Introduction
Ovarian cancer represents the fifth most common
malignancy in women and it is the leading cause of cancer deaths
from gynecological cancer in the western societies (1). The poor survival from epithelial
ovarian cancer that is still reported nowadays is largely
attributed to the high percentage of patients that are diagnosed at
an advanced stage and that often develop resistance to combined
chemotherapy approaches (2). In
light of this imperfect state of clinical management, a better
understanding of the molecular bases of ovarian cancer, coupled
with improved screening tools, is urgently required. In this
context, an increasing number of independent studies have recently
suggested that microRNAs (miRNAs) might play a crucial role in
ovarian cancer pathogenesis (3–5).
Indeed, several miRNA profiling reports have
identified a consensus of aberrantly expressed miRNAs in both
ovarian cancer-derived tumor samples and cell lines (3), suggesting that these small RNAs might
be functionally involved in ovarian cancer development and
progression. Moreover, both gain-of-function and loss-of-function
assays have further validated the role played by several miRNAs in
ovarian cancer pathogenesis (4).
MicroRNAs represent a large family of small
endogenous non-coding RNAs endowed with the ability to regulate
gene expression post-transcriptionally (6). The widely recognized role assigned to
miRNAs in cancer biology is a natural consequence of their
involvement in several biological processes that are closely linked
to cancer development and progression, such as cell proliferation,
differentiation and apoptosis (7).
Indeed, the list of genes that have been reported to be regulated
by miRNAs includes several examples of established tumor suppressor
genes and oncogenes, such as BRCA1, BRCA2,
PTEN, KIT, RAS, E2F3, MET and
MYCN, to name a few (8).
Therefore, substantial experimental evidence has been gathered in
support of the hypothesis that derangement of a complex
cross-regulatory network between miRNAs and transcripts from
cancer-related genes might represent a critical event in the
development and progression of several cancer types. However, due
to the widely acknowledged complexity and heterogeneity of each
cancer type, the current knowledge of the miRNA regulatory networks
involved in cancer development is still far from providing an
exhaustive picture.
The RNASET2 gene encodes a highly conserved
extracellular ribonuclease belonging to the T2/Rh/S family of
endoribonucleolytic enzymes and it has long been ranked as an
ovarian cancer-related tumor suppressor gene (9). Moreover, several independent studies
have consistently reported the oncosuppressive role of
RNASET2 (both in vitro and in vivo) in a
broader range of cancer types, such as malignant melanoma and
colorectal cancer (10,11). In the context of ovarian cancer,
our group has recently described a marked non-cell autonomous
oncosuppressive function for RNASET2. Indeed, in vivo
xenograft studies in immunocompromised mice showed that
RNASET2-mediated tumor suppression was associated with a
significant recruitment of cancer-suppressive macrophages within
the tumor mass in two independent human ovarian cancer-derived cell
lines (12,13). Strikingly, the catalytic activity
of this enzyme was found to be dispensable for its non-cell
autonomous oncosuppressive role, suggesting that RNASET2 might
represent a multitasking or ‘moonlightning’ protein. In agreement
with this speculation, we reported more recently a clear
pleiotropic mode of action for the RNASET2 gene, whereby
gene knock-down approaches coupled with a panel of in vitro
assays unveiled the ability of RNASET2 to affect several
cancer related-parameters in a cell-autonomous manner (14). Of note, these data were reported
for the same experimental model (the human OVCAR3 ovarian cancer
cell line) where previous in vivo studies had shown the
occurrence of a non-cell autonomous oncosuppresive role for this
gene (13). However, the genes and
signaling networks by which RNASET2 carries out such
cell-autonomous oncosuppressive role are largely unknown.
In an attempt to address this issue, we report here
gene profiling data on RNASET2-silenced OVCAR3 cells and
show that RNASET2 expression levels affects the cell
transcriptome at both mRNAs and miRNAs levels. Moreover, our data
suggest that the observed changes in the expression levels of some
mRNAs represent a likely consequence of RNASET2-mediated
regulation of a handful of miRNAs that can directly target these
transcripts.
Materials and methods
Cell lines and clones
Human ovarian cancer-derived OVCAR3 cell clones,
stably transfected with empty pSicoR vector, control scrambled
shRNAs and two RNASET2-targeting shRNAs were previously described
(13). OVCAR3, Hey3Met2 and HeLa
cells were cultured in DMEM-F12 medium (Sigma-Aldrich, St. Louis,
MO, USA) with 10% fetal bovine serum (FBS; Sigma-Aldrich) and 1%
glutamine (Sigma-Aldrich).
RNA extraction
Total RNA was purified from OVCAR3 cells using the
RNeasy Plus kit (Qiagen, Venlo, Limburg, The Netherlands). RNA was
quantified using an ND-1000 UV-Vis spectrophotometer (Thermo Fisher
Scientific, Wilmington, DE, USA), and the integrity of the RNA was
assessed with the Agilent 2100 Bioanalyzer (Agilent Technologies,
Inc., Santa Clara, CA, USA) according to the manufacturer’s
instructions. All of the RNA samples used in this study exhibited a
260/280 ratio above 1.9 and an RNA integrity number (RIN) above
9.0.
Microarray expression profiling
Microarray hybridization assays included two
biological replicates per treatment. All sample-labelling,
hybridization, washing, and scanning steps were carried out
according to the manufacturer’s specifications. Briefly,
Cy3-labelled cRNA was generated from 50 ng input total RNA using
the One Color Quick Amp Labelling kit (Agilent Technologies). For
every sample, 600 ng cRNA from each labelling reaction (with a
specific activity above 9.0) was hybridized using the Gene
Expression Hybridization kit (Agilent Technologies) to the Agilent
Whole Human Genome Oligo Microarray (Agilent Technologies). After
hybridization, the slides were washed and then scanned with the
Agilent G2565BA microarray scanner (Agilent Technologies). The
fluorescence intensities of the scanned images were extracted and
pre-processed using Agilent Feature Extraction software
(10.7.3.1).
Gene expression data analysis
Quality control and array normalization were
performed in the R statistical environment using the
Agi4×44PreProcess (v.1.18.0) package, which was downloaded from the
Bioconductor website (15). The
normalization and filtering steps were based on those described in
the Agi4×44PreProcess reference manual. Briefly, the
Agi4×44PreProcess options were set to use the Mean signal and the
BG Median signal as foreground and background signals,
respectively. The data were normalized between arrays using the
quantile method. Genes with a fold change >1 log2
were designated as modulated. All of the above computations were
conducted using the R statistical programming environment. The
expression analysis systematic explorer (EASE) biological theme
analysis of the regulated genes was conducted online using DAVID
(16).
MicroRNA expression profiling
RNA was extracted using the mirVana kit (Ambion,
Inc., Carlsbad, CA, USA) according to the manufacturer’s
instructions. Concentration and quality of RNA samples were
determined with NanoDrop (Thermo Fisher Scientific). Total RNA was
reverse transcribed with TaqMan MicroRNA reverse transcription kit
using Megaplex RT Primers (Applied Biosystems, Foster City, CA,
USA). Real-time PCR reactions were carried out on preconfigured
microfluidic cards (TaqMan Array MicroRNA Cards, set A, V2.2 and
set B, V3; Applied Biosystems) allowing the detection of ~754
unique specific assays and 4 candidate endogenous control assays.
Two biological replicates for control and two for RNASET2-silenced
cells were carried out. Experimental data were then analyzed by SDS
2.3 software (Applied Biosystems) and the relative expression
values were calculated using U6 miRNA as endogenous control. miRNAs
with a threshold cycle <33 that showed a log2-fold
change greater than one in RNASET2-silenced samples compared
to control samples were considered as induced. To derive a list of
predicted target genes for miRNAs of interest, the selected miRNAs
were uploaded into the miRecords database (http://miRecords.umn.edu/miRecords), in which the
relationship between target genes and miRNAs are predicted by 11
different algorithms. The genes were identified as target when
predicted by at least 5 algorithms. The predicted targets of the
downregulated miRNAs were then crossed with the genes identified as
upregulated by the microarray analysis.
Real-time PCR analysis
Total RNA extracted from OVCAR3 cells was reverse
transcribed with random primers using the ‘High capacity cDNA
reverse transcription kit’ (Life Technologies). Primers for
real-time qPCR were selected within the same region of the mRNA
used to design the probes for microarray experiments. The sequences
of the primers used for qPCR are as follows: 5′-AACTCCAGCAACTTCTT
CTCCATCC-3′ and 5′-AGAGAGGCCAGCTCAAGAGAA AC-3′ for FOSL1;
5′-TCCCAGGACATGGCGAGGAGTA-3′ and 5′-TCACTGGGAACCCGCAGGAAG-3′ for
NOTCH3; 5′-GTTCACTGTGTTTCTGCCGCTGTC-3′ and 5′-CAGTTC
TACATCCATGCCCAAGAAG-3′ for DDIT4L; 5′-GAGTTC
AGGCCTCTGGGATCAAC-3′ and 5′-CGTTATCCTCCAA CCGGGCACAATA-3′ for
NFIB; 5′-TAGCTGCACAAGAAG CAAGAACCAG-3′ and
5′-GCTGCCACCATGCACGAGG TTT-3′ for PDE1A;
5′-GTAATGCTGCACCTCCCTCTC CT-3′ and 5′-AACAAGCGTCTGGATCTCTGCAGG-3′
for LFNG; 5′-CGCGAGAAGATGACCCAGAT-3′ and 5′-ACAG
CCTGGATAGCAACGTACA-3′ for β-actin.
Real-time RT-PCR reactions were performed on ABI
PRISM 7000 (Applied Biosystems) in triplicate in a 25 μl volume
containing target cDNA (25 ng), 40 nM primers, 12.5 μl of Power
SYBR-Green Master Mix and water. Samples were denatured at 95°C for
15 sec and annealed/extended at 60°C for 1 min, for 40 cycles.
Fluorescent signals generated during PCR amplification were
monitored and analyzed with ABI PRISM 7000 SDS software (Applied
Biosystems). Comparison of the amount of each gene transcript among
different samples was made by using β-actin as a reference. The
amount of target RNA, normalized to the endogenous reference gene,
was calculated by means of the difference in threshold cycle
parameter (ΔΔCq) (17). The fold
change in gene expression was calculated using this method and
considering OVCAR3 parental cell line with a level of expression
equal to one.
Cloning of 3′UTR regions from candidate
target genes in pmirGLO
To test whether candidate target genes were
regulated by the corresponding miRNAs, the 3′UTR regions of these
genes were cloned in pmirGLO reporter vector (Promega). The
selected regions were amplified with RT-PCR with specific primers,
XhoI digested and ligated to the plasmid vector.
Cell transfection and reporter gene
expression assays
pmirGLO recombinant constructs carrying the 3′UTR
regions of candidate genes and the corresponding miRNAs were
co-transfected in target cells with Lipofectamine 2000 reagent
(Invitrogen, Carlsbad, CA, USA) following the manufacturer’s
instructions. A total of 100 ng of vector and 1 pmol of the
corresponding miRNA were transfected in a 100 μl final volume in
96-wells white plates with clear bottom. Twelve hours after the
transfection, the medium was completely replaced and each well was
filled with 75 μl of fresh medium. Firefly and Renilla
luciferases were activated using the Dual-Glo Luciferase assay
system (Promega, Fitchburg, WI, USA) following the manufacturer’s
instructions and a luminometer was used to detect luminescence.
Fluorescence staining and cell size
analysis of OVCAR3 cells
OVCAR3 cells were grown on coverslips for 24 h, then
processed for fluorescent staining of the actin cytoskeleton.
Briefly, cells were fixed in 3% paraformaldehyde, washed
extensively in phosphate-buffered saline (PBS) and permeabilized
with 0.1% Triton X-100 in PBS. After blocking (3% BSA in PBS),
cells were stained with 5 μg/ml Phalloidin-TRITC (Phalloidin
tetramethylrhodamine B isothiocyanate conjugated; Sigma-Aldrich).
Coverslips were mounted on microscope slides and images were taken
using a Leica TCS SP8 X confocal laser scanning microscope. Cell
size analysis was performed using the Analyze Particle tool of
ImageJ software.
Western blot analysis
Cultured OVCAR3 cells were mechanically scraped in
PBS + 5 mM EDTA and resuspended in lysis buffer (0.5% Igepal, 0.5%
Triton X-100 in PBS + 5 mM EDTA) supplemented with a protease
inhibitors and a phosphatase inhibitor cocktail. For SDS-PAGE
analysis, 100 μg of intracellular lysates were loaded for each gel
lane. Immunoblot analysis was performed using standard procedures,
using selected primary antibodies [rabbit anti-RNASET2 polyclonal
antibody, rabbit anti-S6K1 (phospho T229) polyclonal antibody or
mouse anti-α-tubulin monoclonal antibody] followed by the
appropriate horseradish peroxidase-conjugated anti-mouse or -rabbit
IgG secondary antibody. Membranes were then processed with a
chemiluminescence assay (SuperSignal West Dura; Thermo Fisher
Scientific). The anti-S6K1 antibody was kindly gifted by Dr Nadia
Zaffaroni (IRCCS Foundation, Milan, Italy).
Statistical analysis
Statistical analysis was performed using two-tailed
Student’s t-test, assuming P<0.05 as a threshold value to
discard the null hypothesis.
Results
Since a cell-autonomous oncosuppressive role for
RNASET2 was previously established in a human ovarian
cancer-derived cell line (OVCAR3) in which endogenous expression of
this gene was silenced by RNA interference (13), we decided to exploit this
experimental model to define genes and signaling networks involved
in RNASET2-mediated tumor suppression.
In a previous study, we addressed the intracellular
distribution pattern of the RNASET2 protein in order to define the
putative sites where its function is carried out. In that study, a
partial re-localization of the RNASET2 protein in processing bodies
(P-bodies) was observed in OVCAR3 cells under stress condition
(18). Moreover, the number of
intracellular P-bodies turned out to be affected in
RNASET2-silenced OVCAR3 cells (18), thus, suggesting a putative role for
this protein in P-bodies function or assembly. Since they represent
intracellular sites where RNA turnover takes place (19), the detection of RNASET2 in P-bodies
led us to ask whether this enzyme plays a critical role in these
subcellular structures. Of note, P-bodies represent sites where
short-interfering RNAs (siRNAs) and microRNAs (miRNAs) carry out
mRNA decay and/or translational repression (19). This fact, coupled to the
established role for several miRNAs in the regulation of a wide
range of cancer-related genes, prompted us to investigate the
putative effects of RNASET2 on both miRNAs and mRNA transcriptional
profiles.
To this end, the gene expression profiles of
parental (wt), RNASET2-silenced (shT2) and scrambled
short-hairpin RNA (scrb)-transfected OVCAR3 cell clones were
assessed by means of both quantitative PCR and microarray
hybridization assays. As a first task, we assessed the expression
profile of miRNAs by qPCR on a TaqMan array platform. From this
assay, a few miRNAs turned out to be downregulated in
RNASET2-silenced clones (15 miRNAs), whereas the others were
upregulated (6 miRNAs) (Table
I).
 | Table IDifferentially expressed
microRNAs. |
Table I
Differentially expressed
microRNAs.
| MicroRNA | Fold change |
|---|
| hsa-miR-1227 | −3.3 |
| hsa-miR-200c | −2.2 |
| hsa-miR-577 | −1.9 |
| hsa-miR-183# | −1.8 |
| hsa-miR-23b | −1.7 |
| hsa-miR-141 | −1.7 |
| hsa-miR-766 | −1.5 |
| hsa-miR-193b | −1.5 |
| hsa-miR-628-5p | −1.4 |
| hsa-miR-183 | −1.3 |
| hsa-miR-24 | −1.3 |
| hsa-miR-1260 | −1.2 |
| hsa-miR-454# | −1.2 |
| hsa-miR-301a | −1.1 |
| hsa-miR-342-3p | −1.0 |
| hsa-miR-1233 | 1.1 |
| hsa-miR-21 | 1.1 |
| hsa-miR-935 | 1.5 |
| hsa-miR-20b | 1.9 |
| hsa-miR-449a | 3.6 |
| hsa-miR-206 | 3.9 |
Based on these results, we decided to extend our
transcriptome investigation on RNASET2-silenced and control
OVCAR3 cells by addressing their mRNA expression profile by
microarray hybridization. A preliminary analysis of the mRNA
expression profile from scrb-transfected OVCAR3 cell clones showed
that the expression of a few mRNAs was unexpectedly affected with
respect to the parental OVCAR3 cells, likely due to the occurrence
of unpredicted off-target effects from the scrambled shRNA vector
used. We therefore decided to remove from further analysis those
genes identified as differentially expressed (2-fold regulation)
when (scrb)-transfected OVCAR3 were compared to parental (wt)
OVCAR3 cells. Genes that were upregulated or downregulated in shT2
but not in scrb clones with respect to wt OVCAR3 cells were
considered as interesting targets.
A total of 299 mRNAs were found to show a 2-fold or
greater change in expression level in RNASET2-silenced
OVCAR3 cells when compared to wt cells. Among these genes, 184 were
downregulated, whereas 115 were upregulated (the gene list is
available on request). Significantly, cross-checking of the two
sets of expression profiling data (miRNAs vs. mRNAs) showed that,
for a few miRNAs that turned out to be downregulated in
RNASET2-silenced clones, some of their putative target mRNAs
were found to be upregulated in the subsequent gene expression
profiling assay. In particular, 6 potential target mRNAs for some
of these downregulated miRNAs turned out to be upregulated in
RNASET2-silenced cells: LFNG, FOSL1,
NFIB, DDIT4L, NOTCH3 and PDE1A. The
inversely correlated expression pattern between these mRNAs and one
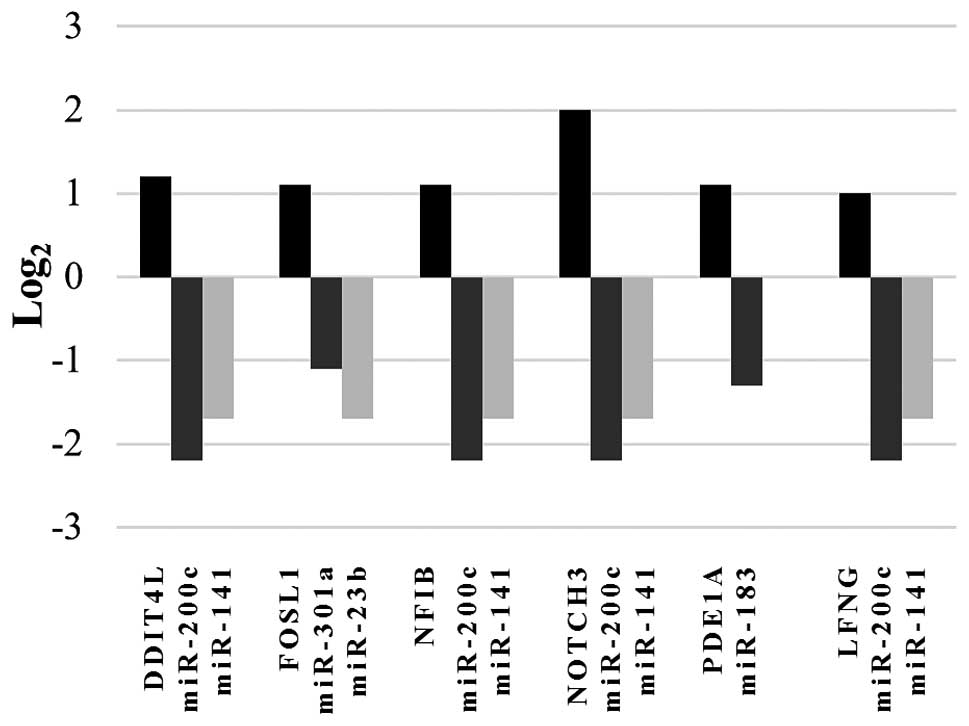
or more of the selected miRNAs is shown in Fig. 1. Thus, these upregulated
transcripts might represent potential targets for regulation by
specific miRNAs whose expression is affected by RNASET2. Of note,
annotation data from all 6 upregulated mRNAs showed a role for the
corresponding genes in cancer-related processes such as cell
growth, proliferation, apoptosis, differentiation and cell
transformation (Table II).
Moreover, a role in microenvironment-mediated control of cancer
growth was reported for 2 of the 6 genes (DDIT4L and
FOSL1) (Table II). To
validate the mRNAs expression changes observed by microarray
hybridization, a real-time RT-PCR assay was carried out with primer
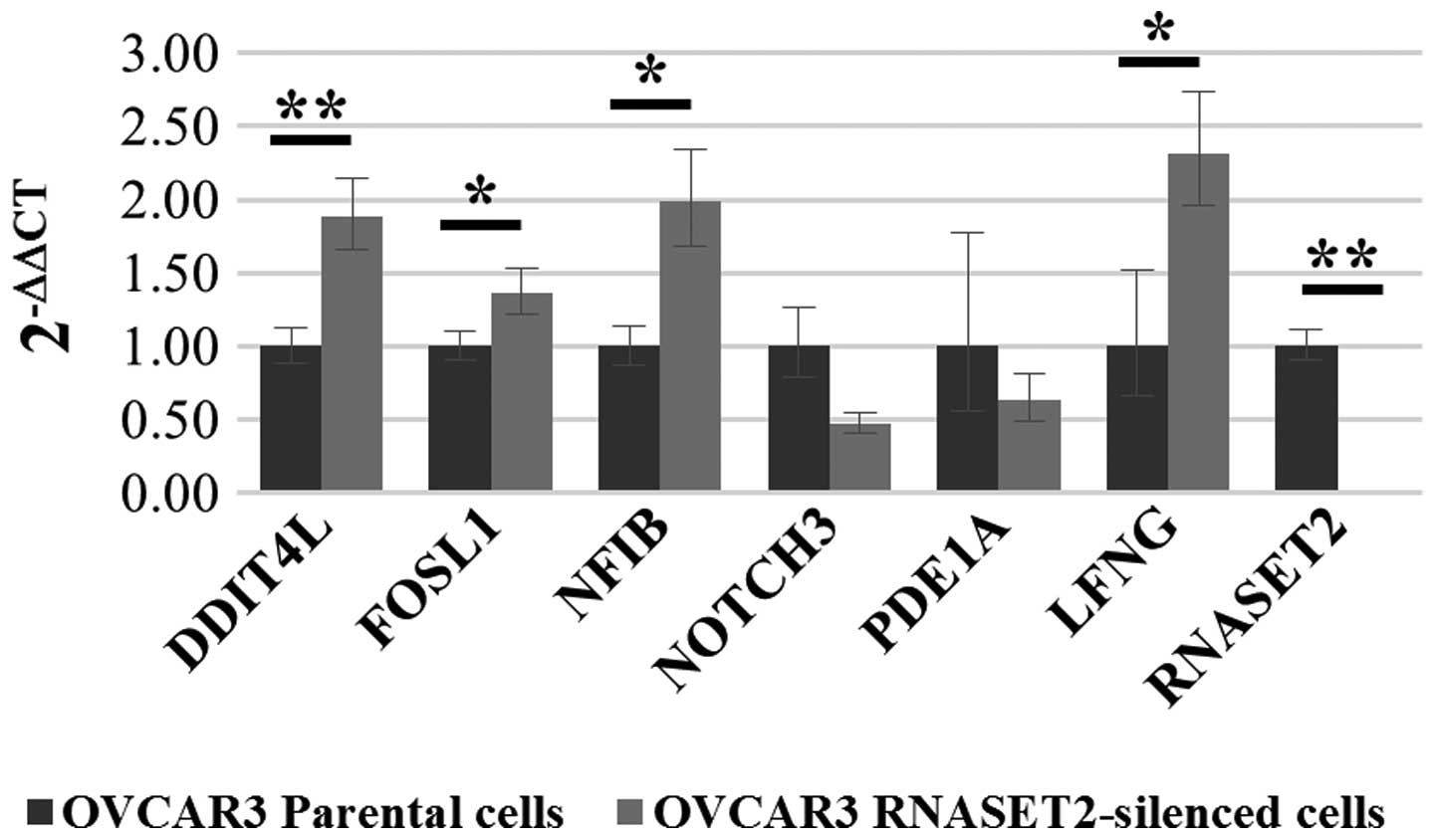
pairs specific for each transcript. As shown in Fig. 2, upregulation in
RNASET2-silenced cells was confirmed only for 4 out of 6
genes (FOSL1, NFIB, LFNG and DDIT4L)
which were therefore selected for further analysis.
 | Table IIList of the genes selected. |
Table II
List of the genes selected.
| Gene ID and
description | Functions | Cancer-related
roles | Location | miRNA |
|---|
| NFIB | | | | |
| (Nuclear factor
I/B) | Binds the
palindromic sequence 5′-TTGGCNNNNNGCCAA-3′ in viral and cellular
promoters and in the origin of replication of adenovirus type
2. | The gene is
rearreanged in several cancer types and overexpressed in
triple-negative breast cancer and small cell lung cancer. Partner
with MYB oncogene in cancer-associated translocations | 9p24.1 | miR-200c,
miR-141 |
| NOTCH3
(Notch3) | Functions as a
receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to
regulate cell-fate determination. Affects the implementation of
differentiation, proliferation and apoptotic programs | Involved in
proliferation of ERBB2-negative breast cancer cells. Overexpressed
in serous ovarian carcinoma and non-small cell lung cancer. | 19p13.2-p13.1 | miR-200c,
miR-141 |
| DDIT4L | | | | |
|
(DNA-damage-inducible transcript
4-like) | Inhibits cell
growth by regulating the TOR signaling pathway. | Disregulated in
breast cancer stromal tissue | 4q23 | miR-200c,
miR-141 |
| LFNG | | | | |
| (LFNG
O-fucosylpeptide 3-beta-N-acetylgluco-saminyltransferase) | Regulator of NOTCH
signalling | Downregulated in
basal-like breast cancer. | 7p22.3 | miR-200c,
miR-141 |
| FOSL1 | | | | |
| (FOS-like antigen
1) | FOS proteins have
been implicated as regulators of cell proliferation,
differentiation, and transformation | Induces pro-tumoral
M2 macrophage polarization pattern | 11q13 | miR-301a,
miR-23b |
| PDE1A | | | | |
| (Phosphodiesterase
1A, calmodulin-dependent) | Cyclic nucleotide
phosphodiesterase with a dual-specificity for the second messengers
cAMP and cGMP, which are key regulators of many important
physiological processes. | Decreased PDE1A
expression induces growth inhibition, cell cycle arrest and
apoptosis in Jurkat cells | 2q32.1 | miR-183 |
We next turned to functional tests to verify whether
the four selected genes are indeed regulated by the corresponding
selected miRNAs. To this end, the whole 3′UTR regions from the
corresponding genes were cloned in pmirGLO reporter vector as
described in Material and methods. For unknown reasons, the 3′UTR
region from the LFNG gene proved to be resistant to cloning
even following several attempts, therefore, we could not include
this gene in subsequent investigations. Due to the same problem,
only the first half from the 3′UTR region of the DDIT4L gene
could be cloned in pmirGLO. Finally, due to its large size (6487
bp), the 3′UTR regions from the NFIB gene was split into two
sub-fragments that were independently cloned into the reporter
vector.
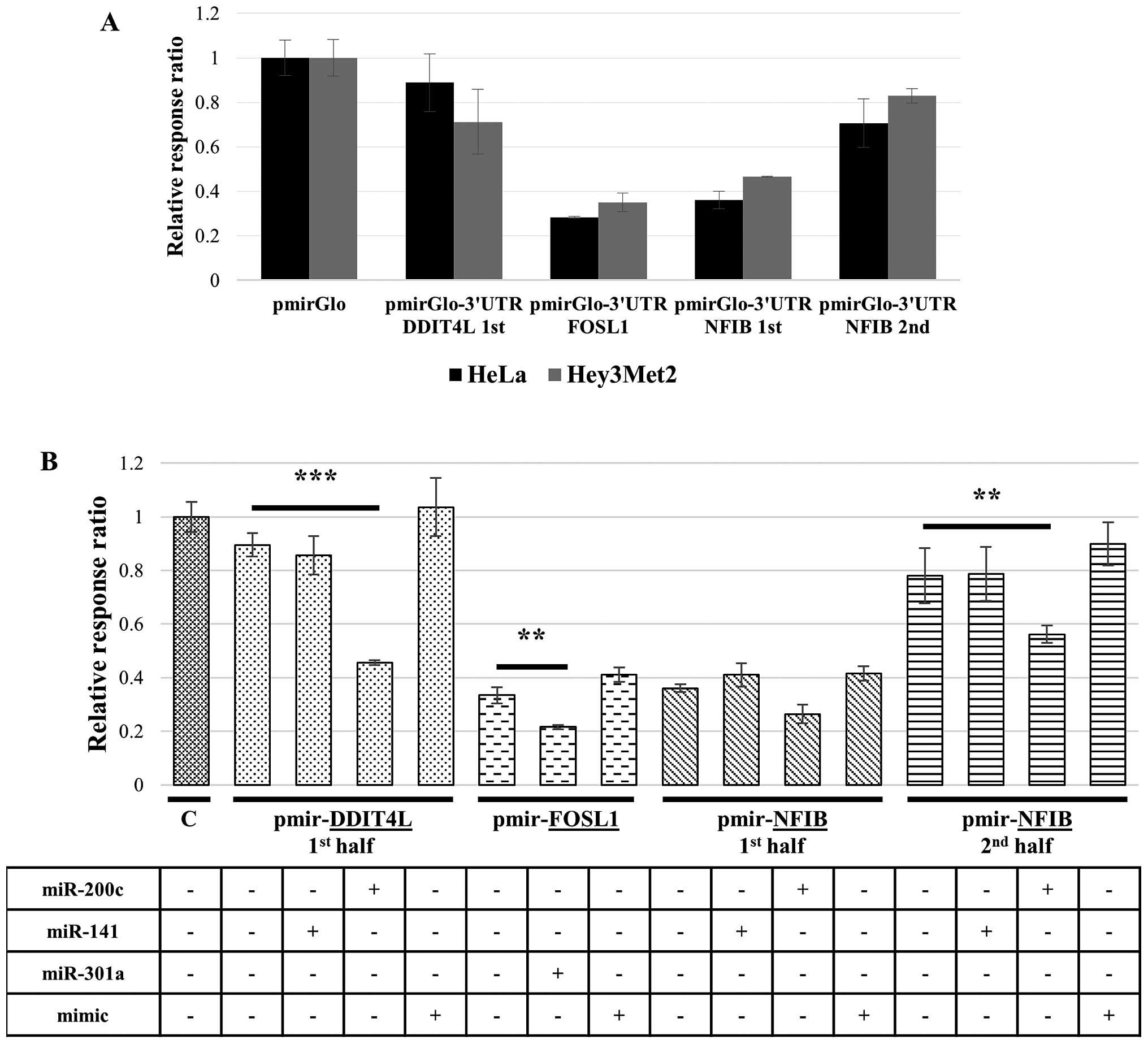
Since parental OVCAR3 cells express high levels of
endogenous RNASET2 protein (13),
the recombinant reporter constructs carrying the different 3′UTR
regions were first transfected in two cell lines (HeLa and
Hey3Met2) known to express low levels of endogenous RNASET2. A
subsequent luciferase gene-based assay showed a high expression
level of the luciferase gene (similar to those observed for the
control pmirGLO empty vector) in both cell lines for reporter
constructs carrying the DDIT4L 3′UTR cloned subregion and
the second half of the NFIB 3′UTR, suggesting that in the
presence of very low RNASET2 expression the selected 3′UTR
regions of the endogenous transcripts from these genes do not
undergo miRNA-mediated regulation in either cell line (Fig. 3A). By contrast, a partial
repression of reporter gene expression was observed following
transfection of the 3′UTR regions from the FOSL1 and the
first half of the NFIB 3′UTR regions only, suggesting that
putative endogenous miRNAs different from those selected in this
study might possibly target some of the relevant 3′UTR regions in
these two cell lines.
To verify that the selected miRNAs were indeed
capable of repressing these putative target genes via binding to
their 3′UTR regions, the same reporter constructs were
co-transfected in HeLa cells together with the corresponding
miRNAs, and luciferase expression assays were again carried out.
Significantly, a clear effect was found for miR-200c on
reporter gene expression driven by recombinant constructs carrying
DDIT4L (first 3′UTR half) and NFIB (second 3′UTR
half) regions (Fig. 3B). A less
pronounced, but detectable effect on reporter gene expression was
observed for miR-200c on the NFIB-carrying construct
(first 3′UTR half) and for miR-301a on the
FOSL1-carrying construct (Fig.
3B). The observed effects were shown to be specific for the
selected miRNAs, since transfection with unrelated miRNAs for which
no binding sites were present in the 3′UTR regions under
investigation (‘mimic’ miRNAs) had no effect on luciferase
expression. Taken together, these data confirmed that some of the
downregulated miRNAs in RNASET2-silenced cells are actually
able to repress the expression of potential target mRNAs by means
of interaction with the corresponding 3′UTR regions.
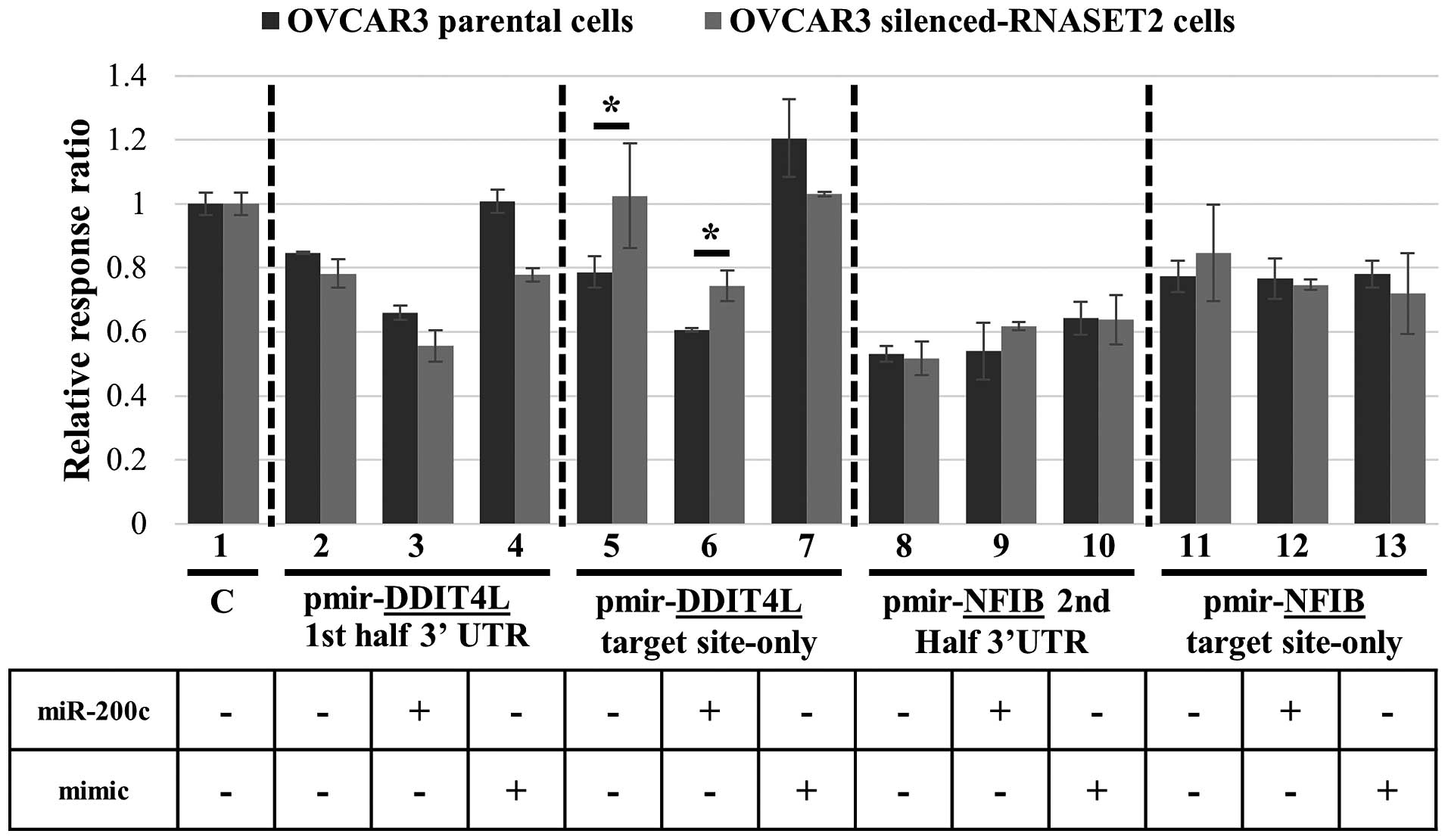
To further verify the role of RNASET2 in
miRNA-mediated regulation of these potential target genes, we
turned back to our OVCAR3 experimental model. The 3′UTR-luciferase
reporter plasmids from the two genes shown to be strongly repressed
by miR-200c (DDIT4L-first 3′UTR half and
NFIB-second 3′UTR half) were transiently transfected in
RNASET2-silenced (shT2) and control OVCAR3 cells. In order
to avoid putative nonspecific effects from unrelated endogenous
miRNAs, for both genes we used pmirGLO reporter constructs bearing
either the whole 3′UTR region under investigation or a shorter DNA
sequence representing just the predicted miR-200c target
sequence within the 3′UTRs (labeled ‘target site-only’).
Strikingly, the RNASET2 expression status was
indeed associated with the level of luciferase expression when the
reporter vector bearing just the miR-200c target site in the
3′UTR of DDIT4L was used, since expression of the luciferase
reporter was increased in RNASET2-silenced cells, as
expected (Fig. 4, lane 5).
Moreover, co-transfection of miR-200c in both control and
RNASET2-silenced cells led to a decrease in reporter
expression, again supporting a role for miR-200c in
targeting the 3′UTR region of DDIT4L (Fig. 4, lane 6). By contrast, the reporter
vectors carrying the whole DDIT4L 3′UTR did not show a
significant effect (Fig. 4, lanes
2 and 3). Once again, dowregulation of luciferase by
miR-200c through DDIT4L 3′UTR binding was shown to be
specific, since a mimic miRNA did not trigger a decrease in
reporter gene expression with respect to the control (Fig. 4, lane 7). These results are in
agreement with the notion that miR-200c-mediated repression
of DDIT4L is somehow impaired following
RNASET2-dowregulation in OVCAR3 cells. By contrast, no
effect of RNASET2 expression status was observed following
transfection of recombinant constructs bearing the 3′UTR of
NFIB in both forms (‘2nd half 3′UTR’ and ‘target site-only’)
(Fig. 4, lanes 8, 9, 11 and
12).
Since the gene expression profile data were derived
from pools of transfected OVCAR3 cells, to further validate these
results we performed the same assay in several independent OVCAR3
clones that were not used in the microarray analysis. Again, an
inverse correlation between RNASET2 expression status and
reporter gene expression was observed when the ‘target site-only’
construct for DDIT4L 3′UTR region was used (data not shown).
Moreover, a slight effect was observed also for NFIB (second
3′UTR half) in this experiment (data not shown). Altogether, the
data show a consistent trend for a role of RNASET2
expression status on miR-200c-mediated silencing of
DDIT4L.
The DDIT4L gene (also known as REDD2
or RTP801L) is involved in the cell growth control by
regulating the mTOR signaling pathway (21). Since expression of this gene turned
out to be affected in RNASET2-silenced OVCAR3 cells, likely
via mir-200c downregulation, we wondered whether
RNASET2 silencing in OVCAR3 cells might be associated with a
change in cell size due to DDIT4L upregulation. Notably,
following fluorescent staining of OVCAR3 cells with a marker for
the actin cytoskeleton (phalloidin-TRITC) a clear change in average
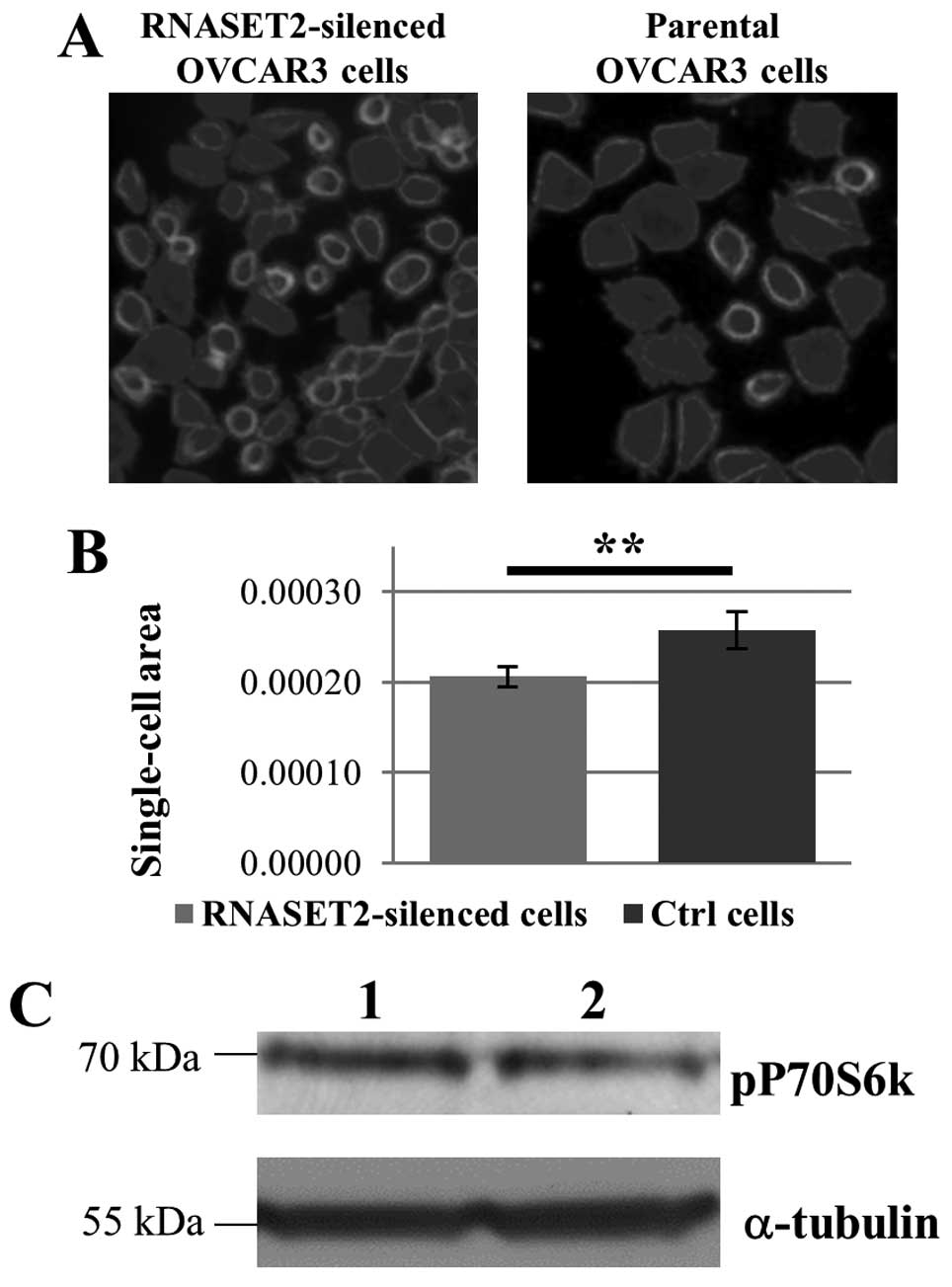
cell size in RNASET2-silenced OVCAR3 cells was observed with
respect to control cells (Fig. 5A and
B), suggesting that RNASET2-mediated increase in
DDIT4L expression in these cells might indeed affect cell
growth. To further validate the involvement of DDIT4L in the
observed RNASET2-mediated changes in cell size, the activation
state of the mTOR signaling pathway was compared in control and
RNASET2-silenced OVCAR3 cells by evaluating the
phosphorylation status of the mTOR downstream effector S6K1 kinase.
As shown in Fig. 5C,
phosphorylation of S6K1 turned out to be slightly decreased in
OVCAR3 cells following RNASET2 silencing, thus, lending support to
the notion that RNASET2-mediated changes in the transcript
levels of DDIT4L gene might affect the activation state of
mTOR pathway.
Discussion
The human RNASET2 gene has been recently
added to the growing list of ribonuclease-encoding genes endowed
with a tumor suppressor activity (22). Indeed, detailed investigations on
the role played by RNASET2 in vivo based on murine xenograft
models unveiled a strong non-cell autonomous oncosuppressive role
for this gene in two independent human ovarian cancer models
(12,13). Remarkably, the observed tumor
suppressive role of RNASET2 was linked to the stimulation of
a marked innate immune response involving cells of the
monocyte-macrophage lineage and was shown to be independent of the
enzyme catalytic activity (23).
On the other hand, several biological functions of T2 family
members are dependent on their ability to process or degrade RNA
substrates, and some of these functions might be related to tumor
suppression as well (22). In this
context, it is likely that the putative catalysis-dependent role(s)
of RNASET2 are carried out within the same cells expressing it and,
as such, they could reveal themselves in a cell-autonomous manner.
Indeed, a recent study carried out in the same experimental model
(the OVCAR3 human ovarian cancer cell line) used to report the
non-cell autonomous oncosuppressive role of RNASET2 allowed
us to show that this gene can also affect several cancer-related
parameters in a cell-autonomous manner (14). Significantly, RNASET2 was
shown to call into play these cell-autonomous roles in response to
a wide range of cellular stresses, some of which are known to be
frequently observed in both pre-cancerous lesions and advanced
cancer (14). Moreover, the recent
discovery that the RNASET2 protein co-localizes to P-bodies under
stress conditions strongly suggests that the stress response role
played by this enzyme might at least in part rely on its catalytic
activity, since P-bodies are widely known to represent
intracellular sites of RNA processing and degradation (19).
Building on this evidence, in the present study we
addressed the effects of RNASET2 silencing on the miRNA
transcriptome, since the latter is known to come into play in
P-bodies. Indeed, RNASET2 silencing in OVCAR3 cells was
shown to significantly affect the expression level of several
miRNAs, some of which have been previously involved in ovarian
cancer pathogenesis (3,20). Moreover, the two miRNAs showing the
strongest impact on the gene expression profile of
RNASET2-silenced OVCAR3 cells (miR-200c and
miR-141; Fig. 1) have been
reported to be disregulated in several human cancers, including
ovarian carcinoma (24,25), thus, lending support to the
hypothesis that the observed tumor suppressive role played by
RNASET2 might at least in part be carried out by means of
miRNA genes deregulation. To the best of our knowledge, this is the
first report on the effects of a mammalian T2 RNase family member
on the miRNA transcriptome. Moreover, by addressing the mRNA
profile in the same cells we found a much more extended effect of
RNASET2 silencing on the global cell transcriptome, with
approximately 300 transcript being affected. Taken together, these
data suggest that the expression levels of RNASET2 seem to
have a deep impact on the cell’s global transcriptome in our
experimental model.
Contrary to our expectations, a great proportion of
transcripts (both miRNAs and mRNAs) turned out to be downregulated
rather than upregulated in the absence of RNASET2, thus, suggesting
that the mechanism(s) by which this ribonuclease affects the
expression level of several intracellular transcripts might not
involve a direct cleavage of target RNAs. Of note, in a previous
report microarray-based gene expression profiling of the Hey3Met2
ovarian cancer cell line showed that overexpression of a
catalytically-dead RNASET2 protein could nevertheless affect the
cell transcriptome significantly, suggesting that this protein can
affect the cellular gene expression profile independently of its
ribonucleolytic activity (26).
These data are in keeping with several recent reports showing that
proteins belonging to the T2 family members can affect several
biological processes in a catalytically-independent manner
(22).
Although miRNAs are known to individually regulate
several independent mRNAs, evidence for a miRNA-mediated mechanism
of gene regulation could not be found for most transcripts whose
expression was dependent on the RNASET2 expression status.
However, among the mRNAs whose expression levels where changed in
RNASET2-silenced cells, a handful was shown to represent
potential target genes for a few ovarian-cancer related,
RNASET2-sensitive miRNAs, thus, suggesting that regulation
of these mRNAs by RNASET2 might involve changes in the expression
levels of their cognate miRNAs.
In support of this hypothesis, we found that two
candidate miRNAs (miR-200c and miR-301a) were indeed
able to affect reporter gene expression levels when the 3′UTR
regions of the corresponding candidate target genes (DDIT4L,
NFIB and FOSL1) were cloned downstream of a
luciferase reporter construct. The role of RNASET2 in
miRNA-mediated silencing of one of these genes (DDIT4L) was
further suggested by showing an inverse correlation between the
expression levels of RNASET2 and the luciferase gene when
the corresponding reporter constructs were assessed in OVCAR3
cells. Finally, since one of the candidate RNASET2 target
genes (DDIT4L, whose expression was shown to be regulated by
miR-200c) is functionally involved in the regulation of the
mTOR pathway (21) the effect of
RNASET2 expression levels on cell growth (a cell phenotype
related to cancer and modulated by this pathway) was evaluated in
our experimental model. Indeed, RNASET2 expression levels in
OVCAR3 cells were shown to affect two parameters downstream of
DDIT4L, i.e. cell size and the activation of the mTOR
pathway. In this regard it is worth noting that, whereas a slight
decrease in mTOR activation pathway was observed in relation to
RNASET2 expression levels, a much more evident effect was
observed on cell size. We considered that such discrepancy might be
attributed to the established pleiotropic roles of the RNASET2
protein, which has been previously shown to significantly affect
cell shape in OVCAR3 cells by remodeling the actin cytoskeleton
(14).
Of note, RNASET2 expression has been
previously shown to be increased in several cell lines (including
OVCAR3 cells) under hypoxic conditions (14) and expression of DDIT4L is
known to be HIF-dependent (27)
suggesting that, under hypoxic conditions, upregulation of
RNASET2 expression might affect cell growth in part via
DDIT4L. Further investigations will be focused on this
topic. Of note, a cancer cell-related parameter (cell proliferation
rate) has been recently shown to be affected by RNASET2 expression
levels in OVCAR3 cells under hypoxic conditions (14).
Though preliminary, these results provide to the
best of our knowledge the first evidence that a member of the T2
RNAse family might affect the expression pattern of selected miRNAs
in mammalian cells. The reason why, among the many miRNAs
potentially expressed in ovarian cancer cells, only a few are
actually affected by RNASET2 silencing has not been addressed in
this study and will require further investigations. To date, T2
ribonucleases have been described as rather aspecific
ribonucleolytic enzymes and only a few specific targets (such as
rRNA) have been reported for this class of RNases (28). Therefore, although targeting of
RNASET2 to P-bodies might suggest a rather general role in
stress-induced RNA processing for this RNase, the results of this
study suggest the occurrence of a sort of selectivity in target
selection by this protein, although the mechanism ruling such
putative selectivity are largely unknown. Our results thus, further
confirm that the effects of RNASET2 on the cell transcriptome are
more complex than expected, as already observed in previous
investigations on the catalytically-dead RNASET2 protein (26).
Finally, although some members of the
miR-200 gene family that were found to be downregulated in
RNASET2-silenced cells have been reported by some authors to
be upregulated in advanced ovarian cancer samples (29,30),
it should be acknowledged that the role played by the
miR-200 gene family in cancer pathogenesis is still rather
controversial. For instance, high expression of miR-200c was
correlated with a decreased risk of disease recurrence in patients
with serous ovarian carcinoma (31) and re-expression of miR-200c
in aggressive ovarian cancer cell lines was shown to trigger a wide
range of oncosuppressive effects both in vitro and in
vivo (32,33). Furthermore, miR-200c was
reported to be downregulated in a subpopulation of cell expressing
the cancer stem cell marker CD133 in the same ovarian cancer cell
line (OVCAR3) used in this study (34). Therefore, it is likely that the
dowregulation of miR-200c observed in RNASET2-silenced
OVCAR3 cells might contribute to the cell-autonomous
oncosuppressive role ascribed to this gene.
In conclusion, the present study shed further light
on the pleiotropic roles played by the human RNASET2 gene in
the context of its oncosuppressive role. The accumulating evidence
suggesting the role of RNASET2 as a stress-inducible gene,
which orchestrates a tumor suppressive response in a both
cell-autonomous and non-cell autonomous manners, is compatible with
a model whereby increased expression of this gene might represent a
key tool in mammalian anticancer response. Our results suggest that
this response might in part entail the regulation of cancer-related
genes by means of RNASET2-mediated modulation of miRNA
expression.
Acknowledgements
The present study is in memory of our friend and
colleague Giovanna Turconi, who is no longer with us. We all miss
her so much. We are also very grateful to Giovanna’s relatives and
friends for their support to the present work. Marco Fabbri is a
student of the PhD program in Biotechnology, School of Biological
and Medical Sciences, University of Insubria (Italy). Francesco
Acquati was supported by Giovanna Turconi’s Memorial Funds.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sopik V, Iqbal J, Rosen B and Narod SA:
Why have ovarian cancer mortality rates declined? Part I Incidence
Gynecol Oncol. 138:741–749. 2015. View Article : Google Scholar
|
|
3
|
Dahiya N and Morin PJ: MicroRNAs in
ovarian carcinomas. Endocr Relat Cancer. 17:F77–F89. 2010.
View Article : Google Scholar :
|
|
4
|
Li SD, Zhang JR, Wang YQ and Wan XP: The
role of microRNAs in ovarian cancer initiation and progression. J
Cell Mol Med. 14:2240–2249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Katz B, Tropé CG, Reich R and Davidson B:
MicroRNAs in Ovarian Cancer. Hum Pathol. 46:1245–1256. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Acquati F, Morelli C, Cinquetti R, Bianchi
MG, Porrini D, Varesco L, Gismondi V, Rocchetti R, Talevi S,
Possati L, et al: Cloning and characterization of a senescence
inducing and class II tumor suppressor gene in ovarian carcinoma at
chromosome region 6q27. Oncogene. 20:980–988. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Monti L, Rodolfo M, Lo Russo G, Noonan D,
Acquati F and Taramelli R: RNASET2 as a tumor antagonizing gene in
a melanoma cancer model. Oncol Res. 17:69–74. 2008.PubMed/NCBI
|
|
11
|
Smirnff P, Roiz L, Algelkovitch B,
Schwartz B and Shoseyov O: A recombinant human RNASET2 glycoprotein
with antituorigenic and antiangiogenic characteristics. Cancer.
107:2760–2769. 2006. View Article : Google Scholar
|
|
12
|
Acquati F, Bertilaccio S, Grimaldi A,
Monti L, Cinquetti R, Bonetti P, Lualdi M, Vidalino L, Fabbri M,
Sacco MG, et al: Microenvironmental control of malignancy exerted
by RNASET2, a widely conserved extracellular RNase. Proc Natl Acad
Sci USA. 108:1104–1109. 2011. View Article : Google Scholar :
|
|
13
|
Acquati F, Lualdi M, Bertilaccio S, Monti
L, Turconi G, Fabbri M, Grimaldi A, Anselmo A, Inforzato A,
Collotta A, et al: Loss of function of Ribonuclease T2, an ancient
and phylogenetically conserved RNase, plays a crucial role in
ovarian tumorigenesis. Proc Natl Acad Sci USA. 110:8140–8145. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lualdi M, Pedrini E, Rea K, Monti L,
Scaldaferri D, Gariboldi M, Camporeale A, Ghia P, Monti E,
Tomassetti A, et al: Pleiotropic modes of action in tumor cells of
RNASET2, an evolutionary highly conserved extracellular RNase.
Oncotarget. 6:7851–7865. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huber W, Carey VJ, Gentleman R, Anders S,
Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al:
Orchestrating high-throughput genomic analysis with bioconductor.
Nat Methods. 12:115–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Vidalino L, Monti L, Haase A, Moro A,
Acquati F, Taramelli R and Macchi P: Intracellular trafficking of
RNASET2, a novel component of P-bodies. Biol Cell. 104:13–21. 2012.
View Article : Google Scholar
|
|
19
|
Eulalio A, Behm-Ansmant I and Izaurralde
E: P bodies: At the crossroads of post-transcriptional pathways.
Nat Rev Mol Cell Biol. 8:9–22. 2007. View Article : Google Scholar
|
|
20
|
Wang L, Zhu MJ, Ren AM, Wu HF, Han WM, Tan
RY and Tu RQ: A ten-microRNA signature identified from a
genome-wide microRNA expression profiling in human epithelial
ovarian cancer. PLoS One. 9:e964722014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Corradetti MN, Inoki K and Guan KL: The
stress-inducted proteins RTP801 and RTP801L are negative regulators
of the mammalian target of rapamycin pathway. J Biol Chem.
280:9769–9772. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luhtala N and Parker R: T2 Family
ribonucleases: Ancient enzymes with diverse roles. Trends Biochem
Sci. 35:253–259. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Acquati F, Possati L, Ferrante L,
Campomenosi P, Talevi S, Bardelli S, Margiotta C, Russo A,
Bortoletto E, Rocchetti R, et al: Tumor and metastasis suppression
by the human RNASET2 gene. Int J Oncol. 26:1159–1168.
2005.PubMed/NCBI
|
|
24
|
Kumar S, Nag A and Mandal CC: A
comprehensive review on miR-200c, a promising cancer biomarker with
therapeutic potential. Curr Drug Targets. 16:1381–1403. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li XY, Li H, Bu J, Xiong L, Guo HB, Liu LH
and Xiao T: Prognostic role of microRNA-200c-141 cluster in various
human solid malignant neoplasms. Dis Markers. 2015:9356262015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Acquati F, Monti L, Lualdi M, Fabbri M,
Sacco MG, Gribaldo L and Taramelli R: Molecular signature induced
by RNASET2, a tumor antagonizing gene, in ovarian cancer cells.
Oncotarget. 2:477–484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cuaz-Pérolin C, Furman C, Larigauderie G,
Legedz L, Lasselin C, Copin C, Jaye M, Searfoss G, Yu KT, Duverger
N, et al: REDD2 gene is upregulated by modified LDL or hypoxia and
mediates human macrophage cell death. Arterioscler Thromb Vasc
Biol. 24:1830–1835. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Haud N, Kara F, Diekmann S, Henneke M,
Willer JR, Hillwig MS, Gregg RG, Macintosh GC, Gärtner J, Alia A,
et al: rnaset2 mutant zebrafish model familial cystic
leukoencephalopathy and reveal a role for RNase T2 in degrading
ribosomal RNA. Proc Natl Acad Sci USA. 108:1099–1103. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Y, Zhang L and Hao Q: Candidate
microRNA biomarkers in human epithelial ovarian cancer: Systematic
review profiling studies and experimental validation. Cancer Cell
Int. 13:86–94. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leskelä S, Leandro-García LJ, Mendiola M,
Barriuso J, Inglada-Pérez L, Muñoz I, Martínez-Delgado B, Redondo
A, de Santiago J, Robledo M, et al: The miR-200 family controls
beta-tubulin III expression and is associated with paclitaxel-based
treatment response and progression-free survival in ovarian cancer
patients. Endocr Relat Cancer. 18:85–95. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cochrane DR, Howe EN, Spoelstra NS and
Richer JK: Loss of miR-200c: A marker of aggressiveness and
chemoresistance in female reproductive cancers. J Oncol.
2010:8217172010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen D, Zhang Y, Wang J, Chen J, Yang C,
Cai K, Wang X, Shi F and Dou J: MicroRNA-200c overexpression
inhibits tumorigenicity and metastasis of
CD117+CD44+ ovarian cancer stem cells by
regulating epithelial-mesenchymal transition. J Ovarian Res.
6:50–60. 2013. View Article : Google Scholar
|
|
34
|
Guo R, Wu Q, Liu F and Wang Y: Description
of the CD133+ subpopulation of the human ovarian cancer
cell line OVCAR3. Oncol Rep. 25:141–146. 2011.
|