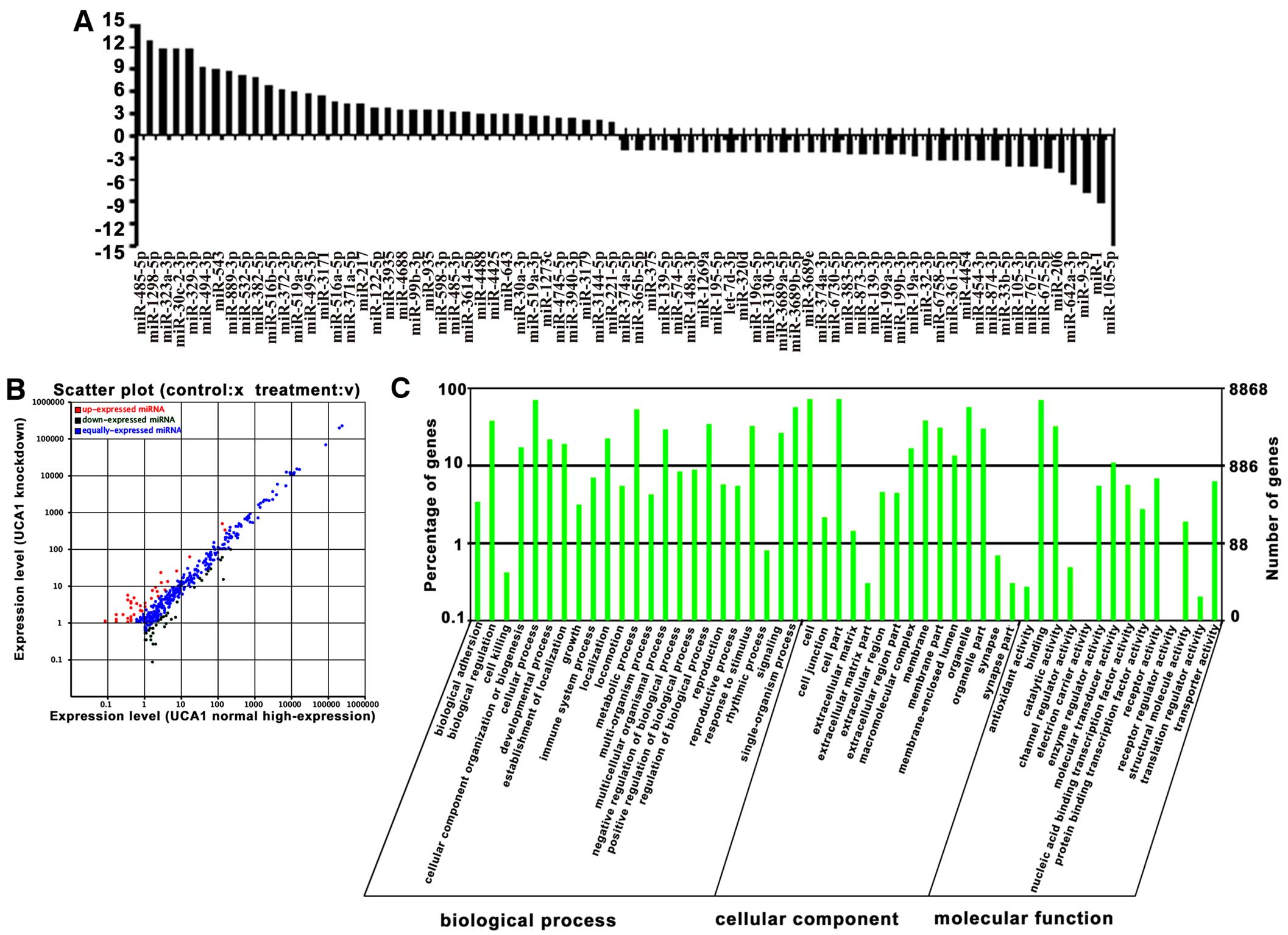
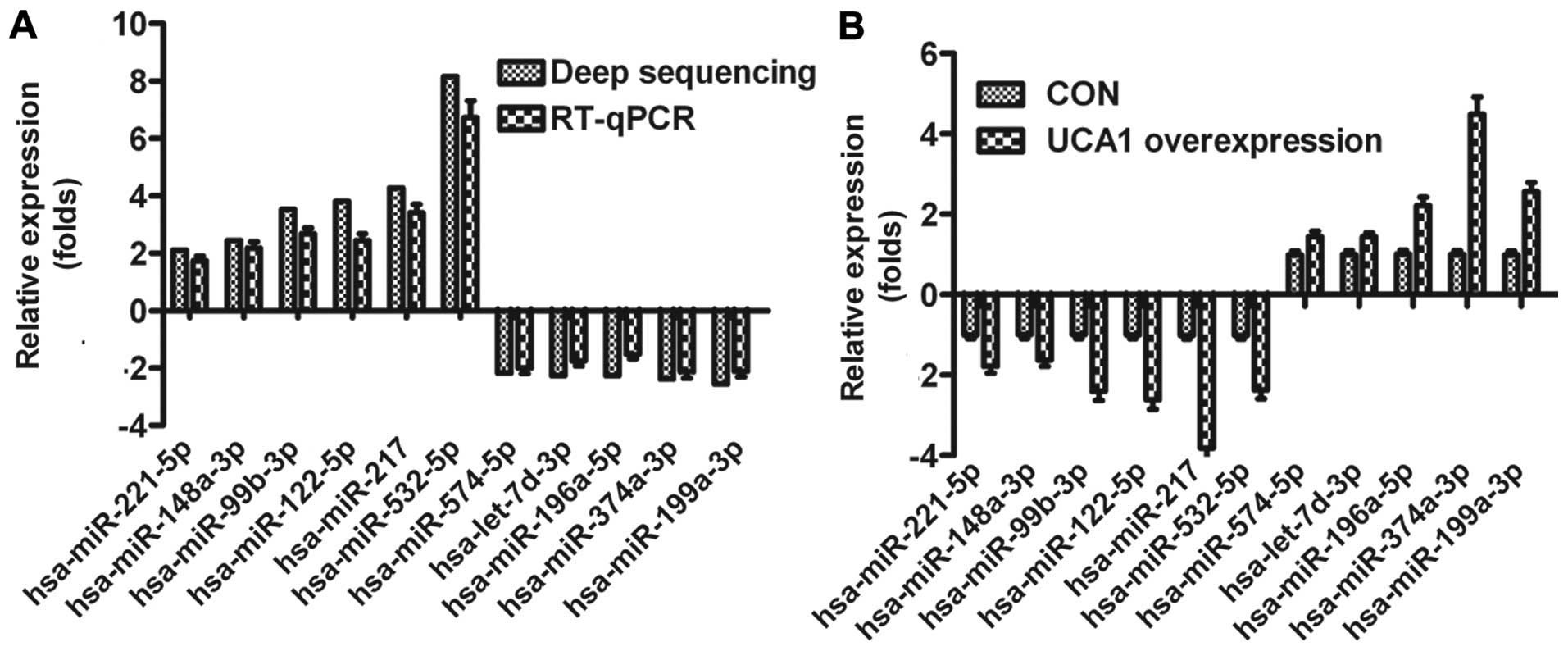
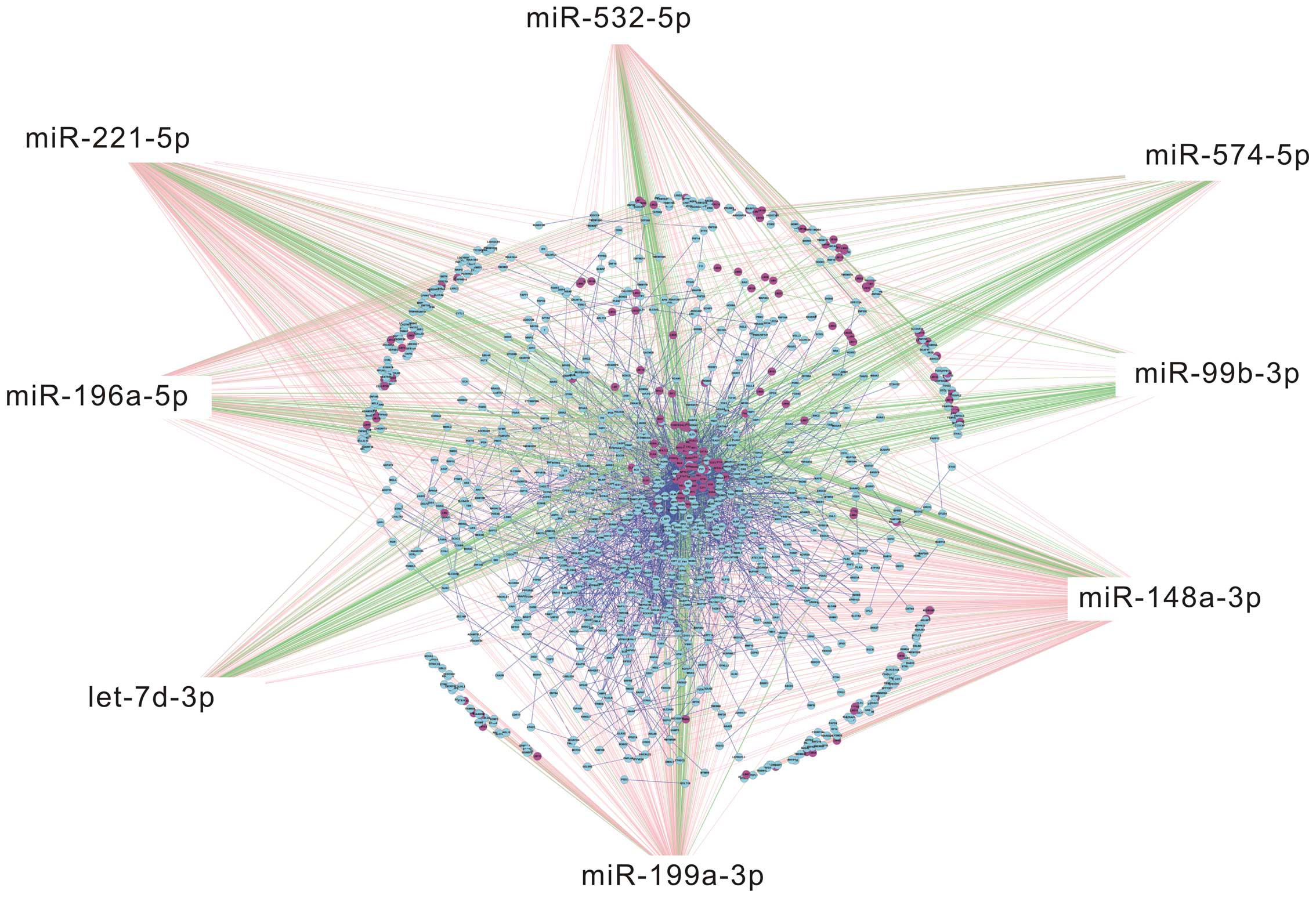
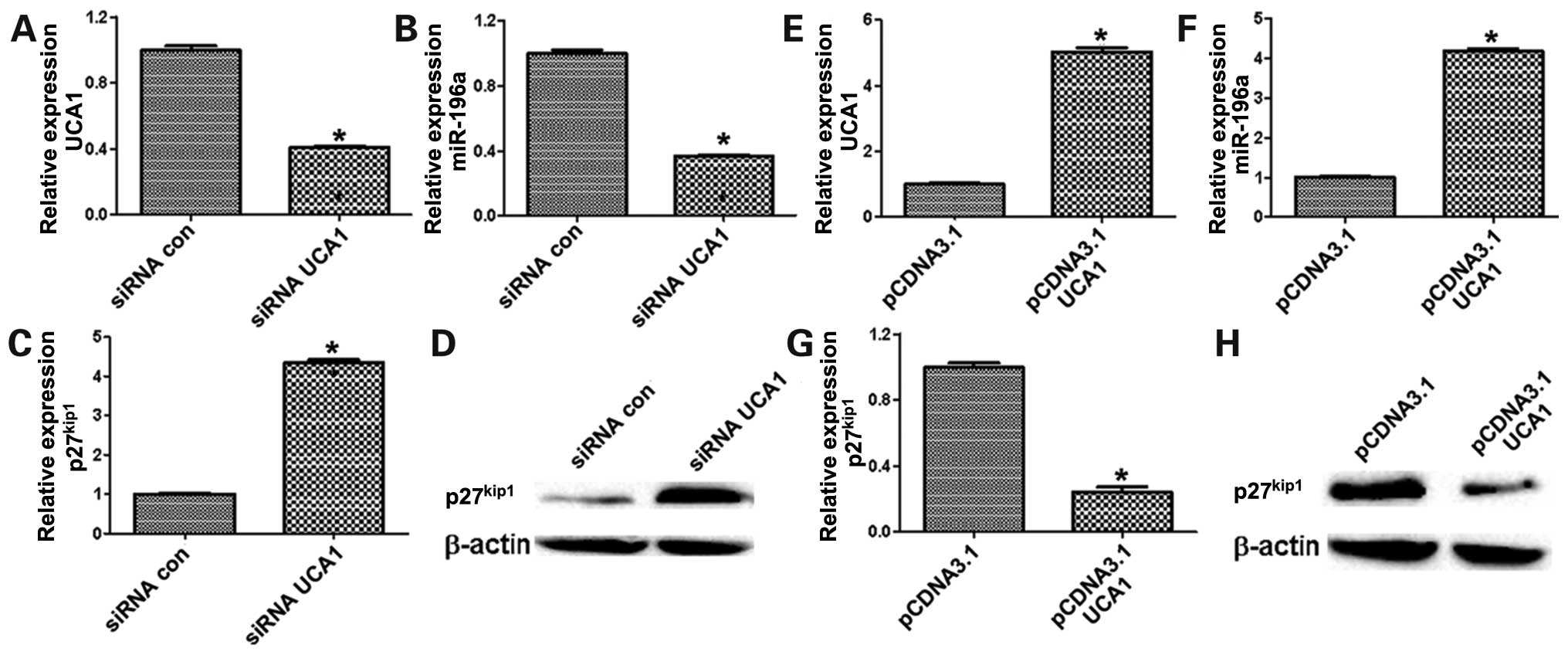
|
1
|
Volders PJ, Helsens K, Wang X, Menten B,
Martens L, Gevaert K, Vandesompele J and Mestdagh P: LNCipedia: A
database for annotated human lncRNA transcript sequences and
structures. Nucleic Acids Res. 41:D246–D251. 2013. View Article : Google Scholar :
|
|
2
|
ENCODE Project Consortium. An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, Song X, Glass CK and Rosenfeld MG:
The long arm of long noncoding RNAs: Roles as sensors regulating
gene transcriptional programs. Cold Spring Harb Perspect Biol.
3:a0037562011. View Article : Google Scholar
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thomas M, Lieberman J and Lal A:
Desperately seeking microRNA targets. Nat Struct Mol Biol.
17:1169–1174. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mattick JS: Non-coding RNAs: The
architects of eukaryotic complexity. EMBO Rep. 2:986–991. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mattick JS and Gagen MJ: The evolution of
controlled multitasked gene networks: The role of introns and other
noncoding RNAs in the development of complex organisms. Mol Biol
Evol. 18:1611–1630. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kasinski AL and Slack FJ: Epigenetics and
genetics. MicroRNAs en route to the clinic: Progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stahlhut C and Slack FJ: MicroRNAs and the
cancer phenotype: Profiling, signatures and clinical implications.
Genome Med. 5:111–122. 2013. View
Article : Google Scholar
|
|
18
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dvinge H, Git A, Gräf S, Salmon-Divon M,
Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, et al:
The shaping and functional consequences of the microRNA landscape
in breast cancer. Nature. 497:378–382. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim TM, Huang W, Park R, Park PJ and
Johnson MD: A developmental taxonomy of glioblastoma defined and
maintained by MicroRNAs. Cancer Res. 71:3387–3399. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Manterola L, Guruceaga E, Gállego
Pérez-Larraya J, González-Huarriz M, Jauregui P, Tejada S,
Diez-Valle R, Segura V, Samprón N, Barrena C, et al: A small
noncoding RNA signature found in exosomes of GBM patient serum as a
diagnostic tool. Neuro-oncol. 16:520–527. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang F, Li X, Xie X, Zhao L and Chen W:
UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma
and embryo, influencing cell growth and promoting invasion. FEBS
Lett. 582:1919–1927. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang C, Li X, Wang Y, Zhao L and Chen W:
Long non-coding RNA UCA1 regulated cell cycle distribution via CREB
through PI3-K dependent pathway in bladder carcinoma cells. Gene.
496:8–16. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Metzker ML: Sequencing technologies - the
next generation. Nat Rev Genet. 11:31–46. 2010. View Article : Google Scholar
|
|
27
|
Buermans HP, Ariyurek Y, van Ommen G, den
Dunnen JT and ‘t Hoen PA: New methods for next generation
sequencing based microRNA expression profiling. BMC Genomics.
11:716–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Anselmo A, Flori L, Jaffrezic F,
Rutigliano T, Cecere M, Cortes-Perez N, Lefèvre F, Rogel-Gaillard C
and Giuffra E: Co-expression of host and viral microRNAs in porcine
dendritic cells infected by the pseudorabies virus. PLoS One.
6:e173742011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stark MS, Tyagi S, Nancarrow DJ, Boyle GM,
Cook AL, Whiteman DC, Parsons PG, Schmidt C, Sturm RA and Hayward
NK: Characterization of the melanoma miRNAome by deep sequencing.
PLoS One. 5:e96852010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li M, Liu Y, Wang T, Guan J, Luo Z, Chen
H, Wang X, Chen L, Ma J, Mu Z, et al: Repertoire of porcine
microRNAs in adult ovary and testis by deep sequencing. Int J Biol
Sci. 7:1045–1055. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang R, Wang ZX, Yang JS, Pan X, De W and
Chen LB: MicroRNA-451 functions as a tumor suppressor in human
non-small cell lung cancer by targeting ras-related protein 14
(RAB14). Oncogene. 30:2644–2658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guan Y, Mizoguchi M, Yoshimoto K, Hata N,
Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, et al:
MiRNA-196 is upregulated in glioblastoma but not in anaplastic
astrocytoma and has prognostic significance. Clin Cancer Res.
16:4289–4297. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schimanski CC, Frerichs K, Rahman F,
Berger M, Lang H, Galle PR, Moehler M and Gockel I: High miR-196a
levels promote the oncogenic phenotype of colorectal cancer cells.
World J Gastroenterol. 15:2089–2096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luthra R, Singh RR, Luthra MG, Li YX,
Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, et al:
MicroRNA-196a targets annexin A1: A microRNA-mediated mechanism of
annexin A1 downregulation in cancers. Oncogene. 27:6667–6678. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun M, Liu XH, Li JH, Yang JS, Zhang EB,
Yin DD, Liu ZL, Zhou J, Ding Y, Li SQ, et al: MiR-196a is
upregulated in gastric cancer and promotes cell proliferation by
downregulating p27(kip1). Mol Cancer Ther. 11:842–852. 2012.
View Article : Google Scholar : PubMed/NCBI
|