Introduction
Osteosarcoma (OS), the frequent sites of which are
the femur, the tibia and the humerus, is the most common primary
malignant bone tumor in the world, representing approximately 56%
of all bone cancers (1). By
combining surgery with multi-agent chemotherapy, the 5-year
cumulative survival rate of primary OS has significantly improved
to 60–90% in the past three decades (2,3).
Unfortunately, approximately 80% of patients eventually developed
metastasis following surgical treatment and pulmonary metastasis in
OS patients is a major cause of fatal outcome (4,5).
Therefore, it is crucical to identify effector molecules and
signaling pathways which exhibit a close relationship with tumor
progression and metastasis. Novel findings may improve the existing
OS treatment.
HLA-F locus adjacent transcript 10 (FAT10) is a
member of the ubiquitin-like protein family, which covalently
modifies target substrates by binding via their C-termini
containing conserved Gly-Gly motifs (6,7) and
is well known as a signal for proteasomal degradation (8). FAT10 has been confirmed to be related
with cell growth and survival; the ectopic expression of FAT10
could induce abnormal alterations in the cell cycle, apoptosis, and
immune response (7,9,10).
Recently, the role of FAT10 in malignant tumors has been widely
studied. Overexpression of FAT10 was examined in various cancers,
such as gastrointestinal cancer (11), hepatocellular carcinoma (HCC)
(12,13), pancreatic ductal adenocarcinoma
(14), and human glioma (15). FAT10 has been proved to
significantly promote invasion and metastasis in mutltiple cancers
(11,14,16).
However, the role of FAT10 in OS was not illuminated. Homeobox B9
(HOXB9), a member of the class I homeobox (HOX) genes, played a
critical role in embryonic segmentation and limb patterning
(17). Recently, biologic function
of HOXB9 in tumor metastasis has been confirmed. It has been
reported that the overexpression of HOXB9 in breast cancer could
increase angiogenesis and distal metastasis, and the knockdown of
HOXB9 significantly decreased the ability of lung cancer cells to
form bone and brain metastases (18,19).
Moreover, silencing FAT10 could inhibit HOXB9 expression in HCC
cells. Thus, we presumed that HOXB9 may be the regulatory target of
FAT10 in invasion and metastasis.
In this study, we found that FAT10 was aberrantly
upregulated in OS, especially in metastatic OS. High expression of
FAT10 was correlated with poor prognosis of OS patients.
Furthermore, silencing FAT10 significantly inhibit invasion and
metastasis of OS cells by regulating HOXB9.
Materials and methods
Clinical specimens and cell culture
This study was conducted with the approval of
Institutional Ethical Review Board of the Fourth Affiliated
Hospital, China Medical University. A total of 65 pairs of
specimens and their adjacent tissue samples frozen in liquid
nitrogen were obtained from the Fourth Affiliated Hospital. No
patients had received any antitumor treatments before biopsy. All
cell lines are cultured in adaptive culture medium according to
ATCC and cultured at 37°C in 5% CO2.
Establishment of cell lines
Human shRNA of FAT10 were cloned into pSuper-puro
vector. Retrovirus supernatants were pSuper and pSuper-sh. PKM2
were produced in Phoenix packaging cells. We respectively
transfected the indicated cell lines with these different viral
supernatant containing 4 μg/ml polybrene (Sigma). Then cells were
selected by puromycin (2 μg/ml). The methods were as described
(20).
RNA extraction, reverse transcription,
and real-time RT-PCR
Total RNA was extracted from freshly-frozen samples
or cells with TRIzol reagent (Invitrogen). Total RNA was
reverse-transcribed with First Strand cDNA Synthesis kit
(Invitrogen). Real-time PCR reactions were conducted using Platinum
SYBR Green qPCR SuperMix-UDG reagents (Invitrogen) on the PRISM
7900HT system (Applied Biosystems). All reactions were done in
triplicate and reactions without reverse transcriptase were used as
negative controls. The GAPDH were used as the endogenous controls
and the 2−ΔΔCT equation was used to calculate the
relative expression levels.
Western blot analysis
Western blot analysis was conducted using
anti-FAT10, anti-β-catenin and anti-β-actin. All antibodies were
purchased from Cell Signaling Technology.
Cell migration and invasion assays
The effects of FAT10 or HOXB9 expression on cell
migration and invasion were assessed using the Transwell and
Matrigel assays as previously described (21).
Immunofluorescence
Cells silencing FAT10 were cultured on glass
coverslips (NEST, 801007) in 24-well plate and fixed by 4%
paraformaldehyde for 15 min at 50–60% density followed by washing
in PBS. After blocking in goat serum (1:10 in PBS) for 30 min, the
coverslips were incubated with primary antibodies (diluted in
primary stain diluting buffer, Beyotime, P0103) overnight at 4°C
and secondary antibodies 1 h at 37°C. Nuclei were visualized by
4′,6-diamidine-2-phenylindole staining (DAPI, Solarbio, D8200). The
coverslips touched face down a drop of Anti-fade Mounting Medium
(Beyotime, P0126) on a slide and the fluorescence was captured by
laser scanning confocal microscopy.
In vivo metastasis
Nude mice were purchased from Shanghai Slac
Laboratory Animal Co. Ltd. and maintained in microisolator cages.
All animals were used in accordance with institutional guidelines
and the current experiments were approved by the Use Committee for
Animal Care. For metastasis assays, cells were resuspended in PBS
at a concentration of 3×107 cells/ml. Cell suspension
(0.1 ml) was injected into tail veins of nude mice. All of the mice
were sacrificed by CO2 8 weeks after inoculation.
Statistical analysis
Statistical analysis data are described as the mean
± SD. Survival percent was estimated using the Kaplan-Meier method.
The relationship between survival period and each of the variables
was analyzed using the log-rank test for categorical variables.
Comparisons between different groups were undertaken using the
Student's two-tailed t-test. The criterion of statistical
significance was P<0.05. Statistical analysis was done with
SPSS/Win11.0 software (SPSS Inc.).
Results
Ectopic expression of FAT10 was
correlated with prognosis of OS patients
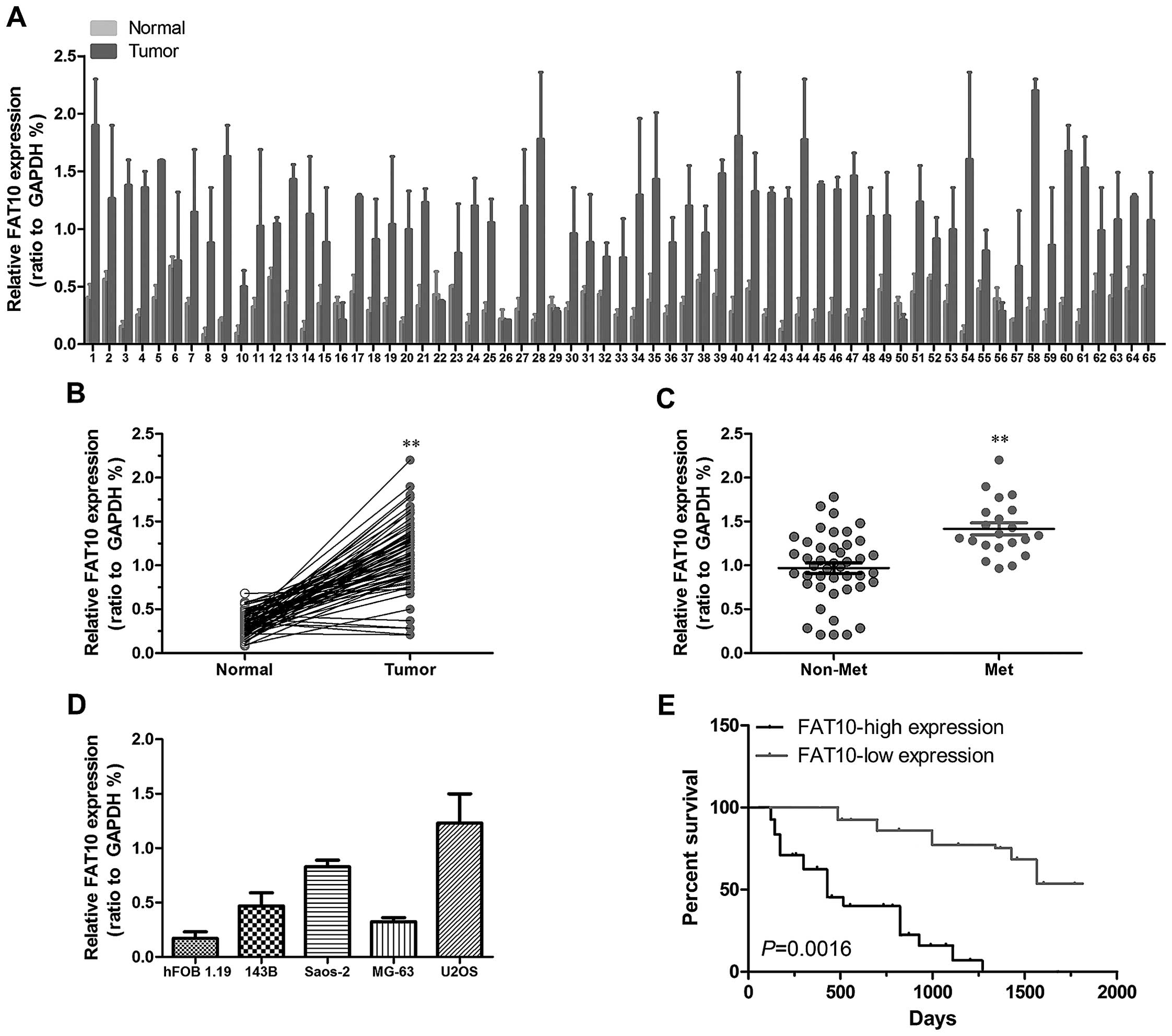
In order to confirm whether FAT10 was upregulated in
OS, we first examined expression levels of FAT10 in OS by qRT-PCR.
As shown in Fig. 1A and B, higher
expression level of FAT10 was examined in 90.7% OS samples which
suggested that FAT10 was correlated with the progression of OS. As
show in the report, metastasis has been proved to be the key cause
of death in cancers. To further confirm the role of FAT10 in OS, we
studied the relationship between FAT10 and metastasis. We first
found that FAT10 was significantly upregulated in metastatic OS
(Fig. 1C) as well as invasive OS
cell lines (Fig. 1D). Moreover,
the analysis of clinical features of OS patients revealed that
higher expression of FAT10 suggested more distant metastasis
(Table I). These data suggested
that FAT10 promoted the metastasis of OS.
 | Table IAssociation of FAT10 expression in
human osteosarcoma tissues with clinicopathological features. |
Table I
Association of FAT10 expression in
human osteosarcoma tissues with clinicopathological features.
| | FAT10 expression | |
|---|
| |
| |
|---|
| Clinicopathological
features | No. of patients | Low (n, %) | High (n, %) | P-value |
|---|
| Gender |
| Male | 35 | 16 (45%) | 19 (55%) | NS |
| Female | 30 | 17 (57%) | 13 (43%) | |
| Age |
| <25 years | 26 | 14 (54%) | 12 (46%) | NS |
| ≥25 years | 39 | 18 (55%) | 19 (45%) | |
| Tumor size |
| >8 cm | 34 | 11 (32%) | 23 (68%) | 0.012 |
| ≤8 cm | 31 | 17 (55%) | 14 (45%) | |
| Serum level of
alkaline phosphatase |
| Elevated | 49 | 31 (63%) | 18 (37%) | NS |
| Normal | 16 | 8 (50%) | 8 (50%) | |
| Anatomic
location |
| Tibia/femur | 44 | 21 (48%) | 23 (52%) | NS |
| Elsewhere | 21 | 11 (52%) | 10 (48%) | |
| Distant
metastasis |
| Absent | 48 | 32 (67%) | 16 (33%) | <0.001 |
| Present | 17 | 5 (29%) | 12 (71%) | |
| Response to
chemotherapy |
| Good | 24 | 18 (75%) | 6 (25%) | <0.001 |
| Poor | 41 | 13 (32%) | 28 (68%) | |
| Clinical stage |
| IIA | 29 | 23 (79%) | 6 (21%) | <0.001 |
| IIB/III | 36 | 7 (19%) | 29 (81%) | |
| Serum level of
lactate dehydrogenase |
| Elevated | 43 | 25 (58%) | 17 (42%) | NS |
| Normal | 22 | 10 (45%) | 12 (55%) | |
Metastasis is closely related with the
prognosis of cancer patients
To evaluate whether FAT10 is related with prognosis
of OS patients, we carried out bioinformatics analysis of the
dataset. As shown in Fig. 1E,
patients with higher FAT10 mRNA level in OS tissues had poorer
survival than those with lower FAT10 expression level suggesting
that FAT10 expression significantly correlated with the prognosis
of OS patients.
Silencing FAT10 inhibits migration and
invasion of OS cells
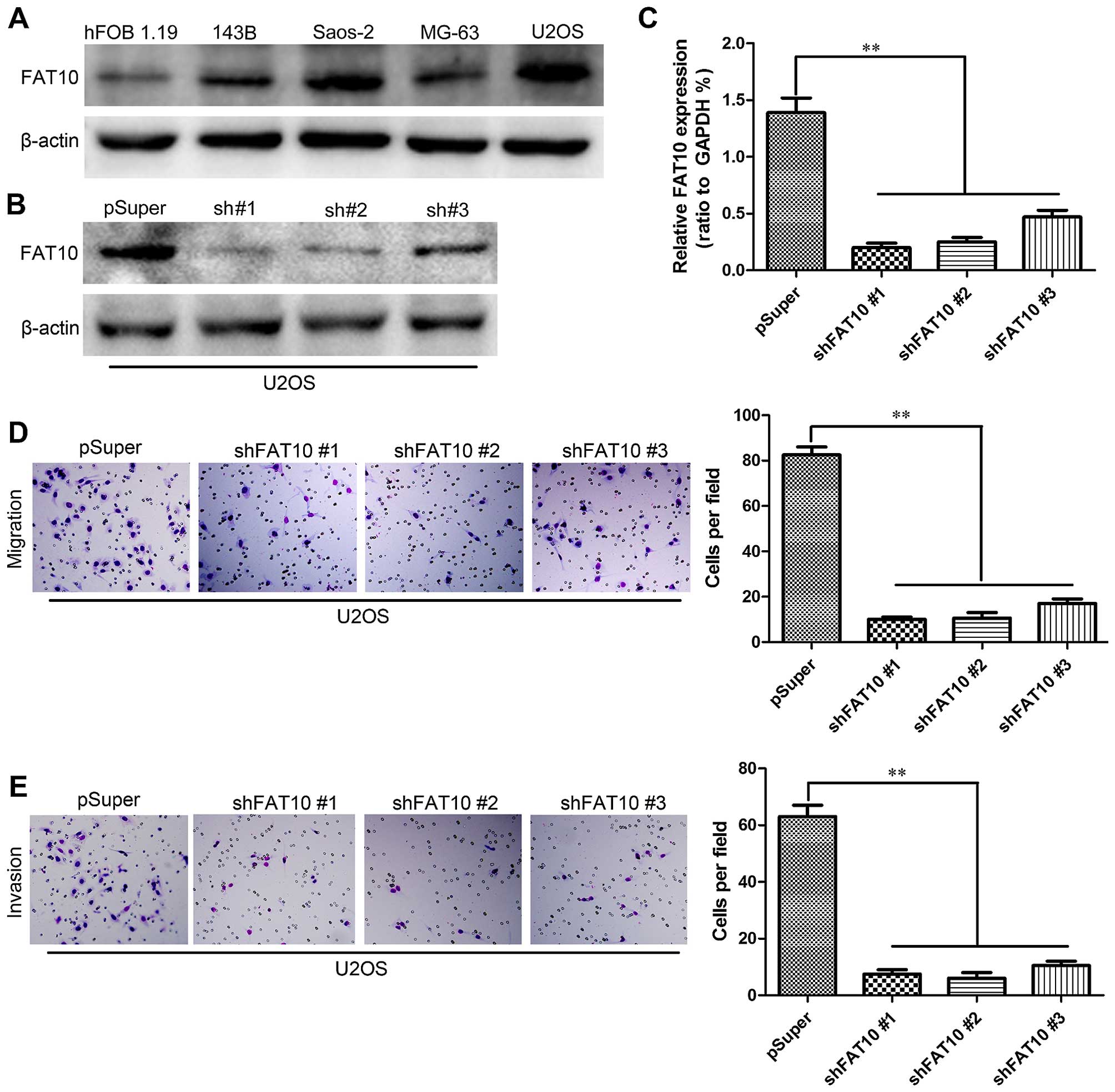
To further confirm the role of FAT10 in metastasis,
we first examined the expression levels of FAT10 in several OS cell
lines by western blotting (Fig.
2A) and then we silenced the expression of FAT10 in U2OS with
high expression of FAT10 by shRNA. The subsequent levels of FAT10
were examined by western blotting (Fig. 2B) and qRT-PCR (Fig. 2C). We then evaluated the effects of
FAT10 on migration and invasion of OS cells. Transwell assay
revealed that silencing FAT10 significantly inhibit the numbers of
U2OS cells migrated through the membrane to the bottom of the
aperture (Fig. 2D). Moreover,
Matrigel assay was used to evaluate the invasive potential of OS
cells with altered FAT10 expression. As shown Fig. 2E, silencing FAT10 decreased the
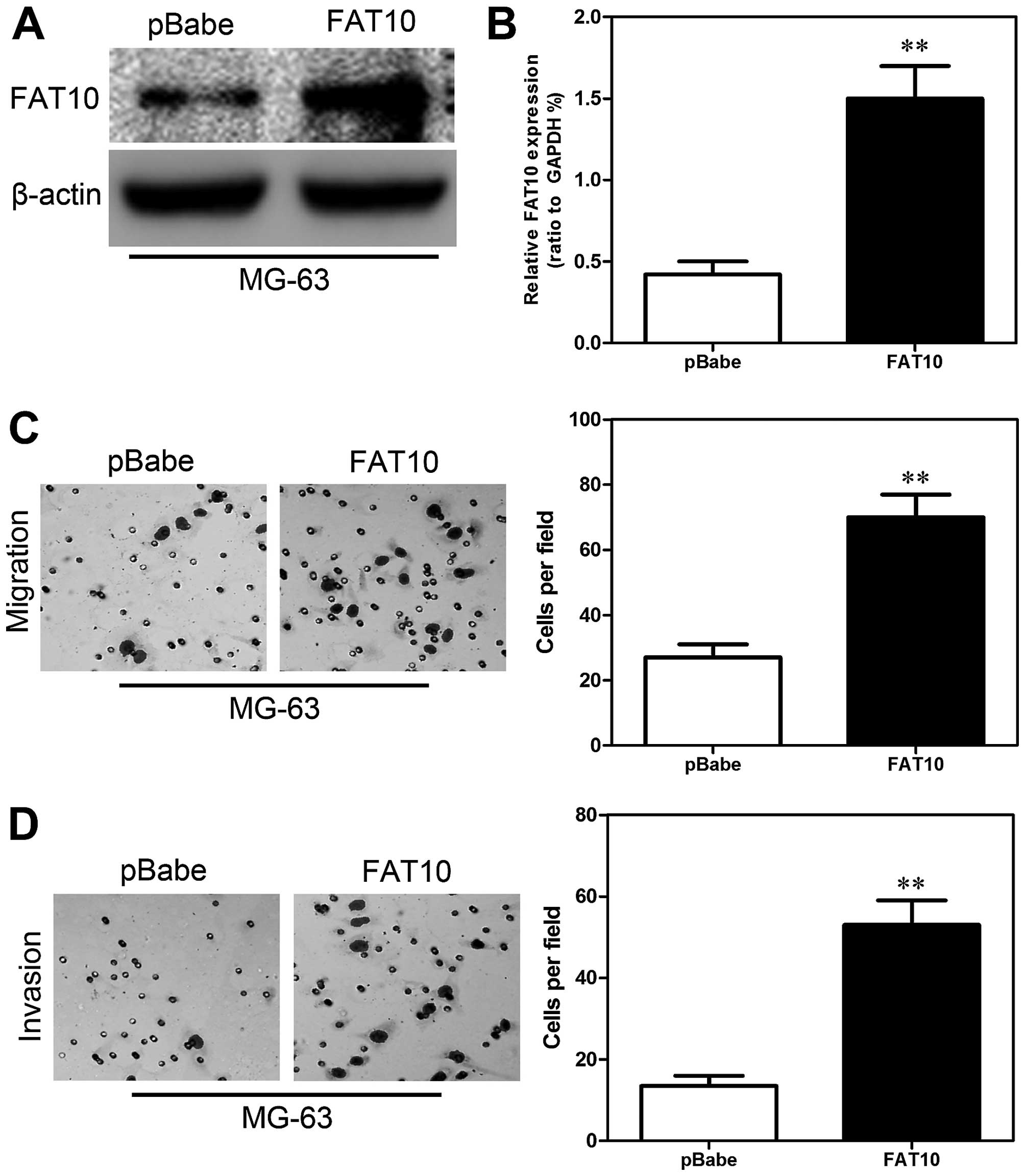
numbers of invaded cells. In order to further confirm the role of
FAT10 in migration and invasion, we overexpressed FAT10 in MG-63
cells. The expression of FAT10 was confirmed by western blotting
(Fig. 3A) and qRT-PCR (Fig. 3B). Then we examined the migratory
and invasive abilities of MG-63-FAT10 cells. Transwell assay
revealed that overexpression of FAT10 significantly increased the
number of migrated cells (Fig. 3C)
and Matrigel assay also showed similar results (Fig. 3D). These results revealed that
FAT10 was essential for the migration and invasion of OS cells.
Silencing FAT10 inhibits osteosarcoma
cell metastasis in vivo
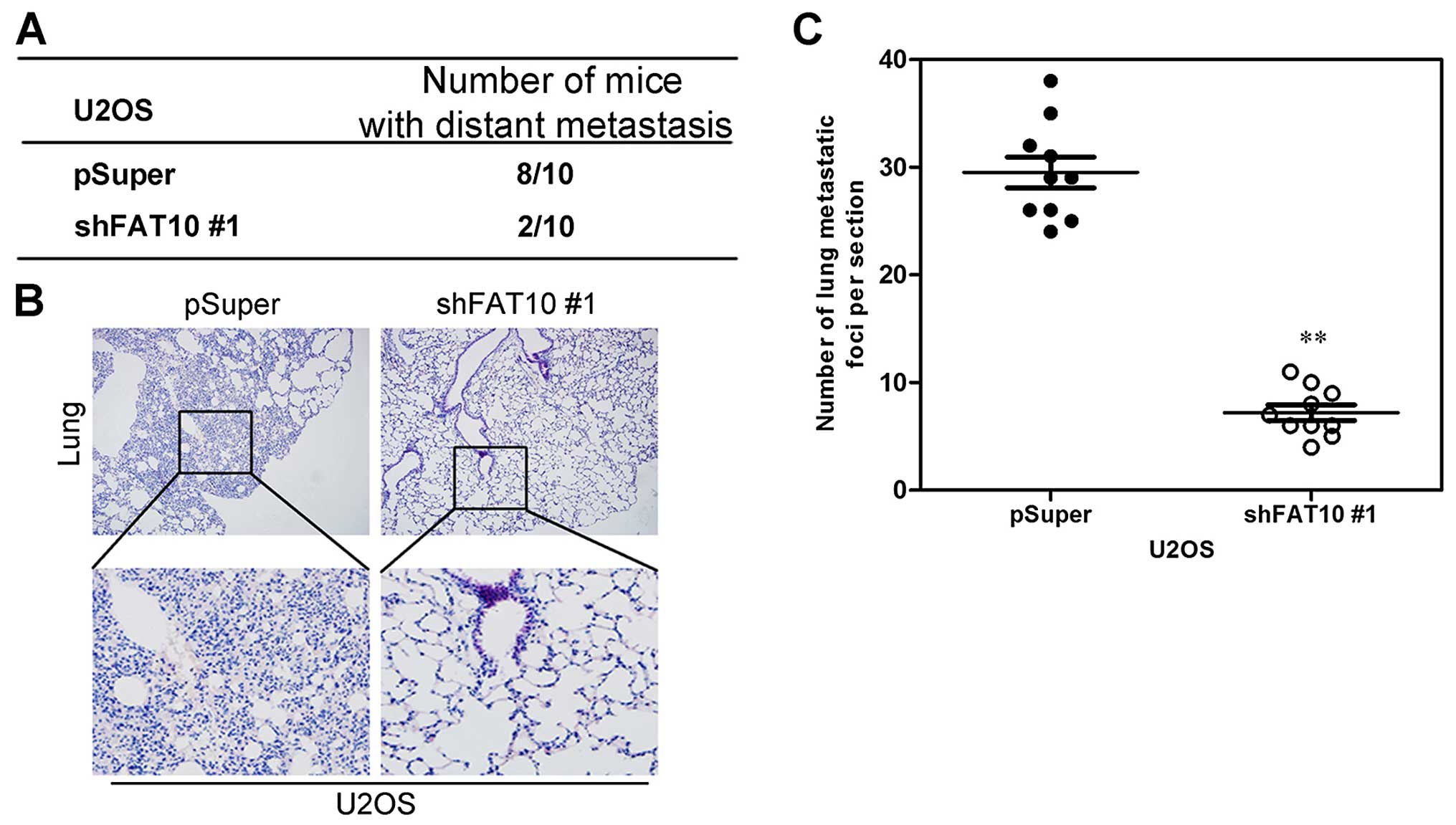
To examine whether the function of FAT10 in
migration and invasion in vitro was relevant to metastasis
of OS cells in vivo, U2OS cells with silent FAT10 was
inoculated into tail vein of BALB/C athymic mice. After 8 weeks, we
observed that less mice, injected with OS cells with silent FAT10,
had distant metastasis (Fig. 4A).
In addition, less metastatic foci in lungs were counted in each
mouse injected with OS cells with silent FAT10 (Fig. 4B and C).
Collectively, both in vivo and in vitro
data strongly showed the biological role of FAT10 as an inducer of
metastasis in OS
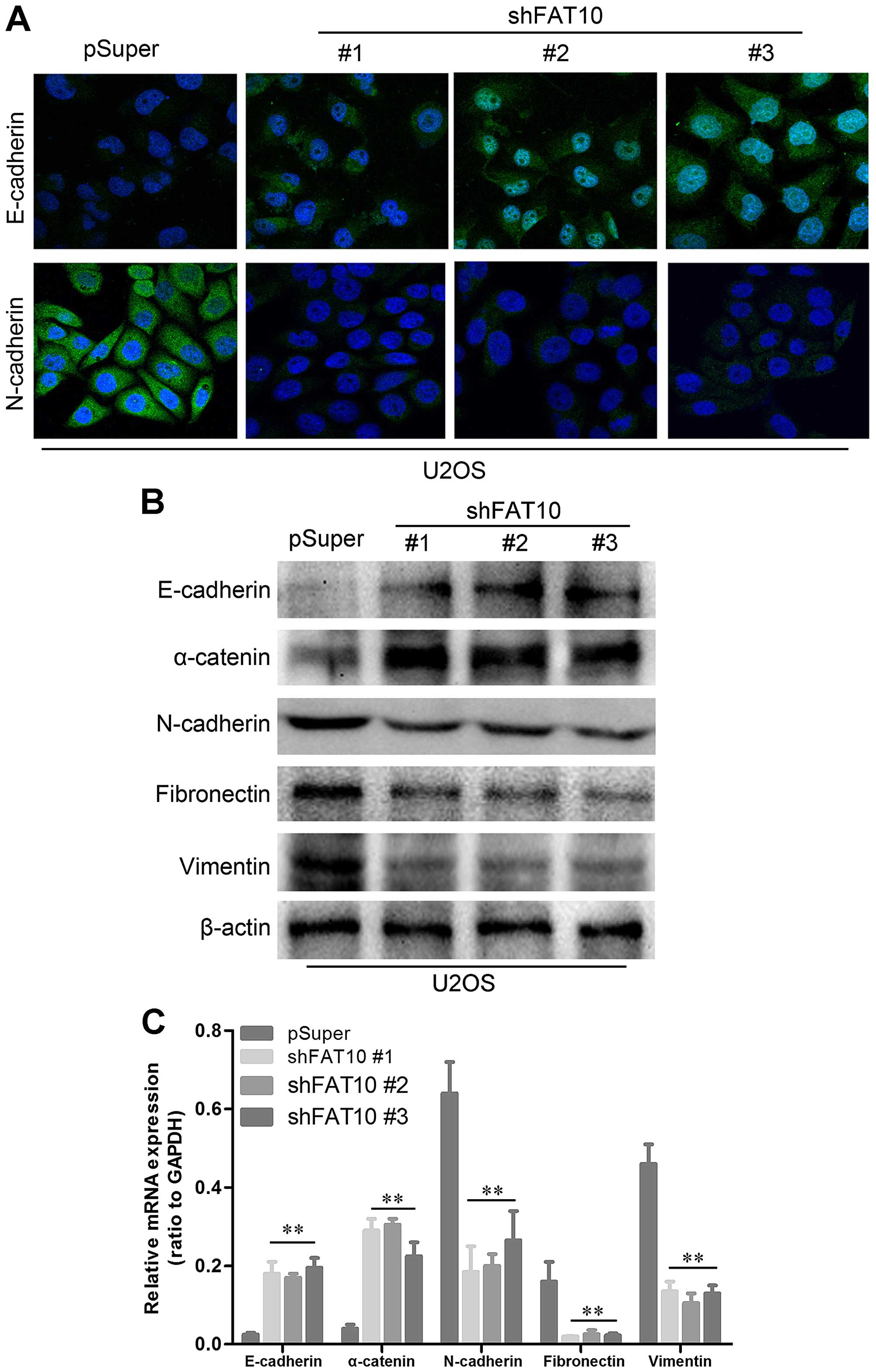
Silencing FAT10 inhibited EMT of OS cells.
Epithelial mesenchymal transformation (EMT) has been proved to be
the key step of metastasis. To evaluate the role of FAT10 in EMT,
we examined the expression levels of some EMT markers. As shown in
Fig. 5A, upregulated E-cadherin
and loss of N-cadherin were found in U2OS with silent FTA10 by
immunofluorescence. Moreover, western blotting revealed upregulated
levels of epithelial markers and loss of mesenchymal markers
(Fig. 5B). We also revealed that
all the changes of EMT-related proteins were based on the ectopic
expressions of EMT-related genes suggesting that FAT10 may regulate
the expression of EMT markers at DNA levels (Fig. 5C).
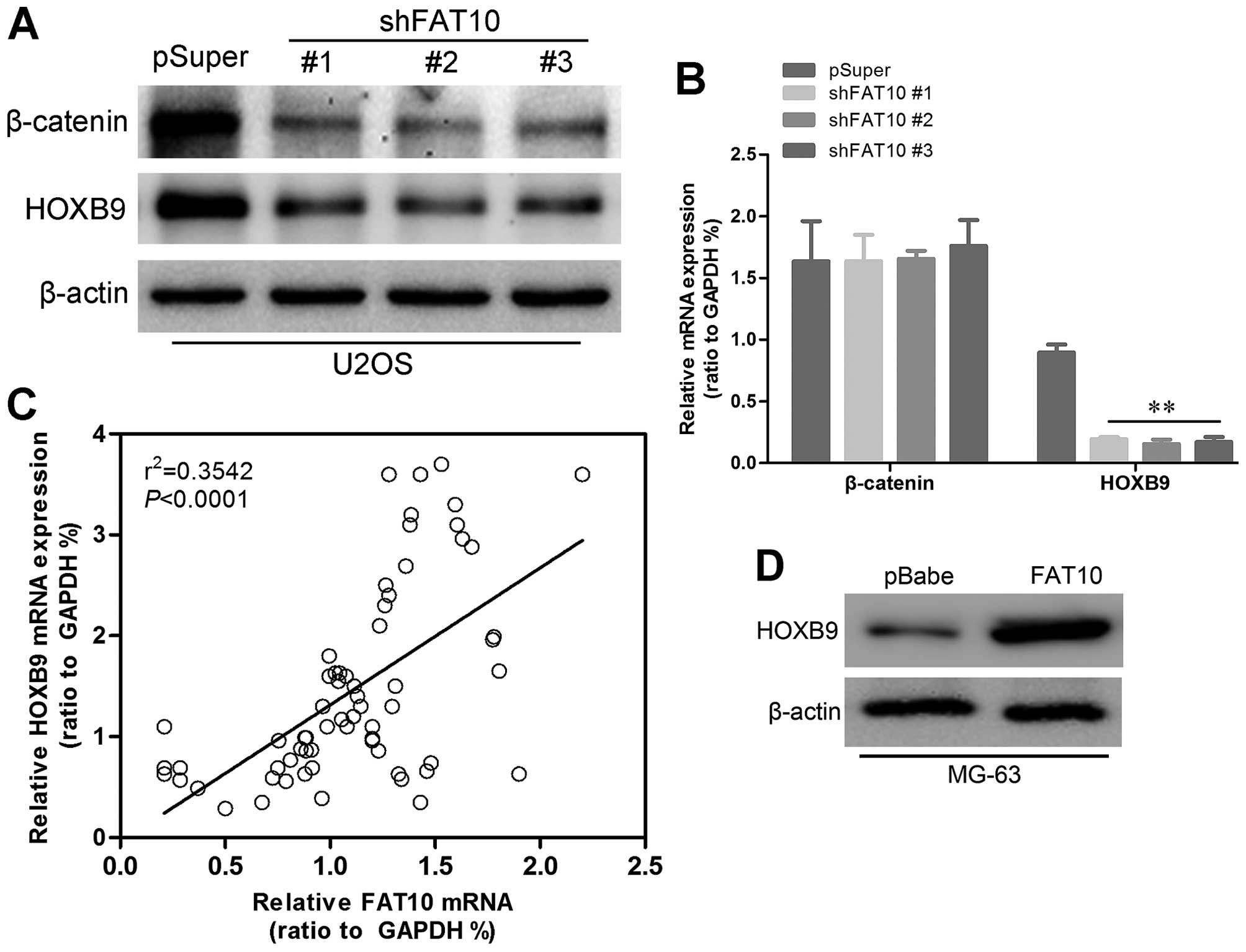
HOXB9 may be the key mediator of
FAT10-induced metastasis of OS patients
HOXB9 has been reported to be a regulatory target of
FAT10, here we also found downregulated HOXB9 in U2OS with silent
FAT10 (Fig. 6A) while upregulated
HOXB9 was examined in MG-63-FAT10 cells (Fig. 6D) suggesting that FAT10 may
regulate expression of HOXB9. Western blotting also revealed that
β-catenin which activated expression of HOXB9 decreased in
U2OS-shFAT10 cells. Moreover, we found a decrease in RNA level of
HOXB9 but no changes in RNA level of β-catenin (Fig. 6B) which indicated that
post-transcriptional modification occurred in β-catenin. In order
to further confirm the relationship between FAT10 and HOXB9, we
performed linear correlated analysis. As shown in Fig. 6C, the expression levels of HOXB9
were positively correlated with the expression levels of FAT10. We
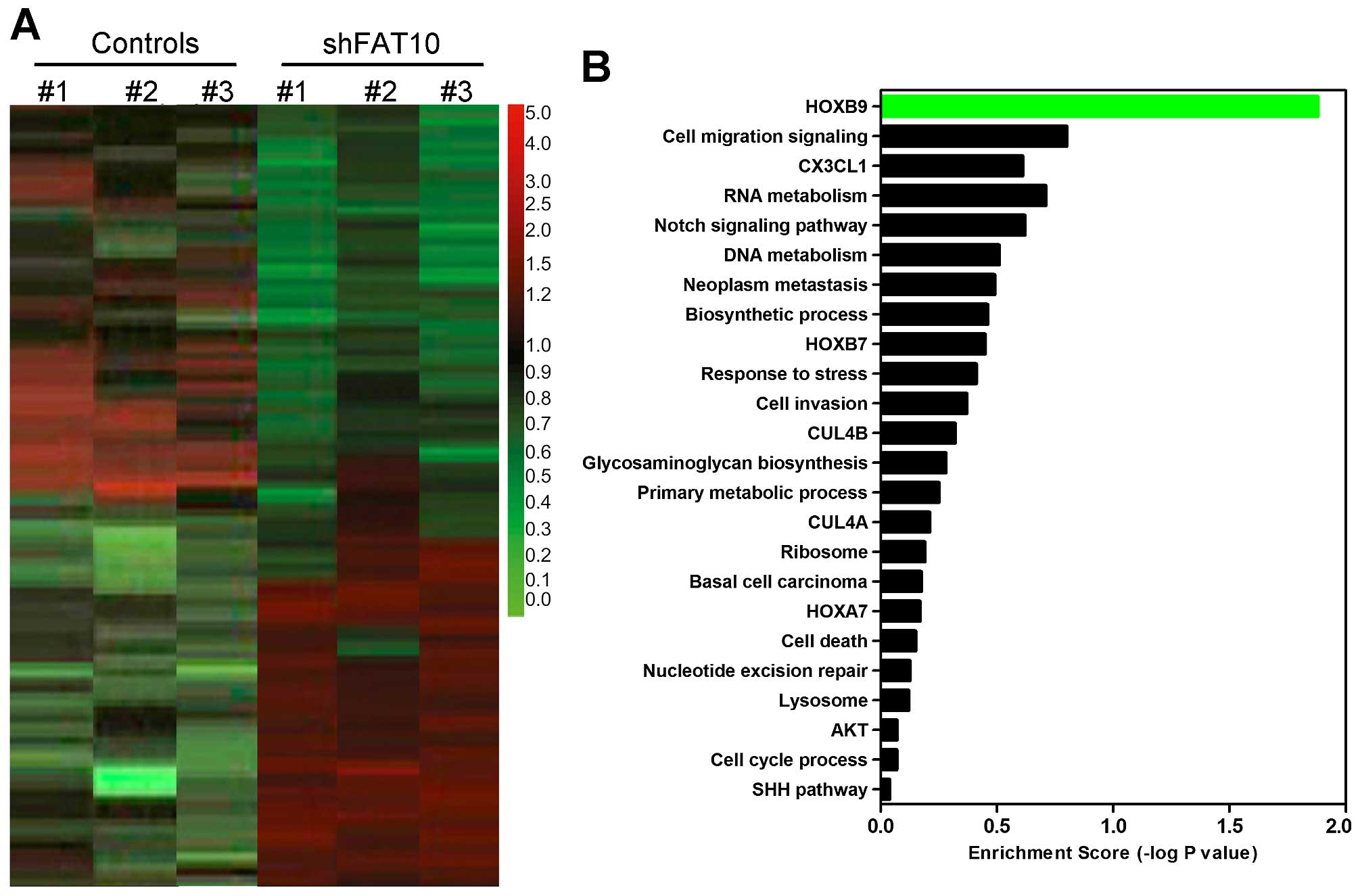
confirmed the relationship between FAT10 and HOXB9 by microarray
assay. As shown in Fig. 7A,
silencing FAT10 induced, respectively, increasing expression (red)
and decreasing expression (green) at DNA levels. HOXB9 was
significantly inhibited by FAT10 in gene enrichment assay (Fig. 7B). These data suggested that FAT10
regulated HOXB9 possibly through β-catenin pathway.
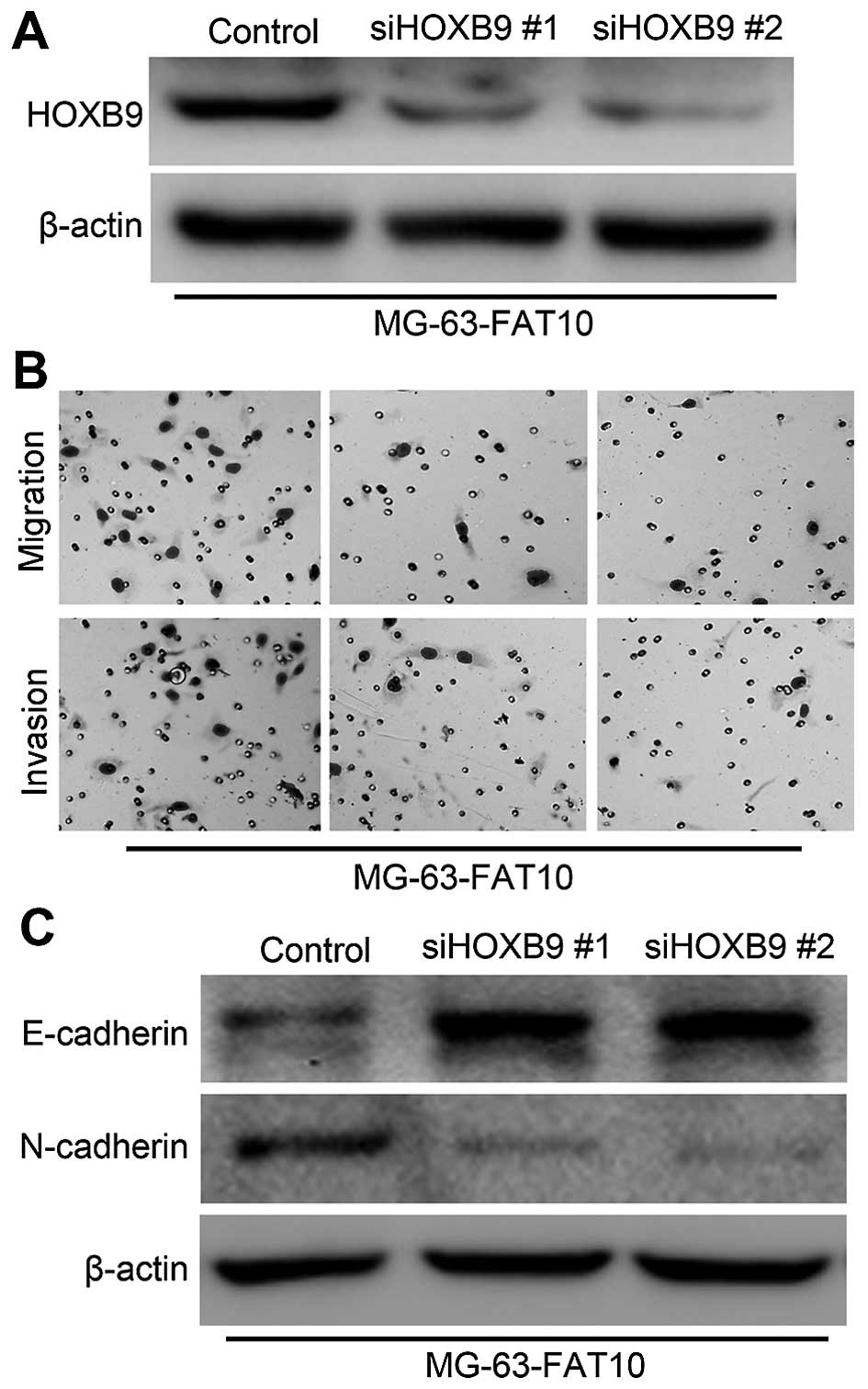
In order to confirm the role of HOXB9, we silenced
the expression of HOXB9 in MG-63 cells with overexpression of
FAT10. The expression levels of HOXB9 were confirmed by western
blotting (Fig. 8A). Then we
examined the invasiveness and migration MG-63 cells. As shown in
Fig. 8B, Transwell and Matrigel
assays revealed that silencing HOXB9 significantly decreased the
number of migrated and invaded cells in MG-63-FAT10 cells. We also
found that silencing HOXB9 promoted the expression of epithelial
marker E-cadherin but inhibited the expression of mesenchymal
marker N-cadherin (Fig. 8C). The
data above suggested that HOXB9 was essential in the FAT10-induced
migration and invasion.
Discussion
FAT10 is a member of the ubiquitin-like protein
family. Herein, we first revealed that FAT10 could induce EMT in OS
cells and promoted migration and invasion of OS cells in
vitro as well as metastasis in vivo. Moreover, FAT10 was
upregulated in OS, especially in metastatic OS and higher
expression level of FAT10 was correlated with poorer prognosis of
OS patients. In this study, we revealed that FAT10 promoted
metastasis of OS possibly by regulating β-catenin pathway in which
HOXB9 played a critical role.
In recent years, FAT10 has been shown to participate
in tumor migration and invasion. In a report, FAT10 promoted cells
invasion and migration through regulating NF-κB-CXCR4/7 pathway
(22). In another study, FAT10
promoted the invasion of HCC cells via the Akt/GSK3b pathway
(23). Moreover, HOXB9 has been
shown to be upregulated by FAT10 in invasion and metastasis of
hepatocarcinoma cells (16). In
this study, we suggested a similar mechanism in which FAT10
promoted OS cell invasion and metastasis by upregulating HOXB9.
First, FAT10 inhibition reduced HOXB9 expression and decreased the
invasion and metastasis of OS cells in vitro and in
vivo. Second, the logistic regression results revealed that
co-overexpression of FAT10 and HOXB9. These data suggested a
possible mechanism in which FAT10 promoted metastasis of OS
patients.
In addition, previous studies have shown that HOXB9
is regulated by the Wnt/β-catenin/TCF4 pathway (18,24).
The Wnt/β-catenin signaling plays a critical role in promoting
progression and metastasis of multiple cancers (25–27).
It has been reported that β-catenin is the critical effector of the
Wnt signaling pathway and played the key role in the transduction
of Wnt signal (28,29). It is also known that β-catenin is
targeted to ubiquitin proteasome-mediated degradation (30). FAT10 is a ubiquitin-like protein,
and it functions similarly to poly-ubiquitin as a tag for
proteasome targeting (8). In this
study, we revealed that silencing FAT10 could decrease the
expression of β-catenin at protein level, but not RAN level, based
on which we speculate that FAT10 may regulate β-catenin at its
protein level. This finding suggested a novel possible mechanism of
degradation of β-catenin.
Collectively, we revealed for the first time that
FAT10 promoted invasion and metastasis of OS cells. Higher level of
FAT10 indicated poor prognosis of OS patients suggesting that FAT10
may be a potential therapeutic target for OS patients.
References
|
1
|
Schwab JH, Springfield DS, Raskin KA,
Mankin HJ and Hornicek FJ: What's new in primary bone tumors. J
Bone Joint Surg Am. 94:1913–1919. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cho Y, Jung GH, Chung SH, Kim JY, Choi Y
and Kim JD: Long-term survivals of stage IIb osteosarcoma: A
20-year experience in a single institution. Clin Orthop Surg.
3:48–54. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lewis VO: What's new in musculoskeletal
oncology. J Bone Joint Surg Am. 91:1546–1556. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wada T, Isu K, Takeda N, Usui M, Ishii S
and Yamawaki S: A preliminary report of neoadjuvant chemotherapy
NSH-7 study in osteosarcoma: Preoperative salvage chemotherapy
based on clinical tumor response and the use of granulocyte
colony-stimulating factor. Oncology. 53:221–227. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dikic I, Wakatsuki S and Walters KJ:
Ubiquitin-binding domains - from structures to functions. Nat Rev
Mol Cell Biol. 10:659–671. 2009. View
Article : Google Scholar
|
|
7
|
Raasi S, Schmidtke G and Groettrup M: The
ubiquitin-like protein FAT10 forms covalent conjugates and induces
apoptosis. J Biol Chem. 276:35334–35343. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hipp MS, Kalveram B, Raasi S, Groettrup M
and Schmidtke G: FAT10, a ubiquitin-independent signal for
proteasomal degradation. Mol Cell Biol. 25:3483–3491. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren J, Wang Y, Gao Y, Mehta SB and Lee CG:
FAT10 mediates the effect of TNF-α in inducing chromosomal
instability. J Cell Sci. 124:3665–3675. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Merbl Y, Refour P, Patel H, Springer M and
Kirschner MW: Profiling of ubiquitin-like modifications reveals
features of mitotic control. Cell. 152:1160–1172. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji F, Jin X, Jiao CH, Xu QW, Wang ZW and
Chen YL: FAT10 level in human gastric cancer and its relation with
mutant p53 level, lymph node metastasis and TNM staging. World J
Gastroenterol. 15:2228–2233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee CG, Ren J, Cheong IS, Ban KH, Ooi LL,
Yong Tan S, Kan A, Nuchprayoon I, Jin R, Lee KH, et al: Expression
of the FAT10 gene is highly upregulated in hepatocellular carcinoma
and other gastrointestinal and gynecological cancers. Oncogene.
22:2592–2603. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lukasiak S, Schiller C, Oehlschlaeger P,
Schmidtke G, Krause P, Legler DF, Autschbach F, Schirmacher P,
Breuhahn K and Groettrup M: Proinflammatory cytokines cause FAT10
upregulation in cancers of liver and colon. Oncogene. 27:6068–6074.
2008. View Article : Google Scholar
|
|
14
|
Sun GH, Liu YD, Yu G, Li N, Sun X and Yang
J: Increased FAT10 expression is related to poor prognosis in
pancreatic ductal adenocarcinoma. Tumour Biol. 35:5167–5171. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yuan J, Tu Y, Mao X, He S, Wang L, Fu G,
Zong J and Zhang Y: Increased expression of FAT10 is correlated
with progression and prognosis of human glioma. Pathol Oncol Res.
18:833–839. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yuan R, Wang K, Hu J, Yan C, Li M, Yu X,
Liu X, Lei J, Guo W, Wu L, et al: Ubiquitin-like protein FAT10
promotes the invasion and metastasis of hepatocellular carcinoma by
modifying β-catenin degradation. Cancer Res. 74:5287–5300. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abate-Shen C: Deregulated homeobox gene
expression in cancer: Cause or consequence? Nat Rev Cancer.
2:777–785. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nguyen DX, Chiang AC, Zhang XH, Kim JY,
Kris MG, Ladany M, Gerald WL and Massagué J: WNT/TCF signaling
through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis.
Cell. 138:51–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hayashida T, Takahashi F, Chiba N,
Brachtel E, Takahashi M, Godin-Heymann N, Gross KW, Vivanco M,
Wijendran V, Shioda T, et al: HOXB9, a gene overexpressed in breast
cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad
Sci USA. 107:1100–1105. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wen M, Kwon Y, Wang Y, Mao JH and Wei G:
Elevated expression of UBE2T exhibits oncogenic properties in human
prostate cancer. Oncotarget. 6:25226–25239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Guessous F, Johnson EB, Eberhart CG,
Li XN, Shu Q, Fan S, Lal B, Laterra J, Schiff D, et al: Functional
and molecular interactions between the HGF/c-Met pathway and c-Myc
in large-cell medulloblastoma. Lab Invest. 88:98–111. 2008.
View Article : Google Scholar
|
|
22
|
Gao Y, Theng SS, Zhuo J, Teo WB, Ren J and
Lee CG: FAT10, an ubiquitin-like protein, confers malignant
properties in non-tumorigenic and tumorigenic cells.
Carcinogenesis. 35:923–934. 2014. View Article : Google Scholar
|
|
23
|
Liu L, Dong Z, Liang J, Cao C, Sun J, Ding
Y and Wu D: As an independent prognostic factor, FAT10 promotes
hepatitis B virus-related hepatocellular carcinoma progression via
Akt/GSK3β pathway. Oncogene. 33:909–920. 2014. View Article : Google Scholar
|
|
24
|
Hatzis P, van der Flier LG, van Driel MA,
Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE,
Welboren W, et al: Genome-wide pattern of TCF7L2/TCF4 chromatin
occupancy in colorectal cancer cells. Mol Cell Biol. 28:2732–2744.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miki T, Yasuda SY and Kahn M:
Wnt/β-catenin signaling in embryonic stem cell self-renewal and
somatic cell reprogramming. Stem Cell Rev. 7:836–846. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lai TY, Su CC, Kuo WW, Yeh YL, Kuo WH,
Tsai FJ, Tsai CH, Weng YJ, Huang CY and Chen LM: β-catenin plays a
key role in metastasis of human hepatocellular carcinoma. Oncol
Rep. 26:415–422. 2011.PubMed/NCBI
|
|
27
|
Lin SY, Xia W, Wang JC, Kwong KY, Spohn B,
Wen Y, Pestell RG and Hung MC: Beta-catenin, a novel prognostic
marker for breast cancer: Its roles in cyclin D1 expression and
cancer progression. Proc Natl Acad Sci USA. 97:4262–4266. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Polakis P: The oncogenic activation of
beta-catenin. Curr Opin Genet Dev. 9:15–21. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rubinfeld B, Robbins P, El-Gamil M, Albert
I, Porfiri E and Polakis P: Stabilization of beta-catenin by
genetic defects in melanoma cell lines. Science. 275:1790–1792.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Aberle H, Bauer A, Stappert J, Kispert A
and Kemler R: beta-catenin is a target for the ubiquitin-proteasome
pathway. EMBO J. 16:3797–3804. 1997. View Article : Google Scholar : PubMed/NCBI
|