Introduction
Gene silencing by small double-stranded RNA (dsRNA)
was first discovered in Caenorhabditis elegans in 1998
(1) and it quickly become an
important method of genetic analysis and it has potential in gene
therapy. mRNA is generally considered a target for RNA-mediated
gene silencing. In the past ten years, studies have shown that
dsRNA targeted to gene promoter regions can regulate gene
expression at transcription level (2–5).
Although the exact mechanisms by which this is regulated are not
well understood, these findings demonstrate that dsRNA have more
diverse roles in gene regulation than previously recognized. Gene
activation by dsRNA is a relatively newly discovered method of
inducing gene overexpression (2,4,6).
Several studies have shown that some exogenously synthetic dsRNAs
can activate a particular gene in a sequence specific manner rather
than silencing it (2,4,7,8).
This phenomenon has been termed RNA activation (RNAa) and this kind
of dsRNA is called small activating RNA (saRNA) (4). Although many mysteries remain,
increasing amounts of evidence suggest that RNAa is not only a
novel method of studying gene function, but also holds exciting
potential for cancer therapeutics (9).
E-cadherin is a cell adhesion molecule. It can
mediate cell-cell adhesion and plays a pivotal role in epithelial
cell behavior and tissue morphogenesis and remodeling (10,11).
Downregulation of E-cadherin is closely associated with tumor
invasiveness, metastasis, and poor patient prognosis (12,13).
Restoration of E-cadherin by RNAa at the transcriptional level may
be a suitable therapy for cancer. Previous studies have shown that
some saRNAs can induce E-cadherin gene transcription in a
sequence-specific manner (4,14).
It is of great interest to identify new saRNA that can upregulate
E-cadherin expression efficiently. It is equally important to
understand the features and mechanism underlying saRNA function on
E-cadherin expression, which can help improve its therapeutic
application.
This report describes the design of several new
dsRNAs targeted to gene promoter regions and identification of a
new saRNA that can significantly enhance E-cadherin expression. The
functional strand of the newly discovered dsRNAs targeted to the
promoter regions of E-cadherin was also investigated. These
findings further revealed functional features of saRNA that may be
used in mechanistic studies and facilitate its medicinal
development for clinical application.
Materials and methods
dsRNA design and synthesis
Promoter sequence (1 kb) was picked away from the
CpG island and scanned for dsRNA targets based on rational siRNA
design rules (9). Four candidates
adhered to the rules of functional dsRNA design. Another two saRNA
(dsEcad-640 and dsEcad-215) were also synthesized as previously
described (4,14). Control dsRNA (dsCon) was
specifically designed to have no homology to any known human
sequences. Asymmetric dsRNA with 2 or 4 base losses in 5′ ends of
either sense or antisense strands were also designed. The
asymmetric dsRNA was designed as previously decribed (15,16).
All of the dsRNA were chemically synthesized by GenePharma
(Shanghai, China) (Table I).
 | Table IThe sequence of synthesized dsRNA and
the PCR primer sequence for E-cadherin and GAPDH. |
Table I
The sequence of synthesized dsRNA and
the PCR primer sequence for E-cadherin and GAPDH.
| dsRNA name | Sequence
(5′-3′) |
|---|
| dsE-661 (−661 to
−642) | Sense: |
CUCAGUGGCUCAUGGCUCA[dT][dT] |
| Antisense: |
UGAGCCAUGAGCCACUGAG[dT][dT] |
| dsE-604 (−604 to
−585) | Sense: |
GGAUCGCUUCAGCCCAGGA[dT][dT] |
| Antisense: |
UCCUGGGCUGAAGCGAUCC[dT][dT] |
| dsE-454 (−454 to
−435) | Sense: |
CAGCUACUAGAGAGGCUGG[dT][dT] |
| Antisense: |
CCAGCCUCUCUAGUAGCUG[dT][dT] |
| dsE-382 (−382 to
−363) | Sense: |
CACCACUGCACUCCAGCUU[dT][dT] |
| Antisense: |
AAGCUGGAGUGCAGUGGUG[dT][dT] |
| dsE-623 (−623 to
−604) | Sense: |
UUGGGAGGCCAAGGCAGGA[dT][dT] |
| Antisense: |
UCCUGCCUUGGCCUCCCAA[dT][dT] |
| dsE-680 (−680 to
−661) | Sense: |
CAAAAAAUUAGGCUGCUAG[dT][dT] |
| Antisense: |
CUAGCAGCCUAAUUUUUUG[dT][dT] |
| dsE-648 (−648 to
−629) | Sense: |
GGCUCACACCUGAAAUCCU[dT][dT] |
| Antisense: |
AGGAUUUCAGGUGUGAGCC[dT][dT] |
| dsControl | Sense: | UUC UCC GAA CGU GUC
ACG U[dT][dT] |
| Antisense: | ACG UGA CAC GUU CGG
AGA A[dT][dT] |
|
| PCR primer | Sequence
(5′-3′) |
|
| E-cadherin | Sense: |
CGCCGAGAGCTACACGTTCA |
| Antisense: |
TGTCGACCGGTGCAATCTTC |
| GAPDH | Sense: |
CGCTCTCTGCTCCTCCTGTT |
| Antisense: |
CCATGGTGTCTGAGCGATGT |
Cell culture and transfection
PC-3 and DU 145 were cultured in RPMI medium-1640
supplemented with 10% FBS, 2 mM L-glutamine, penicillin (100 U/ml),
and streptomycin (100 μg/ml) in a humidified atmosphere of 5%
CO2 maintained at 37°C. A hepatocellular carcinoma cell
line (MHCC-97L, MHCC-97H, and MHCCLM3) was also cultured using DMEM
medium supplemented with 10% FBS, penicillin (100 U/ml), and
streptomycin (100 μg/ml). Transfections of dsRNA were carried out
using Lipofectamine 2000 (Invitrogen, USA) according to the
manufacturer’s protocol and lasted for 72 h.
Real-time quantitative PCR (qPCR)
Total RNA was extracted from cells transfected for
72 h (mock, 50 nM dsCon, 50 nM dsEcad-661, dsEcad-604, dsEcad-454,
dsEcad-382, dsEcad-680, dsEcad-648, dsEcad-623, dsEcad-640 and
dsEcad-215). Reverse transcription was performed with a PrimeScript
RT Reagent kit (Takara Biotechnology, Dalian, China). qPCR was
performed with SYBR Green PCR Reagent kits (Toyobo Co, Osaka,
Japan) at a constant annealing temperature (64°C) according to the
manufacturer’s protocol. Specific primer sets were designed for
qPCR directed against human E-cadherin and GAPDH and synthesized by
Takara Biotechnology Co. (Table
I). Data were recorded and analyzed using qPCR analysis
software Bio-Rad iQ5. Endogenous gene expression was normalized to
GAPDH levels in the cells.
Western blot analysis
The expression of E-cadherin protein was estimated
using western blotting. Cells were harvested 72 h after dsRNA
treatment as described above and then washed and lysed with M-PER
extraction buffer (Pierce Biotechnology) containing protease
inhibitors. A BCA assay (Sangon Biotech Co., Ltd., Shanghai, China)
was used to measure the concentration of protein lysates. Protein
was separated on reducing SDS-polyacrylamide gels and transferred
to polyvinyl difluoride membranes (PVDF, Millipore). The membranes
were blocked with 3% BSA buffer for 1 h at room temperature and
incubated with primary antibodies (anti-E-cadherin rabbit antibody,
Cell Signaling, Beverly, MA, USA, 1:1,000 dilutions; anti-β-actin
antibody, Cell Signaling, 1:1,000 dilutions) overnight at 4°C.
β-actin levels were used to normalize loading. Then the membranes
were incubated with goat anti-rabbit secondary antibody (Cell
Signaling) 1:5,000 at 25 °C for 2 h. Antigen-antibody complexes
were visualized using an enhanced chemiluminescence detection
method (ECL kit, Thermo, Waltham, MA, USA).
Fluorescence microscopy analysis
Labeling of dsRNAs by Cy3 was performed by Guangzhou
RiboBio Co. (Guangzhou, China). Cells were fixed with
paraformaldehyde (PFA) at 6 h after dsRNAs treatment. Staining of
the DNA with 4′, 6-diamidin-2′-phenylindol-dihydrochlorid (DAPI)
was conducted after fixation. A Leica TCS SP5 confocal laser
scanning microscope was used for fluorescence imaging.
Statistical analysis
All experiments were performed at least three times.
Statistical analysis of all data was performed using Student’s
t-test. A P-value <0.05 was considered to indicate statistical
significance.
Results
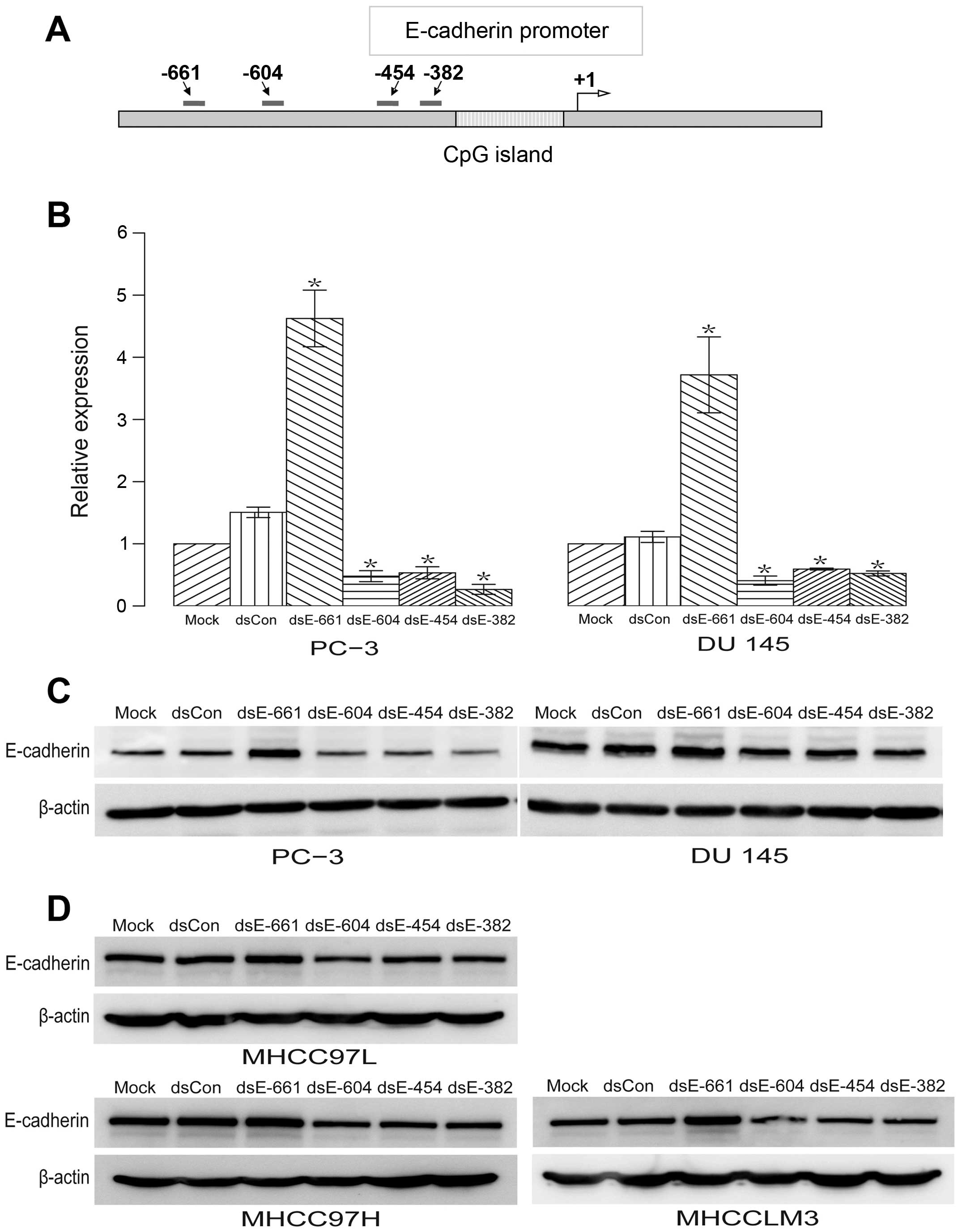
New dsRNA targeting the promoter and
enhancement of E-cadherin expression
Four 21-nt dsRNAs targeting the E-cadherin promoter
at sequence positions −661 (dsEcad-661), −604 (dsEcad-604), −454
(dsEcad-454), and −382 (dsEcad-382) were designed and synthesized
(Fig. 1A). Regions with either
high GC content or low sequence complexity were excluded as dsRNA
targets. These included the CpG island and the Alu repeat
element in the E-cadherin promoter (Fig. 1A). A 21-nt dsCon without
significant homology to any known human sequence was also
synthesized as described previously (4). These dsRNAs were transfected into two
human prostate cancer cell lines: PC-3 and DU 145. The expression
of E-cadherin mRNA and protein was evaluated using qPCR and western
blotting, respectively. We observed a profound induction of
E-cadherin mRNA (Fig. 1B) and
protein expression (Fig. 1C) by
dsEcad-661. While dsEcad-604, dsEcad-454 and dsEcad-382
significantly reduce the expression of E-cadherin mRNA and protein
(Fig. 1B and C). To determine
whether these effect of dsRNAs are specific to prostate cancer cell
lines, these dsRNAs were transfected into hepatocellular carcinoma
cell line MHCC-97L, MHCC-97H, and MHCCLM3. The results showed that
these dsRNAs have similar regulatory effects on the expression of
E-cadherin protein in MHCC-97L, MHCC-97H, and MHCCLM3 cell lines
(Fig. 1D). Overall, no changes in
E-cadherin expression were detected in cells transfected with mock
or dsCon (Fig. 1B–D).
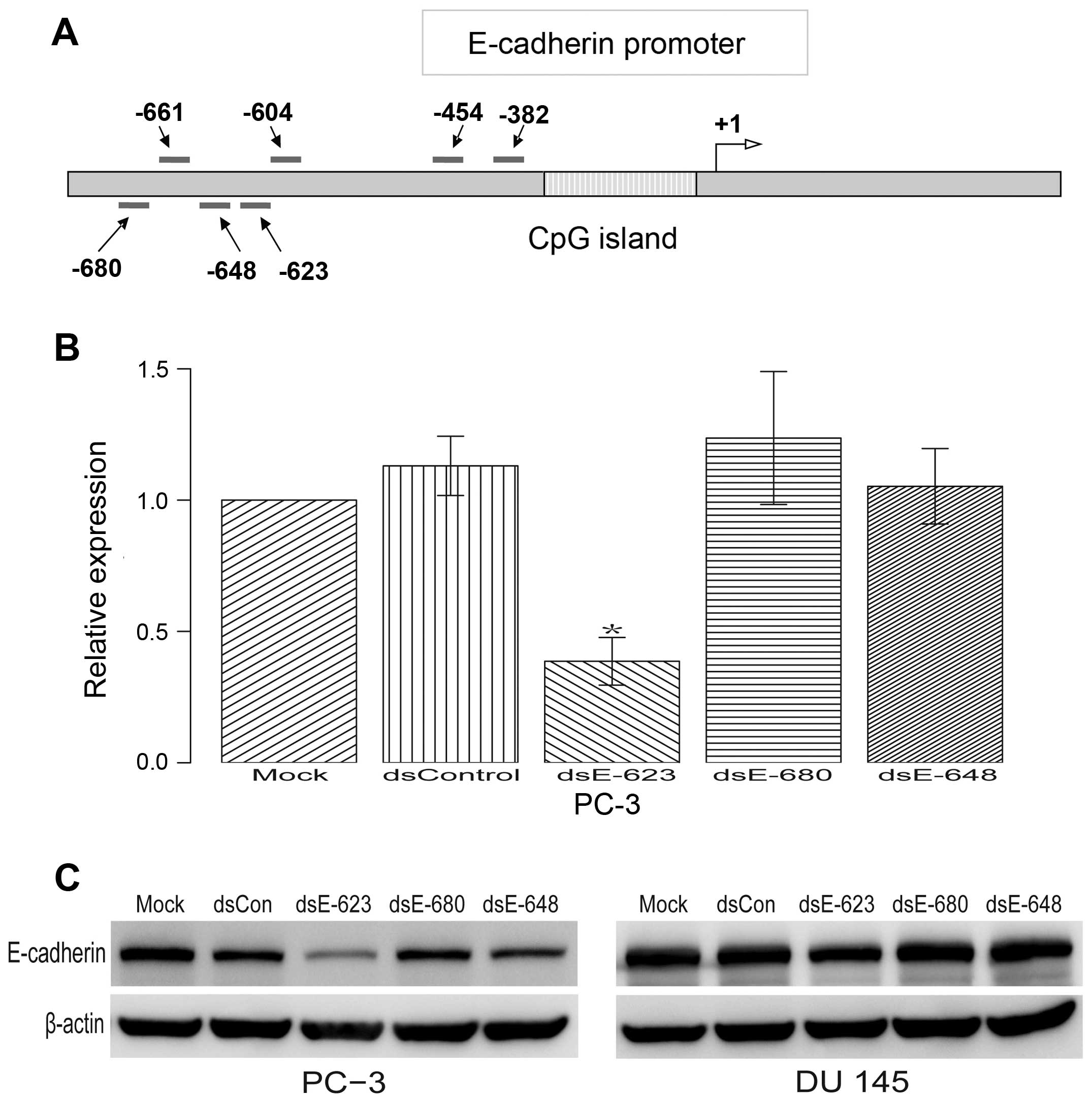
Previous studies have shown that a 21-nt dsRNA
targeting the E-cadherin promoter at sequence positions −640
(dsEcad-640) can also induce E-cadherin expression (14). To further confirm whether there is
any other target near the position −640 and −661 that can also
induce the expression of E-cadherin, another three dsRNAs
(dsEcad-680, dsEcad-648 and dsEcad-623) were tested. The results
indicated that dsEcad-623 can significantly decrease E-cadherin
expression. Neither dsEcad-680 nor dsEcad-648 was capable of
regulating E-cadherin expression (Fig.
2B and C).
Identification of guide strand
Although the RNAa phenomenon was identified almost a
decade ago, the precise mechanism underlying the function still
remains unclear. There is some question regarding the saRNA
targets: DNA or non-coding RNA transcripts (17–19).
Growing evidence suggests that only one strand may be required to
guide activity of the dsRNA (20).
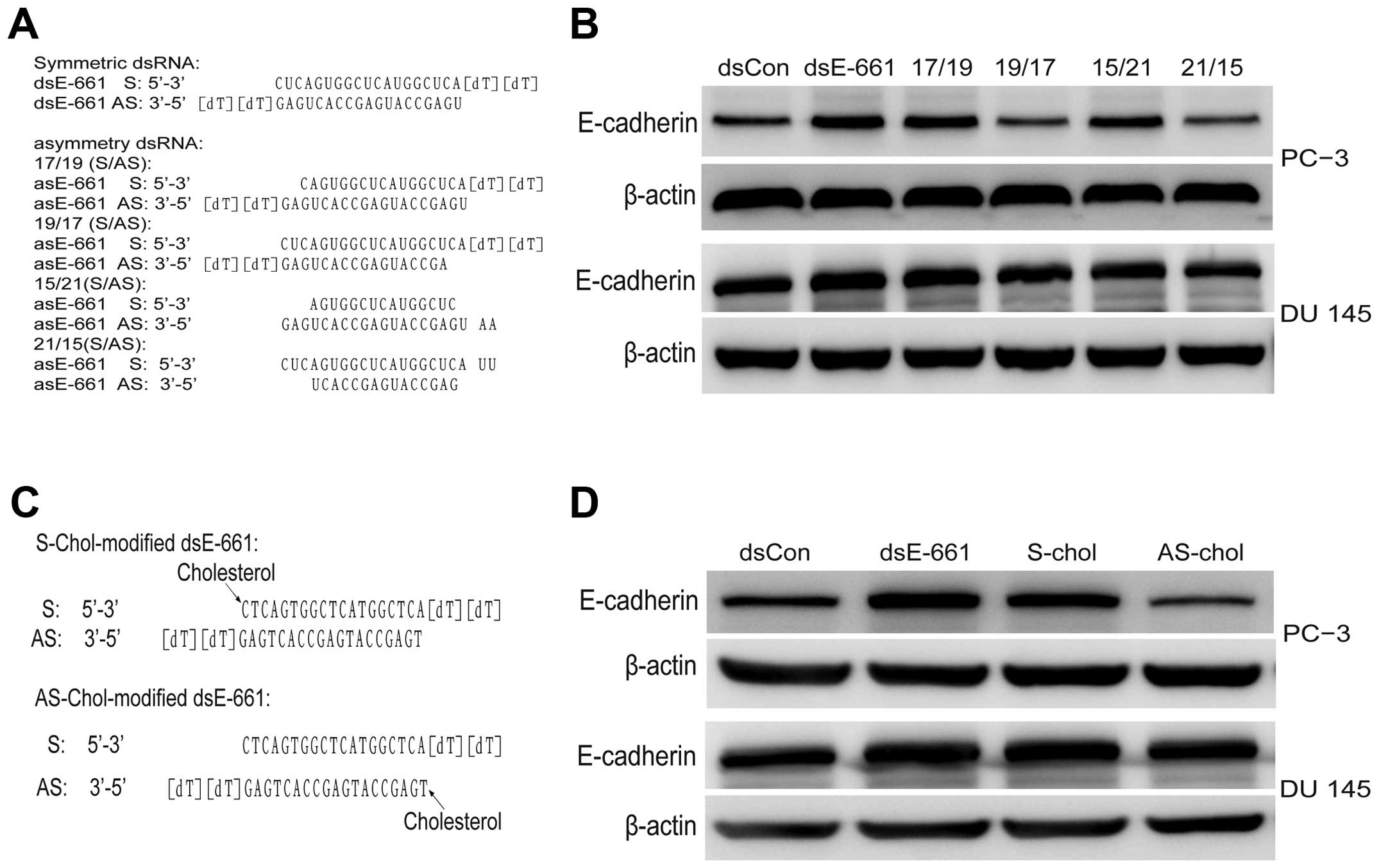
To identify the target of saRNA, which strand of saRNA guides gene
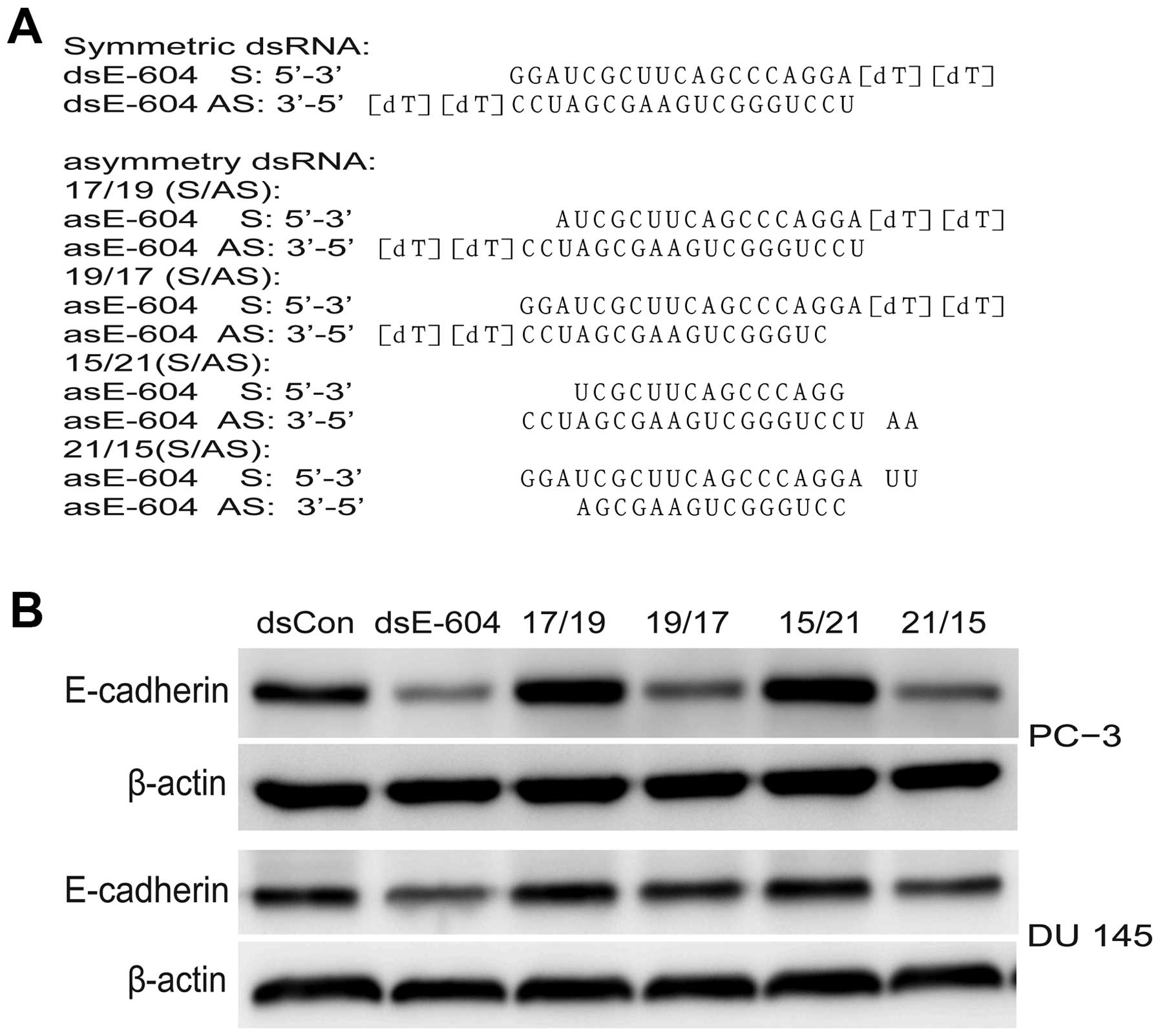
activation must first be determined. Hence, asymmetric saRNA
molecules were synthesized without 2 or 4 base in the 5′ end of
either the sense or antisense strand, which has been reported to
have fewer off-target effects (15). dsEcad-661 (17/19),
dsEcad-661(19/17), dsEcad-661(15/21), and dsEcad-661(21/15) were
transfected to PC-3 and DU 145 cell line and their activity was
analyzed, respectively, using western blot analysis. The results
showed asymmetric dsEcad-661(17/19) and dsEcad-661(15/21) to have
the same E-cadherin activating ability as symmetric dsEcad-661
while asymmetric dsEcad-661(19/17) and dsEcad-661(21/15) lost their
gene induction activity (Fig. 3).
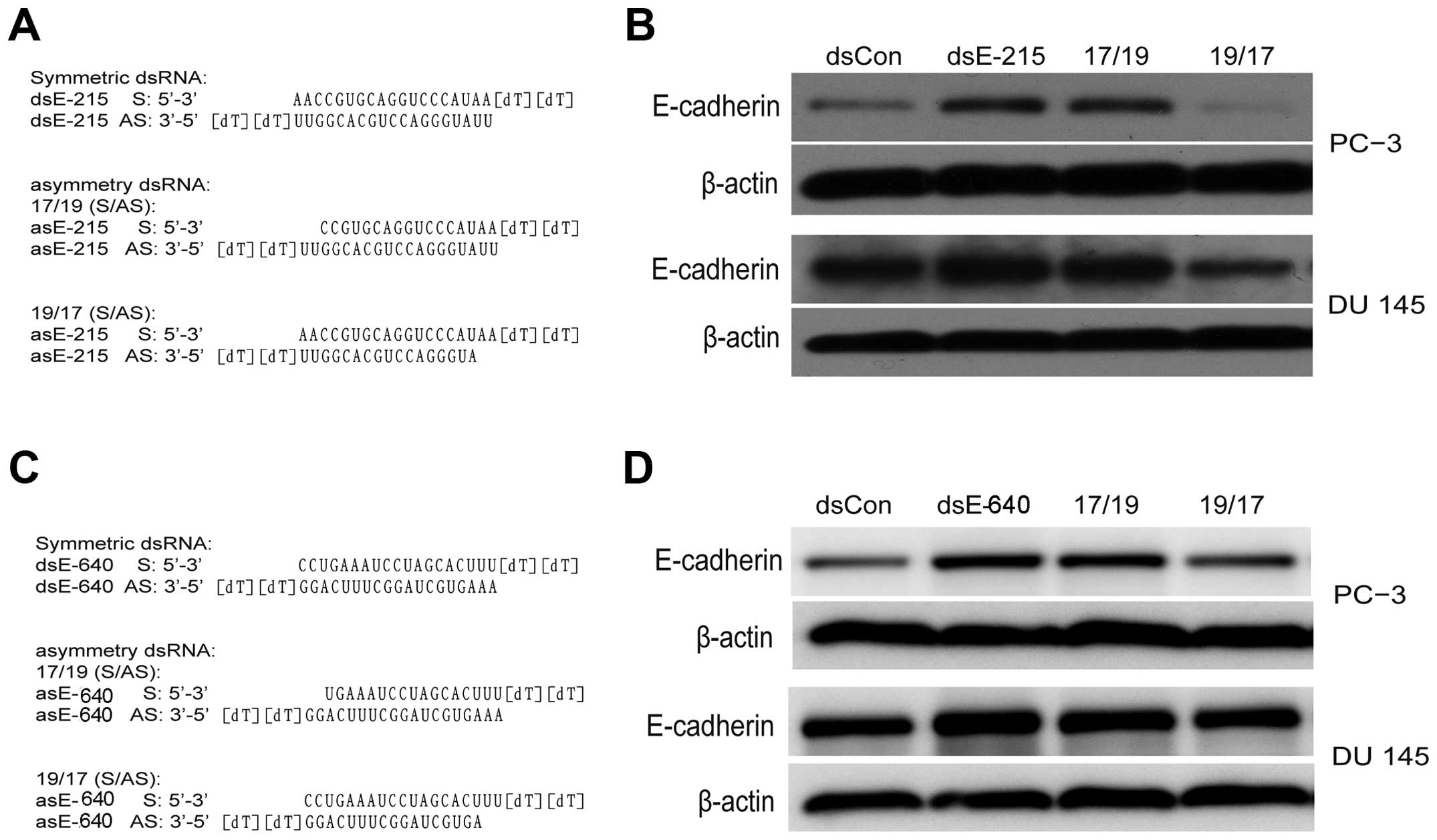
Another two E-cadherin saRNA (dsE-640 and dsE-215) were also tested
as described previously (4,14).
The asymmetric dsEcad-640(17/19) and dsE-215(17/19) can induce
E-cadherin expression while dsEcad-640(19/17) and dsEcad-215(19/17)
lost gene regulating activity (Fig.
4). This evidence indicated that the antisense strand in saRNA
duplexes was responsible for RNAa activity. The target of saRNA
therefore is expected to be in the sense strand of E-cadherin DNA
or the corresponding non-coding RNA transcript.
To further confirm whether the antisense strand is
the guide strand of saRNA, the antisense stand of dsEcad-661 and
the sense strand of dsEcad-661 was modified with cholesterol
respectively, and transfected into PC-3 and DU. The
cholesterol-conjugated saRNA at the sense stand of dsEcad-661
showed similar gene induction activity while cholesterol-conjugated
saRNA at the antisense stand of dsEcad-661 did not show any
regulatory effect on E-cadherin expression (Fig. 3).
To determine whether the promoter targeting dsRNA
that can downregulate E-cadherin expression share the same target
strand as saRNA, another four asymmetric dsRNA were synthesized:
dsEcad-604 (17/19), dsEcad-604 (15/21), dsEcad-604 (19/17), and
dsEcad-604 (21/15). The symmetric dsEcad-604 (21/21) and asymmetric
dsEcad-604 were transfected into PC-3 and DU 145 cell lines as
described previously. Results showed that dsEcad-604 (19/17) and
dsEcad-604 (21/15) had gene silencing activity similar to that of
symmetric dsEcad-604 while dsEcad-604 (17/19) and dsEcad-604
(15/21) lost gene silencing activity (Fig. 5). These results indicated that the
sense strand of interfering dsRNA might be the guide strand.
Identification of nucleus entering
strand
Previous studies have shown that small interfering
dsRNA may, in addition to eliminating specific pools of mRNA in the
cytoplasm, also work by epigenetic regulation of gene expression in
the nucleus (21,22). To develop a preliminary
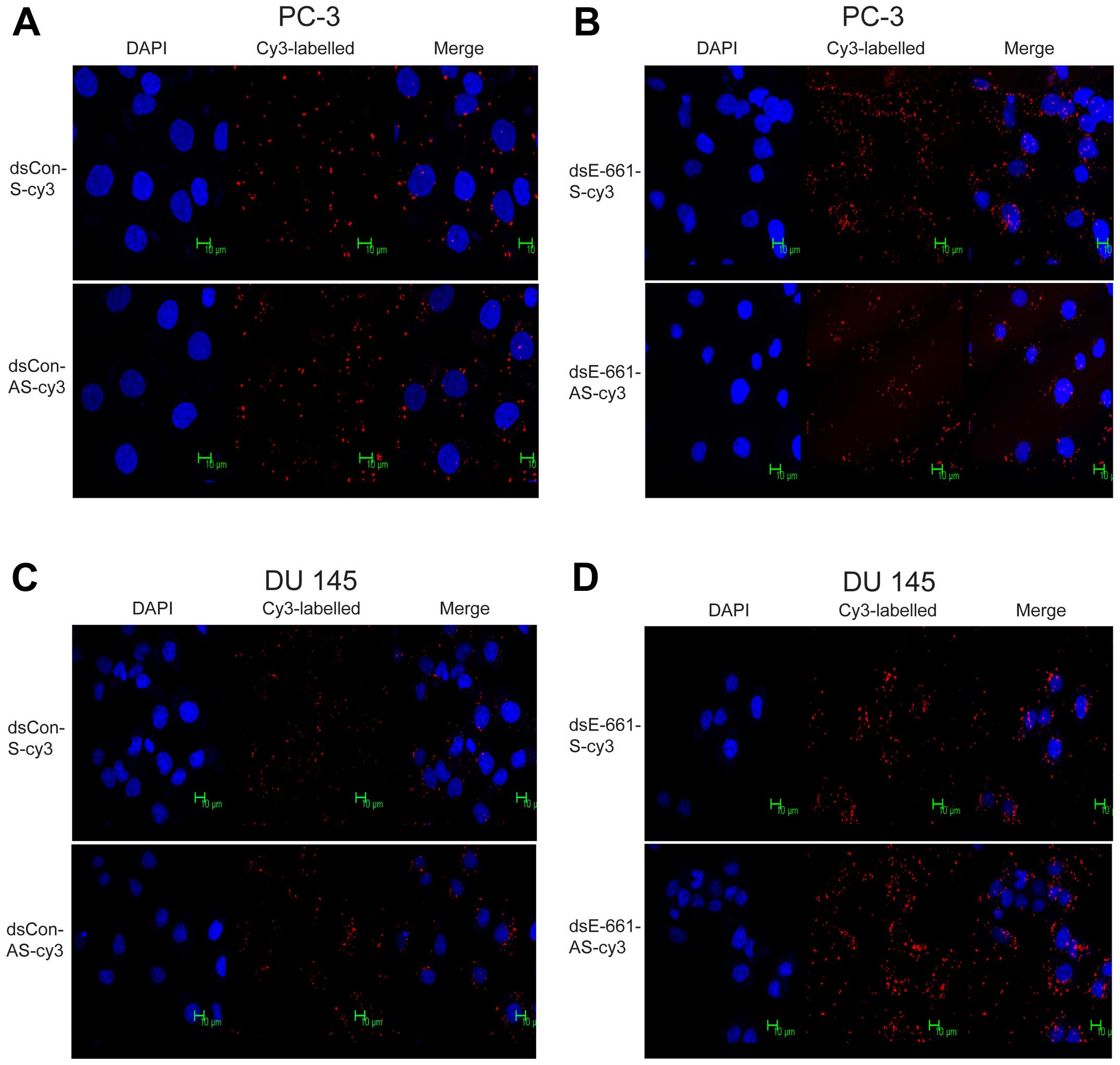
understanding of the position where saRNA exerts its effects and
confirm which strand of saRNA enters the nucleus, the sense and
antisense strands of dsEcad-661 were labeled with Cy3 and
transfected using the procedure described above. Sense and
antisense strands of dsCon served as the control group.
Immunofluorescence analysis indicated that both sense and antisense
strands of dsEcad-661 were present in the cytoplasm and nucleus,
which indicated that both strands of dsEcad-661 can enter the
nucleus (Fig. 6). The control
dsRNA without homology to any known human sequence can also enter
the nucleus of PC-3 and DU 145 cell (Fig. 6).
Discussion
Downregulated E-cadherin is a key event during
epithelial-mesenchymal transition (EMT) and is associated with
enhanced tumor invasion and poor prognosis in variety of solid
tumors (23–25). RNAa is a recently discovered
mechanism of gene regulation. It is mediated by dsRNA that targets
gene regulatory sequences. saRNA can activate the downregulation of
gene expression and so provide a new therapeutic tool for treatment
of cancer. In this study, several potential dsRNAs targeting the
promoter sequence of E-cadherin were designed and a new saRNA that
can induce significant E-cadherin expression was discovered.
Previous studies have reported three saRNA (dsEcad-302, dsEcad-215
and dsEcad-640) can induce E-cadherin expression (4,14).
These results and the newly discovered dsEcad-661 suggest that
E-cadherin expression can be activated by a wide array of saRNA
targeting the promoter sequence. saRNA targeting the E-cadherin
promoter may serve as a useful tool for further elucidation of the
roles of E-cadherin in cancer and make it possible to develop new
gene therapies for cancer. In this study, several dsRNAs targeting
the promoter area close to the target of saRNA were found to
downregulate E-cadherin expression. These findings suggest that
dsRNA have more diverse roles in gene regulation than previously
thought. The mechanism by which dsRNA targets the promoter regions
and so mediates both gene silencing and gene activation may be very
complicated.
Although the RNAa phenomenon has been known for
almost a decade, the use of this technique as a therapeutic method
has not yet been widely used. Poor understanding of the mechanisms
by which it exerts its effects may be the main reason (7). One of these unclear mechanisms is the
manner in which synthesized saRNA enters the nucleus and exerts its
effect. Previous studies have demonstrated that exogenous dsRNAs
may be processed by removing one strand of dsRNA to form active
argonaute protein-RNA complexes containing a selected guide strand
RNA and then enter the nucleus (26–28).
The active complex is guided to its target region by the guide
strand, which may bind to complementary targets on either promoter
DNA or nascent promoter RNA (9,27). A
recent study also found the mechanism that saRNA-loaded Argonaut
protein 2 facilitates the assembly of an RNA-induced
transcriptional activation (RITA) complex, which interacts with RNA
polymerase II to stimulate transcription initiation and productive
elongation (29). In this study,
fluorescently labeled dsRNA was used as a direct indicator of which
strand can enter the nucleus. Results showed that both strands of
dsRNA can enter the nucleus, which suggests that the unwinding
process may take place after the dsRNA enters the nucleus. The
active argonaute protein-RNA complex may form inside the nucleus.
Further studies on this mechanism seem warranted.
It is still not clear how saRNAs activate genes. The
activation mechanism of RNAa may involve both transcriptional and
epigenetic alterations. The target of saRNA may be either promoter
DNA or nascent promoter RNA transcript (18,19).
It is also not clear which strand guides RNAa activity when
targeting DNA or nascent promoter. Recent studies have provided
evidence that RNAa activity was achieved by its antisense strand
with the 5′ region playing a pivotal role (30). However, there still are
controversies on the functional strands of saRNA. Some reports have
suggested that the sense strand plays a pivotal role (18). As such, we designed and synthesized
several asymmetric dsRNA including our newly discovered saRNA
(dsEcad-640), previously described saRNA (dsEcad-640 and
dsEcad-215) and interfering dsRNA (dsEcad-604). Using asymmetric
dsRNA which has one inactivated strand, we showed that the
antisense strand in saRNA duplexes was responsible for the RNAa
activity of E-cadherin while the sense strand of interfering dsRNA
was responsible for E-cadherin gene silence. To further confirm our
results, we designed the cholesterol-conjugated saRNA to block the
binding function of each strand selectively. The result also
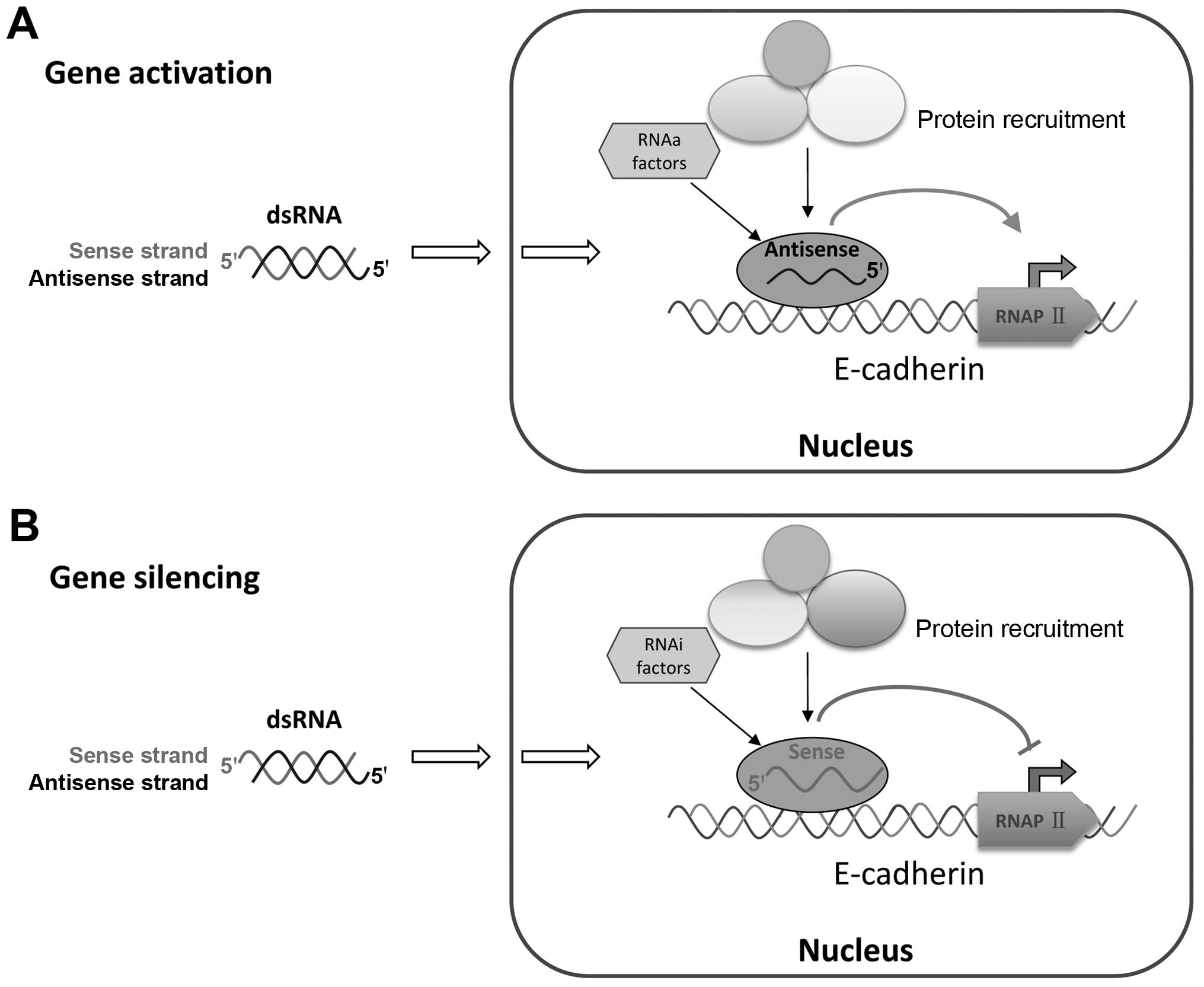
demonstrated that the antisense strand of saRNA was responsible for
RNAa activity. This result suggested that the targets of activating
dsRNA and silencing dsRNA are located on different strands of DNA
or nascent promoter RNA. Previous studies on RNA-directed
transcriptional gene regulation have provided a working model for
the mechanism of action, which requires the recruitment of several
proteins (argonaute-1 and argonaute-2), small RNAs and other
factors (31–34). This study brought up fresh evidence
that the antisense strand of the activating dsRNA is responsible
for RNAa activity and that the sense strand in silencing dsRNA
guides gene activation (Fig.
7).
Recent studies showed the gene silencing mediated by
asymmetric small interfering RNA is efficacious, durable, and
associated with reduced off-target silencing by the sense strand
(15). Asymmetric small
interfering RNA structure has several advantages over the
conventional 19+2 siRNA structures (16,35).
This study found that asymmetric saRNA also has gene activating
capacity similar to that of conventional 19+2 siRNA structures. It
provides a new structural scaffold for designing activating RNA
duplex and may facilitate RNAa applications in functional genomics
and therapeutics.
RNAa offers a promising new therapeutic strategy for
the treatment of diseases with abnormal gene downregulation.
Currently, developing RNAa technology against diseases is still at
the initial stage. Many of the challenges facing RNA interference
based strategies are also applicable to RNAa. Further clarifying
the mechanism of the saRNA action is critical for the clinical
application of saRNA. In conclusion, a new target of dsRNA was here
found to enhance E-cadherin expression and several new dsRNA
targets that can downregulate E-cadherin expression. This study
also established which strand of saRNA is the guide strand. Further
studies are still needed to gain insight into the details of the
mechanisms and biology of RNAa.
Acknowledgements
This study was supported by Shanghai Municipal
Commission of Health and family planning (no. 201540372).
Abbreviations:
|
dsRNAs
|
double-stranded RNAs
|
|
saRNA
|
small activating RNA
|
|
RNAa
|
RNA activation
|
|
EMT
|
epithelial-mesenchymal transition
|
|
qPCR
|
real-time quantitative PCR
|
References
|
1
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Janowski BA, Younger ST, Hardy DB, Ram R,
Huffman KE and Corey DR: Activating gene expression in mammalian
cells with promoter-targeted duplex RNAs. Nat Chem Biol. 3:166–173.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Elbashir SM, Harborth J, Lendeckel W,
Yalcin A, Weber K and Tuschl T: Duplexes of 21-nucleotide RNAs
mediate RNA interference in cultured mammalian cells. Nature.
411:494–498. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li LC, Okino ST, Zhao H, Pookot D, Place
RF, Urakami S, Enokida H and Dahiya R: Small dsRNAs induce
transcriptional activation in human cells. Proc Natl Acad Sci USA.
103:17337–17342. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ting AH, Schuebel KE, Herman JG and Baylin
SB: Short double-stranded RNA induces transcriptional gene
silencing in human cancer cells in the absence of DNA methylation.
Nat Genet. 37:906–910. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Place RF, Noonan EJ, Földes-Papp Z and Li
LC: Defining features and exploring chemical modifications to
manipulate RNAa activity. Curr Pharm Biotechnol. 11:518–526. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pushparaj PN, Aarthi JJ, Kumar SD and
Manikandan J: RNAi and RNAa - the yin and yang of RNAome.
Bioinformation. 2:235–237. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang T, Li M, Yuan H, Zhan Y, Xu H, Wang
S, Yang W, Liu J, Ye Z and Li LC: saRNA guided iNOS up-regulation
improves erectile function of diabetic rats. J Urol. 190:790–798.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng L, Wang L, Gan J and Zhang H: RNA
activation: Promise as a new weapon against cancer. Cancer Lett.
355:18–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Roy F and Berx G: The cell-cell
adhesion molecule E-cadherin. Cell Mol Life Sci. 65:3756–3788.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Repetto O, De Paoli P, De Re V, Canzonieri
V and Cannizzaro R: Levels of soluble E-cadherin in breast,
gastric, and colorectal cancers. Biomed Res Int. 2014:4080472014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Canel M, Serrels A, Frame MC and Brunton
VG: E-cadherin-integrin crosstalk in cancer invasion and
metastasis. J Cell Sci. 126:393–401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schmalhofer O, Brabletz S and Brabletz T:
E-cadherin, beta-catenin, and ZEB1 in malignant progression of
cancer. Cancer Metastasis Rev. 28:151–166. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Place RF, Li LC, Pookot D, Noonan EJ and
Dahiya R: MicroRNA 373 induces expression of genes with
complementary promoter sequences. Proc Natl Acad Sci USA.
105:1608–1613. 2008. View Article : Google Scholar
|
|
15
|
Sun X, Rogoff HA and Li CJ: Asymmetric RNA
duplexes mediate RNA interference in mammalian cells. Nat
Biotechnol. 26:1379–1382. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sano M, Sierant M, Miyagishi M, Nakanishi
M, Takagi Y and Sutou S: Effect of asymmetric terminal structures
of short RNA duplexes on the RNA interference activity and strand
selection. Nucleic Acids Res. 36:5812–5821. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morris KV, Santoso S, Turner AM, Pastori C
and Hawkins PG: Bidirectional transcription directs both
transcriptional gene activation and suppression in human cells.
PLoS Genet. 4:e10002582008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schwartz JC, Younger ST, Nguyen NB, Hardy
DB, Monia BP, Corey DR and Janowski BA: Antisense transcripts are
targets for activating small RNAs. Nat Struct Mol Biol. 15:842–848.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu J, Chen Z, Xia D, Wu J, Xu H and Ye ZQ:
Promoter-associated small double-stranded RNA interacts with
heterogeneous nuclear ribonucleoprotein A2/B1 to induce
transcriptional activation. Biochem J. 447:407–416. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Portnoy V, Huang V, Place RF and Li LC:
Small RNA and transcriptional upregulation. Wiley Interdiscip Rev
RNA. 2:748–760. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morris KV, Chan SW, Jacobsen SE and Looney
DJ: Small interfering RNA-induced transcriptional gene silencing in
human cells. Science. 305:1289–1292. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo D, Barry L, Lin SS, Huang V and Li LC:
RNAa in action: From the exception to the norm. RNA Biol.
11:1221–1225. 2014. View Article : Google Scholar
|
|
23
|
Nakagawa H, Hikiba Y, Hirata Y,
Font-Burgada J, Sakamoto K, Hayakawa Y, Taniguchi K, Umemura A,
Kinoshita H, Sakitani K, et al: Loss of liver E-cadherin induces
sclerosing cholangitis and promotes carcinogenesis. Proc Natl Acad
Sci USA. 111:1090–1095. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Corso G, Carvalho J, Marrelli D, Vindigni
C, Carvalho B, Seruca R, Roviello F and Oliveira C: Somatic
mutations and deletions of the E-cadherin gene predict poor
survival of patients with gastric cancer. J Clin Oncol. 31:868–875.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang H, Stephens LC and Kumar R:
Metastasis tumor antigen family proteins during breast cancer
progression and metastasis in a reliable mouse model for human
breast cancer. Clin Cancer Res. 12:1479–1486. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yue X, Schwartz JC, Chu Y, Younger ST,
Gagnon KT, Elbashir S, Janowski BA and Corey DR: Transcriptional
regulation by small RNAs at sequences downstream from 3′ gene
termini. Nat Chem Biol. 6:621–629. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiao AL and Slack FJ: RNA-mediated gene
activation. Epigenetics. 9:27–36. 2014. View Article : Google Scholar :
|
|
28
|
Li LC: Chromatin remodeling by the small
RNA machinery in mammalian cells. Epigenetics. 9:45–52. 2014.
View Article : Google Scholar :
|
|
29
|
Portnoy V, Lin SH, Li KH, Burlingame A, Hu
ZH, Li H and Li LC: saRNA-guided Ago2 targets the RITA complex to
promoters to stimulate transcription. Cell Res. 26:320–335. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meng X, Jiang Q, Chang N, Wang X, Liu C,
Xiong J, Cao H and Liang Z: Small activating RNA binds to the
genomic target site in a seed-region-dependent manner. Nucleic
Acids Res. 44:2274–2282. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Janowski BA, Huffman KE, Schwartz JC, Ram
R, Nordsell R, Shames DS, Minna JD and Corey DR: Involvement of
AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct
Mol Biol. 13:787–792. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim DH, Saetrom P, Snøve O Jr and Rossi
JJ: MicroRNA-directed transcriptional gene silencing in mammalian
cells. Proc Natl Acad Sci USA. 105:16230–16235. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Morris KV: RNA-directed transcriptional
gene silencing and activation in human cells. Oligonucleotides.
19:299–306. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gagnon KT, Li L, Chu Y, Janowski BA and
Corey DR: RNAi factors are present and active in human cell nuclei.
Cell Rep. 6:211–221. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chang CI, Yoo JW, Hong SW, Lee SE, Kang
HS, Sun X, Rogoff HA, Ban C, Kim S, Li CJ, et al: Asymmetric
shorter-duplex siRNA structures trigger efficient gene silencing
with reduced nonspecific effects. Mol Ther. 17:725–732. 2009.
View Article : Google Scholar : PubMed/NCBI
|