Introduction
Breast cancer, the most frequently diagnosed cancer
and the leading cause of cancer deaths in women worldwide (1), is a heterogeneous disease with
different biological hallmarks, and thereby varying prognostic and
therapeutic characteristics. Tumors are classified into different
subtypes based on the immunohistochemical expression of estrogen
receptor α (ERα), progesterone receptor (PR), human epidermal
growth factor receptor-2 (HER-2) and Ki67 which also provides
prognostic and predictive information on response to hormonal or
targeted therapies (2).
Approximately 20–30% of all breast cancers overexpress HER-2
leading to an uncontrolled growth of cancer cells (3). Estrogen and progesterone are steroid
hormones that play an essential role for normal mammary gland
growth and development as well as for breast cancer progression.
Most of their effects are mediated by ERs and PR which are
intracellular receptors which constitute members of the nuclear
receptor superfamily of transcription factors. Approximately 75% of
malignant breast tumors are ERα positive and more than half of
these tumors also express PR (4,5). For
decades, ERα was thought to be the only ER present in mammary
epithelial cells until the identification of a second estrogen
receptor, ERβ, in 1996 (6). ERβ is
the most widely expressed ER in normal, mammary tissue. Five
different isoforms (ERβ1-ERβ5) exist in humans, though ERβ1 is
considered the only fully functional isoform. The role for ERβ1 and
other splice variants in breast cancer is still being investigated
and may not be consistent among different breast cancer subtypes
(7,8).
A solid, malignant tumor consists of cancer cells
within a tumor stroma of essentially fibroblasts, endothelial
cells, smooth muscle cells and immune cells (9). Of the latter, macrophages appear to
play a significant role in carcinogenesis (10). Thus, the association between
inflammation and cancer is well established (11,12).
An inflammatory microenvironment influences every hallmark of
cancer e.g. cell proliferation, invasion, angiogenesis and
metastasis (9). Macrophages are
innate immune cells originating from peripheral blood monocytes
with important roles in normal tissue homeostasis such as primary
response to pathogens, resolution of inflammation and wound
healing. They also constitute the most abundant immune cell present
in the tumor stroma and display extensive diversity and plasticity
(13–15). Within the tumor microenvironment
monocytes differentiate into tumor-associated macrophages (TAMs),
mainly due to tumor-derived chemo-tactic factors. TAMs are active
in the progression of tumors and may, in response to various
signals, exhibit dual roles in the microenvironment such as
facilitate tumor growth, or in contrast, contribute to destruction
of tumors (16,17). Simplified, human macrophages can be
classified into two phenotypically extremes, the classically
activated M1 phenotype considered to exhibit tumoricidal activities
arising from stimulation of macrophages with Th1-cytokines such as
interferon-γ (IFNγ) and/or microbial products like
lipopolysaccharide (LPS). This phenotype is characterized by high
levels of pro-inflammatory cytokines such as interleukin (IL)12,
IL23, IL6, tumor necrosis factor-α (TNF-α) and reactive
oxygen/nitrogen intermediates (ROI/RNI) (10,15).
On the contrary, the alternatively activated M2
macrophage is polarized by Th2-cytokines such as IL4 and IL13, and
releases high levels of anti-inflammatory cytokines such as IL10
and transforming growth factor-β. In most tumors the infiltrating
macrophages are polarized towards the M2 phenotype and show a
pro-tumoral role (10,15,16).
CD163 is a scavenger receptor that is regarded as highly specific
for M2 macrophages, while CD68 is a pan-macrophage marker and
stains both M1 and M2 phenotypes (18,19).
Furthermore, TAMs produce growth promoting and sustaining
cytokines, including epithelial growth factor, vascular endothelial
growth factor and matrix metalloproteinases (MMPs) (20). Activation of (cell-signaling)
urokinase receptor (uPAR) through binding of urokinase-type
plasminogen activator (uPA) triggers the conversion of plasminogen
to plasmin, which in turn, initiates a cascade of extracellular
proteases, e.g. MMPs. MMPs degrade components of extracellular
matrix leading to/promoting tumor cell invasion and metastatic
progression (21,22).
In human breast cancer, the inflammatory cells,
mainly lymphocytes and macrophages, can constitute as much as half
of the tumor mass and several studies suggest that high density of
TAMs is associated with high vascularity, high tumor grade,
increased tumor size, nodal metastasis and reduced overall survival
(23–25). Previously published studies have
also associated high infiltration of CD68+ and/or
CD163+ macrophages with ERα and PR-negative tumors and
high Ki67 proliferative index, whereas there are inconsistent
results of infiltration of macrophages, and HER-2 positivity
(25–29).
The aim of the current study, undertaken with human
breast cancer tissue, as well as with the human breast cancer cell
line T47D was to examine, in human breast cancer, the relationship
between infiltrating macrophages and their phenotype(s), hormone
receptor status comprising PR, ERα and ERβ1, the expression of
HER-2, MMP-9, uPAR and the proliferation marker Ki67. Furthermore,
we investigated how conditioned media (CM) from macrophages of the
M1 and M2 phenotypes, respectively, may influence the proliferation
and the expression of the markers mentioned above.
Materials and methods
Immunohistochemistry
Breast cancer specimens being analyzed in this study
were archival material stored in paraffin blocks, having been taken
for diagnostic purpose prior to any treatment at the Department of
Clinical Pathology and Cytology, Karlstad Central Hospital
(Karlstad, Sweden). The study included tumor specimen from all
patients (n=19) selected for neoadjuvant therapy at Karlstad
Central Hospital between 2009 and 2012, two samples were excluded
because of too little materials left. All samples were
de-identified prior to analysis. Serial sections of 4 µm
were cut from each sample and were mounted on IHC microscope glass
slides (Dako, Glostrup, Denmark). The sections were de-paraffinized
followed by antigen retrieval using PT-link at 97°C for 20 min in
EnVision FLEX Target Retrieval Solution (Dako). The sections were
incubated for 30 min with either of the following primary
antibodies: Monoclonal rabbit anti-human estrogen receptor (ER)α
(clone EP1, ready-to-use), monoclonal mouse anti-human progesterone
receptor (PR; clone PgR 636, ready-to-use), monoclonal mouse
anti-human-CD68 antibody (clone Kp1, ready-to-use), HercepTest™
polyclonal rabbit anti-human HER2, monoclonal mouse anti-human
estrogen receptor β1 (clone PPG5/10, dilution 1:40), monoclonal
mouse anti-human Ki-67 (clone MIB1, ready-to-use), polyclonal
rabbit anti-human MMP-9 (1:50), monoclonal mouse anti-human uPAR
(clone R4, 1:50) (all from Dako) and monoclonal mouse
anti-human-CD163 antibody (clone 10D6, 1:200, Novocastra, Leica
Microsystems, Newcastle, UK). The monoclonal mouse anti-human
estrogen receptor β1 (clone PPG5/10, 1:40) (Dako) demanded an
additional step of incubating sections with EnVision FLEX/mouse
linker (Dako) for 15 min prior to addition of the secondary
antibody. Immunohistochemical EnVision visualization system was
performed with the standard method of horseradish peroxidase and 3,
3′-diamino-benzidine incubating the sections with secondary
anti-mouse/anti-rabbit (ready-to-use) for 20 min and substrate
working solution FLEX DAB sub-chromophore 5 min in Autostainer Link
48 according to the manufacturer (Dako).
Benign human cervix tissue was used as control for
ERα and PR antibodies, benign cervix and breast carcinoma for ERβ1
and human tonsil for CD68, CD163, uPAR, MMP-9 and Ki67 antibodies.
After immunostaining, slides were counter-stained with Mayer's
haematoxylin, dehydrated, cleared and mounted using Tissue-Tek
coverslipping film (Sakura Finetek, Torrence, CA, USA). Assessments
of all immunostainings were done by a senior pathologist (A.S.).
Positive immunoreactivity (IR) for ERα, ERβ1, PR and Ki67 were
denoted as percentage of positive breast carcinoma cells while
positive IR for HER-2 was scored from 0 to 3+ according
to current clinical guidelines in Sweden. Staining for CD68 and
CD163 were scored as 1–3 where 1 (1–10%, 'low'), 2 (10–30%,
'moderate') and 3 (>30%, 'high') indicating percentage of
positive cells in the intratumoral and stromal area. The MMP-9 or
uPAR immunoreactivity was determined by counting the total number
of positive tumor (T) cells, and macrophage like stroma (S) cells,
respectively. Cells were counted in five randomly selected 320×250
µm areas and ranged from 0 (negative), score 1 (1–10
positive cells/five areas), score 2 (10–30 positive cells/five
areas) and score 3 (>30 positive cells/five areas). All scoring
were performed at ×400 magnification and a resolution of 6.24
pixels/µm. Images at ×400 magnification were captured using
a Leica DMD108 light microscope with an integrated camera.
Isolation of human monocytes and their
differentiation to M1 or M2 macro phage phenotype, and collection
of macrophage conditioned media
The generation of human monocyte-derived macrophages
was conducted as previously described (30). Briefly, 45 ml of buffy coat
obtained from healthy blood donors at Clinical Immunology and
Transfusion Medicine, Akademiska University Hospital (Uppsala,
Sweden) was mixed with an equal volume of PBS containing 3 mM EDTA
(Sigma-Aldrich, St. Louis, MO, USA) and was then gradient
centrifuged with Ficoll Paques PLUS (GE Healthcare, Little
Chalfont, UK). The separated band of peripheral blood mononuclear
cells (PBMC) was collected, and pelleted cells were washed by
repeated centrifugation steps. Monocytes were purified by adherence
to the cell culture dishes and macrophages were generated by
culturing monocytes for 6 days in RPMI-1640 (RPMI) (Life
Technologies, Carlsbad, CA, USA) with 20% heat-inactivated fetal
calf serum (FCS) (Thermo Scientific, Waltham, MA, USA) and 20 ng/ml
macrophage colony-stimulating factor (M-CSF; R&D Systems,
Minneapolis, MN, USA).
For further differentiation of the macrophages 100
ng/ml LPS (Sigma-Aldrich) and 20 ng/ml IFNγ (R&D Systems) were
added to generate the M1 phenotype. Conversely, 20 ng/ml IL4 and 20
ng/ml IL13 (both from R&D Systems) were added to generate the
M2 phenotype. M0-macrophages were cultured in RPMI +5% FCS without
any additions. After 48 h, the differentiated macrophages were
washed twice with PBS and were, furthermore, cultured in RPMI with
5% FCS for another 48 h. Thereafter the conditioned media (CM) from
M1 and M2 macrophages, containing neither LPS plus IFNγ nor IL4
plus IL13, was collected, centrifuged to remove cellular debris and
then stored in aliquots at −20°C. Macrophages of either M0, M1 or
M2 phenotype were also lysed for RNA isolation and reverse
transcriptase quantitative PCR (RT-qPCR) at two different time
points, first directly after washing and removal of the prior
addition of LPS, IFNγ, IL4, IL13 and second, after the 48 h
incubation in RPMI with 5% FCS.
Cell culture and cell cycle analysis
The human ductal breast epithelial tumor cell line
T47D was purchased from American Type Culture Collection (Manassas,
VA, USA). The cells were cultured at 37°C with 5% CO2 in
RPMI medium supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml
of penicillin, 100 µg/ml of streptomycin (Life
Technologies). For treatment with macrophage CM, cells were seeded
at 25,000 cells/cm2 onto cell culture plates (Greiner
Bio-One, Frickenhausen, Germany) and were allowed to adhere for 48
h before treatment with M1 and M2 CM for another 48 h. Cells that
were exposed to RPMI +5% FCS only, served as untreated controls.
After the respective treatment, RNA extraction was undertaken with
fractions of the cells. Alternatively, in order to investigate
possible epigenetic effect of M1 and M2 CM, respectively, on the
hormone receptor expression of T47D cell line, cells were re-seeded
and cultured for another 72 and 140 h prior to RNA extraction.
To investigate the effect of CM from M1 and M2
phenotypes on cell growth of the T47D cell line, cells were
cultured and treated for 48 h as described above. Next, they were
detached by trypsinization and counted in a hemocytometer.
Approximately 250,000 cells from each treatment (including
untreated controls) were collected for cell cycle analysis. These
cells were washed with PBS containing 1% bovine serum albumin
(BSA), centrifuged 10 min at 200 × g and resuspended in 450
µl ice-cold PBS/BSA prior to the addition of 5 ml ice-cold
70% ethanol. Samples were stored at −20°C until analysis, prior to
which Triton X-100 (Sigma-Aldrich) was added to a final
concentration of 0.1% and samples were incubated for 5 min at 6°C.
Thereafter, the cells were centrifuged 10 min at 200 × g and
resuspended in 1 ml PBS/BSA and this procedure was repeated once.
The cells were then resuspended in PBS/BSA and 0.1% Triton X-100,
200 µg/ml RNaseA and 50 µg/ml propidium iodide (the
latter two items were obtained from Sigma-Aldrich) were added
followed by incubation in the dark at room temperature for 45 min.
Cell cycle analysis was performed on FACSCalibur flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA) and the results was
calculated using ModFit LT v3.1 (Verity Software House, Inc.,
Topsham, ME, USA).
RNA extraction, cDNA synthesis and
quantification of mRNA (RT-qPCR)
Total RNA was extracted from M0, M1 and M2
macrophages as well as from cultured T47D cells treated as
described above using RNeasy Plus Mini kit (Qiagen, Hilden,
Germany) in accordance with the manufacturer's instructions. For
purity and quantification of the extracted RNA absorbance was
measured at wavelengths 260 and 280 nm using a NanoQuant plate with
the M200 Pro plate reader (Tecan, Männedorf, Switzerland).
Synthesis of cDNA was performed from 0.2 µg of total RNA
using High Capacity cDNA Reverse Transcription kit (Applied
Biosystems, Foster City, CA, USA) with a total reaction volume of
20 µl in accordance with the manufacturer's instructions.
Expression of CCL2, CCL17, CCL18, CXCL9, IL6, IL8, IL10, IL12, NFκB
and TNFα mRNA in M0, M1 and M2 macrophages and ERα, ERβ1, PR,
HER-2, p21 and p27 mRNA in T47D cells was evaluated by RT-qPCR,
using StepOnePlus real-time PCR with Power SYBR-Green Master Mix
(both from Applied Biosystems) in a total volume of 25 µl
containing 4 µl of cDNA (diluted 5×) and 200 nM of each
primer. All primer sequences are listed in Table I. Samples were run in duplicates
with appropriate negative controls and gene expression was
normalized to the housekeeping genes POLR2F and GAPDH. The
efficiency of the primers was calculated using LinRegPCR software
(31) and the size of the
amplified PCR products were validated using agarose
gel-electrophoresis. Fold changes were calculated using the ΔΔCq
method.
 | Table IPrimer sequences used for qPCR. |
Table I
Primer sequences used for qPCR.
| Gene name | Forward primer
sequence 5′-3′ | Reverse primer
sequence 5′-3′ | Genebank accession
no. |
|---|
| CCL2 |
CAGCCAGATGCAATCAATGCC |
TGGAATCCTGAACCCACTTCT | NM_002982.3 |
| CCL17 |
CTTCTCTGCAGCACATCCAC |
AGTACTCCAGGCAGCACTCC | NM_002987.2 |
| CCL18 |
CTCCTTGTCCTCGTCTGCAC |
TCAGGCATTCAGCTTCAGGT | NM_002988.3 |
| CXCL9 |
CCAGTAGTGAGAAAGGGTCGC |
AGGGCTTGGGGCAAATTGTT | NM_002416.2 |
| ERα |
GGGAAGTATGGCTATGGAATCTG |
TGGCTGGACACATATAGTCGTT | NM_000125.3 |
| ERβ1 |
TCCATCGCCAGTTATCACATCT |
CTGGACCAGTAACAGGGCTG | NM_001437.2 |
| GAPDH |
CAACAGCGACACCCACTCCT |
CACCCTGTTGCTGTAGCCAAA | NM_002046.4 |
| HER-2 |
TGTGACTGCCTGTCCCTACAA |
CCAGACCATAGCACACTCGG | NM_001005862.2 |
| IL6 |
GATCCAAAAACCACCCCTGACCC |
CAATCTGAGGTGCCCATGCTAC | NM_000600.3 |
| IL8 |
CATGACTTCCAAGCTGGCCGTG |
CCACTCTCAATCACTCTCAGTTC | NM_000584.3 |
| IL10 |
CTGGGGGAGAACCTGAAGA |
GGCCTTGCTCTTGTTTTCAC | NM_000572.2 |
| IL12 |
CAGCCTGGGAAACATAACAAGAC |
CTCCTGCCTCATCCTCCTGAA | NM_002187.2 |
| MMP-9 |
GGGACGCAGACATCGTCATC |
TCGTCATCGTCGAAATGGGC | NM_004994.2 |
| NFκB |
CCAACAGATGGCCCATACCT |
AACCTTTGCTGGTCCCACAT | NM_001165412.1 |
| p21 |
TTAGCAGCGGAACAAGGAGT |
AGCCGAGAGAAAACAGTCCA | NM_000389.4 |
| P27 |
TAATTGGGGCTCCGGCTAACT |
TGCAGGTCGCTTCCTTATTCC | NM_004064.3 |
| POLR2F |
ATGTCAGACAACGAGGACAATTT |
TTCGGCATTCTCCAAGTCATC | NM_001301129.1 |
| PR |
ACCCGCCCTATCTCAACTACC |
AGGACACCATAATGACAGCCT | NM_000926.4 |
| TNF-α |
CCTCTCTCTAATCAGCCCTCTG |
GAGGACCTGGGAGTAGATGAG | NM_000594.3 |
| uPAR |
GAGCTATCGGACTGGCTTGAA |
CGGCTTCGGGAATAGGTGAC | NM_002659.3 |
Immunocytochemistry
Approximately 200,000 cells of T47D treated as
described in the cell culture section were detached by
trypsinization and centrifuged for 10 min at 300 × g. Cells were
resuspended in PBS and spun onto positively charged microscopic
glass slides (Thermo Scientific). Slides were allowed to dry and
fixed in 4% formaldehyde solution for 10 min prior to
immunostaining using monoclonal rabbit anti-human estrogen receptor
α (clone EP1, ready-to-use), monoclonal mouse anti-human
progesterone receptor (clone PgR 636, ready-to-use,) mouse
anti-human estrogen receptor β1 (clone PPG5/10, 1:40) monoclonal
mouse anti-human Ki-67 (clone MIB1, ready-to-use) and monoclonal
mouse anti-human uPAR (clone R4, 1:50) (all from Dako) as
previously described.
Ethics
The study has been approved by the Uppsala Ethics
Committee (license 2014/498).
Statistics
Data are presented as mean ± SEM. A paired Student's
t-test was used for all cell counting experiments comparing treated
samples vs. untreated controls and also for the RT-qPCR mRNA
expression data comparing ΔCt values for treated samples vs.
untreated controls. Basal mRNA expression levels and cell cycle
distributions were compared using an unpaired Student's t-test. A
Jonckheere-Terpstra test was used to analyze significant
associations between increasing CD68+ mononuclear cell
infiltration in the biopsies with increasing tumor cell expression
of Ki67, uPAR, HER2 or MMP-9, or decreasing tumor cell expression
of ERα, ERβ or PR.
Results
The number of macrophages in human breast
cancer tissue have a positive association with the expression of
uPAR or Ki67 as well as an inverse association with ERα or PR
To investigate association between infiltration of
macrophages and selected markers in human breast cancer tissue, 17
breast cancer biopsies were immunohistochemically stained for CD68,
CD163, ERα, PR, HER-2, ERβ1, Ki67, MMP-9 and uPAR. The
immunoreactivity (IR) of the selected antigens was evaluated and is
presented in Table II.
Representative images of the IR obtained by staining of CD68,
CD163, ERβ1 and uPAR are shown in Fig.
1. The score for CD68 and CD163 were equal in 15/17 cases
strongly indicating that the M2 macrophage phenotype is the
dominant one being present in the currently investigated biopsies.
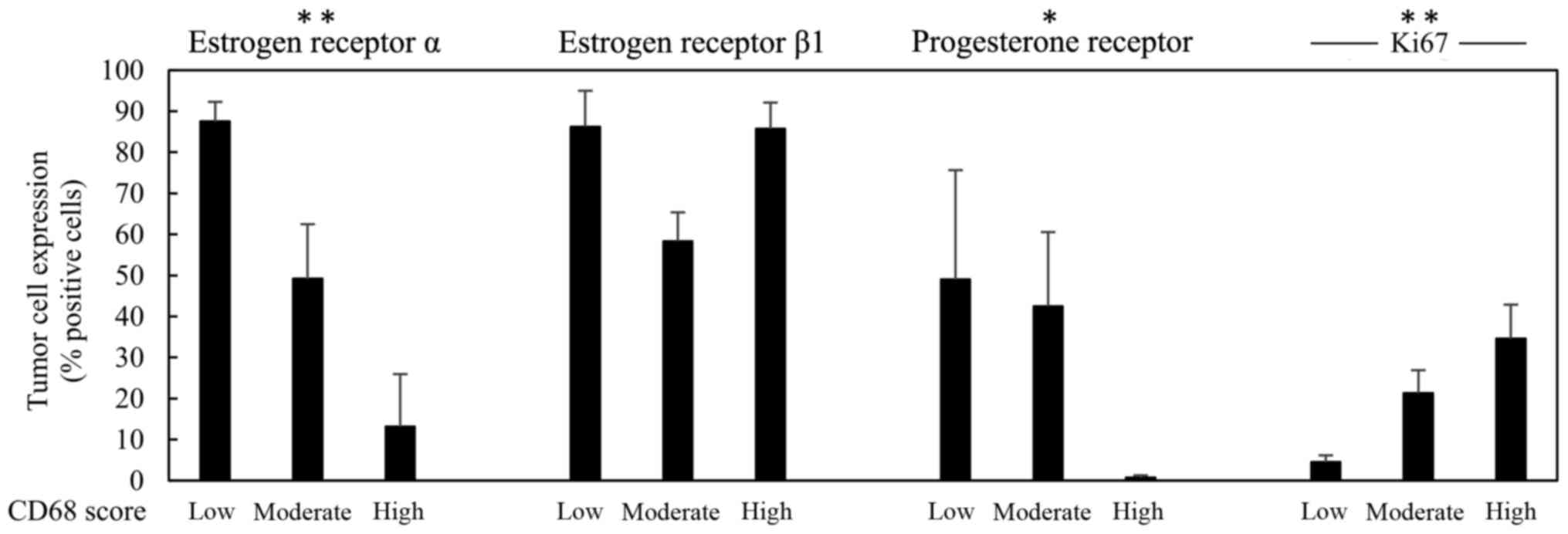
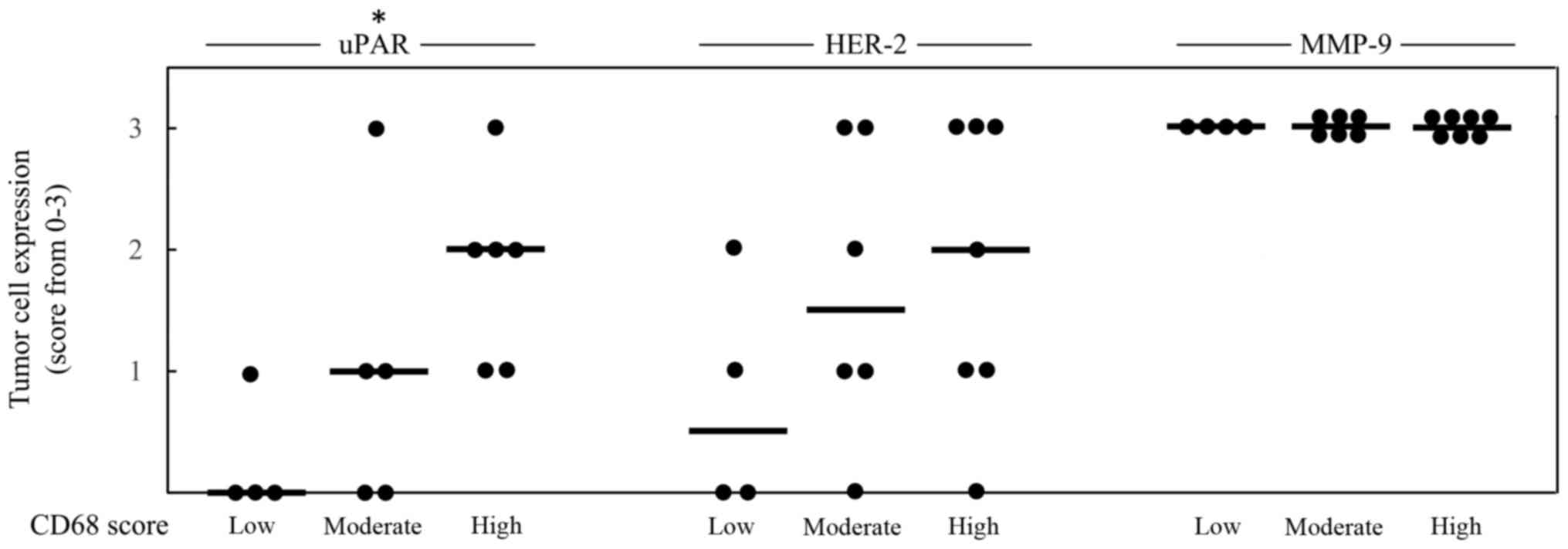
In Figs. 2 and 3, the score 1–3 of CD68 is positively
associated with higher expression of Ki67 or uPAR, respectively. No
statistical significant association could be seen between CD68 and
the expression of HER-2, ERβ1 or MMP-9, while an inverse
association between CD68 and ERα as well as between CD68 and PR,
could be noted (Fig. 2).
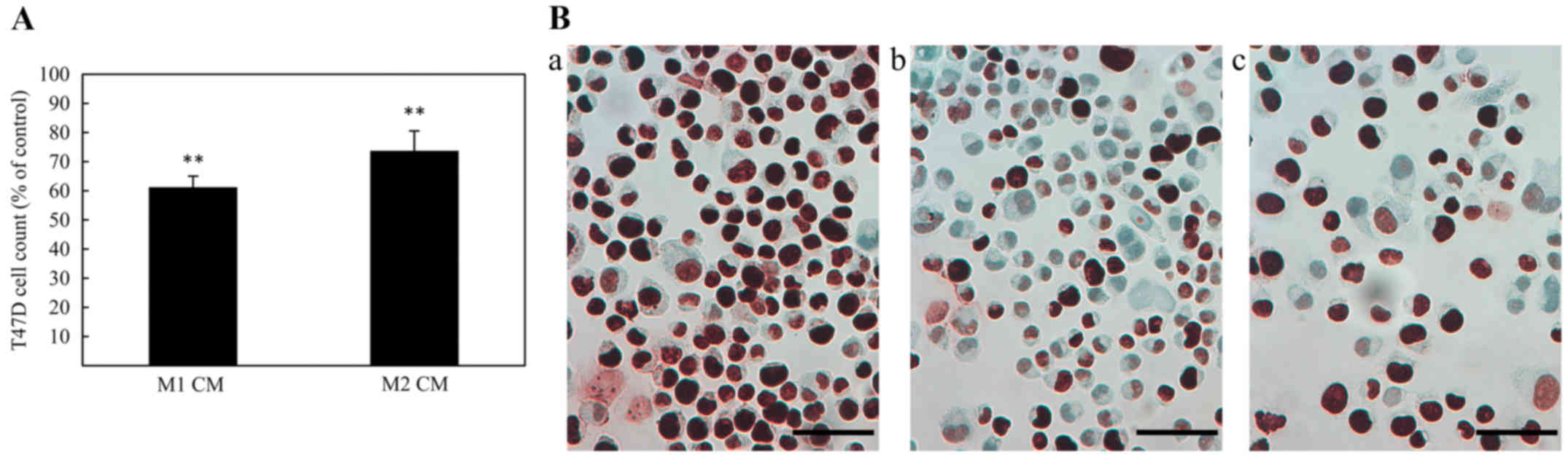
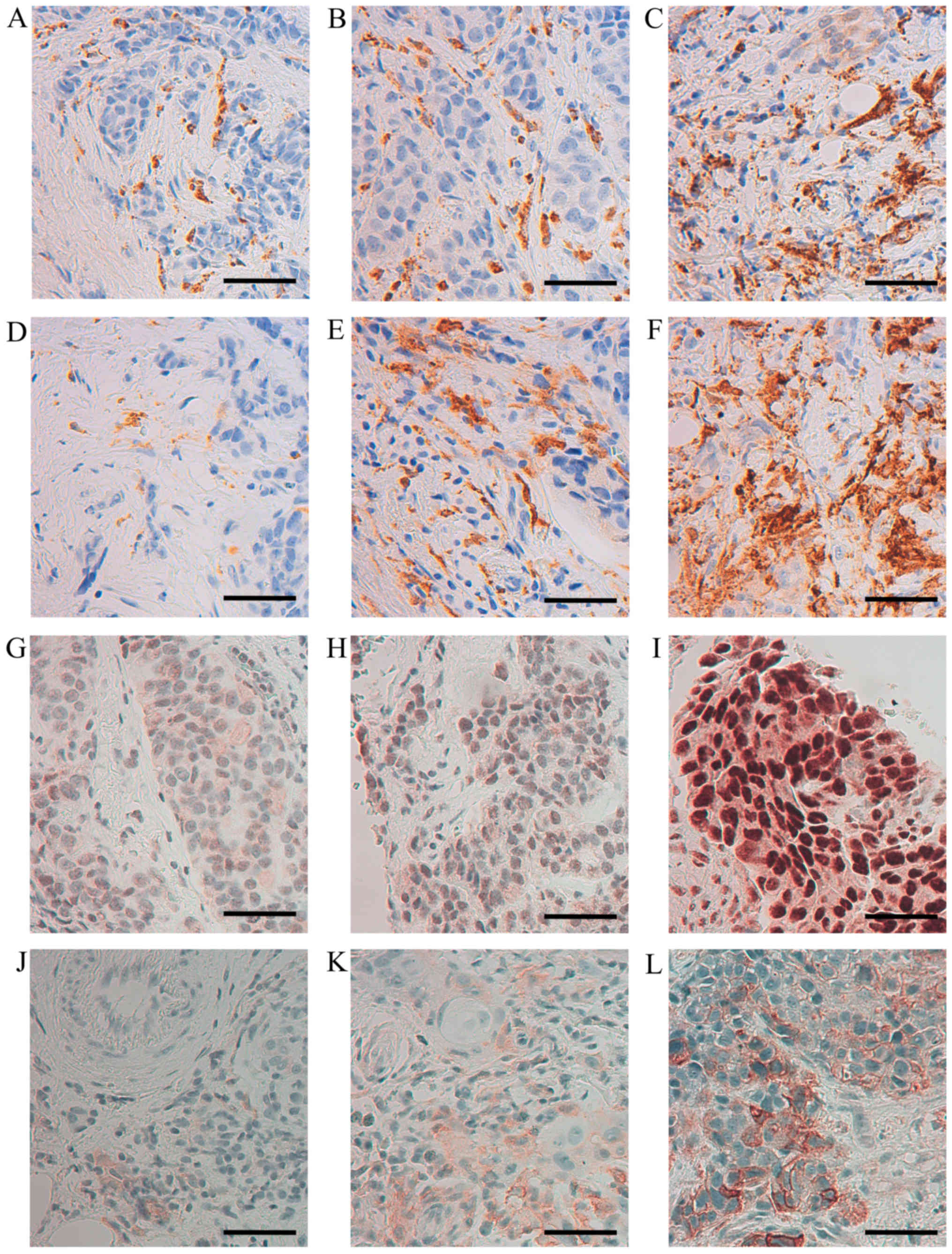 | Figure 1Immunohistochemical staining of CD68,
CD163, ERβ1 and uPAR in human, breast cancer biopsies.
Representative images of CD68 (A) score 1, (B) score 2, (C) score
3; CD163 (D) score 1, (E) score 2, (F) score 3; ERβ1 (G) 30%
positive tumor cells, (H) 50% positive tumor cells (I) 100%
positive tumor cells; uPAR (J) score 1, (K) score 2, (L) score 3
(×400 magnification, calibration bar is 50 µm in all
micrographs). |
 | Table IIImmunohistochemical staining of 17
breast cancer biopsies. |
Table II
Immunohistochemical staining of 17
breast cancer biopsies.
| Sample | CD68 | CD163 | ERα (%) | ERβ1 (%) | PR (%) | HER-2 | Ki-67 (%) | MMP-9 (T/S) | uPAR (T/S) |
|---|
| 1 | 3 | 3 | 90 | 100 | 0 | 3+ | 15 | 3 | 3 | 2 | 0 |
| 2 | 1 | 1 | 100 | 95 | 100 | 2+ | 0 | 3 | 3 | 0 | 0 |
| 3 | 2 | 2 | 75 | 30 | 75 | 0 | 25 | 3 | 2 | 1 | 0 |
| 4 | 3 | 3 | 0 | 95 | 0 | 3+ | 50 | 3 | 3 | 1 | 0 |
| 5 | 2 | 2 | 0 | 70 | 0 | 3+ | 25 | 3 | 3 | 0 | 0 |
| 6 | 1 | 1 | 80 | 95 | 5 | 1+ | 5 | 3 | 3 | 0 | 0 |
| 7 | 2 | 2 | 30 | 60 | 10 | 3+ | 8 | 3 | 2 | 0 | 0 |
| 8 | 1 | 1 | 80 | 60 | 90 | 0 | 5 | 3 | 2 | 0 | 0 |
| 9 | 1 | 2 | 90 | 95 | 1 | 0 | 8 | 3 | 3 | 1 | 0 |
| 10 | 3 | 3 | 0 | 90 | 0 | 0 | 70 | 3 | 3 | 3 | 3 |
| 11 | 3 | 3 | 1 | 90 | 1 | 2+ | 50 | 3 | 2 | 2 | 0 |
| 12 | 3 | 3 | 0 | 95 | 0 | 3+ | 25 | 3 | NA | 1 | NA |
| 13 | 2 | 1 | 40 | 60 | 0 | 2+ | 10 | 3 | 2 | NA | NA |
| 14 | 2 | 2 | 60 | 80 | 70 | 1+ | 45 | 3 | 2 | 3 | 2 |
| 15 | 3 | 3 | 1 | 80 | 4 | 1+ | 15 | 3 | 3 | 2 | 0 |
| 16 | 3 | 3 | 0 | 50 | 0 | 2+ | 17 | 3 | 3 | NA | NA |
| 17 | 2 | 2 | 90 | 50 | 100 | 0 | 15 | 3 | 1 | 1 | 0 |
Conditioned media from cultured human
macrophages decrease cell proliferation, reduce protein expression
of Ki-67, and mRNA expression of ERα and PR, and increase mRNA
expression of ERβ1 in T47D
For investigation of the effect of CM (48 h
challenge) from M1 or M2 macrophages on the proliferation of T47D,
cells were counted in a hemocytometer and immunocytochemically
stained for the proliferation marker, Ki67. As shown in Fig. 4A and B, both M1 and M2 macrophage
CM caused a decrease in cell number as well as a reduced Ki67
protein expression, when compared with untreated controls. In
addition, cell cycle distribution analysis for T47D cells treated
with macrophage CM indicated that treatment with M1 CM caused an
accumulation of cells in G0/G1 phase
(Table III).
 | Table IIICell cycle analysis of T47D cells
treated with macrophage CM. |
Table III
Cell cycle analysis of T47D cells
treated with macrophage CM.
| Treatment | Cells in
G1/G0-phase (%) | Cells in S-phase
(%) | Cells in
G2/M-phase (%) |
|---|
| RPMI 5% FCS | 70 | 20 | 10 |
| M1 CM | 85 | 5 | 10 |
| M2 CM | 75 | 15 | 10 |
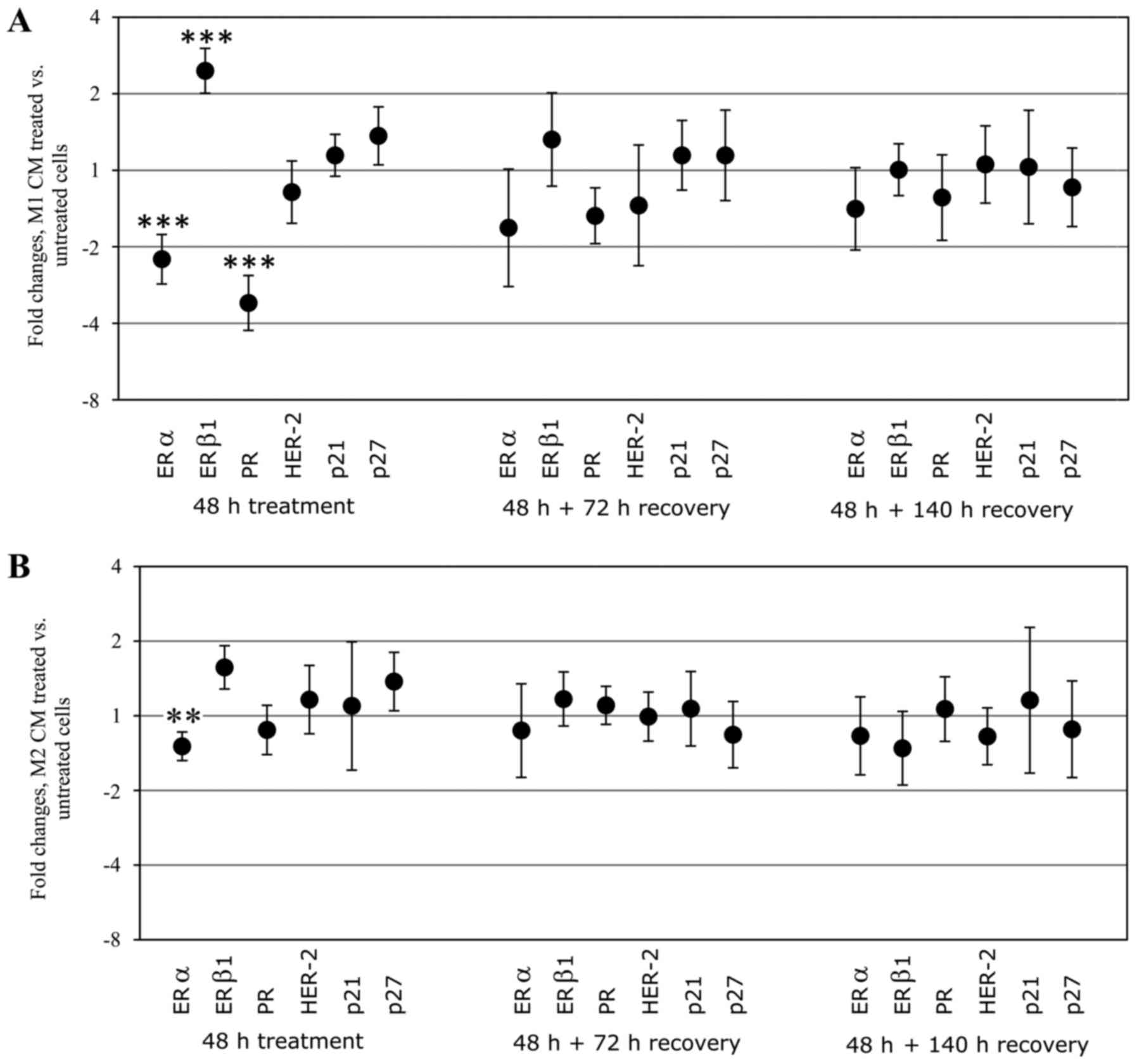
The mRNA expression of ERα, ERβ1, PR, HER-2, p21,
p27, MMP-9 and uPAR, respectively, was analyzed in treated T47D
cells and was compared with untreated controls. In cells treated
with M1 CM there was a significant downregulation of ERα and PR
mRNA. In contrast, the expression of ERβ1 was significantly
upregulated with M1 CM. Treatment with CM from M2 macrophages also
significantly downregulated the mRNA expression of ERα, however, to
a lesser extent than treatment with M1 CM. When the CM was removed,
normal expression of hormone receptors in T47D was restored after
140 h. No significant change in HER-2 or the cell cycle regulatory
genes p21 and p27 expression in mRNA could be observed in response
to either M1 or M2 CM treatment (Fig.
5). There was no detectable level of MMP-9 mRNA in T47D cells
(data not shown). Gene expression of uPAR indicated downregulation
at the mRNA level, however, no immunoreactivity for the uPAR
protein could be confirmed in the T47D cells making this mRNA data
less relevant (data not shown).
Conditioned media from cultured human
macrophages reduce ERα protein expression in T47D
Immunocytochemical staining for ERα, ERβ1, PR and
uPAR in T47D cells treated with CM from M1 or M2 macrophages was
performed, and were compared with untreated control cells. In
accordance with the downregulation of ERα mRNA in cells treated
with M1 CM (above), reduced immunoreactivity for ERα protein could
be observed in T47D treated with conditioned media from either M1
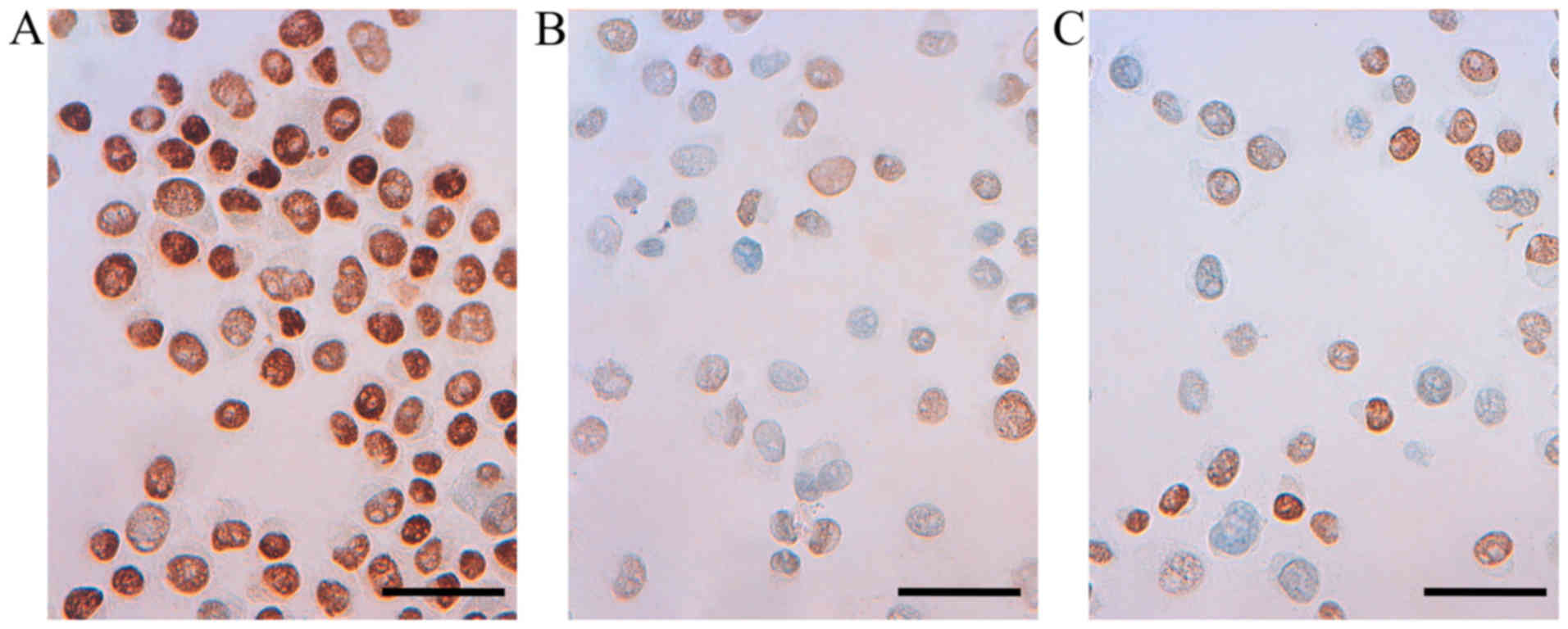
or M2 macrophages (Fig. 6).
Immunocytochemical staining for PR in untreated and
treated T47D cells demonstrated a very strong nuclear staining with
only a slightly weaker positivity in a few of the T47D cells
treated with M1 CM. Staining for ERβ1 revealed a very strong
nuclear staining of both untreated T47D and T47D cells treated with
M1 or M2 CM. No immunoreactivity for the uPAR protein could be
confirmed in the T47D cells (Table
IV).
 | Table IVImmunoreactivity of T47D cells
treated with CM from M1 or M2 macrophages. |
Table IV
Immunoreactivity of T47D cells
treated with CM from M1 or M2 macrophages.
| Treatment | ERα | ERβ1 | PR | Ki-67 | uPAR |
|---|
| RPMI 5% FCS | +++ | +++ | +++ | +++ | − |
| M1 CM | + | +++ | ++ | + | − |
| M2 CM | + | +++ | +++ | ++ | − |
Characterization of cultured macrophages
of M0, M1 and M2 phenotypes using quantitative PCR
CM generated from the cultured human M1 and M2
macrophages exhibited different effects on the breast carcinoma
cell line T47D. To study possible differences in the CM used,
RT-qPCR was performed on cultured macrophages of M0, M1 and M2
phenotype. Macrophages were terminated for RNA extraction at two
different time points, first directly after the washing and removal
of the additives used for differentiation of the M1 and M2
macrophages (LPS, IFNγ, IL4 and IL13), i.e. at the start of
collection of CM (0 h) and second at the end of collection of CM
(48 h incubation). The M1 macrophage phenotype was found to express
significantly lower mRNA level of IL10 and significantly higher
mRNA levels of IL6, IL8, IL12, CXCL9, TNFα and NFκB, these
upregulated genes had a higher significant upregulation at the
start of media collection (0 h) than at the end (48 h) (Table V). Expression of uPAR and MMP-9
mRNA in M0, M1 and M2 macrophages has been previously investigated
(32) showing in that study no
difference between either phenotype.
 | Table VmRNA expression levels in macrophages
of M1 and M2 phenotypes in comparison to M0. |
Table V
mRNA expression levels in macrophages
of M1 and M2 phenotypes in comparison to M0.
| Target mRNA | M0 0 h | M1 0 h | M2 0 h | M0 48 h | M1 48 h | M2 48 h |
|---|
| IL6 | 1 | U 190-foldb | U 10-fold | ND | ND | ND |
| IL8 | 1 | U 1700-foldc | D 2-folda | 1 | U 26-foldc | U 2-folda |
| IL10 | 1 | D 5-folda | 1 | 1 | U 5-fold | 1 |
| IL12 | 1 | U 5-foldc | 1 | D 2-fold | 1 | D 3-fold |
| CCL2 | 1 | D 2-fold | 1 | D 2-fold | 1 | D 8-fold |
| CCL17 | 1 | D 10-fold | U 4-fold | D 5-fold | ND | U 3-fold |
| CCL18 | 1 | U 26-fold | U 115-fold | D 2-fold | U 30-folda | U 30-fold |
| CXCL9 | 1 | U
14,000-foldc | 1 | D 5-fold | U 190-folda | D 3-fold |
| TNF-α | 1 | U 95-foldc | 1 | D 2-fold | U 7-folda | D 5-folda |
| NFκB | 1 | U 9-foldb | 1 | 1 | U 4-folda | 1 |
Discussion
Breast cancer, being the most common malignancy in
women, constitute a heterogeneous disease in which the status of
hormone receptors, HER-2 and Ki67 are routinely used to categorize
tumors and to provide predictive information of response to
hormonal therapy or targeted treatments. Adenocarcinoma of the
breast are frequently infiltrated with TAMs and the aim of the
current study was to investigate a possible relationship between
such infiltration and the expression of receptors for various
hormones as well as for HER-2, Ki67, MMP-9 and uPAR in 17 breast
tumor biopsies chosen for neoadjuvant therapy, and in addition in a
human breast cancer cell line.
It has been demonstrated that high number of M2
macrophages correlate with poor outcome in breast cancer (27) and that all histological locations
of TAM have prognostic value (29). In our current study we noted both
intratumoral and stromal dominance of M2 macrophages indicated by a
large proportion of CD68+/CD163+ macrophages
in all 17 breast tumor biopsies. Effects of macrophages on the
markers analyzed in the current study could contribute to the
understanding of the underlying mechanisms of TAMs in tumor
progression. With regard to uPAR, there are indications that tumor
cells which express this receptor may stimulate macrophage
polarization towards a more tumor permissive M2 macrophage
phenotype with the ability to promote tumor invasion and metastasis
(22). uPAR is of major interest
in breast cancer, ligation of uPAR has been shown to elicit an
activation of uPA a validated biomarker for a worse outcome in
breast cancer and also to activate MMP-9 that correlate with poor
prognosis in breast cancer (21,33,34).
In the current study, we could demonstrate a
significant positive association between the level of uPAR
expressed on the surface of the tumor cell and the macrophage
score. The number of MMP-9-positive tumor cells was high in all the
17 tumor biopsies selected for neoadjuvant therapy and therefore no
association with the level of macrophages or uPAR could be
demonstrated. Moreover, a significant positive association between
the extent of macrophage infiltration and the expression of Ki67
was found, Ki67 is a proliferation marker and high expression is
associated with a more aggressive tumor growth and higher risk of
developing recurrent disease (35). Numerous studies have revealed that
the expression of the growth factor HER-2 is associated with poor
prognosis in breast cancer, however the link between macrophage
infiltrates and HER-2 status is inconsistent (25–29).
We could not demonstrate a significant positive association between
the extent of macrophage infiltration and the expression of HER-2.
Loss of ERα or PR expression in breast cancer gives a worse
prognosis and exclude the possibility to treat these patients with
hormone blocking therapy (36). We
found that the amount of M2 macrophage infiltration is inversely
associated with the expression of ERα as well as of PR. These
findings support previous reports which suggest that high
infiltration of macrophages expressing either CD68 or CD163 is
associated with ERα and PR-negative tumors (25–29),
and demonstrate another possible route for TAMs in breast cancer to
act in an unfavorable manner.
Moreover, we demonstrate that CM from macrophages of
M1 and M2 phenotype could decrease the amount of ERα at the mRNA
levels in vitro in the breast carcinoma cell line T47D. The
downregulation of ERα mRNA was accompanied by an apparent decrease
in ERα immunoreactivity, as demonstrated by
immunocytochemistry.
Our current findings are in concert with those by
Stossi et al (37). Thus,
these authors also reported that conditioned media from THP-1
macrophages induced a loss of expression of ERα mRNA and protein in
MCF-7 breast cancer cell line via the involvement of MAPK and
c-Jun. There are previous reports of high contents of inflammatory
cytokines and of infiltrating leukocytes in ERα-negative tumors
(38) and several
macrophage-derived cytokines have been implicated in the
downregulation of ERα (39). We
observed that IL6, IL8, IL12, CXCL9, TNFα and NFκB were upregulated
in the M1 macrophages in comparison with the M2 macrophages,
although expressed also by M2. TNF-α as well as NFκB have been
associated with suppression of ERα in vitro (39–41).
Moreover, IL8 was overexpressed in ER-negative breast cancer cells
(42). A previous observation that
IL6 elicited a loss of ERα mRNA expression and caused methylation
of the promoter for ERα in MCF-7 cells (43), could not be confirmed in our study.
Thus, when the CM was removed and cells were re-cultured in RPMI
the expression of hormone receptors was restored after 140 h.
Moreover, the effect of CM from M1 or M2 macrophage
phenotypes on cell proliferation, and expression of HER-2, Ki67 and
hormone receptors, was investigated in the T47D cell line. After 48
h of treatment, both M1 and M2 macrophage CM caused a decrease in
the cell number of T47D compared with untreated controls, with the
highest effect elicited by the M1 macrophage. This was accompanied
by a reduced protein expression of Ki67 in T47D. However, this
finding could not be confirmed in the tumor biopsies where a strong
infiltration of macrophages was associated with high Ki67
proliferation index. It is most likely that TAM contribute to tumor
growth in vivo by means not taken into account in our in
vitro experiments, for instance by contributing to sustained
angiogenesis (23,24).
The prognostic role of the hormone receptor ERβ1 in
breast cancer is less clear (7,8) and
whether an association between macrophages and expression of ERβ1
in breast cancer exists is not known. The expression of ERβ1 was
high in the 17 breast tumor biopsies analyzed and we could not
demonstrate any association between the amount of macrophage and
the tumor cell expression of ERβ1. However, the mRNA level of ERβ1
in T47D cells was upregulated when treated with macrophage CM.
Unlike ERα, ERβ has a putative anti-proliferative effect when
binding to its ligand (7). M1 CM
caused an accumulation of T47D cells in G0/G1
phase and a decrease of cells in S-phase, indicating a cell cycle
arrest in G0/G1. This inhibitory effect of M1
CM on T47D cells is in agreement with previous studies on the colon
cancer cell line HT-29 and the lung cancer cell line H520 (30,32,44)
and suggests that macrophages of the M1 phenotype in breast cancer
tumor stroma might have an inhibitory effect and reduce the growth
of breast cancer cells. However, it was also stated that CM from M1
macrophage phenotype attenuated the effect of chemotherapy for
cells which responds with a G0/G1 cell cycle
arrest (44). In the current
study, we could not observe any changes on the cell cycle
inhibitory gene p21 and this has been observed previously in the
small cell lung cancer cell line H69 (32).
In conclusion, our in vivo and in
vitro results confirm the potential of the macrophages alone to
influence the expression of PR and ERα. We also demonstrated in
vivo and in vitro differences in the influence of Ki67,
HER-2 and ERβ1, though; our in vivo results demonstrated a
significant positive association of macrophages and the tumor cell
expression of uPAR and Ki67. Our result support previous studies
suggesting that macrophages are involved in impairing the prognosis
for breast cancer patients and that there could be a reason to
control the level of macrophages in some breast carcinoma
patients.
Acknowledgments
The present study was supported financially by the
County Council of Värmland, Karlstad University and Örebro
University.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldhirsch A, Wood WC, Coates AS, Gelber
RD, Thürlimann B and Senn HJ: Panel members: Strategies for
subtypes - dealing with the diversity of breast cancer: Highlights
of the St. Gallen International Expert Consensus on the Primary
Therapy of Early Breast Cancer 2011. Ann Oncol. 22:1736–1747. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferretti G, Felici A, Papaldo P, Fabi A
and Cognetti F: HER2/neu role in breast cancer: From a prognostic
foe to a predictive friend. Curr Opin Obstet Gynecol. 19:56–62.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thomas C and Gustafsson JA: The different
roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer.
11:597–608. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Diep CH, Daniel AR, Mauro LJ, Knutson TP
and Lange CA: Progesterone action in breast, uterine, and ovarian
cancers. J Mol Endocrinol. 54:R31–R53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kuiper GG, Enmark E, Pelto-Huikko M,
Nilsson S and Gustafsson JA: Cloning of a novel receptor expressed
in rat prostate and ovary. Proc Natl Acad Sci USA. 93:5925–5930.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Haldosén LA, Zhao C and Dahlman-Wright K:
Estrogen receptor beta in breast cancer. Mol Cell Endocrinol.
382:665–672. 2014. View Article : Google Scholar
|
|
8
|
Leung YK, Lee MT, Lam HM, Tarapore P and
Ho SM: Estrogen receptor-beta and breast cancer: Translating
biology into clinical practice. Steroids. 77:727–737. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Biswas SK, Allavena P and Mantovani A:
Tumor-associated macrophages: Functional diversity, clinical
significance, and open questions. Semin Immunopathol. 35:585–600.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Balkwill F and Mantovani A: Inflammation
and cancer: Back to Virchow? Lancet. 357:539–545. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sica A and Mantovani A: Macrophage
plasticity and polarization: In vivo veritas. J Clin Invest.
122:787–795. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hao NB, Lü MH, Fan YH, Cao YL, Zhang ZR
and Yang SM: Macrophages in tumor microenvironments and the
progression of tumors. Clin Dev Immunol. 2012:9480982012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Allavena P, Sica A, Solinas G, Porta C and
Mantovani A: The inflammatory micro-environment in tumor
progression: The role of tumor-associated macrophages. Crit Rev
Oncol Hematol. 66:1–9. 2008. View Article : Google Scholar
|
|
16
|
Sica A, Schioppa T, Mantovani A and
Allavena P: Tumour-associated macrophages are a distinct M2
polarised population promoting tumour progression: Potential
targets of anti-cancer therapy. Eur J Cancer. 42:717–727. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sica A, Allavena P and Mantovani A: Cancer
related inflammation: The macrophage connection. Cancer Lett.
267:204–215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lau SK, Chu PG and Weiss LM: CD163: A
specific marker of macrophages in paraffin-embedded tissue samples.
Am J Clin Pathol. 122:794–801. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Holness CL and Simmons DL: Molecular
cloning of CD68, a human macrophage marker related to lysosomal
glycoproteins. Blood. 81:1607–1613. 1993.PubMed/NCBI
|
|
20
|
Matsumoto H, Koo SL, Dent R, Tan PH and
Iqbal J: Role of inflammatory infiltrates in triple negative breast
cancer. J Clin Pathol. 68:506–510. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Noh H, Hong S and Huang S: Role of
urokinase receptor in tumor progression and development.
Theranostics. 3:487–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu J, Jo M, Eastman BM, Gilder AS, Bui JD
and Gonias SL: uPAR induces expression of transforming growth
factor β and interleukin-4 in cancer cells to promote
tumor-permissive conditioning of macrophages. Am J Pathol.
184:3384–3393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lewis CE, Leek R, Harris A and McGee JO:
Cytokine regulation of angiogenesis in breast cancer: The role of
tumor-associated macrophages. J Leukoc Biol. 57:747–751.
1995.PubMed/NCBI
|
|
24
|
Leek RD, Hunt NC, Landers RJ, Lewis CE,
Royds JA and Harris AL: Macrophage infiltration is associated with
VEGF and EGFR expression in breast cancer. J Pathol. 190:430–436.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mahmoud SM, Lee AH, Paish EC, Macmillan
RD, Ellis IO and Green AR: Tumour-infiltrating macrophages and
clinical outcome in breast cancer. J Clin Pathol. 65:159–163. 2012.
View Article : Google Scholar
|
|
26
|
Mohammed ZM, Going JJ, Edwards J,
Elsberger B, Doughty JC and McMillan DC: The relationship between
components of tumour inflammatory cell infiltrate and
clinicopathological factors and survival in patients with primary
operable invasive ductal breast cancer. Br J Cancer. 107:864–873.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tiainen S, Tumelius R, Rilla K, Hämäläinen
K, Tammi M, Tammi R, Kosma VM, Oikari S and Auvinen P: High numbers
of macrophages, especially M2-like (CD163-positive), correlate with
hyaluronan accumulation and poor outcome in breast cancer.
Histopathology. 66:873–883. 2015. View Article : Google Scholar
|
|
28
|
Medrek C, Pontén F, Jirström K and
Leandersson K: The presence of tumor associated macrophages in
tumor stroma as a prognostic marker for breast cancer patients. BMC
Cancer. 12:3062012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gwak JM, Jang MH, Kim DI, Seo AN and Park
SY: Prognostic value of tumor-associated macrophages according to
histologic locations and hormone receptor status in breast cancer.
PLoS One. 10:e01257282015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Engström A, Erlandsson A, Delbro D and
Wijkander J: Conditioned media from macrophages of M1, but not M2
phenotype, inhibit the proliferation of the colon cancer cell lines
HT-29 and CACO-2. Int J Oncol. 44:385–392. 2014.
|
|
31
|
Ruijter JM, Ramakers C, Hoogaars WM,
Karlen Y, Bakker O, van den Hoff MJ and Moorman AF: Amplification
efficiency: Linking baseline and bias in the analysis of
quantitative PCR data. Nucleic Acids Res. 37:e452009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hedbrant A, Wijkander J, Seidal T, Delbro
D and Erlandsson A: Macrophages of M1 phenotype have properties
that influence lung cancer cell progression. Tumour Biol.
36:8715–8725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duffy MJ, O'Donovan N, McDermott E and
Crown J: Validated biomarkers: The key to precision treatment in
patients with breast cancer. Breast. 29:192–201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao S, Ma W, Zhang M, Tang D, Shi Q, Xu
S, Zhang X, Liu Y, Song Y, Liu L, et al: High expression of CD147
and MMP-9 is correlated with poor prognosis of triple-negative
breast cancer (TNBC) patients. Med Oncol. 30:3352013. View Article : Google Scholar
|
|
35
|
Dai X, Xiang L, Li T and Bai Z: Cancer
hallmarks, biomarkers and breast cancer molecular subtypes. J
Cancer. 7:1281–1294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dunnwald LK, Rossing MA and Li CI: Hormone
receptor status, tumor characteristics, and prognosis: A
prospective cohort of breast cancer patients. Breast Cancer Res.
9:R62007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stossi F, Madak-Erdoğan Z and
Katzenellenbogen BS: Macrophage-elicited loss of estrogen
receptor-α in breast cancer cells via involvement of MAPK and c-Jun
at the ESR1 genomic locus. Oncogene. 31:1825–1834. 2012. View Article : Google Scholar
|
|
38
|
Chavey C, Bibeau F, Gourgou-Bourgade S,
Burlinchon S, Boissière F, Laune D, Roques S and Lazennec G:
Oestrogen receptor negative breast cancers exhibit high cytokine
content. Breast Cancer Res. 9:R152007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee SH and Nam HS: TNF alpha-induced
down-regulation of estrogen receptor alpha in MCF-7 breast cancer
cells. Mol Cells. 26:285–290. 2008.PubMed/NCBI
|
|
40
|
Bhat-Nakshatri P, Campbell RA, Patel NM,
Newton TR, King AJ, Marshall MS, Ali S and Nakshatri H: Tumour
necrosis factor and PI3-kinase control oestrogen receptor alpha
protein level and its transrepression function. Br J Cancer.
90:853–859. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Oida K, Matsuda A, Jung K, Xia Y, Jang H,
Amagai Y, Ahn G, Nishikawa S, Ishizaka S, Jensen-Jarolim E, et al:
Nuclear factor-ĸB plays a critical role in both intrinsic and
acquired resistance against endocrine therapy in human breast
cancer cells. Sci Rep. 4:40572014. View Article : Google Scholar
|
|
42
|
Freund A, Chauveau C, Brouillet JP, Lucas
A, Lacroix M, Licznar A, Vignon F and Lazennec G: IL-8 expression
and its possible relationship with estrogen-receptor-negative
status of breast cancer cells. Oncogene. 22:256–265. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
D'Anello L, Sansone P, Storci G, Mitrugno
V, D'Uva G, Chieco P and Bonafé M: Epigenetic control of the
basal-like gene expression profile via Interleukin-6 in breast
cancer cells. Mol Cancer. 9:3002010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hedbrant A, Erlandsson A, Delbro D and
Wijkander J: Conditioned media from human macrophages of M1
phenotype attenuate the cytotoxic effect of 5-fluorouracil on the
HT-29 colon cancer cell line. Int J Oncol. 46:37–46. 2015.
|