Introduction
Colorectal cancer is one of the most common
malignancies causing mortality in the world (1). However, current knowledge of
molecular carcinogenesis in the development of colorectal cancer is
still limited. Although significant advances have been achieved,
more effective therapeutic options for advanced colorectal cancer
are still needed, and many efforts have been made to develop novel
treatments for targeting tumor-specific genes.
Emerging evidence suggests that specific
sub-populations of cancer cells with stem cell characteristics
within the bulk of tumors are implicated in the pathogenesis of
heterogeneous malignant tumors (2–4). To
study the behavior of cancer stem cells (CSCs), markers for
prospective isolation of CSCs are crucial. CD44 has been proposed
as one of the CSC markers of colorectal cancer (2,4,5).
CD44 is a transmembrane glycoprotein acting as a cell adhesion
molecule through the binding to hyaluronic acid, and plays a key
role in remodeling and degradation of hyaluronic acid (4). Furthermore, CD44 is involved in
fundamental aspects of cancer cell biology such as tumor stem cell
phenotype, cell adhesion, invasion, and metastasis (6). Several studies have shown that CD44
expression was associated with tumor progression, metastasis, and
poor prognosis (7–10).
Recent studies have reported that knockdown of CD44
resulted in the inhibition of tumor growth and metastasis (11–13).
In our previous studies, we have shown that CD44 enhanced the
epithelial-mesenchymal transition, which is associated with colon
cancer invasion (14), and that
knockdown of CD44 expression using inducible short hairpin RNA
(shRNA) significantly reduced cell proliferation, invasion, and
migration (13). The therapeutic
effect of RNA interference depends on the stability and tissue
specificity of small interference RNA (siRNA) and the efficiency of
siRNA transduction. We have previously used plasmids to suppress
CD44 expression (13); however,
the efficiency of plasmid delivery remains poor. Therefore, more
efficient means of delivering therapeutic siRNA are necessary. One
promising therapeutic modality is the use of oncolytic viruses,
which have cancer specificity and also act as a vector for stable
introduction of siRNA (15,16).
Here, we developed an in vitro model using
CD44-shRNA recombinant adenovirus, and evaluated the impact of CD44
knockdown adenovirus on proliferation, invasion, migration, and
apoptosis of colon cancer cells.
Materials and methods
Cell culture
The HCT116 human colon cancer cells were obtained
from the American Type Culture Collection (ATCC, Manassas, VA,
USA). The cells were routinely maintained in complete medium (DMEM;
Lonza, Walkersville, MN, USA) supplemented with 10% fetal bovine
serum (FBS; Biowest, Nuaillé, France), 50 U/ml penicillin, and 50
µg/ml streptomycin (Lonza) at 37°C in a humidified incubator
with 5% CO2.
Construction of shRNA-CD44 plasmid
The shRNA with vector was purchased from OriGene
(OriGene Technologies, Inc., Rockville, MD, USA). The shRNA-CD44
(sense: GACAGAAAGCCAAGTGGACTCAACGGAGA) and pGFP-V-RS vectors, the
latter of which contained an ineffective shRNA cassette against
GFP, were used for knockdown of CD44 expression.
Construction of recombinant
adenovirus
The human shRNA targeting the CD44 sequence and a
negative control scrambled sequence were each amplified by
polymerase chain reaction (PCR) from plasmids containing the 29-mer
shRNA construct using primers containing KpnI and
XbaI restriction sites (Enzynomics, Daejeon, Korea).
Purified (Qiagen, Valencia, CA, USA) PCR products and adenovirus
shuttle plasmids (Agilent Technologies, Palo Alto, CA, USA) were
digested with KpnI and XbaI, ligated with T4 DNA
ligase (Promega, Madison, WI, USA), and then transformed into DH5a
chemically competent E. coli. Miniprepped DNA of the
different clones was analyzed on a 0.8% agarose gel and the correct
clones were confirmed by DNA sequencing.
Clones carrying the correct target sequences were
selected, linearized with PmeI, subcloned into the pAdEasy-1
backbone, and transformed into BJ5183 bacteria. Recombinant
adenoviral plasmids were selected against kanamycin and screened by
diagnostic digestions. These plasmids were then digested by
PacI, and the larger fragments were transfected using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) into AD293 cells
(Stratagene, La Jolla, CA, USA). After 11–13 days, the recombinant
adenoviruses were collected after freeze-thaw lysis of the AD293
cells. The primary viral stock was used to infect new AD293 cells
for 2–3 days to produce a bulk viral stock. Infected cells
exhibiting cytopathic effects were lysed for collection of the
virus.
Viral particles were purified and concentrated using
the Vivapure AdenoPACK™20 RT (Sartorius Stedim Biotech, Göttingen,
Germany) kit. The viral particle concentration was determined by
measuring absorbance at 260 nm, and a standard TCID50
(50% tissue culture infective dose) assay was performed on AD293
cells to determine the infectious virus titer (17). Purified viral particles were stored
at −70°C until use.
Adenoviral infection
The HCT116 colon cancer cells were seeded onto a
6-well plate at a density of 0.1×106 cells/ml, cultured
overnight, and infected with serially diluted concentrations of
recombinant adenovirus. After a 24-h incubation, the previous
growth medium was removed and fresh complete growth medium was
added, and treated with adenoviral aliquots with or without
shRNA-CD44.
Reverse transcription PCR (RT-PCR)
analysis
Total RNA was isolated from cells with TRIzol
reagent (Invitrogen) according to the manufacturer's instructions.
The quantity and purity of total RNA were determined by measuring
absorbance at 260 and 280 nm using the Nanodrop ND-1000
spectrophotometer (BCM, Houston, TX, USA). Next, cDNA was
synthesized from 3 µg total RNA using Oligo(dT) (Promega)
and reverse transcriptase (Beams Biotech, Seongnam, Korea). PCR
amplification of cDNA was performed using gene-specific primers
(Table I) and nTaq DNA polymerase
(Enzynomics, Daejeon, Korea). PCR products were separated on a 1%
agarose gel, visualized and photographed under UV light.
 | Table IPrimers for RT-PCR. |
Table I
Primers for RT-PCR.
| Protein | Primers | Sequences |
|---|
| CD44 | Forward | 5′-GAA TAT AAC CTG
CCG CTT TG-3′ |
| Reverse | 5′-CTG AAG TGC TGC
TCC TTT CAC-3′ |
| GAPDH | Forward | 5′-ACC ACA GTC CAT
GCC ATC AC-3′ |
| Reverse | 5′-TCC ACC ACC CTG
TTG CTG TA-3′ |
Western blotting
Total cell extracts were lysed in cell lysis buffer
(Cell Signaling Technology, Inc., Danvers, MA, USA) with a protease
inhibitor cocktail (Roche, Basel, Switzerland). Protein
concentrations were determined by a BCA protein assay (Thermo
Fisher Scientific, Rockford, IL, USA). The protein was separated by
10% SDS-PAGE and transferred onto a PVDF (polyvinylidene fluoride)
membrane (Millipore, Billerica, MA, USA). The membranes were
incubated for 1 h in blocking solution [5% skim milk in TBS with
0.1% Tween-20 (TBST)] and sequentially blotted with the following
primary antibodies: anti-CD44 (R&D Systems, Minneapolis, MN,
USA), anti-GAPDH (Aviva Systems Biology, San Diego, CA, USA),
anti-AKT, anti-phospho-AKT (Ser/Thr), anti-phospho-GSK-3β,
anti-β-catenin, anti-Bax, anti-Bcl-2, anti-Bcl-xL, anti-caspase-3,
anti-phospho-caspase-3, anti-cleaved-caspase-3, anti-caspase-9,
anti-phospho-caspase-9, anti-cleaved-caspase-9, anti-PARP
poly(ADP-ribose) polymerase, and anti-cleaved-PARP (Cell Signaling
Technology) at 4°C overnight. After rinsing in TBST (0.1%),
membranes were incubated with horseradish peroxidase-labeled
anti-rabbit (Thermo Fisher Scientific) or anti-mouse IgG secondary
antibodies (Cell Signaling Technology) at room temperature for 1 h.
The blot was detected by ECL (enhanced chemiluminescence) of an HRP
substrate (Millipore) on an image reader (Ras4000, Fujifilm, Tokyo,
Japan).
Cell viability assay
The viability of treated cells was measured using a
Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). Cells were
plated into 96-well plate at 5,000 cells/well one day prior to the
viral transduction. Then, cells were infected with the recombinant
adenoviruses for 1 day, followed by medium replacement. Cell growth
and viability were assayed 4–5 days post-infection. For cell
viability assay, the cells are incubated with reagents from the
CCK-8 kit for 1 h and the absorbance was measured at 450 nm in a
microplate reader (BioTek, Winooski, VT, USA). Each sample was
assayed in triplicate, and each experiment was repeated at least
twice.
Flow cytometry analysis
Apoptosis was quantified using flow cytometry after
being stained with APC (allophycocyanin)-labeled Annexin V and
7-amino-dactinomycin (BD Biosciences, San Diego, CA, USA). We
analyzed for intact cells (Annexin V/7AAD-double-negative), early
apoptotic cells (Annexin V-positive), and late apoptotic cells or
necrotic cells (Annexin V/7AAD-double-positive). The cells were
plated in 6-well plates at of 200,000 cells/well prior to infected
with the recombinant adenoviruses for 4–5 days. Both uninfected and
infected HCT116 cells were trypsinized, washed twice with cold PBS,
and resuspended in 1X binding buffer (BD Biosciences). Analysis of
400 µl of this cell resuspension was performed on a
fluorescence active cell sorting (FACS)Calibur flow cytometer
(Becton-Dickinson, San Jose, CA, USA) using the CellQuest version
3.3 software (Becton-Dickinson).
Cell migration assay
Cells were cultured in 6-well plates and infected
with the recombinant adenoviruses for 24–48 h. The infected HCT116
cells were then seeded in culture-inserts (2×0.22 cm2;
IBIDI GmbH, Martinsried, Germany) at 5×104 cells/well.
To create a cell-free gap, Culture-Inserts were gently removed
using sterile tweezers after a 24-h incubation. The progress of
cell migration into the cell-free gap was photographed at 0, 20,
and 40 h using an inverted microscope. The distance between gaps
was measured using the Focus Lite Ver 2.90 (Focus, Daejeon, Korea)
software after three random sites were photographed.
Cell invasion assay
Cell invasion assays were carried out using 24-well
Transwell filters with 8-µM pores (Coring Inc., NY, USA).
Transwell filters were coated with 500 µg Matrigel/DMEM for
3–4 h and unbound material was aspirated at room temperature. Cells
infected with the recombinant adenoviruses were resuspended at a
density of 2.5×105 cells in 120 µl 0.2% BSA
medium and then seeded into the upper chamber. Then 400 µl
of 0.2% BSA medium containing 50 µg/ml human plasma
fibronectin (Calbiochem, La Jolla, CA, USA) as a chemoattractant
was loaded into the lower chamber. After a 24-h incubation, invaded
cells on the bottom surface of the Transwell were stained with
Diff-Quick solution (Sysmex, Kobe, Japan) and quantified in five
selected fields (1 mm2 each) using a hematocytometer
under a light microscope.
Soft-agar colony formation assay
Soft agar assays were constructed in 6-well plates.
The foundation layer of each well consisted of 1.5 ml of 0.6% agar
solution in 1X media. The HCT116 cells were transduced
(1.5×104 cells/well) for 1 day, and then mixed with 0.6%
soft agar (1:1) and seeded onto the bottom. An additional 3 ml of
1X media without agarose was poured on top of the growth layer.
After a 2-week incubation, the colonies were stained with 0.05%
crystal violet and photographed using an inverted microscope
camera. The number of colonies was counted at ×40
magnification.
Statistical analysis
All statistical analyses were performed using a
t-test with SPSS 21.0 (IBM Inc., Armonk, NY, USA) software.
Results
Expression of CD44
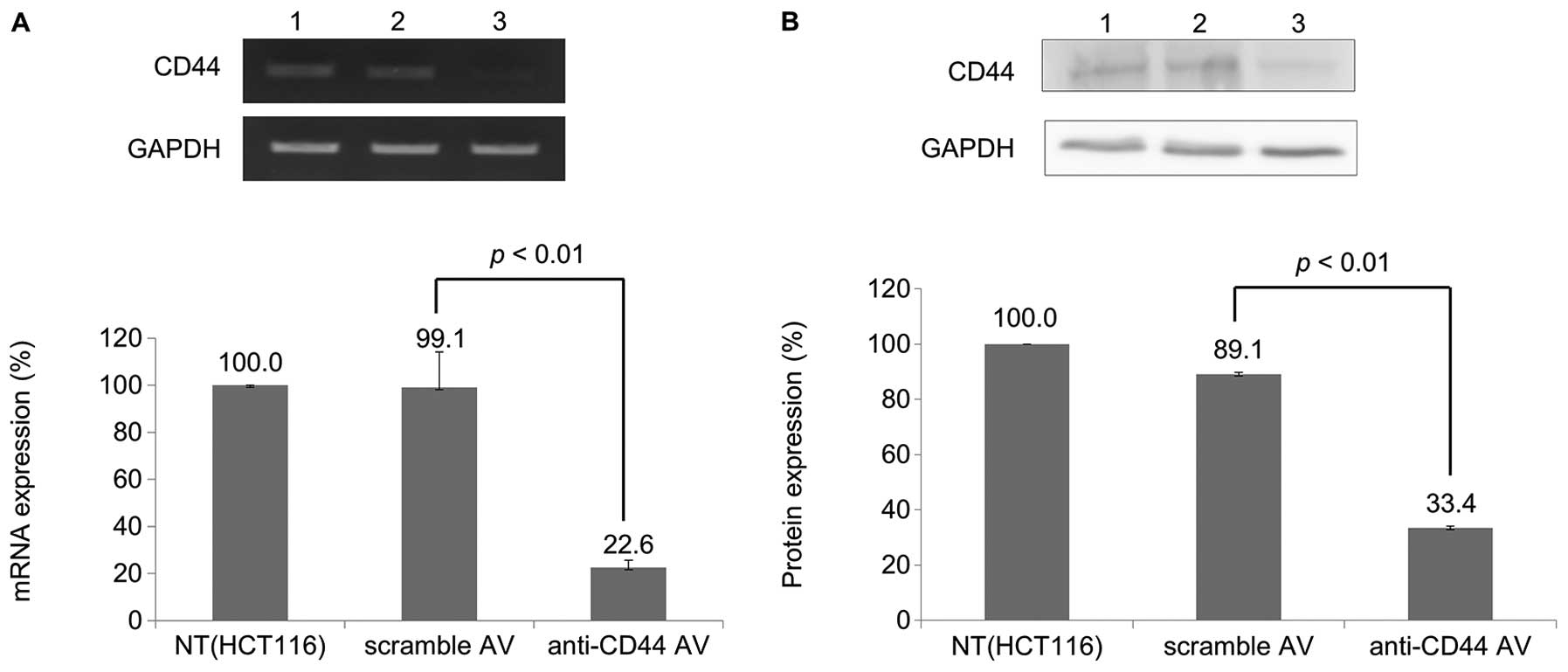
The level of expression of CD44 mRNA was
evaluated by RT-PCR; GAPDH served as an internal control. As shown
in Fig. 1A, the level of
expression of CD44 mRNA in cells infected with Ad-CD44-shRNA
was significantly downregulated compared with parental (HCT116) and
scramble-Ad-infected cells (p<0.01). A significant reduction in
CD44 protein was also detected in Ad-CD44-shRNA-infected cells
compared with scramble-Ad-infected cells (p<0.01).
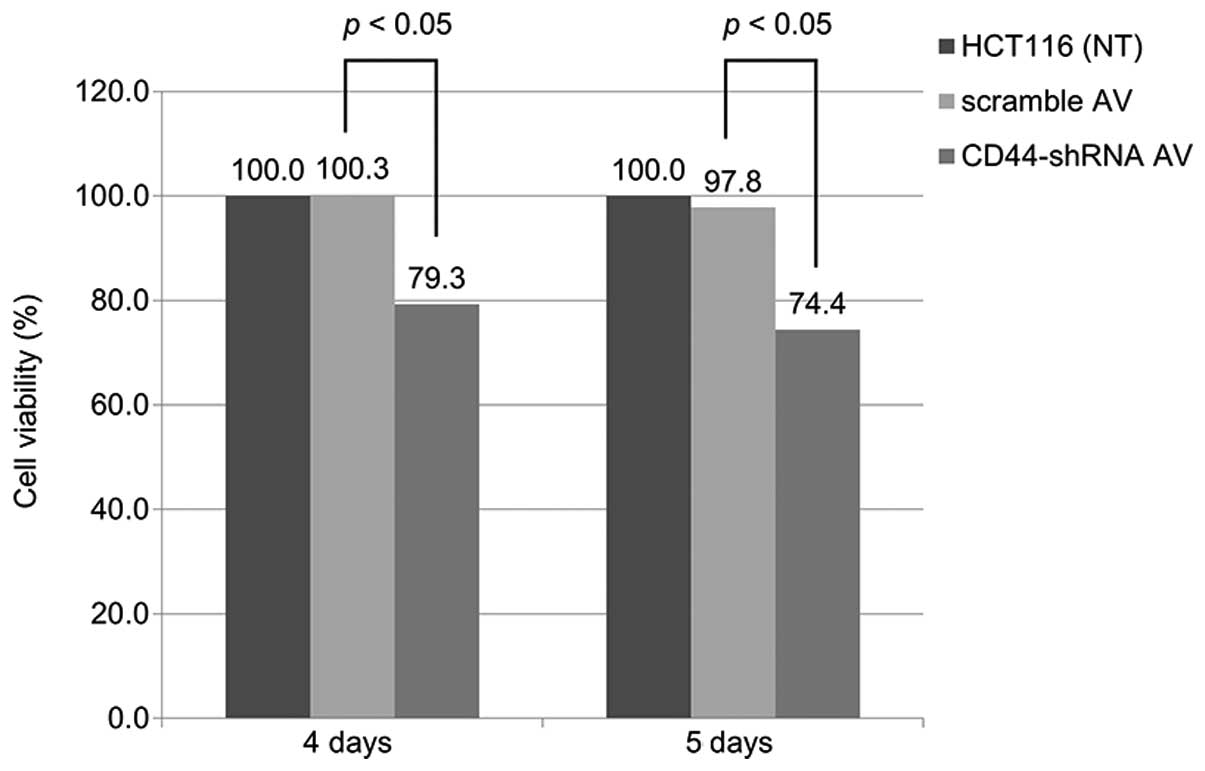
Cell viability
The results of the cell viability assay are shown in
Fig. 2. Whereas the scramble-Ad
showed little cytotoxicity, Ad-CD44-shRNA suppressed cell viability
(p<0.05) 4–5 days post-infection.
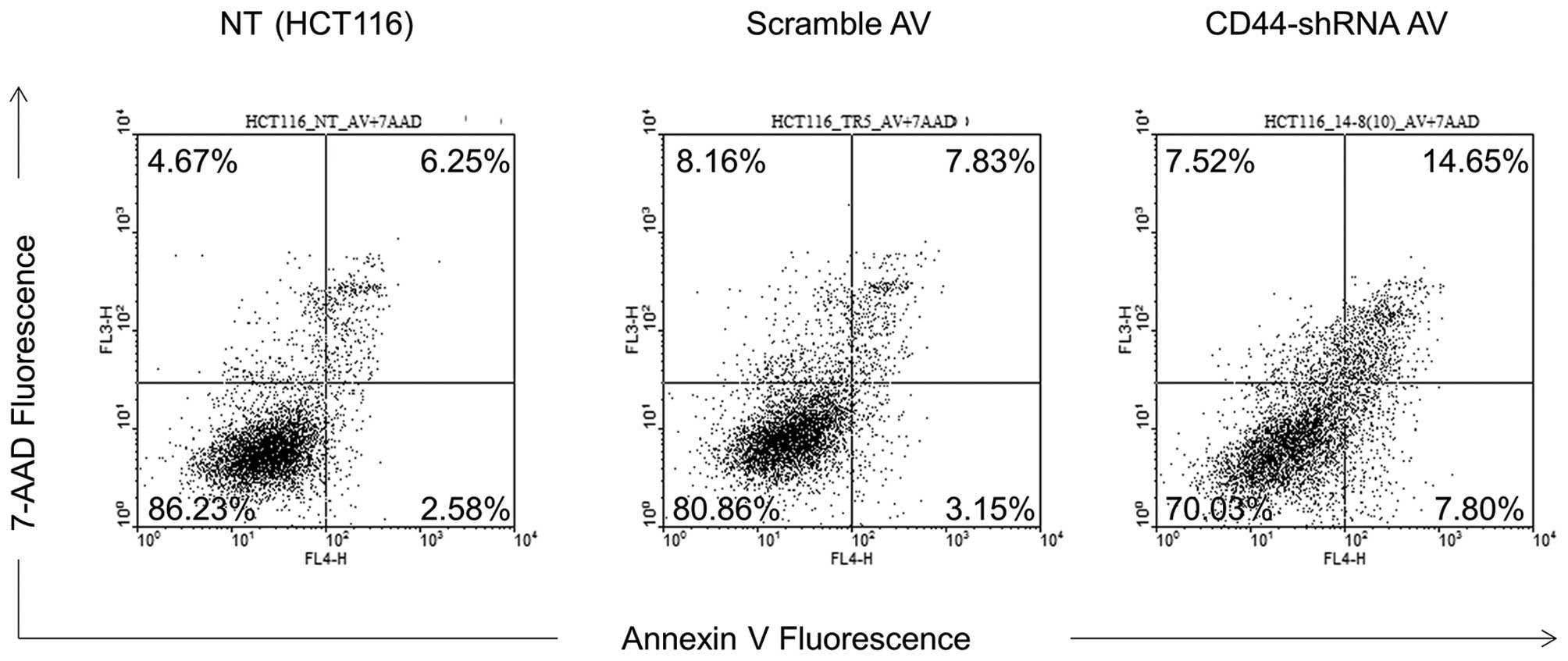
Flow cytometry analysis
The early (7.80%) and late (14.65%) apoptotic rate
of Ad-CD44-shRNA-infected cells was increased compared with
parental (2.58 and 6.25%) and scramble Ad-infected (3.15 and 7.83%)
cells (p<0.01) (Fig. 3).
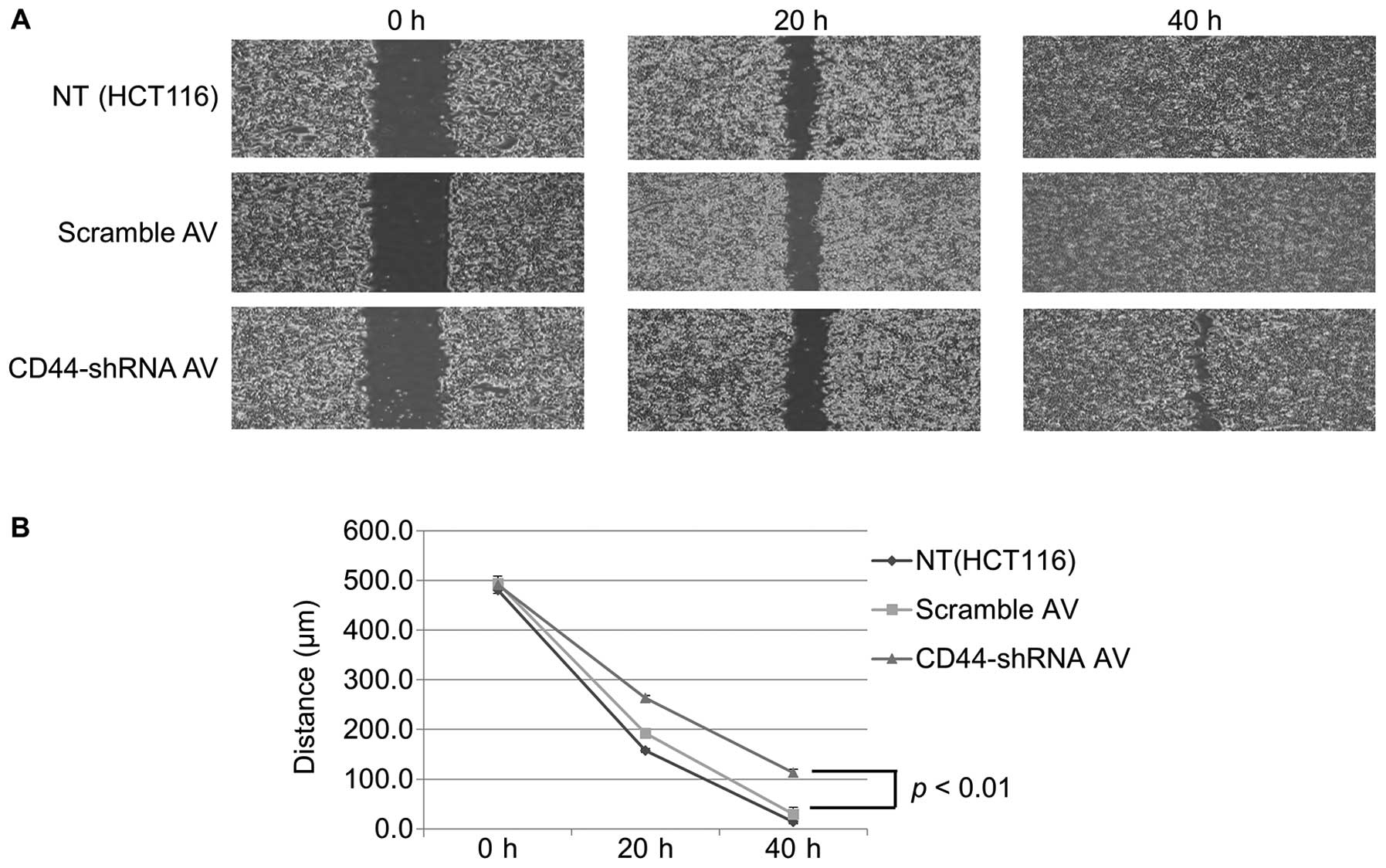
Cell migration assay
The Ad-CD44-shRNA-infected cells showed much lower
migratory capacity than scramble-Ad-infected cells at 40 h after
plating (p<0.01) (Fig. 4).
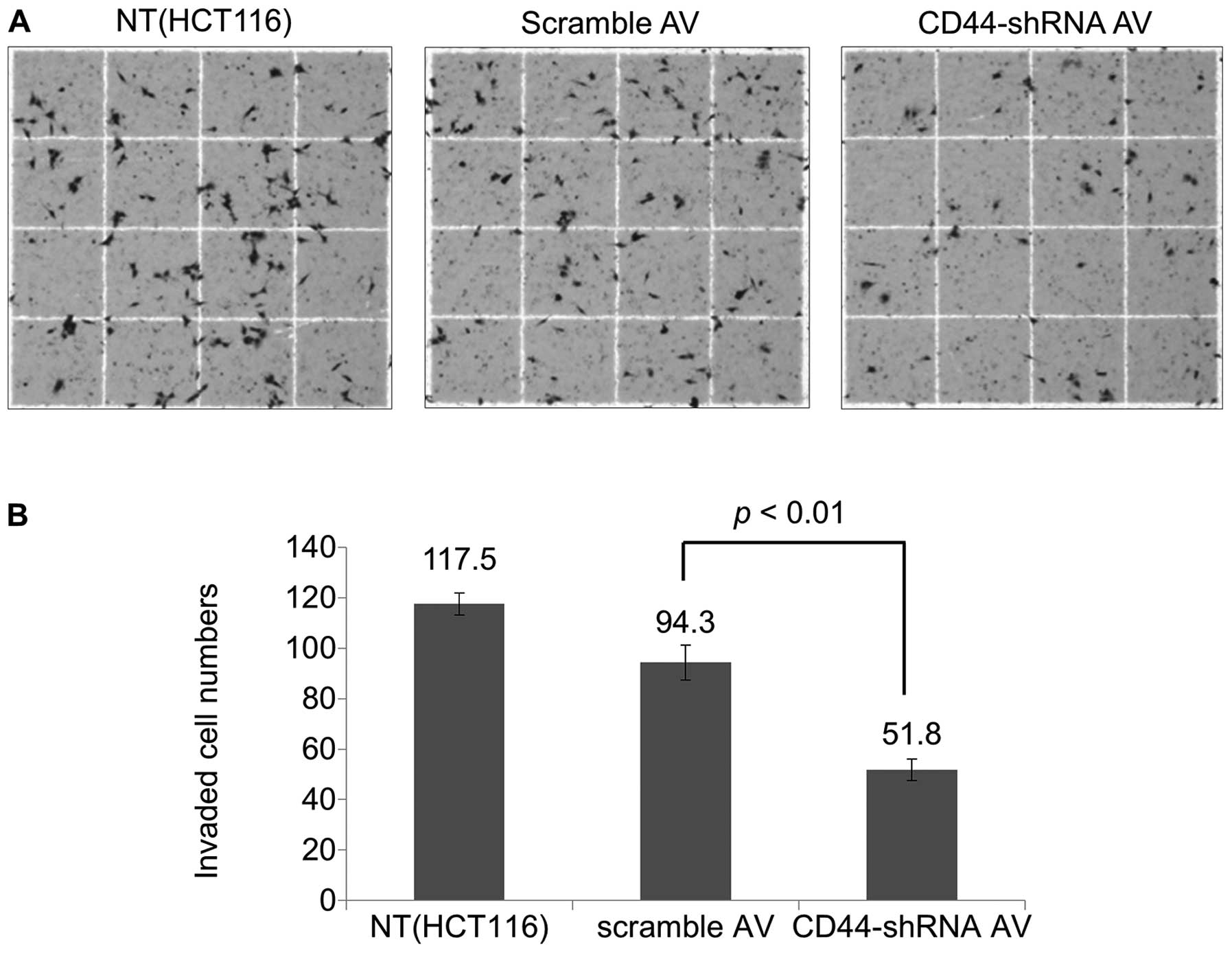
Cell invasion assay
The invasion activity of Ad-CD44-shRNA-infected
cells was significantly decreased compared with
scramble-Ad-infected cells (51.8 vs. 94.3, p<0.01) (Fig. 5).
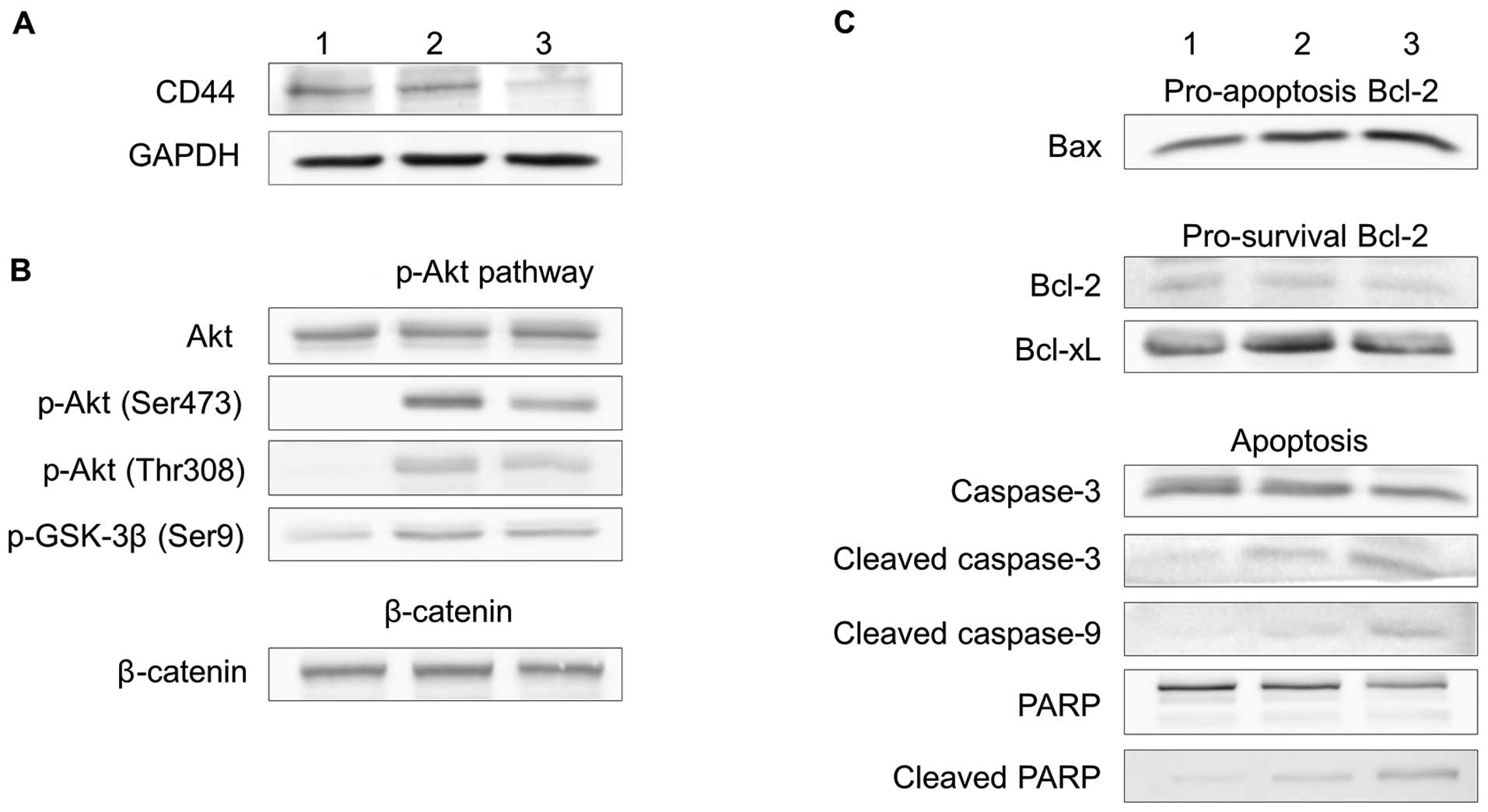
PI3-Akt signaling and apoptosis
Western blot analysis for expression of PI3-Akt
signaling and apoptotic molecules is shown in Fig. 6. The Ad-CD44-shRNA resulted in a
decrease in the expression of phospho-Akt and phospho-GSK-3β
(Fig. 6B). In contrast, there was
minimal change of β-catenin expression (Fig. 6B). The Ad-CD44-shRNA also resulted
in a decrease in the expression of Bcl-2 and Bcl-xL, but an
increase in the expression of Bax, and promoted the cleavage of
caspase-3, -9 and PARP (Fig.
6C).
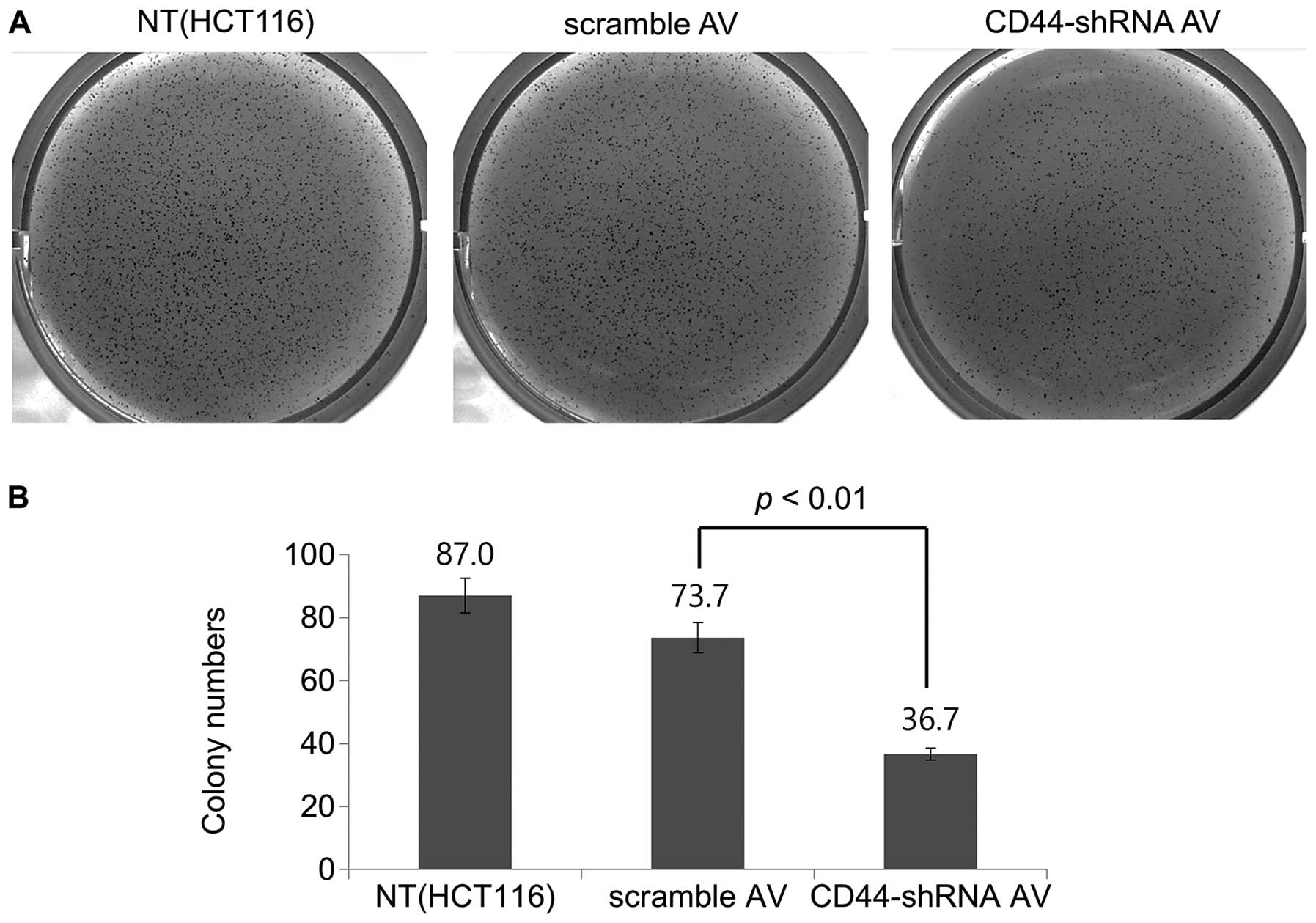
Soft-agar colony formation assay
The Ad-CD44-shRNA-infected cells showed a marked
decrease in colony formation (Fig.
7). When quantified, there was a 50.2% decrease in colony
formation units in the presence of Ad-CD44-shRNA compared with
scramble-Ad (Fig. 7B), suggesting
a significant tumorigenic inhibition of HCT116 colon cancer cells
by Ad-CD44-shRNA.
Discussion
In this study, we have constructed a recombinant
adenoviral model to reduce the expression of CD44. We showed that
Ad-CD44-shRNA inhibited cell proliferation, migration, and invasion
in HCT116 colon cancer cells, which supports the feasibility of an
adenovirus-mediated RNA interference therapy targeting colon cancer
via the CD44 antigen.
Colorectal cancer is the result of genetic
alterations that lead to a transformation of normal colonic
epithelial cells into cancer cells. Currently, radical surgery
followed by adjuvant chemotherapy is recommended to high risk
patients for management of colon cancer (18). However, this standard treatment is
not ideally effective because of the recurrence of the cancer and
toxicity of the chemotherapeutic agents. CSCs are believed to be
the reason of resistance to the conventional chemotherapy and
radiotherapy that targets the bulk of cancer, leaving the stem
cells unaffected (2–4). In the traditional stochastic model,
every cancer cell from the bulk tumor has a carcinogenic potential.
According to the hierarchical model, however, only a small
proportion of tumor cells are actually cancer stem cells (4). In contrast with the stochastic model,
slowly proliferating CSCs displaying multipotency and self-renewal
are only responsible for tumor initiation, maintenance, and
metastasis (4). These CSCs are
hypothesized to be spared from the chemotherapy that interferes
with the ability of rapidly growing cells to divide (2).
To identify and isolate CSCs, there have been many
efforts to identify specific CSC markers. Well-known CSC markers
for colorectal cancer include CD44, CD133, EpCAM, CD24, and CD29
(4). Above all, CD44, a
transmembrane glycoprotein functioning as a cell adhesion protein
and a signaling receptor (6), is
one of the most well-studied CSC surface markers. CD44 enhances the
epithelial-mesenchymal transition, which is related to cancer cell
migration and invasion (14), and
therefore is associated with tumor progression, metastasis, and
poor prognosis in colon cancer (9,10,19,20).
Importantly, inhibition of these CSC surface markers may result in
the inhibition of tumor cell proliferation, invasion, and
metastasis (3). We have previously
developed a CD44 knockdown model using plasmids for RNA
interference and reported that shRNA against CD44 inhibited cell
proliferation, invasion, and migration (13). However, as mentioned above, more
efficient means of delivering therapeutic siRNA are still needed
because of the limited efficiency of delivery via plasmid.
In this study, we successfully constructed a
recombinant adenoviral model to knock down CD44 using adenoviruses,
which are among the most widely used vectors for gene therapy
(21). Oncolytic virotherapy using
recombinant adenoviruses has a number of potential advantages. It
can be used to specifically target cancer cells while leaving
normal tissue stem cells unharmed, thus minimizing systemic
toxicity (15,22). There is also a low possibility of
resistance because of the diverse ways it induces oncolysis
(22). Above all, therapeutic
genes, such as inhibitory RNA against specific oncogenes, can be
delivered using recombinant adenoviral vectors (22). Because of the high efficiency of
transduction in vivo, the adenoviral system has been used
for virus-based therapies (15,23).
The result of this study demonstrated that reduced
cell proliferation, migration, and invasion, and enhanced apoptosis
were likely to be a result of the Ad-CD44-shRNA infection. Also, we
showed differential expression of PI3-Akt signaling and apoptotic
molecules in colon cancer cells treated with Ad-CD44-shRNA. Tumor
proliferation, differentiation, and apoptosis are known to be under
the control of several signaling pathways such as the Wnt signaling
pathway (24,25). We demonstrated that Ad-CD44-shRNA
infection inhibited Akt phosphorylation (Fig. 6B), which is one of the most
important Wnt-target genes for the survival of cancer cells
(26,27). We also showed the downregulation of
GSK-3β (Fig. 6B), the target of
PDK1/Akt signal transduction, inactivates various proteins involved
in cell proliferation and survival such as β-catenin, cyclin D1,
c-jun, and c-myc (28). β-catenin
is a downstream molecule in the Wnt signaling pathway and plays an
important role in cell-to-cell adhesion, tumor invasion, and
metastasis (25). In addition, we
showed decreased expression of Bcl-2 and Bcl-xL; increased
expression of Bax; and cleavage of caspase-3, -9, and PARP
(Fig. 6C). The Bcl-2 family
proteins are key regulators of apoptosis, with a pro-survival
subfamily including Bcl-2, Bcl-xL, Bcl-w, Mcl-1, and A1; and a
pro-apoptotic subfamily including Bax, Bak, and Bok (29–31).
Apoptosis is precipitated by the activation of cysteine proteases
of the caspase family, including caspase-3, -8 and -9, and their
cleavage is considered the primary hallmark of apoptosis (32,33).
The results of our study also demonstrated that Ad-CD44-shRNA
infection induces apoptosis in HCT116 colon cancer cells,
suggesting reduced clonogenic ability.
We also utilized the soft agar colony formation
assay, or 3D culture, as a novel modality to identify the
inhibition of tumorigenesis by Ad-CD44-shRNA. Because of the
intrinsic difficulties in investigating the tumor progression in
vivo, the soft agar colony formation assay, which is a close
mimicry of the 3D cellular environment in vivo, has recently
been used (34). With this assay,
we assessed the effects of Ad-CD44-shRNA on cell proliferation and
migration. The result of the assay provided us with a
straightforward and intuitive result, as well as a quantitative
assessment of the inhibitory potential of Ad-CD44-shRNA.
Until now, there have been several clinical trials
using oncolytic adenoviruses (22); however, regarding colorectal
cancer, it is rarely reported (35–38).
Although there is an increasing demand for novel therapeutic
modalities, such as non-pathogenic viruses in the treatment of
colorectal cancer, clinical evidence of oncolytic virotherapy is
still lacking (22). Our results
support the feasibility of an adenovirus-mediated RNA interference
therapy targeting colon cancer via the CD44 antigen, which can be
used as a therapeutic intervention with the
anti-survival/pro-apoptotic machinery in human colon cancer. This
study is also meaningful as a cornerstone to potential future gene
therapies using oncolytic adenoviruses against colorectal cancer.
Oncolytic adenoviral therapy, despite its limited efficacy as a
single agent, has a potential role in combination therapy with
conventional chemotherapy (22).
Further translational studies and clinical trials focusing on the
administration of cancer virotherapy in combination with
conventional chemotherapy are needed.
Acknowledgments
This study was supported by a grant from Chonnam
National University 2010, Research Institute of Medical Sciences,
Chonnam National University (2011-CURIMS-DR007) and a Research
Grant 0720570 from the National Cancer Center, Korea.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Todaro M, Francipane MG, Medema JP and
Stassi G: Colon cancer stem cells: Promise of targeted therapy.
Gastroenterology. 138:2151–2162. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Puglisi MA, Tesori V, Lattanzi W,
Gasbarrini GB and Gasbarrini A: Colon cancer stem cells:
Controversies and perspectives. World J Gastroenterol.
19:2997–3006. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fanali C, Lucchetti D, Farina M, Corbi M,
Cufino V, Cittadini A and Sgambato A: Cancer stem cells in
colorectal cancer from pathogenesis to therapy: Controversies and
perspectives. World J Gastroenterol. 20:923–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dalerba P, Dylla SJ, Park IK, Liu R, Wang
X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponta H, Sherman L and Herrlich PA: CD44:
From adhesion molecules to signalling regulators. Nat Rev Mol Cell
Biol. 4:33–45. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ropponen KM, Eskelinen MJ, Lipponen PK,
Alhava E and Kosma VM: Expression of CD44 and variant proteins in
human colorectal cancer and its relevance for prognosis. Scand J
Gastroenterol. 33:301–309. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fernández JC, Vizoso FJ, Corte MD, Gava
RR, Corte MG, Suárez JP, García-Muñíz JL and García-Morán M: CD44s
expression in resectable colorectal carcinomas and surrounding
mucosa. Cancer Invest. 22:878–885. 2004. View Article : Google Scholar
|
|
9
|
Huh JW, Kim HR, Kim YJ, Lee JH, Park YS,
Cho SH and Joo JK: Expression of standard CD44 in human colorectal
carcinoma: Association with prognosis. Pathol Int. 59:241–246.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lugli A, Iezzi G, Hostettler I, Muraro MG,
Mele V, Tornillo L, Carafa V, Spagnoli G, Terracciano L and Zlobec
I: Prognostic impact of the expression of putative cancer stem cell
markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer.
Br J Cancer. 103:382–390. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Harada N, Mizoi T, Kinouchi M, Hoshi K,
Ishii S, Shiiba K, Sasaki I and Matsuno S: Introduction of
antisense CD44S CDNA down-regulates expression of overall CD44
isoforms and inhibits tumor growth and metastasis in highly
metastatic colon carcinoma cells. Int J Cancer. 91:67–75. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Du L, Wang H, He L, Zhang J, Ni B, Wang X,
Jin H, Cahuzac N, Mehrpour M, Lu Y, et al: CD44 is of functional
importance for colorectal cancer stem cells. Clin Cancer Res.
14:6751–6760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Park YS, Huh JW, Lee JH and Kim HR: shRNA
against CD44 inhibits cell proliferation, invasion and migration,
and promotes apoptosis of colon carcinoma cells. Oncol Rep.
27:339–346. 2012.
|
|
14
|
Cho SH, Park YS, Kim HJ, Kim CH, Lim SW,
Huh JW, Lee JH and Kim HR: CD44 enhances the epithelial-mesenchymal
transition in association with colon cancer invasion. Int J Oncol.
41:211–218. 2012.PubMed/NCBI
|
|
15
|
Short JJ and Curiel DT: Oncolytic
adenoviruses targeted to cancer stem cells. Mol Cancer Ther.
8:2096–2102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
St George JA: Gene therapy progress and
prospects: Adenoviral vectors. Gene Ther. 10:1135–1141. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McDougal JS, Cort SP, Kennedy MS,
Cabridilla CD, Feorino PM, Francis DP, Hicks D, Kalyanaraman VS and
Martin LS: Immunoassay for the detection and quantitation of
infectious human retrovirus, lymphadenopathy-associated virus
(LAV). J Immunol Methods. 76:171–183. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang GJ, Kaiser AM, Mills S, Rafferty JF
and Buie WD; Standards Practice Task Force of the American Society
of Colon and Rectal Surgeons: Practice parameters for the
management of colon cancer. Dis Colon Rectum. 55:831–843. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jing F, Kim HJ, Kim CH, Kim YJ, Lee JH and
Kim HR: Colon cancer stem cell markers CD44 and CD133 in patients
with colorectal cancer and synchronous hepatic metastases. Int J
Oncol. 46:1582–1588. 2015.PubMed/NCBI
|
|
20
|
Bendardaf R, Algars A, Elzagheid A,
Korkeila E, Ristamäki R, Lamlum H, Collan Y, Syrjänen K and
Pyrhönen S: Comparison of CD44 expression in primary tumours and
metastases of colorectal cancer. Oncol Rep. 16:741–746.
2006.PubMed/NCBI
|
|
21
|
Cerullo V, Koski A, Vähä-Koskela M and
Hemminki A: Chapter eight - Oncolytic adenoviruses for cancer
immunotherapy: Data from mice, hamsters, and humans. Adv Cancer
Res. 115:265–318. 2012. View Article : Google Scholar
|
|
22
|
Bourke MG, Salwa S, Harrington KJ,
Kucharczyk MJ, Forde PF, de Kruijf M, Soden D, Tangney M, Collins
JK and O'Sullivan GC: The emerging role of viruses in the treatment
of solid tumours. Cancer Treat Rev. 37:618–632. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hawkins LK, Lemoine NR and Kirn D:
Oncolytic biotherapy: A novel therapeutic plafform. Lancet Oncol.
3:17–26. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Logan CY and Nusse R: The Wnt signaling
pathway in development and disease. Annu Rev Cell Dev Biol.
20:781–810. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Medema RH, Kops GJ, Bos JL and Burgering
BM: AFX-like Forkhead transcription factors mediate cell-cycle
regulation by Ras and PKB through p27kip1. Nature.
404:782–787. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao X, Gan L, Pan H, Kan D, Majeski M,
Adam SA and Unterman TG: Multiple elements regulate
nuclear/cytoplasmic shuttling of FOXO1: Characterization of
phosphorylation- and 14-3-3-dependent and -independent mechanisms.
Biochem J. 378:839–849. 2004. View Article : Google Scholar
|
|
28
|
Downward J: PI 3-kinase, Akt and cell
survival. Semin Cell Dev Biol. 15:177–182. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Czabotar PE, Lessene G, Strasser A and
Adams JM: Control of apoptosis by the BCL-2 protein family:
Implications for physiology and therapy. Nat Rev Mol Cell Biol.
15:49–63. 2014. View
Article : Google Scholar
|
|
30
|
Zheng JH, Viacava Follis A, Kriwacki RW
and Moldoveanu T: Discoveries and controversies in BCL-2
protein-mediated apoptosis. FEBS J. 283:2690–2700. 2016. View Article : Google Scholar
|
|
31
|
Um HD: Bcl-2 family proteins as regulators
of cancer cell invasion and metastasis: A review focusing on
mitochondrial respiration and reactive oxygen species. Oncotarget.
7:5193–5203. 2016.
|
|
32
|
Galluzzi L, López-Soto A, Kumar S and
Kroemer G: Caspases connect cell-death signaling to organismal
homeostasis. Immunity. 44:221–231. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Man SM and Kanneganti TD: Converging roles
of caspases in inflammasome activation, cell death and innate
immunity. Nat Rev Immunol. 16:7–21. 2016. View Article : Google Scholar :
|
|
34
|
Horibata S, Vo TV, Subramanian V, Thompson
PR and Coonrod SA: Utilization of the soft agar colony formation
assay to identify inhibitors of tumorigenicity in breast cancer
cells. J Vis Exp. e527272015.PubMed/NCBI
|
|
35
|
Nemunaitis J, Cunningham C, Buchanan A,
Blackburn A, Edelman G, Maples P, Netto G, Tong A, Randlev B, Olson
S, et al: Intravenous infusion of a replication-selective
adenovirus (ONYX-015) in cancer patients: Safety, feasibility and
biological activity. Gene Ther. 8:746–759. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Reid TR, Freeman S, Post L, McCormick F
and Sze DY: Effects of Onyx-015 among metastatic colorectal cancer
patients that have failed prior treatment with 5-FU/leucovorin.
Cancer Gene Ther. 12:673–681. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kemeny N, Brown K, Covey A, Kim T,
Bhargava A, Brody L, Guilfoyle B, Haag NP, Karrasch M,
Glasschroeder B, et al: Phase I, open-label, dose-escalating study
of a genetically engineered herpes simplex virus, NV1020, in
subjects with metastatic colorectal carcinoma to the liver. Hum
Gene Ther. 17:1214–1224. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fong Y, Kim T, Bhargava A, Schwartz L,
Brown K, Brody L, Covey A, Karrasch M, Getrajdman G, Mescheder A,
et al: A herpes oncolytic virus can be delivered via the
vasculature to produce biologic changes in human colorectal cancer.
Mol Ther. 17:389–394. 2009. View Article : Google Scholar
|