Introduction
Osteosarcoma is a common malignant bone tumor in
children and adolescents. This disease presents as a bimodal age
peak distribution, that is, 0–24 years and >60 years. Incidence
in youth (0–24 years) is similar across the world at ~3.4 patients
per million (1). Current
treatment, based on surgery and chemotherapy, provides a 5-year
survival rate of ~60%. However, that survival rate seems to have
reached a plateau since 1980 (2).
In order to improve treatment outcomes, the challenge is to develop
a better understanding of tumorigenesis and to search for new
targets or strategies of treatment.
The pathophysiology of disease initiation and
progression in osteosarcoma is still unclear. There is no specific
risk factor predisposing for this disease (3), the peak incidence has been shown to
be related to secondary growth (1). No specific genetic abnormality
patterns for disease predilection have been identified, although
there is a high incidence of osteosarcoma related to the mutation
of p53, RB and the RECQ-Helicase family (4). The epigenetics and protein regulatory
controls has become a focus for identifying common pathways in the
various steps of pathological events. Some agents which related to
these have been studied as potential targets in experimental and
clinical trials scale (5).
Proteomics describes global changes in the
expression patterns of proteins under different biological
conditions. Notably, there is still a lack of information regarding
the proteomic profile of osteosarcoma since the preparation of
proteins under properly matched condition is limited. Heterogeneous
tissue, consisting of various cellular and matrix content, causes
uncertain results of proteomic analysis. Furthermore, there is a
lack of validated information on some potential proteins from
proteomic studies in clinical samples due to the disease being
relatively rare.
In order to determine pathogenesis-related proteins
in osteosarcoma, proteomics serves as a valuable tool for
identifying differentially expressed proteins between primary
osteosarcoma and osteoblast cells, which were derived from primary
biopsy of osteosarcoma patients and normal cancellous bone of
non-cancer participants, respectively. Expression levels of
candidate protein was further validated in both osteosarcoma cell
culture system and tissue specimens. Importantly, we also explored
biological roles of candidate protein with progressive behavior of
osteosarcoma. This study reveals a possibly therapeutic target of
osteosarcoma from proteomics study of well-characterized primary
cells.
Materials and methods
Study design and participant recruitment
protocol
Proteomics study was carried out from primary cell
osteosarcoma and osteoblasts in order to identify the proteins
involving in pathogenesis. Primary cells were extracted from
chemo-naïve osteosarcoma tissue of 7 patients (osteosarcoma cell),
and normal cancellous bone of 7 participants (osteoblast cells).
Using 2-DE based proteomics approach, there were 49 biologically
different sets in this experiment. A fold-change of more than two
times the initial threshold for screening protein spots of all 49
match sets. A consistently up- or downregulated match was
determined when the spots presented in the same direction (up or
down) in >70% of all match sets. The consistently up-or
downregulated spots were subjected to tryptic In-gel digestion and
LC-MS/MS analysis. Candidate spots were determined when they had
the statistically significant difference of percent volume between
osteosarcoma and osteoblast. The selected protein was validated and
tested for the possibility to be the potential therapeutic targets.
Patients, or parent of patients, and participants gave consent
before being recruited into the study. All research protocols were
approved by the Ethics committee of the Faculty of Medicine, Chiang
Mai University, Thailand, EC no. ORT-11-09-16A-14 (ID#757).
Primary osteosarcoma and osteoblastic
cell isolation, and cell characterizations
The tissue (naïve chemotherapy) was immediately
subjected to primary cell extraction after biopsy. Primary
osteosarcoma cells were extracted by incubating minced tissue in 5
mg/ml collagenase type I solution (Gibco, MA, USA) at 37°C for 18
h. The cells were then isolated by centrifugation and cultured in
10% FBS-DMEM (Gibco). The healthy participants were patients who
had been diagnosed with other, non-cancer, orthopedic conditions,
and required the use of an autologous bone grafts for substitution
procedure. Some bone graft was immediately subjected to perform
osteoblast extraction after harvested. Osteoblasts were isolated
with sequential collagenase type I-trypsin digestion. The cells
were isolated and expanded in 10% FBS-DMEM (6,7). The
cells from the 2–4 passages were used for cellular
characterization, proteomics study, and protein validation.
Doubling time of primary cells was estimated using proliferation
analysis by using a hemocytometer with time-course measurement.
Molecular marker expression was monitored by quantitative real-time
polymerase chain reaction (RT-PCR) (8). Collagen type I, osteonectin,
bonesialoprotein were selected as the osteogenic markers.
MMP-9, collagen type X were selected as the cancer markers
(7). Alkaline phosphatase (ALP)
activity assay was conducted kinetically by monitoring the
conversion of p-nitrophenyl phosphate (pNPP) to
p-nitrophenol (9). Alizarin
red-S histochemistry was performed to measure mineralization
ability (10). Activity of MMP-9
in the culture media was measured by gelatin zymographic assay
(11).
Real-time polymerase chain reaction
Total RNA was isolated using an Ilustra RNAspin Mini
kit (GE Healthcare, UK), and cDNA was synthesized from total RNA
using a Bioline SensiFAST™ cDNA synthesis kit (Bioline, UK).
Real-time PCR was performed using cDNA as a template under standard
conditions with Taq DNA polymerase (Bioline). The RT-PCR was
carried out using a Bioline SensiFAST™ SYBR® No-ROX kit
(Bioline). A Chromo4™ Real-Time PCR Detection system (Bio-Rad, CA,
USA) was used to detect cDNA amplification levels. Relative
expression levels were normalized to the expression of GAPDH
or 18S-RNA by the 2−ΔCT method (8). The primer sequences used in this
experiment were follows: GAPDH, 18S-RNA, Collagen type I,
osteonectin, bonesialoprotein, MMP-9, collagen type X and
KSRP, respectively. F, 5′-GAAGGTGAAGGTCGGAGTC-3′ and R,
5′-GAAGATGGTGATGGGATTTC-3′; F, 5′-CTT AGAGGGACAAGTGGCG-3′ and R,
5′-ACGCTGAGC CAGTCAGTGTA-3′; F, 5′-CAGCCGCTTCACCTACAGC-3′ and R,
5′-TTTTGTATTCAATCACTGTCTTGCC-3′; F, 5′-TCTATGTTAGCACCTTGTCTCCAG-3′
and R, 5′-CAG CCGCTTCACCTACAGC-3′; F, 5′-GCAGTAGTGACT CATCCGAAGA-3′
and R, 5′-GCCTCAGAGTCTTCATCTT CATTC-3′; F,
5′-TGAGAACCAATCTCACCGACAG-3′ and R, 5′-TGCCACCCGAGTGTAACCAT-3′; F,
5′-AGCCAG GGTTGCCAGGACCA-3′ and R, 5′-TTTTCCCACTCCAG GAGGGC-3′; F,
5′-CCGCCTACTACTCACACTACTA-3′ and R, 5′-TCTTCCCAGGCCTTAGTGTA-3′.
Protein extraction, 2-DE and
visualization of spots, and In-gel Tryptic digestion
Primary osteosarcoma and osteoblast cells were
cultured until reaching their 90% confluence. Cells were washed
with PBS, scraped in 5 ml of 0.25 M sucrose containing protease
inhibitor cocktail 1:500 (Sigma, MO, USA) and spun down. Three
hundred microliters of lysis buffer was added into the cell pellet,
then, cell suspension was incubated on ice and separated by a
sonicator. Cellular debris was removed, and the protein
concentration in supernatant was quantified by Bradford assay kits
(Bio-Rad) (12).
Protein analysis was performed with 2-DE, and the
setup conditions including the first dimension electrophoresis, the
second dimension electrophoresis, gel scanning and software
analysis were conducted as described briefly (12). For the first dimension, Immobiline™
drystrip, 7 cm, non-linear, pH 3.0–10.0 gradient and immobilize pH
gradient (IPG) gel strips were used. The first dimension of gel
electrophoresis was run on Ettan IPGphor III system. Total 150 μg
proteins with 75 μl of rehydration buffer was loaded into IPG strip
and incubation in Immobiline Drystrip Re-swelling Tray. The current
application was 50 mA per IGP gel strip and the four running
conditions as shown in literature (12). Two equilibration steps were
performed before subjecting to the second dimension. For the second
dimension, the proteins were separated on 12.5% SDS gel with 4%
stacking SDS gel. For the running program, i) current 10 mA/gel for
15 min, and ii) current 20 mA/gel until the bromophenol blue
reached the end of the gel. Finally, the gels were submerged in the
coomassie blue solution overnight and de-stained with 50% (v/v)
methanol, 10% (v/v) acetic acid (12). Gels were scanned and the background
adjusted by using Ettan labscan 5.0. Next, the images were analyzed
using ImageMaster 2D Platinum 7.0. Protein spots were considered as
candidates when the percent volume (% volume) of consistent spots
was significantly different between osteosarcoma and osteoblast
cells (Student’s t-test, p<0.05). For Tryptic In-gel digestion,
the candidate spots were cut to small pieces and repeatedly
incubated in 0.1 M NH4HCO3 in 50%
acetonitrite (ACN) to remove Coomassie dye. Reduction was performed
by re-swelling the gel pieces in 50 μl buffer solution and
incubated at 60°C for 45 min. After cooling, the excess liquid was
removed and quickly replaced by the same volume of freshly prepared
100 mM iodoacetamide in 0.1 M NH4HCO3
solution. The reaction was incubated in the dark at room
temperature for 30 min. The iodoacetamide solution was removed and
the gel pieces were washed with 50% ACN in water for 10 min. Gel
pieces were completely dried by centrifugation under vacuum. One
spot of gel piece requires 30 μl of trypsin digestion buffer. After
incubating the reaction mixture at 37°C overnight, the digestion
buffer was removed and saved in a new tube (peptide tube). The
protein from the gel pieces were then extracted by adding 60 μl of
2% freshly prepared trifluoroacetic acid and incubating for 30 min
at 60°C. The remaining solution was again saved in peptide tube
(12). Extracted proteins within
the peptide solution was finally pooled and dried by vacuum
centrifugation. Samples were kept in −20°C before being analyzed by
LC-MS/MS.
LC-MS/MS analysis
Mass spectrometry, nanoflow liquid chromatography
coupled with the AmaZon speed ion trap mass spectrometry (Bruker,
MA, USA) was utilized to identify protein spots. A 75 μm id × 100
mm C18 EASY-nLC™ column (Thermo Scientific, MA, USA) was used.
Gradient separation was performed using 0.1% formic acid in water
(solution A) and 0.1% formic acid in ACN (solution B) followed by
MS/MS equipped with CaptiveSpray™ source. Parent mass peaks with
ranges from 500 to 3,000 m/z were picked out for MS/MS analysis.
The collision energy was fixed at 3,000 V. The MS/MS data were
processed using Burker compass 1.4 software. Peptides were searched
with MASCOT search engine (http://www.matrixscience.com). The search parameters
were set as follows: NCBInr 20130918 (October 2013; 32611672
sequences; 11345269536 residues) and SwissProt (October 2013;
540958 sequences; 192206270 residues) database, taxonomy Homo
sapiens, enzyme set for trypsin, allowance of one missed
cleavage, variable modifications were carbamidomethylation at
cysteine residues and oxidation at methionine residues, peptide and
fragment mass tolerance were 1.2 and 0.6 Da, respectively, the
limit of peptide charges was 1+, 2+, and 3+. Significant hits were
defined by the MASCOT probability analysis (p<0.05) with MASCOT
scores >48.
Western blot analysis
Cells were lysed in RIPA lysis buffer (radioimmuno
precipitation assay lysis). Extracted protein (10 μg) was separated
on a 10% SDS-PAGE gel and then transferred to Immobilon-P
polyvinylidene difluoride membranes (PVDF) (Millipore). Protein
bands were visualized by chemiluminescence using ECL-Advance
Western Blotting Detection kit (GE Healthcare) (13). The antibody concentrations used for
KSRP were 1:5,000, ab150393 (Abcam, UK), 1:1,000 for
immunohistochemistry, β-actin 1:7,500, ab8227 (Abcam), goat
anti-rabbit IgG H&R 1:6,000 (Abcam).
Immunohistochemistry analysis
The sections, 3-micron formalin-fixed,
paraffin-embedded tissue, were deparaffinized and hydrated. Heat
mediated antigen retrieval was done in citrate buffer pH 6.0 in a
pressure cooker. Automated immunohistochemical staining was
performed using a Ventana Benchmark XT automated stainer (Ventana
Medical Systems, AZ, USA). The detection system was a Ventana
Ultraview DAB detection kit (Ventana Medical Systems).
KSRP knock-down by small interference RNA
(siRNA)
Osteosarcoma cell line (MMNG-HOS) catalog no. CLS
300289 passage 24 and U2OS catalog no. CLS 300364 passage 24 were
used as representative osteosarcoma cells. The siRNAs were diluted
in serum-free OptiMEM media and transfected using Lipofectamine
RNAiMAX reagent according to the manufacturer’s instructions
(Invitrogen, MA, USA). Short interference RNAs (siRNAs) against
KSRP were synthesized using Ambion by Life Technologies (14). The sequences (sense/antisense) for
the siRNAs were as follows: siRNA1
(5′-CAGGCUCAAUGAAUCGAAUtt-3′/5′-AUUCGAUUCAU UGAGCCUGct-3′); siRNA2
(5′-GGAUUCAGGCUGCAAA GUAtt-3′/5′-UACUUUGCAGCCUGAAUCCtg-3′). Cells
were treated with KSRP siRNA (10 and 30 nM), non-targeting siRNA
controls (10 and 30 nM) and transfection reagent only.
Wound healing assay
The MNNG-HOS and U2OS were transfected with 20 nM
KSRP siRNA and 20 nM negative control siRNA for 48 and 72 h. The
cells were trypsinized and plated in 24-well plates at 90%
confluence and incubated for 18 h. The monolayer cell culture was
scratched with a 200-μl yellow tip to create a wound line. The
width of the wound gaps was measured using AxioVision Analytic
Software and the percentage of migration ability was calculated.
The formula was: % migration = 100% − [(width at X hour)/(width at
0 hour) ×100] (15).
Tumor implantation in chick
chorioallantoic membrane assay (CAM)
Fertilized native crossbreed chicken eggs were
obtained from the Department of Animal and Aquatic Science, Faculty
of Agriculture, Chiang Mai University, and were incubated at 37.5°C
and 60% relative humidity (16).
On day 8, the shells were cut at the blunt end of the egg, a 1×1-cm
hole was created and the shell membrane was detached. A sterile
plastic ring was placed on the CAM and 5×105 cells/20 μl
of transfected cell solution was deposited after gentle laceration
of the CAM surface (16). Only
embryos surviving at day 15 were included in the analysis; tumors
were collected. Tumor formation was observed and tumor size was
estimated based on the tumor volume calculation V=4/3 πr3, with r =
1/2 square root (d1 × d2) (16).
Statistical analysis
Gene expression levels, enzymatic activity levels,
doubling time, protein intensity levels and tumor volume were
compared between groups with the Mann-Whitney U test or the
Kruskal-Wallis test. The statistical significance was defined as
p<0.05.
Results
Osteosarcoma and osteoblast
characterization
The demographic characterizations and survival rate
of patients with osteosarcoma (n=7) and participants (n=7) are
presented in Table I. The average
doubling time of osteosarcoma and osteoblast cells was 75.6±6.0 and
148.6±26.0 h, respectively (p<0.05). Both types of primary cells
expressed osteogenic markers (collagen type-I,
bonesialoprotein and osteonectin), but the osteoblasts
were expressed at higher levels than osteosarcomas. On the other
hand, collagen type-X and MMP-9 were significantly
upregulated in the osteosarcomas (data not shown). The high
activity of MMP-9 in osteosarcoma cells was confirmed by
zymographic assay (data not shown). Osteoblasts showed more
mineralization ability than osteosarcoma cells on days 21 and 28
(data not shown). When cells were activated with osteogenic media,
alkaline phosphatase activity was significantly increased in
osteoblast cells compared to osteosarcoma cells (data not
shown).
 | Table ICharacteristics of patients with
osteosarcoma and non-cancer participants. |
Table I
Characteristics of patients with
osteosarcoma and non-cancer participants.
| Case ID | Diagnosis
(site) | Gender | Age | Tissue from | Enneking stage | % Tumor
necrosisa | Clinical status
(follow-up month) |
|---|
| OS 1 | OS (femur) | F | 9 | Femur | IIB | 90 | Alive (36) |
| OS 2 | OS (femur) | M | 21 | Femur | III | 15 | Alive (28)b |
| OS 3 | OS (femur) | F | 12 | Femur | IIB | 90 | Alive (36) |
| OS 4 | OS (femur) | F | 5 | Femur | IIB | 10 | Dead (15)c |
| OS 5 | OS (femur) | F | 15 | Femur | IIB | 90 | Alive (30) |
| OS 6 | OS (tibia) | F | 16 | Tibia | IIB | 5 | Alive (18)b |
| OS 7 | OS (femur) | F | 18 | Femur | IIB | 10 | Alive (26) |
| OB 1 | Fracture
(femur) | F | 23 | Iliac crest | - | - | Alive (36) |
| OB 2 | Fracture
(femur) | M | 24 | Iliac crest | - | - | Alive (36) |
| OB 3 | Fracture
(tibia) | M | 35 | Iliac crest | - | - | Alive (30) |
| OB 4 | Chondroblastoma
(humerus) | F | 11 | Iliac crest | - | - | Alive (30) |
| OB 5 | Bone cyst
(femur) | M | 12 | Iliac crest | - | - | Alive (30) |
| OB 6 | Fracture
(tibia) | F | 17 | Iliac crest | - | - | Alive (28) |
| OB 7 | Scoliosis
(spine) | F | 15 | Iliac crest | - | - | Alive (26) |
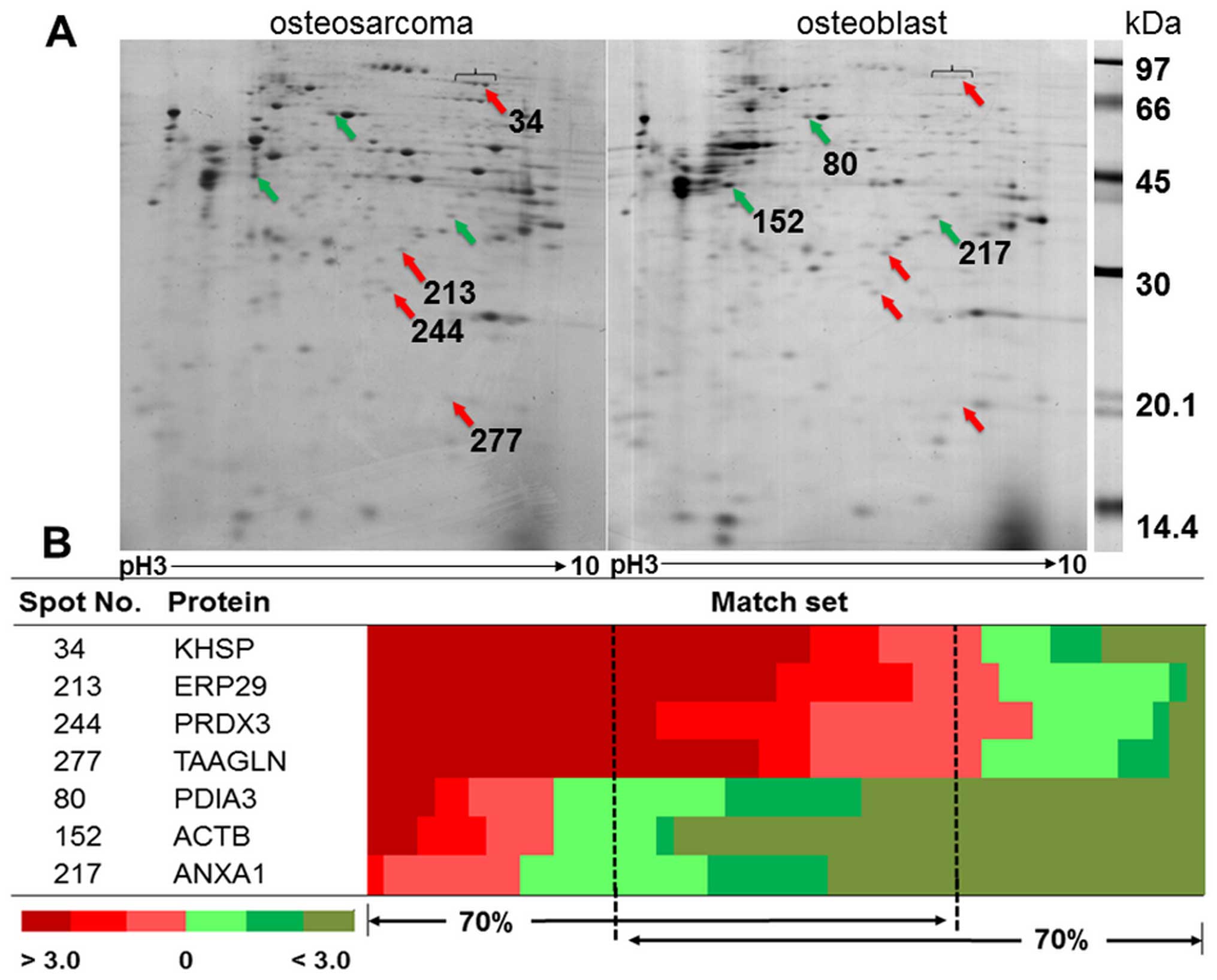
Proteomic analysis and candidate protein
selection
The representative gels from osteosarcoma and
osteoblast cells are shown in Fig.
1A. The average number of protein spots from osteosarcoma and
osteoblast cells was 348±30 and 415±74, respectively. There were
257±62 matching spots from all 49 matched sets, but only 33 spots
had consistent presentation of either up- or downregulation.
Thirty-three spots were categorized as follows; 29% of all
consistent proteins involved in ribonucleic metabolism, 14% were
cytoskeletal and cytoskeletal related proteins, 14% involved in
redox reaction related proteins, 11% involved in cellular
metabolism, 6% were protease proteins, and 26% were either signal
transduction related proteins or other proteins. There were four
statistically significant upregulated proteins: KH-type splicing
regulatory protein (KSRP), endoplasmic reticulum resident protein
29 (ERP29), thioredoxin-dependent peroxidase reductase (PRDX3), and
transgelin (TAGL). There were three statistically significant
downregulated proteins: protein disulfide-isomerase A3 (PDIA3),
actin, cytoplasmic1 (ACTB), and Annexin A1 (ANXA1) (Fig. 1B). The details of protein
identification from LC-MS/MS are shown in Table II.
 | Table IISummary of significant altered
proteins in primary osteosarcoma cells identified by LC-MS/MS. |
Table II
Summary of significant altered
proteins in primary osteosarcoma cells identified by LC-MS/MS.
| | | | | | | % Volume intensity
(mean ± SD)d |
|---|
| | | | | | |
|
|---|
| ID | Accession
no.a | Name | Gene | MW/pI
(kDa/pI)b | MASCOT score | Peptide
matchc/% coverage | Osteosarcoma | Osteoblast |
|---|
| Upregulated
protein | | | | | | | | |
| 34 | gi|2055427 | KH-type splicing
regulatory protein | KSRP | 73.1/6.84 | 255 | 6/7 | 0.156±0.11 | 0.049±0.03 |
| 213 | gi|5803013 | Endoplasmic
reticulum resident protein 29 | ERP29 | 30.0/6.77 | 270 | 5/18 | 0.411±0.49 | 0.131±0.02 |
| 244 | gi|5802974 |
Thioredoxin-dependent peroxide reductase,
mitochondrial | PRDX3 | 27.7/7.67 | 239 | 5/18 | 0.217±0.09 | 0.110±0.06 |
| 277 | gi|48255905 | Transgelin | TAGLN | 22.6/8.87 | 197 | 4/21 | 0.662±0.59 | 0.307±0.24 |
| Downregulated
protein | | | | | | | | |
| 80 | gi|2507461 | Protein
disulfide-isomerase A3 | PDIA3 | 56.7/5.98 | 279 | 6/10 | 0.178±0.14 | 0.332±0.16 |
| 152 | gi|4501885 | Actin, cytoplasmic
1 | ACTB | 41.71/5.29 | 160 | 7/21 | 0.992±1.07 | 2.715±1.98 |
| 217 | gi|4502101 | Annexin A1 | ANXA1 | 38.7/6.57 | 359 | 7/19 | 0.113±0.05 | 0.264±0.12 |
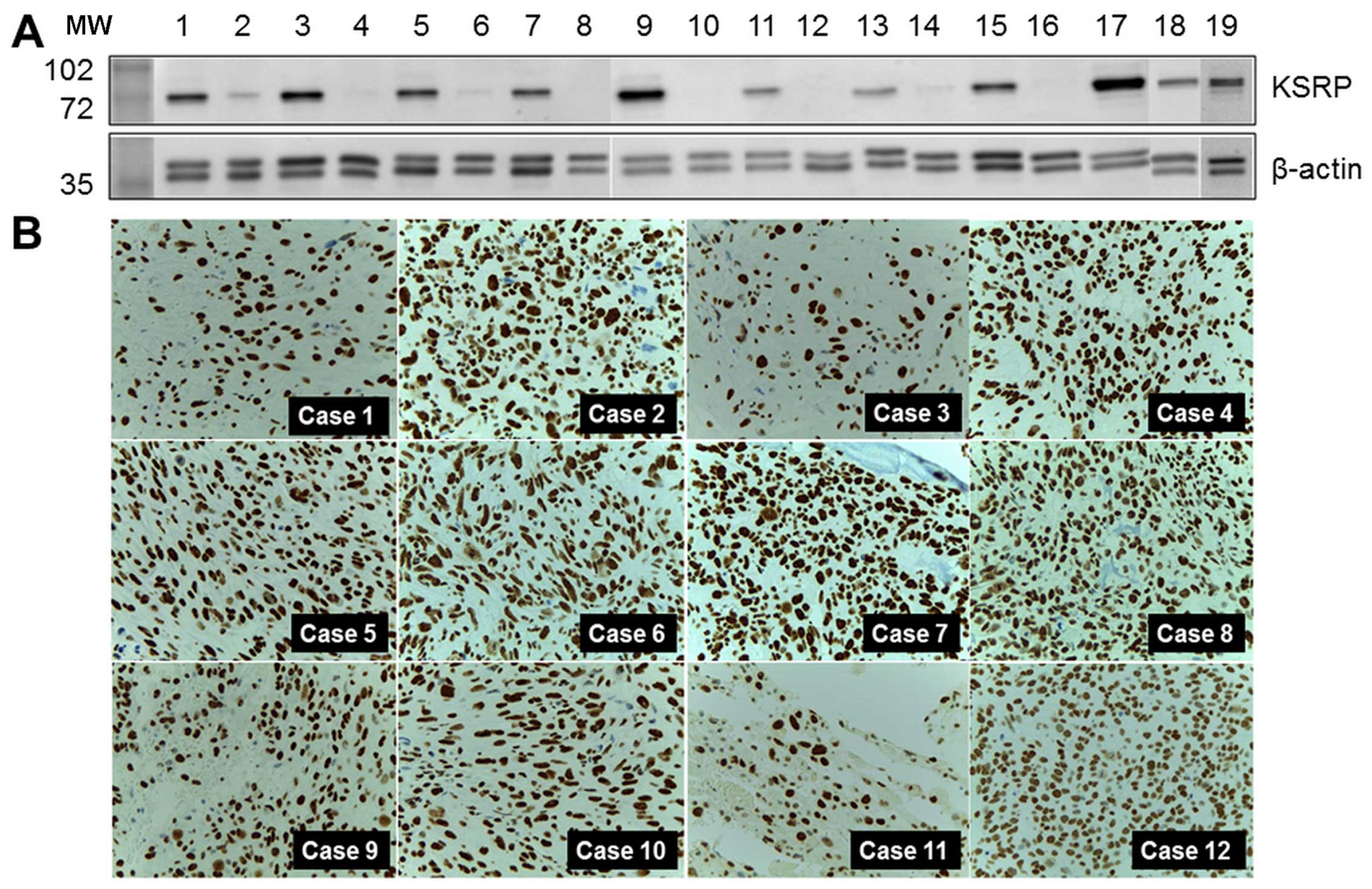
Levels of KSRP expression in samples,
cell lines, and other cases of the experimental group
KH-type splicing regulatory protein (far upstream
element-binding protein 2: FUBP2) was the protein of interest, and
was selected for further detailed exploration. Western blot
analysis was performed in both pooled samples and crude protein
extracted from individual samples. Relatively high regulation was
indicated by the intensity of the western blots as shown in
Fig. 2A. KSRP was also observed in
MNNG-HOS and U2OS, human osteosarcoma cell lines. KSRP expression
in primary biopsies of 12 representative osteosarcoma cases, the
separate cases from the proteomics study, are demonstrated by
immunohistochemistry staining in Fig.
2B. All cases revealed diffuse and strong nuclear staining in
tumor cells.
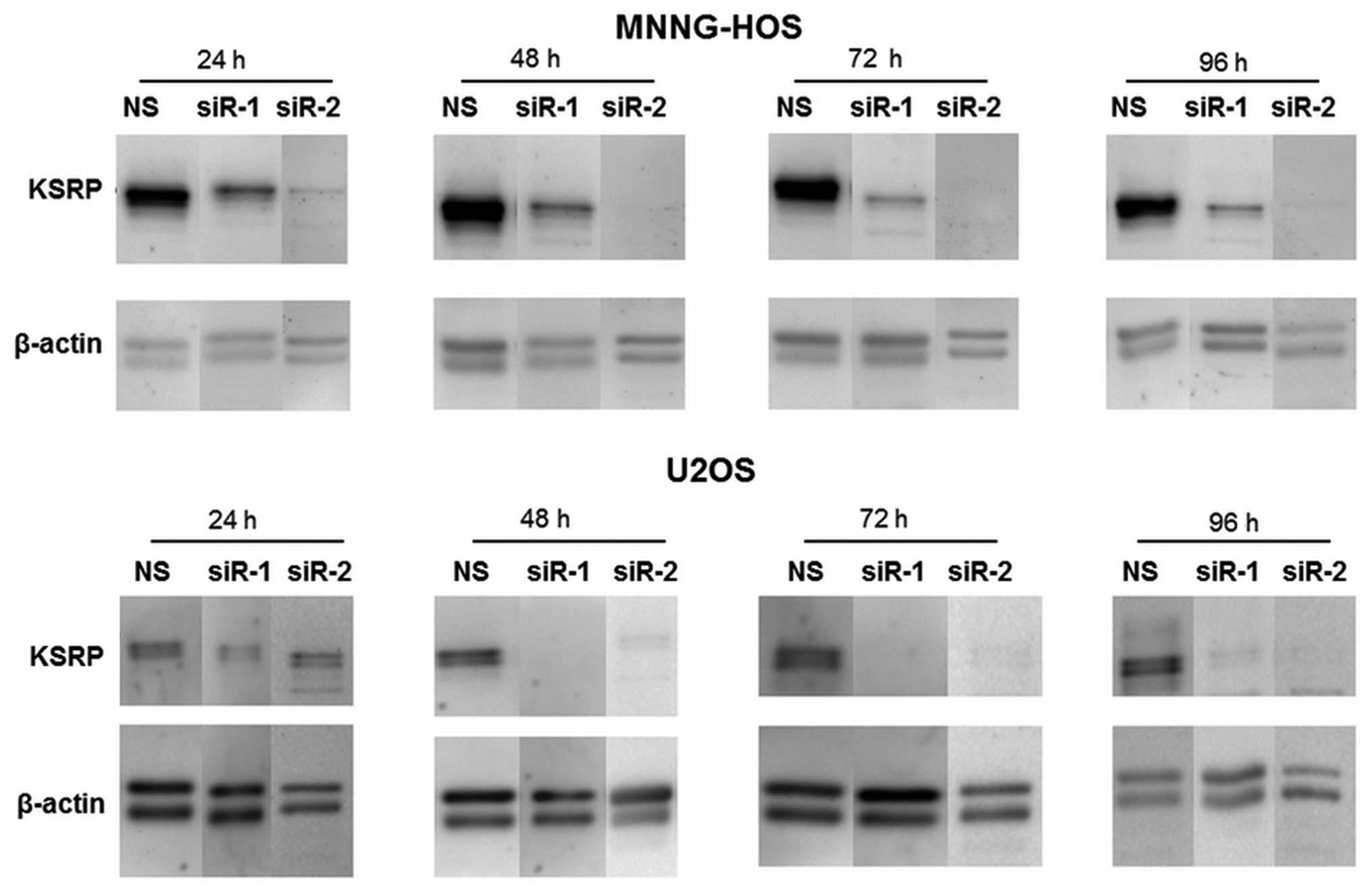
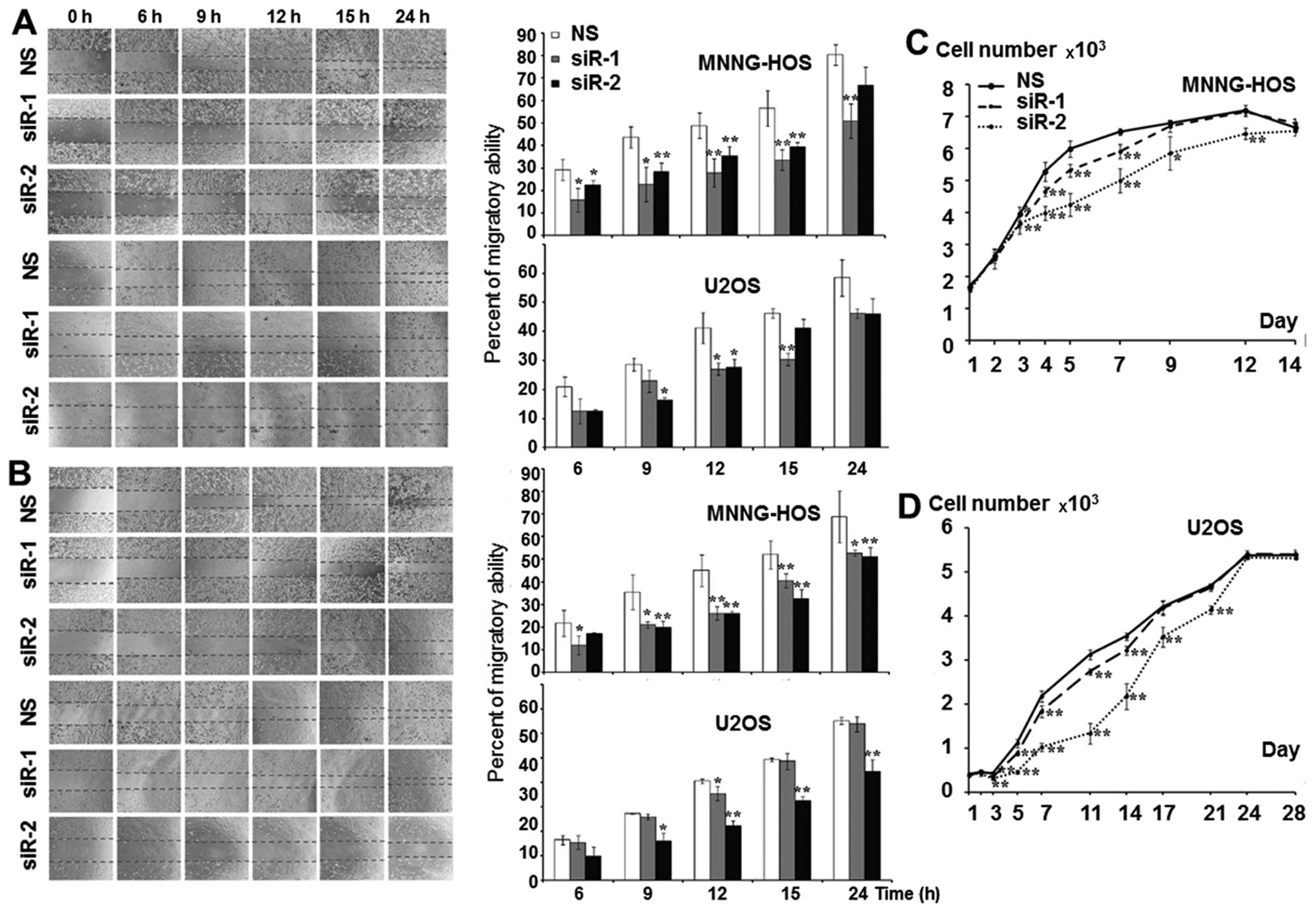
KSRP silencing decreases migration
ability, and proliferation rate of osteosarcoma cells
The KSRP silencing conditions were optimized at 24,
48, 72 and 96 h. Both siRNA-1 and siRNA-2 compromised KSRP
expression from 24 h comparing to non-sense from both cell lines
and persisted >96 h (Fig. 3).
The KSRP silencing at 48 and 72 h was subjected to wound healing
assay. KSRP silencing decreased the migratory rate of both cell
lines. Silencing by siRNA-2 presented a stronger inhibitory effect
in both MNNG-HOS and U2OS than siRNA-1 as shown in Fig. 4A and B with both 48 and 72 h of
incubation. The proliferative rates were significantly reduced in
both cell lines and both types of siRNA. Silencing by siRNA-2
presented a stronger proliferative inhibition in both MNNG-HOS and
U2OS compared to siRNA-1 as shown in Fig. 4C and D, respectively.
KSRP silencing impairs implant and growth
ability of osteosarcoma cells in CAM model
MNNG-HOS cell lines exhibited good ability for
successful implantation in the CAM model (16). This study used MNNG-HOS to
represent osteosarcoma growth in the CAM model. KSRP silencing with
siRNA-1 and siRNA-2 decreased rates of tumor implantation 70 and
25%, respectively, shown in Table
III. Mass-forming tumors and non-mass-forming tumors (membrane)
are shown in Fig. 5A. The average
tumor size of the non-sense group was significantly larger, and
more number of mass-forming tumors than that presented by KSRP
silencing with siRNA-1 and siRNA-2, respectively. The
representative histology of mass-forming tumor showed a dense
population of cancer cells (Fig.
5B) and the histology of non-mass-forming tumors showed small
loose colonies of tumors embedded in chorioallantoic membrane
(Fig. 5C).
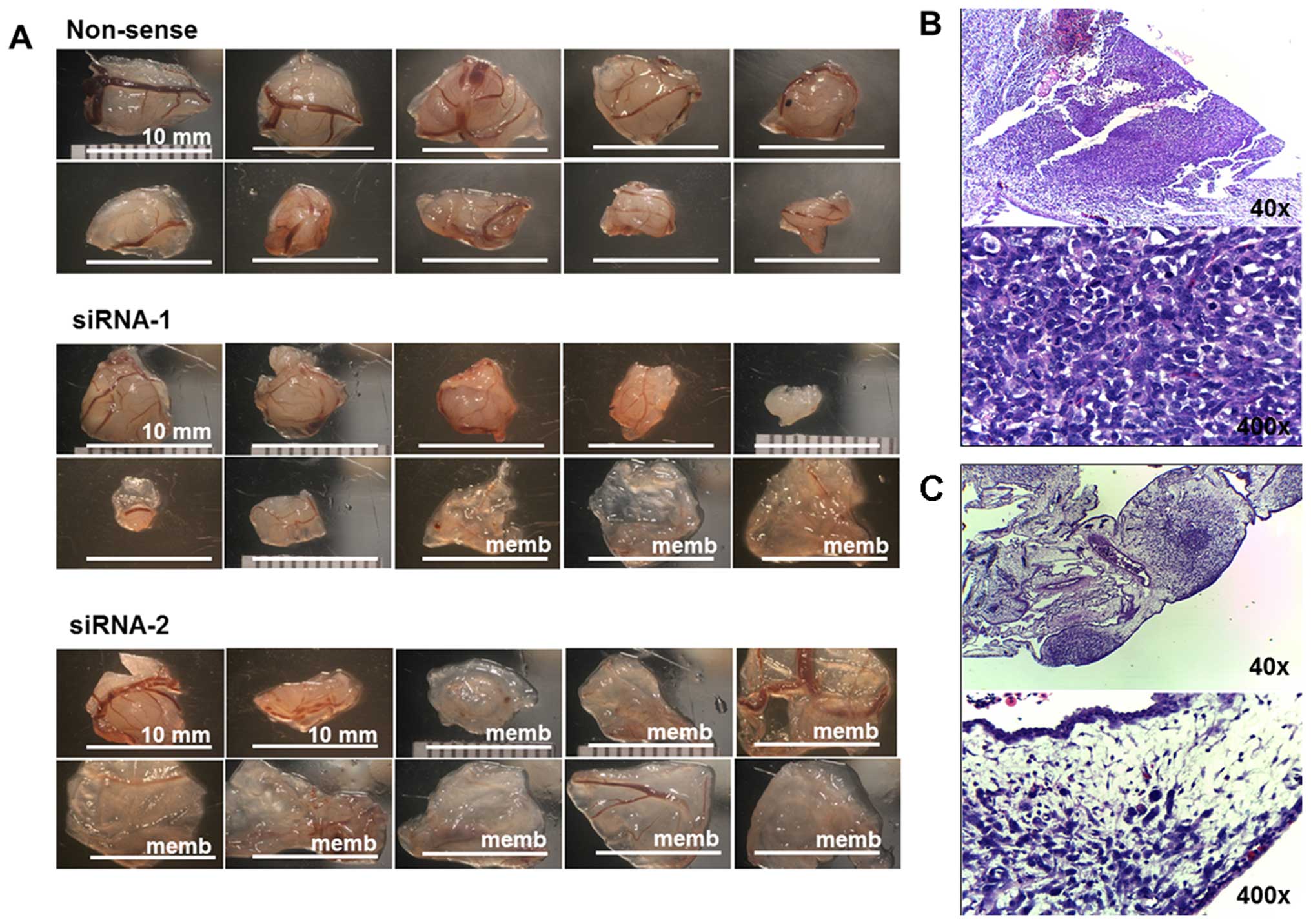 | Figure 5(A) The ten largest implanted tumors
of non-sense, siRNA-1, and siRNA-2 of KSRP knocked down MNNG-HOS in
CAM assay. (B) The representative histology of mass-forming tumors
was observed as a proliferative nodular architecture with high
cellularity, dense deep blue colony, embedded in chorioallantoic
membrane (H&E, ×40). Plump spindle malignant cells which are
anisonucleosis, nuclear pleomorphism, hyperchromatin patterns, and
prominent nucleoli supplied by chick vasculature. Mitotic figures
were frequently observed (H&E, ×400). (C) The representative
histology of non-mass-forming tumor (membrane) showed small loose
colonies of tumors embedded in chorioallantoic membrane (H&E,
×40), and hypocellular cancer cells loosely in extracellular matrix
within edematous chick membrane (H&E, ×400). |
 | Table IIIThe comparative data of implanted
tumor in CAM model. |
Table III
The comparative data of implanted
tumor in CAM model.
| Non-sense | siRNA-1 | siRNA-2 |
|---|
| Number of
implantation | 15 | 15 | 15 |
| Embryo survive | 14 | 10 | 12 |
| Tumor forming:
membrane | 13:1 | 7:3 | 2:10 |
| Tumor forming
(%)a | 93 | 70 | 17 |
| Ten selected
presentation (membrane)b | 10 | 7 (3) | 2 (8) |
| Mean tumor volume ±
SD (ml) | 1.14 (0.65) | 0.36 (0.40) | 0.15 (0.34) |
| p-value | | 0.005 | <0.001 |
Discussion
There have been few studies of bone analysis using
proteomic technologies comparing other types of tissues because the
preparation of samples with different physical properties was
limited, thus restricting the opportunity for comparison. There are
thirty-one studies including the keywords ‘osteosarcoma’ ‘human’
and ‘proteomics’ which were listed on the PubMed database until the
year 2015. Twenty-one studies were conducted in human osteosarcoma
cell lines, four were investigated in serum or plasma from
patients, and six were based on patient tissues. Of those six, one
compared proteomics of formalin-fixed paraffin-embedded tissues
between osteosarcoma and desmoid tumors (benign fibrous tumors)
(17). Another study compared the
proteomic profiles of osteosarcoma tissue and benign bone tumors
(18). Three of six research
efforts studied biopsy samples from both good and poor response
chemotherapy cases (19–21). Peroxiredoxin 2 (PRDX2) was
expressed at higher levels in the poor responders and siRNA-induced
silencing of PRDX2 resulted in a decrease of proliferation,
invasion and migration in osteosarcoma cells (20,21).
Heterogeneous tissue of sample and control, for which it is
difficult to identify an exact zone of pathogenesis, might alter
the protein outcome during the analysis process. Folio et al
used a different approach, extracting the primary cells from a
biopsy sample of osteosarcoma tissue and osteoblast cells from
cancellous bone from patients. Crystallin α/β and ezrin were
selected for study in greater detail (22). Ezrin has been widely studied for
its role of metastasis behavior in osteosarcoma, and is becoming a
potential target for metastasis control (23).
Significant upregulation of KH type splicing
regulatory protein (KSRP or FUBP2) was found in proteomics study of
our experiment, and the strong evidence of KSRP existence was
demonstrated by western blot analysis of experimental samples, and
immunohistochemistry from primary biopsy sample of osteosarcoma
tissue (n=12 cases) out of the experimental group. Previous studies
showed important roles of KSRP in pathogenesis of some cancers
including hepatocellular carcinoma (HCC), colon cancer, and some
hematologic malignancies upon diverse molecular mechanisms.
Upregulated KSRP was found in HCC comparing to normal liver tissue
via 2D-DIGE profiling (24).
Overexpression related to proliferation and dissemination behavior
of HCC, and correlated with a poor survival rate in patients
(25). Overexpression of KSRP was
able to inactivate β-catenin/Lymphoid-enhancer factor/T-cell
factor(Lef/Tcf)-sensitive transcription in colon carcinoma
(26). KSRP is involved in
maturation of several miRNAs which were dysregulated in leukemia
and lymphoma (27).
KSRP is a member of the far upstream element (FUSE)
binding protein family. The far upstream binding element is an AT
rich DNA element located 1.7 kb upstream of the c-myc
oncogene promoter. Three members of the family include the proteins
FUBP1, FUBP2, and FUBP3, all of which are able to bind to the FUSE
and to upregulate the expression of the c-myc (28,29).
It is possible that KSRP is involved in pathogenesis of
osteosarcoma through c-Myc regulation. c-Myc amplification was also
present in up to 78% of cases in later studies which used
conventional cytogenetic and comparative genomic hybridization
(30). Using immunohistochemistry,
expression of c-Myc was observed in up to 85.7% of osteosarcoma
tissue samples and was significantly correlated with the survival
time (31). A recent study using a
mouse model suggested that c-Myc amplification was the key
regulator and was sufficient to drive both bone marrow mesenchymal
stem cells and lineage-committed immature osteoblasts into becoming
osteosarcoma (32).
KSRP has other roles in post-translational
regulation by controlling mRNA level and miRNA maturation process.
KSRP is one of the AU-rich element-binding proteins (ARE-BP)
controlling the target mRNA level. The stability of β-catenin mRNA
level was maintained through phosphorylation of KSRP activated by
PI3-kinase/AKT signaling (33).
Suppression of KSRP level increased 5-fold accumulation of
β-catenin, and provoked activation of β-catenin transcription
targets. On the other hand, overexpression of KSRP level attenuated
Wnt3a-induced β-catenin accumulation (34). The inactivation of the
Wnt/β-catenin pathway was also found to relate with tumorigenesis
of osteosarcoma (35).
KSRP controls the maturation of miRNA by interacting
with the terminal loop of miRNA precursors, and it is also able to
interact with Drosha and Dicer in the miRNA maturation process
(36). Therefore, KSRP presents
its diversity function involving many biological processes
including the stem cell differentiation and cancer pathogenesis.
KSRP has demonstrated a role in controlling mesenchymal stem cells
during the cell fate differentiation between myoblast and
osteoblast differentiation. The presence of KSRP plays a critical
role in blocking mesenchymal stem cell differentiation toward
osteoblast by enhancing the BMP2-dependent myogenic miRNA
maturation process (37).
KSRP controls maturation of the Let-7 miRNA family
which is known as a tumor suppressive miRNA (38). KSRP knock-down limited Let-7 miRNA
maturation and increased mRNA levels of Let-7 targets including
c-Myc (39). Decreasing maturation
of Let-7a miRNA after KSRP knockdown was also demonstrated in the
U2OS, osteosarcoma cell line, and resulted in increased cell
proliferation (36). On the other
hand, overexpression of Lin-28 plays an opposite role to the KSRP
controlling levels of Let-7 maturation. Lin-28 is able to bind to
Let-7 miRNA at the terminal loop, at the same site of KSRP, and
leading to suppression of Let-7 maturation (40). There is still only limited data on
the indirect regulation control between KSRP and Lin-28 and Let-7
family maturation process for the pathogenesis of osteosarcoma.
This study has not covered in detail the intensities
of signals from both promoter control of c-Myc and other
mechanisms, downstream effects, or crosstalk with other related
proteins. However, results of our study supported that KSRP would
be one of the candidate proteins for osteosarcoma control. KSRP and
related cascades also have been studied as potential new targets
for cancer treatment. Curcumin analogue (GO-Y086) is able to bind
with nuclear protein KSRP by covalent modification and markedly
suppresses c-Myc protein expression. GO-Y086 has been studied for
the control of c-Myc expression in colon cancer cell lines (HCT116)
(41).
In conclusion, our study supports overexpression of
KSRP to be involved in pathogenesis of osteosarcoma and related to
the aggressive phenotypes of osteosarcoma cells. KSRP might be a
potential target for further studies at a higher level for disease
control. Up- and down-stream regulation of KSRP overexpression are
appropriate for further exploration to identify more potential
biomarkers and therapeutic targets for osteosarcoma.
Acknowledgements
This study received funding from the National
Science and Technology Development Agency (NSTDA), code P-12-01469,
and the Faculty of Medicine, Chiang Mai University. Additional
support was provided by the Excellence Center in Osteology Research
and Training Center (ORTC), Chiang Mai University, and The Thailand
National Research University (NRU) Fund. The authors would also
like to express their sincere thanks to Dr G. Lamar Robert for
editing the English manuscript.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allison DC, Carney SC, Ahlmann ER,
Hendifar A, Chawla S, Fedenko A, Angeles C and Menendez LR: A
meta-analysis of osteosarcoma outcomes in the modern medical era.
Sarcoma. 2012:7048722012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gelberg KH, Fitzgerald EF, Hwang S and
Dubrow R: Growth and development and other risk factors for
osteosarcoma in children and young adults. Int J Epidemiol.
26:272–278. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ottaviani G and Jaffe N: The etiology of
osteosarcoma. Cancer Treat Res. 152:15–32. 2009. View Article : Google Scholar
|
|
5
|
Zhou W, Hao M, Du X, Chen K, Wang G and
Yang J: Advances in targeted therapy for osteosarcoma. Discov Med.
17:301–307. 2014.PubMed/NCBI
|
|
6
|
Jonsson KB, Frost A, Nilsson O, Ljunghall
S and Ljunggren O: Three isolation techniques for primary culture
of human osteoblast-like cells: A comparison. Acta Orthop Scand.
70:365–373. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pautke C, Schieker M, Tischer T, Kolk A,
Neth P, Mutschler W and Milz S: Characterization of osteosarcoma
cell lines MG-63, Saos-2 and U-2 OS in comparison to human
osteoblasts. Anticancer Res. 24:3743–3748. 2004.
|
|
8
|
Alameh M, Jean M, Dejesus D, Buschmann MD
and Merzouki A: Chitosanase-based method for RNA isolation from
cells transfected with chitosan/siRNA nanocomplexes for real-time
RT-PCR in gene silencing. Int J Nanomed. 5:473–481. 2010.
|
|
9
|
Martins AM, Pham QP, Malafaya PB, Sousa
RA, Gomes ME, Raphael RM, Kasper FK, Reis RL and Mikos AG: The role
of lipase and alpha-amylase in the degradation of
starch/poly(epsilon-caprolactone) fiber meshes and the osteogenic
differentiation of cultured marrow stromal cells. Tissue Eng Part
A. 15:295–305. 2009. View Article : Google Scholar
|
|
10
|
Gregory CA, Gunn WG, Peister A and Prockop
DJ: An Alizarin red-based assay of mineralization by adherent cells
in culture: Comparison with cetylpyridinium chloride extraction.
Anal Biochem. 329:77–84. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kleiner DE and Stetler-Stevenson WG:
Quantitative zymography: Detection of picogram quantities of
gelatinases. Anal Biochem. 218:325–329. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pruksakorn D, Lirdprapamongkol K,
Chokchaichamnankit D, Subhasitanont P, Chiablaem K, Svasti J and
Srisomsap C: Metabolic alteration of HepG2 in scaffold-based 3-D
culture: Proteomic approach. Proteomics. 10:3896–3904. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bolt MW and Mahoney PA: High-efficiency
blotting of proteins of diverse sizes following sodium dodecyl
sulfate-polyacrylamide gel electrophoresis. Anal Biochem.
247:185–192. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weil D, Garçon L, Harper M, Duménil D,
Dautry F and Kress M: Targeting the kinesin Eg5 to monitor siRNA
transfection in mammalian cells. Biotechniques. 33:1244–1248.
2002.PubMed/NCBI
|
|
15
|
Ridley AJ, Schwartz MA, Burridge K, Firtel
RA, Ginsberg MH, Borisy G, Parsons JT and Horwitz AR: Cell
migration: Integrating signals from front to back. Science.
302:1704–1709. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Balke M, Neumann A, Kersting C,
Agelopoulos K, Gebert C, Gosheger G, Buerger H and Hagedorn M:
Morphologic characterization of osteosarcoma growth on the chick
chorioallantoic membrane. BMC Res Notes. 3:582010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rao UN, Hood BL, Jones-Laughner JM, Sun M
and Conrads TP: Distinct profiles of oxidative stress-related and
matrix proteins in adult bone and soft tissue osteosarcoma and
desmoid tumors: A proteomics study. Hum Pathol. 44:725–733. 2013.
View Article : Google Scholar
|
|
18
|
Li Y, Liang Q, Wen YQ, Chen LL, Wang LT,
Liu YL, Luo CQ, Liang HZ, Li MT and Li Z: Comparative proteomics
analysis of human osteosarcomas and benign tumor of bone. Cancer
Genet Cytogenet. 198:97–106. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kawai A, Kondo T, Suehara Y, Kikuta K and
Hirohashi S: Global protein-expression analysis of bone and soft
tissue sarcomas. Clin Orthop Relat Res. 466:2099–2106. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kikuta K, Tochigi N, Saito S, Shimoda T,
Morioka H, Toyama Y, Hosono A, Suehara Y, Beppu Y, Kawai A, et al:
Peroxiredoxin 2 as a chemotherapy responsiveness biomarker
candidate in osteosarcoma revealed by proteomics. Proteomics Clin
Appl. 4:560–567. 2010.PubMed/NCBI
|
|
21
|
Kubota D, Mukaihara K, Yoshida A, Tsuda H,
Kawai A and Kondo T: Proteomics study of open biopsy samples
identifies peroxiredoxin 2 as a predictive biomarker of response to
induction chemotherapy in osteosarcoma. J Proteomics. 91:393–404.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Folio C, Mora MI, Zalacain M, Corrales FJ,
Segura V, Sierrasesúmaga L, Toledo G, San-Julián M and
Patiño-García A: Proteomic analysis of chemonaive pediatric
osteosarcomas and corresponding normal bone reveals multiple
altered molecular targets. J Proteome Res. 8:3882–3888. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ren L and Khanna C: Role of ezrin in
osteosarcoma metastasis. Adv Exp Med Biol. 804:181–201. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zubaidah RM, Tan GS, Tan SB, Lim SG, Lin Q
and Chung MC: 2-D DIGE profiling of hepatocellular carcinoma
tissues identified isoforms of far upstream binding protein (FUBP)
as novel candidates in liver carcinogenesis. Proteomics.
8:5086–5096. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Malz M, Weber A, Singer S, Riehmer V,
Bissinger M, Riener MO, Longerich T, Soll C, Vogel A, Angel P, et
al: Overexpression of far upstream element binding proteins: A
mechanism regulating proliferation and migration in liver cancer
cells. Hepatology. 50:1130–1139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sinner D, Kordich JJ, Spence JR, Opoka R,
Rankin S, Lin SC, Jonatan D, Zorn AM and Wells JM: Sox17 and Sox4
differentially regulate beta-catenin/T-cell factor activity and
proliferation of colon carcinoma cells. Mol Cell Biol.
27:7802–7815. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baou M, Norton JD and Murphy JJ: AU-rich
RNA binding proteins in hematopoiesis and leukemogenesis. Blood.
118:5732–5740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Davis-Smyth T, Duncan RC, Zheng T,
Michelotti G and Levens D: The far upstream element-binding
proteins comprise an ancient family of single-strand DNA-binding
transactivators. J Biol Chem. 271:31679–31687. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu J, Kouzine F, Nie Z, Chung HJ,
Elisha-Feil Z, Weber A, Zhao K and Levens D: The FUSE/FBP/FIR/TFIIH
system is a molecular machine programming a pulse of c-myc
expression. EMBO J. 25:2119–2130. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Squire JA, Pei J, Marrano P, Beheshti B,
Bayani J, Lim G, Moldovan L and Zielenska M: High-resolution
mapping of amplifications and deletions in pediatric osteosarcoma
by use of CGH analysis of cDNA microarrays. Genes Chromosomes
Cancer. 38:215–225. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu X, Cai ZD, Lou LM and Zhu YB:
Expressions of p53, c-MYC, BCL-2 and apoptotic index in human
osteosarcoma and their correlations with prognosis of patients.
Cancer Epidemiol. 36:212–216. 2012. View Article : Google Scholar
|
|
32
|
Shimizu T, Ishikawa T, Sugihara E,
Kuninaka S, Miyamoto T, Mabuchi Y, Matsuzaki Y, Tsunoda T, Miya F,
Morioka H, et al: c-MYC overexpression with loss of Ink4a/Arf
transforms bone marrow stromal cells into osteosarcoma accompanied
by loss of adipogenesis. Oncogene. 29:5687–5699. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gherzi R, Trabucchi M, Ponassi M, Ruggiero
T, Corte G, Moroni C, Chen CY, Khabar KS, Andersen JS and Briata P:
The RNA-binding protein KSRP promotes decay of beta-catenin mRNA
and is inactivated by PI3K-AKT signaling. PLoS Biol. 5:e52006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bikkavilli RK and Malbon CC:
Dishevelled-KSRP complex regulates Wnt signaling through
post-transcriptional stabilization of beta-catenin mRNA. J Cell
Sci. 123:1352–1362. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cai Y, Mohseny AB, Karperien M, Hogendoorn
PC, Zhou G and Cleton-Jansen AM: Inactive Wnt/beta-catenin pathway
in conventional high-grade osteosarcoma. J Pathol. 220:24–33. 2010.
View Article : Google Scholar
|
|
36
|
Trabucchi M, Briata P, Garcia-Mayoral M,
Haase AD, Filipowicz W, Ramos A, Gherzi R and Rosenfeld MG: The
RNA-binding protein KSRP promotes the biogenesis of a subset of
microRNAs. Nature. 459:1010–1014. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pasero M, Giovarelli M, Bucci G, Gherzi R
and Briata P: Bone morphogenetic protein/SMAD signaling orients
cell fate decision by impairing KSRP-dependent microRNA maturation.
Cell Rep. 2:1159–1168. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nicastro G, García-Mayoral MF,
Hollingworth D, Kelly G, Martin SR, Briata P, Gherzi R and Ramos A:
Noncanonical G recognition mediates KSRP regulation of let-7
biogenesis. Nat Struct Mol Biol. 19:1282–1286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sampson VB, Rong NH, Han J, Yang Q, Aris
V, Soteropoulos P, Petrelli NJ, Dunn SP and Krueger LJ: MicroRNA
let-7a downregulates MYC and reverts MYC-induced growth in Burkitt
lymphoma cells. Cancer Res. 67:9762–9770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Viswanathan SR, Powers JT, Einhorn W,
Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA,
Lockhart VL, et al: Lin28 promotes transformation and is associated
with advanced human malignancies. Nat Genet. 41:843–848. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yamakoshi H, Kanoh N, Kudo C, Sato A, Ueda
K, Muroi M, Kon S, Satake M, Ohori H, Ishioka C, et al: KSRP/FUBP2
is a binding protein of GO-Y086, a cytotoxic curcumin analogue. ACS
Med Chem Lett. 1:273–276. 2010. View Article : Google Scholar : PubMed/NCBI
|