1. Introduction
miR-22, primitively cloned from HeLa cells, is an
evolutionarily-conserved gene located in chromosome 17p13, its cDNA
catalyzed by RNA polymerase II is ~1.3 kb and promoter TSS
(transcription start site) lacks TATA box (1). Recently, increasing numbers of
studies have confirmed that miR-22, to a large extent, determines
the destiny of many cancers, to die soon or survive, by the
complicated known or unknown mechanisms through targeting and
suppressing downstream transcription factors. Frequently, since
miR-22 in different contexts are aberrantly expressed upregulation
or downregulation in various cancers, such as prostatic cancer,
esophageal squamous cell carcinoma, breast cancer, and gastric
cancers, thus miR-22 shows different effects in these cancers
(2–5), that is, it served not only as a
tumor-suppressive miRNA, but also as an oncogenic miRNA to encumber
or aggravate cancer formation and malignant transformation
(3,6). Besides, reports also showed that
miR-22 may prominently influence cancer biological behaviors, such
as proliferation, invasion and metastasis (7,8), and
it genetically alters expression of numerous related genes
(9), which unveils the intrinsic
mechanisms of miR-22 in regulating cancer formation by means of
multi-approaches and multi-layers, indicating the central roles of
miR-22 in manipulating the occurrence and development of different
cancers. Since the underlying regulatory mechanisms of miR-22 are
complicated and remain poorly expounded, we concentrated on the
potential mechanisms and clinical applications of miR-22 in
modulating cancer progression.
2. miR-22 works as suppressor gene in tumor
malignant development
miR-22 inhibits tumor proliferation,
invasion and metastasis by accelerating cell senescence, inhibiting
energy metabolism and angiogenesis
Considering that tumor progression, including
proliferation, invasion and metastasis, are intimately involved in
tumor growth status and energy supply, miR-22 could interrupt these
processes by mediating tumor growth status and energy supply. For
instance, miR-22 induced p53 expression and concurrently targeted
SIRT1, CDK6 and Sp1 to activate pRb signaling pathway, thereby
hastening senescence, retarding cellular growth, invasion and
metastasis in cervical cancer and breast cancer, which shows
evident anticancer effects (8)
(Fig. 1).
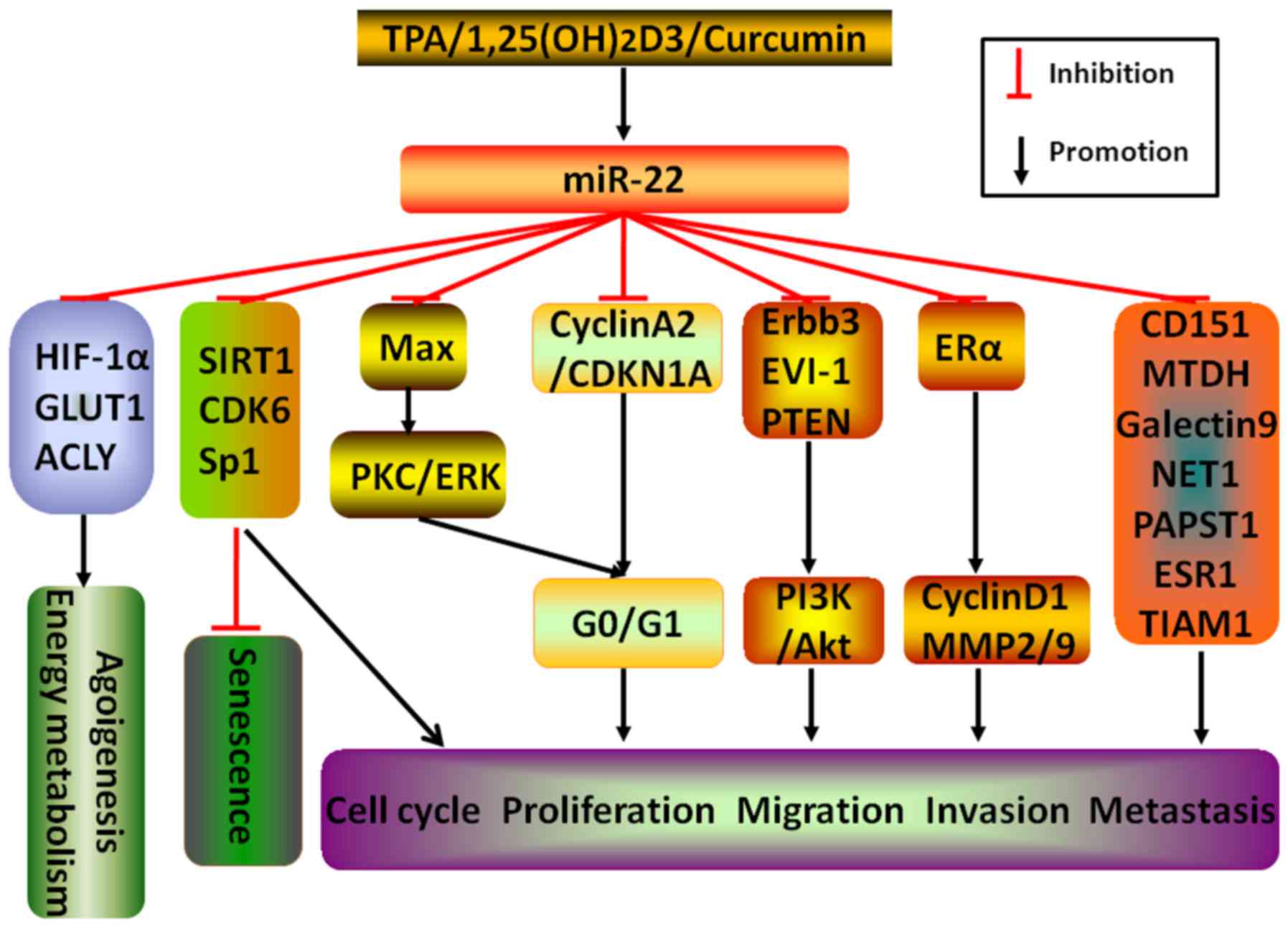 | Figure 1miR-22 inhibits tumor malignant
progressions. Different stimulators, including TPA,
1,25(OH)2D3 and curcumin, induce miR-22
expression. Through targeting various downstream related molecules,
such as HIF-1α, GLUT1, ACLY, SIRT1, CDK6, Sp1, CD151, MTDH,
Galectin-9, NET1, PAPST1, ESR1, TIAM1, Max, Cyclin A2/CDKN1A,
Erbb3, EVI-1, PTEN and ERα, miR-22 is capable of directly or
indirectly abrogate the process of tumor malignancy, including
acceleration of senescence and the abruption of angiogenesis,
energy metabolism, cell cycle, proliferation, migration, invasion
and metastasis. |
Glucose is one of the crucial energy sources, and
angiogenesis conveys energy and nutrient for rapid cancer growth
beyond the restrictions of original blood supply. Congruently,
miR-22 could cut off energy metabolism by directly silencing GLUT1
(glucose transporter protein type 1), a protein unidirectionally
transferring glucose into the cytoplasm to promote energy
metabolism, and ACLY (ATP citrate lyase), an enzyme accelerating
lipid synthesis and elevated expression in cancers, restraining
cancer proliferation, migration and invasion and inducing
apoptosis, which is negatively linked to TNM stage, metastasis,
recurrence and survival rates of breast, prostate, osteosarcoma,
lung and cervical cancers (10,11).
Moreover, miR-22 has low expression in colorectal cancer (CRC), and
increased expression of miR-22 to silence HIF-1α (hypoxia inducible
factor 1α) may severely repress VEGF (vascular endothelial growth
factor) expression to block angiogenesis, leading to the disruption
of cancer progression (12)
(Fig. 1).
Considering the above, one of the feasible methods
to effectively disrupt tumor formation may be by elevating miR-22
expression to hasten senescence, cut energy supplies and block
angiogenesis.
miR-22 inhibits tumor proliferation,
invasion and metastasis via repressing tumor cell cycle and
promoting apoptosis
It is universally known that numerous cancer cells
continuously enter proliferation and division through G0/G1
checkpoint along with reducing apoptosis, eventually causing rapid
cancer growth and enlargement in size. Recently, extensive evidence
has demonstrated that miR-22 could repress tumor malignant process
by inhibition of the cell cycle. For example, miR-22 may
post-transcriptionally target cyclin A2 and CDKN1A
(cyclin-dependent kinase inhibitor 1A) to arrest the cell cycle in
G0/G1 phage in CRC and liver cancer, respectively (13,14).
Besides, augmenting expression of miR-22 in ER (estrogen receptor)
α-positive endometrioid adenocarcinoma where miR-22 expression is
usually low could downregulate ERα expression to further decrease
the expression of cyclin D1 and member matrix metalloproteinase 2/9
(15). Furthermore, carcinogen TPA
(12-O-tetradecanoylphorbol-13-acetate)-induced miR-22 may
inversely regulate PKC/ERK pathway via dramatically downregulating
Max (a transcription factor binding to and activate c-Myc)
expression, thus resulting in G0/G1 arrest in lung, breast and
prostate cancer cells (16). These
findings confirmed that the ultimate effects of miR-22 by cell
cycle arrest in different ways may lead to the attenuation of
cancer growth and invasion and the disruption of tumor malignant
progression, indicating that miR-22 in response to different
carcinogens may serve as a tumor suppressor miRNA to mitigate or
block cancer occurrence and development.
Several emerging studies have validated that miR-22
could influence tumor proliferation by regulating hormone-related
signaling pathway. For instance, the proliferation and migration of
CRC cells and ERα-positive breast cancer cells may be attenuated by
1,25(OH)2D3 (the in vivo metabolite of
vitamin D3)-induced miR-22 and by ectopic introduction of miR-22,
respectively (17,18), insinuating that the regulation of
miR-22 participating in hormone signal transduction pathway should
not be ignored as critical underlying mechanism for tumorigenesis
and progression.
Furthermore, miR-22 may also contribute to the
cessation of cancer aggression through post-transcriptionally
regulating downstream molecules with respect to cellular migration
and adhesion, a case in point is that miR-22 in gastric cancer may
separately silence CD151, a molecule promoting cellular migration,
and MTDH (metadherin), a molecule involved in cellular adhesion, to
effectively interfere with cancer cellular proliferation and
metastatic dissemination (19,20).
Additionally to the above, miR-22 has been reported
to tightly repress cellular immune escape and proliferation and
trigger apoptosis by targeting a variety of downstream molecules,
such as Galectin-9, NET1 (neuro-epithelial transforming gene 1) and
PAPST1, in liver cancer, chronic myeloid leukemia (CML) and
medulloblastoma, respectively (21–23).
In addition, not only could curcumin-induced miR-22
post-transcriptionally degrade oncogene Erbb3 in retinoblastoma,
but also ectopic elevated miR-22 expression could directly target
Erbb3 or EVI-1, subsequently causing the repression of PI3K/Akt
cascade, the end result is inhibition of cellular proliferation,
migration, invasion and metastasis in lung cancer and breast
cancer, respectively (24–26). Moreover, miR-22-mediated silencing
of ESR1 and TIAM1 may directly repress cancer cellular migration
and invasion without exerting effects on cellular viability and
apoptosis in metastatic ovarian cancer (27).
To summarize, miR-22 shows considerable antitumor
effects via various and synthetic rather than only a single
mechanism to intervene in multi-step processes of tumorigenesis
(Fig. 1).
3. miR-22 functions as an oncogene to
promote tumor proliferation, migration and invasion
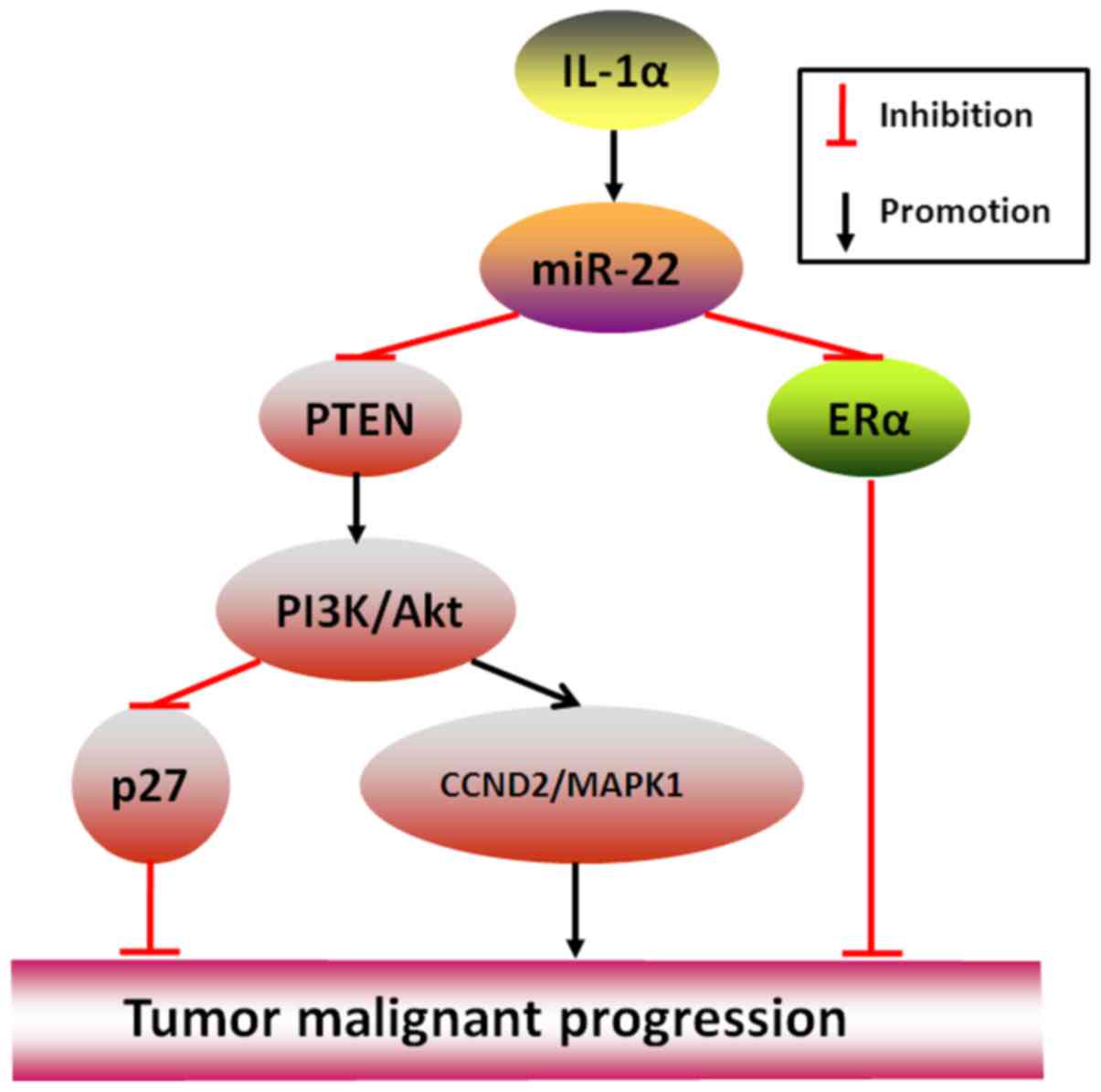
Conversely, in some cancers, miR-22 may serve as a
crucial driver to promote rather than inhibit cancer
aggressiveness. An example is that IL-1α-stimulated miR-22 may
initiate HBV-related liver cancer by suppressing ERα (28), and another example is that by
directly targeting PTEN (phosphatase and tensin homolog, a gene
usually regarded as tumor suppressor factor), miR-22 in clear cell
renal cell carcinoma where its expression is frequently
downregulated has been shown to abolish cancer proliferation,
migration and invasion but in prostate cancer where its expression
is high to stimulate (7,29), the most important reason may be
explained by a further well-performed study in CLL (chronic
lymphocytic leukemia) that miR-22-targeted PTEN silencing may
spontaneously activate PI3K/AKT pathway to the downregulated
expression of p27 (-Kip1) and upregulated expression of Survivin,
CCND2 (Cyclin D2) and MAPK1 (mitogen-activated protein kinase 1),
thereby resulting in the end telling effects of tumor formation
(30) (Fig. 2).
Collectively, adequate access to tumorigenesis of
miR-22 in some tumors is achieved by modulating tumor suppressor
molecules to initiate oncogene-related signal cascade events,
therefore, in these tumors, repressing miR-22 expression to
persistently inactivate downstream interactive oncogenic molecules
may be an effective means of preventing tumor proliferation,
migration and invasion.
4. The functions of miR-22 participating in
the feedback loops
Intriguingly, in the form of positive or negative
feedback loops, numerous miRNAs, such as miR-200, miR-203 and
miR-183/96/182, play pivotal roles in intimately inhibiting or
promoting cancer occurrence and development (31–34),
and so does miR-22. For instance, in cervical cancer and breast
cancer, miR-22 has been revealed to act as an onco-miRNA to
directly target PTEN and subsequently initiate PI3K/AKT/FoxO1
pathway. Nevertheless, the activated AKT unexpectedly induces
miR-22 expression, eventually forming a positive feedback loop and
continuously simulate PI3K/AKT/FoxO1 cascade to promote tumor
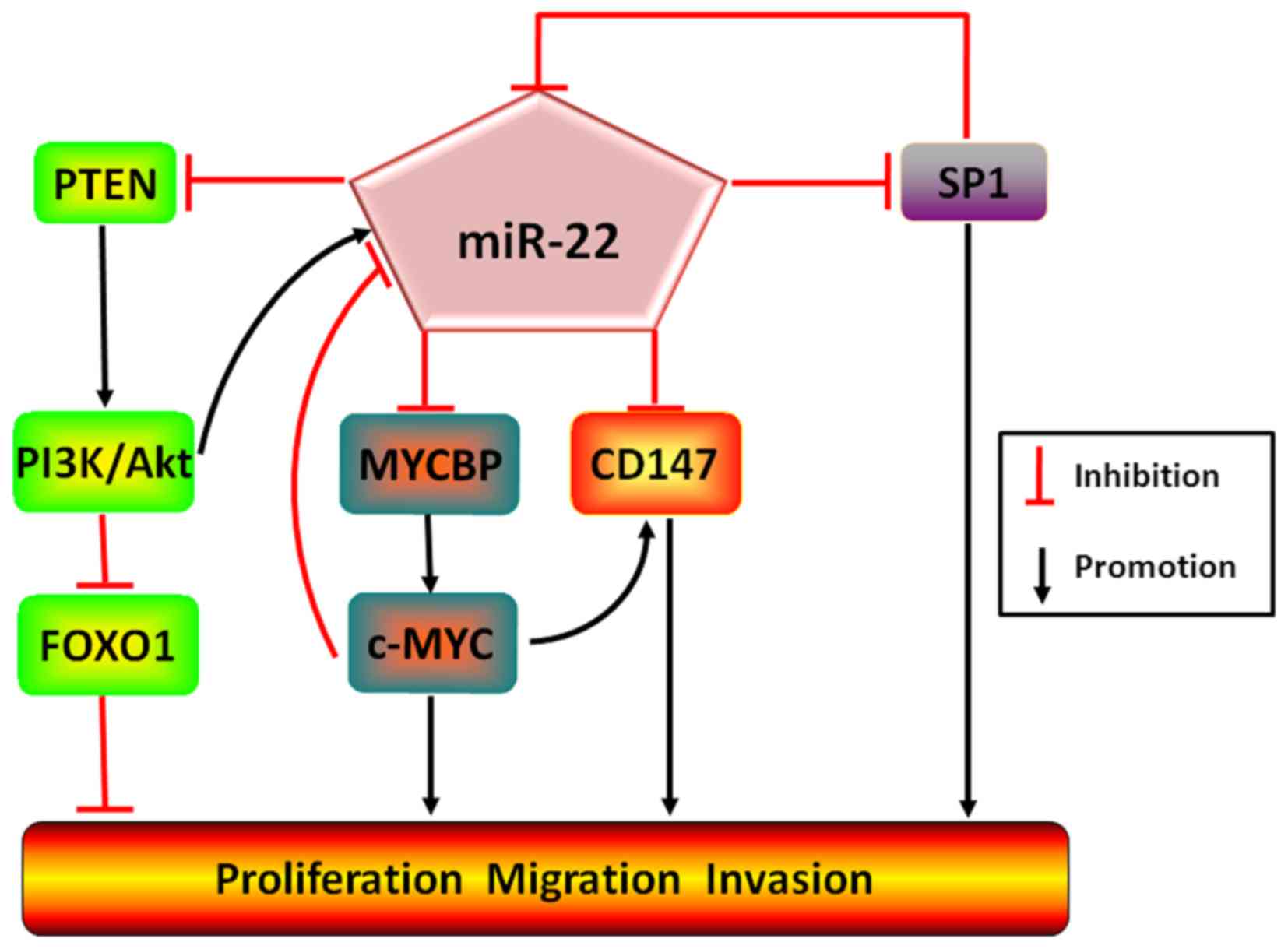
malignant transformation (6). In
addition, binding of MYCBP (c-Myc-binding protein) to inactive
c-Myc which is capable of repressing miR-22 expression, miR-22
forms a strongly positive feedback loop in favor of the inhibition
of breast cancer growth (35).
Moreover, additional experimental results have hinted that besides
straightly degrading transcription factor SP1 to retard the
migration and invasion of gastric cancer (36), miR-22 may as well concurrently
target and inactive CD147 (an inducible factor of extracellular
matrix metalloproteinase) and SP1, but both SP1 and c-Myc are
capable of binding to the promoter region of CD147 and subsequently
enhancing CD147 expression, and simultaneously of miR-22 and
inhibiting miR-22 expression, which constitutively promotes
expression of SP1 and c-Myc to upregulate CD147 expression, finally
facilitating the proliferation, migration, invasion and metastasis
of breast cancer. Oppositely, the reverse could be observed as
miR-22 expression was increased (37) (Fig.
3).
Taking the data collectively, miR-22 undertakes the
kernel role in controlling the proliferation, migration, invasion
and metastasis of different cancers by closely intertwining with
multiple tumor suppressor genes or oncogenes of upstream or
downstream molecules to form positive or negative feedback loops.
However, since miR-22 has dual (inhibitory or promoting) functions
in different cancers, especially breast cancer, in different
experiments, it is therefore necessary to further elucidate the
underlying mechanisms of miR-22 in regulating feedback loops,
correspondingly augmenting or reducing miR-22 expression in
different cancers, particularly breast cancer, will be maximally
instrumental for amplifying its anticancer effects or restricting
its accelerated effects.
5. miR-22 plays a critical role in EMT
process in cancer
Surprisingly, increasing findings have documented a
fascinating and usually ignored mechanism of miR-22 with reference
to the regulation of EMT, a process expediting cancer invasion and
metastasis and shifting cells from an epithelial status to a
mesenchymal status accompanied by morphological loss of
cohesiveness and an increased motility and genetically the
downregulated expression of epithelial adhesive molecules,
including E-cadherin and ZO-1, and upregulated expression of
mesenchymal molecules, including Zeb1/2, snail1/2, Vimentin, Twist
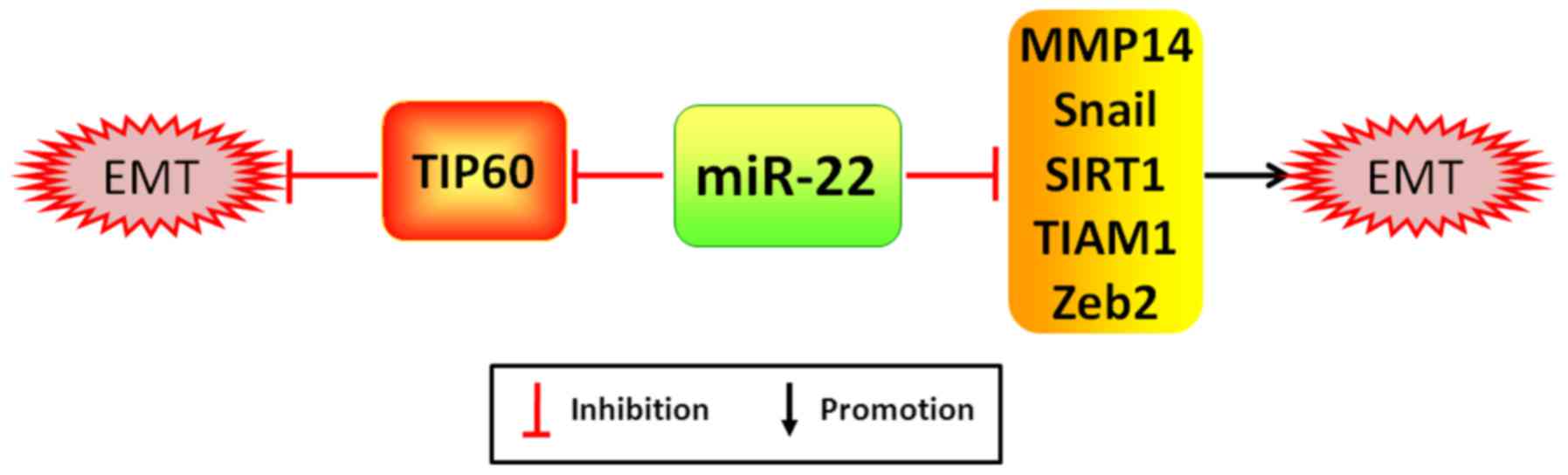
(38). As examples, miR-22 is
capable of promoting cancer proliferation, migration and incursion
by potently silencing acetylase TIP60, a gene inhibiting cancer
formation, which is significantly correlated with the worse
prognosis of patients with breast cancer (39). Notwithstanding, miR-22 has been
reported to markedly attenuate EMT process and cancer distant
metastasis by directly targeting TIAM1 (T-cell lymphoma invasion
and metastasis 1), a gene augmenting expression of MMP2/9 (member
matrix metalloproteinase 2/9) to exacerbate cancer invasion and
metastasis, MMP14 (member matrix metalloproteinase 14), Snail and
SIRT1 in CRC, gastric cancer and RCC, respectively (40–42)
(Fig. 4). Moreover, the
downexpression of miR-22 in folate deficiency HCC cells may lead to
the upexpression of its target gene Zeb2, which may be related to
the initiation of EMT process (43).
Given miR-22 could directly target either
EMT-associated tumor suppressors or oncogenes to induce or
debilitate EMT progression and metastasis, sustained and targeted
upregulation or downregulation of miR-22 in various cancer types
may forcefully cease the EMT process and distant metastasis,
thereby displaying optimal therapeutic effects on patients with
malignant cancers.
6. Molecular regulatory mechanisms of miR-22
at the genetic level in regulating tumorigenesis and malignant
transformation
Strikingly, accumulating findings have revealed the
link between epigenetic abnormalities and miR-22 (Fig. 5). The gene CNAs and SNPs may
significantly impact on the modulation of miR-22 to different
cancers. As an example, 2/21 cases of acute lymphoblastic leukemia
patients have miR-22 copy number deletions at 17p13.3 (44), and in cervical cancer, the failed
binding of miR-22 to rs11064 variant GG allelic genotype of TNFAIP8
(tumor necrosis factor-α-induced protein 8, a target of miR-22)
will distinctively result in the overexpression of TNFAIP8 and
subsequently against apoptosis and facilitate unimpeded tumor
formation, which is highly pertinent to platinum resistance
(45), evincing that the persons
suffering copy number deletions of miR-22 gene or SNP alterations
of miR-22 target genes may be much more vulnerable to malignant
tumors than the normal ones.
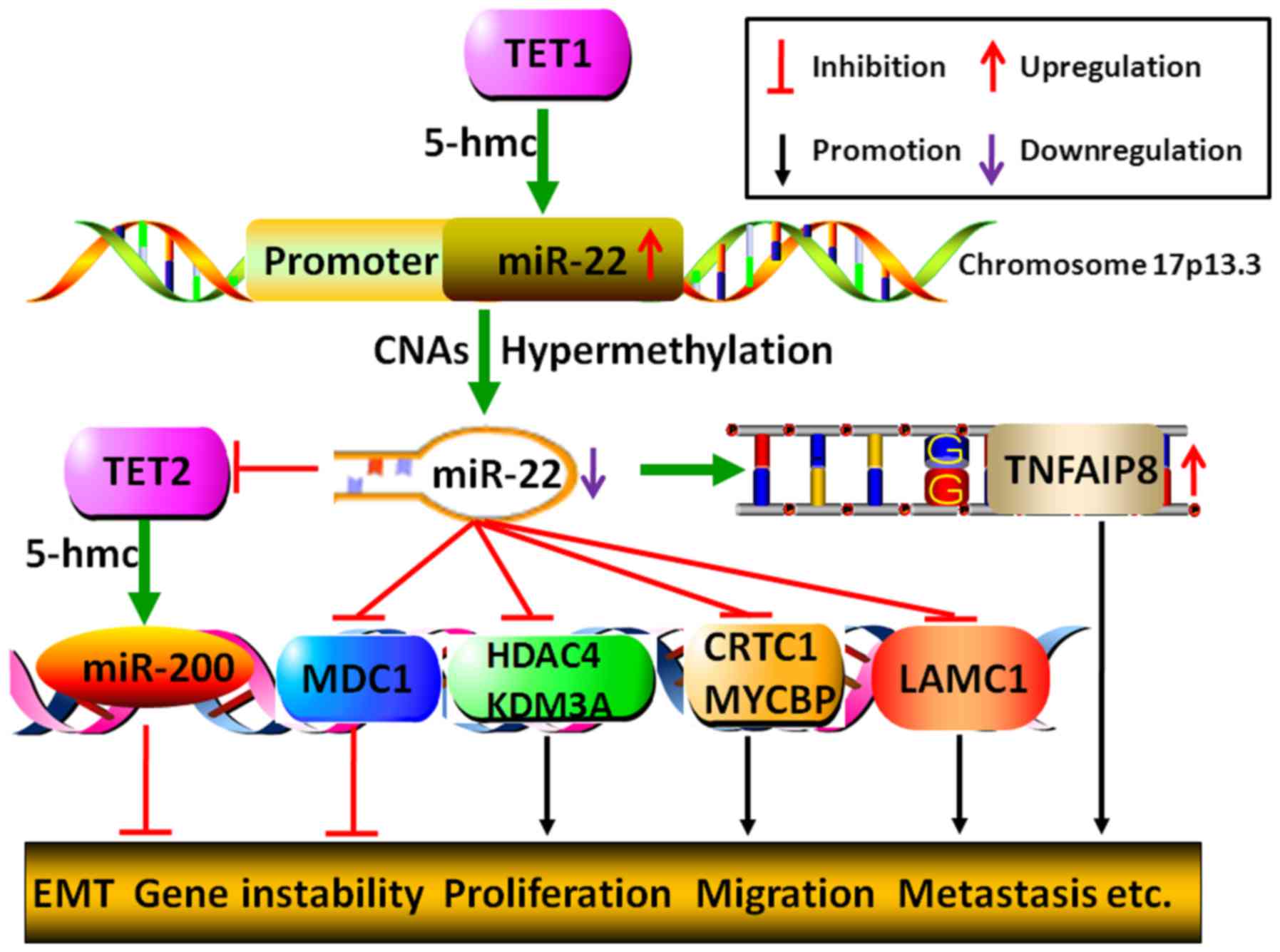 | Figure 5Genetical or epigenetical mechanisms
by which miR-22 regulates tumor malignant progression. miR-22 gene
5-hmc catalyzed by TET1 may lead to its increased expression,
whereas, either CNAs or hypermethylation of miR-22 promoter may
bring about its decreased expression. Moreover, miR-22 could
restrain miR-200 gene 5-hmc to initiate EMT process by directly
degrading TET2. Additionally, miR-22 may fail to inhibit cancer
process due to its target gene TNFAIP8 SNPs-GG allelic genotype,
which results in the upregulation of TNFAIP8. Furthermore, by
modulating histone acetylation, DNA methylation and gene
repair-related molecules, MDC, HDAC4, KDM3A, CRTC1, MYCBP and
LAMC1, miR-22 could either inhibit or promote tumor malignant
transformation, including gene instability, proliferation,
migration, invasion and metastasis. |
Growing number of mechanistic experiments documented
that miR-22 may control cancer proliferation and growth through
epigenetically modulating histone acetylation, DNA methylation and
gene repair. In hepatocellular carcinoma and Ewing sarcoma,
externally induced expression of miR-22 has been shown to repress
cellular proliferation and tumorigenesis via post-transcriptionally
silencing histone deacetylase HDAC4 (histone deacetylase 4) and
KDM3A (lysine (K)-specific demethylase 3A), respectively (46,47).
Generally, promoter hypermethylation implies the silence of gene
expressions, inversely, promoter demethylation hints at promotion
of gene expressions. In androgen receptor-positive cancer cells of
prostate cancer, miR-22 and miR-29 promoters are frequently
hypermethylated which makes their expression low. Whereas, cancer
cells will obviously undergo restrained migration and stimulated
apoptosis by upregulated miR-22 and miR-29 to separately target
LAMC1 (laminin γ1, a gene promoting cell migration) and MCL1
(myeloid cell leukemia 1, a gene against apoptosis) (9). Additionally, AKT-induced miR-22 in
CRC HCT116 has been reported to directly target MDC1 (mediator of
DNA damage checkpoint 1), giving rise to the aberrant repair of
damaged DNA and instability of genes, thus, causing probable
susceptibility to aging and cancer (48).
Extremely absorbing, miR-22 has been shown to
participate in tumorigenesis via regulating 5-hmC
(5-hydroxymethylcytosine), usually called the six base and
originating from 5-mC (5-methylcytosine) catalyzed by TET
(ten-eleven translocation) enzymes TET1, TET2 and TET3. 5-hmC of
transcription factor binding sites commonly initiates gene
expression, and the down-expression of TET often fails to activate
gene transcription owing to the low level of 5-hmC (49,50).
For instance, analysis of patients with refractory cytopenia of
childhood revealed that the high expression of miR-22 has a closely
converse relationship with the low expression of TET and 5-hmC
(51). Furthermore, recent
investigations have confirmed in miR-22 transgenic mice that by
directly targeting TET2, not only is miR-22 against methylation of
tumor suppressor miR-200 promoter, causing the downregulation of
miR-200 and the initiation of EMT process and distant metastasis
for breast cancer stem cells, which is closely correlated with the
poor prognosis of patients (52),
but also it contributes to the low levels of other 5-hmC of
downstream genes, leading to self-renewal and malignant
transformation of blood stem cells in these mice, which eventually
undergo MDS and malignant blood diseases. Instead, miR-22
depression may lead to the inhibitory proliferation of leukemia
cells in mice and human (53).
Nonetheless, the latest investigation hinted that in AML (acute
myeloid leukemia) cells, the expression of TET2 has a positive
rather than negative relation with the expression of miR-22, but
upregulated TET1 has a negative association with the expression of
miR-22. Although TET1 is conducive to the hypomethylation of miR-22
promoter, it, strangely, does not induce miR-22 expression, the
primary reason may be that both copy number deletions of miR-22
gene and binding of upstream cofactors GFI1/EZH2/SIN3A and TET1,
especially TET1, to the region of miR-22 promoter result in the
inhibition of miR-22 expression. However, restoration of miR-22 may
markedly abate the CREB and MYC pathways by directly targeting
CRTC1 (CREB-regulated transcription coactivator 1) and MYCBP,
bringing about the inhibition of tumor formation and malignant
transformation (54). Briefly,
these data suggest that miR-22 may directly be controlled by
hydroxymethylation, but it does not necessarily mean that the
hypomethylation of miR-22 promoter leads to the upregulation of
miR-22, to some extent, many other factors, including copy number
deletions of miR-22 gene and the direct inhibition of miR-22
promoter by hydroxymethylation-related genes, are prominently
correlated with suppressing miR-22 expression, miR-22 may turn off
the expression of downstream suppressor genes with respect to the
hydroxy-methylation-related pathways, thereby playing an oncogenic
role in the process of tumor initiation and malignant
transformation.
Taken together, at the genetic level, on the one
hand, copy number deletions or hypermethylation of miR-22 gene may
result in its dysfunction to facilitate tumor uncontrolled
malignant progression, indicating that miR-22 likely serves as a
tumor suppressor to design epigenetic drugs in some cancers. On the
other hand, miR-22 could bring about genomic instability to promote
tumor malignant transformation via manipulating epigenetic
modification to turn off the expression of tumor suppressors,
suggesting that miR-22 probably functions as an internal engine for
some cancers. As so many are underlying mechanisms involved in
miR-22 participating in cancer formation that it is necessary to
genetically explore and unveil the intricate mechanisms of miR-22
for epigenetic therapy in different cancers.
7. miR-22 influences cancer progression via
collaborating with other miRNAs
Tantalizingly, increasing evidence has recently
corroborated that the combined effects of miR-22 complexing with
many other miRNAs may play a crucial role in controlling cancer
differentiation, proliferation, migration and invasion in some
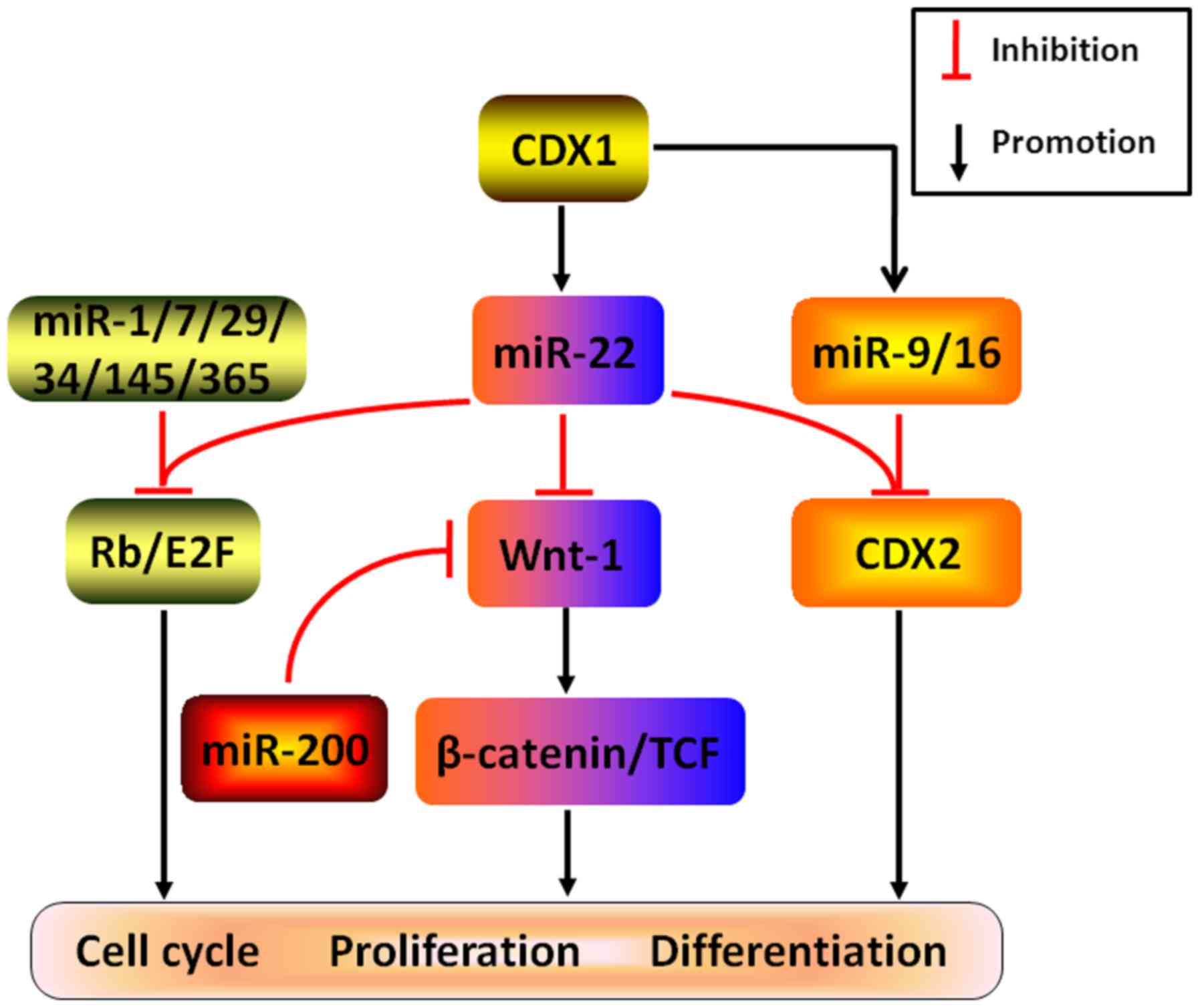
cancers (Fig. 6). Considering
several examples, experimental validation demonstrated that both
CDX1-induced miR-22, miR-9 and miR-16 may directly target CDX2 to
abrogate cell differentiation in CRC (55). In addition, cooperating with
several other miRNAs, such as miR-1, Let-7, miR-29, miR-34,
miR-145, and miR-365, miR-22 was found to attenuate Rb/E2F
signaling pathway by post-transcriptionally silencing Rb to
remarkably arrest cell cycle and DNA replication, ultimately
resulting in the restriction of cancer growth (56). Likewise, by coordinating with tumor
suppressor gene miR-200 and directly targeting Wnt-1, an oncogene
positively modifying Wnt/β-catenin pathway, miR-22 is capable of
repressing the expression of β-catenin and TCF to potently restrain
cancer colony-forming, which may be responsible for the anticancer
effects in gastric cancer (57).
Overall, these findings indicate that it is perhaps
a feasible way to force expression of the miR-22 combining with
many other miRNAs regarding the repression of carcinoma-related
pathways to strengthen anticancer effects in some cancers, opening
a new window for unearthing the intrinsic underlying mechanisms of
miR-22 in modulating the cancer progression.
8. miR-22 functions as a sensitizer in
cancer treatments
Clinically, it is very common that the same
chemotherapeutic drugs show extraordinarily different therapeutic
effects on different cancers even on the same cancers owing to
chemo-resistance which is one of the most imperative reasons for
the failure of treatment, the toughest challenges and seemingly
insurmountable obstacles. Utterly inspiring is the good news that
miR-22 in several cancers, to some extent, displays the capability
of increasing chemosensitivity to different anticarcinogens by
directly targeting and activating or inactivating various
downstream genes (Fig. 7).
Findings from recent experiments have confirmed that p53-wild
rather than p53-mutant type CRC cells display chemoresensitivity to
paclitaxel partly in that enhanced miR-22 binding to and activating
PTEN can counteract a cascade of PI3K/Akt events and stimulate
apoptosis, leading to resensitization to paclitaxel (58). Further research revealed that via
binding to the 5-upstream regions and intron regions of C17orf91, a
gene in which miR-22 locates, adriamycin-triggered p53 in
p53-wild-type CRC cells is capable of augmenting miR-22 expression
to target downstream molecule p21 to subsequently cease the cell
cycle and induce cell apoptosis, ultimately resulting in
intensifying the chemosensitivity to adriamycin (59). In addition, there were several
interesting and important mechanisms, including autophagy pathway
and apoptosis pathway, with respect to miR-22 in enhancing
sensitivity to several chemotherapeutics. Through separately
targeting BTG1 (B-cell translocation gene 1) and HMGB1
(high-mobility group box 1), miR-22 could inhibit autophagy and
enhance apoptosis against proliferation, migration and invasion,
eventually contributing to the reverse chemo-resistance to 5-FU,
cisplatin and doxorubicin in CRC and osteosarcoma (60,61).
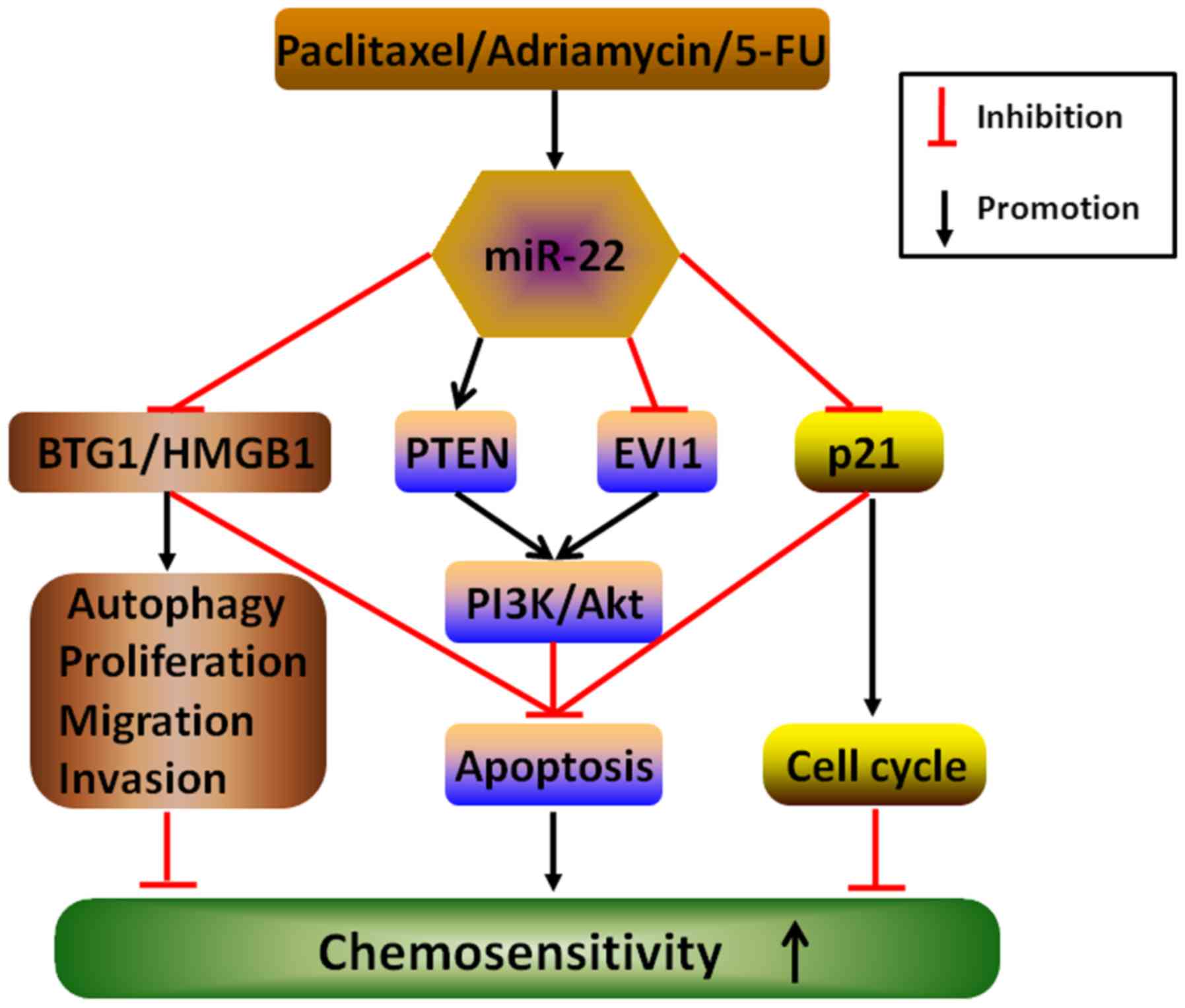 | Figure 7Underlying mechanisms by which miR-22
enhances chemosensitivity to therapeutic drugs in cancers. The
elevated expression of miR-22 stimulated by anticancer drugs,
including paclitaxel, adriamycin and 5-FU, may directly target and
activate or inactivate multiple downstream molecules, such as BTG1,
HMGB1, PTEN, EVI1 and p21, to counteract autophagy, proliferation,
migration and invasion and cell cycle and induce apoptosis, thereby
enhancing chemo-sensitivity in different cancers. |
Moreover, miR-22 may profoundly enhance
chemosensitivity to anticancer drugs by intimately working with
many other miRNAs. In clear cell ovarian cancer, PI3K/Akt/mTOR
pathway was predominately repressed due to the combined effects
through directly targeting both FGFR3 (fibroblast growth factor
receptor 3) and mTOR by miR-100 and simultaneously targeting EVI1
by miR-22, allowing for the subsequent inhibition of proliferation,
growth and survival, which, in the end, increases chemosensitivity
to anticancer drug everolimus (62).
Taken together, these data hint that miR-22 may
notably enhance chemosensitivity or reverse chemoresistance to
corresponding cancer drugs by a variety of underlying mechanisms,
and that multiple anticancer drugs with predominately therapeutic
effects may further maximize their advantages or minimize
disadvantages via augmenting miR-22 expression to exert the
inhibition of cancer proliferation, migration and invasion.
Therefore, to obtain the optimal treatment effects for miR-22 as an
adjuvant therapeutic intervention in the near future, illuminating
the potential mechanisms of miR-22 in regulating chemosensitivity
or chemoresistance will be beneficial for patients with various
cancers in precision medicine.
9. miR-22 is an independent biomarker for
cancer diagnosis, surveillance and prognosis
Early diagnosis is one of the primary challenges of
cancers and would be beneficial for effectively prolonging the
survival time and improving the patient life quality. As miR-22
expression is different in different cancers and plays a central
role in cancer cell proliferation, migration, invasion and
metastasis, it is reflected in cancer diagnosis, monitoring, and
prognosis. For instance, the low expression of miR-22 in the serum
of patients with ICC (intrahepatic cholangio-carcinoma) and
hepatocellular cancer with hepatitis C virus and the malignant
pleural effusion of patients with lung adenocarcinoma may well be
promising as an independent early diagnostic biomarker for these
cancers (63–65). On the contrary, the sustained high
expression of miR-22 in the serum of patients with esophageal
squamous cell carcinoma, pancreatic cancer and metastatic prostatic
cancer may well be a reliable serum biomarker for cancer diagnosis,
along with the desirable diagnosis of sensitivity and specificity
(2,66,67)
(Table I).
 | Table IClinical applications of miR-22 in
tumor diagnosis, surveillance and prognosis. |
Table I
Clinical applications of miR-22 in
tumor diagnosis, surveillance and prognosis.
| Tumor | Body
fluids/tissues | miR-22 levels | Diagnosis | Treatment
response/prognosis | Refs. |
|---|
| ICC | Serum | ↓ | √ | N | (63) |
| Hepatocellular
carcinoma | Serum | ↓ | √ | Poor | (46,64) |
| Lung
adenocarcinoma | Pleural
effusion | ↓ | √ | N | (65) |
| ESCC | Serum | ↑ | √ | N | (66) |
| Pancreatic
cancer | Serum | ↑ | √ | N | (67) |
| Prostatic
cancer | Serum | ↑ | √ | N | (2) |
| NSCLC | Serum | ↑ | N | Poor | (68) |
| Gastric cancer | Cancer tissue | ↓ | N | Poor | (69) |
| CRC | Cancer tissue | ↓ | N | Poor | (70) |
Additionally, the alteration of miR-22 expression in
body fluids may, to a certain extent, directly mirror the
therapeutic effects. The elevated expression of miR-22 in the serum
of NSCLC (non-small cell lung cancer) is conspicuously associated
with the cancer aggression and the unresponsiveness to the
chemotherapeutic drug pemetrexed (68), implying that miR-22 may be a
telling serum predictor for monitoring the chemotherapeutic effects
in NSCLC. Instead, the reduced expression of miR-22 frequently
portend poor prognosis in patients with certain cancers. An example
of this is that the downregulated miR-22 expression shows shorter
overall survival time and is more likely to have a tendency of
distant metastasis in cancer tissues than normal adjacent tissues
in patients with hepatocellular carcinoma, gastric cancer and CRC
(46,69,70)
(Table I).
Collectively, as for some cancers, monitoring the
fluctuation of miR-22 expression in the serum or body fluids or the
cancer tissues may be of great latent significance for cancer
diagnosis, particularly early diagnosis, assessing therapeutic
effects and prognosis, therefore, it is very promising that miR-22
may be eventually utilized as a predictive cancer biomarker for
early accurate diagnosis, monitoring treatment responses in
real-time and prognosis of outcomes and as an effective strategy
for supplementary or even principal treatment in special
cancers.
10. Conclusions and prospects
Numerous studies have revealed that miR-22 functions
as either a tumor suppressor miRNA or an onco-miRNA to inhibit or
promote tumor formation and malignant transformation from genetic
to post-transcription level via intricate mechanisms, in which
miR-22 could stimulate or turn off different cascades of events
concerning pathways by directly or indirectly interacting with
upstream or downstream molecules/pathways, either synergistically
or antagonistically. Also, as the formation of miR-22-related
positive or negative feedback loops extraordinarily amplify the
inhibitory or promoting effects of miR-22 in a variety of cancers,
therefore, miR-22 and countless related molecules constitute
complex signaling networks where miR-22 is at the core of events
(Table II). Indicating that
miR-22 may serve as a hopeful therapeutic target for precision
treatments in diverse cancers to inhibit proliferation, migration,
invasion and metastasis, thus weakening or reversing
chemoresistance to anticancer drugs. Besides, miR-22 expression in
cancer cells and body fluids may fluctuate in different cancers and
different growth stages in the same cancer, which makes it possible
for miR-22 to be a potential and complementary or even independent
biomarker in cancer diagnosis, monitoring treatment effects and
prognosis.
 | Table IIMolecular mechanisms of miR-22
regulating tumor progression. |
Table II
Molecular mechanisms of miR-22
regulating tumor progression.
| Tumor | miR-22 level | Target genes | Pathways | Effect | Refs. |
|---|
| GC | ↓ | MMP14 Snail | EMT | − | (19,20,36,41,57) |
| | MTDH | Wnt/β-catenin/ | | |
| | CD151 Wnt-1 | TCF | | |
| | SP1 | | | |
| PC | ↑ or ↓ | PTEN Max | PI3K/AKT | + or − | (7,9,11,16) |
| | LAMC1 | MAPK/ERK | | |
| | ACLY | PKC/ERK | | |
| BC | N or ↓ | TIP60 GLUT1 | EMT | + or − | (6,8,10,16,26,35,37,39,52) |
| | CD147 SIRT1 | pRB pathway | | |
| | CDK6 Sp1 | PI3K/Akt | | |
| | Erbb3 EVI-1 | ER pathway | | |
| | ERα MYCBP | PKC/ERK | | |
| | PTEN Max | | | |
| | TET2 | | | |
| RCC | ↓ | SIRT1 PTEN | Apoptosis | − | (29,42) |
| | | EMT | | |
| Liver cancer | ↓ | Galectin-9 | Apoptosis | − | (13,21,28,43) |
| | CCNA2 | Cell cycle | | |
| | CDKN1A | | | |
| | ERα HDAC4 | | | |
| | Zeb2 | | | |
| CRC | ↓ | MDC1 | PI3K/AKT | + or − | (12,13,40,48,55,58,59,60) |
| | CCNA2 BTG1 | Cell cycle | | |
| | TIAM1 CDX2 | Autophagy | | |
| | HIF-1α p21 | Apoptosis | | |
| | PTEN | Hypoxia | | |
| Lung cancer | ↓ or ↑ | Erbb3 Max | PKC/ERK | − | (11,16,25) |
| | ACLY | | | |
| AML | ↓ | CRTC1 | CREB/MYC | – | (35,54) |
| | MYCBP | | | |
| CLL | ↑ | PTEN | PI3K/AKT/FOXO1 | + | (30) |
| MDS | ↑ | TET2 | 5-hmc | + | (53) |
| CML | | NET1 | Cell cycle | − | (22) |
| Osteosarcoma | ↓ | HMGB1 | Autophagy | − | (11,61) |
| | ACLY | | | |
| EEC | ↓ | ERα | ER pathway | − | (15) |
|
Medulloblastoma | ↓ | PAPST1 | Cell
proliferation | − | (23) |
| | | Apoptosis | | |
| Ewing sarcoma | ↓ | KDM3A | Histone
demethylation | − | (47) |
| Cervical
cancer | ↓ or ↑ | TNFAIP8 | Apoptosis | + or − | (6,8,11,45) |
| | SIRT1 CDK6 | pRB pathway | | |
| | Sp1 PTEN | PI3K/AKT | | |
| | ACLY | | | |
| Retinoblastoma | ↓ | Erbb3 | Cell
proliferation | − | (24) |
| Ovarian cancer | ↓ | TIAM1 ESR1
EVI1 | PI3K/Akt/mTOR | − | (27,62) |
However, there will be still many problems to be
settled in the future due to the intricate and intrinsic mechanisms
of miR-22 regulating cancer formation. For instance, the same
cancer has different expression of miR-22 at the different growth
steps, in which step does miR-22 play a primary or secondary role?
Or inhibitory or promoting role or both? How does it interact with
many other molecules? In particular, it is seemly paradoxical that
miR-22 may show distinctively opposite effects (inhibition or
promotion) on the biological behavior of different cancers by
post-transcriptionally targeting the same transcription factors,
such as PTEN. Therefore, further excavating the underlying
mechanisms of miR-22 with many other molecules in manipulating
tumor malignant progression may, to some degree, be very valuable
for cancer diagnosis, treatment, and prognosis in precision
medicine in the coming years.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (81672730), the Zhejiang
Provincial Natural Science Foundation (LY15H160067), the Jiaxing
Municipal Science and Technology Project (2015AY23012, 2016AY23043)
and Medical Key Discipline of Jiaxing (Pathology, 04-Z-01).
References
|
1
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Knyazev EN, Samatov TR, Fomicheva KA,
Nyushko KM, Alekseev BY and Shkurnikov MY: MicroRNA hsa-miR-4674 in
hemolysis-free blood plasma is associated with distant metastases
of prostatic cancer. Bull Exp Biol Med. 161:112–115. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang C, Ning S, Li Z, Qin X and Xu W:
miR-22 is down-regulated in esophageal squamous cell carcinoma and
inhibits cell migration and invasion. Cancer Cell Int. 14:1382014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Damavandi Z, Torkashvand S, Vasei M,
Soltani BM, Tavallaei M and Mowla SJ: Aberrant expression of breast
development-related microRNAs, miR-22, miR-132, and miR-212, in
breast tumor tissues. J Breast Cancer. 19:148–155. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang M, Jiang N, Cao QW and Sun Q: EDD1
predicts prognosis and regulates gastric cancer growth in vitro and
in vivo via miR-22. Biol Chem. Apr 28–2016.Epub ahead of print.
View Article : Google Scholar
|
|
6
|
Bar N and Dikstein R: miR-22 forms a
regulatory loop in PTEN/AKT pathway and modulates signaling
kinetics. PLoS One. 5:e108592010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Budd WT, Seashols-Williams SJ, Clark GC,
Weaver D, Calvert V, Petricoin E, Dragoescu EA, O'Hanlon K and
Zehner ZE: Dual action of miR-125b as a tumor suppressor and
oncomiR-22 promotes prostate cancer tumorigenesis. PLoS One.
10:e01423732015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu D, Takeshita F, Hino Y, Fukunaga S,
Kudo Y, Tamaki A, Matsunaga J, Takahashi RU, Takata T, Shimamoto A,
et al: miR-22 represses cancer progression by inducing cellular
senescence. J Cell Biol. 193:409–424. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pasqualini L, Bu H, Puhr M, Narisu N,
Rainer J, Schlick B, Schäfer G, Angelova M, Trajanoski Z, Börno ST,
et al: miR-22 and miR-29a are members of the androgen receptor
cistrome modulating LAMC1 and Mcl-1 in prostate cancer. Mol
Endocrinol. 29:1037–1054. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen B, Tang H, Liu X, Liu P, Yang L, Xie
X, Ye F, Song C, Xie X and Wei W: miR-22 as a prognostic factor
targets glucose transporter protein type 1 in breast cancer. Cancer
Lett. 356:410–417. 2015. View Article : Google Scholar
|
|
11
|
Xin M, Qiao Z, Li J, Liu J, Song S, Zhao
X, Miao P, Tang T, Wang L, Liu W, et al: miR-22 inhibits tumor
growth and metastasis by targeting ATP citrate lyase: Evidence in
osteosarcoma, prostate cancer, cervical cancer and lung cancer.
Oncotarget. 7:44252–44265. 2016.PubMed/NCBI
|
|
12
|
Yamakuchi M, Yagi S, Ito T and Lowenstein
CJ: MicroRNA-22 regulates hypoxia signaling in colon cancer cells.
PLoS One. 6:e202912011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang F, Hu Y, Liu HX and Wan YJ:
miR-22-silenced cyclin A expression in colon and liver cancer cells
is regulated by bile acid receptor. J Biol Chem. 290:6507–6515.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi C and Xu X: MicroRNA-22 is
down-regulated in hepatitis B virus-related hepatocellular
carcinoma. Biomed Pharmacother. 67:375–380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li S, Hu R, Wang C, Guo F, Li X and Wang
S: miR-22 inhibits proliferation and invasion in estrogen receptor
α-positive endometrial endometrioid carcinomas cells. Mol Med Rep.
9:2393–2399. 2014.PubMed/NCBI
|
|
16
|
Ting Y, Medina DJ, Strair RK and Schaar
DG: Differentiation-associated miR-22 represses Max expression and
inhibits cell cycle progression. Biochem Biophys Res Commun.
394:606–611. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Alvarez-Díaz S, Valle N, Ferrer-Mayorga G,
Lombardía L, Herrera M, Domínguez O, Segura MF, Bonilla F, Hernando
E and Muñoz A: MicroRNA-22 is induced by vitamin D and contributes
to its antiproliferative, antimigratory and gene regulatory effects
in colon cancer cells. Hum Mol Genet. 21:2157–2165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pandey DP and Picard D: miR-22 inhibits
estrogen signaling by directly targeting the estrogen receptor
alpha mRNA. Mol Cell Biol. 29:3783–3790. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang X, Yu H, Lu X, Zhang P, Wang M and Hu
Y: miR-22 suppresses the proliferation and invasion of gastric
cancer cells by inhibiting CD151. Biochem Biophys Res Commun.
445:175–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang Y, Liu X, Su B, Zhang Z, Zeng X, Lei
Y, Shan J, Wu Y, Tang H and Su Q: microRNA-22 acts as a metastasis
suppressor by targeting metadherin in gastric cancer. Mol Med Rep.
11:454–460. 2015.
|
|
21
|
Yang Q, Jiang W, Zhuang C, Geng Z, Hou C,
Huang D, Hu L and Wang X: microRNA-22 downregulation of galectin-9
influences lymphocyte apoptosis and tumor cell proliferation in
liver cancer. Oncol Rep. 34:1771–1778. 2015.PubMed/NCBI
|
|
22
|
Ahmad HM, Muiwo P, Ramachandran SS, Pandey
P, Gupta YK, Kumar L, Kulshreshtha R and Bhattacharya A: miR-22
regulates expression of oncogenic neuroepithelial transforming gene
1, NET1. FEBS J. 281:3904–3919. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu QF, Pan YW, Li LC, Zhou Z, Huang QL,
Pang JC, Zhu XP, Ren Y, Yang H, Ohgaki H, et al: miR-22 is
frequently downregulated in medulloblastomas and inhibits cell
proliferation via the novel target PAPST1. Brain Pathol.
24:568–583. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sreenivasan S, Thirumalai K, Danda R and
Krishnakumar S: Effect of curcumin on miRNA expression in human Y79
retinoblastoma cells. Curr Eye Res. 37:421–428. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ling B, Wang GX, Long G, Qiu JH and Hu ZL:
Tumor suppressor miR-22 suppresses lung cancer cell progression
through post-transcriptional regulation of ErbB3. J Cancer Res Clin
Oncol. 138:1355–1361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Patel JB, Appaiah HN, Burnett RM,
Bhat-Nakshatri P, Wang G, Mehta R, Badve S, Thomson MJ, Hammond S,
Steeg P, et al: Control of EVI-1 oncogene expression in metastatic
breast cancer cells through microRNA miR-22. Oncogene.
30:1290–1301. 2011. View Article : Google Scholar
|
|
27
|
Li J, Liang S, Yu H, Zhang J, Ma D and Lu
X: An inhibitory effect of miR-22 on cell migration and invasion in
ovarian cancer. Gynecol Oncol. 119:543–548. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang R, Deng L, Zhao L, Li X, Zhang F,
Xia Y, Gao Y, Wang X and Sun B: miR-22 promotes HBV-related
hepatocellular carcinoma development in males. Clin Cancer Res.
17:5593–5603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fan W, Huang J, Xiao H and Liang Z:
MicroRNA-22 is down-regulated in clear cell renal cell carcinoma,
and inhibits cell growth, migration and invasion by targeting PTEN.
Mol Med Rep. 13:4800–4806. 2016.PubMed/NCBI
|
|
30
|
Palacios F, Abreu C, Prieto D, Morande P,
Ruiz S, Fernández-Calero T, Naya H, Libisch G, Robello C, Landoni
AI, et al: Activation of the PI3K/AKT pathway by microRNA-22
results in CLL B-cell proliferation. Leukemia. 29:115–125. 2015.
View Article : Google Scholar
|
|
31
|
Tang J, Li Y, Wang J, Wen Z, Lai M and
Zhang H: Molecular mechanisms of microRNAs in regulating
epithelial-mesenchymal transitions in human cancers. Cancer Lett.
371:301–313. 2016. View Article : Google Scholar
|
|
32
|
Lu M, Jolly MK, Levine H, Onuchic JN and
Ben-Jacob E: MicroRNA-based regulation of
epithelial-hybrid-mesenchymal fate determination. Proc Natl Acad
Sci USA. 110:18144–18149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moes M, Le Béchec A, Crespo I, Laurini C,
Halavatyi A, Vetter G, Del Sol A and Friederich E: A novel network
integrating a miRNA-203/SNAI1 feedback loop which regulates
epithelial to mesenchymal transition. PLoS One. 7:e354402012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ding X, Park SI, McCauley LK and Wang CY:
Signaling between transforming growth factor β (TGF-β) and
transcription factor SNAI2 represses expression of microRNA miR-203
to promote epithelial-mesenchymal transition and tumor metastasis.
J Biol Chem. 288:10241–10253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiong J, Du Q and Liang Z:
Tumor-suppressive microRNA-22 inhibits the transcription of
E-box-containing c-Myc target genes by silencing c-Myc binding
protein. Oncogene. 29:4980–4988. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo MM, Hu LH, Wang YQ, Chen P, Huang JG,
Lu N, He JH and Liao CG: miR-22 is down-regulated in gastric
cancer, and its overexpression inhibits cell migration and invasion
via targeting transcription factor Sp1. Med Oncol. 30:5422013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kong LM, Liao CG, Zhang Y, Xu J, Li Y,
Huang W, Zhang Y, Bian H and Chen ZN: A regulatory loop involving
miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer
invasion and metastasis. Cancer Res. 74:3764–3778. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Choi JH, Hwang YP, Kim HG, Khanal T, Do
MT, Jin SW, Han HJ, Lee HS, Lee YC, Chung YC, et al: Saponins from
the roots of Platycodon grandiflorum suppresses TGFβ1-induced
epithelial-mesenchymal transition via repression of PI3K/Akt,
ERK1/2 and Smad2/3 pathway in human lung carcinoma A549 cells. Nutr
Cancer. 66:140–151. 2014. View Article : Google Scholar
|
|
39
|
Pandey AK, Zhang Y, Zhang S, Li Y,
Tucker-Kellogg G, Yang H and Jha S: TIP60-miR-22 axis as a
prognostic marker of breast cancer progression. Oncotarget.
6:41290–41306. 2015.PubMed/NCBI
|
|
40
|
Li B, Song Y, Liu TJ, Cui YB, Jiang Y, Xie
ZS and Xie SL: miRNA-22 suppresses colon cancer cell migration and
invasion by inhibiting the expression of T-cell lymphoma invasion
and metastasis 1 and matrix metalloproteinases 2 and 9. Oncol Rep.
29:1932–1938. 2013.PubMed/NCBI
|
|
41
|
Zuo QF, Cao LY, Yu T, Gong L, Wang LN,
Zhao YL, Xiao B and Zou QM: MicroRNA-22 inhibits tumor growth and
metastasis in gastric cancer by directly targeting MMP14 and Snail.
Cell Death Dis. 6:e20002015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang S, Zhang D, Yi C, Wang Y, Wang H and
Wang J: MicroRNA-22 functions as a tumor suppressor by targeting
SIRT1 in renal cell carcinoma. Oncol Rep. 35:559–567. 2016.
|
|
43
|
Su YH, Huang WC, Huang TH, Huang YJ, Sue
YK, Huynh TT, Hsiao M, Liu TZ, Wu AT and Lin CM: Folate deficient
tumor microenvironment promotes epithelial-to-mesenchymal
transition and cancer stem-like phenotypes. Oncotarget.
7:33246–33256. 2016.PubMed/NCBI
|
|
44
|
Ninomiya S, Tyybäkinoja A, Borze I, Räty
R, Saarinen-Pihkala UM, Usvasalo A, Elonen E and Knuutila S:
Integrated analysis of gene copy number, copy neutral LOH, and
microRNA profiles in adult acute lymphoblastic leukemia. Cytogenet
Genome Res. 136:246–255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shi TY, Cheng X, Yu KD, Sun MH, Shao ZM,
Wang MY, Zhu ML, He J, Li QX, Chen XJ, et al: Functional variants
in TNFAIP8 associated with cervical cancer susceptibility and
clinical outcomes. Carcinogenesis. 34:770–778. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu
S, Wu M, Pan Z and Zhou W: microRNA-22, downregulated in
hepatocellular carcinoma and correlated with prognosis, suppresses
cell proliferation and tumourigenicity. Br J Cancer. 103:1215–1220.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Parrish JK, Sechler M, Winn RA and
Jedlicka P: The histone demethylase KDM3A is a
microRNA-22-regulated tumor promoter in Ewing Sarcoma. Oncogene.
34:257–262. 2015. View Article : Google Scholar :
|
|
48
|
Lee JH, Park SJ, Jeong SY, Kim MJ, Jun S,
Lee HS, Chang IY, Lim SC, Yoon SP, Yong J, et al: MicroRNA-22
suppresses DNA repair and promotes genomic instability through
targeting of MDC1. Cancer Res. 75:1298–1310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Madzo J, Liu H, Rodriguez A, Vasanthakumar
A, Sundaravel S, Caces DB, Looney TJ, Zhang L, Lepore JB, Macrae T,
et al: Hydroxymethylation at gene regulatory regions directs
stem/early progenitor cell commitment during erythropoiesis. Cell
Rep. 6:231–244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shen L, Wu H, Diep D, Yamaguchi S,
D'Alessio AC, Fung HL, Zhang K and Zhang Y: Genome-wide analysis
reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics.
Cell. 153:692–706. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Coutinho DF, Monte-Mór BC, Vianna DT,
Rouxinol ST, Batalha AB, Bueno AP, Boulhosa AM, Fernandez TS,
Pombo-de-Oliveira MS, Gutiyama LM, et al: TET2 expression level and
5-hydroxymethylcytosine are decreased in refractory cytopenia of
childhood. Leuk Res. 39:1103–1108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Song SJ, Poliseno L, Song MS, Ala U,
Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al:
MicroRNA-antagonism regulates breast cancer stemness and metastasis
via TET-family-dependent chromatin remodeling. Cell. 154:311–324.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Song SJ, Ito K, Ala U, Kats L, Webster K,
Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J,
Avigan DE, et al: The oncogenic microRNA miR-22 targets the TET2
tumor suppressor to promote hematopoietic stem cell self-renewal
and transformation. Cell Stem Cell. 13:87–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jiang X, Hu C, Arnovitz S, Bugno J, Yu M,
Zuo Z, Chen P, Huang H, Ulrich B, Gurbuxani S, et al: miR-22 has a
potent anti-tumour role with therapeutic potential in acute myeloid
leukemia. Nat Commun. 7:114522016. View Article : Google Scholar
|
|
55
|
Tagawa T, Haraguchi T, Hiramatsu H,
Kobayashi K, Sakurai K, Inada K and Iba H: Multiple microRNAs
induced by Cdx1 suppress Cdx2 in human colorectal tumour cells.
Biochem J. 447:449–455. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Marzi MJ, Puggioni EM, Dall'Olio V, Bucci
G, Bernard L, Bianchi F, Crescenzi M, Di Fiore PP and Nicassio F:
Differentiation-associated microRNAs antagonize the Rb-E2F pathway
to restrict proliferation. J Cell Biol. 199:77–95. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tang H, Kong Y, Guo J, Tang Y and Xie X,
Yang L, Su Q and Xie X: Diallyl disulfide suppresses proliferation
and induces apoptosis in human gastric cancer through Wnt-1
signaling pathway by up-regulation of miR-200b and miR-22. Cancer
Lett. 340:72–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li J, Zhang Y, Zhao J, Kong F and Chen Y:
Overexpression of miR-22 reverses paclitaxel-induced
chemoresistance through activation of PTEN signaling in p53-mutated
colon cancer cells. Mol Cell Biochem. 357:31–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tsuchiya N, Izumiya M, Ogata-Kawata H,
Okamoto K, Fujiwara Y, Nakai M, Okabe A, Schetter AJ, Bowman ED,
Midorikawa Y, et al: Tumor suppressor miR-22 determines
p53-dependent cellular fate through post-transcriptional regulation
of p21. Cancer Res. 71:4628–4639. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang H, Tang J, Li C, Kong J, Wang J, Wu
Y, Xu E and Lai M: miR-22 regulates 5-FU sensitivity by inhibiting
autophagy and promoting apoptosis in colorectal cancer cells.
Cancer Lett. 356:781–790. 2015. View Article : Google Scholar
|
|
61
|
Guo S, Bai R, Liu W, Zhao A, Zhao Z, Wang
Y, Wang Y, Zhao W and Wang W: miR-22 inhibits osteosarcoma cell
proliferation and migration by targeting HMGB1 and inhibiting
HMGB1-mediated autophagy. Tumour Biol. 35:7025–7034. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Nagaraja AK, Creighton CJ, Yu Z, Zhu H,
Gunaratne PH, Reid JG, Olokpa E, Itamochi H, Ueno NT, Hawkins SM,
et al: A link between miR-100 and FRAP1/mTOR in clear cell ovarian
cancer. Mol Endocrinol. 24:447–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kawahigashi Y, Mishima T, Mizuguchi Y,
Arima Y, Yokomuro S, Kanda T, Ishibashi O, Yoshida H, Tajiri T and
Takizawa T: MicroRNA profiling of human intrahepatic
cholangiocarcinoma cell lines reveals biliary epithelial
cell-specific microRNAs. J Nippon Med Sch. 76:188–197. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zekri AN, Youssef AS, El-Desouky ED, Ahmed
OS, Lotfy MM, Nassar AA and Bahnassey AA: Serum microRNA panels as
potential biomarkers for early detection of hepatocellular
carcinoma on top of HCV infection. Tumour Biol. 37:12273–12286.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shin YM, Yun J, Lee OJ, Han HS, Lim SN, An
JY, Lee KH, Lee KM and Choe KH: Diagnostic value of circulating
extracellular miR-134, miR-185, and miR-22 levels in lung
adenocarcinoma-associated malignant pleural effusion. Cancer Res
Treat. 46:178–185. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang C, Wang C, Chen X, Yang C, Li K,
Wang J, Dai J, Hu Z, Zhou X, Chen L, et al: Expression profile of
microRNAs in serum: A fingerprint for esophageal squamous cell
carcinoma. Clin Chem. 56:1871–1879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ganepola GA, Rutledge JR, Suman P,
Yiengpruksawan A and Chang DH: Novel blood-based microRNA biomarker
panel for early diagnosis of pancreatic cancer. World J
Gastrointest Oncol. 6:22–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Franchina T, Amodeo V, Bronte G, Savio G,
Ricciardi GR, Picciotto M, Russo A, Giordano A and Adamo V:
Circulating miR-22, miR-24 and miR-34a as novel predictive
biomarkers to pemetrexed-based chemotherapy in advanced non-small
cell lung cancer. J Cell Physiol. 229:97–99. 2014.
|
|
69
|
Wang W, Li F, Zhang Y, Tu Y, Yang Q and
Gao X: Reduced expression of miR-22 in gastric cancer is related to
clinicopathologic characteristics or patient prognosis. Diagn
Pathol. 8:1022013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang G, Xia S, Tian H, Liu Z and Zhou T:
Clinical significance of miR-22 expression in patients with
colorectal cancer. Med Oncol. 29:3108–3112. 2012. View Article : Google Scholar : PubMed/NCBI
|