Introduction
Mesothelioma, primarily associated with exposure to
asbestos, causes ~2,500 deaths per year in the United States alone
(1). There is a recent trend of
decreasing incidence of mesothelioma in the USA and Canada
(2), but its incidence is still
rising in Europe, Japan (3) and in
developing countries. Vital Statistics Data published yearly by
Ministry of Health, Labor and Welfare, Japan indicated that there
is an increasing trend in the death due to mesothelioma, from 500
cases in 1995 to 710 in 2000, 911 in 2005, 1109 in 2010 and 1,376
in 2014. It is a highly aggressive cancer with poor prognosis
(4) and refractory to currently
available therapeutic regimes (5,6). In
population-based studies, survival times ranged from 5 to 13.2
months (7). Although surgery is an
option for patients with early stage mesothelioma, most patients
present with advanced invasive stage are not amenable to surgical
resection and development of new therapeutic regimes is needed.
MicroRNAs (miRNAs) are regulators of developmental
and cellular pathways and most commonly silence the genes by
binding to the 3′-untranslated region of target mRNA. This binding
induces either mRNA degradation or inhibition of translation
(8). More than 50% of miRNA genes
are found in cancer-associated genomic regions and their altered
expression in numerous cancers supports the notion that these small
RNAs serve as a novel class of oncogenes or tumor suppressors
(9). Recently, Truini et al
reviewed the aberrant expression of miRNAs in mesothelioma for
their role in carcinogenesis, diagnosis and prognosis (10). miRNAs such as miR-31 (11), miR-34b/c (12) as potential therapeutic target,
miR-29c* (13) as
prognostic biomarker, the miR-200 family (14) as diagnostic marker, miR-126
(15), miR-625-3p (16) as biomarker for early detection have
been investigated.
We analyzed the miRNA expression in mesothelioma
cell lines and found significant number of differentially expressed
miRNAs between mesothelioma cell lines and non-neoplastic cells by
TaqMan Low Density miRNA expression PCR array profiling. We focused
on two highly expressed miRNAs, the miR-182 and miR-183, and two
negligibly expressed miRNAs, miR-1 and miR-214, in mesothelioma
cell lines and analyzed their functional role in mesothelioma cell
lines.
Materials and methods
Mesothelioma cell lines and pleural
tissue
Seven mesothelioma cell lines were included in the
study. Five cell lines (CRL-2081, -5820, -5826, -5915, -5946) were
purchased from the American Type Culture Collection (ATCC)
(Manassas, VA, USA) and two cell lines (ACC-Meso-1, ACC-Meso-4)
(17) from the RIKEN BioResearch
Center (Tsukuba, Ibaraki, Japan). All cell lines were maintained in
Roswell Park Memorial Institute 1640 medium with GlutaMAX
(RPMI-1640) with 1% kanamycin, 1% fungizone and 10% fetal bovine
serum (all purchased from Gibco/Life Technologies, Tokyo, Japan).
Non-neoplastic mesothelial cells were retrieved from the strip of
pleura of patients who underwent autopsy for causes other than
neoplastic lesions, and were snap-frozen immediately in liquid
nitrogen and processed for total RNA extraction. The use of these
samples conformed to the statement of Declaration of Helsinki
Ethical principles for Medical Research.
RNA isolation
Total RNA containing small non-coding RNA was
extracted from mesothelioma cell lines and non-neoplastic
mesothelial cells using mirVana miRNA isolation kit (Ambion/Life
Technologies) according to the manufacturer's recommended protocol.
The extracted RNA was directly quantified by the absorbance ratio
at 260 or 280 nM with a NanoVue Plus spectrophotometer (GE
Healthcare BioSciences, Tokyo, Japan) and with fluorometer-based
RNA Assay kit using Qubit 2.0 (Life Technologies). The integrity of
total RNA was evaluated by capillary electrophoresis on RNA
HighSens analysis kit using Experion automated electrophoresis
system (Bio-Rad, Tokyo, Japan).
miRNA expression profiling
Total RNA (1 μg) in 7.5 μl reaction volume was
reverse-transcribed using Megaplex RT Primers, Human Pool set v3.0
using GeneAmp PCR systems 9700 (Applied Biosystems/Life
Technologies) and 6 ml of Megaplex RT product was used for miRNA
expression profiling in TaqMan Low Density Array Human miRNA Cards
v3.0 using 7900HT Fast Real-time PCR system (Applied
Biosystems/Life Technologies) according to the manufacturer's
instructions. The array card set consisted of 2 cards of 384 low
density designed for analysis of 754 human miRNAs based in Sanger
miRbase v14. Relative expression was determined using the
ΔΔCt method and a ≥40 Ct value was interpreted as
amplification too low to quantify. miRNA expressions were analyzed
after normalization to RNU6B (U6 snRNA, an endogenous control assay
designed in both cards) using DataAssist software version 3.0 (Life
Technologies) to calculate the fold change and hierarchical
clustering of significant genes using JMP 9 software.
miRNA expression validation by TaqMan
microRNA assays
Total RNA from mesothelioma cells (5 μl containing
10 ng total RNA) was reverse transcribed with the TaqMan MicroRNA
Reverse Transcription kit and 3 μl of TaqMan MicroRNA assays
specific for hsa-miR-182, hsa-miR-183, hsa-miR-1, hsa-miR-214 and
endogenous controls (Life Technologies) using Eppendorf
Mastercycler (Eppendorf AG, Hamburg, Germany). The resulting cDNA
(1.33 μl) was used as template in a total reaction volume of 20 μl
for real-time quantitative PCR analysis (qPCR) which was performed
in triplicates with TaqMan microRNA assay primer/probes specific
for hsa-miR-182, hsa-miR-183, hsa-miR-1, hsa-miR-214 and endogenous
controls in TaqMan Universal PCR Master Mix (Life Technologies)
using 7900HT Fast Real-time PCR system (Applied Biosystems/Life
Technologies).
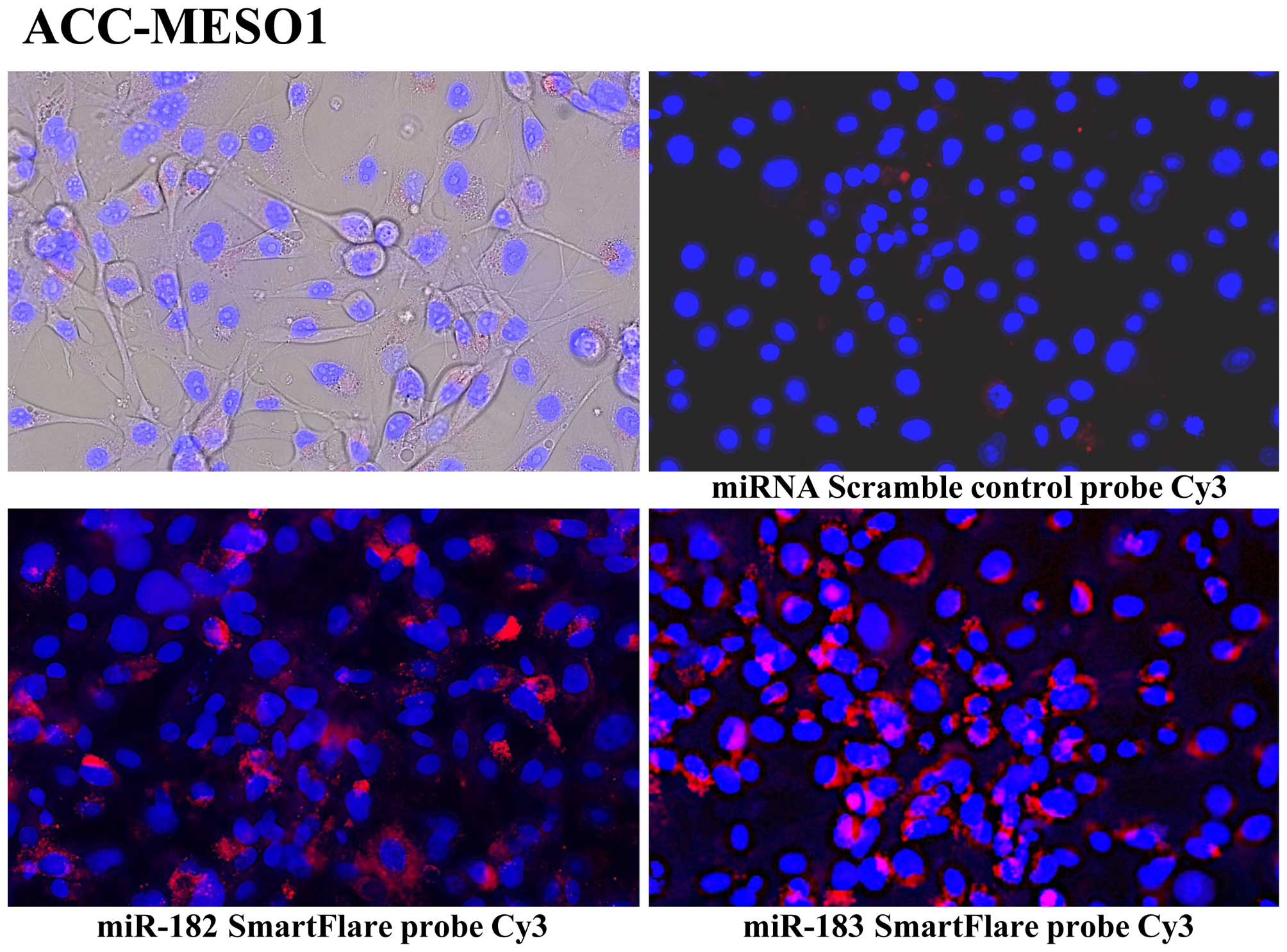
Live cell miRNA validation by SmartFlare
probes
SmartFlare technology detects RNA or miRNA in live
cells. Mesothelioma cells were cultured in collagen-coated 24-well
glass-bottom plates at a concentration of 1×104 cells
per well, in 1 ml of RPMI-1640 for 12 h. SmartFlare probes (3 μl)
(Cy3 labeled miR-182-5p, miR-183-5p, and scramble control detection
probes, purchased from Millipore) prediluted into 50 μl of PBS were
added to the each well in triplicates. Cells were incubated
overnight (~16 h) at 37°C and 5% CO2 and analyzed with
fluorescent microscope and digital picture was taken with similar
light exposure for expression of miRNA.
Transient transfections of mesothelioma
cells with miRNA mimics
All transfection studies were carried out in
triplicates with 10 nM of the each mirVana miRNA mimics by the
Lipofectamine RNAiMax reagent (Lipofectamine) in OptiMEM media
(Life Technologies) using collagen-plated 6-well plates for 2 or 3
days with exchange of miRNA mimics and Lipofectamine every 24 h.
Mesothelioma cell lines, ACC-Meso-1 and CRL 5915, were transfected
with mirVana miRNA mimics specific for hsa-miR-1, hsa-miR-214, and
negative control mimic (Life Technologies). After transient
transfection with miRNA mimics, the cells were analyzed by various
cell based assays.
Cell proliferation, cell cycle, migration
and invasion assays
Cell proliferation assay was analyzed with Guava
ViaCount reagent using Guava EasyCyte Mini Flow Cytometer (Guava
Technologies/Millipore, Hayward, CA, USA). miRNA mimic transfected
cells were incubated in RPMI-1640 media with 10% FBS in
collagen-coated 24-well plates in triplicates. After 24, 48 or 72
h, cells were harvested by trypsinization with TrypLE Express (Life
Technologies), stained for 5 min with ViaCount reagent, and total
numbers of viable cells were determined with flow cytometer using
Cytosoft viacount module software (Guava Technologies).
Cell cycle phase analysis was performed with Guava
Cell Cycle reagent using Guava Easycyte Mini Flow cytometer. miRNA
mimic transfected cells were incubated in RPMI-1640 media with 10%
FBS in collagen-coated 24-well plates in triplicates. After 48 or
72 h, cells were harvested by trypsinization with TrypLE Express
and slowly fixed with 10 ml of 70% ethanol for ≥2 h. After removal
of ethanol by centrifuging and washing cells with PBS (−), cells
were stained with Guava Cell Cycle reagent for 30 min and flow
cytometric data were collected with flow cytometer using Cytosoft
cell cycle module software (Guava Technologies). Later, flow
cytometric raw data were analyzed using FCS Express 5 Pro software
(DeNovo Software, Glendale, CA, USA).
Cell migration was analyzed using a wound/scratch
assay. miRNA mimic transfected cells incubated in RPMI-1640 media
with 10% FBS in collagen-coated 24-well plates overnight. The wound
was created in cell monolayer by scratching the cells with 1 ml
micropipette tips and floating cells were removed by washing with
fresh media. Real-time microscopic images were taken at 0, 6, 12,
18 and 24 h post-scratching by incubating the cells in stage-top
incubator on an inverted microscope with automatic picture
acquisition at given time interval (Olympus IX81 with cellSens
software). The area of the wound gap that was not covered by cell
migration (percentage) was determined using TScrach software
(18).
Cell invasion was performed using BD FluoroBlok
culture Inserts with 8-μm pores (BD Biosciences, Franklin Lakes,
NJ, USA) coated with matrix Matrigel (Life Technologies) according
to the manufacturer's protocol. miRNA mimics transfected cells were
stained with cell permeant nuclear fluorescent dye Hoechst 33342
(Life Technologies) for 10 min and fluorescent image (16
consecutive images with 10X objective) was captured with DP80 CCD
camera using an inverted fluorescent microscope IX81. The total
numbers of invading cells were calculated by analyzing fluorescent
images with CellProfiler cell imaging software (19).
Cell culture in chamber-slide or
cell-block preparation for immunocytochemistry
Approximately 1×105 cells were cultured
in collagen coated chamber glass slide (Millicell EZ Slide:
Millipore, EMD Millipore Corp., Billerica, MA, USA) overnight and
fixed with 10% formalin. Each of mesothelioma cell lines was
cultured for 2–3 days on 100-mm collagen-coated culture dish
(Corning Life Sciences, Corning, NY, USA) and were subjected to
cell-block preparation using glucomannan (HoldGel 110, EBIS1
cell-block construction kit, Asia Kizai, Tokyo, Japan) (20). Three-μm-sections were prepared from
formalin-fixed paraffin-embedded cell-blocks in Platinum Pro
Adhesive Glass slides (Matsunami Glass, Osaka, Japan). Fully
automatic immunohistochemical staining of anti-PIM1 antibody
(rabbit monoclonal, Cat #ab75776, Abcam, Cambridge, MA, USA, 1:250
dilution filled in antibody dispenser) was performed with Ventana
BenchMark GX using ultraView Universal DAB Detection kit (Ventana,
Roche Diagnostics, Tokyo, Japan). Similar immunohistochemical
procedure was carried out with the omission of the primary antibody
as a negative control.
Western blot analysis
Cell lysates were obtained from miRNA mimic
transfected cells using CytoBuster Protein Extraction reagent
(Novagen, Madison, WI, USA) with Halt Protease Inhibitor Cocktail
(Thermo Scientific Fisher, Tokyo, Japan). Total protein (20 μg) was
separated on the Bolt 4–12% Bis-Tris Plus Gels (Life Technologies)
with lithium dodecyl sulfate (LDS) electrophoresis at 165 V for 35
min and transferred onto a polyvinylidene difluoride (PVDF)
membrane using iBlot transfer stack (Life Technologies) at 20 V for
7 min. The protein-transferred membrane was processed on BenchPro
4100 Western System (Life Technologies) with primary antibodies,
anti-PIM1 antibody (1:500, rabbit monoclonal, ab75776, Abcam) and
anti-GAPDH antibody (1:500, rabbit polyclonal sc-25778, Santa Cruz
Biotechnology, Dallas, TX, USA) and biotin labeled secondary
antibody [Biotin-XX goat anti-rabbit IgG (H+L) (Life
Technologies)]. The membrane was washed and stained with Qdot 625
streptavidin conjugate (Life Technologies) for 30 min and bands
were detected using E-Gel Imager (Life Technologies).
Statistical analysis
The TaqMan Low Density microRNA Array data were
analyzed with DataAssist software and JMP 9. All other statistical
analysis was performed with JMP 9 software (SAS Institute, Inc.,
Cary, NC, USA).
Results
Differential miRNA expression in
mesothelioma cells and its validation
Out of 754 analyzable miRNAs by TaqMan Low Density
Array Human miRNA Cards, significant number of miRNA were found to
be differentially expressed between mesothelioma cell lines
(CRL-2081, -5820, -5826, -5915, -5946, ACC-Meso-1, ACC-Meso-4) and
non-neoplastic mesothelial tissue. We found increased expression of
18 miRNAs and reduced or absent expression of 20 miRNAs in
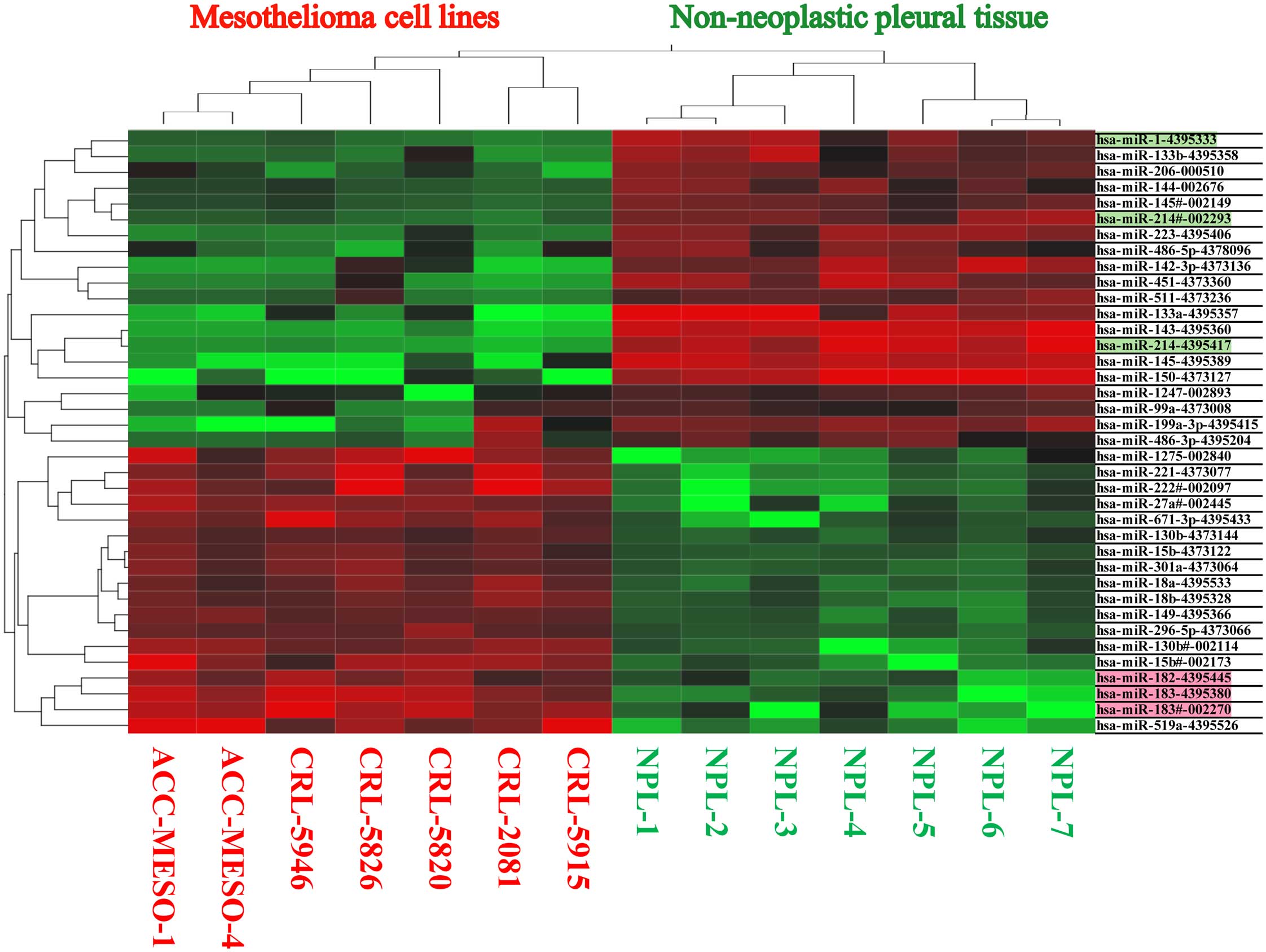
mesothelioma cell lines (Fig. 1
and Table I). Using SmartFlare
miRNA probes and scrambled negative control probe, we found all of
these 7 mesothelioma cell lines have increased miR-182, miR-183
(Fig. 2). We also validated miRNA
expression in these mesothelioma cell lines by individual microRNA
expression assay (data not shown).
 | Table IExpression fold change, average CT,
and average dCT value in mesothelioma cell lines (MESO) and
non-neoplastic pleural tissue (NPL). |
Table I
Expression fold change, average CT,
and average dCT value in mesothelioma cell lines (MESO) and
non-neoplastic pleural tissue (NPL).
| Expression fold
change | Average CT | Average dCT |
|---|
|
|
|
|
|---|
| Assay | MESO | NPL | P-value | MESO | NPL | MESO | NPL |
|---|
|
hsa-miR-519a-4395526 | 135.1 | | 0.119 | 32.5 | 39.1 | 15.1 | 21.6 |
|
hsa-miR-222#-002097 | 54.9 | | 0.057 | 27.9 | 33.7 | 10.5 | 16.2 |
|
hsa-miR-183-4395380 | 42.6 | | 0.011 | 30.9 | 37.2 | 13.5 | 19.8 |
|
hsa-miR-221-4373077 | 41.7 | | 0.095 | 25.1 | 29.9 | 7.6 | 12.4 |
|
hsa-miR-15b#-002173 | 31.8 | | 0.020 | 28.3 | 33.7 | 10.9 | 16.2 |
|
hsa-miR-1275-002840 | 21.2 | | 0.040 | 31.1 | 36.4 | 13.6 | 19.0 |
|
hsa-miR-183#-002270 | 20.2 | | 0.002 | 29.4 | 36.2 | 11.9 | 18.7 |
|
hsa-miR-130b#-002114 | 15.7 | | 0.018 | 31.7 | 36.3 | 14.3 | 18.8 |
|
hsa-miR-671-3p-4395433 | 15.0 | | 0.005 | 30.6 | 35.2 | 13.2 | 17.7 |
|
hsa-miR-27a#-002445 | 10.2 | | 0.012 | 28.5 | 32.8 | 11.1 | 15.4 |
|
hsa-miR-149-4395366 | 8.0 | | 0.033 | 25.1 | 27.9 | 7.7 | 10.5 |
|
hsa-miR-18a-4395533 | 7.8 | | 0.042 | 28.1 | 30.8 | 10.6 | 13.4 |
|
hsa-miR-182-4395445 | 7.4 | | 0.010 | 28.6 | 32.4 | 11.2 | 14.9 |
|
hsa-miR-301a-4373064 | 7.1 | | 0.055 | 28.6 | 31.2 | 11.1 | 13.8 |
|
hsa-miR-18b-4395328 | 7.0 | | 0.011 | 32.2 | 35.1 | 14.7 | 17.7 |
|
hsa-miR-15b-4373122 | 6.3 | | 0.026 | 25.9 | 28.2 | 8.4 | 10.7 |
|
hsa-miR-296-5p-4373066 | 6.1 | | 0.021 | 31.0 | 33.4 | 13.5 | 15.9 |
|
hsa-miR-130b-4373144 | 3.6 | | 0.014 | 29.6 | 31.3 | 12.1 | 13.9 |
|
hsa-miR-99a-4373008 | 3.4 | | 0.249 | 26.3 | 26.0 | 8.8 | 8.5 |
|
hsa-miR-486-3p-4395204 | | 26.3 | 0.016 | 38.0 | 29.4 | 20.6 | 11.9 |
|
hsa-miR-199a-3p-4395415 | | 41.2 | 0.000 | 35.5 | 23.5 | 18.0 | 6.1 |
|
hsa-miR-511-4373236 | | 134.6 | 0.002 | 39.1 | 28.9 | 21.6 | 11.4 |
|
hsa-miR-1247-002893 | | 171.0 | 0.001 | 34.2 | 25.1 | 16.8 | 7.7 |
|
hsa-miR-142-3p-4373136 | | 352.0 | 0.000 | 38.2 | 25.8 | 20.7 | 8.3 |
|
hsa-miR-206-000510 | | 490.7 | 0.020 | 37.1 | 26.8 | 19.6 | 9.4 |
|
hsa-miR-486-5p-4378096 | | 588.6 | 0.038 | 35.8 | 25.9 | 18.3 | 8.5 |
|
hsa-miR-451-4373360 | | 759.8 | 0.009 | 39.1 | 26.6 | 21.6 | 9.2 |
|
hsa-miR-144-002676 | | 1,223.0 | 0.040 | 40.0 | 30.4 | 22.6 | 12.9 |
|
hsa-miR-145#-002149 | | 1,340.9 | 0.014 | 40.0 | 29.9 | 22.6 | 12.5 |
|
hsa-miR-133b-4395358 | | 1,643.8 | 0.112 | 39.2 | 28.1 | 21.7 | 10.7 |
|
hsa-miR-214#-002293 | | 2,788.9 | 0.005 | 39.9 | 28.7 | 22.4 | 11.3 |
|
hsa-miR-223-4395406 | | 2,977.7 | 0.001 | 32.7 | 20.6 | 15.2 | 3.1 |
|
hsa-miR-1-4395333 | | 6,705.6 | 0.049 | 40.0 | 28.3 | 22.6 | 10.8 |
|
hsa-miR-150-4373127 | | 10,404.8 | 0.001 | 37.3 | 21.7 | 19.9 | 4.2 |
|
hsa-miR-145-4395389 | | 12,025.2 | 0.004 | 37.7 | 22.6 | 20.2 | 5.2 |
|
hsa-miR-133a-4395357 | | 13,032.1 | 0.050 | 37.9 | 23.6 | 20.4 | 6.2 |
|
hsa-miR-214-4395417 | | 32,285.8 | 0.001 | 40.0 | 25.0 | 22.5 | 7.5 |
|
hsa-miR-143-4395360 | | 55,283.3 | 0.002 | 39.7 | 24.0 | 22.3 | 6.5 |
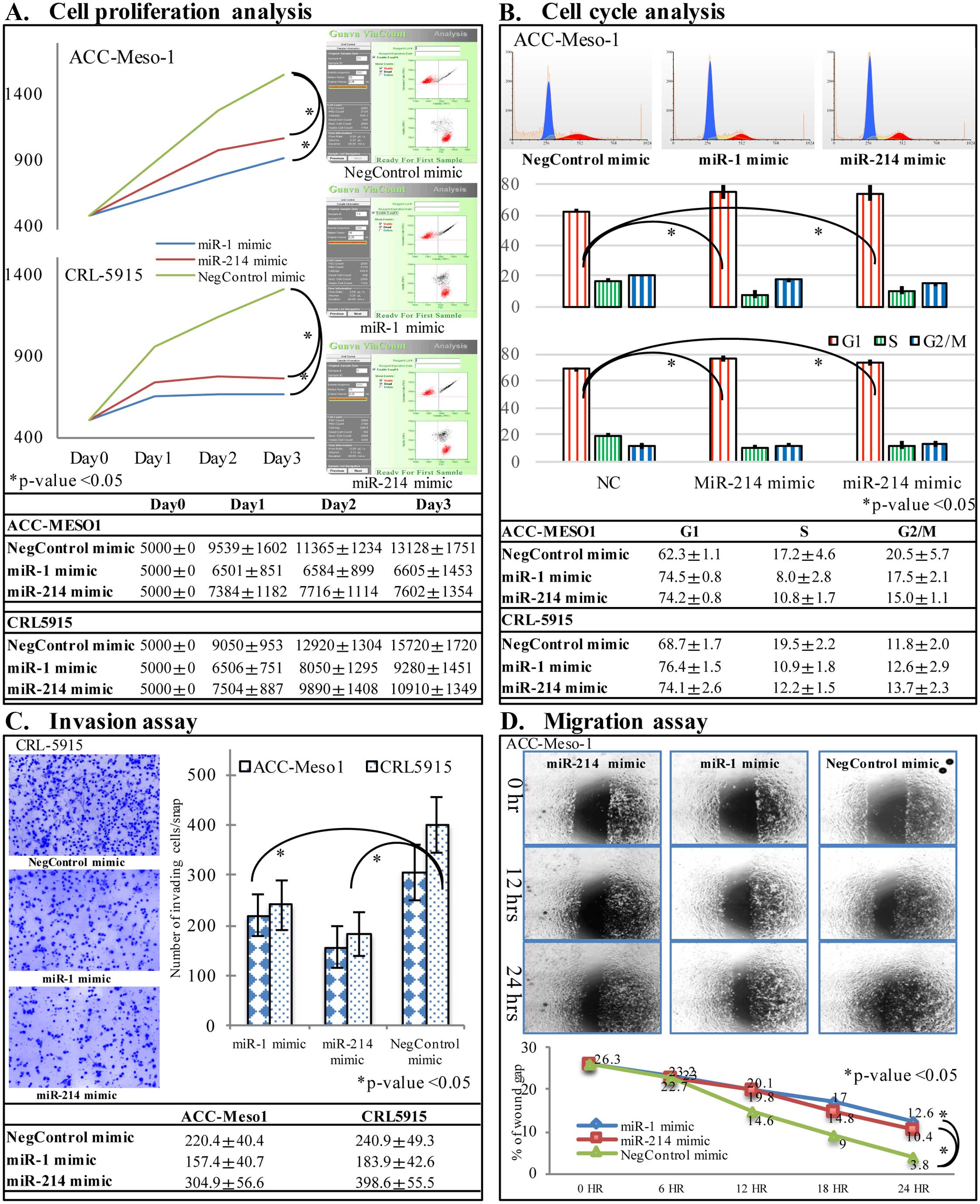
miR-1 and miR-214 mimic transfection
inhibits cell growth, cell cycle progression, migration and
invasion of mesothelioma cells
Two mesothelioma cell lines, ACC-Meso-1 and CRL-5915
were transfected with hsa-miR-1, and hsa-miR-214 miRNA mimics, and
negative control miRNA mimic. Cell proliferation was significantly
suppressed in mesothelioma cells transfected with miR-1 and miR-214
mimic compared to negative control group at 48 and 72 h after
transfection by flow cytometric analysis of viable cells
(p<0.05, Fig. 3A). Furthermore,
flow cytometric analysis indicated that proportion of cells in
G0/G1 phase was increased significantly, whereas cells in S and
G2/M phase was significantly decreased in miR-1 and miR-214 mimic
transfected cells compared to control group (p<0.05, Fig. 3B). The cell migration analysis by
wound scratch assay showed decreased speed of cell migration with
hsa-miR-1 and hsa-miR-214 miRNA mimics compared to negative control
miRNA mimic (Fig. 3C). Matrigel
invasion chamber assay showed less number of cell invasion through
the Matrigel in cells transfected with hsa-miR-1 and hsa-miR-214
mimics (p<0.05, Fig. 3D).
Target prediction database of miR-1 and
miR-214 shows PIM1 as possible target in mesothelioma
To analyze the underlying mechanism of miR-1 and
miR-214 in mesothelioma cell carcinogenesis, online miRTarBase
(http://mirtarbase.mbc.nctu.edu.tw/php/search.php)
was used to search for potential targets of miR-1 and miR-214, and
we noted PIM1 was the target gene of both miR-1, miR-214. We also
found similar result from TargetScan (www.targetscan.org) and miRanda (www.microrna.org). Thus, we investigated whether the
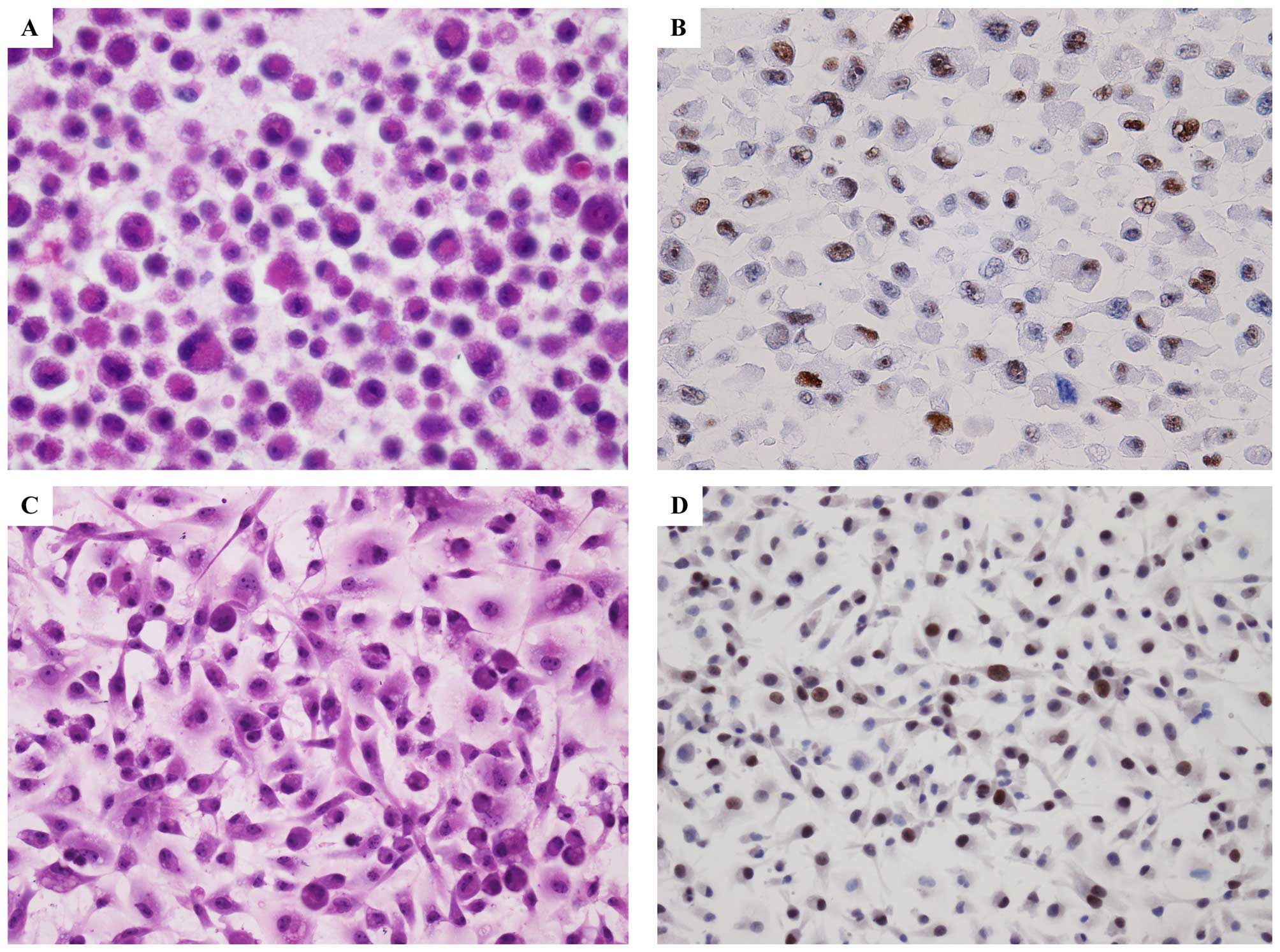
PIM1 was expressed in mesothelioma cell lines or not. By
immunocytochemical study of cell-blocks prepared from mesothelioma
cell lines, we found PIM1 was highly expressed in all of the seven
mesothelioma cells lines (Fig.
4B). In addition, we also investigated the expression of PIM1
in mesothelioma cell line cultured directly in chamber-slide
(Fig. 4D).
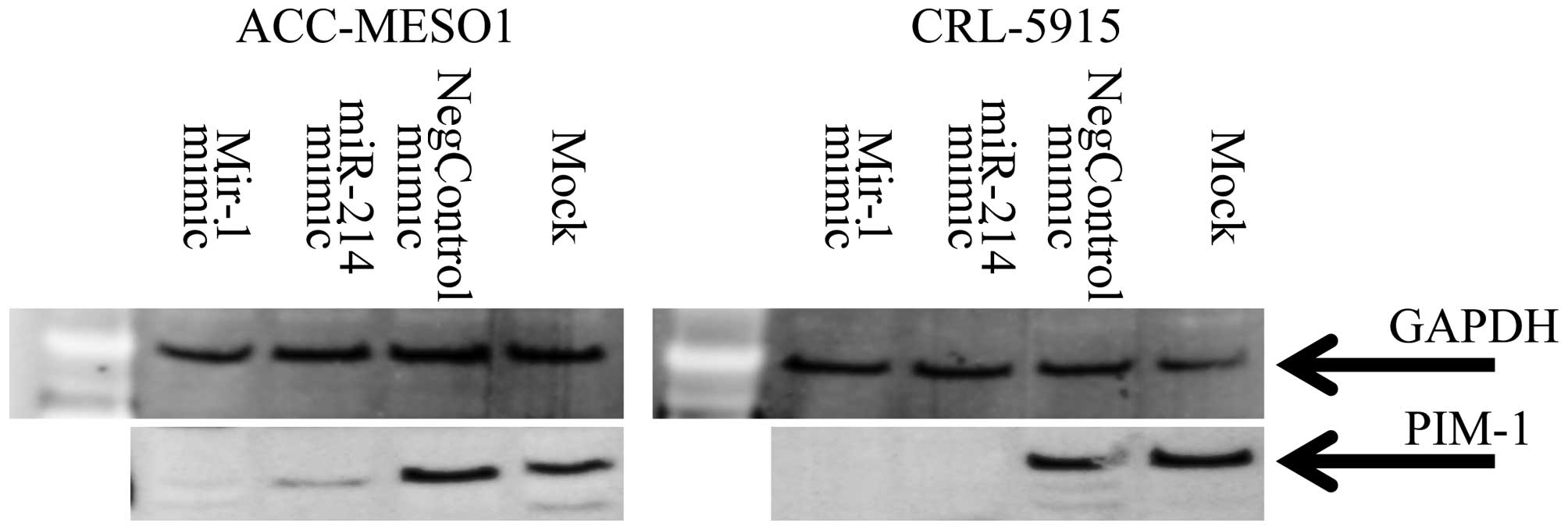
Transfection of miR-1 and miR-214
downregulates PIM1 in mesothelioma cell lines
To determine if miR-1 and miR-214 target PIM1 in
mesothelioma, the miR-1, miR-214 mimic or a control were
transfected into ACC-Meso-1 and CRL5915 cells. Both of the cell
lines used in this study, showed PIM1 expression. The serum starved
cells transfected with miR-1 and miR-214 mimic showed reduced PIM1
protein, respectively, after 2 days and 3 days of transfection
compared to control miRNA mimic (Fig.
5). These results confirmed that miR-1 and miR-214 mimic
transfection downregulated PIM1 expression in mesothelioma
cells.
Discussion
Malignant pleural mesothelioma is an aggressive
cancer as current treatment modalities do not improve patient
survival (21). Present
understanding of molecular mechanism is not yet sufficient to
improve the management of these patients. miRNAs for their powerful
regulatory potential are recently considered as potential
therapeutic targets to various human malignancies, including
malignant mesothelioma miR-31 (11), miR-34b/c (12). In addition, the investigation of
the target genes involved in various malignancies are also possible
by analyzing aberration of the miRNA expression. Many reports,
summarized by Truini et al (10), have identified various miRNAs
associated with mesothelioma. Here we add miR-1, miR-214 and its
prediction target PIM1 gene as possible target of therapeutic
potential in mesothelioma.
Previous reports have shown downregulation of miR-1
and mir-214 in various human malignancies. miR-1 expression is
reported to be downregulated in colon cancer (22), gastric cancer (23), lung cancer (24), prostate cancer (25), renal cancer (26), and esophageal cancer (27). Xu et al (28) have shown low miR-1 expression in
mesothelioma and transfection of precursor miR-1 been shown to
suppress cell growth and induce apoptosis. In our study, we found
negligible miR-1 in mesothelioma cell line by real-time RT PCR
method and transaction of miR-1 mimic showed reduced cell
proliferation with cell cycle G1 arrest, reduced cell migration and
invasive nature by in vitro experiments. Aberrant expression
of miR-214 has been reported in the past with its downregulation in
liver cancers (29,30), cervical cancer (31), prostate cancer (32), and upregulation in lung cancer
(33), bladder cancer (34) and esophageal cancer (35). In malignant mesothelioma, we found
no expression of miR-1 and miR-214 in mesothelioma cell lines.
The loss of expression of miR-1 and miR-214 may have
potential as the diagnostic biomarker of malignant mesothelioma.
The transfection of miR-1 or miR-214 both inhibits cell
proliferation, cell migration and invasion. The histopathological
differentiation of early malignant mesothelioma from reactive
mesothelial proliferation has been performed even in an
immunohistochemical study (36).
The assessment of miR-1 and miR-214 expression may be utilized as
differentiation marker of mesothelioma from reactive mesothelial
proliferation. In situ hybridization with miR-1 and/or
miR-214 probes could be utilized to differentiate mesothelioma from
reactive mesothelial tissue. However, further analysis of the miRNA
expression in human mesothelioma and reactive mesothelial
hyperplasia tissue is necessary. We intend to perform such analysis
using human tissue in future.
By target prediction analysis using online
miRTarBase release 6.1 (37), we
analyzed the common target of miR-1 and miR-214 and we primarily
focused on PIM1 proto-oncogene. We analyzed whether the
mesothelioma cell line express PIM1 or not. We investigated PIM1
expression by immunohistochemistry of cell lines cultured directly
on collagen coated chamber-slide and cell-block. We found that PIM1
was highly expressed in all seven mesothelioma cell lines that we
analyzed in our miRNA profiling study. In addition, by transfecting
miR-1 and miR-214 mimic to two cell lines, ACC-MESO-1, and
CRL-5915, we also found the downregulation of PIM1 protein
expression suggesting the role of miR-1, and miR-214 in inhibition
of PIM1 expression. The expression of PIM1 gene in human cancer has
been studied in great depth and was found to be overexpressed in a
wide range of human tumors. PIM expression level has shown
favorable prognosis in prostate cancer (38), pancreatic ductal carcinoma
(39), and non-small cell lung
cancer (40), and unfavorable
prognosis in gastric cancer (41),
and squamous cell carcinoma of the head and neck (42). As yet, there is no published report
on PIM1 expression in malignant mesothelioma. This is the first
report on increased PIM1 expression in mesothelioma. Further, novel
molecules inhibiting PIM kinases have been evaluated in preclinical
studies, demonstrating to be effective and with a favorable
toxicity profile. Given the promising results, some of these
compounds are currently under investigation in clinical trials
(43). However, its application
and therapeutic potential in mesothelioma needs additional
investigation to assess the significance in mesothelioma.
In conclusion, we have shown that loss of miR-1 and
miR-214 expression and high expression of their target gene, PIM1,
in malignant mesothelioma. By miRNA mimic transfection study we
consider the loss of miR-1 and miR-214 play a role in cell
proliferation, invasion, and migration in mesothelioma, probably by
downregulation of PIM1. It remains feasible that loss of miR-1 and
miR-214 expression may behave as an oncogene in mesothelioma and
have potential as an oncogenic target. miR-1 and miR-214 may be
attractive targets for future mesothelioma therapeutic studies.
Acknowledgements
Part of this study was carried out at the Analysis
Center of Life Science, Hiroshima University.
References
|
1
|
Weill H, Hughes JM and Churg AM: Changing
trends in US mesothelioma incidence. Occup Environ Med. 61:438–441.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Centers for Disease Control and Prevention
(CDC). Malignant mesothelioma mortality - United States, 1999–2005.
MMWR Morb Mortal Wkly Rep. 58:393–396. 2009.
|
|
3
|
Takeshima Y, Inai K, Amatya VJ, Gemba K,
Aoe K, Fujimoto N, Kato K and Kishimoto T: Accuracy of pathological
diagnosis of mesothelioma cases in Japan: Clinicopathological
analysis of 382 cases. Lung Cancer. 66:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amatya VJ, Takeshima Y, Aoe K, Fujimoto N,
Okamoto T, Yamada T, Kishimoto T, Morimoto C and Inai K: CD9
expression as a favorable prognostic marker for patients with
malignant mesothelioma. Oncol Rep. 29:21–28. 2013.
|
|
5
|
Pass HI, Vogelzang N, Hahn S and Carbone
M: Malignant pleural mesothelioma. Curr Probl Cancer. 28:93–174.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Robinson BW and Lake RA: Advances in
malignant mesothelioma. N Engl J Med. 353:1591–1603. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Montanaro F, Rosato R, Gangemi M, Roberti
S, Ricceri F, Merler E, Gennaro V, Romanelli A, Chellini E,
Pascucci C, et al: Survival of pleural malignant mesothelioma in
Italy: A population-based study. Int J Cancer. 124:201–207. 2009.
View Article : Google Scholar
|
|
8
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Drakaki A and Iliopoulos D: MicroRNA gene
networks in Oncogenesis. Curr Genomics. 10:35–41. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Truini A, Coco S, Alama A, Genova C, Sini
C, Dal Bello MG, Barletta G, Rijavec E, Burrafato G, Boccardo F, et
al: Role of microRNAs in malignant mesothelioma. Cell Mol Life Sci.
71:2865–2878. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Busacca S, Germano S, De Cecco L, Rinaldi
M, Comoglio F, Favero F, Murer B, Mutti L, Pierotti M and Gaudino
G: MicroRNA signature of malignant mesothelioma with potential
diagnostic and prognostic implications. Am J Respir Cell Mol Biol.
42:312–319. 2010. View Article : Google Scholar
|
|
12
|
Kubo T, Toyooka S, Tsukuda K, Sakaguchi M,
Fukazawa T, Soh J, Asano H, Ueno T, Muraoka T, Yamamoto H, et al:
Epigenetic silencing of microRNA-34b/c plays an important role in
the pathogenesis of malignant pleural mesothelioma. Clin Cancer
Res. 17:4965–4974. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pass HI, Goparaju C, Ivanov S, Donington
J, Carbone M, Hoshen M, Cohen D, Chajut A, Rosenwald S, Dan H, et
al: hsa-miR-29c* is linked to the prognosis of malignant
pleural mesothelioma. Cancer Res. 70:1916–1924. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gee GV, Koestler DC, Christensen BC,
Sugarbaker DJ, Ugolini D, Ivaldi GP, Resnick MB, Houseman EA,
Kelsey KT and Marsit CJ: Downregulated microRNAs in the
differential diagnosis of malignant pleural mesothelioma. Int J
Cancer. 127:2859–2869. 2010. View Article : Google Scholar :
|
|
15
|
Santarelli L, Strafella E, Staffolani S,
Amati M, Emanuelli M, Sartini D, Pozzi V, Carbonari D, Bracci M,
Pignotti E, et al: Association of miR-126 with soluble
mesothelin-related peptides, a marker for malignant mesothelioma.
PLoS One. 6:e182322011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kirschner MB, Cheng YY, Badrian B, Kao SC,
Creaney J, Edelman JJ, Armstrong NJ, Vallely MP, Musk AW, Robinson
BW, et al: Increased circulating miR-625-3p: A potential biomarker
for patients with malignant pleural mesothelioma. J Thorac Oncol.
7:1184–1191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Usami N, Fukui T, Kondo M, Taniguchi T,
Yokoyama T, Mori S, Yokoi K, Horio Y, Shimokata K, Sekido Y, et al:
Establishment and characterization of four malignant pleural
mesothelioma cell lines from Japanese patients. Cancer Sci.
97:387–394. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gebäck T, Schulz MM, Koumoutsakos P and
Detmar M: TScratch: A novel and simple software tool for automated
analysis of monolayer wound healing assays. Biotechniques.
46:265–274. 2009.PubMed/NCBI
|
|
19
|
Lamprecht MR, Sabatini DM and Carpenter
AE: CellProfiler: Free, versatile software for automated biological
image analysis. Biotechniques. 42:71–75. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Amatya VJ, Takeshima Y, Kushitani K,
Yamada T, Morimoto C and Inai K: Overexpression of CD26/DPPIV in
mesothelioma tissue and mesothelioma cell lines. Oncol Rep.
26:1369–1375. 2011.PubMed/NCBI
|
|
21
|
Milano MT and Zhang H: Malignant pleural
mesothelioma: A population-based study of survival. J Thorac Oncol.
5:1841–1848. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Furukawa S, Kawasaki Y, Miyamoto M,
Hiyoshi M, Kitayama J and Akiyama T: The miR-1-NOTCH3-Asef pathway
is important for colorectal tumor cell migration. PLoS One.
8:e806092013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han C, Zhou Y, An Q, Li F, Li D, Zhang X,
Yu Z, Zheng L, Duan Z and Kan Q: MicroRNA-1 (miR-1) inhibits
gastric cancer cell proliferation and migration by targeting MET.
Tumour Biol. 36:6715–6723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nasser MW, Datta J, Nuovo G, Kutay H,
Motiwala T, Majumder S, Wang B, Suster S, Jacob ST and Ghoshal K:
Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression
of tumorigenic property of lung cancer cells and their
sensitization to doxorubicin-induced apoptosis by miR-1. J Biol
Chem. 283:33394–33405. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Karatas OF, Guzel E, Suer I, Ekici ID,
Caskurlu T, Creighton CJ, Ittmann M and Ozen M: miR-1 and miR-133b
are differentially expressed in patients with recurrent prostate
cancer. PLoS One. 9:e986752014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kawakami K, Enokida H, Chiyomaru T,
Tatarano S, Yoshino H, Kagara I, Gotanda T, Tachiwada T, Nishiyama
K, Nohata N, et al: The functional significance of miR-1 and
miR-133a in renal cell carcinoma. Eur J Cancer. 48:827–836. 2012.
View Article : Google Scholar
|
|
27
|
Du YY, Zhao LM, Chen L, Sang MX, Li J, Ma
M and Liu JF: The tumor-suppressive function of miR-1 by targeting
LASP1 and TAGLN2 in esophageal squamous cell carcinoma. J
Gastroenterol Hepatol. Sep 28–2015.(Epub ahead of print).
View Article : Google Scholar
|
|
28
|
Xu Y, Zheng M, Merritt RE, Shrager JB,
Wakelee H, Kratzke RA and Hoang CD: miR-1 induces growth arrest and
apoptosis in malignant mesothelioma. Chest. 144:1632–1643. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xia H, Ooi LL and Hui KM: miR-214 targets
β-catenin pathway to suppress invasion, stem-like traits and
recurrence of human hepatocellular carcinoma. PLoS One.
7:e442062012. View Article : Google Scholar
|
|
30
|
Li B, Han Q, Zhu Y, Yu Y, Wang J and Jiang
X: Down-regulation of miR-214 contributes to intrahepatic
cholangiocarcinoma metastasis by targeting Twist. FEBS J.
279:2393–2398. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang F, Liu M, Li X and Tang H: miR-214
reduces cell survival and enhances cisplatin-induced cytotoxicity
via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett.
587:488–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Srivastava A, Goldberger H, Dimtchev A,
Ramalinga M, Chijioke J, Marian C, Oermann EK, Uhm S, Kim JS, Chen
LN, et al: MicroRNA profiling in prostate cancer - the diagnostic
potential of urinary miR-205 and miR-214. PLoS One. 8:e769942013.
View Article : Google Scholar :
|
|
33
|
Long H, Wang Z, Chen J, Xiang T, Li Q,
Diao X and Zhu B: microRNA-214 promotes epithelial-mesenchymal
transition and metastasis in lung adenocarcinoma by targeting the
suppressor-of-fused protein (Sufu). Oncotarget. 6:38705–38718.
2015.PubMed/NCBI
|
|
34
|
Wang J, Zhang X, Wang L, Yang Y, Dong Z,
Wang H, Du L and Wang C: MicroRNA-214 suppresses oncogenesis and
exerts impact on prognosis by targeting PDRG1 in bladder cancer.
PLoS One. 10:e01180862015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Phatak P, Byrnes KA, Mansour D, Liu L, Cao
S, Li R, Rao JN, Turner DJ, Wang JY and Donahue JM: Overexpression
of miR-214-3p in esophageal squamous cancer cells enhances
sensitivity to cisplatin by targeting survivin directly and
indirectly through CUG-BP1. Oncogene. View Article : Google Scholar : [Epub ahead of
print].
|
|
36
|
Tsukiji H, Takeshima Y, Amatya VJ,
Kushitani K and Inai K: Myogenic antigen expression is useful for
differentiation between epithelioid mesothelioma and non-neoplastic
mesothelial cells. Histopathology. 56:969–974. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res.
42D:D78–D85. 2014. View Article : Google Scholar
|
|
38
|
Dhanasekaran SM, Barrette TR, Ghosh D,
Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA and Chinnaiyan
AM: Delineation of prognostic biomarkers in prostate cancer.
Nature. 412:822–826. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Reiser-Erkan C, Erkan M, Pan Z, Bekasi S,
Giese NA, Streit S, Michalski CW, Friess H and Kleeff J:
Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in
pancreatic ductal adenocarcinoma. Cancer Biol Ther. 7:1352–1359.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Warnecke-Eberz U, Bollschweiler E, Drebber
U, Pohl A, Baldus SE, Hoelscher AH and Metzger R: Frequent
down-regulation of pim-1 mRNA expression in non-small cell lung
cancer is associated with lymph node metastases. Oncol Rep.
20:619–624. 2008.PubMed/NCBI
|
|
41
|
Warnecke-Eberz U, Bollschweiler E, Drebber
U, Metzger R, Baldus SE, Hölscher AH and Mönig S: Prognostic impact
of protein overexpression of the proto-oncogene PIM-1 in gastric
cancer. Anticancer Res. 29:4451–4455. 2009.PubMed/NCBI
|
|
42
|
Peltola K, Hollmen M, Maula SM, Rainio E,
Ristamäki R, Luukkaa M, Sandholm J, Sundvall M, Elenius K, Koskinen
PJ, et al: Pim-1 kinase expression predicts radiation response in
squamocellular carcinoma of head and neck and is under the control
of epidermal growth factor receptor. Neoplasia. 11:629–636. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mondello P, Cuzzocrea S and Mian M: Pim
kinases in hematological malignancies: Where are we now and where
are we going? J Hematol Oncol. 7:952014. View Article : Google Scholar : PubMed/NCBI
|