Introduction
It is reported that there were 1.8 million new cases
of lung cancer in 2012 (12.9% of the total), 58% of which occurred
in less developed regions (1).
Lung cancer is estimated to be responsible for nearly one in five
cancer related deaths worldwide (1.59 million deaths, 19.4% of the
total), which makes it a major public health problem (2).
In most Western countries, lung cancer incidence and
death rates are decreasing in men and plateauing in women (3,4);
however, both incidence and mortality rates of lung cancer are
still increasing in China and there were ~652,800 new cases and
597,200 deaths in China in 2015, accounting for 35.78% and 37.56%
of the whole world (5).
Approximately 85% of patients diagnosed with lung cancer are
non-small cell lung cancer (NSCLC) and two in three of these cases
are diagnosed in metastatic or advanced stages (6). NSCLC patients with early stage could
have better opportunity of admirable survival (7). Therefore minimal damage, effective
and convenient detection for early stage of non-small cell lung
cancer is particularly necessary.
Currently there are several methods to detect NSCLC,
such as tissue biopsy, which is the golden standard of diagnosis.
However, it is often difficult to obtain biopsy samples, and it is
very challenging or time-consuming to acquire samples especially in
early-stage cases from different medical centers. Low-dose computed
tomography (LDCT) and X-ray examination are two other main
approaches. LDCT is an ever emerged National Lung Screening Trial
(NLST) in US; however, LDCT screening has a relatively low
specificity (73.4%), which results a high false-positive findings
rate (96.4%) (8). LDCT screening
is simple and was shown to confer a 20% reduction in lung cancer
mortality and a higher proportion of early NSCLC diagnosis in a
high-risk population (9), but its
utility and validity are still under debate (10–12).
Recently the U.S. Preventive Services Task Force recommendation
statement (USPSTF) recommended annual LDCT-screening for lung
cancer in high-risk individuals and stressed the need for more
research into the use of biomarkers to complement LDCT
screening.
CfDNA corresponds to cell-free DNA fragments
circulating in the bloodstream which can be extracted from plasma
or serum. CfDNA is mostly composed of constitutive genomic DNA
(13). One of the most immediate
applications of circulating cfDNA has been termed ‘liquid biopsy’
in research studies as well as in clinical practices (14,15).
Detection of genetic and specific mutations will be the most
promising tool for large-scale population-based lung cancer
screening when considering its safety, availability and accuracy
(11). Sun et al found that
compared with other non-invasive approaches to monitor EGFR-TKI
treatment in NSCLC patients, cfDNA displayed many advantages:
moderate sensitivity, high specificity, feasible on small-amount
samples, rapid and low cost, and high reproducibility (16). Identified mutations in cfDNA have
also been found to be potential prognostic biomarkers of NSCLC
(17,18). Here we expanded the increasing
interest in this approach to explore potential application in early
stage screening and diagnosis of NSCLC patients.
Materials and methods
Patient information and ethics
statement
Tumor and blood samples from 10 NSCLC patients were
analyzed in this study. All patients, including 8 males and 2
females with an average age of 57.8±8.88, were diagnosed with stage
IA, IB, IIA, and IIB NSCLC, of these 5 were adenocarcinoma, 4 were
squamous cell carcinoma (SCC), and 1 was sarcomatoid carcinoma.
Three of these patients have direct relatives diagnosed with
cancer. Four of these patients have long term smoking history
(>20 years). 20 healthy controls, including 13 elderly people
and 7 middle-age smokers with an average age of 66.8±15.46, were
also recruited from noncancerous out-patients of Hebei Medical
University Fourth Hospital. The study was approved by Ethics
Committees of Hebei Medical University Fourth Hospital. All
participants, including NSCLC patients and healthy controls,
provided written informed consent for this study.
Sample DNA handling
Fresh tumor tissue, peripheral blood lymphocytes
(PBLs), and plasma were collected for analysis for each patient.
Tubes (10 ml) containing blood samples with EDTA added were
centrifuged at 1000 g for 10 min. The cell pellets containing
peripheral blood lymphocytes were stored at −20°C. The supernatants
were centrifuged again at 10,000 g for 10 min, and plasma was
collected and stored at −80°C. Tiangen tissue DNA kit (Tiangen,
Beijing, China) and Tiangen whole blood DNA kit (Tiangen) were used
to extracted DNA from fresh tumor tissue and peripheral blood
lymphocytes, respectively. QIAamp Circulating Nucleic Acid kit
(Qiagen, Germany) was used to extract cfDNA form plasma. All kits
were used according to the manufacturer’s instructions.
Library preparation and sequencing
For each sample, DNA was quantified with the Qubit
dsDNA HS Assay kit (Life Technologies, USA) according to the
manufacturer’s instructions. Targeted amplification and Illumina
adapter-ligated library preparation was performed using Amplicon
Sequencing-Illumina Compatible kit following the manufacturer’s
instructions (Genecrab, Beijng, China). All samples were subjected
to Illumina HiSeq X-Ten for paired-end sequencing (150 bp each
end). The AmpliSeq Cancer Panel covers 92 continuous region with
10,235 bp in 22 cancer-associated genes (KRAS, EGFR, BRAF, PIK3CA,
AKT1, ERBB2, PTEN, NRAS, STK11, MAP2K1, ALK, DDR2, CTNNB1, MET,
TP53, SMAD4, FBX7, FGFR3, NOTCH1, ERBB4, FGFR1 and FGFR2), which
developed by Life Technologies Co.
Variant calling
Initial data from HiSeq X-Ten were evaluated by
using fastQC (v0.11.3). Raw reads were mapped to reference genome
hg19 by using BWA (0.7.12-r1039). Program Samtools and VarScan
(v2.4.1) was used for variant calling: i) the average total
coverage depth was defined as >1,000 and each variant coverage
as >10; for called variant, at least one sample with variant
frequency >1%, variant frequency of each sample >0.5%, and
P-value <0.01; ii) visual examination of the mutations was
performed using Samtools software (http://samtools.sourceforge.net) and possible errors
specific to one DNA strand were filtered out. Software ANNOVAR
(v2015-06-17) and snpEff (v4.2) was used for variant
annotation.
Statistical analysis
For variant frequency <0.5%, 0 was replaced. R
(hclust, v3.2.4) was used for variant frequency clustering analysis
to show the types of samples from cancer patients that are more
similar. Student’s t-test was applied for comparison of cfDNA
concentration and p<0.05 was considered statistically
significant.
Results
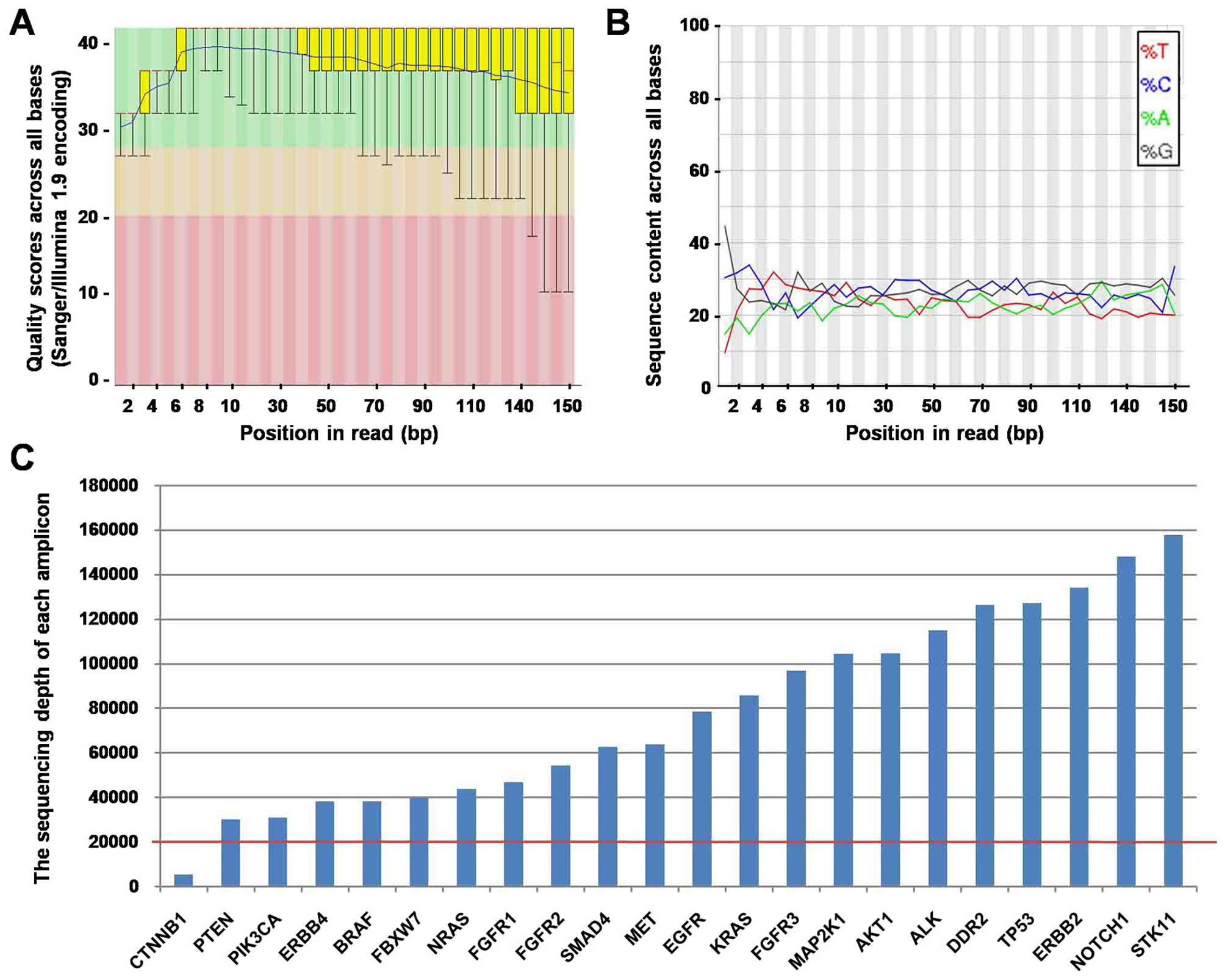
Sequence QC with the Illumina X10
For the 10 NSCLC patients, tumor DNA, matched blood
DNA, and plasma cfDNA were all subjected to sequencing. Of all 30
sequenced samples, sequence lengths were all set to 150 bp. The
quality scores were ~40, indicated that the accuracy is very good
and error rate was ~0.01% (Fig.
1A). The GC content was ~50% besides the first 1–15 bp, which
was removed before further analysis (Fig. 1B). The sequencing depth in all
samples ranged between 10,000× and 750,000x, and most amplications
were >20,000× (Fig. 1C).
Concordance of tumor DNA and matched
plasma cfDNA sample
All detected mutations are listed in Table I. Considering possible systematic
error, we defined mutations with >0.5% percentages as positive
mutations. Comparing each plasma cfDNA with its matched tumor DNA,
concordant mutations were identified in all 10 patients.
 | Table IMutations detected in tumor DNA and
plasma cfDNA of NSCLC patients. |
Table I
Mutations detected in tumor DNA and
plasma cfDNA of NSCLC patients.
| Patient ID | Position | Gene | Mutation | Mutation type | Blood mutation rate
(reads) | cfDNA mutation rate
(reads) | Tumor mutation rate
(reads) |
|---|
| NSCLC1 | chr10:89624218 | PTEN | p.L171V | SUB | 9.01 (11538) | 0.07 (108648) | 0.02 (21955) |
| NSCLC1 | chr8:38285913 | FGFR1 | p.D133D | DEL | 5.60 (1340) | 2.22 (13695) | 2.59 (9729) |
| NSCLC1 | chr9:139399408 | NOTCH1 | p.L1579L | DEL | 1.81 (22782) | 0.70 (153196) | 1.03 (34967) |
| NSCLC2 | chr10:89720705 | PTEN | p.T459A | SUB | 0.13 (3197) | 1.04 (220526) | 0.14 (2876) |
| NSCLC2 | chr19:1221293 | STK11 | p.Y272Y | SUB | 0.05 (11956) | 2.54 (1434088) | 0.38 (11433) |
| NSCLC2 | chr2:29443617 | ALK | p.A1200A | SUB | 0 (107957) | 0 (1566735) | 49.29 (69422) |
| NSCLC2 | chr7:116339672 | MET | p.S178S | SUB | 0.09 (4232) | 0.58 (504813) | 0.56 (3416) |
| NSCLC2 | chr7:55259515 | EGFR | p.L858R | SUB | 0.01 (43374) | 0.01 (421187) | 24.72 (34533) |
| NSCLC2 | chr8:38285913 | FGFR1 | p.D133D | DEL | 2.60 (12201) | 3.46 (170139) | 3.10 (7457) |
| NSCLC2 | chr9:139399408 | NOTCH1 | p.L1579L | DEL | 0.94 (33155) | 1.11 (806291) | 1.05 (22104) |
| NSCLC3 | chr17:7577538 | TP53 | p.R209L | SUB | 0.01 (65412) | 0.01 (668018) | 3.61 (62283) |
| NSCLC3 | chr17:7579476 | TP53 | p.P32S | SUB | 20.06 (325417) | 16.12 (112725) | 3.75 (266676) |
| NSCLC3 | chr8:38285913 | FGFR1 | p.D133D | DEL | 2.92 (20513) | 2.22 (14258) | 2.96 (20809) |
| NSCLC3 | chr9:139399408 | NOTCH1 | p.L1579L | DEL | 0.94 (46337) | 0.67 (162970) | 1.05 (46751) |
| NSCLC4 | chr10:89624218 | PTEN | p.L171V | SUB | 12.84 (14043) | 51.14 (3154) | 53.31 (12299) |
| NSCLC4 | chr19:1223125 | STK11 | p.F354L | SUB | 3.02 (48133) | 56.19 (46898) | 34.46 (40610) |
| NSCLC4 | chr3:178938796 | PIK3CA | p.V680L | SUB | 0.44 (4980) | 2.13 (47) | 0.24 (13656) |
| NSCLC4 | chr8:38285913 | FGFR1 | p.D133D | DEL | 4.97 (3017) | 3.14 (8777) | 2.72 (7789) |
| NSCLC4 | chr9:139399408 | NOTCH1 | p.L1579L | DEL | 1.68 (33691) | 0.67 (150) | 1.05 (17506) |
| NSCLC5 | chr17:7577565 | TP53 | p.N200S | SUB | 0.05 (27965) | 0.10 (386509) | 15.33 (46046) |
| NSCLC5 | chr2:29443617 | ALK | p.A1200A | SUB | 47.93 (63513) | 51.31 (335544) | 42.54 (104652) |
| NSCLC5 | chr3:178936091 | PIK3CA | p.E545K | SUB | 0 (0) | 0.73 (33218) | 21.64 (5084) |
| NSCLC5 | chr8:38285913 | FGFR1 | p.D133D | DEL | 3.04 (7144) | 2.34 (36438) | 3.35 (15840) |
| NSCLC6 |
chr10:123279651 | FGFR2 | p.G172R | SUB | 0.03 (22113) | 0.02 (146276) | 2.02 (30992) |
| NSCLC6 | chr12:25398285 | KRAS | p.G12C | SUB | 0.01 (34200) | 0.01 (144676) | 5.74 (37623) |
| NSCLC6 | chr17:7578406 | TP53 | p.R136H | SUB | 0.07 (27351) | 0.05 (164157) | 2.64 (37926) |
| NSCLC6 | chr8:38285913 | FGFR1 | p.D133D | DEL | 2.90 (10253) | 2.39 (30748) | 3.01 (16766) |
| NSCLC7 | chr8:38285913 | FGFR1 | p.D133D | DEL | 2.44 (6968) | 2.63 (53550) | 3.03 (7382) |
| NSCLC7 | chr9:139399408 | NOTCH1 | p.L1579L | DEL | 0.82 (33227) | 0.82 (149833) | 1.01 (28019) |
| NSCLC8 | chr17:7578272 | TP53 | p.H154Y | SUB | 0.04 (26414) | 0.24 (330592) | 28.81 (17584) |
| NSCLC8 | chr7:116339672 | MET | p.S178S | SUB | 49.17 (7269) | 50.86 (181222) | 43.71 (4985) |
| NSCLC8 | chr7:116340262 | MET | p.N375S | SUB | 50.06 (21006) | 50.47 (213064) | 44.18 (13086) |
| NSCLC8 | chr8:38285913 | FGFR1 | p.D133D | DEL | 2.74 (21644) | 3.26 (188537) | 2.76 (13826) |
| NSCLC8 | chr9:139399408 | NOTCH1 | p.L1579L | DEL | 0.96 (53785) | 1.10 (543910) | 0.96 (42236) |
| NSCLC9 | chr10:89624218 | PTEN | p.L171V | SUB | 9.05 (4155) | 0.01 (297185) | 0.06 (16938) |
| NSCLC9 | chr3:178938796 | PIK3CA | p.V680L | SUB | 1.14 (9864) | 0.23 (304805) | 0.21 (21326) |
| NSCLC9 | chr8:38285913 | FGFR1 | p.D133D | DEL | 5.11 (2054) | 3.32 (229921) | 2.79 (12710) |
| NSCLC9 | chr9:139399408 | NOTCH1 | p.L1579L | DEL | 1.65 (23599) | 1.14 (439058) | 0.84 (31467) |
| NSCLC10 | chr17:7578534 | TP53 | p.K93N | SUB | 0.05 (13254) | 0.04 (279526) | 14.22 (8318) |
| NSCLC10 | chr7:55259515 | EGFR | p.L858R | SUB | 0.02 (46073) | 0.17 (368684) | 18.84 (36517) |
| NSCLC10 | chr8:38285913 | FGFR1 | p.D133D | DEL | 3.02 (14564) | 3.37 (146536) | 2.59 (9555) |
| NSCLC10 | chr9:139399408 | NOTCH1 | p.L1579L | DEL | 1.10 (34408) | 1.09 (532945) | 0.77 (25075) |
For all concordant mutations identified in the 10
patients, mutation percentages were higher in plasma cfDNA (average
12.04%) than in tumor DNA (average 10.80%) (Table I). Mutations of 26 alleles located
in 7 genes (PTEN, STK11, FGFR1, TP53, NOTCH1, ALK and MET) were
identified in both tumor DNA and plasma cfDNA. Mutations in EGFR
(24.72% and 18.84%), KRAS (5.74%), and FGFR2 (2.02%) were only
identified in tumor DNA while no mutated genes found in plasma
cfDNA but not in tumor DNA, indicated that plasma cfDNA could
partially reflect the genetic condition of NSCLC tumors.
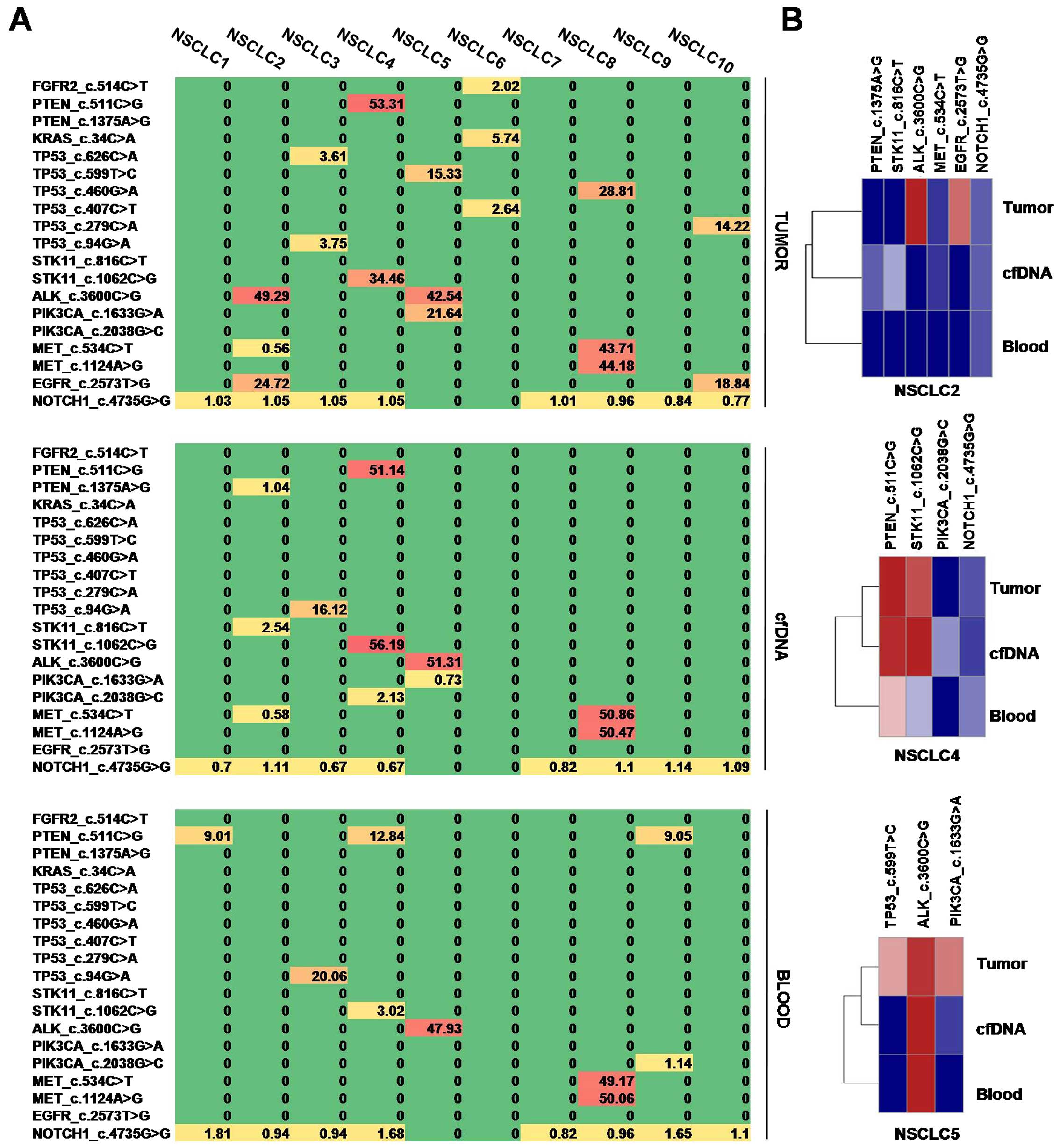
Differences in mutation pattern among
tumor tissue, cfDNA and blood of NSCLC
To reveal the association between mutation pattern
of tumor tissue and cfDNA, unclustered heatmap diagrams were
displayed for mutation patterns of tumor tissue, cfDNA and blood
(Fig. 2A). It is clearly showed
that cfDNA have a moderate mutation pattern, which is weaker than
that of tumor tissue and stronger than that of blood. Mutation
patterns of different tissues from the same patients were clustered
by hierarchical clustering algorithm (Fig. 2B). For each representative patient,
plasma cfDNA was closely clustered with tumor DNA not blood DNA,
indicated that plasma cfDNA was associated with tumor
genetically.
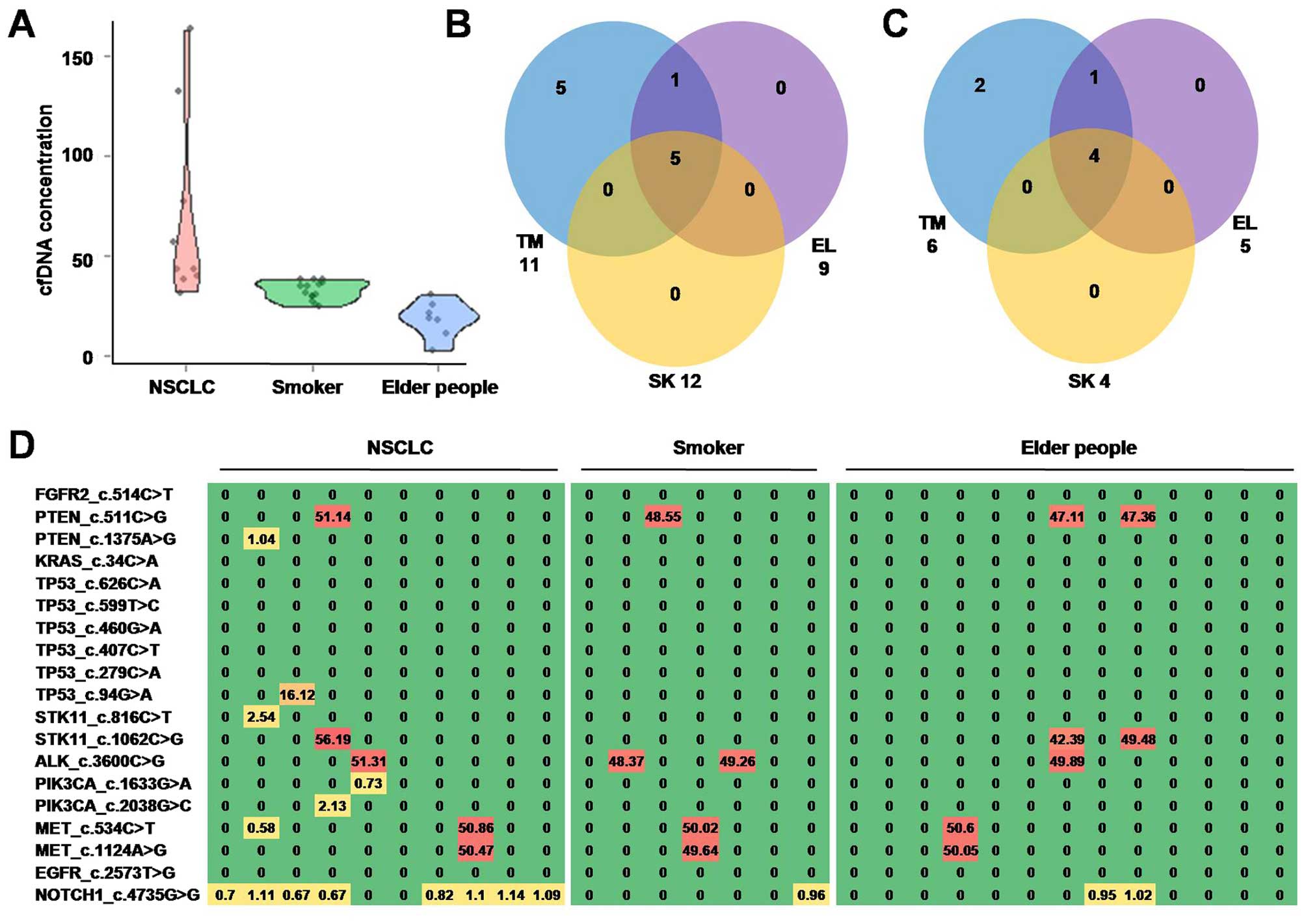
Mutations in plasma cfDNA sample of
healthy people
It is widely known that mutations in NSCLC patients
could be caused by senescence and smoking. To further investigate
the source and specificity of mutations in NSCLC plasma cfDNA, we
recruited 13 elderly people and 7 middle-age smokers and subjected
their plasma cfDNA samples to sequencing.
The plasma cfDNA concentration of NSCLC patients
(69.2±46.9 ng/ml) is significantly higher than that of elderly
people (32.5±5.2 ng/ml, t=2.96, p=0.007) and middle-aged smokers
(17.9±9.1 ng/ml, t=2.83, p=0.013). Violin plot of plasma cfDNA
concentration is shown in Fig.
3A.
Nearly half of mutations (PTEN_c.511C>G,
STK11_c.1062C>G, ALK_c.3600C>G, MET_c.534C>T,
MET_c.1124A>G, NOTCH1_c.4735G>G) were identified in both
NSCLC patients and healthy controls (elderly people and smokers).
However, five mutations (PTEN_c.1375A>G, TP53_c.94G>A,
STK11_c.816C>T, PIK3CA_c.1633G>A, PIK3CA_c.2038G>C) only
identified in NSCLC patients, indicated that cfDNA mutation pattern
was largely different between NSCLC patients and non-cancerous
people.
Discussion
The 5-year survival rate following surgical
resection for NSCLC at early stage is significantly higher than
that at late stage (2,19). Unfortunately, NSCLC is always
diagnosed at advanced stages because the symptoms are not apparent
initially and the detection is difficult at stage I or II (20,21).
Thus, exploring an effective approach to detect early-stage
patients can observably increase survival. Even though biopsy has
been the golden diagnostic method, in many cases of NSCLC, it is
always difficult to obtain tissue samples in early-stage or even in
advanced or metastatic stage (18). Additionally, the procedure of
biopsy may increase the risk of cancer ‘seeding’ to other sites
(22). In recent years, ‘liquid
biopsy’ receives more and more attention but most studies involved
in mutations of NSCLC are about late stages. Therefore the
detection of liquid biopsy biomarkers in early-stage NSCLC patients
is necessary and can provide a non-invasive way to gain genotypic
information and will have broad application prospects in the
large-scale in the future (18).
Several hypotheses have been proposed to explain the
mechanism of cellular DNA release into the circulation. Wu et
al proposed that tumor DNA was released into the circulation
and was enriched in the plasma and serum and the increased cfDNA in
plasma of cancer patients may be due to the derivation from tumor
DNA (23). Lee and colleagues
hypothesized that the cfDNA was composed of small fragments of
nucleic acid that are not associated with cells or cell fragments
and should be from a leakage after tumor necrosis or apoptosis
(24).
Jamal-Hanjani et al found there were
ubiquitous and heterogeneous single-nucleotide variants (SNVs)
between tumor DNA and cfDNA in lung cancer, incidence of which are
68% and 32%, respectively (25).
Couraud et al reported that not all tumor mutations could be
detected in cfDNA (50 mutations were identified in tumor DNA, while
only 26 were detected in cfDNA) (26). Further detection of mutations in
tumor DNA and cfDNA was required to define its potential use in
clinical practice. In this study, we recruited 10 NSCLC patients of
early stage (includes IA, IB, IIA and IIB); collected matched DNA
samples and conducted further research. We found that tumor DNA and
its matched plasma cfDNA samples showed high concordance in their
mutation patterns.
Notably, some specific mutations were identified in
tumor cfDNA, locating in genes of EGFR, KRAS and FGFR2, which is
not common in lung cancer tumor samples. These results are also
coherent with precious studies (17,27).
However, the incidence of mutations in plasma cfDNA was roughly
higher than that in tumor DNA in some previous studies (18,28).
In this study, plasma cfDNA had a moderate mutation pattern, which
was weaker than that of tumor tissue and stronger than that of
PBLs. Generally, plasma cfDNA was closely clustered with tumor DNA
not blood DNA, indicating that plasma cfDNA was associated with
tumor genetically.
Many studies proposed that cfDNA concentration can
reflect the tumor burden in the cases of cancer (26). In cancer patients, a relatively
high concentration of cfDNA (range 0–1,000 ng/ml, average 180
ng/ml) were reported (29)
compared to that of health people (range 0–100 ng/ml, average 30
ng/ml) (30), which may be
predominantly originated from apoptotic cells, inflammation, tissue
trauma and so on (31). Some
studies even revealed that cfDNA concentration was different
between early and late stages (26).
Association between mutations of cfDNA with those of
tumor tissue DNA was also observed in prostate cancer and breast
cancer (32,33). cfDNA is not only associated with
carcinogenesis. Its concentration may also be elevated in
inflammatory, infectious and other health-related conditions such
as senescence and smoking status (34). Thus further studies need to
establish a standard to distinguish potential lung cancer patients
with healthy subjects.
Our results suggested that the concentrations of
cfDNA in NSCLC patients showed significant differences when
compared with that of elderly people (t=2.96, p=0.007) and that of
middle-aged smokers (t=2.83, p=0.013) although the median
concentration of cfDNA can vary with senescence and smoking among
healthy subjects (35). Therefore
our data may suggest that the quantification of cfDNA from plasma
may be a useful non-invasive technique for diagnosis and dynamic
monitoring of lung cancer at early stage.
In some circumstances cfDNA alterations are
detectable ahead of cancer diagnosis, raising the possibility of
exploiting them as biomarkers for monitoring cancer occurrence
(36). Although detection of
mutations in cfDNA is difficult due to the low amount of mutant
alleles in a background of wild-type DNA (37), there are still five mutations
(PTEN_c.1375A>G, TP53_c.94G>A, STK11_c.816C>T,
PIK3CA_c.1633G>A, PIK3CA_c.2038G>C) identified in NSCLC
patients specifically, suggesting that cfDNA mutation pattern could
distinguish early NSCLC patients with non-cancerous individuals.
Stankovic et al reported that coexistence of aberrant p53
and PTEN was the most frequent marker and significantly associated
with poor survival of NSCLC patients (38). Stjernström et al also found
when analyzing all PI3K pathway related genes together, NSCLC
patients would have at least one alteration (39). STK11 mutation was first found in
Peutz-Jeghers syndrome patients, which is significantly associated
with KRAS and EGFR (40). Mutation
in STK11 was also found associated with prognosis (41).
In conclusion, we find that cfDNA has a similar
mutation pattern with its matched tumor tissue DNA. Specific
mutations or elevated concentration of plasma cfDNA could be useful
non-invasive biomarkers through liquid biopsy in prevention and
diagnosis of NSCLC. However, future investigation in utility and
perspectives of cfDNA still need to be performed in large
prospective cohorts.
Acknowledgements
This study was supported by grants from the National
Natural Scientific Foundation of China (81272682) and the Financial
department of Hebei Province (no. [2014]1257).
References
|
1
|
Catarino R, Coelho A and Medeiros R:
Circulating DNA and NSCLC: Old findings with new perspectives. J
Thorac Dis. 4:442–443. 2012.PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Islami F, Torre LA and Jemal A: Global
trends of lung cancer mortality and smoking prevalence. Transl Lung
Cancer Res. 4:327–338. 2015.PubMed/NCBI
|
|
4
|
Torre LA, Siegel RL, Ward EM and Jemal A:
International variation in lung cancer mortality rates and trends
among women. Cancer Epidemiol Biomarkers Prev. 23:1025–1036. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reade CA and Ganti AK: EGFR targeted
therapy in non-small cell lung cancer: Potential role of cetuximab.
Biologics. 3:215–224. 2009.PubMed/NCBI
|
|
7
|
Agarwal M, Brahmanday G, Chmielewski GW,
Welsh RJ and Ravikrishnan KP: Age, tumor size, type of surgery, and
gender predict survival in early stage (stage I and II) non-small
cell lung cancer after surgical resection. Lung Cancer. 68:398–402.
2010. View Article : Google Scholar
|
|
8
|
Aberle DR, Adams AM, Berg CD, Black WC,
Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM and Sicks
JD; National Lung Screening Trial Research Team: Reduced
lung-cancer mortality with low-dose computed tomographic screening.
N Engl J Med. 365:395–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Church TR, Black WC, Aberle DR, Berg CD,
Clingan KL, Duan F, Fagerstrom RM, Gareen IF, Gierada DS, Jones GC,
et al; National Lung Screening Trial Research Team. Results of
initial low-dose computed tomographic screening for lung cancer. N
Engl J Med. 368:1980–1991. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lai Y, Shen C, Wang X, Du H, Chen D, Tian
L, Zhou X and Che G: Status and perspectives of detection by
low-dose computed tomography or computed radiography in surgical
patients with lung cancer, based on a five-year study. Thorac
Cancer. 7:111–117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao SJ and Wu N: Early detection of lung
cancer: Low-dose computed tomography screening in China. Thorac
Cancer. 6:385–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Silva M, Galeone C, Sverzellati N,
Marchianò A, Calareso G, Sestini S, La Vecchia C, Sozzi G, Pelosi G
and Pastorino U: Screening with low-dose computed tomography does
not improve survival of small cell lung cancer. J Thorac.
11:187–193. 2016. View Article : Google Scholar
|
|
13
|
Tissot C, Toffart AC, Villar S, Souquet
PJ, Merle P, Moro-Sibilot D, Pérol M, Zavadil J, Brambilla C,
Olivier M, et al: Circulating free DNA concentration is an
independent prognostic biomarker in lung cancer. Eur Respir J.
46:1773–1780. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moyer VA; U.S. Preventive Services Task
Force. Screening for lung cancer: U.S. Preventive Services Task
Force recommendation statement. Ann Intern Med. 160:330–338.
2014.PubMed/NCBI
|
|
15
|
Humphrey L, Deffebach M, Pappas M, et al:
U.S. Preventive Services Task Force Evidence Syntheses, formerly
Systematic Evidence Reviews. Screening for Lung Cancer: Systematic
Review to Update the US Preventive Services Task Force
Recommendation. Agency for Healthcare Research and Quality (US);
Rockville, MD: 2013
|
|
16
|
Sun W, Yuan X, Tian Y, Wu H, Xu H, Hu G
and Wu K: Non-invasive approaches to monitor EGFR-TKI treatment in
non-small-cell lung cancer. J Hematol Oncol. 8:952015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nygaard AD, Garm Spindler KL, Pallisgaard
N, Andersen RF and Jakobsen A: The prognostic value of KRAS mutated
plasma DNA in advanced non-small cell lung cancer. Lung Cancer.
79:312–317. 2013. View Article : Google Scholar
|
|
18
|
Guo K, Zhang Z, Han L, Han J, Wang J, Zhou
Y, Liu H, Tong L, Li X and Yan X: Detection of epidermal growth
factor receptor mutation in plasma as a biomarker in Chinese
patients with early-stage non-small cell lung cancer. Onco Targets
Ther. 8:3289–3296. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rossi A, Maione P, Colantuoni G, Gaizo FD,
Guerriero C, Nicolella D, Ferrara C and Gridelli C: Screening for
lung cancer: New horizons? Crit Rev Oncol Hematol. 56:311–320.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brambilla C, Fievet F, Jeanmart M, de
Fraipont F, Lantuejoul S, Frappat V, Ferretti G, Brichon PY and
Moro-Sibilot D: Early detection of lung cancer: Role of biomarkers.
Eur Respir J. (Supplement 39): S36–S44. 2003. View Article : Google Scholar
|
|
22
|
Murtaza M, Dawson SJ, Pogrebniak K, Rueda
OM, Provenzano E, Grant J, Chin SF, Tsui DW, Marass F, Gale D, et
al: Multifocal clonal evolution characterized using circulating
tumour DNA in a case of metastatic breast cancer. Nat Commun.
6:87602015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu TL, Zhang D, Chia JH, Tsao K, Sun CF
and Wu JT: Cell-free DNA: Measurement in various carcinomas and
establishment of normal reference range. Clin Chim Acta. 321:77–87.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee YJ, Yoon K-A, Han J-Y, Kim HT, Yun T,
Lee GK, Kim HY and Lee JS: Circulating cell-free DNA in plasma of
never smokers with advanced lung adenocarcinoma receiving gefitinib
or standard chemotherapy as first-line therapy. Clin Cancer Res.
17:5179–5187. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jamal-Hanjani M, Wilson GA, Horswell S,
Mitter R, Sakarya O, Constantin T, Salari R, Kirkizlar E,
Sigurjonsson S, Pelham R, et al: Detection of ubiquitous and
heterogeneous mutations in cell-free DNA from patients with
early-stage non-small-cell lung cancer. Ann Oncol. 27:862–867.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Couraud S, Vaca-Paniagua F, Villar S,
Oliver J, Schuster T, Blanché H, Girard N, Trédaniel J,
Guilleminault L, Gervais R, et al: Noninvasive diagnosis of
actionable mutations by deep sequencing of circulating free DNA in
lung cancer from never-smokers: A proof-of-concept study from
BioCAST/IFCT-1002. Clin Cancer Res. 20:4613–4624. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Duan H, Lu J, Lu T, Gao J, Zhang J, Xu Y,
Wang M, Wu H, Liang Z and Liu T: Comparison of EGFR mutation status
between plasma and tumor tissue in non-small cell lung cancer using
the Scorpion ARMS method and the possible prognostic significance
of plasma EGFR mutation status. Int J Clin Exp Pathol.
8:13136–13145. 2015.
|
|
28
|
Jung K, Fleischhacker M and Rabien A:
Cell-free DNA in the blood as a solid tumor biomarker - a critical
appraisal of the literature. Clin Chim Acta. 411:1611–1624. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shapiro B, Chakrabarty M, Cohn EM and Leon
SA: Determination of circulating DNA levels in patients with benign
or malignant gastrointestinal disease. Cancer. 51:2116–2120. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Anker P and Stroun M: Circulating DNA in
plasma or serum. Medicina (B Aires). 60:699–702. 2000.
|
|
31
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Feng J, Gang F, Li X, Jin T, Houbao H, Yu
C and Guorong L: Plasma cell-free DNA and its DNA integrity as
biomarker to distinguish prostate cancer from benign prostatic
hyperplasia in patients with increased serum prostate-specific
antigen. Int Urol Nephrol. 45:1023–1028. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang ZH, Li LH and Hua D: Quantitative
analysis of plasma circulating DNA at diagnosis and during
follow-up of breast cancer patients. Cancer Lett. 243:64–70. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ulivi P and Silvestrini R: Role of
quantitative and qualitative characteristics of free circulating
DNA in the management of patients with non-small cell lung cancer.
Cell Oncol (Dordr). 36:439–448. 2013. View Article : Google Scholar
|
|
35
|
Huang R, Wei Y, Hung RJ, Liu G, Su L,
Zhang R, Zong X, Zhang ZF, Morgenstern H, Brüske I, et al:
Associated links among smoking, chronic obstructive pulmonary
disease, and small cell lung cancer: A pooled analysis in the
International Lung Cancer Consortium. EBioMedicine. 2:1677–1685.
2015. View Article : Google Scholar
|
|
36
|
Gormally E, Caboux E, Vineis P and Hainaut
P: Circulating free DNA in plasma or serum as biomarker of
carcinogenesis: Practical aspects and biological significance.
Mutat Res. 635:105–117. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sherwood JL, Corcoran C, Brown H, Sharpe
AD, Musilova M and Kohlmann A: Optimised pre-analytical methods
improve KRAS mutation detection in circulating tumour DNA (ctDNA)
from patients with non-small cell lung cancer (NSCLC). PLoS One.
11:e01501972016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stankovic T, Milinkovic V, Bankovic J,
Dinic J and Tanic N, Dramicanin T and Tanic N: Comparative analyses
of individual and multiple alterations of p53, PTEN and p16 in
non-small cell lung carcinoma, glioma and breast carcinoma samples.
Biomed Pharmacother. 68:521–526. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Stjernström A, Karlsson C, Fernandez OJ,
Söderkvist P, Karlsson MG and Thunell LK: Alterations of INPP4B,
PIK3CA and pAkt of the PI3K pathway are associated with squamous
cell carcinoma of the lung. Cancer Med. 3:337–348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ylikorkala A, Avizienyte E, Tomlinson IP,
Tiainen M, Roth S, Loukola A, Hemminki A, Johansson M, Sistonen P,
Markie D, et al: Mutations and impaired function of LKB1 in
familial and non-familial Peutz-Jeghers syndrome and a sporadic
testicular cancer. Hum Mol Genet. 8:45–51. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pécuchet N, Laurent-Puig P, Mansuet-Lupo
A, Legras A, Alifano M, Pallier K, Didelot A, Gibault L, Danel C,
Just PA, et al: Different prognostic impact of STK11 mutations in
non-squamous non-small-cell lung cancer. Oncotarget. Nov
25–2015.(Epub ahead of print). PubMed/NCBI
|