Introduction
Gastric carcinoma is a highly aggressive tumor with
approximately 50% of cases occurring in Eastern Asia, mainly China
(1,2). Although the current operation
techniques, chemo- and radio therapies are continuously improving,
the 5-year survival rate remains low due to the highly invasive and
metastatic properties of gastric carcinoma. Therefore,
understanding the crucial events underlying gastric tumorigenesis
and progression is urgently needed, which could pave the way for
the future anti-anticancer drug discovery and efficacious
therapeutic strategies.
It is well known that tumors are composed of
heterogeneous cell populations (2–5),
which are inconsistent in tumor-propagating ability. Cancer stem
cells, also referred to as tumor initiating cells (TICs), which
exhibit the self-renewal capacity and highly tumorigenic ability
are responsible for tumor invasion, relapse and therapy resistance
(6–9). Increasing evidence has suggested that
high aldehyde dehydrogenase activity can be used to characterize
cells with CSC properties (10–22).
The human ALDH enzymes are a family comprised of 19 isoforms
(23,24), mainly functioning in oxidizing
aldehydes to their corresponding carboxylic acid and converting
retinol to retinoic acid (RA) which would activate the RA signaling
pathway. Among the various ALDH isoforms, ALDH-1A1 is the most
extensively investigated isoform and has always been considered to
be the cause of aldefluor activity (18). However, recent studies have shown
that not all tumors exhibit the correlation between ALDH-1A1
prevalence and tumor progression, therapeutic resistance or
prognosis estimation, that is, ALDH activity is not necessarily
ALDH-1A1 specific, but could be attributed to other ALDH isoforms
(17,25–30).
Therefore, identifying the individual responsible ALDH isoforms in
each cancer is of vital importance and can be the foundation of the
following research.
ALDHbright gastric cancer cells have
recently been demonstrated to possess certain CSC properties
(31–33). However, the existing research all
regarded ALDH-1 as being responsible for aldefluor activity in
gastric cancer, although ALDH-1 families contain 6 members
(23,24,34).
Few reports identified the specific isoforms responsible for ALDH
activity in gastric carcinoma. We therefore investigated the
expression of 19 human ALDH isoforms in ALDHbright
gastric cancer cells which have been proved possessing certain
stemness properties. Our study shows that ALDH-3A1, but not
ALDH-1A1 is robustly upregulated in gastric cancer stem-like cells.
Moreover, high levels of ALDH-3A1 expression associates with poorly
differentiation degree in gastric cancer tissue which suggests that
certain level of dedifferentiation may have occurred. Collectively,
our research reveals that the aldefluor activity in gastric cancer
stem-like cells is primarily due to ALDH-3A1 and its expression
correlates well with gastric cancer progression.
Materials and methods
Cell lines and culture conditions
The human gastric cancer cell lines MKN-45 and AGS
were obtained from American Type Culture Collection. The other four
human gastric cancer cell lines SGC-7901, BGC-823, MGC-803 and
HGC-27 were obtained from Cell Bank of Shanghai Institute of Cell
Biology, Chinese Academy of Sciences. MKN-45, SGC-7901, BGC-823,
MGC-803 were cultured in RPMI-1640 medium (Gibco) supplemented with
10% fetal bovin serum (FBS, Invitrogen). HGC-27 and AGC were
cultured in Dulbecco's modified Eagle's medium (Gibco) and F-12K
(Boster), respectively supplemented with 10% FBS. All cell lines
were maintained at 37°C in a humidified atmosphere containing 5%
CO2. Images of cell lines were taken by using a Nikon
Eclipse TS100 inverted microscope.
Patients and clinical samples
Gastric cancer specimens used in this study were
obtained from the 93 cases of gastric cancer patients, with written
informed consent, who underwent a surgical resection from 2013 to
2014 at Sir Run Run Shaw Hospital. The samples of patients who
underwent preoperative radiotherapy or chemotherapy were excluded.
The clinical characteristics of the patients are presented in
Table III. The study was
approved by the Clinical Research Ethics Committee of Sir Run Run
Shaw Hospital of Zhejiang University (no. 20150227–13).
 | Table IIIRelationship between ALDH-3A1 and
clinicopathological charateristics of gastric cancer. |
Table III
Relationship between ALDH-3A1 and
clinicopathological charateristics of gastric cancer.
| | Expression of
ALDH-3A1 | |
|---|
| |
| |
|---|
| Clinical
factors | Nos. | High | Low | P-value |
|---|
| Age (years) |
| <60 | 47 | 29 | 18 | 0.527 |
| >60 | 46 | 25 | 21 | |
| Gender |
| Male | 63 | 38 | 25 | 0.237 |
| Female | 30 | 15 | 15 | |
| T stage |
| Tis, T1 | 1 | 1 | 0 | 0.452 |
| T2 | 16 | 8 | 8 | |
| T3 | 69 | 38 | 31 | |
| T4 | 7 | 6 | 1 | |
| Lymph node
metastasis |
| Presence | 31 | 23 | 8 | 0.015 |
| Absence | 62 | 30 | 32 | |
| Tumor stage |
| I | 12 | 7 | 5 | 0.011 |
| II | 51 | 22 | 29 | |
| III | 20 | 17 | 3 | |
| IV | 10 | 7 | 3 | |
|
Differentiation |
| Well | 12 | 5 | 7 | <0.001 |
| Moderate | 40 | 15 | 25 | |
| Poor | 41 | 33 | 8 | |
Aldefluor assay and FACS isolation of
cells
The ALDH activity of tumor cells was evaluated by
Aldefluor kit (Stem Cell Technologies) according to the
manufacturer's instructions. Briefly, 106 harvested
gastric cancer cells were resuspended in 500 μl aldefluor buffer
containing 2.5 μl ALDH substrate and incubated for 45 min at 37°C.
The specific ALDH inhibitor diethylaminobenzaldehye (DEAB) served
as a negative control. For FACS sorting, cells were labeled using
Aldefluor kit and the desired populations were sorted by a FACS
Aria cell sorter (BD Bioscience).
Tumorsphere formation assay
For sphere formation assay, cells were cultured in
100-mm ultralow attachment plates (Corning) at a density of 2,000
cells/ml in DMEM with nutrient mixture F-12 (Invitrogen),
supplemented with 1% N2 supplement (Gibco), 2% B27 supplement
(Gibco), 100 ng/ml epidermal growth factor (Pepro Tech), 20 ng/ml
basic fibroblast growth factor (Pepro Tech) at 37°C in humidified
air 5% CO2. To passage tumorspheres, spheres cultured
for 7 days were collected, disaggregated with 0.05% trypsin/EDTA
(Solarbio), sieved through a 40-μm filter and replated as described
above.
Colony formation assay
For colony formation analysis, two hundred viable
sorted cells were plated in each well of 6-well plates and cultured
in RPMI-1640 containing 10% FBS. After incubation for 2 weeks at
37°C, colonies containing >50 cells were counted with Giemsa
staining.
Immunofluorescence confocal
microscopy
To analyze cell differentiation, the sorted
ALDHbright cells were seeded on glass coverslips and
cultured in DMEM containing 10% FBS for 7–10 days. The cells were
then fixed in 4% paraformaldehyde for 10 min at room temperature,
permeabilized with 0.1% Triton X-100 for 1 h, blocked in 10% goat
serum in PBS for 1 h and labeled overnight at 4°C with primary
mouse anti-human CK-18 antibodies (Zhongshan Gold Bridge
Biotechnology) and mouse anti-human CD44 antibodies (Thermo)
respectively. After washing the cells with PBS, the cells were
incubated for an additional 1 h at room temperature with goat
anti-mouse IgG conjugated Alexa 594 (Invitrogen) and goat
anti-mouse IgG conjugated FITC (Invitrogen) respectively. Cell
nuclei were counterstained with DAPI. Samples were then observed
and photographed under immunofluorenscence confocal microscope
(Zeiss).
In vivo tumorigenicity assay
The sorted cells were resuspended in 150 μl of 1:1
PBS/Matrigel (BD Bioscience) and injected subcutaneously into the
inguen of 6-week-old NOD/SCID mice with 1×103 or
5×103 cells per mouse. Xenograft tumors were removed at
the end of 6th week and measured according to the
formula: Tumor volume = (length × width2)/2. A portion
of each tumor tissue was fixed in 4% formaldehyde for
immunohistochemical (IHC) analysis.
Lung metastasis assay
For generation of lung metastasis, 1×106
sorted ALDHbright and ALDHlow were
resuspended into 300 μl PBS and injected through the lateral tail
vein of 6-week-old NOD/SCID mice. All mice were sacrificed at the
16th week, and the lungs were harvested and examined for
tumor nodules.
RNA extraction and qRT-PCR analysis
The total RNA of cells was extracted with RNAiso
reagent (Takara). cDNA was synthesized using equivalent amount of
total RNA (1 μg) with primers in a 20-μl reverse transcriptase
reaction mixture (Takara). The primers were designed and purchased
from Invitrogen, the sequences of each primer pair were listed in
Tables IV and V. qRT-PCR was performed using
LightCycler® 480 Real-Time PCR system. The expression of
each gene was calculated using 2−ΔΔCT method. Results
were normalized against the level of GAPDH. All assays were
performed in triplicates and results were plotted as the mean ±
SD.
 | Table IVReal-time PCR primers used for all 19
human ALDH isoforms and GAPDH. |
Table IV
Real-time PCR primers used for all 19
human ALDH isoforms and GAPDH.
| Genes | Sequences |
|---|
| ALDH1A1 | F:
5′-ACTGCTCTCCACGTGGCATCTTTA-3′
R: 5′-TGCCAACCTCTGTTGATCCTGTGA-3′ |
| ALDH1A2 | F:
5′-AGGGCAGTTCTTGCAACCATGGAA-3′
R: 5′-CACACACTCCAATGGGTTCATGTC-3′ |
| ALDH1A3 | F:
5′-ACCTGGAGGTCAAGTTCACCAAGA-3′
R: 5′-ACGTCGGGCTTATCTCCTTCTTCC-3′ |
| ALDH1B1 | F:
5′-TGCTGCAGAGTGTCAGCAT-3′
R: 5′-GGTGGTAGGGTTGACCGTCG-3′ |
| ALDH1L1 | F:
5′-ATCTTTGCTGACTGTGACCT-3′
R: 5′-GCACCTCTTCTACCACTCTC-3′ |
| ALDH1L2 | F:
5′-GCCTGGTCTCGTTACCAAAA-3′
R: 5′-GCCACTTTCACCTCTTCAGC-3′ |
| ALDH2 | F:
5′-CCAACCAGCAGCCCGAGGTC-3′
R: 5′-AAGGCCTTGTCCCCTTCAGCTACC-3′ |
| ALDH3A1 | F:
5′-TGTGTCAAAGGCGCCATGAGCAAG-3′
R: 5′-GGCGTTCCATTCATTCTTGTGCAG-3′ |
| ALDH3A2 | F:
5′-TGCACTTCACGCTCAACTCT-3′
R: 5′-GACTGGCTGTTGGGAGGATA-3′ |
| ALDH3B1 | F:
5′-ACAAGTCAGCCTTCGAGTCGG-3′
R: 5′-AGCACCACACAGTTCCCTGC-3′ |
| ALDH3B2 | F:
5′-ACAGAGAAGGTCCTGGCTGA-3′
R: 5′-CATGACAATCTTGCCCACAC-3′ |
| ALDH4A1 | F:
5′-TGCAGTACCAAGTGTCGCCTTT-3′
R: 5′-AATCTCCGCTTGGATCACGGTCTT-3′ |
| ALDH5A1 | F:
5′-ACCAATTCTTGGTGCAAAGG-3′
R: 5′-GTTGGTGTCGTTTTCCACCT-3′ |
| ALDH6A1 | F:
5′-GGCTCTTTCAACAGCAGTCC-3′
R: 5′-ATGGAAGCTCCCTCCTTTGT-3′ |
| ALDH7A1 | F:
5′-CGAGCCAATAGCAAGAGTCC-3′
R: 5′-CTTCACCCACACCTTCCACT-3′ |
| ALDH8A1 | F:
5′-TGGTGAGCATAGGTGCTCTG-3′
R: 5′-GTTATCACCGTGGGAAGCAT-3′ |
| ALDH9A1 | F:
5′-CACTCATCAACCGACCACAC-3′
R: 5′-GGACATAACAGGCCCAAAGA-3′ |
| ALDH16A1 | F:
5′-GCTCCTCATCCAGGAGTCTG-3′
R: 5′-AAGGTTGGGGGATAGAATGG-3′ |
| ALDH18A1 | F:
5′-CTGAGTATGGGGACCTGGAA-3′
R: 5′-GCGGTAACCATCAGAAAAGC-3′ |
 | Table VReal-time PCR primers used for EMT
associated transcription factors and stem cell related markers. |
Table V
Real-time PCR primers used for EMT
associated transcription factors and stem cell related markers.
| Genes | Sequences |
|---|
| Snail | F:
5′-GACCACTATGCCGCGCTCTT-3′
R: 5′-TCGCTGTAGTTAGGCTTCCGATT-3′ |
| Slug | F:
5′-AGCGAACTGGACACACATAC-3′
R: 5′-TCTAGACTGGGCATCGCAG-3′ |
| Twist-1 | F:
5′-CACTGAAAGGAAAGGCATCA-3′
R: 5′-GGCCAGTTTGATCCCAGTAT-3′ |
| Zeb-1 | F:
5′-CGAGTCAGATGCAGAAAATGAGCAA-3′
R: 5′-ACCCAGACTGCGTCACATGTCTT-3′ |
| Zeb-2 | F:
5′-GAGTTGATGCCTCGGCTATTGC-3′
R: 5′-CTGGACATTGAGCTGCTTCGATC-3′ |
| E-cadherin | F:
5′-TACACTGCCCAGGAGCCAGA-3′
R: 5′-TGGCACCAGTGTCCGGATTA-3′ |
| Vimentin | F:
5′-TGAGTACCGGAGACAGGTGCAG-3′
R: 5′-TAGCAGCTTCAACGGCAAAGTTC-3′ |
| OCT-4 | F:
5′-GCAGCGACTATGCACAACGA-3′
R: 5′-CCAGAGTGGTGACGGAGACA-3′ |
| NANOG | F:
5′-GACTTGACCACCGAACC- CAT-3′
R: 5′-CTGGATGTTCTGGGTCTGGT-3′ |
| BMI-1 | F:
5′-TCGTTCTTGTTATTACGCTGTTTT-3′
R: 5′-CGGTAGTACCCGCTTTTAGGC-3′ |
| GAPDH | F:
5′-GGTGTGAACCATGAGAAGTATG-3′
R: 5′-GATGGCATGGACTGTGGTCAT-3′ |
| Snail | F:
5′-GACCACTATGCCGCGCTCTT-3′
R: 5′-TCGCTGTAGTTAGGCTTCCGATT-3′ |
| Slug | F:
5′-AGCGAACTGGACACACATAC-3′
R: 5′-TCTAGACTGGGCATCGCAG-3′ |
| Twist-1 | F:
5′-CACTGAAAGGAAAGGCATCA-3′
R: 5′-GGCCAGTTTGATCCCAGTAT-3′ |
| Zeb-1 | F:
5′-CGAGTCAGATGCAGAAAATGAGCAA-3′
R: 5′-ACCCAGACTGCGTCACATGTCTT-3′ |
| Zeb-2 | F:
5′-GAGTTGATGCCTCGGCTATTGC-3′
R: 5′-CTGGACATTGAGCTGCTTCGATC-3′ |
| E-cadherin | F:
5′-TACACTGCCCAGGAGCCAGA-3′
R: 5′-TGGCACCAGTGTCCGGATTA-3′ |
| Vimentin | F:
5′-TGAGTACCGGAGACAGGTGCAG-3′
R: 5′-TAGCAGCTTCAACGGCAAAGTTC-3′ |
| OCT-4 | F:
5′-GCAGCGACTATGCACAACGA-3′
R: 5′-CCAGAGTGGTGACGGAGACA-3′ |
Western blot analysis
Cultured cells were lysed with 200 μl of ice-cold
RIPA buffer (Beyotime) containing 1 mM PMSF. Proteins were resolved
by SDS-polyarylamide gel and transferred to PVDF membranes
(Millipore). The membranes were then blocked with 5% non-fat milk
and were then incubated with primary antibodies overnight at 4°C.
The membranes were washed and incubated with appropriate
HPR-conjugated secondary antibodies for 1 h at room temperature.
Protein bands were detected by using Pierce ECL Western blotting
substrate (Thermo). GAPDH or TUBLIN were used as a loading control.
The primary antibodies used for western blot analyses were as
follows: NANOG (Abcam), OCT-4 (Abcam), BMI-1 (Abcam), ALDH-1A1 (BD
Bioscience), ALDH-3A1 (Santa Cruz Biotechnology), GAPDH (Abcam),
TUBLIN (Cwbiotech).
TMA and image analysis
TMAs were either constructed by author (Key
Laboratory of Biotherapy of Zhejing University) or purchased from
Alenabio Biotech Co.. Staining of tissue microarray slides was
carried out according to the manufacturer's protocol. Briefly, IHC
staining was performed on 10% phosphate-buffered formalin fixed and
paraffin-embedded gastric cancer sections (4 μm) using xylene. The
slides were hydrated by a graded series of ethanol washes and
incubated in 0.3% H2O2. After incubation with
blocking solution for 30 min at room temperature, the slides were
incubated with primary antibodies at 4°C overnight. The secondary
antibodies were added for incubation at 37°C. TMA slides were
scanned by using Aperio Slide Scanner and analyzed by Image Scope
software (Aperio). The IHC staining was scored independently by two
pathologists blinded to the clinical data as follows: Score =
percentage of immunoreactive cells (0, <10%; 1, 11–25%; 2,
26–50%; 3, 51–75%; 4, >75%) × mean stain intensity (0–3).
Statistical analysis
Statistical analysis was performed using SPASS
statistical analysis software Version 17.0 (SPSS). Student's t-test
was used to examine the statistical significance when two groups
were compared. To determine differences among 3 groups, an ANOVA
analysis was performed. P-value <0.05 was regarded as
statistically significant.
Results
ALDHbright gastric cancer
cells exibit cancer stem-like cell properties
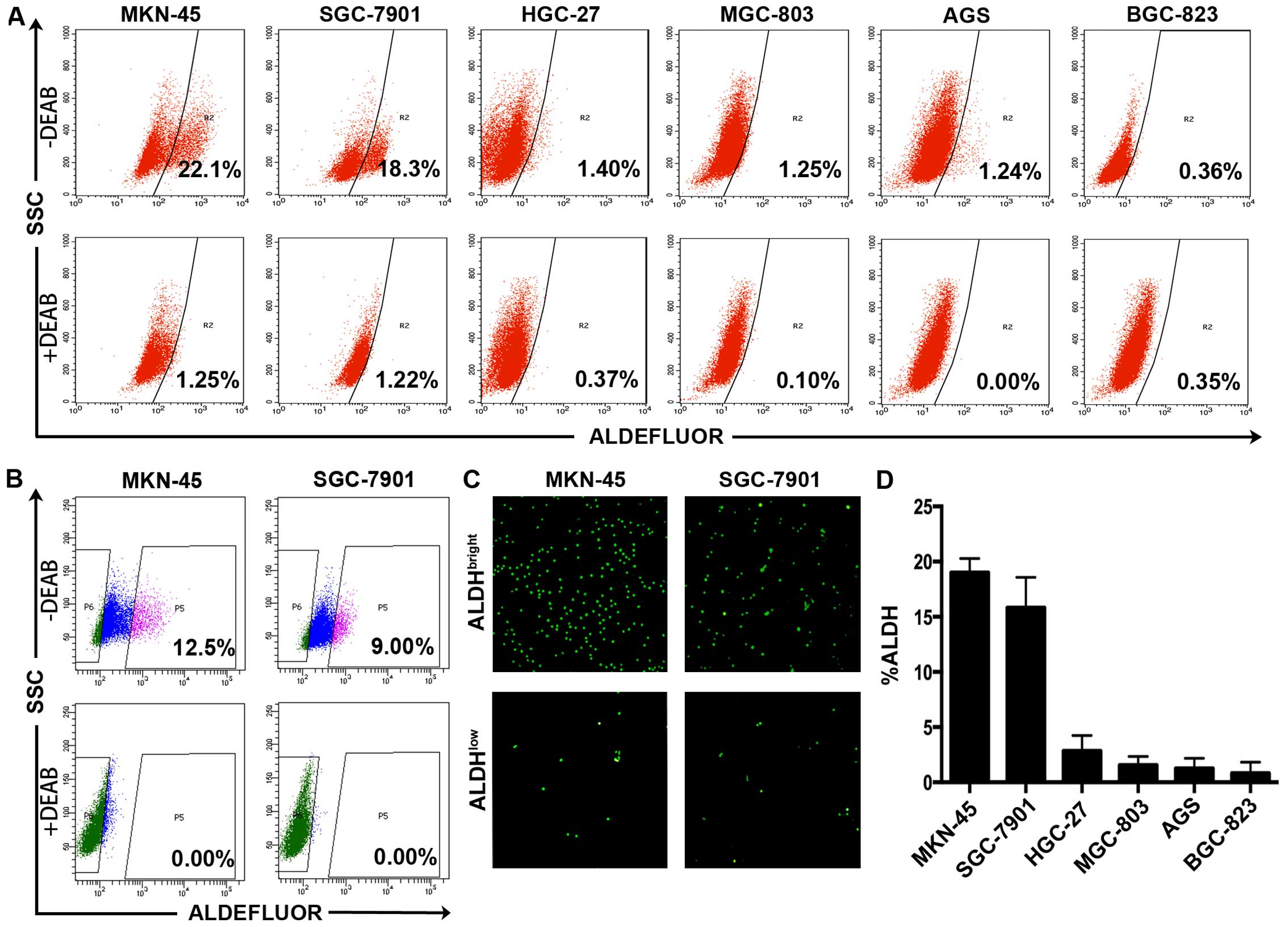
We first examined the proportion of
ALDHbright cells in the six human gastric cancer cell
lines using Aldefluor kit and found that each of the 6 gastric
cancer cell lines (MKN-45, SGC-7901, HGC-27, AGS, BGC-823, MGC-803)
contained ALDH-positive cells (Fig.
1A). Compared with the DEAB-treated control groups, high ALDH
activity was detected in 22.1% of the MKN-45 cells, 18.3% of the
SGC-7901 cells, 1.40% of the HGC-27 cells, 1.25% of the MGC-803
cells, 1.24% of the AGS cells, 0.36% of the BGC-823 cells (Fig. 1D). We therefore chose MKN-45 and
SGC-7901 for the follow-up research taking advantage that both cell
lines contained prominent ALDHbright subpopulations
(indicated by the bright green fluorescence), making the ALDH
activity based separation more efficient and convincing (Fig. 1B and C). We present here that
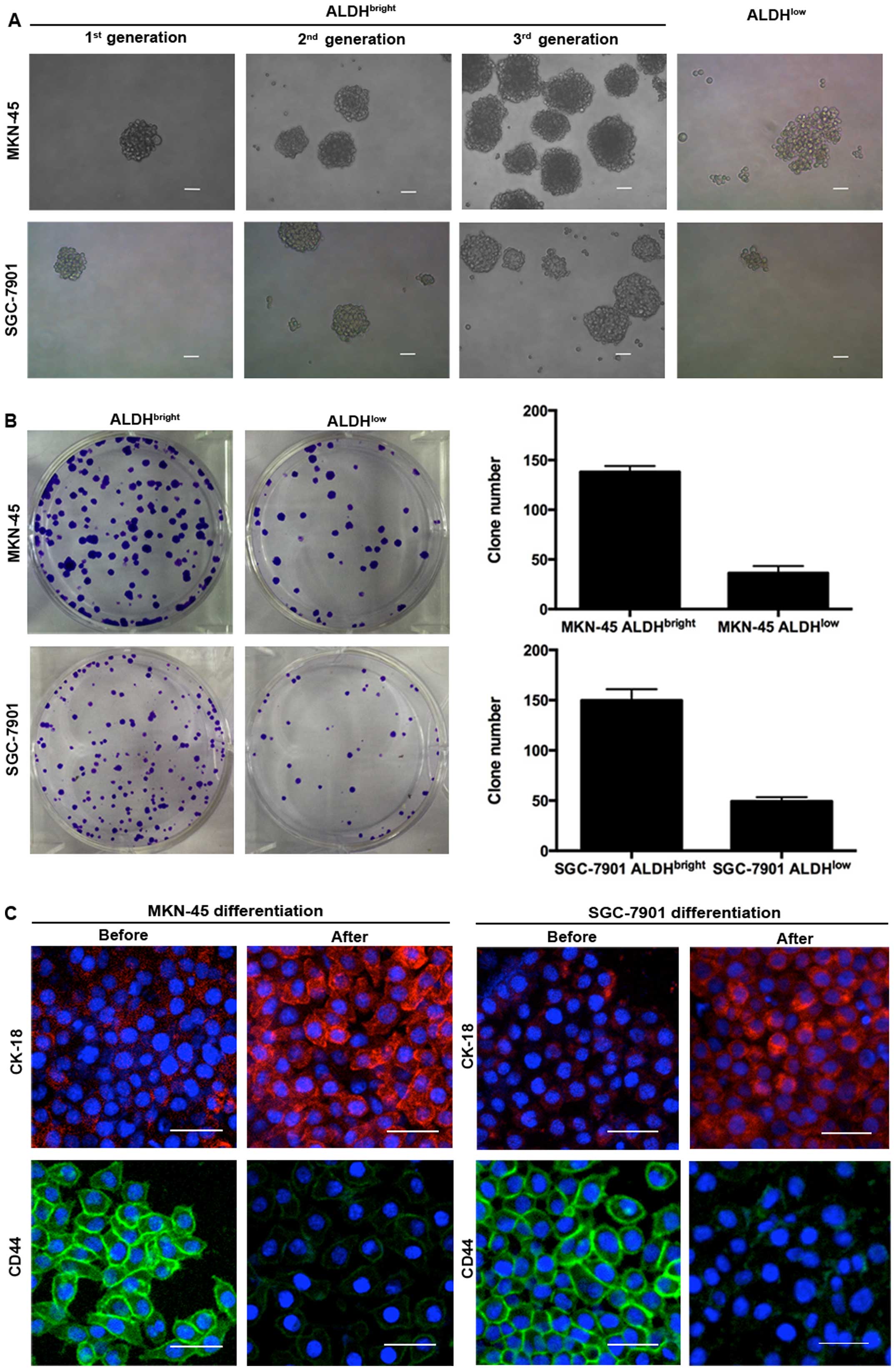
ALDHbright cells isolated from the 2 gastric cancer cell
lines showed higher capabilities of tumorsphere formation compared
with ALDHlow cells. These spheres could be passaged for
at least 3 consecutive generations with an increasing forming
efficiency, while the ALDHlow cells did not form typical
tumorspheres, only a few loose cell aggregates (Fig. 2A). The number of colonies formed by
ALDHbright MKN-45 cells and ALDHbright
SGC-7901 cells were higher than those formed by ALDHlow
cells (138 versus 36, 150 versus 49, respectively) (Fig. 2B). The ALDHbright cells
from MKN-45 and SGC-7901 also showed multi-potent capacity of
differentiation by expressing decreased level of CD44 but increased
level of CK-18 after induction of differentiation (Fig. 2C). ALDHbright MKN-45 and
SGC-7901 were also found to express higher levels of stem
cell-related markers including OCT-4, BMI-1, NANOG than
ALDHlow cells (Fig.
2D). The tumorigenicity assay with isolated
ALDHbright and ALDHlow gastric cancer cells
in NOD/SCID mice showed that ALDHbright MKN-45 and
SGC-7901 cells have enhanced tumor initiation capacity as compared
to ALDHlow cells at each cell dose (103, 5/6
versus 0/6, 6/6 versus 0/6; 5×103, 6/6 versus 1/6, 6/6
versus 2/6, respectively) (Fig. 2E
and Table I). Also, the volume of
ALDHbright cell derived xenografts were larger than
those of ALDHlow cell-derived (Fig. 2E).
 | Table ITumorigenicity of
ALDHbright and ALDHlow cells from MKN-45 and
SGC-7901. |
Table I
Tumorigenicity of
ALDHbright and ALDHlow cells from MKN-45 and
SGC-7901.
| | Cell dose |
|---|
| |
|
|---|
| Cell line | Subpopulation |
1×103 |
5×103 |
|---|
| MKN-45 |
ALDHbright | 5/6 | 6/6 |
|
ALDHlow | 0/6 | 1/6 |
| SGC-7901 |
ALDHbright | 6/6 | 6/6 |
|
ALDHlow | 0/6 | 2/6 |
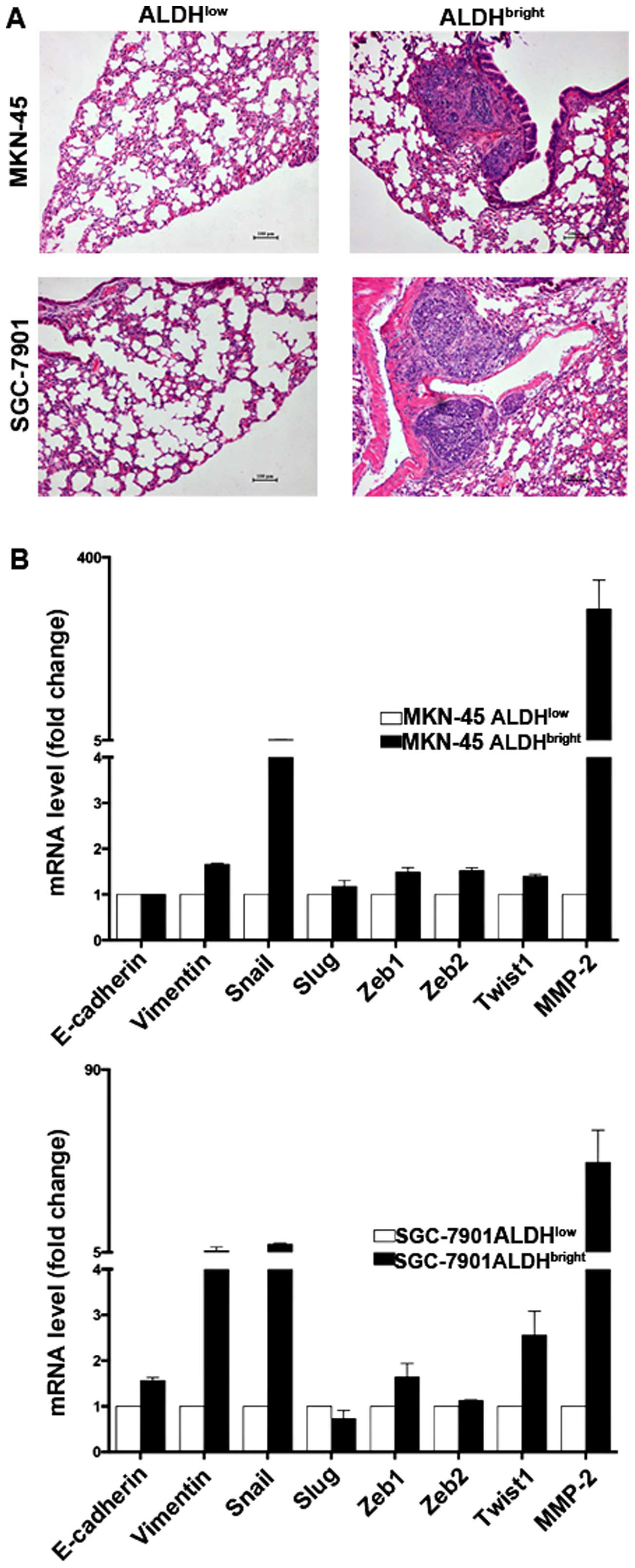
ALDHbright cells display
enhanced metastatic ability correlating with a mesenchymal
transition phenotype
Accumulating evidence suggests that CSCs are
responsible for tumor relapse, we therefore examined the metastatic
capabilities and the expression of related molecules in the sorted
ALDHbright cells. The ALDHbright cells
derived from MKN-45 and SGC-7901 formed lung metastasis in four of
six mice, and five of six mice, respectively, whereas few
metastasis was formed by ALDHlow cells (Fig. 3A and Table II). Gene expression analysis
further showed that MKN-45 ALDHbright cells express
higher levels of Vimentin and Snail but lower level of E-cadherin,
similar results were also obtained in SGC-7901
ALDHbright cells (Fig.
3B), suggesting EMT might be responsible for their enhanced
metastatic capabilities.
 | Table IIGeneration of lung metastasis of
ALDHbright and ALDHlow cells from MKN-45 and
SGC-7901. |
Table II
Generation of lung metastasis of
ALDHbright and ALDHlow cells from MKN-45 and
SGC-7901.
| Cell line | Subpopulation | Lung metastasis
(yes/no) |
|---|
| MKN-45 |
ALDHbright | 4/2 |
|
ALDHlow | 0/6 |
| SGC-7901 |
ALDHbright | 5/1 |
|
ALDHlow | 1/5 |
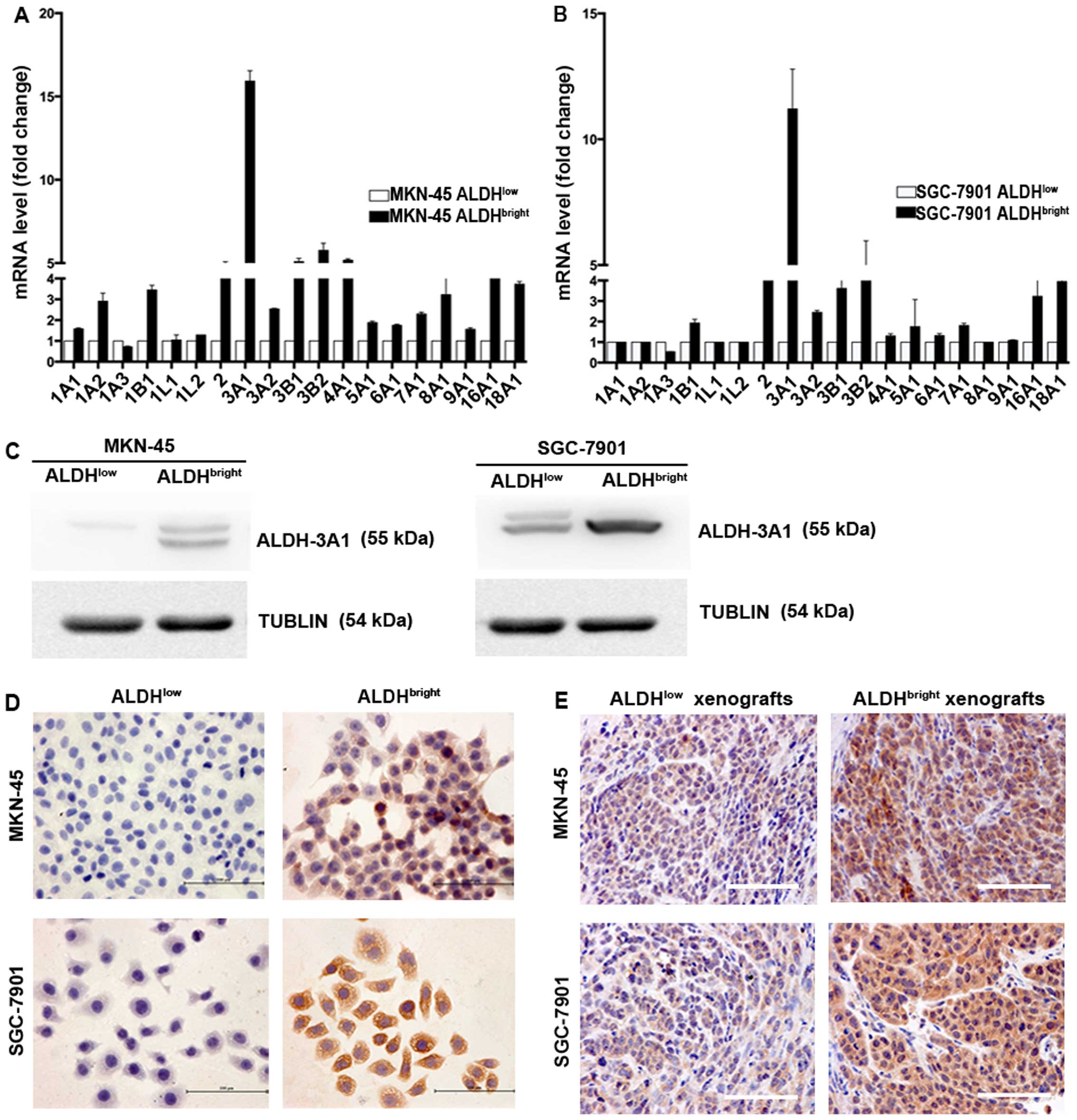
Specific increase in ALDH-3A1 expression
in ALDHbright subpopulations
Since the ALDH isoform responsible for Aldefluor
activity may vary depending on cancer type and tissue origin, we
characterized the gene expression of all 19 ALDH isoforms between
ALDHbright and ALDHlow cell populations
sorted from MKN-45 and SGC-7901. Our results showed that among all
the various human ALDH isoforms, ALDH-3A1 was the most elevated
isoform in the sorted MKN-45 and SGC-7901 cells (15.9- and
11.2-fold, respectively) (Fig. 4A and
B). In addition, several other isoforms were also found
upregulated including ALDH-16A1, ALDH-2, ALDH-3B1, ALDH-4A1,
ALDH-3B2 in MKN-45 derived ALDHbright cells, ALDH-18A1,
ALDH-2, ALDH-3B2 in SGC-7901 derived ALDHbright cells,
the elevated folds of which ranges from 4.4 to 5.8, 4.0 to 4.6,
respectively. Notably, no fold change was detected in ALDH-1A1 in
ALDHbright MKN-45 and SGC-7901 cells. Moreover, the mRNA
levels of ALDH-1A1 were undetectable until cycle 35 for two cell
lines. The expression of ALDH-3A1 was subsequently confirmed by
western blot analysis (Fig. 4C),
immunocytochemical (ICC) staining (Fig. 4D) and IHC staining (Fig. 4E).
ALDH-3A1 expression associates with
gastric cancer evolution
To understand the possible role of ALDH-3A1 in the
development and progression of gastric cancer, we examined the
expression of ALDH-3A1 in a set of primary gastric tumors of
different severity and metastatic gastric tumors. We were able to
show here that ALDH-3A1 could not be detected or was only slightly
expressed in the basal layer in normal gastric epithelia, but was
detectable at low levels in the gastric dysplastic lesions, then
the staining intensity of ALDH-3A1 progressed from weak to strong
with increasing grades of dysplasia and carcinoma (Fig. 5B and C). The expression of ALDH-3A1
was associated with histological degrees (P<0.001), N stage
(P=0.015) and cancer stage (P=0.011) of gastric cancer (Table III). Considering that CSCs play
critical roles in tumor relapse, we subsequently examined the level
of ALDH-3A1 in several pairs of local and lymph node metastatic
gastric cancer and found higher levels of ALDH-3A1 in metastatic
lesions compared with the local carcinomas (Fig. 5D). Collectively, these results
indicate that the expression level of ALDH-3A1 correlates with the
increasing severity of gastric cancer.
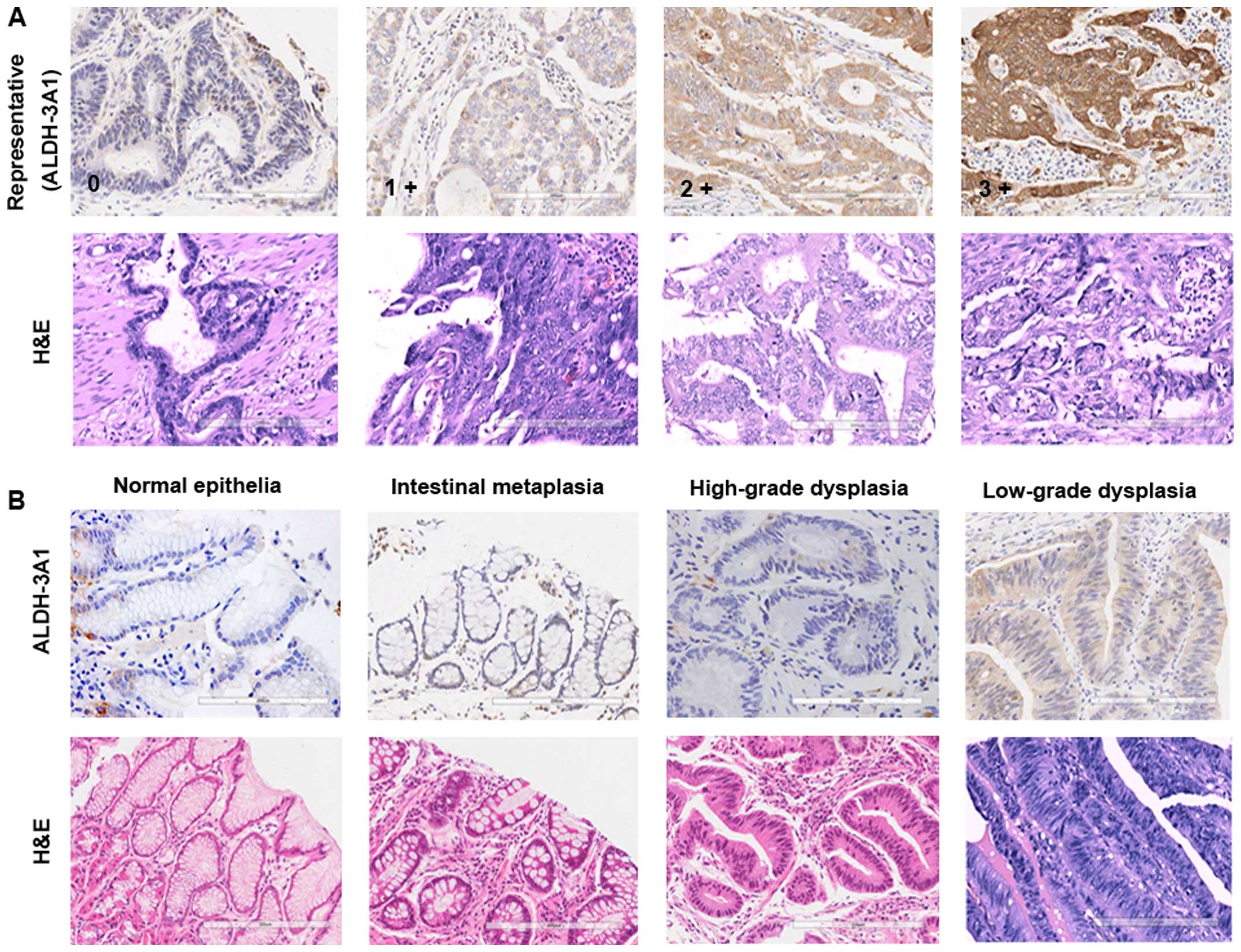 | Figure 5IHC analysis of ALDH-3A1-positive
cells in normal gastric mucosa and gastric carcinoma samples with
different severity. (A) Representative IHC staining intensity of
ALDH-3A1 from patients with gastric adenocarcinoma. In all cases,
scores were assigned to a scale of 0–3; 0, no staining; 1+, mild;
2+, moderate; and 3+, strong intensity of staining. (B) Expression
for ALDH-3A1 in normal gastric epithelia, gastric dysplastic
lesions and gastric epithelia dysplasia. (C) Representative IHC
images of ALDH-3A1-positive cells in gastric cancer tissue with
different histological grade. (D) Expression for ALDH-3A1 in local
gastric carcinoma and corresponding lymph node metastasis lesions.
G1, G2 and G3, tumor histological grade I, II and III,
respectively. |
Discussion
In this study, we provide strong evidence that
ALDHbright gastric cancer cells possess cancer stem-like
properties and revealed for the first time that ALDH-3A1 is the
highly expressed isoform in gastric cancer stem-like cells.
Together with IHC analysis in tissue level, we show that
ALDH-3A1-expressing cells play a vital role in the initiation and
progression of gastric cancer. We convincingly demonstrate that
ALDH-3A1 functions as a pivotal enzyme in regulating gastric cancer
stem-like population, ALDH-3A1 prevalence therefore might be used
as a novel indicator to predict gastric cancer patient outcome.
The human ALDH enzymes are a family comprised of 19
isoforms, mainly functioning in oxidizing various aldehydes to
their corresponding carboxylic acids (27,34).
High ALDH activity was originally identified as a marker for
hematopoietic stem cells (25,35,36)
and is thought to play a vital role in the self-renewal and
differentiation of hematopoietic progenitor cells via converting
retinal to retinoic acid (37).
Recent studies suggest that high levels of ALDH activity
characterize a subpopulation of cells with CSC properties in
several malignancies (14,19–21,38,39).
Moreover, enhanced ALDH activity predicts poor patients outcome in
breast cancer (11,22,40–42),
lung cancer (15,40), and esophageal squamous cell
carcinoma (43), which is
consistent with CSCs being responsible for cancer evolution and
metastasis. Katsuno et al (32) have reported that
ALDHbright cells derived from HSC-39 and OCUM-2ML
gastric cancer cell lines exhibited certain stem cell traits but
little is known about traits of ALDHbright cells in
other gastric cancer cell lines. In this study, we isolated
ALDHbright and ALDHlow subpopulations from 2
different gastric cancer cell lines MKN-45 and SGC-7901. Our data
show that these sorted cells have capabilities of self-renewal,
multilineage differential potential, high tumorigenicity and
express high levels of stemness associated hallmarks, thereby
representing a more reliable cancer stem-like cell marker in
gastric cancer. We also found that the ALDHbright cells
are more invasive and exhibit a phenotype of EMT by expressing
increased level of Vimentin and Snail but decreased level of
E-cadherin, in agreement with EMT being able to endow tumor cells
with stem cell-like properties. We have previously reported that
Snail is required for the maintenance of stem cell-like phenotype
in pancreatic carcinoma (44), as
such, it would be interesting to examine its contributions to
gastric cancer stem-like compartment, which might provide novel
therapeutic strategies.
With our discoveries that ALDHbright
gastric cancer cells exhibit stemness charateristics and may
contribute to tumor invasion, we hypothesized that the activity of
ALDH in tumor might associate with gastric tumorigenesis and
provide clinical prognostic values in gastric cancer patients.
Generally, CSCs quantification in clinic requires
immunohistological methods for detection of the specific protein
expression in fixed tumor tissue. Therefore, the prognostic
application by evaluating ALDH prevalence requires detection and
quantification of ALDH expression at protein level in gastric
tissue, but not activity. Although the Aldefluor assay is able to
identify the celluar ALDH activity, it is not practical for
clinical evaluation of ALDH at protein level. More importantly, the
Aldefluor assay has been proved to be not merely ALDH-1A1-specific.
Increasing evidence has demonstrated that the ALDH isoforms
responsible for Aldefluor assay may vary depending on the cell
origin and tumor types, the reason of which could be attributed to
the cross-reactivity between the similar amino acid sequence
sharing among 19 ALDH isoforms. Van den Hoogen et al
(45) reported high expression of
ALDH-7A1 in both prostate cancer cell lines and malignant tissues.
Yan et al (46)
demonstrated the association of ALDH-3A1 with prostate cancer
progression. These findings suggest that more than one of these
isoforms could be responsible for the Aldefluor activity. Chen
et al (25) compared the
expression levels of ALDH-1A1 and ALDH-3A1 in colon adenocarcinoma
and reported that 98% of the tumor samples were ALDH-1B1 positive.
Marcato et al (26)
characterized the Aldefluor positive cells isolated from breast
cancer patient tumor samples and found a better correlation with
ALDH-1A3. Similar studies have successively been published and
attracted broad attention in defining the responsible ALDH isoforms
in different tumors. So far, the specific ALDH isoform responsible
for Aldefluor activity in gastric cancer is still unclear.
Furthermore, staining with ALDH-1A1 has shown no prognostic impact
in gastric cancer patients (33),
suggesting that ALDH-1A1 might not be attributed to ALDH activity,
thus unable to provide clinical prognostic value. We show here the
obvious upregulation of ALDH-3A1 in both MKN-45 and SGC-7901
derived ALDHbright cells compared with
ALDHlow cells. In the tissue level, the staining
intensity of ALDH-3A1 elevates along with increasing severity of
gastric cancer, suggesting the expression of ALDH-3A1 may have
resulted in certain level of dedifferentiation in which process
gastric cancer stem-like cells may have been produced and
contributed to gastric tumorigensis. Particularly, enhanced
expression of ALDH-3A1 was observed in gastric cancer metastatic
lesions compared to the matched primary carcinoma. This fits well
with ALDHbright cells possessing increased metastatic
capabilities with EMT phenotype. In addition, ALDH-1A1 expression
could not be detected in sorted gastric cancer stem-like cells,
suggesting ALDH-1A1 might not function in gastric cancer stem cell
biology, thereby could not be deemed as a cancer stem cell related
marker in gastric cancer. We speculate this possibility could
account for the negative follow-up results of applying ALDH-1A1
prevalence to predict gastric cancer patients outcome.
ALDH-3A1 is an important isoform of ALDH family
located in the cytoplasm (27) and
shows multiple catalysis including serving as a corneal crystalline
to protect eyes from UV radiation, contributing to the maintainance
of hematopoietic stem cell population, regulating cell
proliferation and apoptosis and conducing to the chemotherapeutic
drug resistance (47–54). All these functions in particular
have been linked to tumor evolution. We demonstrate here that
ALDH-3A1 is involved in the process of malignant transformation,
tumor relapse in gastric cancer. These findings are consistent with
those of Yan et al and Calderaro et al, who have
shown upregulation of ALDH-3A1 correlated with tumorigenesis in
hepatocellular carcinoma and protate cancer, respectively (46,55).
It is necessary to clarify the mechanisms underlying ALDH-3A1
prevalence and gastric cancer development. With the recent
discovery that EMT transcription factors, such as TWIST could
promote cancer stem-like cells by directly binding to E-box
sequences in promoter region of CD24 (56), it would be intriguingly to clarify
whether the elevated EMT transcription factors would directly act
on the ALDH-3A1 promoter region thereby contributing to the
maintainance of ALDHbright gastric cancer stem-like
subpopulation.
In conclusion, our discoveries identify ALDH-3A1 as
a critical CSC marker with potential clinical prognostic
application, and demonstrate a clear association between ALDH-3A1
prevalence and gastric cancer evolution.
Acknowledgements
This study was supported by grants by Natural
Science Foundation of Zhejiang Province (CN) (grant no. LY15H160030
and no. LY15H160031) and Chinese Medicine Research Program of
Zhejiang Province (CN) (grant no. 2012ZA087).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Jones RJ, Matsui WH and Smith BD: Cancer
stem cells: Are we missing the target? J Natl Cancer Inst.
96:583–585. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fidler IJ and Hart IR: Biological
diversity in metastatic neoplasms: Origins and implications.
Science. 217:998–1003. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heppner GH and Miller BE: Tumor
heterogeneity: Biological implications and therapeutic
consequences. Cancer Metastasis Rev. 2:5–23. 1983. View Article : Google Scholar
|
|
5
|
Nowell PC: Mechanisms of tumor
progression. Cancer Res. 46:2203–2207. 1986.PubMed/NCBI
|
|
6
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Greaves M: Cancer stem cells as ‘units of
selection’. Evol Appl. 6:102–108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Clarke MF, Dick JE, Dirks PB, Eaves CJ,
Jamieson CH, Jones DL, Visvader J, Weissman IL and Wahl GM: Cancer
stem cells - perspectives on current status and future directions:
AACR Workshop on cancer stem cells. Cancer Res. 66:9339–9344. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Luo Y, Dallaglio K, Chen Y, Robinson WA,
Robinson SE, McCarter MD, Wang J, Gonzalez R, Thompson DC, Norris
DA, et al: ALDH1A isozymes are markers of human melanoma stem cells
and potential therapeutic targets. Stem Cells. 30:2100–2113. 2012.
View Article : Google Scholar :
|
|
11
|
Landen CN Jr, Goodman B, Katre AA, Steg
AD, Nick AM, Stone RL, Miller LD, Mejia PV, Jennings NB, Gershenson
DM, et al: Targeting aldehyde dehydrogenase cancer stem cells in
ovarian cancer. Mol Cancer Ther. 9:3186–3199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rahadiani N, Ikeda J, Mamat S, Matsuzaki
S, Ueda Y, Umehara R, Tian T, Wang Y, Enomoto T, Kimura T, et al:
Expression of aldehyde dehydrogenase 1 (ALDH1) in endometrioid
adenocarcinoma and its clinical implications. Cancer Sci.
102:903–908. 2011. View Article : Google Scholar
|
|
13
|
Clay MR, Tabor M, Owen JH, Carey TE,
Bradford CR, Wolf GT, Wicha MS and Prince ME: Single-marker
identification of head and neck squamous cell carcinoma cancer stem
cells with aldehyde dehydrogenase. Head Neck. 32:1195–1201. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim MP, Fleming JB, Wang H, Abbruzzese JL,
Choi W, Kopetz S, McConkey DJ, Evans DB and Gallick GE: ALDH
activity selectively defines an enhanced tumor-initiating cell
population relative to CD133 expression in human pancreatic
adenocarcinoma. PLoS One. 6:e206362011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sullivan JP, Spinola M, Dodge M, Raso MG,
Behrens C, Gao B, Schuster K, Shao C, Larsen JE, Sullivan LA, et
al: Aldehyde dehydrogenase activity selects for lung adenocarcinoma
stem cells dependent on notch signaling. Cancer Res. 70:9937–9948.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qin J, Liu X, Laffin B, Chen X, Choy G,
Jeter CR, Calhoun-Davis T, Li H, Palapattu GS, Pang S, et al: The
PSA(−/lo) prostate cancer cell population harbors self-renewing
long-term tumor-propagating cells that resist castration. Cell Stem
Cell. 10:556–569. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
van den Hoogen C, van der Horst G, Cheung
H, Buijs JT, Lippitt JM, Guzmán-Ramírez N, Hamdy FC, Eaton CL,
Thalmann GN, Cecchini MG, et al: High aldehyde dehydrogenase
activity identifies tumor-initiating and metastasis-initiating
cells in human prostate cancer. Cancer Res. 70:5163–5173. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Su Y, Qiu Q, Zhang X, Jiang Z, Leng Q, Liu
Z, Stass SA and Jiang F: Aldehyde dehydrogenase 1 A1-positive cell
population is enriched in tumor-initiating cells and associated
with progression of bladder cancer. Cancer Epidemiol Biomarkers
Prev. 19:327–337. 2010. View Article : Google Scholar
|
|
19
|
Chu P, Clanton DJ, Snipas TS, Lee J,
Mitchell E, Nguyen ML, Hare E and Peach RJ: Characterization of a
subpopulation of colon cancer cells with stem cell-like properties.
Int J Cancer. 124:1312–1321. 2009. View Article : Google Scholar
|
|
20
|
Carpentino JE, Hynes MJ, Appelman HD,
Zheng T, Steindler DA, Scott EW and Huang EH: Aldehyde
dehydrogenase-expressing colon stem cells contribute to
tumorigenesis in the transition from colitis to cancer. Cancer Res.
69:8208–8215. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang EH, Hynes MJ, Zhang T, Ginestier C,
Dontu G, Appelman H, Fields JZ, Wicha MS and Boman BM: Aldehyde
dehydrogenase 1 is a marker for normal and malignant human colonic
stem cells (SC) and tracks SC overpopulation during colon
tumorigenesis. Cancer Res. 69:3382–3389. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ginestier C, Hur MH, Charafe-Jauffret E,
Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG,
Liu S, et al: ALDH1 is a marker of normal and malignant human
mammary stem cells and a predictor of poor clinical outcome. Cell
Stem Cell. 1:555–567. 2007. View Article : Google Scholar
|
|
23
|
Vasiliou V and Nebert DW: Analysis and
update of the human aldehyde dehydrogenase (ALDH) gene family. Hum
Genomics. 2:138–143. 2005.PubMed/NCBI
|
|
24
|
Marchitti SA, Brocker C, Stagos D and
Vasiliou V: Non-P450 aldehyde oxidizing enzymes: The aldehyde
dehydrogenase superfamily. Expert Opin Drug Metab Toxicol.
4:697–720. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Y, Orlicky DJ, Matsumoto A, Singh S,
Thompson DC and Vasiliou V: Aldehyde dehydrogenase 1B1 (ALDH1B1) is
a potential biomarker for human colon cancer. Biochem Biophys Res
Commun. 405:173–179. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marcato P, Dean CA, Pan D, Araslanova R,
Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, et al:
Aldehyde dehydrogenase activity of breast cancer stem cells is
primarily due to isoform ALDH1A3 and its expression is predictive
of metastasis. Stem Cells. 29:32–45. 2011. View Article : Google Scholar
|
|
27
|
Vasiliou V, Thompson DC, Smith C, Fujita M
and Chen Y: Aldehyde dehydrogenases: From eye crystallins to
metabolic disease and cancer stem cells. Chem Biol Interact.
202:2–10. 2013. View Article : Google Scholar
|
|
28
|
Levi BP, Yilmaz OH, Duester G and Morrison
SJ: Aldehyde dehydrogenase 1a1 is dispensable for stem cell
function in the mouse hematopoietic and nervous systems. Blood.
113:1670–1680. 2009. View Article : Google Scholar :
|
|
29
|
Rovira M, Scott SG, Liss AS, Jensen J,
Thayer SP and Leach SD: Isolation and characterization of
centroacinar/terminal ductal progenitor cells in adult mouse
pancreas. Proc Natl Acad Sci USA. 107:75–80. 2010. View Article : Google Scholar :
|
|
30
|
Marchitti SA, Orlicky DJ, Brocker C and
Vasiliou V: Aldehyde dehydrogenase 3B1 (ALDH3B1):
Immunohistochemical tissue distribution and cellular-specific
localization in normal and cancerous human tissues. J Histochem
Cytochem. 58:765–783. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nishikawa S, Konno M, Hamabe A, Hasegawa
S, Kano Y, Ohta K, Fukusumi T, Sakai D, Kudo T, Haraguchi N, et al:
Aldehyde dehydrogenase high gastric cancer stem cells are resistant
to chemotherapy. Int J Oncol. 42:1437–1442. 2013.PubMed/NCBI
|
|
32
|
Katsuno Y, Ehata S, Yashiro M, Yanagihara
K, Hirakawa K and Miyazono K: Coordinated expression of REG4 and
aldehyde dehydrogenase 1 regulating tumourigenic capacity of
diffuse-type gastric carcinoma-initiating cells is inhibited by
TGF-β. J Pathol. 228:391–404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wakamatsu Y, Sakamoto N, Oo HZ, Naito Y,
Uraoka N, Anami K, Sentani K, Oue N and Yasui W: Expression of
cancer stem cell markers ALDH1, CD44 and CD133 in primary tumor and
lymph node metastasis of gastric cancer. Pathol Int. 62:112–119.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Marcato P, Dean CA, Giacomantonio CA and
Lee PW: Aldehyde dehydrogenase: Its role as a cancer stem cell
marker comes down to the specific isoform. Cell Cycle.
10:1378–1384. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Storms RW, Trujillo AP, Springer JB, Shah
L, Colvin OM, Ludeman SM and Smith C: Isolation of primitive human
hematopoietic progenitors on the basis of aldehyde dehydrogenase
activity. Proc Natl Acad Sci USA. 96:9118–9123. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kastan MB, Schlaffer E, Russo JE, Colvin
OM, Civin CI and Hilton J: Direct demonstration of elevated
aldehyde dehydrogenase in human hematopoietic progenitor cells.
Blood. 75:1947–1950. 1990.PubMed/NCBI
|
|
37
|
Collins SJ: Retinoic acid receptors,
hematopoiesis and leukemogenesis. Curr Opin Hematol. 15:346–351.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bosch FX, Lorincz A, Muñoz N, Meijer CJ
and Shah KV: The causal relation between human papillomavirus and
cervical cancer. J Clin Pathol. 55:244–265. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu SY and Zheng PS: High aldehyde
dehydrogenase activity identifies cancer stem cells in human
cervical cancer. Oncotarget. 4:2462–2475. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jiang F, Qiu Q, Khanna A, Todd NW, Deepak
J, Xing L, Wang H, Liu Z, Su Y, Stass SA, et al: Aldehyde
dehydrogenase 1 is a tumor stem cell-associated marker in lung
cancer. Mol Cancer Res. 7:330–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Charafe-Jauffret E, Ginestier C, Iovino F,
Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci
F, Jacquemier J, et al: Aldehyde dehydrogenase 1-positive cancer
stem cells mediate metastasis and poor clinical outcome in
inflammatory breast cancer. Clin Cancer Res. 16:45–55. 2010.
View Article : Google Scholar
|
|
42
|
Deng S, Yang X, Lassus H, Liang S, Kaur S,
Ye Q, Li C, Wang LP, Roby KF, Orsulic S, et al: Distinct expression
levels and patterns of stem cell marker, aldehyde dehydrogenase
isoform 1 (ALDH1), in human epithelial cancers. PLoS One.
5:e102772010. View Article : Google Scholar :
|
|
43
|
Yang L, Ren Y, Yu X, Qian F, Bian BS, Xiao
HL, Wang WG, Xu SL, Yang J, Cui W, et al: ALDH1A1 defines invasive
cancer stem-like cells and predicts poor prognosis in patients with
esophageal squamous cell carcinoma. Mod Pathol. 27:775–783. 2014.
View Article : Google Scholar
|
|
44
|
Zhou W, Lv R, Qi W, Wu D, Xu Y, Liu W, Mou
Y and Wang L: Snail contributes to the maintenance of stem
cell-like phenotype cells in human pancreatic cancer. PLoS One.
9:e874092014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
van den Hoogen C, van der Horst G, Cheung
H, Buijs JT, Pelger RC and van der Pluijm G: The aldehyde
dehydrogenase enzyme 7A1 is functionally involved in prostate
cancer bone metastasis. Clin Exp Metastasis. 28:615–625. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yan J, De Melo J, Cutz JC, Aziz T and Tang
D: Aldehyde dehydrogenase 3A1 associates with prostate
tumorigenesis. Br J Cancer. 110:2593–2603. 2014. View Article : Google Scholar :
|
|
47
|
Singh S, Brocker C, Koppaka V, Chen Y,
Jackson BC, Matsumoto A, Thompson DC and Vasiliou V: Aldehyde
dehydrogenases in cellular responses to oxidative/electrophilic
stress. Free Radic Biol Med. 56:89–101. 2013. View Article : Google Scholar :
|
|
48
|
Gasparetto M, Sekulovic S, Brocker C, Tang
P, Zakaryan A, Xiang P, Kuchenbauer F, Wen M, Kasaian K, Witty MF,
et al: Aldehyde dehydrogenases are regulators of hematopoietic stem
cell numbers and B-cell development. Exp Hematol. 40:318–329.e2.
2012. View Article : Google Scholar
|
|
49
|
Moreb JS, Mohuczy D, Ostmark B and Zucali
JR: RNAi-mediated knockdown of aldehyde dehydrogenase class-1A1 and
class-3A1 is specific and reveals that each contributes equally to
the resistance against 4-hydroperoxycyclophosphamide. Cancer
Chemother Pharmacol. 59:127–136. 2007. View Article : Google Scholar
|
|
50
|
Muzio G, Trombetta A, Maggiora M,
Martinasso G, Vasiliou V, Lassen N and Canuto RA: Arachidonic acid
suppresses growth of human lung tumor A549 cells through
down-regulation of ALDH3A1 expression. Free Radic Biol Med.
40:1929–1938. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pappa A, Brown D, Koutalos Y, DeGregori J,
White C and Vasiliou V: Human aldehyde dehydrogenase 3A1 inhibits
proliferation and promotes survival of human corneal epithelial
cells. J Biol Chem. 280:27998–28006. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Muzio G, Trombetta A, Martinasso G, Canuto
RA and Maggiora M: Antisense oligonucleotides against aldehyde
dehydrogenase 3 inhibit hepatoma cell proliferation by affecting
MAP kinases. Chem Biol Interact. 143–144:37–43. 2003. View Article : Google Scholar
|
|
53
|
Wang JS, Fang Q, Sun DJ, Chen J, Zhou XL,
Lin GW, Lu HZ and Fei J: Genetic modification of hematopoietic
progenitor cells for combined resistance to
4-hydroperoxycyclophosphamide, vincristine, and daunorubicin. Acta
Pharmacol Sin. 22:949–955. 2001.PubMed/NCBI
|
|
54
|
Canuto RA, Muzio G, Ferro M, Maggiora M,
Federa R, Bassi AM, Lindahl R and Dianzani MU: Inhibition of
class-3 aldehyde dehydrogenase and cell growth by restored lipid
peroxidation in hepatoma cell lines. Free Radic Biol Med.
26:333–340. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Calderaro J, Nault JC, Bioulac-Sage P,
Laurent A, Blanc JF, Decaens T and Zucman-Rossi J: ALDH3A1 is
overexpressed in a subset of hepatocellular carcinoma characterised
by activation of the Wnt/β-catenin pathway. Virchows Arch.
464:53–60. 2014. View Article : Google Scholar
|
|
56
|
Vesuna F, Lisok A, Kimble B and Raman V:
Twist modulates breast cancer stem cells by transcriptional
regulation of CD24 expression. Neoplasia. 11:1318–1328. 2009.
View Article : Google Scholar : PubMed/NCBI
|