Introduction
p53 is one of the most frequently mutated tumor
suppressor genes (1, 2). In response to various cellular
stresses, ATM-Chk2 cascade stabilizes p53 protein through the
phosphorylation of its N-terminal domain (3). Activated p53 functions as a
transcription factor and exerts its tumor suppressive effects such
as apoptosis, cell cycle arrest, and senescence through the
induction of its target genes (1,2). In
addition to genes related with cell proliferation, regulation of
glycolysis (4), energy metabolism,
antioxidant effect (5), autophagy
(6), and respiration with
mitochondria are reported as novel functions of p53. Thus, p53
regulates not only tumor cell growth but also pathways related with
cellular homeostasis. Since inactivation of p53 is the most common
feature of cancer cells, the elucidation of p53 signaling pathways
would contribute to the understanding of tumor cells as well as for
drug development.
Myo-inositol is water-soluble vitamin found in a
variety of food products, and are also synthesized in cells
(7). Previous studies indicated
that myo-inositol has various functions including glucose and lipid
metabolism (8,9), neurotropic effect (10), and tumor suppression (11–13).
However, the regulation of myo-inositol biosynthesis in cancer
tissues has not been disclosed yet. Through a cDNA microarray
screening using mRNAs isolated from HCT116 p53+/+
and HCT116 p53−/− cells, we identified
ISYNA1 which encodes an enzyme essential for myo-inositol
biosynthesis as a novel p53 target.
Materials and methods
cDNA microarray
Gene expression analysis was performed using
SurePrint G3 Human GE 8×60K microarray (Agilent, Santa Clara, CA,
USA) according to the manufacturer's protocol. Briefly, HCT116
p53+/+ or HCT116 p53−/− cells
were treated with 2 μg/ml of adriamycin (ADR) for 2 h and incubated
at 37°C until harvest. At 12, 24 and 48 h after treatment, total
RNA was isolated from the cells using standard protocols. Each RNA
sample was labeled and hybridized to array slides.
Cell culture and treatment
Human embryonic kidney cells HEK293T were obtained
from Riken Cell Bank. Human cancer cell lines U373MG (astrocytoma),
HepG2 (hepatocellular carcinoma), and HCT116 (colorectal
adenocarcinoma) were purchased from American Type Culture
Collection. HCT116 p53+/+ and HCT116
p53−/− cells lines were gifts from B. Vogelstein
(Johns Hopkins University, Baltimore, MD, USA). HEK293T, HCT116,
and HepG2 cells were transfected with plasmids using FuGENE 6
(Promega, Madison, WI, USA). U373 MG cells were transfected with
plasmids using FuGENE 6 or Lipofectamine LTX (Invitrogen, Carlsbad,
CA, USA). Small interfering RNA (siRNA) oligonucleotides,
commercially synthesized by Sigma Genosys, were transfected with
Lipofectamine RNAiMAX reagent (Invitrogen). Sequences of siRNA
oligonucleotides are shown in Table
I. We generated and purified replication-deficient recombinant
viruses expressing p53 (Ad-p53) or LacZ (Ad-LacZ) as described
previously (14). U373MG
(p53-mutant) cells were infected with viral solutions at
various amounts of multiplicity of infection (MOI) and incubated at
37°C until the time of harvest. For treatment with genotoxic
stress, cells were incubated with 2 μg/ml of ADR for 2 h.
 | Table ISequence of primers and
oligonucleotides. |
Table I
Sequence of primers and
oligonucleotides.
| siRNA | Sense | Antisense |
|---|
| siISYNA1-A |
GCGCUUCUGUGAGGUGAUUTT |
AAUCACCUCACAGAAGCGCTT |
| siISYNA1-B |
GCCUCAAGACCAUGUCCAUTT |
AUGGACAUGGUCUUGAGGCTT |
| siISYNA1-C |
UCAAGUCAGGCCAGACCAATT |
UUGGUCUGGCCUGACUUGATT |
| sip53 |
GACUCCAGUGGUAAUCUACTT |
GUAGAUUACCACUGGAGUCTT |
| siEGFP |
GCAGCACGACUUCUUCAAGTT |
CUUGAAGAAGUCGUGCUGCTT |
|
| Expression
vecter | Forward | Reverse |
|
| ISYNA1 isoform
1 |
CCCGATATCGCCGCGATGGAGGCCGCCGC |
CCCCTCGAGGGTGGTGGGCATTGGGGGC |
| ISYNA1 isoform
4 |
CCCGATATCCTGCCCATGGTGGCGCCC |
CCCCTCGAGGGTGGTGGGCATTGGGGGC |
|
| Quantitative
real-time PCR | Forward | Reverse |
|
| hISYNA1 |
AGTCCGTGCTTGTGGACTTC |
CCGATAGGTTCTCCCCATC |
| hGAPDH |
ACCATGGGGAAGGTGAAG |
AATGAAGGGGTCATTGATGG |
| mIsyna1 |
CCTTGGTGCTCCATAATACCTG |
AGTCTGTGCAGAAGCTCACG |
| mGapdh |
AATGTGTCCGTCGTGGATCTGA |
GATGCCTGCTTCACCACCTTCT |
|
| Gene reporter
assay | Forward | Reverse |
|
| RE1 |
CCCCTCGAGTGTATTGAGACGGGGTTTCC |
CCCAAGCTTCACCCAGTCTGCTCCCTTTAAG |
| RE2 |
CCCCTCGAGGCCTTCCTCAATGGGTCTC |
CCCAAGCTTTCACGATGGACATGGTCTGTG |
| mRE |
CCCCTCGAGATACCCAAGTGCTGGAGGTG |
CCCAAGCTTACCTTGTGTGTGGTCCCTTC |
|
| Chromatin
immunoprecipitation assay | Forward | Reverse |
|
| RE2 |
GATGACTTCAAGTCAGGCCAGAC |
CCCACGCACCTTGAGGCCGG |
|
| Mutagenesis | Forward | Reverse |
|
| ISYNA1 isoform
2 |
AGGCCAACTACTACGGCTCGCTGA |
CCTTGAGAACGCCACCCTCGCGGC |
| RE1mt |
CACTGTTCCTGGCTGACTGCCTATTTTTCG |
GCTAATACCTGTAATCCTAGCACTTTGGGAG |
| RE2mt |
GTGTTTTTGGACTTCCTCATTGGCTCCGGC |
GGAATTAACTTTGGTCTGGCCTGACTTGAAG |
| mREmt |
GGGTTATTTACCACCACTGATGCCGTGACC
AGGAATACTGGCCCTGTACACCCGTGCTTG |
Plasmid construction
The entire coding sequence of ISYNA1 isoforms
1 and 4 were amplified by PCR using KOD-Plus DNA polymerase
(Toyobo, Osaka, Japan), and inserted into the EcoRV and
XhoI sites of pCAGGS vector. ISYNA1 isoform 2 expression
vector was constructed by site-directed mutagenesis using ISYNA1
isoform 1 as a template. The construct was confirmed by DNA
sequence analysis. Primers are shown in Table I.
Quantitative real-time PCR
Total RNA was isolated from human cells and mouse
tissues using RNeasy Plus Mini kits (Qiagen, Valencia, CA, USA) and
RNeasy Plus Universal Mini kits (Qiagen) according to the
manufacturer's instructions. Complementary DNAs were synthesized
using Super Script III reverse transcriptase (Invitrogen).
Quantitative real-time PCR (qPCR) was conducted using SYBR Green
Master Mix on a Light Cycler 480 (Roche, Basel, Switzerland).
Primer sequences are shown in Table
I.
Western blot analysis
To prepare whole cell extracts, cells were collected
and lysed in chilled RIPA buffer (50 mmol/l Tris-HCl at pH 8.0, 150
mmol/l NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% NP40)
containing 1 mM phenyl-methylsulphonyl fluoride (PMSF), 0.1 mM DTT
and 0.1% Calbiochem Protease Inhibitor Cocktail Set III, EDTA-Free
(EMD Chemicals Inc., Merck KGaA, Darmstadt, Germany). Samples were
sonicated for 15 min with a 30 sec on/30 sec off cycle using
Bioruptor UCD-200 (Cosmobio, Tokyo, Japan). After centrifugation at
16,000 × g for 15 min, supernatants were collected and boiled in
SDS sample buffer (Bio-Rad, Hercules, CA, USA). SDS-polyacrylamide
gel electrophoresis (SDS-PAGE) was performed for each sample, and
the proteins were then transferred to a nitrocellulose membrane
(Hybond™ ECL™, Amersham, Piscataway, NJ, USA). Protein bands on
western blots were visualized by chemiluminescent detection (ECL,
Amersham and Immobilon, Millipore). Anti-β-actin monoclonal
antibody (AC-15) was purchased from Abcam (Cambridge, UK).
Anti-ISYNA1 monoclonal antibody (sc-271830) and anti-p53 monoclonal
antibody were purchased from Santa Cruz Biotechnology (Santa Cruz,
CA, USA). Anti-p21WAF1 monoclonal antibody (OP64) was
purchased from Merck Millipore (Darmstadt, Germany).
Immunocytochemical analyses
Cells were seeded on coverslips in 24-well plates.
After each treatment indicated in the text, cells were washed in
phosphate-buffered saline (PBS) before fixation in 4%
paraformaldehyde. Cells were immunostained overnight with primary
antibodies followed by incubation with Alexa Fluor 488-conjugated
secondary IgG (Molecular Probes) for 1 h. Cells were subjected to
4′-6-diamidino-2-phenylindole (DAPI) staining to visualize cell
nuclei. Immunofluorescence was visualized and recorded on an
Olympus FV1000D laser confocal microscope. Images were processed
using Olympus FV10-ASW software and Adobe Photoshop CS3.
Gene reporter assay
DNA fragments, including the potential p53-response
elements (REs), were amplified and subcloned into the pGL4.24
vector (Promega). Point mutations ‘T’ were inserted at the 4th and
the 14th nucleotide ‘C’ and the 7th and the 17th nucleotide ‘G’ of
each RE by site-directed mutagenesis. Reporter assays were
performed using the Dual Luciferase assay system (Promega) as
described previously (15).
Primers for amplification and mutagenesis are shown in Table I.
Chromatin immunoprecipitation (ChIP)
assay
ChIP assay was performed using EZ-Magna ChIP G
Chromatin Immunoprecipitation kit (Merck Millipore, Darmstadt,
Germany) following the manufacturer's protocol. In brief, U373MG
cells infected with Ad-p53- or Ad-LacZ at a MOI of 10 were
cross-linked with 1% formaldehyde for 10 min, washed with PBS, and
lysed in nuclear lysis buffer. The lysate was then sonicated using
Bioruptor UCD-200 (CosmoBio) to shear DNA to ~200–1,000 bp.
Supernatant from 1×106 cells was used for each
immunoprecipitation with anti-p53 antibody (OP140, Merck Millipore)
or normal mouse IgG (sc-2025, Santa Cruz). Column-purified DNA was
quantified by qPCR. Primer sequences are shown in Table I.
Myo-inositol (MI) assay
To prepare cell homogenate, cells were collected and
suspended in PBS. Samples were sonicated for 15 min with a 30 sec
on/30 sec off cycle using Bioruptor UCD-200 (Cosmobio). After
centrifugation at 16,000 × g for 5 min, myo-inositol content in
supernatants was measured using myo-inositol assay kit (K-INOSL,
Megazyme International Ireland, Bray, Wicklow, Ireland) according
to the manufacturer's instructions.
Colony formation assay
HCT116 cells and HepG2 cells were seeded on 6-well
flat bottomed microplates. At 24 h after seeding, cells were
transfected with pCAGGS (mock) vector or pCAGGS/ISYNA1 isoform 1.
HCT116 and HepG2 cells were cultured with 0.5 or 1.2 mg/ml of G418,
respectively. After 2 or 3 weeks of drug selection, colonies were
washed in phosphate-buffered saline and stained with 0.1% crystal
violet for 24 h.
ATP assay
HCT116 p53+/+ cells were
transfected with siRNAs and seeded on 24-well plates. At 24 h after
transfection, cells were treated with 2 μg/ml of ADR for 2 h. At 48
h after ADR treatment, cell viability was evaluated by Cell
Titer-Glo Luminescent Cell Viability assay (Promega). After removal
of culture medium, cells were incubated with 100 μl of Cell
Titer-Glo reagent and 100 μl of culture medium for 10 min and
lysed. The luminescence of cell lysate was measured by ARVO X3
plate reader (Perkin-Elmer, Waltham, MA, USA) according to the
manufacturer's protocol.
Animal models
p53−/− mice were provided by RIKEN
BioResource Center (Ibaraki, Japan) (16). The mice were maintained under
specific pathogen-free conditions and were handled in accordance
with the Guidelines for Animal Experiments of the University of
Tokyo. p53+/+ and p53−/− mice
at 6 weeks of age were irradiated with 10 Gy of X-ray. At 24 h
after irradiation, mice were sacrificed for liver extraction.
Database analysis
ISYNA1 expression and p53 mutation status in
clinical samples were obtained from the TCGA project via data
portal on 15 May 2015 (17). The
association between ISYNA1 expression and the presence of
the p53 gene mutation was determined by using the Student's
t-test.
Results
p53 regulates genes related with
myo-inositol metabolism
To screen novel p53 target genes, we conducted cDNA
micro-array analysis using mRNAs isolated from HCT116
p53+/+ and HCT116 p53−/− cells
that were treated with 2 μg/ml of adriamycin (ADR). Fig. 1A shows a schematic representation
of inositol phosphate metabolism pathway. The result of cDNA
microarray analysis indicated that five genes related with
myo-inositol metabolism were induced by p53 (Fig. 1B). We selected inositol 3-phosphate
synthase (ISYNA1) for further analysis, because
ISYNA1 showed the highest expression among the five
genes.
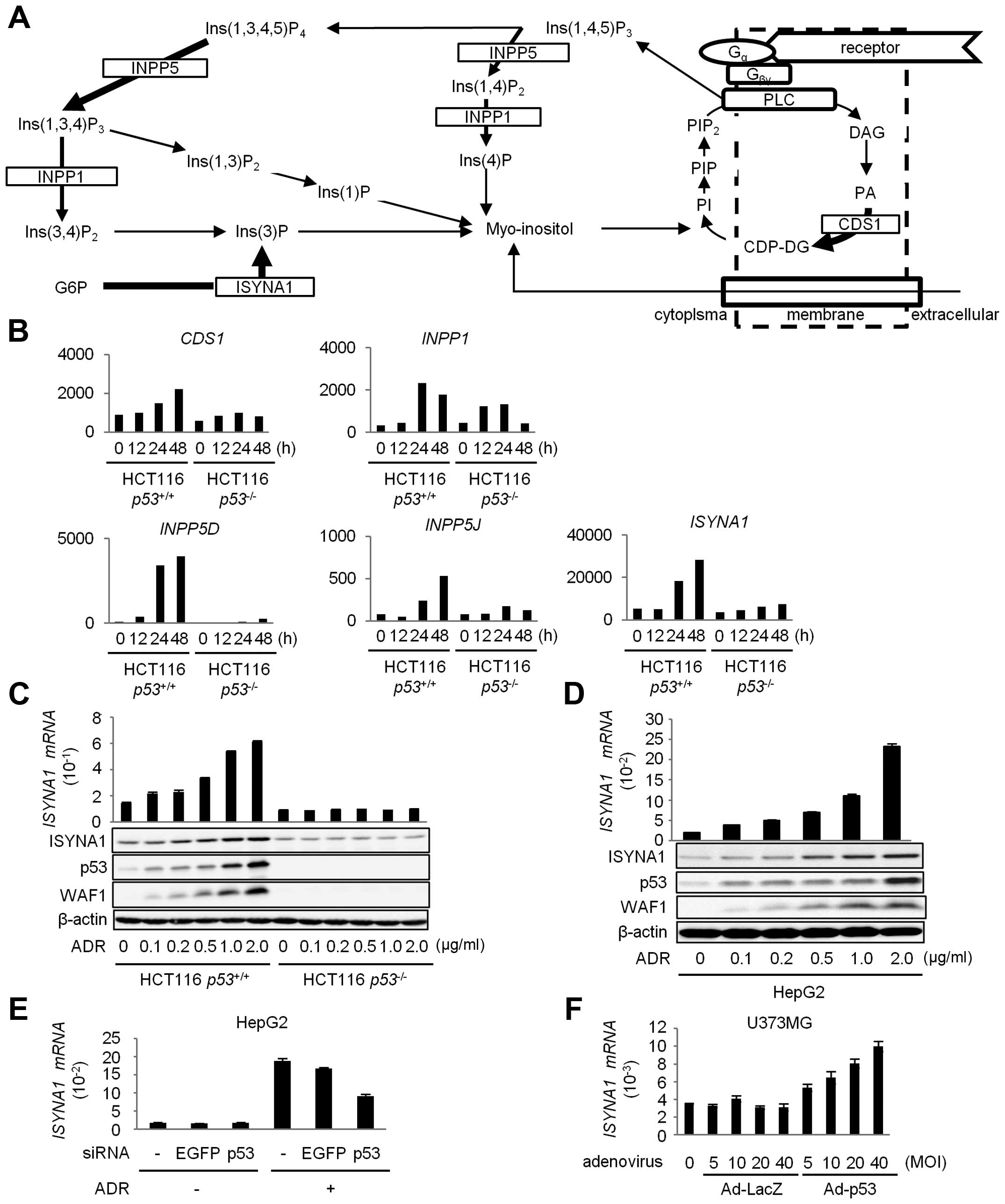 | Figure 1Regulation of ISYNA1 by p53. (A)
Schematic representation of the inositol phosphate metabolism
pathway. PI, phosphatidylinositol; PIP, phosphatidylinositol
4-phosphate; PIP2, phosphatidylinositol
4,5-bisphosphate; Ins(1,4,5)P3, inositol
1,4,5-trisphosphate; Ins(1,4)P2, inositol
1,4-bisphosphate; Ins(4) P, inositol 4-phosphate;
Ins(1,3,4,5)P4, inositol 1,3,4,5-tetrakisphosphate;
Ins(1,3,4)P3, inositol 1,3,4-trisphosphate;
Ins(3,4)P2, inositol 3,4-phosphtate; Ins(3)P, inositol
3-phosphate; Ins(1,3)P2, inositol 1,3-bisphosphate;
Ins(1)P, inositol 1-phosphate; G6P, glucose 6-phosphate; DAG,
diacylglycerol; PA, phosphatidate; CDP-DAG, CDP-diacylglycerol. (B)
Induction of genes related with myo-inositol biosynthesis by p53.
HCT116 p53+/+ and HCT116 p53−/−
cells were treated with 2 μg/ml of adriamycin (ADR) for 2 h. mRNAs
isolated from these cells were subjected to microarray analysis.
Five genes related with inositol phosphate metabolism were shown to
be induced by p53. (C) qPCR analysis (upper) and western blotting
(lower) of ISYNA1, p53, and WAF1 in HCT116 p53+/+
and HCT116 p53−/− cells at 36 h after treatment
with ADR for 2 h. Glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) and β-actin were used for the normalization of
expression levels. Error bars represent SD (n=3). (D) qPCR analysis
(upper) and western blotting (lower) of ISYNA1, p53, and WAF1 in
HepG2 cells at 36 h after treatment with ADR for 2 h. GAPDH
and β-actin were used for the normalization of expression levels.
Error bars represent SD (n=3). (E and F) qPCR analysis of
ISYNA1 mRNA in HepG2 (E) or U373MG (F) cells. At 24 h after
transfection of each siRNA, HepG2 cells were treated with 2 μg/ml
of ADR for 2 h. At 40 h after treatment, cells were harvested for
qPCR analysis. U373MG cells were harvested at 36 h after infection
with Ad-p53. siEGFP or Ad-LacZ were used as controls. GAPDH
was used for the normalization of expression levels. Error bars
represent SD (n=3). |
To validate the result of cDNA microarray analysis,
we performed quantitative real-time PCR (qPCR) analysis and western
blotting of ISYNA1 using HCT116 p53+/+ and HCT116
p53−/− cells treated with ADR. As a result, we
found dose-dependent induction of ISYNA1 mRNA and protein only in
HCT116 p53+/+ cells in response to ADR treatment
(Fig. 1C). We also confirmed the
induction of ISYNA1 mRNA and protein by ADR treatment in HepG2
(Fig. 1D). Moreover, transfection
with siRNA against p53 remarkably inhibited the induction of
ISYNA1 (Fig. 1E).
p53-mediated induction of ISYNA1 was also observed in U373MG
glioblastoma cells that were infected with adenovirus designed to
express wild-type p53 (Ad-p53) (Fig.
1F). These results clearly indicated that ISYNA1 was
regulated by p53.
Expression and subcellular localization
of ISYNA1
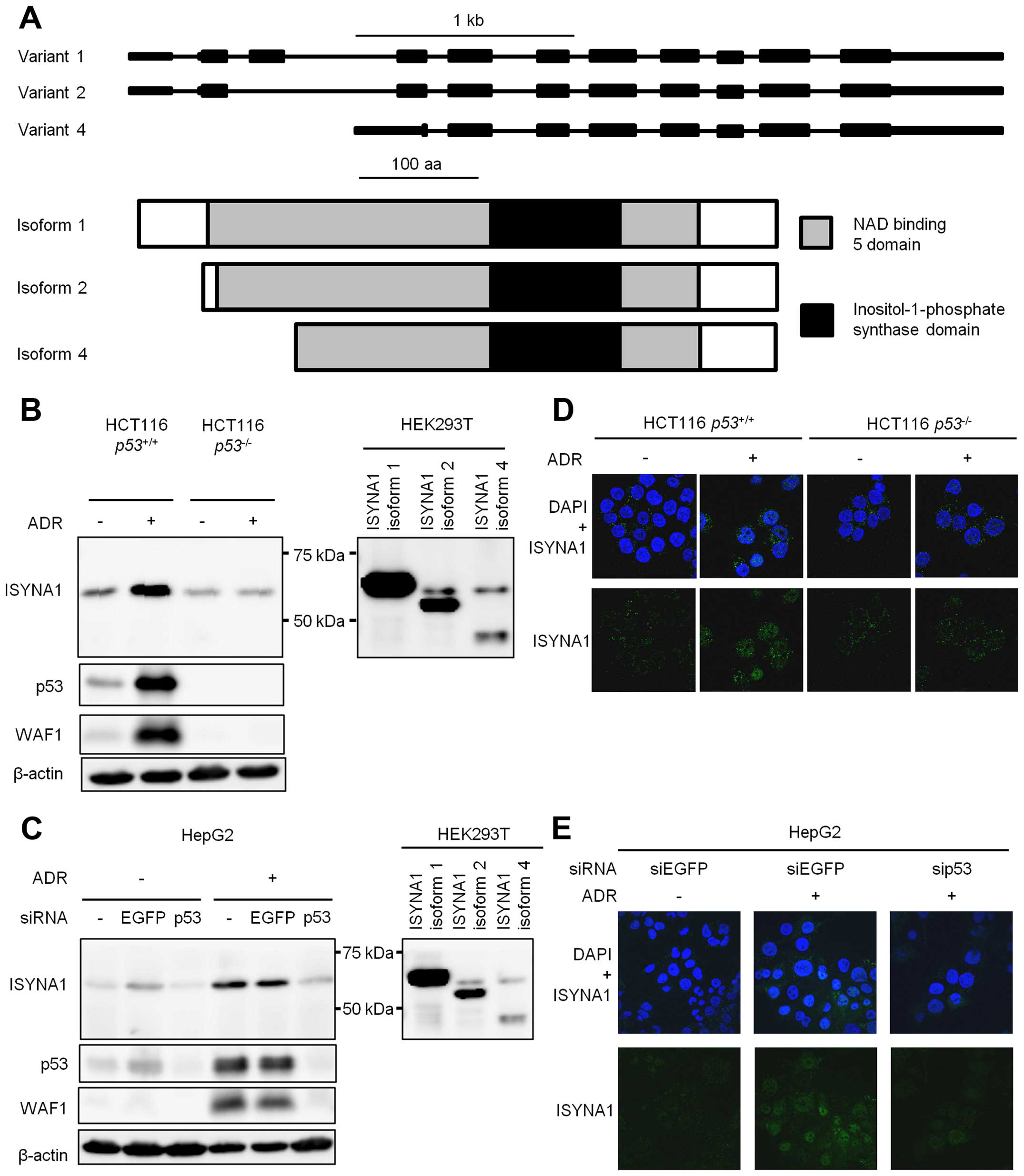
There are three major variants of human ISYNA1,
namely isoform 1, 2, and 4. All isoforms are similar in domain
structure as shown in Fig. 2A. We
constructed plasmids expressing each isoform. Result of western
blotting indicated that isoform 1 is the major ISYNA1 isoform that
was expressed in HCT116 and HepG2 cells treated with ADR (Fig. 2B and C).
Then we performed immunocytochemical analysis using
HCT116 p53+/+, HCT116 p53−/−
cells, or HepG2 cells (Fig. 2D and
E). ADR treatment increased ISYNA1 protein in the cytoplasm and
the nucleus of HCT116 p53+/+ and HepG2 cells, but
ISYNA1 expression was very low in HCT116 p53−/−
cells or HepG2 cells treated with sip53.
Identification of ISYNA1 as a novel p53
target
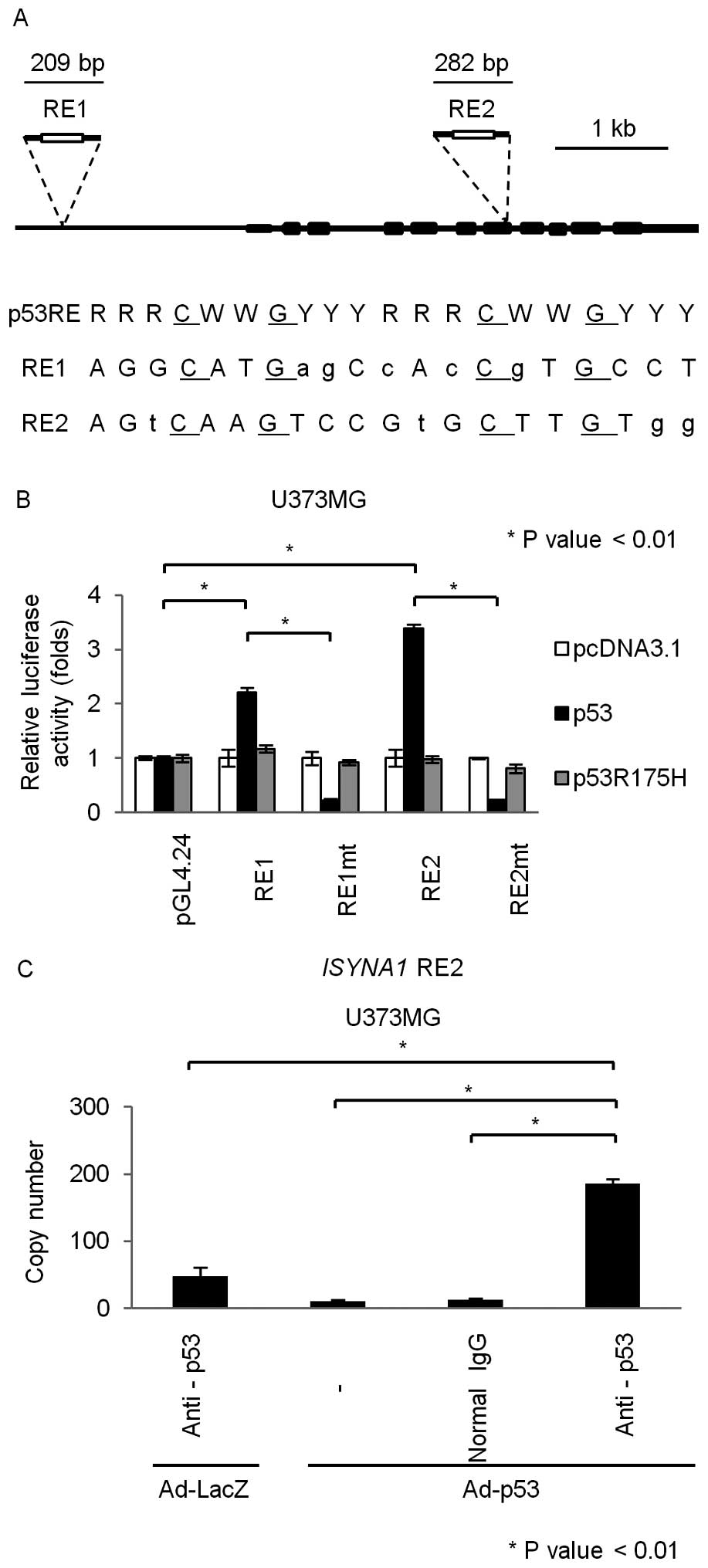
To investigate whether ISYNA1 is a direct target of
p53, we searched for p53 response element (RE) (18) within the ISYNA1 genomic region
which is located on chromosome 19p13. We found putative p53 RE in
the promoter region (RE1) and the seventh exon (RE2) (Fig. 3A). We subcloned DNA fragments
including the RE1 or RE2 into pGL4.24 vector (pGL4.24/RE1 and
pGL4.24/RE2) and performed gene reporter assay using U373MG cells.
As a result, U373MG cells transfected with pGL4.24/RE1 or
pGL4.24/RE2 showed enhanced luciferase activity only in the
presence of plasmid expressing wild-type p53 (Fig. 3B). In addition, base substitutions
within the RE1 and RE2 (pGL4.24/RE1mt and pGL4.24/RE2mt) completely
abolished the enhancement of luciferase activity (Fig. 3B). To investigate whether p53 could
directly bind to RE2 which showed higher transcriptional
activatity, we performed chromatin immunoprecipitation (ChIP) assay
using U373MG cells that were infected with Ad-p53 or Ad-LacZ. qPCR
analysis of the immunoprecipitated DNA indicated that the p53
protein bound to the genomic fragment that included the RE2
(Fig. 3C). Taken together, p53
directly regulated ISYNA1 expression through binding to the
RE2 in the seventh exon.
Growth suppressive effect of ISYNA1
ISYNA1 is the rate-limiting enzyme of myo-inositol
de novo synthesis (7) which
is conserved among eukaryotes (19–25).
To evaluate the biosynthesis of myo-inositol by ISYNA1, we
performed myo-inositol (MI) assay using HEK293T cells that were
transfected with mock or plasmid expressing mock or ISYNA1 isoform
1 (Fig. 4A). The results showed
that intracellular myo-inositol content in cells expressing ISYNA1
isoform 1 was significantly higher than those in control cells. In
addition, DNA damage significantly increased intracellular
myo-inositol content in HCT116 p53+/+ cells, but
did not affect the myo-inositol content in HCT116
p53−/− cells (Fig.
4B). Thus, our results indicated that p53 could regulate
intracellular myo-inositol levels in response to DNA damage.
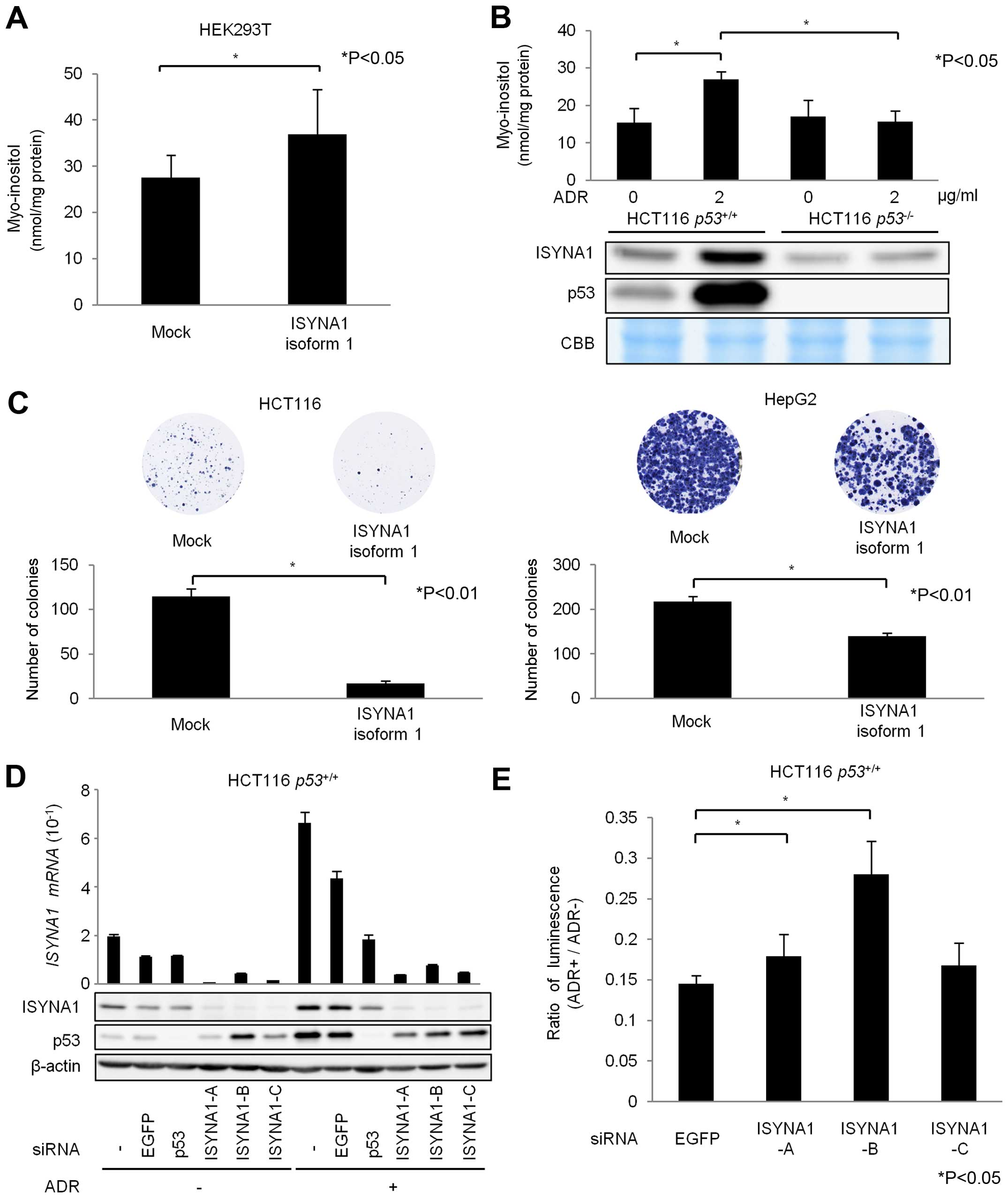 | Figure 4Regulation of myo-inositol synthesis
and cell growth by p53-ISYNA1 pathway. (A) At 36 h after
transfection with mock vector or plasmid expressing ISYNA1 isoform
1, the amounts of myo-inositol were evaluated. Total protein
content was used for normalization. Error bars, SD (n=3). (B)
Upper, myo-inositol assay at 36 h after treatment with 2 μg/ml of
ADR in HCT116 p53+/+ and HCT116
p53−/− cells. Total protein content was used for
normalization. Error bars, SD (n=3). Lower, expression of ISYNA1
and p53 protein. (C) HCT116 and HepG2 cells were transfected with
mock or plasmid expressing ISYNA1 isoform 1. The number of colonies
was quantified by ImageJ software. Error bar, SD (n=3). (D) At 24 h
after transfection of each siRNA, HCT116 p53+/+
cells were treated with 2 μg/ml of ADR for 2 h. At 48 h after
treatment, qPCR (upper) and western blot (lower) analyses were
performed to evaluate the expression of ISYNA1 and p53. siEGFP was
used as a control. GAPDH and β-actin were used for the
normalization of expression levels. Error bars represent SD (n=3).
(E) At 24 h after transfection of each siRNA, HCT116
p53+/+ cells were treated with 2 μg/ml of ADR for
2 h. At 48 h after treatment, ATP assay was performed. Relative
cell viability was calculated by dividing the luminescence of
ADR-treated cells by that of untreated cells. Error bars represent
SD (n=3). |
We also evaluated the effect of p53-ISYNA1 pathway
on cancer cell growth. The result of colony formation assay using
HCT116 and HepG2 cells indicated that ISYNA1 overexpression
suppressed cell proliferation (Fig.
4C). We then designed three siRNAs (siA, siB and siC) and found
that siRNAs effectively suppressed ISYNA1 mRNA and protein
(Fig. 4D). We performed ATP assay
using HCT116 p53+/+ cells and found that
ISYNA1-silencing caused resistance to ADR treatment (Fig. 4E). These results indicated ISYNA1
is likely to be one of the key mediators of p53 induced growth
suppression.
Regulation of ISYNA1 by p53 in vivo
Since ISYNA1 is conserved among eukaryotes, we
investigated whether mouse Isyna1 is also regulated by p53. p53
wild-type or p53 knockout mice at 6 weeks of age were irradiated
with 10 Gy of X-ray. At 24 h after irradiation, we isolated total
RNA from liver tissues. qPCR analysis revealed that mouse
Isyna1 mRNA was induced by DNA damage only in p53 wild-type
mice (Fig. 5A). Screening of p53
RE within Isyna1 genomic region identified a putative RE
(mRE) at ~10 kb upstream of the Isyna1 gene (Fig. 5B). We subcloned a DNA fragment
including mRE into the pGL4.24 vector (pGL4.24/mRE) and performed
gene reporter assay using U373MG cells (Fig. 5C). Luciferase activity was strongly
enhanced by co-transfection with wild-type p53, but not with mutant
p53. In addition, base substitutions within mRE diminished the
enhancement of luciferase activity, demonstrating regulation of
Isyna1 by p53 through mRE.
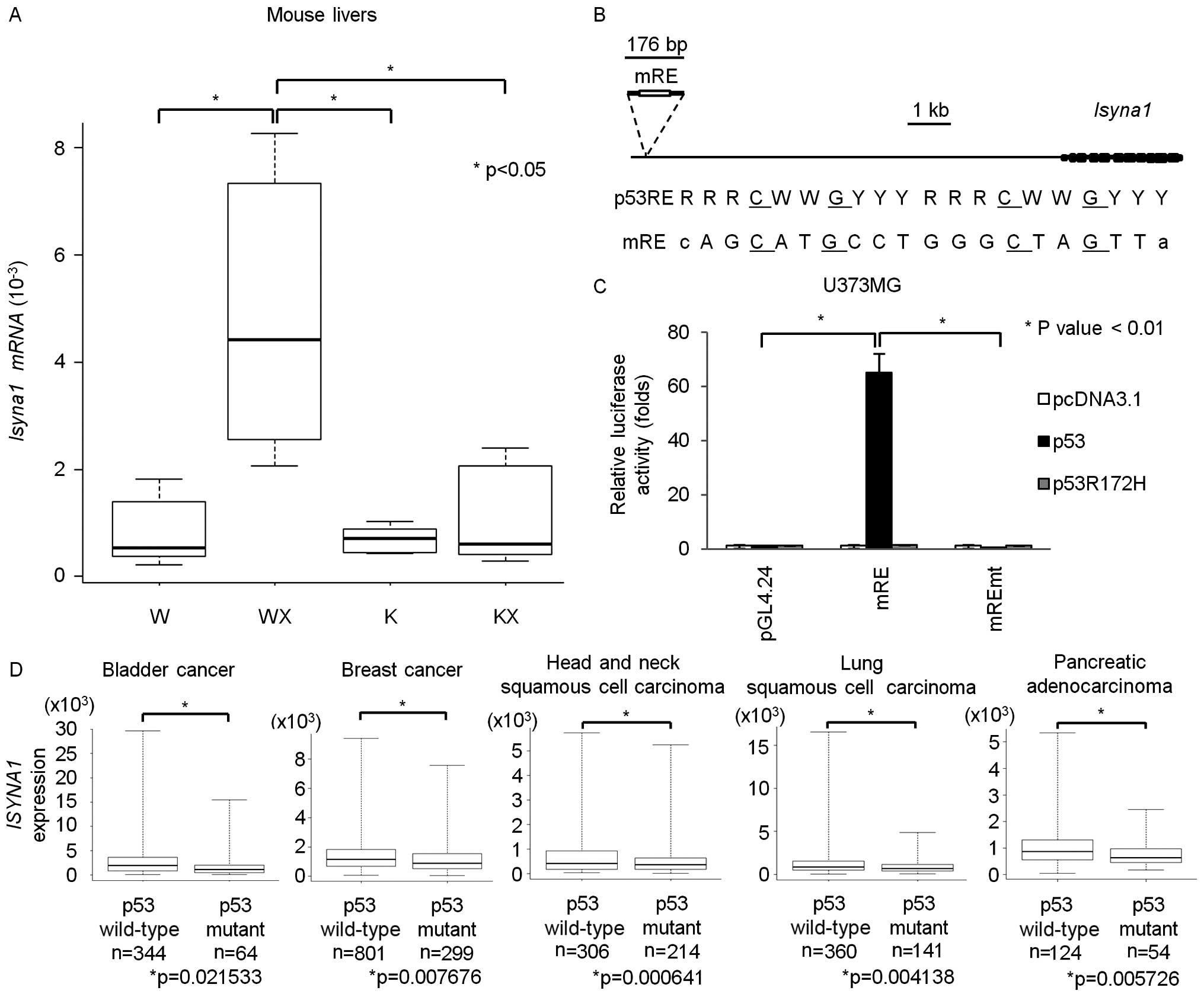 | Figure 5Regulation of ISYNA1 by p53 in
vivo. (A) qPCR analysis of Isyna1 in mouse livers. Mice
were divided into four groups; p53 wild-type mice without
irradiation (W), p53 wild-type mice with irradiation (WX),
p53 knockout mice without irradiation (K), p53
knockout mice with irradiation (KX) (n=6 per group).
Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used
for the normalization of expression level. Top bar represents
maximum observation, lower bar represents minimum observation, the
top side of the box represents the third quartile, and the bottom
side, the first quartile. The middle bar represents the median
value. The P-value was calculated by Student's t-test. (B) Upper,
genomic structure of mouse Isyna1. Black boxes indicate the
locations and relative sizes of exons. The white box indicates the
location of the p53 response element (mRE). Lower, comparison of
mRE to the consensus p53RE. R, purine; W, A or T; Y pyrimidine. (C)
Luciferase assay of mRE in U373MG with or without mutation of the
RE. Luciferase activity is indicated relative to the activity of
the mock vectors. The plasmid expressing mouse p53 carrying a
missense mutation (R172H) served as a negative control. Error bars
represent the SD (n=3). (D) Box plot of ISYNA1 expression in
bladder cancer, breast cancer, head and neck squamous cell
carcinoma, lung squamous cell carcinoma, and pancreatic
adenocarcinoma tissues from the TCGA database. The vertical axis
indicates the normalized expression level of ISYNA1, top bar
represents maximum observation, lower bar represents minimum
observation, the top side of the box represents the third quartile,
and the bottom side, the first quartile. The middle bar represents
the median value. The P-value was calculated by Student's
t-test. |
We also analyzed whether p53 regulates ISYNA1
in human cancer tissues. Correlation between p53 mutation
and ISYNA1 expression was analyzed by using omics data of
various tumor tissues released from the TCGA database (17). Interestingly, ISYNA1 mRNA
expression in bladder cancer, breast cancer, head and neck squamous
cell carcinoma, lung squamous cell carcinoma, and pancreatic
adenocarcinoma was significantly decreased in tumor tissues with
p53 mutation compared with those without p53 mutation
(Fig. 5D). These findings indicate
that p53 regulates ISYNA1 expression in vivo.
Discussion
We identified ISYNA1 as a novel p53 target. ISYNA1
is a key enzyme which affects myo-inositol de novo synthesis
(7,26,27).
In addition, p53 induced INPP1 and INPP5 (28) that are involved in myo-inositol
salvage pathway. Myo-inositol is one of the chemical compounds
which is essential for living organisms (29), and myo-inositol depletion affects
cell survival and growth (30).
Myo-inositol was also reported to suppress tumor growth in
vitro and in vivo (31–38).
Previous studies indicated that myo-inositol suppresses
phosphorylation of Akt and Erk by inhibiting PI3K activity
(12,13). p53 was also shown to suppress
PI3K-Akt pathway by inducing PTEN (39) and Phlda3 (40). Our results suggested a novel
mechanism whereby p53 negatively regulates PI3K-Akt pathway by
inducing ISYNA1.
Epidemiological studies indicate that myo-inositol
prevents progression of dysplasia in smokers (11–13),
and decreases tumorigenesis in chronic hepatitis patients (33). These findings suggested that p53
would suppress tumorigenesis by inducing biosynthesis of
myo-inositol. We also found that ISYNA1 was induced in mouse liver
tissue by DNA damage. To evaluate the chemopreventive effect of
myo-inositol, we fed p53 knockout mice with myo-inositol in
drinking water. However, oral myo-inositol did not suppress tumor
development (data not shown). Although, myo-inositol was shown to
suppress liver cancer (32,33),
liver cancer is relatively rare for p53 knockout mice compared with
lymphoma of thymus or spleen (41). In addition, although induction of
Isyna1 was observed in liver tissues, Isyna1 was not
induced in thymus and spleen (data not shown). Therefore, to
evaluate the chemopreventive effect of myo-inositol or ISYNA1 in
vivo, liver cancer model would be appropriate.
Taken together, ISYNA1 was shown to be a mediator of
p53-dependent growth suppression, and ISYNA1 expression was
reduced in several types of cancers with p53 mutations. Therefore,
myo-inositol could be a potential anticancer agent for cancer cells
with p53 mutation. Our findings revealed a novel role of p53 in
myo-inositol biosynthesis which could be a possible therapeutic
target.
Acknowledgements
We thank Satomi Takahashi and Misato Oshima for
technical assistance. We also thank The Cancer Genome Atlas (TCGA)
project and members of the Cancer Genomics Hub (CGHub) for making
all TCGA data publicly accessible. This study was supported
partially by grant from Japan Society for the Promotion of Science
and Ministry of Education, Culture, Sports, Science and Technology
of Japan to K.M and C.T., grant from Japan Agency for medical
Research and Development to K.M. and C. T., grant from the Ministry
of Health, Labour and Welfare, Japan to K.M., and grant in-Aid from
the Tokyo Biochemical Research Foundation to K.M.
References
|
1
|
Brady CA and Attardi LD: p53 at a glance.
J Cell Sci. 123:2527–2532. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Levine AJ and Oren M: The first 30 years
of p53: Growing ever more complex. Nat Rev Cancer. 9:749–758. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirao A, Kong YY, Matsuoka S, Wakeham A,
Ruland J, Yoshida H, Liu D, Elledge SJ and Mak TW: DNA
damage-induced activation of p53 by the checkpoint kinase Chk2.
Science. 287:1824–1827. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bensaad K, Tsuruta A, Selak MA, Vidal MN,
Nakano K, Bartrons R, Gottlieb E and Vousden KH: TIGAR, a
p53-inducible regulator of glycolysis and apoptosis. Cell.
126:107–120. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu W, Zhang C, Wu R, Sun Y, Levine A and
Feng Z: Glutaminase 2, a novel p53 target gene regulating energy
metabolism and antioxidant function. Proc Natl Acad Sci USA.
107:7455–7460. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kenzelmann Broz D, Spano Mello S, Bieging
KT, Jiang D, Dusek RL, Brady CA, Sidow A and Attardi LD: Global
genomic profiling reveals an extensive p53-regulated autophagy
program contributing to key p53 responses. Genes Dev. 27:1016–1031.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ju S, Shaltiel G, Shamir A, Agam G and
Greenberg ML: Human 1-D-myo-inositol-3-phosphate synthase is
functional in yeast. J Biol Chem. 279:21759–21765. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Croze ML and Soulage CO: Potential role
and therapeutic interests of myo-inositol in metabolic diseases.
Biochimie. 95:1811–1827. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maeba R, Hara H, Ishikawa H, Hayashi S,
Yoshimura N, Kusano J, Takeoka Y, Yasuda D, Okazaki T, Kinoshita M,
et al: Myo-inositol treatment increases serum plasmalogens and
decreases small dense LDL, particularly in hyperlipidemic subjects
with metabolic syndrome. J Nutr Sci Vitaminol (Tokyo). 54:196–202.
2008. View Article : Google Scholar
|
|
10
|
Mukai T, Kishi T, Matsuda Y and Iwata N: A
meta-analysis of inositol for depression and anxiety disorders. Hum
Psychopharmacol. 29:55–63. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lam S, McWilliams A, LeRiche J, MacAulay
C, Wattenberg L and Szabo E: A phase I study of myo-inositol for
lung cancer chemoprevention. Cancer Epidemiol Biomarkers Prev.
15:1526–1531. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han W, Gills JJ, Memmott RM, Lam S and
Dennis PA: The chemopreventive agent myoinositol inhibits Akt and
extracellular signal-regulated kinase in bronchial lesions from
heavy smokers. Cancer Prev Res (Phila). 2:370–376. 2009. View Article : Google Scholar
|
|
13
|
Gustafson AM, Soldi R, Anderlind C,
Scholand MB, Qian J, Zhang X, Cooper K, Walker D, McWilliams A, Liu
G, et al: Airway PI3K pathway activation is an early and reversible
event in lung cancer development. Sci Transl Med.
2:26ra252010.PubMed/NCBI
|
|
14
|
Oda K, Arakawa H, Tanaka T, Matsuda K,
Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et
al: p53AIP1, a potential mediator of p53-dependent apoptosis, and
its regulation by Ser-46-phosphorylated p53. Cell. 102:849–862.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tanikawa C, Matsuda K, Fukuda S, Nakamura
Y and Arakawa H: p53RDL1 regulates p53-dependent apoptosis. Nat
Cell Biol. 5:216–223. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsukada T, Tomooka Y, Takai S, Ueda Y,
Nishikawa S, Yagi T, Tokunaga T, Takeda N, Suda Y, Abe S, et al:
Enhanced proliferative potential in culture of cells from
p53-deficient mice. Oncogene. 8:3313–3322. 1993.PubMed/NCBI
|
|
17
|
Taura M, Eguma A, Suico MA, Shuto T, Koga
T, Komatsu K, Komune T, Sato T, Saya H, Li JD, et al: p53 regulates
Toll-like receptor 3 expression and function in human epithelial
cell lines. Mol Cell Biol. 28:6557–6567. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
el-Deiry WS, Kern SE, Pietenpol JA,
Kinzler KW and Vogelstein B: Definition of a consensus binding site
for p53. Nat Genet. 1:45–49. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Michell RH: Inositol derivatives:
Evolution and functions. Nat Rev Mol Cell Biol. 9:151–161. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Seelan RS, Lakshmanan J, Casanova MF and
Parthasarathy RN: Identification of myo-inositol-3-phosphate
synthase isoforms: Characterization, expression, and putative role
of a 16-kDa gamma(c) isoform. J Biol Chem. 284:9443–9457. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Konarzewska P, Esposito M and Shen CH:
INO1 induction requires chromatin remodelers Ino80p and Snf2p but
not the histone acetylases. Biochem Biophys Res Commun.
418:483–488. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Valluru R and Van den Ende W: Myo-inositol
and beyond - emerging networks under stress. Plant Sci.
181:387–400. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deranieh RM, He Q, Caruso JA and Greenberg
ML: Phosphorylation regulates myo-inositol-3-phosphate synthase: A
novel regulatory mechanism of inositol biosynthesis. J Biol Chem.
288:26822–26833. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Parthasarathy RN, Lakshmanan J, Thangavel
M, Seelan RS, Stagner JI, Janckila AJ, Vadnal RE, Casanova MF and
Parthasarathy LK: Rat brain myo-inositol 3-phosphate synthase is a
phosphoprotein. Mol Cell Biochem. 378:83–89. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Henry SA, Gaspar ML and Jesch SA: The
response to inositol: Regulation of glycerolipid metabolism and
stress response signaling in yeast. Chem Phys Lipids. 180:23–43.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guan G, Dai P and Shechter I: cDNA cloning
and gene expression analysis of human myo-inositol 1-phosphate
synthase. Arch Biochem Biophys. 417:251–259. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chauvin TR and Griswold MD:
Characterization of the expression and regulation of genes
necessary for myo-inositol biosynthesis and transport in the
seminiferous epithelium. Biol Reprod. 70:744–751. 2004. View Article : Google Scholar
|
|
28
|
Ye Y, Jin L, Wilmott JS, Hu WL, Yosufi B,
Thorne RF, Liu T, Rizos H, Yan XG, Dong L, et al: PI(4,5)P2
5-phosphatase A regulates PI3K/Akt signalling and has a tumour
suppressive role in human melanoma. Nat Commun. 4:15082013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ohnishi T, Murata T, Watanabe A, Hida A,
Ohba H, Iwayama Y, Mishima K, Gondo Y and Yoshikawa T: Defective
craniofacial development and brain function in a mouse model for
depletion of intracellular inositol synthesis. J Biol Chem.
289:10785–10796. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eagle H, Oyama VI, Levy M and Freeman AE:
Myo-inositol as an essential growth factor for normal and malignant
human cells in tissue culture. J Biol Chem. 226:191–205.
1957.PubMed/NCBI
|
|
31
|
Hecht SS, Upadhyaya P, Wang M, Bliss RL,
McIntee EJ and Kenney PM: Inhibition of lung tumorigenesis in A/J
mice by N-acetyl-S-(N-2-phenethylthiocarbamoyl)-L-cysteine and
myo-inositol, individually and in combination. Carcinogenesis.
23:1455–1461. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee HJ, Lee SA and Choi H: Dietary
administration of inositol and/or inositol-6-phosphate prevents
chemically-induced rat hepatocarcinogenesis. Asian Pac J Cancer
Prev. 6:41–47. 2005.PubMed/NCBI
|
|
33
|
Nishino H: Phytochemicals in
hepatocellular cancer prevention. Nutr Cancer. 61:789–791. 2009.
View Article : Google Scholar
|
|
34
|
Vucenik I and Shamsuddin AM: Protection
against cancer by dietary IP6 and inositol. Nutr Cancer.
55:109–125. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wattenberg LW and Estensen RD:
Chemopreventive effects of myo-inositol and dexamethasone on
benzo[a]pyrene and
4-(methyl-nitrosoamino)-1-(3-pyridyl)-1-butanone-induced pulmonary
carcinogenesis in female A/J mice. Cancer Res. 56:5132–5135.
1996.PubMed/NCBI
|
|
36
|
Witschi H, Espiritu I and Uyeminami D:
Chemoprevention of tobacco smoke-induced lung tumors in A/J strain
mice with dietary myo-inositol and dexamethasone. Carcinogenesis.
20:1375–1378. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kassie F, Kalscheuer S, Matise I, Ma L,
Melkamu T, Upadhyaya P and Hecht SS: Inhibition of vinyl
carbamate-induced pulmonary adenocarcinoma by indole-3-carbinol and
myo-inositol in A/J mice. Carcinogenesis. 31:239–245. 2010.
View Article : Google Scholar :
|
|
38
|
Kassie F, Matise I, Negia M, Lahti D, Pan
Y, Scherber R, Upadhyaya P and Hecht SS: Combinations of
N-Acetyl-S-(N-2-Phenethylthiocarbamoyl)-L-Cysteine and myo-inositol
inhibit tobacco carcinogen-induced lung adenocarcinoma in mice.
Cancer Prev Res (Phila). 1:285–297. 2008. View Article : Google Scholar
|
|
39
|
Memmott RM and Dennis PA: The role of the
Akt/mTOR pathway in tobacco carcinogen-induced lung tumorigenesis.
Clin Cancer Res. 16:4–10. 2010. View Article : Google Scholar :
|
|
40
|
Kawase T, Ohki R, Shibata T, Tsutsumi S,
Kamimura N, Inazawa J, Ohta T, Ichikawa H, Aburatani H, Tashiro F,
et al: PH domain-only protein PHLDA3 is a p53-regulated repressor
of Akt. Cell. 136:535–550. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jacks T, Remington L, Williams BO, Schmitt
EM, Halachmi S, Bronson RT and Weinberg RA: Tumor spectrum analysis
in p53-mutant mice. Curr Biol. 4:1–7. 1994. View Article : Google Scholar : PubMed/NCBI
|