Introduction
Renal cell carcinoma (RCC) is the most common type
of kidney cancer, >200,000 new cases and 100,000 deaths are
estimated to occur worldwide each year (1). Up to 30% of RCC patients present with
metastases at the time of diagnosis and nearly half of the rest
will subsequently develop metastases in their course. When
metastasis occurs, it is largely incurable, with a very poor 5-year
survival rate (2). RCC is highly
resistant to chemotherapy and radiotherapy, immunotherapies such as
interleukin-2 and interferon α are once used as first-line
treatments for metastatic RCC (mRCC), however, the response rates
are extremely low (3). Newly
developed targeted-therapies based on the understanding of
molecular mechanisms of RCC progression make significant
improvements over immunotherapies for mRCC. Unfortunately, <40%
of patients have response to targeted-therapies and nearly all
patients will eventually develop resistance (4,5). It
appears that a therapeutic ceiling has been reached for mRCC, thus,
it is important to comprehensively study the mechanisms of how RCC
develops metastasis, and explore promising therapeutic approaches
for this disease.
SPOP, a BTB/POZ domain containing speckle-type POZ
protein, was first identified as a component for the E3 ubiquitin
ligase (6). In Drosophila,
D-SPOP (ortholog of human SPOP) has been shown to promote the
ubiquitination and degradation of Cubitus interruptus (Ci) in the
Hedgehog pathway, and JNK phosphatase puckered (Puc) in the tumor
necrosis factor (TNF) pathway, respectively (7–9). In
human, SPOP has been recently shown to mediate ubiquitination of
the death domain-associated protein (Daxx) (10), the polycomb group protein BMI-1,
the histone variant MacroH2A (11), and the transcription factor Gli
(9).
In RCC, hypoxia-inducible factor (HIF) and mammalian
target of rapamycin (mTOR) pathways are considered as the most
predominant pathways controlling RCC development and progression
(12,13), and therapies targeting these two
pathways have brought clinical benefits to mRCC (14). A more recent study shows that SPOP
is a direct target of HIF, and cytoplasmic SPOP promotes RCC
tumorigenesis through the ubiquitination and degradation of
multiple regulators of cellular proliferation and apoptosis,
including the tumor suppressor PTEN, ERK phosphatases DUSP7, the
proapoptotic molecule Daxx, and the Hedgehog pathway transcription
factor Gli2 (15). However, other
potential functions of SPOP in RCC have not been studied. In this
study, we aim to determine whether SPOP promotes invasion and
metastasis in RCC.
Materials and methods
Human RCC specimens and
immunohistochemistry staining
Forty-seven human RCC and 11 matched normal kidney
specimens were obtained from patients who underwent surgical
resection with the approval of the Institutional Review Board
(IRB). Immunohistochemistry was performed as described previously
(16). Briefly, paraffin-embedded
sections were subjected to deparaffinization, rehydration, and
heat-induced antigen retrieval. After blocking of endogenous
peroxidase with 3% hydrogen peroxide, sections were subsequently
incubated with primary SPOP antibody (Santa Cruz Biotechnology),
horseradish-peroxidase-labeled dextran polymer (Dako EnVision™) and
developed with 3,3′-diaminobenzidine chromogen followed by counter
staining with hematoxylin. All stains were assessed by an
independent pathologist according to the histologic scoring system
(H-score) based on the product of staining intensity (0, no
staining; 1, weak; 2, moderate; and 3, strong) and percentage of
stained cells (0, 0%; 1, 1–30%; 2, 31+70%; and 3, 71–100%). The
expression of SPOP in each tissue was considered either negative
(H-score, <2) or positive (H-score, >2).
Cell culture
Human normal kidney cell HK-2, RCC cells 786-0,
A498, RCC4 and 769-P were maintained in RPMI-1640 medium (Gibco)
supplied with 10% fetal bovine serum (FBS), RCC cells ACHN, CAKI-1,
CAKI2 and A498 were maintained in Dulbecco’s modified Eagle’s
medium (DMEM) (Gibco) supplied with 10% FBS. All the cells were
cultured in a humidified incubator containing 5% CO2 at
37°C.
cDNA constructs, siRNA and
transfection
Human SPOP cDNA cloned into pDONR221 vector was
obtained from the DNASU Plasmid Repository. Control siRNA (si-NC)
and siRNA specifically targeting SPOP (si-SPOP) were from Guangzhou
RiboBio Co., Ltd. For transfections, 2×105 cells were
seeded in 6-well plate and cultured overnight, SPOP plasmids or
siRNAs were transfected into cells by Xfect™ transfection reagent
(Clontech) according to the manufacturer’s instructions.
Twenty-four hours (h) after the transfection, cell protein or RNA
was collected for further assays.
Transwell invasion assay
Matrigel-coated Transwell chambers were applied to
examine RCC cell in vitro invasive ability. RCC cells were
pre-transfected with the indicated plasmids or siRNAs, 100 μl 0.5%
FBS medium of 3×104 cell suspension was then planted
into the upper chamber, and 600 μl of 10% FBS medium was supplied
to the lower chamber. Cells were cultured at 37°C in 5%
CO2 for 24 h. Invaded cells onto the lower surface of
the upper chambers were stained with 0.5% crystal violent (Sigma)
and photographed and counted.
Immunofluorescence
Microslide cultured cells were fixed with 4%
paraformaldehyde, permeabilized with 0.3% Triton X-100 and blocked
with 5% bovine serum albumin. Cells were incubated with β-catenin
primary antibody (Cell Signaling Technology) overnight at 4°C and
subsequently incubated with AlexaFluor 488-conjugated secondary
antibody (Sigma) for 1 h at room temperature, followed by nuclear
staining with 4,6-diamidino-2-phenylindole and fluorescence was
visualized by fluorescence microscopy (Olympus Optical Co.)
Reverse transcriptional (RT) real-time
PCR
Cell total RNA was extracted with RNeasy mini kit
(Qiagen) and reverse transcribed with cDNA synthesis kit
(Invitrogen). Real-time PCR analysis was set up with SYBR Green
qPCR Supermix kit (Invitrogen) supplied with commercial primers
specific for the indicated genes, and carried out in the iCycler
thermal cycler (Bio-Rad). The relative level of mRNA expression of
each gene was determined by normalizing with an internal control
gene GAPDH.
Western blotting
Western blotting was performed as previously
described (17). Cells were first
lysed and total proteins were collected, equivalent amounts of
protein were separated on 10% NuPAGE Bis-Tris gels (Invitrogen) and
transferred to nitrocellulose membranes. Membranes were blocked
with 3% skim-milk (w/v), and incubated with primary antibodies
overnight at 4°C. After washing, membranes were incubated with
appropriate secondary antibodies conjugated with horseradish
peroxidase and signals were then detected by chemiluminescence
(Pierce). Primary SPOP, vimentin, pan-cytokeratin (Pan-CK), TCF4
and GAPDH antibodies were purchased from Santa Cruz Biotechnology;
E-cadherin and α-SMA antibodies were from BD Biosciences, ZEB1 and
MMP-2 antibodies were from Cell Signaling Technology.
Bioinformatic and statistical
analyses
The RNA-sequencing-based mRNA expression data for
SPOP, TCF4, ZEB1 genes and the reverse phase protein array-based
protein expression data for β-catenin of human clear cell RCC
samples were all retrieved from The Cancer Genome Atlas (TCGA) Data
Portal (18). Gene microarray data
for SPOP of human normal kidney and clear cell RCC tissues were
retrieved from the GEO datasets (GSE14994, GSE781 and GSE15641).
SPOP gene microarray data and DNA copy number data of multiple
types of cancer cell lines were retrieved from the Cancer Cell Line
Encytopedia (CCLE) datasets. The Kaplan-Meier analysis (long-rank
test) was performed to analyze recurrence-free survival. Pearson’s
correlation coefficient was used to test the association between
genes. Data from in vitro assay are presented as the mean ±
SEM from three independent experiments, and the differences between
two groups were compared by Student’s two-tail t-test. All
statistical analyses were performed by GraphPad Prism6 (GraphPad
Software).
Results
SPOP is highly expressed in clear cell
RCC
Previous studies suggest that overexpression of SPOP
may lead to dysregulation of pathways involved in tumorigenesis
(8,9). We assessed SPOP expression in
urological tumors including prostate cancer, bladder cancer, RCC
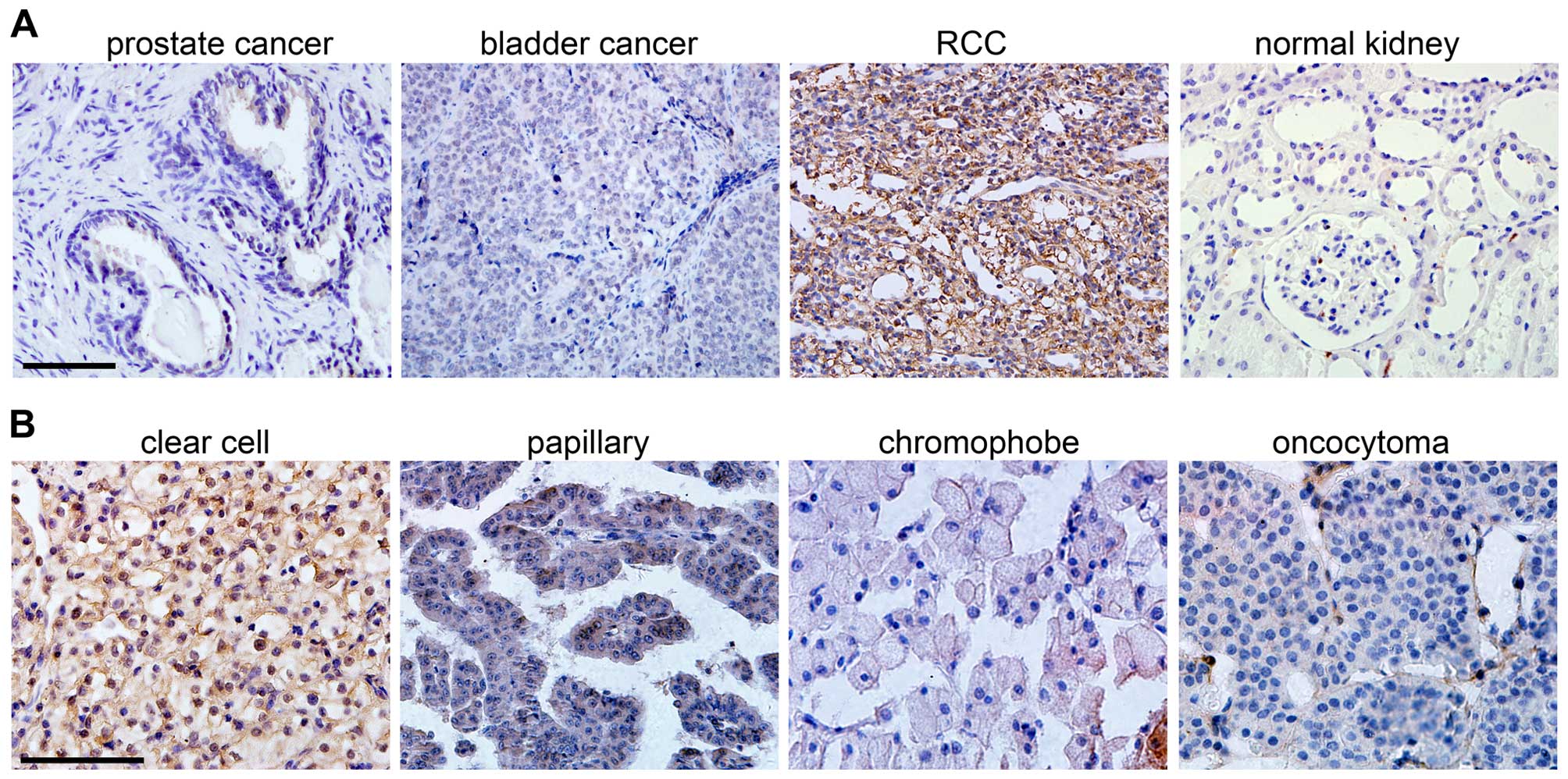
and normal kidney tissues by immunohistochemistry. Interestingly,
we found SPOP was negative in prostate cancer, bladder cancer and
normal kidney tissues, but it was highly expressed in RCC tissues
(Fig. 1A). RCC is a heterogeneous
group of tumors with distinct histological subtypes, including
clear cell, papillary, chromophobe, and other rare subtypes in
addition to oncocytoma (19). When
RCC subtypes were stratified, we found that the papillary,
chromphobe or oncocytoma RCC were weak or negative for SPOP, but
clear cell RCC was positively stained with this protein (Fig. 1B). In total we analyzed 47 clear
cell RCC and matched 11 normal kidney tissues, and the results
showed that 83% clear cell RCC were positive for SPOP, while only
18% normal tissues were positive (Table I). This indicates that SPOP is
highly expressed in clear cell RCC and may serve as a specific
biomarker for this type of RCC.
 | Table ISPOP IHC staining in normal and clear
cell RCC tissues. |
Table I
SPOP IHC staining in normal and clear
cell RCC tissues.
| SPOP | |
|---|
|
| |
|---|
| Positive (%) | Negative (%) | P-value |
|---|
| Normal tissues | 2 (18) | 9 (82) | <0.001 |
| Clear cell RCC | 39 (83) | 8 (17) | |
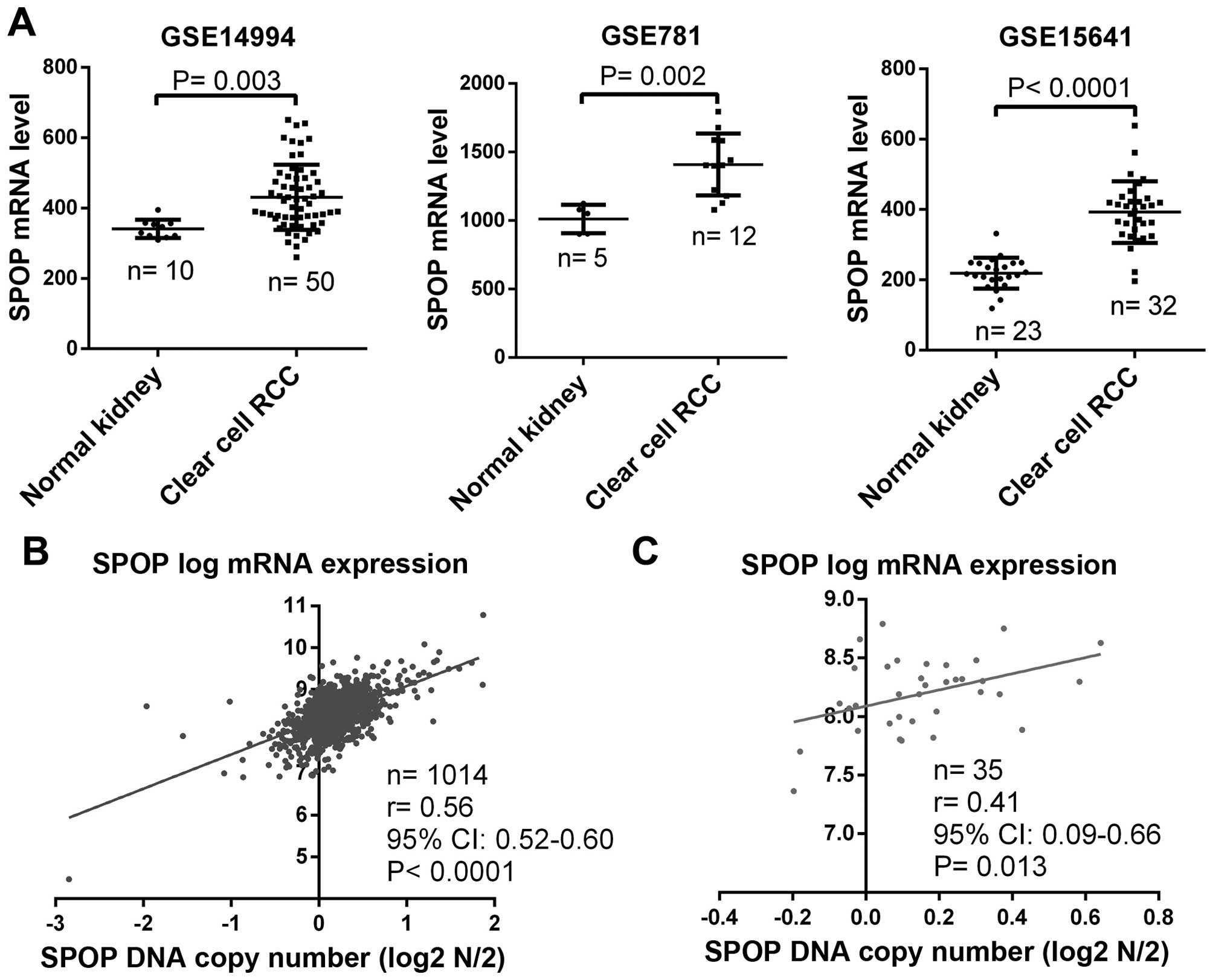
In addition to the determination of SPOP protein
status in clear cell RCC, we checked SPOP mRNA expression by
analysis of gene microarray data of normal kidney and clear cell
RCC from the GEO datasets. Results from three independent datasets
consistently showed that SPOP mRNA was significantly upregulated in
clear cell RCC compared to normal kidney (Fig. 2A). To further explore whether SPOP
upregulation is due to genomic abnormality, we analyzed the
association of SPOP mRNA level and its DNA copy number in multiple
types of cancer cell lines including 1,014 samples from the CCLE
datasets, and found there was a positive correlation between SPOP
mRNA level and its DNA copy number (Fig. 2B), and in RCC cell lines, a
similarly positive correlation was also found (Fig. 2C). These data suggest that the
upregulation of SPOP in RCC may be due to the genomic
variation.
SPOP is associated with progressive clear
cell RCC
Previous studies indicate that SPOP plays important
roles during tumor cell apoptosis and proliferation (7,10,15),
we further investigated the potential functions of SPOP in tumor
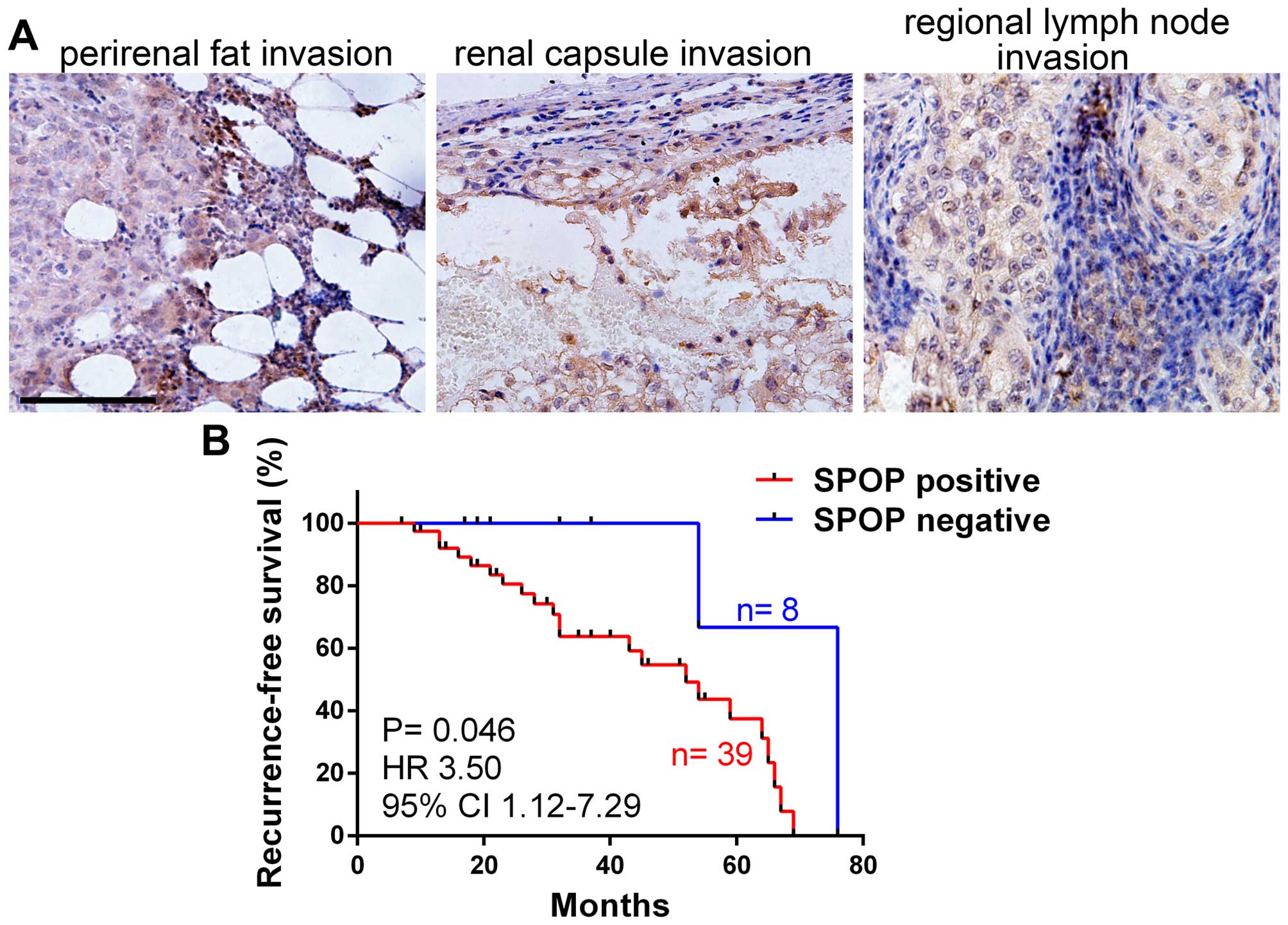
progression. The expression of SPOP in human clear cell RCC with
local invasion (tumor cell invaded into perirenal fat, renal
capsule or regional lymph node) was detected by
immunohistochemistry, notably, the results showed that almost all
the RCC with local invasion were SPOP-positive (Fig. 3A). We compared SPOP expression in
RCC with different pathological stages according to the 2010 AJCC
TNM classfication (20), and found
RCC in T3–4 stages (primary tumors with local invasion) showed much
high frequency of SPOP positive staining compared to RCC in T1–2
stages (without local invasion). Similarly, RCC with lymph node
invasion (N1) or distant metastasis (M1) showed very high frequency
of SPOP positive staining compared to RCC in N0 (without lymph node
invasion) or M0 (without distant metastasis) (Table II). These date suggest SPOP is
associated with clear cell RCC invasion and metastasis. We further
analyzed the association of SPOP expression and tumor
recurrence-free survival, and the result demonstrated that SPOP was
negatively correlated with RCC recurrence-free survival (Fig. 3B), indicating SPOP as novel
prognostic marker for RCC patients.
 | Table IISPOP IHC staining in clear cell RCC
with local invasion/metastasis. |
Table II
SPOP IHC staining in clear cell RCC
with local invasion/metastasis.
| SPOP |
|---|
|
|
|---|
| Positive (%) | Negative (%) |
|---|
| Tumor stage |
| T1–2 | 17 (70) | 7 (30) |
| T3–4 | 22 (96) | 1 (4) |
| Lymph nodes |
| N0 | 28 (80) | 7 (20) |
| N1 | 11 (92) | 1 (8) |
| Metastasis |
| M0 | 31 (79) | 8 (21) |
| M1 | 8 (100) | 0 (0) |
SPOP promotes the invasiveness of
RCC
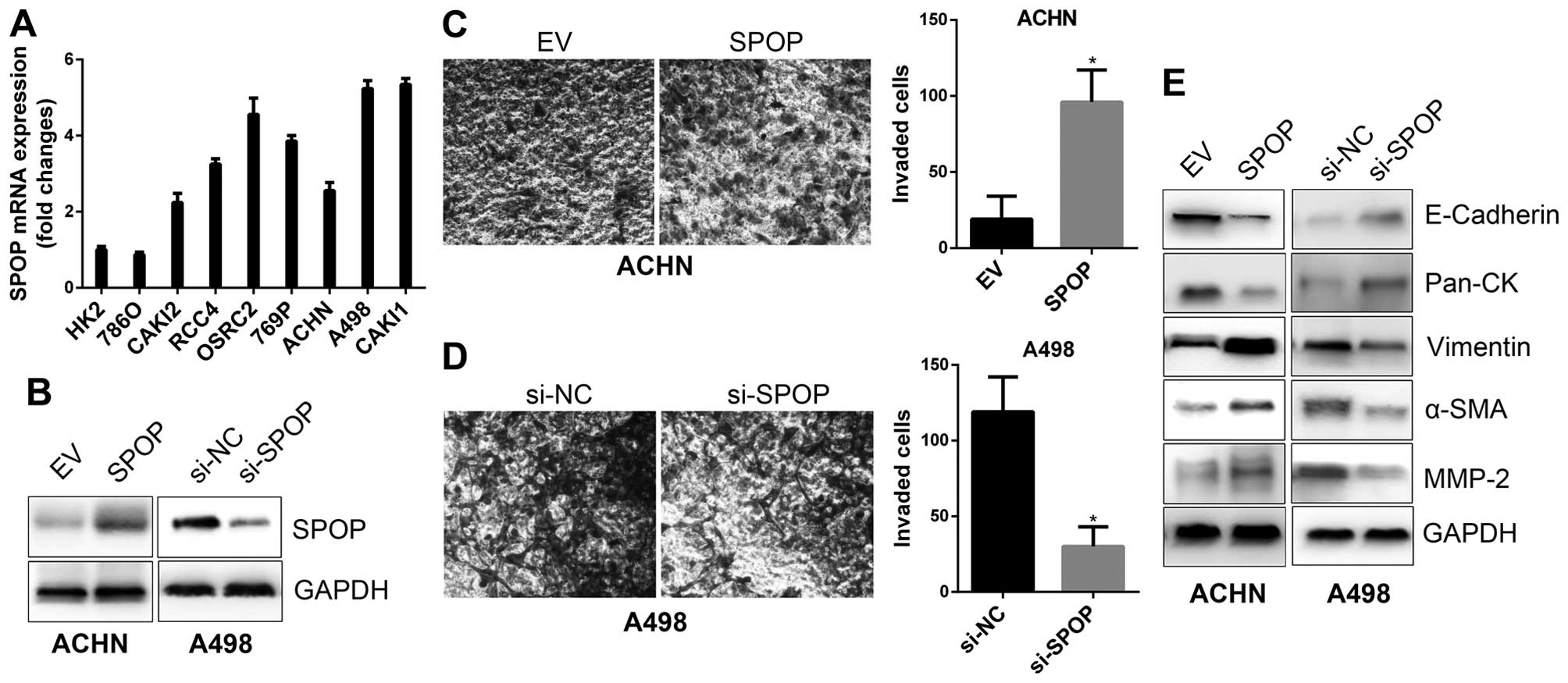
To confirm the biological function of SPOP in RCC
invasion, in vitro cell line based assays were performed.
Profile of SPOP expression in a series of RCC cell lines by RT-PCR
showed ACHN cells with low SPOP expression, thus, it was applied
for SPOP overexpression model, while A498 cells with high SPOP was
applied for SPOP silencing model (Fig.
4A and B). In vitro Transwell invasion assays
demonstrated that overexpression of SPOP promoted ACHN invasion
(Fig. 4C), while silencing of SPOP
by siRNA in A498 cells suppressed cell invasion (Fig. 4D). It has been well documented that
epithelial-mesenchymal transition (EMT) is a process thought to
initiate metastasis, and it is characterized by the gain of
mesenchymal markers (e.g., vimentin, α-SMA, MMP2) and the loss of
epithelial markers (e.g., E-cadherin, cytokeratins), as well as
increased motility and invasion of cancer cells (21,22).
We examined the expression of EMT markers in SPOP overexpressing or
silencing cells, and the results showed that SPOP downregulated
epithelial markers, such as E-cadherin and Pan-CK, and upregulated
mesenchymal makers, such as vimentin, α-SMA and MMP-2 (Fig. 4E). These data indicate SPOP as an
inducer for the invasiveness of RCC cells.
Mechanisms of SPOP in regulation of RCC
invasion
We further dissected the molecular mechanisms of
SPOP in regulation of RCC invasion. Firstly, the associations
between SPOP and a panel of EMT markers and EMT-inducing
transcription factors (EMT-TFs) were analyzed in human clear cell
RCC samples from TCGA datasets by Pearson correlation analyses. The
results showed negative correlations between SPOP and epithelial
marker CDH1 (E-cadherin), and positive correlations between SPOP
and mesenchymal makers VIM (vimentin), ACTA2 (α-SMA) and MMP-2
(Table III). Importantly, SPOP
was positively correlated with many critical EMT-TFs, such as TCF4
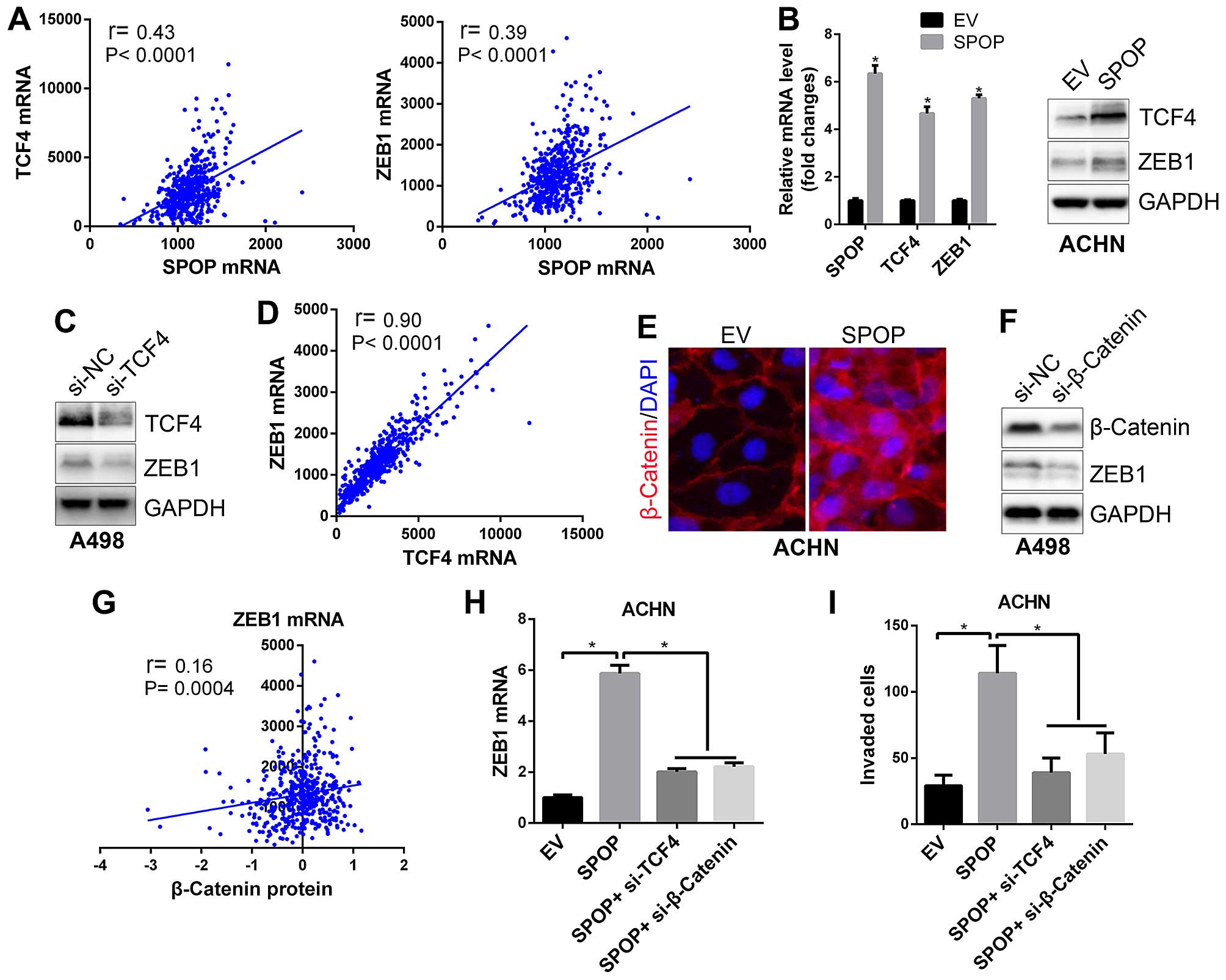
and ZEB1 (Table III and Fig. 5A). Further cell line based assays
confirmed that overexpression of SPOP upregulated TCF4 and ZEB1
expression (Fig. 5B). ZEB1 is well
known as one of the most critical transcriptional factors driving
EMT in many cancer cells (23),
and β-catenin/TCF4 complex has been demonstrated to bind ZEB1 gene
promoter region and promote its transcription (24). Our data showed that silencing of
TCF4 in RCC cells suppressed ZEB1 expression (Fig. 5C), and there was an extremely
positive correlation between TCF4 and ZEB1 mRNA in clear cell RCC
samples (Fig. 5D), these data
again confirmed that TCF4 is an upstream regulator for ZEB1
expression. Additionally, we observed that SPOP could also
upregulate cytosolic β-catenin protein expression and promote its
nuclear translocation (Fig. 5E).
Silencing of β-catenin suppressed ZEB1 expression (Fig. 5F), and there was a positive
correlation between β-catenin protein and ZEB1 expression in clear
cell RCC samples (Fig. 5G),
suggesting β-catenin as upstream regulator for ZEB1. Furthermore,
although overexpression of SPOP upregulated ZEB1 expression as well
as cell invasion, co-transfection of TCF4 siRNA or β-catenin siRNA
could ablate the effects of SPOP on ZEB1 gene expression and cell
invasive ability (Fig. 5H and I).
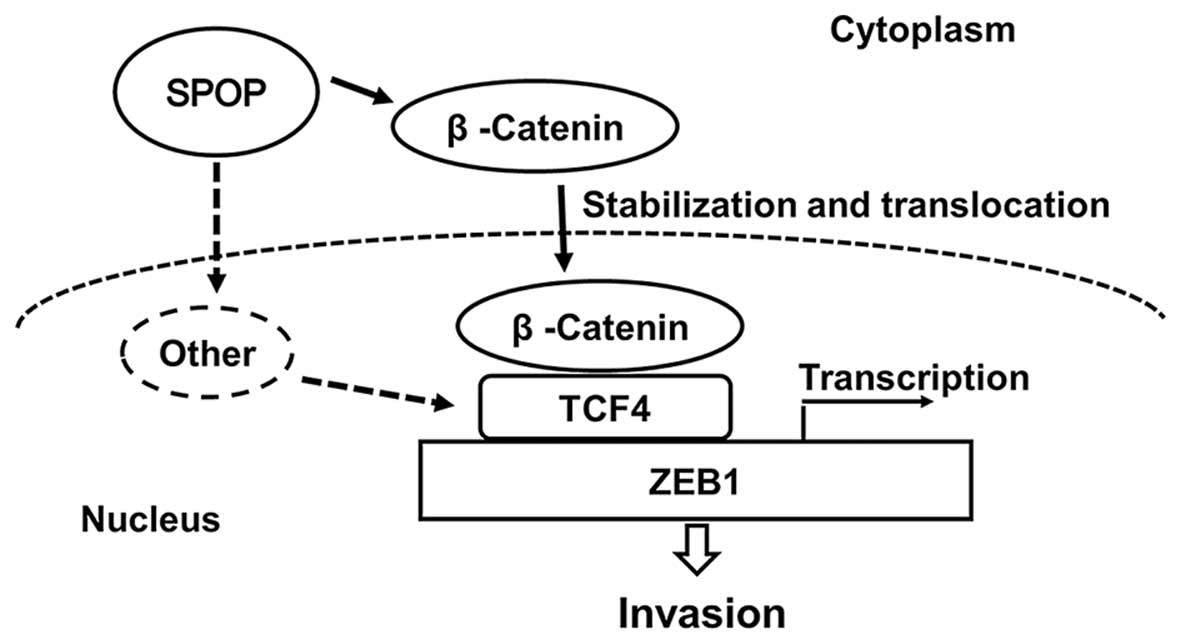
Taken together, our results indicate that SPOP promotes ZEB1 to
drive RCC cell invasion via activating the β-catenin/TCF4 complex
(Fig. 6).
 | Table IIIPearson correlation analyses of the
mRNA expression of SPOP and EMT related genes in human RCC samples
from TCGA dataset (RNA Seq V2 RSEM). |
Table III
Pearson correlation analyses of the
mRNA expression of SPOP and EMT related genes in human RCC samples
from TCGA dataset (RNA Seq V2 RSEM).
| Pearson r | 95% CI | P-value | Significant
(α=0.05) | No. of samples |
|---|
| SPOP vs. CDH1 | −0.1875 | −0.27 to −0.10 | <0.0001 | Yes | 534 |
| SPOP vs. VIM | 0.1393 | 0.06 to 0.22 | 0.0013 | Yes | 534 |
| SPOP vs. ACTA2 | 0.185 | 0.10 to 0.27 | <0.0001 | Yes | 534 |
| SPOP vs. MMP2 | 0.2734 | 0.19 to 0.35 | <0.0001 | Yes | 534 |
| SPOP vs. MMP9 | −0.0442 | −0.13 to 0.04 | 0.3079 | No | 534 |
| SPOP vs. TCF4 | 0.4277 | 0.36 to 0.49 | <0.0001 | Yes | 534 |
| SPOP vs. ZEB1 | 0.3942 | 0.32 to 0.46 | <0.0001 | Yes | 534 |
| SPOP vs. ZEB2 | 0.2884 | 0.21 to 0.36 | <0.0001 | Yes | 534 |
| SPOP vs. SMAD4 | 0.2086 | 0.13 to 0.29 | <0.0001 | Yes | 534 |
| SPOP vs. SNAI1 | 0.2581 | 0.18 to 0.34 | <0.0001 | Yes | 534 |
| SPOP vs. SNAI2 | 0.3063 | 0.23 to 0.38 | <0.0001 | Yes | 534 |
| SPOP vs.
TWIST1 | 0.1092 | 0.025 to 0.19 | 0.0115 | Yes | 534 |
Discussion
RCC is a clinicopathologically heterogeneous disease
with distinct histological subtypes, including clear cell which
accounts for the 70% of cases, and other rare subtypes, such as
papillary, chromophobe, and oncocytoma (25). Although different subtypes of RCC
exhibit certain distinguishing morphology, diagnostic difficulties
arise when one subtype displays morphologic features that overlap
with others. Recent advances are paving the way for seeking
specific molecular abnormalities based on improved knowledge of the
cytogenetics and molecules to recognize distinct molecular
subtypes. A panel of immunohistochemical markers are used to
differentiate the major subtypes of RCC. Unfortunately, these
markers lack specificity and sensitivity. For example, carbonic
anhydrase IX has been proposed as a sensitive marker for clear cell
RCC, but it is not positive for all cases (26). Vimentin, a broad mesenchymal
marker, is expressed in 87–100% clear cell and papillary RCC, but
also in 73% oncocytoma (27). PAX2
is found to be a good marker for kidney cancers, but it is also
positive for normal kidney tissues (28). We find that SPOP is negative in
prostate cancer and bladder cancer but positive in 83% of the clear
cell RCC, and all the cases with local invasion included for this
study are positive. Although a large cohort of samples is required
for further study, results from small number of samples in this
study indicate SPOP may serve as a new marker for clear cell RCC,
especial for metastatic cases.
Epithelial-mesenchymal transition (EMT) is a process
thought to initiate metastasis, and it is characterized by the gain
of mesenchymal markers (e.g., vimentin, α-SMA, MMP2) and the loss
of epithelial markers (e.g., E-cadherin, cytokeratins), as well as
increased motility and invasion of cancer cells. EMT is driven by
many EMT-inducing transcription factors (EMT-TFs) (21,23).
The well documented EMT-TFs include the Zinc-finger factors Snail,
Slug, ZEB2 and ZEB1, and the HLH factors Twist and E12/E47. All of
which directly bind to E-boxes in the promoter of the E-cadherin
gene and repress its expression (29). By analyses of the associations of
SPOP and EMT markers and a panel of EMT-TFs in a large cohort of
clear cell RCC samples, we find SPOP is negatively correlated with
epithelial maker and positively correlated with mesenchymal markers
and all the EMT-TFs, suggesting SPOP indeed plays essential roles
in inducing EMT and promoting RCC progression, which is consistent
with the finding that SPOP predicts a poor recurrence-free survival
of RCC patient.
Within the SPOP associated EMT-TFs, we further
confirm ZEB1 is the critical downstream effector of SPOP to drive
RCC cell invasion. ZEB1 has been reported to be regulated by the
TGF-β signaling pathway (30) and
Wnt/β-catenin signaling pathway (24). The activation of Wnt signaling
inactivates the glycogen synthase kinase-3β, and leads to the
stabilization of β-catenin protein in cytoplasm followed by the
nuclear translocation to complex with TCF4 and enhance the
transcriptional activity of TCF4 (31). β-catenin/TCF4 complex has been
demonstrated to bind the ZEB1 promoter region and promote its
transcription in intestinal tumor cells (24). We confirm that SPOP upregulates
ZEB1 in clear cell RCC by promoting β-catenin protein nuclear
translocation and TCF4 mRNA expression. SPOP, as an E3 ubiquitin
ligase component, and appears to promote RCC tumorigenesis by the
ubiquitination and degradation of PTEN, DUSP7 and Daxx as
previously reported (15).
However, results in this study indicate new actions of SPOP in RCC,
of which SPOP seems to regulate β-catenin in posttranscriptional
level and TCF4 in transcriptional level, further studies are
required to illuminate the details of the mechanism.
Acknowledgements
This study was supported by the Shaanxi Provincial
Key Scientific Foundation (grant no. 2013KTCL03-04 to Y. Xu), the
National Natural Science Foundation of China (grant no.
NSFC81172436 to Y. Sun) and the Shaanxi Provincial Natural Science
Foundation (grant no. 2016JQ8011 to J. Zhou).
References
|
1
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hollingsworth JM, Miller DC, Daignault S
and Hollenbeck BK: Five-year survival after surgical treatment for
kidney cancer: A population-based competing risk analysis. Cancer.
109:1763–1768. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McDermott DF, Regan MM, Clark JI, Flaherty
LE, Weiss GR, Logan TF, Kirkwood JM, Gordon MS, Sosman JA, Ernstoff
MS, et al: Randomized phase III trial of high-dose interleukin-2
versus subcutaneous interleukin-2 and interferon in patients with
metastatic renal cell carcinoma. J Clin Oncol. 23:133–141. 2005.
View Article : Google Scholar
|
|
4
|
Kroeger N, Choueiri TK, Lee JL, Bjarnason
GA, Knox JJ, MacKenzie MJ, Wood L, Srinivas S, Vaishamayan UN, Rha
SY, et al: Survival outcome and treatment response of patients with
late relapse from renal cell carcinoma in the era of targeted
therapy. Eur Urol. 65:1086–1092. 2014. View Article : Google Scholar
|
|
5
|
Koshkin VS and Rini BI: Emerging
therapeutics in refractory renal cell carcinoma. Expert Opin
Pharmacother. 17:1225–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mains PE, Kemphues KJ, Sprunger SA,
Sulston IA and Wood WB: Mutations affecting the meiotic and mitotic
divisions of the early Caenorhabditis elegans embryo. Genetics.
126:593–605. 1990.PubMed/NCBI
|
|
7
|
Kent D, Bush EW and Hooper JE: Roadkill
attenuates Hedgehog responses through degradation of Cubitus
interruptus. Development. 133:2001–2010. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu J, Ghanim M, Xue L, Brown CD, Iossifov
I, Angeletti C, Hua S, Nègre N, Ludwig M, Stricker T, et al:
Analysis of Drosophila segmentation network identifies a JNK
pathway factor overexpressed in kidney cancer. Science.
323:1218–1222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Q, Zhang L, Wang B, Ou CY, Chien CT
and Jiang J: A hedgehog-induced BTB protein modulates hedgehog
signaling by degrading Ci/Gli transcription factor. Dev Cell.
10:719–729. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kwon JE, La M, Oh KH, Oh YM, Kim GR, Seol
JH, Baek SH, Chiba T, Tanaka K, Bang OS, et al: BTB
domain-containing speckle-type POZ protein (SPOP) serves as an
adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase.
J Biol Chem. 281:12664–12672. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hernández-Muñoz I, Lund AH, van der Stoop
P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B,
Marahrens Y and van Lohuizen M: Stable X chromosome inactivation
involves the PRC1 Polycomb complex and requires histone MACROH2A1
and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA.
102:7635–7640. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gordan JD, Lal P, Dondeti VR, Letrero R,
Parekh KN, Oquendo CE, Greenberg RA, Flaherty KT, Rathmell WK,
Keith B, et al: HIF-alpha effects on c-Myc distinguish two subtypes
of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell.
14:435–446. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sato Y, Yoshizato T, Shiraishi Y, Maekawa
S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki
H, et al: Integrated molecular analysis of clear-cell renal cell
carcinoma. Nat Genet. 45:860–867. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Su D, Stamatakis L, Singer EA and
Srinivasan R: Renal cell carcinoma: Molecular biology and targeted
therapy. Curr Opin Oncol. 26:321–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li G, Ci W, Karmakar S, Chen K, Dhar R,
Fan Z, Guo Z, Zhang J, Ke Y, Wang L, et al: SPOP promotes
tumorigenesis by acting as a key regulatory hub in kidney cancer.
Cancer Cell. 25:455–468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou J, Luo J, Wu K, Yun EJ, Kapur P, Pong
RC, Du Y, Wang B, Authement C, Hernandez E, et al: Loss of DAB2IP
in RCC cells enhances their growth and resistance to mTOR-targeted
therapies. Oncogene. Feb 15–2016.(Epub ahead of print). View Article : Google Scholar
|
|
17
|
Zhou J, Zhu G, Huang J, Li L, Du Y, Gao Y,
Wu D, Wang X, Hsieh JT, He D, et al: Non-canonical GLI1/2
activation by PI3K/AKT signaling in renal cell carcinoma: A novel
potential therapeutic target. Cancer Lett. 370:313–323. 2016.
View Article : Google Scholar
|
|
18
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Muglia VF and Prando A: Renal cell
carcinoma: Histological classification and correlation with imaging
findings. Radiol Bras. 48:166–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eggener S: TNM staging for renal cell
carcinoma: Time for a new method. Eur Urol. 58:517–519; discussion
519–521. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tania M, Khan MA and Fu J: Epithelial to
mesenchymal transition inducing transcription factors and
metastatic cancer. Tumour Biol. 35:7335–7342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sánchez-Tilló E, de Barrios O, Siles L,
Cuatrecasas M, Castells A and Postigo A: β-catenin/TCF4 complex
induces the epithelial-to-mesenchymal transition (EMT)-activator
ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci USA.
108:19204–19209. 2011. View Article : Google Scholar
|
|
25
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Al-Ahmadie HA, Alden D, Qin LX, Olgac S,
Fine SW, Gopalan A, Russo P, Motzer RJ, Reuter VE and Tickoo SK:
Carbonic anhydrase IX expression in clear cell renal cell
carcinoma: An immunohistochemical study comparing 2 antibodies. Am
J Surg Pathol. 32:377–382. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hes O, Michal M, Kuroda N, Martignoni G,
Brunelli M, Lu Y, Adley BP, Alvarado-Cabrero I and Yang XJ:
Vimentin reactivity in renal oncocytoma: Immunohistochemical study
of 234 cases. Arch Pathol Lab Med. 131:1782–1788. 2007.PubMed/NCBI
|
|
28
|
Ozcan A, Zhai J, Hamilton C, Shen SS, Ro
JY, Krishnan B and Truong LD: PAX-2 in the diagnosis of primary
renal tumors: Immunohistochemical comparison with renal cell
carcinoma marker antigen and kidney-specific cadherin. Am J Clin
Pathol. 131:393–404. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huber MA, Kraut N and Beug H: Molecular
requirements for epithelial-mesenchymal transition during tumor
progression. Curr Opin Cell Biol. 17:548–558. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peinado H, Quintanilla M and Cano A:
Transforming growth factor beta-1 induces snail transcription
factor in epithelial cell lines: Mechanisms for epithelial
mesenchymal transitions. J Biol Chem. 278:21113–21123. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kypta RM and Waxman J: Wnt/β-catenin
signalling in prostate cancer. Nat Rev Urol. 9:418–428. 2012.
View Article : Google Scholar : PubMed/NCBI
|