Introduction
Colorectal cancer (CRC) is one of the most common
malignancies in the United States and worldwide (1). Genetic alterations, specifically gene
mutations implicated in CRC tumorigenesis, lead to a gain of
oncogene function and loss of tumor suppressor gene function
(2). Epigenetic modifications,
particularly DNA methylation in selected gene promoters, are also
recognized common molecular alterations in human tumors (2,3).
Neurotensin (NTS), a tridecapeptide mainly
distributed along the gastrointestinal (GI) tract, functions to
decrease gastric motility, to increase pancreaticobiliary
secretion, to facilitate fatty acid absorption, and to increase
proliferation of normal intestinal mucosa (4–6). In
addition to these physiologic effects, NTS influences intestinal
inflammation and promotes the growth of cancers, including breast,
prostate, pancreas, lung and cancers of the GI tract (4,7,8). The
effects of NTS are exerted primarily by its G protein-coupled
receptors: the high-affinity NTS receptor 1 (NTSR1), and the
low-affinity NTSR2 (7,8). NTSR3/sortilin, a single transmembrane
receptor, also binds NTS and contributes to the diversity of
effects on multiple tissues (7,8).
The properties of NTS are predominantly mediated
through NTSR1, and this neuropeptide-receptor complex is
deregulated during cancer progression (8,9).
Although increased expression of NTSR1 has been identified in
certain cancer types, such as colon and neuroendocrine tumors
(NETs) (10–16), the molecular mechanisms regulating
NTSRs expression have not been clarified. Recently, we reported
that promoter methylation is an important molecular process that
regulates the differential expression of NTSR1 and silences NTSR2
in NET cells, and that silencing of NTSR1 suppressed the oncogenic
effects of NTS (16). In the
current study, we analyzed the endogenous expression of NTS
signaling components, the transcriptional change of NTSR genes
mediated by a demethylating agent, and the methylation status of
their promoters in CRC cell lines. Importantly, we show that
inhibition of NTSR1 by either gene knockdown or treatment with an
NTSR1 inhibitor decreases the growth and migration of CRC
cells.
Materials and methods
Cell lines, reagents and siRNA
transfection
The human CRC cell lines KM12c, Caco2, DLD1, HT29,
HCT116, and SW480 were used in these studies. Cell lines were
authenticated in February and May 2016 at Genetica DNA Laboratories
(Cincinnati, OH). KM12c cells were kindly provided by Dr Isaiah J.
Fidler (M.D. Anderson Cancer Center, Houston, TX, USA); other cell
lines were obtained from American Type Culture Collection
(Manassas, VA, USA). KM12c cells were cultured in MEM supplemented
with 10% fetal bovine serum (FBS), 1% sodium pyruvate, 1%
non-essential amino acids and 2% MEM essential vitamins. Caco2
cells were incubated in MEM supplemented with 15% FBS, 1% sodium
pyruvate and 1% non-essential amino acids. DLD1 cells were grown in
RPMI-1640 with 10% FBS. HT29 and HCT116 cells were maintained in
McCoy's 5A medium supplemented with 10% FBS. SW480 cells were
cultured in DMEM with 10% FBS. Cells were maintained at 37°C in a
humidified 5% CO2 incubator. The DNA methyltransferase
inhibitor, 5-aza-2′-deoxycytidine (5-aza-CdR), and a selective
inhibitor for NTSR1, SR48692, were purchased from Sigma-Aldrich
(St. Louis, MO, USA) and dissolved in dimethyl sulfoxide (DMSO).
Transfections with non-targeting control and SMARTPool NTSR1 siRNA
(Dharmacon, Lafayette, CO, USA) were performed using Lipofectamine
RNAiMAX (Invitrogen, Carlsbad, CA, USA) as previously described
(16).
RNA isolation, reverse transcription-PCR
(RT-PCR) and quantitative reverse transcription-PCR (RT-qPCR)
analysis
Total RNA isolation, cDNA synthesis and RT-PCR
analysis were carried out as previously described (16,17).
Briefly, RT-PCR reactions for NTS signaling elements were performed
using cDNA synthesized from 1 µg of total RNA from CRC
cells, HotStarTaq DNA polymerase (Qiagen, Valencia, CA, USA) and
primers as follows: NTSR1 F, 5′-TCATCGCCTTTGTGGTCTGCT-3′,
and NTSR1 R, 5′-TGGTTGCTGGACACGCTGTCG-3′, 33 cycles;
NTSR2 F, 5′-GTCTCCTCAGCTTCATCGTAT-3′, and NTSR2 R,
5′-TCCCCAAAGCCTGAAGCTGTA-3′, 40 cycles; NTSR3 F,
5′-AGAATGGTCGAGACTATGTTG-3′, and NTSR3 R,
5′-AAGAGCTATTCCAAGAGGTCC-5′, 33 cycles; NTS F,
5′-GATGATGGCAGGAATGAAAATCCAG-3′, and NTS R,
5′-GTTGAAAAGCCCTGCTGTGACAGA-3′, 40 cycles; β-actin F,
5′-TCACCAACTGGGACGACATG-3′, and β-actin R,
5′-ACCGGAGTCCATCACGATG-3′, 28 cycles; IL-8 F,
5′-CATGACTTCCAAGCTGGCCG-3′, and IL-8 R,
5′-AATTTTTTTATGAATTCTCAGCCCTC-3′, 33 cycles; cyclin D1 F,
5′-ATGTGTGCAGAAGGAGGTCC-3′ and cyclin D1 R,
5′-CTTAGAGGCCACGAACATGC-3′, 38 cycles. Cycling conditions for the
reactions were: initial melting at 95°C for 15 min, followed by the
above described numbers of cycles at 94°C for 30 sec, 55°C for 30
sec and 72°C for 45 sec and a final extension of 10 min at 72°C (MJ
Mini Thermal Cycler, Bio-Rad, Irvine, CA, USA). The PCR products
were analyzed on a 2% agarose gel and visualized with the Alpha
Innotech Imaging system (Alpha Innotech Corp., San Leandro, CA,
USA). RT-qPCR was carried out using TaqMan kits (Applied
Biosystems, Foster City, CA, USA) under StepOnePlus Real-Time PCR
System (Applied Biosystems) as previously described (16), according to the manufacturer's
protocol.
Methylation analysis
The methylation status of the NTSR1 and
NTSR2 promoters was determined by methylation-specific PCR
(MSp) and bisulfite sequencing (BS) analyses as previously
described (16,17). In brief, PCR with 35 cycling
reactions was performed using bisulfite-modified genomic DNA,
HotStarTaq DNA polymerase (Qiagen) and primers as follows:
NTSR1 MSP methyl (M) F, 5′-TTGGAATTCGTGGTAAGC-3′, and
NTSR1 MSP M R, 5′-GTCTCAAACGAAAACCGATA-3′; NTSR1 MSP
unmethyl (U) F, 5′-TATTTGGAATTTGTGGTAAGT-3′, and NTSR1 MSP U
R, 5′-ATCTCAAACAAAAACCAATAAAC-3′; NTSR2 MSP M F,
5′-GTGGAGTTCGGTTTAATTC-3′, and NTSR2 MSP M R
5′-ACTACCCGAAATCTAAACG-3′; NTSR2 MSP U F,
5′-GGTGGAGTTTGGTTTAATTT-3′, and NTSR2 MSP U R,
5′-CACTACCCAAAATCTAAACA-5′; NTSR1 BS F,
5′-TTGTGGATATTTAGGAGTGGG-3′ and NTSR1 BS R,
5′-CTCCAAAAAACCAAAATTCC-3′; NTSR2 BS F,
5′-TGTTGGGAAAGTTTTTTTTAAG-3′ and NTSR2 BS R,
5′-AAACACCTCCTCTTCTCTAAAAA-3′. The PCR products for MSP were
visualized as indicated above. For BS, PCR products were cloned
into the TOPO TA cloning vector (Invitrogen) and the plasmids from
individual bacterial colonies were sequenced.
Cell proliferation
Equal numbers of HCT116 and HT29 cells were seeded
in 24-well plates. Proliferation of cells transfected with siRNA or
treated with SR48692 was assessed at 48 and 96 h after seeding by
direct cell counting using a Beckman Coulter Cell Viability
Analyzer (Beckman-Coulter, Fullerton, CA, USA).
Western blot analysis
Western blot analysis was done as previously
described (16). The antibodies
for rabbit polyclonal NTSR1 (PA3-214, 1:2500 dilution) and rabbit
monoclonal cyclin D1 (2261-1, 1:5000 dilution) were purchased from
Thermo fisher Scientific (Rockford, IL, USA) and Epitomics
(Burlingame, CA), respectively. The rabbit polyclonal IL-8
(ab106350, 1:500 dilution) and mouse monoclonal anti-β-actin
(A5316, 1:5000 dilution) antibodies were obtained from Abcam
(Cambridge, MA, USA) and Sigma-Aldrich, respectively.
Wound-healing migration assay
A wound-healing migration assay was performed, using
the Ibidi Culture Insert (Ibidi, Munich, Germany), with control and
NTSR1 knockdown HCT116 or HT29 cells. The wounded monolayers,
generated by removal of the insert, which provides a cell-free gap,
were maintained for the indicated time periods. Phase-contrast
microscopic images were acquired using a Nikon Eclipse Ti
microscope and NIS Elements software (Nikon, Melville, NY, USA).
The data are the quantified gap distance.
Luciferase reporter assays
HCT116 and HT29 cells, seeded in 24-well plates,
were transiently transfected with the IL-8 reporter (0.4 µg)
and the Renilla luciferase reporter (0.05 µg) using
Lipofectamine 2000 (Invitrogen) according to the manufacturers'
instructions. For the NTSR1 inhibitor treatments, CRC cells were
treated with SR48692 (0, or 10 µM) one day after
transfection. Luciferase activity was measured from cell lysates
using a Dual-Luciferase Reporter Assay System (Promega, Madison,
WI) through Sirius Luminometer (Berthold Detection Systems,
Pforzheim, Germany) according to the manufacturers' protocols.
Transwell migration assay
Cell migration assessments were performed using 8.0
µm pore size Transwell filter inserts (Corning Inc.,
Corning, NY, USA) coated with 15 µg/ml type I collagen in
24-well plates. Briefly, HCT116 cells, serum-starved for 24 h, were
trypsinized, washed twice and resuspended in serum-free media
supplemented with 0.1% BSA. For replicates, equivalent cell numbers
were placed in the upper chamber, while the lower chamber was
filled with the same serum-free media containing SR48692 (either 0
or 10 µM). The cells were incubated in the Transwell
chambers for 20 h at 37°C. After removing the non-migrated cells
from the upper surface of the membrane, cells on the lower surface
were fixed with methanol and stained with 0.5% crystal violet.
Migration was determined by counting cell numbers in four random
fields per membrane under a Nikon Eclipse 80i microscope and NIS
Elements software (Nikon) at ×10.
Statistical analysis
Means and standard deviations for triplicate or
quadruple samples were calculated, and graphic representations
summarize mRNA levels of NTSR1 and NTSR2, number of
counted cells, migration distance, IL-8 luciferase activity and
migrated cell number. Comparisons between groups were performed
using two-sample t-test or analysis of variance, as indicated, with
tests for pairwise comparison or linear trend over dose levels.
Results
Expression analysis of NTS signaling
components in endogenous or 5-aza-CdR treated CRC cell lines
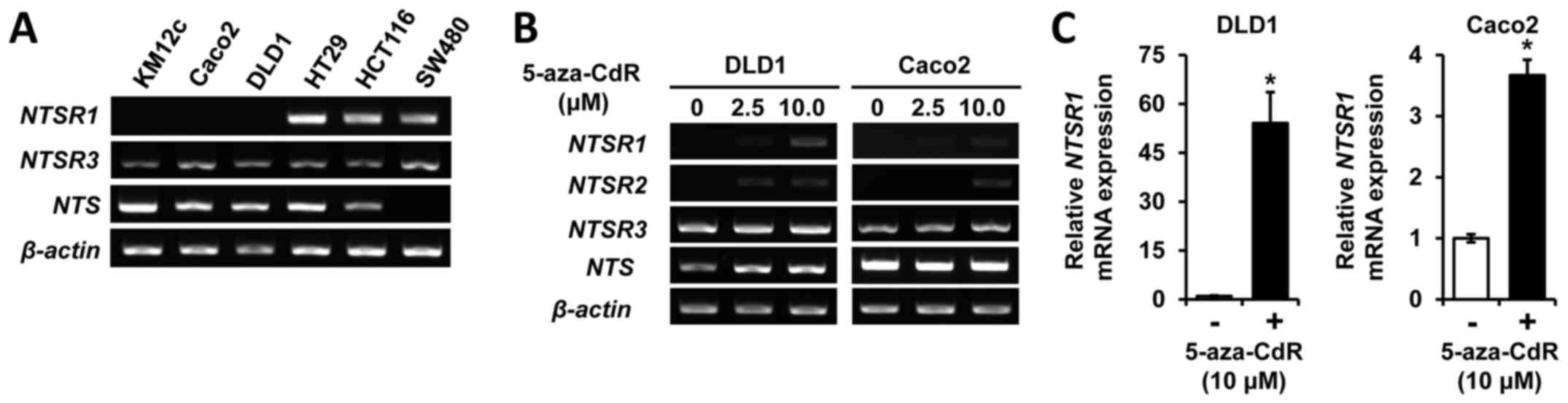
To assess NTS and NTSR expression in
CRC cells, we performed RT-PCR analysis of NTS constituents in six
human CRC cell lines (KM12c, Caco2, DLD1, HT29, HCT116 and SW480).
Whereas NTSR3 mRNA was consistently expressed in all six
cell lines (Fig. 1A), NTSR2
expression was not observed (data not shown). Selective expression
of NTSR1 and NTS was noted; little to no expression
of NTSR1 was noted in KM12c, Caco2 and DLD1 cells, and an
absence of NTS expression was detected in SW480 cells
(Fig. 1A). Recently, we showed
that promoter methylation is a key mechanism regulating the
differential expression of NTSR1 and to silence NTSR2
in NET cells (16). To confirm
whether the altered expression of NTSR1 and NTSR2 in
CRC cells was a result of DNA methylation, we treated DLD1 and
Caco2 cells with the DNA methyltransferase inhibitor, 5-aza-CdR,
and evaluated the expression of the NTSRs by RT-PCR
(Fig. 1B). Treatment with
5-aza-CdR augmented the expression of NTSR1 and NTSR2
in DLD1 and Caco2 cells, respectively. To verify these results, the
level of mRNA expression was also examined by RT-qPCR (Fig. 1C). Treatment with 5-aza-CdR
resulted in an approximate 54-fold induction of NTSR1
expression in DLD1 and an approximate 4-fold induction in Caco2
cells. Together, these results suggest that NTSR1 and
NTSR2 are targets of methylation in CRC cells.
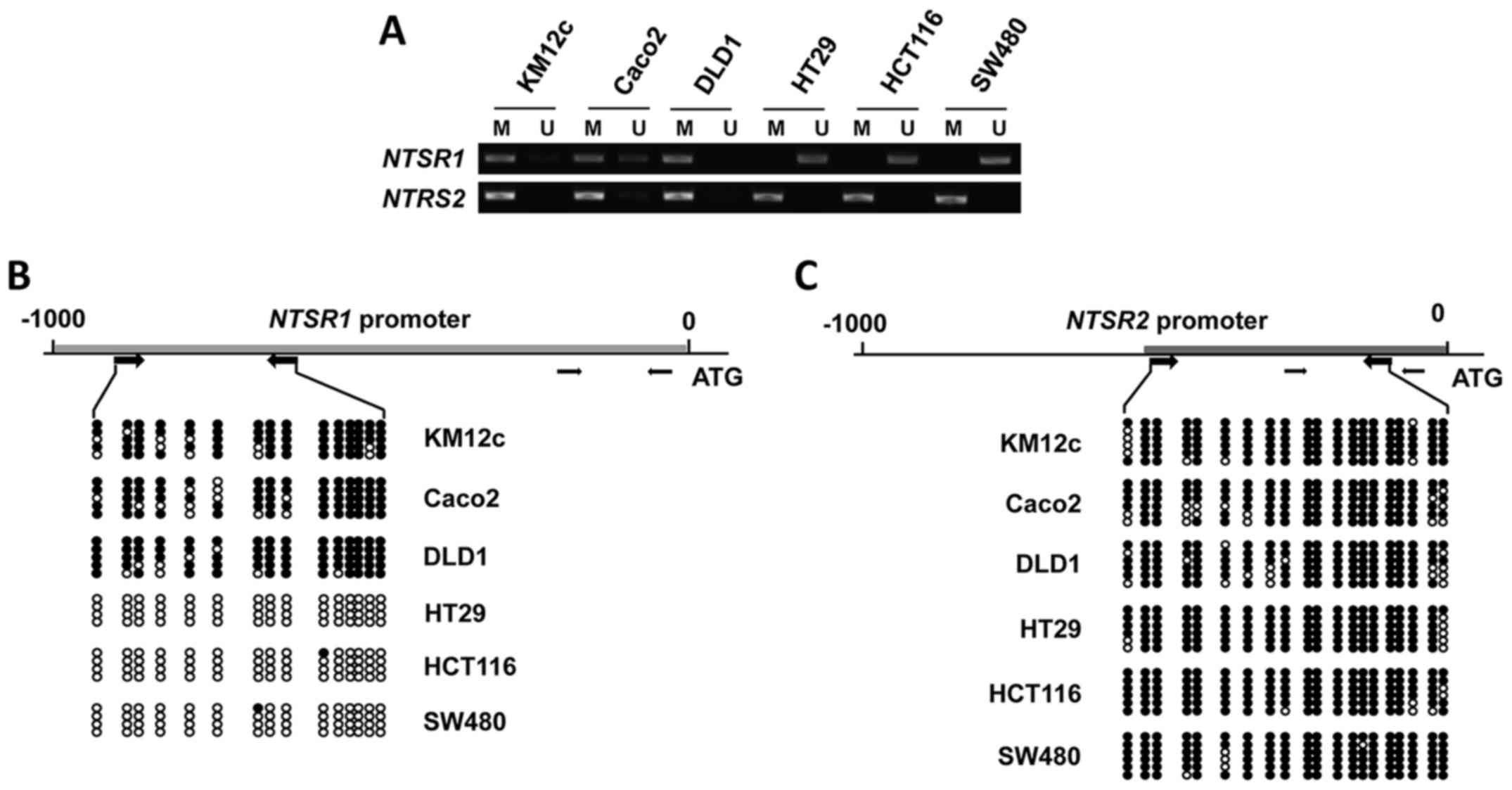
Correlation between gene silencing and
promoter methylation of NTSR1 and NTSR2 in CRC cells
To determine whether induction of NTSR1 and
NTSR2 expression by 5-aza-CdR was due to promoter
methylation, we investigated the methylation status of CpG islands
for these genes using methylation-specific PCR (MSP) and bisulfite
sequencing. While the NTSR1/2 promoters were found to be
almost completely methylated in KM12c, Caco2 and DLD1 cells, the
NTSR1 promoter in HT29, HCT116 and SW480 cells was shown to
be unmethylated (fig. 2A). The
methylation profile of the CpG sites of NTSR1 was further
analyzed by bisulfite sequencing (Fig.
2B). Consistent with the MSP data, the CpG islands of
NTSR1 were not methylated in HT29, HCT116 and SW480 cells.
Moreover, promoter methylation of NTSR2 was noted in all six
CRC cell lines by MSP analysis (fig.
2A) and bisulfite sequencing analyses (Fig. 2C). These data demonstrate that
promoter methylation silences the NTSR1/2 genes.
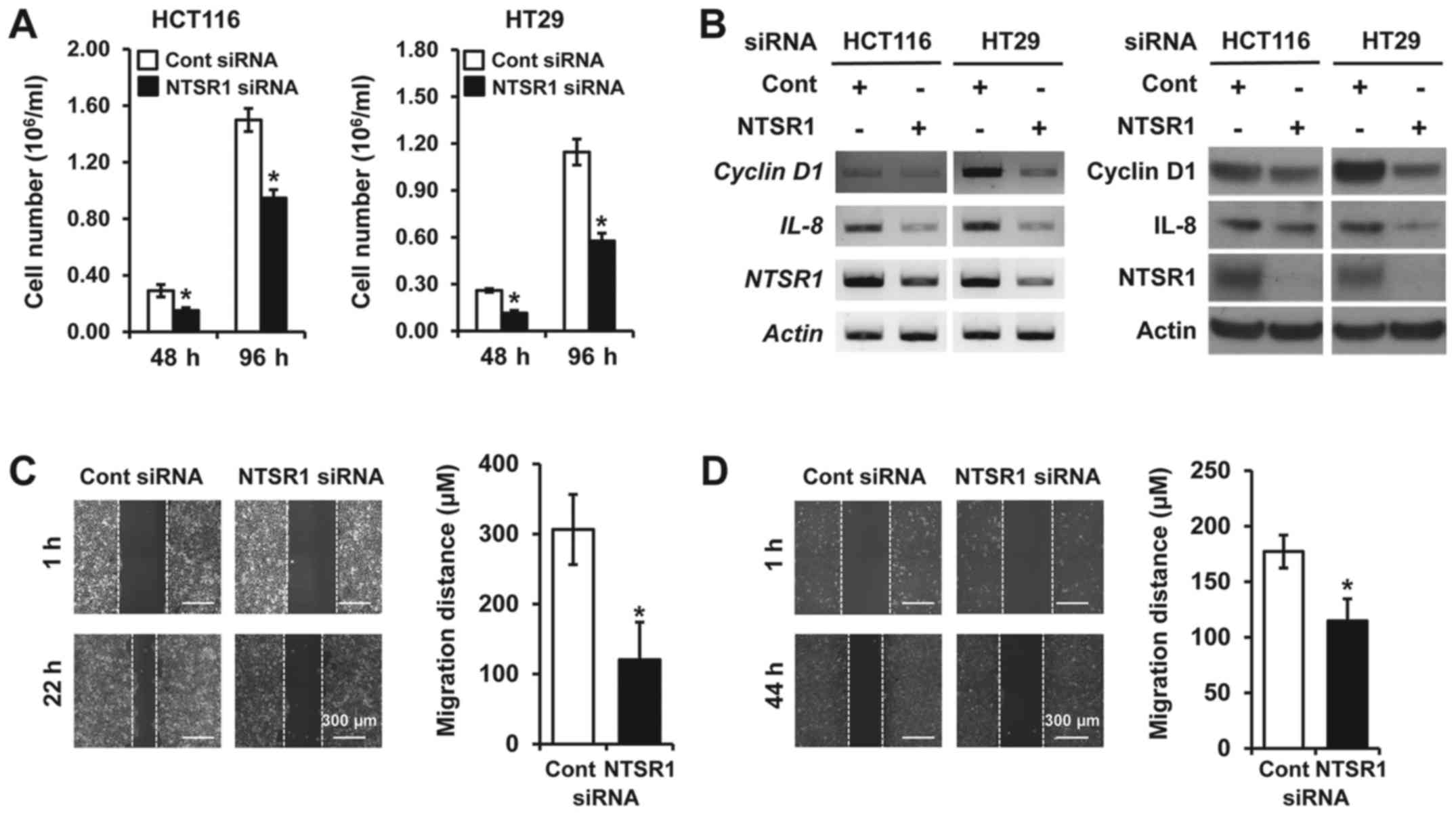
Knockdown of NTSR1 represses cell growth
and migration in CRC cells
We previously reported that inhibition of NTS
signaling components such as NTS and NTSR1 suppressed tumorigenic
functions in NET cells (16,18).
To elucidate the potential oncogenic functions of NTS components in
CRC cells, small interfering RNA (siRNA) directed against
NTSR1, which is inconsistently expressed among the tested
cell lines (Fig. 1A), was used in
HCT116 and HT29 cells since these cell lines do express the
NTS transcript (Fig. 1A),
and release NTS peptide (19).
Knockdown of NTSR1 inhibited cell growth for 48 and 96 h in
both HCT116 and HT29 cells compared with cells transfected with
non-targeting control siRNA (Fig.
3A). Furthermore, silencing NTSR1 significantly
suppressed mRNA expression of IL-8, which can be induced by
NTS signaling (20) and is
associated with tumorigenic functions in CRC (21) and cyclin D1, which is
important for cell growth (18,22)
(Fig. 3B, left). Protein
expressions of IL-8 and cyclin D1 were also decreased in cell lines
with NTSR1 knockdown (Fig.
3B, right). In addition to cell proliferation, NTSR1 activation
influences cell migration and invasion in some cancers including
breast and NET (13, 16, 23). On the basis of these studies, we
next assessed the migratory activity of CRC cells transfected with
NTSR1 siRNA using a wound-healing migration assay. Silencing NTSR1
suppressed cell migration of HCT116 cells (Fig. 3C) and HT29 cells (fig. 3D), respectively. Collectively,
these findings suggest that knockdown of NTSR1 significantly
inhibits the growth and migration of CRC cells.
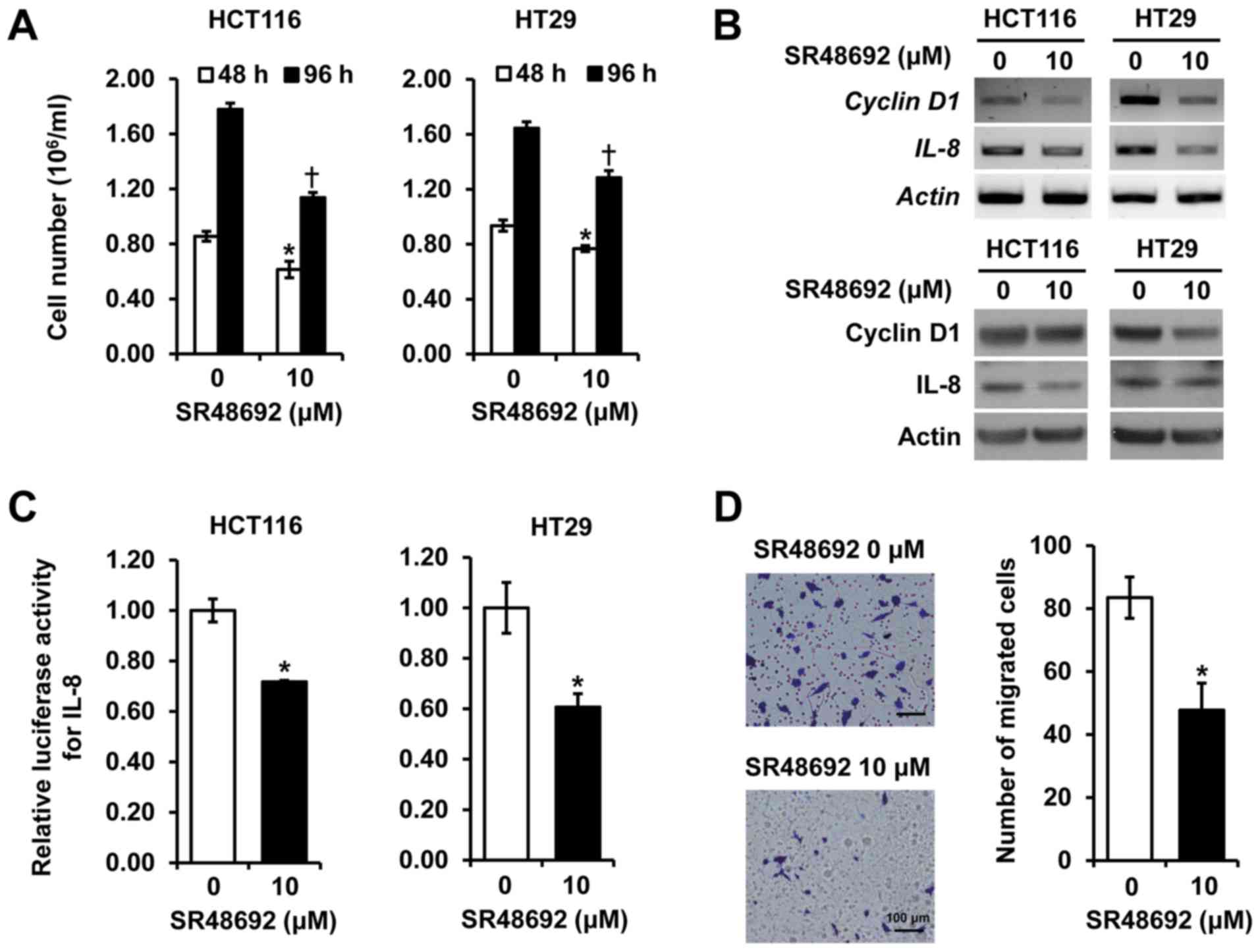
SR48692, an NTSR1 antagonist, suppresses
cell proliferation and migration of CRC cells
To further delineate the role of NTSR1, we examined
the effect of a pharmacologic blockade of NTSR1 using SR48692 in
HCT116 and HT29 cells. Treatment with SR48692 repressed
proliferation in HCT116 and HT29 cells for 48 and 96 h (Fig. 4A) and reduced both mRNA (upper
panels) and protein expressions (lower panels) of IL-8 and cyclin
D1 in the cell lines (fig. 4B).
Additionally, SR48692 inhibited the promoter activity of
IL-8, which promotes tumor growth, metastasis and
angiogenesis in CRC cell lines (21) (Fig.
4C). Moreover, HCT116 cells treated with SR48692 showed
significantly decreased cell migration compared to vehicle-treated
control cells (fig. 4D). These
results confirm that treatment with a selective NTSR1 antagonist
inhibits cell growth and migration of CRC cells, and is consistent
with results obtained with the siRNA experiments outlined above.
These data indicate that inhibition of NTSR1, either through gene
knockdown or receptor blockade, suppresses the growth and migration
of CRC cells, and demonstrates a role for NTSR1 in CRC
tumorigenesis.
Discussion
It has been reported that NTS and/or NTSR1 are over
expressed in some types of cancers and many cancer cell lines, and
that inhibition of NTS signaling can suppress the oncogenic
activities in several cancer cell lines (10–14,
16, 23, 24). Moreover, there is accumulating
evidence that NTSR1 activation is linked with poor prognosis,
cancer progression and a higher incidence of metastases in lung and
breast cancers (13,14). However, the regulatory mechanism
directing the expression of NTS pathway components in CRC cells is
not well-delineated. One regulatory process has been proposed;
activation of Wnt/β-catenin signaling plays an essential role in
the upregulation of NTSR1 in CRC (25) and NTS is a direct target of the
Wnt/β-catenin pathway in NET cells (18). In addition, Dong et al in
our laboratory showed that promoter methylation contributes to the
regulation of NTS expression in human cancer cells including colon
cancer (26,27) and we have also recently
demonstrated that promoter methylation is an important molecular
process regulating the expression of NTSR1 and NTSR2
in NET cells (16). Herein, we
observed diverse expression of NTSR1 and NTS, no
expression of NTSR2, and consistent expression of
NTSR3 in the CRC cells examined in this study. In
particular, we speculate that the varied level of NTS
expression (KM12c, Caco2, DLD1, HT29 and HCT116 versus SW480 cells,
Fig. 1A) is attributed to promoter
methylation on the basis of our current result and previous study
that DNA methylation is associated with NTS expression in
CRC cells (27). We also found
that the gene expression patterns for NTSR1 and NTSR2
in CRC cells are similar to those in NET (16) suggesting that promoter methylation
of NTS signaling constituents is closely associated with silencing
of the genes in human cancer cells.
In NTS pathway genes (especially NTSR1 and
NTSR2) whose expression was demonstrated to be modulated by
promoter methylation in our study, it has been reported that
activation of NTSR1 induces cell proliferation, migration and
invasion in a variety of cancers (8, 11,
24). Therefore, we evaluated the
role of NTSR1 on cell growth and migration in CRC cell lines. In
previous studies, the effects of NTS-mediated NTSR1 activation were
usually assessed through direct treatment of cancer cells with NTS
or an NTS agonist (11, 15, 23,
24). Herein, we determined the
effect of NTSR1 inhibition without NTS treatment on cell growth and
migration in CRC cells. Previously, NTS expression was
reported in some cancer cells including CRC (11–14);
these findings were confirmed in our present study by RT-PCR
analysis (Fig. 1A). In addition,
our group identified NTS peptide secretion in CRC cell lines
including HCT116 and HT29 (19).
Analogous to our previous findings and current study, we find that
endogenous NTS can exert stimulatory properties of cell growth and
migration through NTSR1 in CRC cells through both autocrine and
paracrine pathways.
As aforementioned, it has been reported that a
higher level of NTSR1 expression is observed in several human
cancer tissues such as pancreas, lung, breast, prostate, colon and
NET and that the NTSR1 correlates with tumor progression and
aggressiveness (10–16). These findings suggest that NTSR1
may be a potential diagnostic biomarker and treatment target for
these cancers. However, as noted above, NTSR1 promoter
hypermethylation, which can lead to the gene silencing, was also
found in lung and pancreatic cancers through genome scanning or
global DNA methylation profiling, and was identified as a
predictive marker for the cancers (28–30).
These intricacies imply that additional unrevealed adjusters or
mechanisms may affect the level of NTSR1 expression and that
further work is needed to clarify the detailed machinery between
expression and silencing of NTSR1 in various types of cancer.
In conclusion, our study shows that promoter
methylation is a key regulatory event for NTSR1 and
NTSR2 expression in CRC cells. In addition, we confirmed
that inhibition of NTSR1 suppresses cell proliferation and
migration in these cells. Our findings provide a rationale and
incentive to explore NTSR1 as a therapeutic target of CRC growth
and pathogenesis.
Acknowledgments
We thank Catherine E. Anthony (Markey Cancer
Center's Research Communications Office, University of Kentucky)
for assistance with manuscript preparation. This study was
supported by National Institutes of Health (NIH) grant R01 DK112034
and by the Biostatistical and Bioinformatics shared resource
facility of the University of Kentucky Markey Cancer Center
(supported by National Cancer Institute grant no. P30CA177558).
Abbreviations:
|
CRC
|
colorectal cancer
|
|
NTS
|
neurotensin
|
|
NTSR
|
neurotensin receptor
|
|
GI
|
gastrointestinal
|
|
NET
|
neuroendocrine tumor
|
|
FBS
|
fetal bovine serum
|
|
5-aza-CdR
|
5-aza-2′-deoxycytidine
|
|
RT-(q)PCR
|
(quantitative) reverse
transcription-polymerase chain reaction
|
|
MSP
|
methylation-specific PCR
|
|
BS
|
bisulfite sequencing
|
|
IL-8
|
interleukin-8
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar
|
|
3
|
Ashktorab H and Brim H: DNA methylation
and colorectal cancer. Curr Colorectal Cancer Rep. 10:425–430.
2014. View Article : Google Scholar
|
|
4
|
Evers BM: Neurotensin and growth of normal
and neoplastic tissues. Peptides. 27:2424–2433. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kalafatakis K and Triantafyllou K:
Contribution of neurotensin in the immune and neuroendocrine
modulation of normal and abnormal enteric function. Regul Pept.
170:7–17. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Song J, Zaytseva YY, Liu Y, Rychahou
P, Jiang K, Starr ME, Kim JT, Harris JW, Yiannikouris FB, et al: An
obligatory role for neurotensin in high-fat-diet-induced obesity.
Nature. 533:411–415. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mustain WC, Rychahou PG and Evers BM: The
role of neurotensin in physiologic and pathologic processes. Curr
Opin Endocrinol Diabetes Obes. 18:75–82. 2011. View Article : Google Scholar
|
|
8
|
Wu Z, Martinez-Fong D, Trédaniel J and
Forgez P: Neurotensin and its high affinity receptor 1 as a
potential pharmacological target in cancer therapy. Front
Endocrinol (Lausanne). 3:1842013.
|
|
9
|
Dupouy S, Mourra N, Doan VK, Gompel A,
Alifano M and Forgez P: The potential use of the neurotensin high
affinity receptor 1 as a biomarker for cancer progression and as a
component of personalized medicine in selective cancers. Biochimie.
93:1369–1378. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Friess H, Zhu Z, Graber H,
Zimmermann A, Korc M, Reubi JC and Büchler MW: Neurotensin
receptor-1 mRNA analysis in normal pancreas and pancreatic disease.
Clin Cancer Res. 6:566–571. 2000.PubMed/NCBI
|
|
11
|
Souazé F, Dupouy S, Viardot-Foucault V,
Bruyneel E, Attoub S, Gespach C, Gompel A and Forgez P: Expression
of neurotensin and NT1 receptor in human breast cancer: A potential
role in tumor progression. Cancer Res. 66:6243–6249. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gui X, Guzman G, Dobner PR and Kadkol SS:
Increased neurotensin receptor-1 expression during progression of
colonic adenocarcinoma. Peptides. 29:1609–1615. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dupouy S, Viardot-Foucault V, Alifano M,
Souazé F, Plu-Bureau G, Chaouat M, Lavaur A, Hugol D, Gespach C,
Gompel A, et al: The neurotensin receptor-1 pathway contributes to
human ductal breast cancer progression. PLoS One. 4:e42232009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alifano M, Souazé F, Dupouy S,
Camilleri-Broët S, Younes M, Ahmed-Zaïd SM, Takahashi T,
Cancellieri A, Damiani S, Boaron M, et al: Neurotensin receptor 1
determines the outcome of non-small cell lung cancer. Clin Cancer
Res. 16:4401–4410. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Valerie NC, Casarez EV, Dasilva JO,
Dunlap-Brown ME, Parsons SJ, Amorino GP and Dziegielewski J:
Inhibition of neurotensin receptor 1 selectively sensitizes
prostate cancer to ionizing radiation. Cancer Res. 71:6817–6826.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim JT, Li J, Song J, Lee EY, Weiss HL,
Townsend CM Jr and Evers BM: Differential expression and
tumorigenic function of neurotensin receptor 1 in neuroendocrine
tumor cells. Oncotarget. 6:26960–26970. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim JT, Li J, Jang ER, Gulhati P, Rychahou
PG, Napier DL, Wang C, Weiss HL, Lee EY, Anthony L, et al:
Deregulation of Wnt/β-catenin signaling through genetic or
epigenetic alterations in human neuroendocrine tumors.
Carcinogenesis. 34:953–961. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim JT, Liu C, Zaytseva YY, Weiss HL,
Townsend CM Jr and Evers BM: Neurotensin, a novel target of
Wnt/β-catenin pathway, promotes growth of neuroendocrine tumor
cells. Int J Cancer. 136:1475–1481. 2015. View Article : Google Scholar
|
|
19
|
Evers BM, Ishizuka J, Chung DH, Townsend
CM Jr and Thompson JC: Neurotensin expression and release in human
colon cancers. Ann Surg. 216:423–430; discussion 430–431. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao D, Kuhnt-Moore S, Zeng H, Wu JS,
Moyer MP and Pothoulakis C: Neurotensin stimulates IL-8 expression
in human colonic epithelial cells through Rho GTPase-mediated
NF-kappa B pathways. Am J Physiol Cell Physiol. 284:C1397–C1404.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ning Y, Manegold PC, Hong YK, Zhang W,
Pohl A, Lurje G, Winder T, Yang D, LaBonte MJ, Wilson PM, et al:
Interleukin-8 is associated with proliferation, migration,
angiogenesis and chemosensitivity in vitro and in vivo in colon
cancer cell line models. Int J Cancer. 128:2038–2049. 2011.
View Article : Google Scholar :
|
|
22
|
Kerkhoff E and Rapp UR: Cell cycle targets
of Ras/Raf signalling. Oncogene. 17:1457–1462. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Servotte S, Camby I, Debeir O, Deroanne C,
Lambert CA, Lapière CM, Kiss R, Nusgens B and Decaestecker C: The
in vitro influences of neurotensin on the motility characteristics
of human U373 glioblastoma cells. Neuropathol Appl Neurobiol.
32:575–584. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shimizu S, Tsukada J, Sugimoto T, Kikkawa
N, Sasaki K, Chazono H, Hanazawa T, Okamoto Y and Seki N:
Identification of a novel therapeutic target for head and neck
squamous cell carcinomas: A role for the neurotensin-neurotensin
receptor 1 oncogenic signaling pathway. Int J Cancer.
123:1816–1823. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Souazé F, Viardot-Foucault V, Roullet N,
Toy-Miou-Leong M, Gompel A, Bruyneel E, Comperat E, Faux MC, Mareel
M, Rostène W, et al: Neurotensin receptor 1 gene activation by the
Tcf/beta-catenin pathway is an early event in human colonic
adenomas. Carcinogenesis. 27:708–716. 2006. View Article : Google Scholar
|
|
26
|
Dong Z, Wang X, Zhao Q, Townsend CM Jr and
Evers BM: DNA méthylation contributes to expression of the human
neurotensin/neuromedin N gene. Am J Physiol. 274:G535–G543.
1998.PubMed/NCBI
|
|
27
|
Dong Z, Wang X and Evers BM: Site-specific
DNA méthylation contributes to neurotensin/neuromedin N expression
in colon cancers. Am J Physiol Gastrointest Liver Physiol.
279:G1139–G1147. 2000.PubMed/NCBI
|
|
28
|
Hagihara A, Miyamoto K, Furuta J, Hiraoka
N, Wakazono K, Seki S, Fukushima S, Tsao MS, Sugimura T and
Ushijima T: Identification of 27 5′ CPG islands aberrantly
methylated and 13 genes silenced in human pancreatic cancers.
Oncogene. 23:8705–8710. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan AC, Jimeno A, Lin SH, Wheelhouse J,
Chan F, Solomon A, Rajeshkumar V, Rubio-Viqueira B and Hidalgo M:
Characterizing DNA méthylation patterns in pancreatic cancer
genome. Mol Oncol. 3:425–438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo S, Yan F, Xu J, Bao Y, Zhu J, Wang X,
Wu J, Li Y, Pu W, Liu Y, et al: Identification and validation of
the methylation biomarkers of non-small cell lung cancer (NSCLC).
Clin Epigenetics. 7:32015. View Article : Google Scholar : PubMed/NCBI
|