Introduction
Non-small cell lung cancer (NSCLC) comprises 80–85%
of lung cancer cases and is the leading cause of cancer deaths
worldwide (1). Despite recent
advances in understanding the molecular biology of lung cancer and
the use of combined therapy including surgical resection,
chemotherapy, radiation therapy and targeted therapy, the 5-year
survival rate of NSCLC patients has not substantially changed. Only
15.9% of lung cancer patients survive ≥5 years, and ~50% of
patients die of disease recurrence and metastasis (2).
The cancer stem cell (CSC) hypothesis suggests that
a rare population of cancer cells with self-renewing capacity is
responsible for tumor initiation, metastasis, relapse and
therapeutic resistance (3,4). The existence of cancer stem-like
cells (CSLCs) has been confirmed in hematopoietic malignancies and
in solid tumors (5,6). CSLCs can be identified and isolated
by spheroid formation in serum-free medium and by the expression of
specific cell surface markers. Among the lung CSLC markers are
CD133+, CD326+ (EpCAM), CD44+,
ALDH1+ and Oct-4 (7–12).
Some or all of these markers may be co-expressed to confer CSLCs
properties. In our previous study, we demonstrated that a
subpopulation of CD133+/CD326+ cells
represents CSLC in the A549 cell line (13). CSLCs represent the ultimate target
for cancer therapy.
Epithelial-to-mesenchymal transition (EMT) leads to
the loss of epithelial cell adhesion and the acquisition of the
mesenchymal phenotype (14,15).
EMT plays a key role in the metastasis of NSCLC (16). Cancer cells treated with TGF-β1, an
inducer of EMT, show decreased expression of E-cadherin and
increased expression of vimentin, N-cadherin and matrix
metalloproteinases (MMP), such as MMP-2, MMP-3 and MMP-9. Cells
undergoing EMT also show increased motility and invasiveness.
MicroRNAs are known to be critical regulators of EMT (17). The miR-200 family plays an
essential role in suppressing the EMT through targeting ZEB in
NSCLC and other cancers (18–20).
miR-23a regulates TGF-β-induced EMT by targeting E-cadherin in lung
cancer cells (21). The Snail and
miR-34a-mediated regulation of ZNF281/ZBP99 promotes EMT (22). Accordingly, microRNAs can serve as
novel therapeutic targets as well as diagnostic and prognostic
markers in cancer (23).
To determine whether microRNAs also regulate EMT in
CSLCs, we treated both A549 cells and
CD133+/CD326+ cells with TGF-β1 and compared
microRNA expression profiles. We demonstrated that both in A549
cells and CD133+/CD326+ cells, miR-181b-5p
promotes tumor invasion and metastasis in vitro and in
vivo by targeting the expression of E-cadherin.
Materials and methods
Cell culture, TGF-β1 treatment and
transfections
The human lung cancer cell line A549 was obtained
from the American Type Culture Collection (ATCC) and was cultured
in DMEM (Hyclone, USA) with 10% fetal bovine serum at 37°C, 5%
CO2. The induced CD133+/CD326+
subpopulation cells were suspended in serum-free medium
supplemented with 0.4% BSA (Sigma, USA), insulin (5 ng/ml, Sigma),
bFGF (10 ng/ml, PeproTech, USA), EGF (20 ng/ml, PeproTech), and B27
(20 ng/ml, Invitrogen, USA) at a density of 103 cells/3
ml in ultralow attachment plates (Corning, USA).
To induce the EMT process, adherent A549 cells and
CD133+/CD326+ spheroids were treated with 5
ng/ml TGF-β1 (Sigma) for 72 h. Adherent A549 cells and suspended
CD133+/CD326+ cells were transfected with
agomiR-181b-5p and antagomiR-181b-5p, which were purchased from
RiboBio Co. Ltd. (China). Cells were plated in a 24-well plate at
1×105 cells/plate. Agomirs or antagomirs of miR-181b-5p
were appropriately diluted according to the manufacturer's protocol
and added to the culture medium to transfect the cells. The
concentration of agomiR-181b-5p and antagomiR-181b-5p were 50 nM
and 100 nM, respectively. The expression of 181b-5p was determined
48 h after transfection.
Patients and peripheral blood
samples
Peripheral blood samples were obtained from NSCLC
patients prior to treatment at the Xinqiao Hospital of the Third
Military Medical University between 2014 and 2015 and were stored
at −80°C. This project was approved by the ethics committee of the
Xinqiao Hospital of the Third Military Medical University, and
informed consent was obtained from all the patients.
Flow cytometry
Spheres were dissociated into single cells, washed
and incubated with monoclonal antibodies specific for human
CD133/1-PE and CD326-FITC (Miltenyi, Germany). The appropriate
dilution and procedures were carried out according to the
manufacturer's instructions. After incubation, the samples were
washed with PBS and analyzed by FACSAria II (BD, USA). All
CD133+/CD326+ cells were collected for
subsequent experiments.
Quantitative real-time PCR
Total RNA was isolated using RNAiso Plus (Takara,
Japan). Circulating microRNA in peripheral blood was isolated using
the mirVana PARIS Kit (Ambion, USA). Reverse transcription
reactions were performed using the PrimeScript™ RT reagent kit with
gDNA Eraser (Takara) for mRNA and Bulge-Loop™ miRNA qRT-PCR Starter
kit (RiboBio) for miRNA to make cDNA from total RNA in a MyCycler
PCR system (Bio-Rad, USA). Subsequently, quantitative real-time PCR
was performed using SYBR® Premix Ex Taq II (Takara).
Primer pairs for miR-181b-5p, cel-miR-39 and U6 were purchased from
RiboBio Co. Ltd. Primer pairs for GAPDH and genes associated with
stemness and EMT were designed by Sangon Biotech Co. Ltd. (China).
Each sample was performed in triplicate, and the reaction products
were analyzed using the ABI 7500 Prism Sequence Detection system
(Applied Biosystems, USA). Data analysis was based on the Ct method
(∆∆Ct according to Applied Biosystems). All operations followed the
manufacturer's protocol.
Immunofluorescence assay
Spheres were centrifuged (800 rpm, 5 min) on slides
by cytospin and fixed with 4% paraformaldehyde and 0.1% Triton for
30 min, washed with PBS, blocked with BSA for 30 min at room
temperature, and then incubated with primary antibodies at 4°C
overnight. Primary antibodies were rabbit monoclonal anti-CD133
(Abcam, UK) and goat polyclonal anti-CD326 (Santa Cruz, USA) at a
dilution of 1:300. After washing, the spheroids were incubated with
goat anti-rabbit IgG-FITC (Beyotime, China) and donkey anti-goat
IgG-Cy3 (BioLegend, USA) fluorescent antibodies at a dilution of
1:400 for 30 min and protected from light. After DAPI staining for
the nucleus, the spheres were observed under an Olympus confocal
microscope.
MicroRNA expression profiling array and
data analysis
Adherent A549 cells and
CD133+/CD326+ cells were left untreated or
were treated with TGF-β1 as described above. Cells were lysed using
TRIzol (Life Technologies, USA) according to the manufacturer's
instructions. First, E. coli poly(A) polymerase was used to
generate polyadenylated tails at the 3′-end of all RNA molecules.
Second, after annealing oligo-dT primers, cDNA was synthesized
using the qScript Flex cDNA synthesis kit (Quanta Biosciences, USA)
according to the manufacturer's instructions for gene-specific
priming (a universal tag that would extend from the 3′-end of cDNA
molecules was added during reverse transcription). With the
addition of this universal tag, individual miRNAs were detected
with miRNA-specific forward primers and a reverse universal primer
mix. A SYBR Green-based quantitative real-time PCR method was used
to quantify the relative expression of mature miRNAs. A total of
362 mature miRNAs were evaluated in the microRNA expression
profiling array. miRNA expression was normalized by geometric
mean-based global normalization using the Real-Time StatMiner
(Integromics, Spain) analysis software. The array data were
processed by bioinformatics analysis.
Western blot assay
For western blot analysis, 40 µg of total
protein was resolved by SDS polyacrylamide gel (Boster, China)
electrophoresis, and the proteins were then transferred onto
polyvinylidene difluoride (PVDF) membranes (Millipore, USA). The
membranes were incubated with the Blocking kit (Boster) for 1 h and
then incubated overnight at 4°C with antibodies from the EMT
antibody sampler kit (1:1,000 dilution, Cell Signaling Technology,
USA). The protein levels were normalized against GAPDH from the
same sample (1:3,000 dilution, ZSGB-Bio, China). After three
washes, the membranes were incubated for 1 h at 37°C with
species-specific horseradish peroxidase-conjugated secondary
antibodies. The membranes were developed using an enhanced
chemiluminescence (ECL) detection system followed by exposure to
Hyperfilm ECL (Beyotime).
Transwell invasion assay
A Transwell invasion assay was performed using
24-well Transwell permeable supports with 8-µm pores
(Corning). Cells (2×105) were suspended in 200 µl
serum-free DMEM/F12 medium and seeded into the Transwell inserts
coated with Matrigel (BD Biosciences, USA). The bottoms of the
wells were filled with 500 µl complete medium containing 20%
fetal bovine serum. All cells were incubated at 37°C, 5%
CO2. After 24 h, the cells in the upper chamber were
removed with cotton swabs and the filter membrane was fixed with 4%
paraformaldehyde for 20 min. Subsequently, the remaining cells of
the filter membrane in the lower chamber were stained with 0.1%
crystal violet stain solution. From five randomly selected fields,
the cells that had invaded through the membrane to the lower
surface were counted under a light microscope.
In vivo xenograft experiments
Five-week-old male nude mice were purchased from the
Chinese Academy of Medical Sciences (Beijing, China). To generate
xenografts, serial dilutions (1×106, 1×105,
1×10, and 1×103) of A549 cells and
CD133+/CD326+ cells were injected into the
mice (n=5). Mice were monitored twice a week and sacrificed after 6
weeks.
Tumor metastasis assay
A549 cancer cells at 2×106/100 µl
were injected into the right armpit of the forelimb in all 10 mice.
The mice were observed twice per week. When xenografts reached ~5×5
mm (after nearly 2 weeks), we randomly chose 5 mice and injected 1
nmol micrON™ agomiR-181b-5p every 3 days for 4 weeks. The other 5
mice injected with micrON agomiR-181b-5p negative control were the
control group. After 6 weeks, all mice were sacrificed and
metastases were observed. All mouse experiments were performed in
accordance with local ethics guidelines.
Statistical analysis
The data are expressed as the means ± SD.
Statistical analysis of data was assessed using the Student's
t-test and one-way ANOVA with SPSS 18.0 software. Differences with
p<0.05 were considered statistically significant.
Results
Establishment of non-small lung cancer
stem-like cells from A549 cells
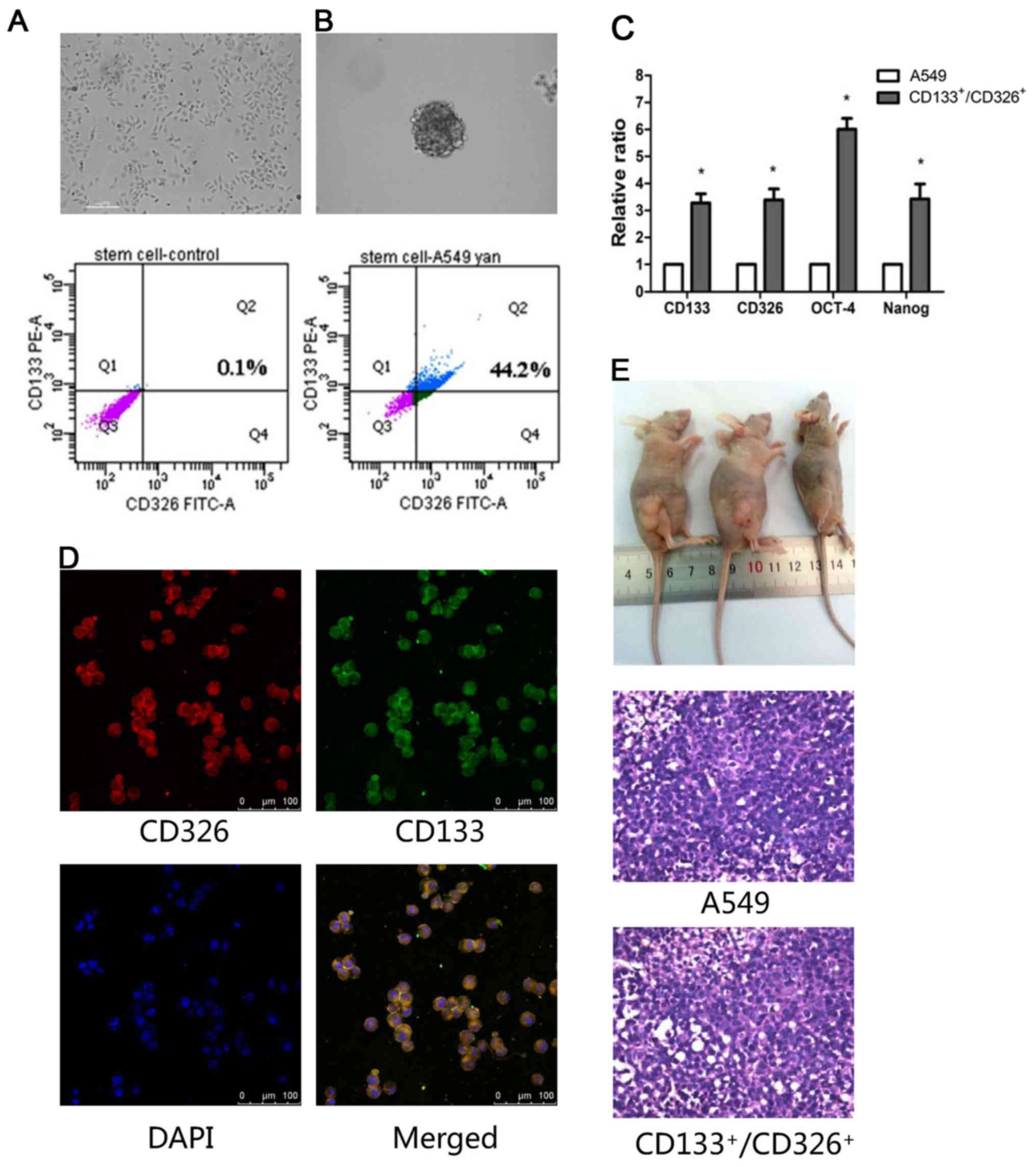
Only 0.1% of adherent A549 cells co-expressed CD133
and CD326 (Fig. 1A), which are
markers of stem cells. To enrich the
CD133+/CD326+ cells, A549 cells were
resuspended in serum-free medium and grown as spheroids. We
confirmed by flow cytometry that 44.2% cells in the spheroids
co-expressed CD133 and CD326 (Fig.
1B). We collected CD133+/CD326+ cells for
the subsequent experiments. Next, we studied the expression of
stemness-associated genes, including CD133, CD326, OCT-4 and Nanog,
using quantitative real-time PCR. We demonstrated that the
expression of these genes was higher in
CD133+/CD326+ cells compared to the adherent
A549 cells (p<0.05, Fig. 1C).
Immunofluorescence assay confirmed that most of the cells in
spheroids co-expressed CD133/CD326 (Fig. 1D). We demonstrated that as few as
1×104 CD133+/CD326+ cells
generated tumor xenografts in nude mice, whereas 1×105
unselected A549 cells failed to do so. All xenografts were
confirmed as adenocarcinoma by histologic examination (Fig. 1E and Table I). According to these results and
our previous study (13), we
concluded that the CD133+/CD326+ cell
subpopulation of A549 cells has the traits of CSLCs.
 | Table IXenografts on nude mice (male, 6
weeks old). |
Table I
Xenografts on nude mice (male, 6
weeks old).
| Cell number |
1×106 |
1×105 |
1×104 |
1×103 |
|---|
|
CD133+/CD326+
cells | – | 5/5 | 5/5 | 0/5 |
| A549 cells | 5/5 | 0/5 | 0/5 | – |
| Latency (day) | 13 | 15 | 18 | – |
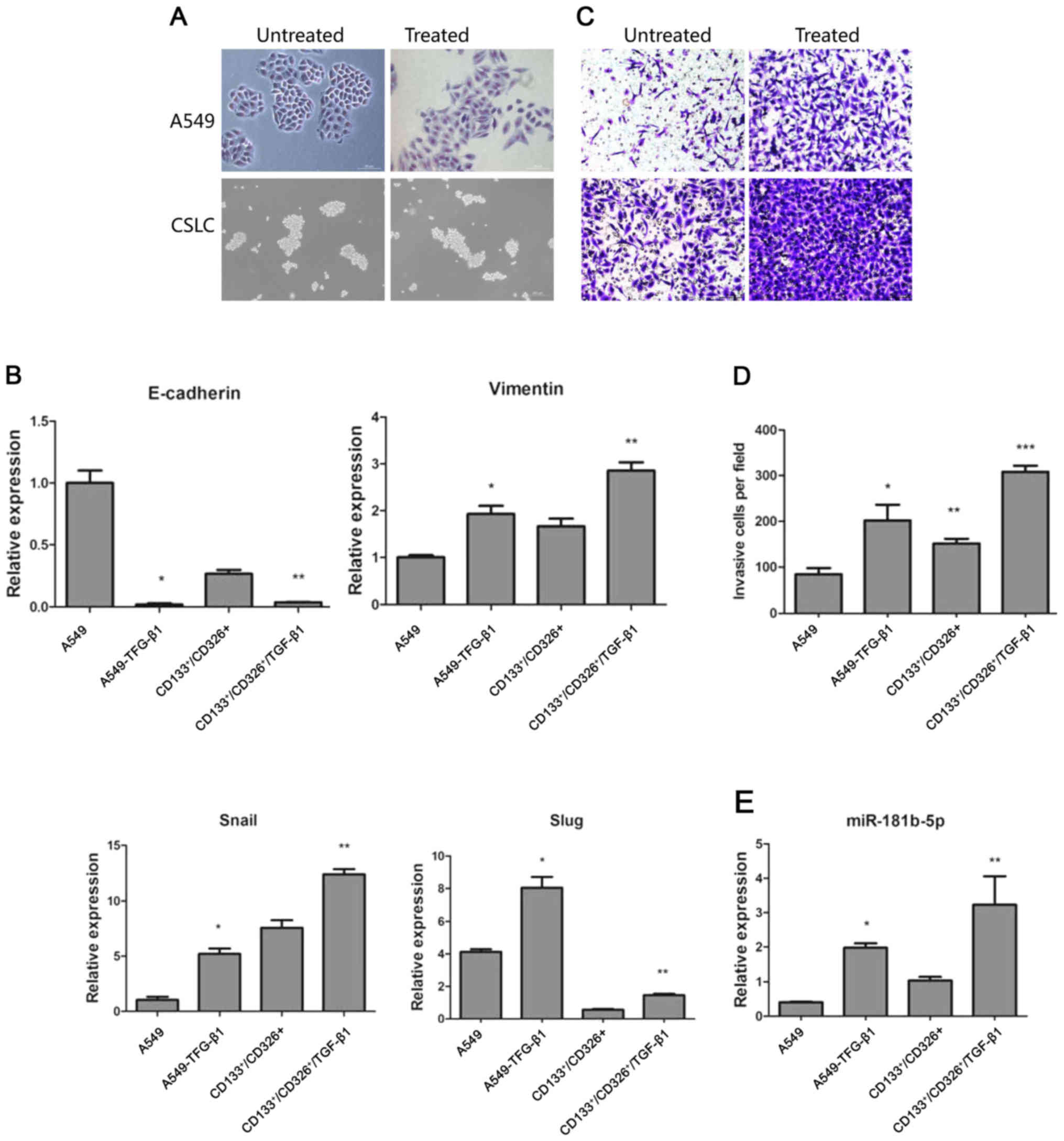
TGF-β1 promotes EMT both in CSLCs and
non-CSLCs
TGF-β1 has been shown to induce EMT in cancer cells
(24). We treated adherent A549
cells and CD133+/CD326+ cells with 5 ng/ml of
TGF-β1 for 72 h. As expected, adherent A549 cells treated with
TGF-β1 lost their epithelial morphology. Their shape had changed
drastically, the junction between cells became loose and cells
acquired a fibroblast-like appearance. In contrast, TGF-β1
treatment did not trigger significant morphological changes in the
CD133+/CD326+ subpopulation (Fig. 2A). We used quantitative real-time
PCR to assess the expression of E-cadherin, vimentin, Snail and
Slug. We demonstrated that TGF-β1 treatment induced the
upregulation of vimentin, Snail and Slug and the downregulation of
E-cadherin in both adherent A549 cells and
CD133+/CD326+ cells (Fig. 2B). This result indicates that
although the distinctive morphological changes of cells undergoing
EMT were not observed in CD133+/CD326+ cells,
the expression of EMT-associated genes indicative of the epithelial
to mesenchymal transition was observed in parental A549 cells and
in CD133+/CD326+ cells. Cancer cells
undergoing EMT are widely known to gain the ability to invade and
metastasize. We used Transwell invasion assay to show that compared
to control A549 cells, A549 cells treated with TGF-β1 have
increased invasion potential through the Matrigel membrane.
CD133+/CD326+ have an increased basal ability
to invade compared to unselected A549 cells and TGF-β1 further
increased their invasive capacity (Fig. 2C and D). Based on these results, we
concluded that TGF-β1 promotes EMT both in CSLCs and non-CSLCs.
MicroRNA profile reveals differential
miRNA expression in EMT and stem cell phenotype
To identify microRNAs involved in EMT and stem cell
phenotype, we performed microRNA expression profiles in A549 cells,
A549 cells treated with TGF-β1, CD133+/CD326+
cells and in CD133+/CD326+ cells treated with
TGF-β1 using a quantitative real-time PCR miRNA expression
profiling array. We identified common microRNAs between two groups
(A549 vs A549/TGF-β1 and CD133+/CD326+ vs
CD133+/CD326+/TGF-β1), their predicted target
genes (n=1180) and their binding sites using the TargetScan and
miRanda databases. We performed GO analysis and pathway analysis
based on the Gene Ontology database and the KEGG database. Finally,
according to the interactions between these genes and their
corresponding microRNAs, we constructed a microRNA-gene network.
The microRNAs in the network were evaluated by the degree, which is
the number of target genes regulated by one microRNA. A high degree
indicates that a large number of target genes is regulated by this
particular miRNA. Similarly, the degree of the gene indicates the
number of microRNAs targeting this gene. If a gene has a high
degree, multiple microRNAs may target it. The key miRNA and gene in
the network always have the highest degrees (Tables II and III). Several miRNAs in Table II have an established role in
cancer; however, the role of miR-181b-5p in EMT of CSLCs has not
been described. We confirmed by quantitative real-time PCR that
miR-181b-5p was significantly upregulated both in A549 and
CD133+/CD326+ cells treated with TGF-β1
(Fig. 2E). We hypothesize that
this may contribute to the invasion and metastasis of CSLCs and
non-CSLCs.
 | Table IIThe key microRNAs in the network
(degree >20). |
Table II
The key microRNAs in the network
(degree >20).
| microRNA | A549/TGF-β1 vs
A549 |
CD133+/CD326+/TGF-β1
vs CD133+/CD326+ | Degree |
|---|
| hsa-miR-497-5p | Down | Up | 69 |
| hsa-let-7e-5p | Up | Down | 62 |
| hsa-miR-671-5p | Up | Up | 42 |
|
hsa-miR-302c-3p | Up | Up | 37 |
|
hsa-miR-181b-5p | Up | Up | 27 |
|
hsa-miR-302a-3p | Down | Up | 25 |
| hsa-miR-25-3p | Up | Up | 23 |
|
hsa-miR-130b-3p | Up | Up | 20 |
 | Table IIIThe target genes regulated mostly in
the network (degree) >3. |
Table III
The target genes regulated mostly in
the network (degree) >3.
| Gene symbol | Description | Degree |
|---|
| PPARA | Peroxisome
proliferator-activated receptor α | 3 |
| TGFA | Transforming growth
factor, α | 3 |
| HDAC4 | Histone deacetylase
4 | 3 |
| ABL2 | v-abl Abelson
murine leukemia viral oncogene homolog 2 | 3 |
| E2F3 | E2F transcription
factor 3 | 3 |
| GABBR2 | γ-aminobutyric acid
(GABA) B receptor, 2 | 3 |
| TPP1 | Tripeptidyl
peptidase I | 3 |
miR-181b-5p is the key upregulator of EMT
and targets E-cadherin in CSLCs and non-CSLCs
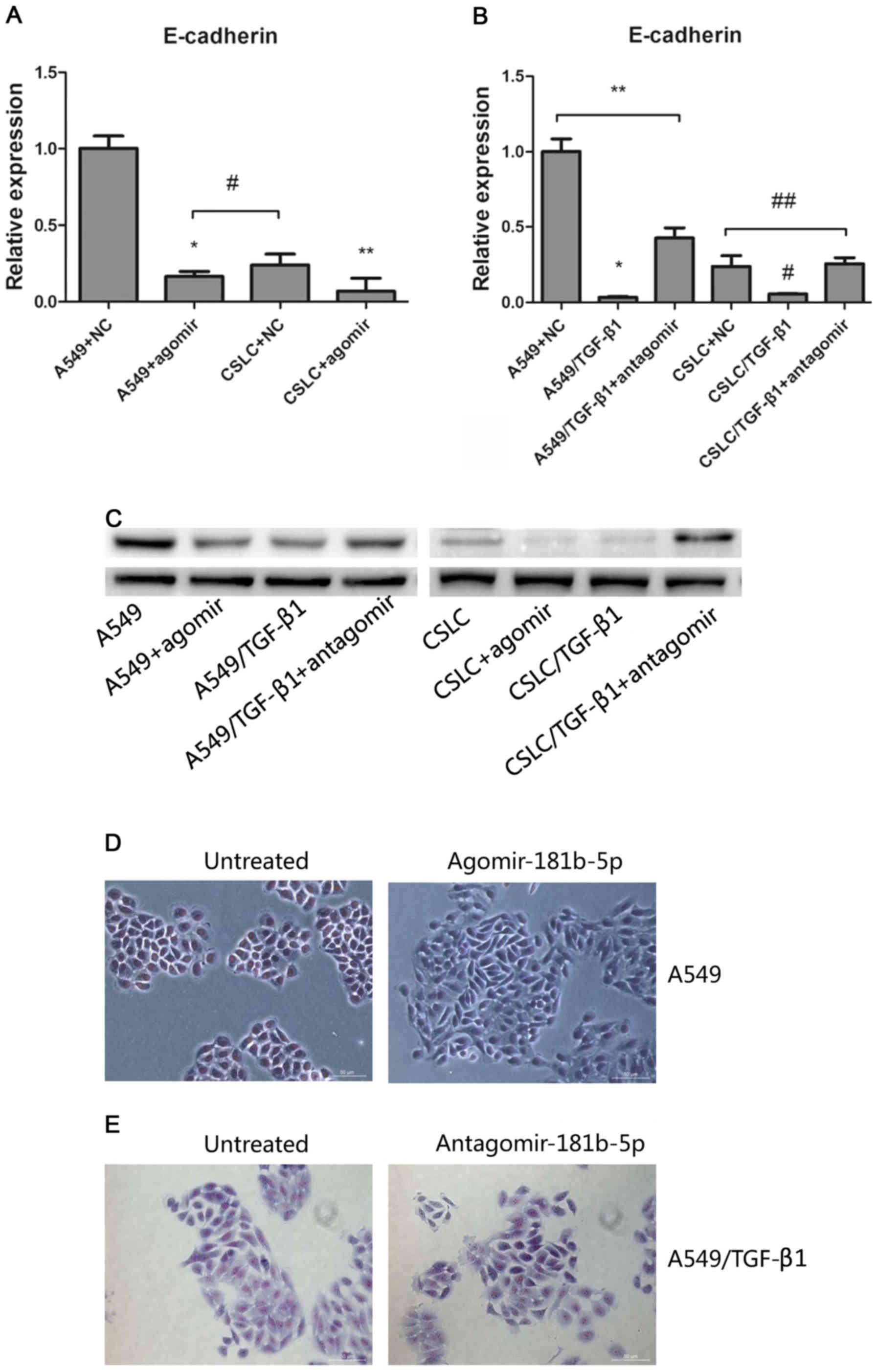
To modulate the expression of miR-181b-5p, we
transfected agomiR-181b-5p into A549 cells and
CD133+/CD326+ cells. In addition, we
transfected antagomiR-181b-5p into A549 cells and
CD133+/CD326+ cells that were treated with
TGF-β1 for 72 h. We confirmed that the expression of miR-181b-5p in
A549 cells and CD133+/CD326+ cells
transfected with agomiR-181b-5p was increased 8.75- and 25.3-fold
(Fig. 3A). Accordingly, the
transfection of antagomiR-181b-5p into A549 cells and
CD133+/CD326+ cells treated with TGF-β1
significantly reduced the expression of miR-181b-5p (Fig. 3B). This result confirmed that
agomiR-181b-5p was the agonist of miR-181b-5p and that
antagomiR-181b-5p was the antagonist of miR-181b-5p.
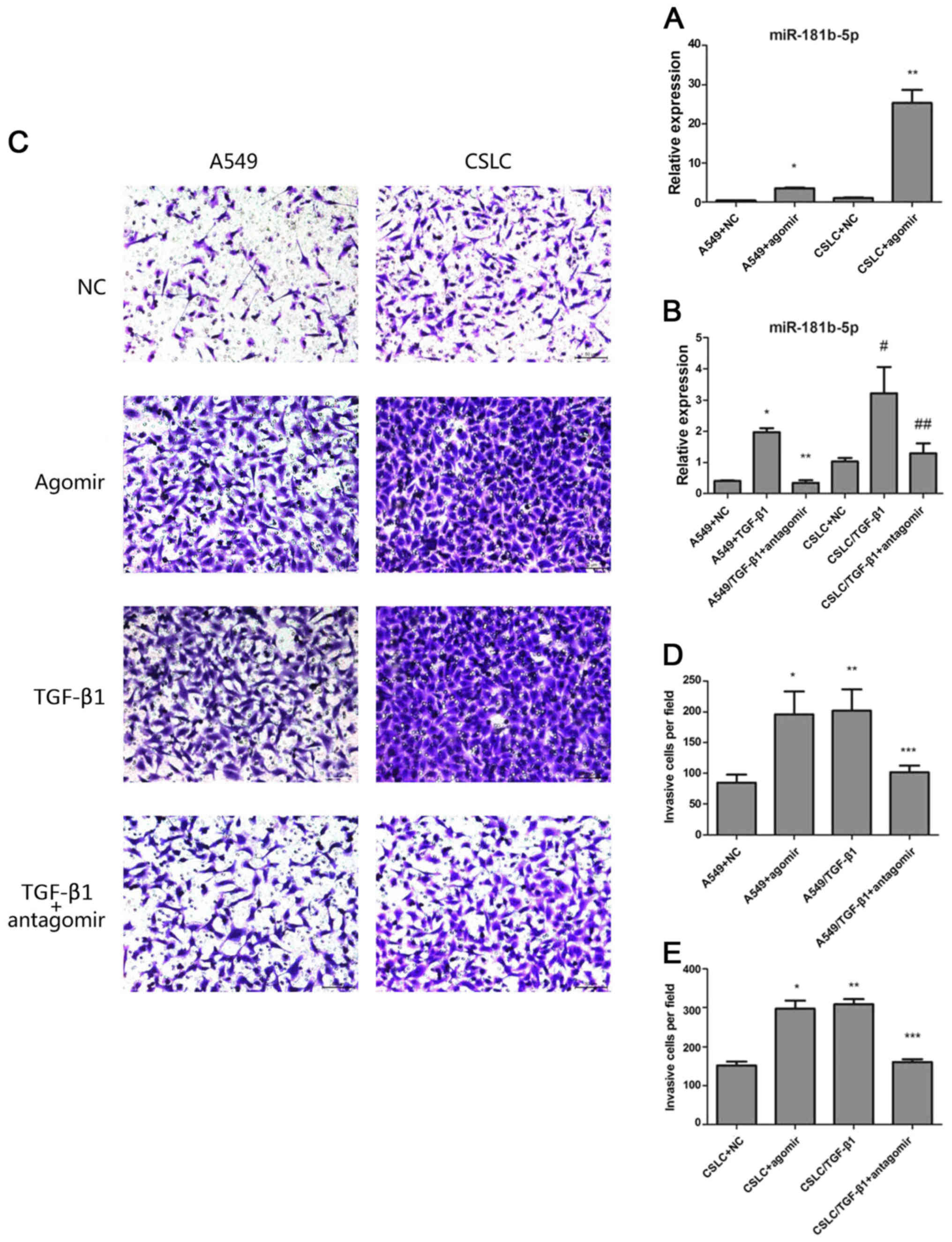 | Figure 3The effects of agomiR-181b-5p and
antagomiR-181b-5p. (A) Quantitative real-time PCR measured the
expression levels of miR-181b-5p in A549 cells and in CSLCs
transfected with agomiR-181b-5p (* vs A549, p<0.05;
** vs CSLC p<0.05). (B) Quantitative real-time PCR
was used to measure the expression levels of miR-181b-5p in
A549/TGF-β1 and CSLC/TGF-β1 transfected with antagomiR-181b-5p
(* vs A549/TGF-β1+antagomir p<0.05; # vs
CSLC/TGF-β1+antagomir, p<0.05; ** vs A549, p=0.2529;
## vs CSLC, p=0.300). (C) Detecting cell invasion by
Transwell invasion assay. (D) The effect of agomiR-181b-5p and
antagomiR-181b-5p on the invasion of A549 cells through the
Matrigel membrane (* vs A549, p<0.05; **
vs A549+agomir, p=0.6443; *** vs A549, p<0.05). (E)
The effect of agomiR-181b-5p and antagomiR-181b-5p on the invasion
CSLCs through the Matrigel membrane (* vs CSLC,
p<0.05; ** vs CSLC+agomir, p=0.0818; ***
vs CSLC, p<0.05). |
Next, we sought to determine whether miR-181b-5p
regulates EMT in vitro and in vivo. Transwell
invasion assay confirmed that agomiR-181b-5p, similar to TGF-β1
treatment, significantly enhanced the invasion of both A549 cells
and CD133+/CD326+ cells. In contrast,
antagomiR-181b-5p reduced the TGF-β1-mediated invasion in A549
cells and CD133+/CD326+ cells but did not
completely eliminate the effect of TGF-β1 (Fig. 3C–E). We treated adherent A549 cells
with 50 nM of agomiR-181b-5p for 7 days and observed that a part of
adherent A549 cells lost their epithelial morphology and acquired a
fibroblast-like appearance (Fig.
5D). These results indicated that the expression of miR-181b-5p
correlates with the ability of cells to invade in both CSLCs and
non-CSLCs and that miR-181b-5p contributes to TGF-β1-induced EMT.
Based on the in vitro studies, we hypothesized that the
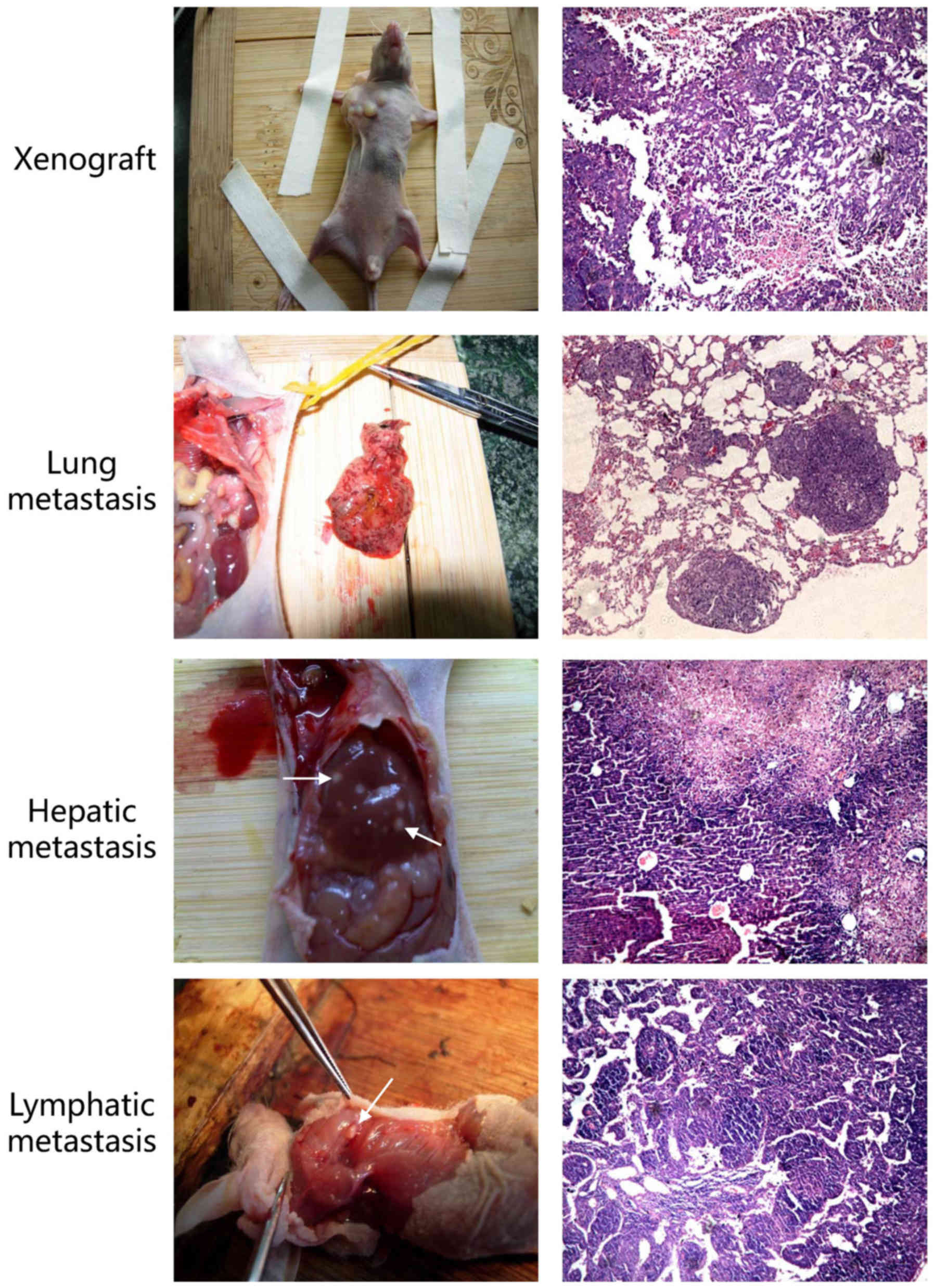
overexpression of miR-181b-5p may promote metastasis in
vivo. To confirm this critical question, we performed a tumor
metastasis assay and observed that the group of mice injected with
agomiR-181b-5p had more metastasis than the negative control
(Fig. 4 and Table IV). Thus, our findings suggest
that miR-181b-5p promotes invasion and metastasis both in CSLCs and
non-CSLCs in vitro and in vivo.
 | Table IVMetastasis on nude mice. |
Table IV
Metastasis on nude mice.
| Metastasis
sites | A549+NC | A549+agomir |
|---|
| Lung | 1 | 3 |
| Liver | 1 | 4 |
| Lymph node | 2 | 4 |
To identify the target gene of miR-181b-5p, we
evaluated the expression of EMT markers (such as E-cadherin,
vimentin, Snail and Slug) after the treatment of miR-181b-5p agomir
and antagomir in untreated or TGF-β1 treated A549 cells and
CD133+/CD326+ cells. After treatment with
agomiR-181b-5p, decreased E-cadherin expression was observed only
in A549 cells and CD133+/CD326+ cells
(Fig. 5A). In contrast,
antagomiR-181b-5p increased the expression of E-cadherin expression
in TGF-β1-treated A549 cells and
CD133+/CD326+ cells (Fig. 5B). Western blot analysis was used
to detect the protein levels of E-cadherin and to confirm our
results (Fig. 5C). These findings
suggested that miR-181b-5p promotes EMT at least in part by
targeting E-cadherin in CSLCs and non-CSLCs.
Circulating miR-181b-5p is a potential
clinical diagnostic marker
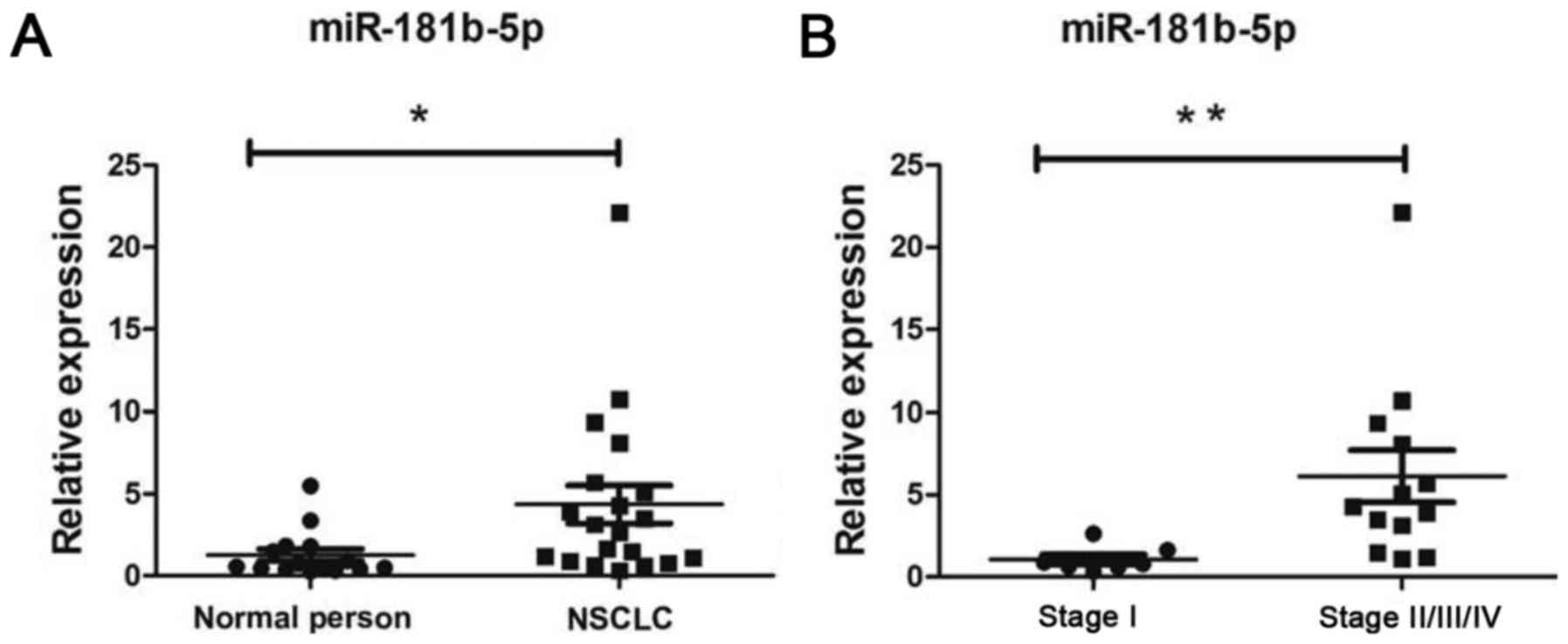
Finally, we compared the expression of miR-181b-5p
in peripheral blood of 15 healthy people and 20 patients with NSCLC
(patient characteristics are indicated in Table V). We found that the levels of
miR-181b-5p in peripheral blood were increased in NSCLC patients
(Fig. 6A) compared to healthy
individuals (p<0.05). miR-181b-5p was upregulated in stage
II/III/IV NSCLC patients (Fig. 6B,
p<0.05). According to this result, miR-181b-5p may be a
potential therapeutic target and a prognostic marker in NSCLC
patients.
 | Table VClinical data statistics of NSCLC
patients. |
Table V
Clinical data statistics of NSCLC
patients.
| Items | No. of patients
(n=20) |
|---|
| Age (year) | |
| ≤60 | 8 |
| >60 | 12 |
| Sex | |
| Male | 14 |
| Female | 6 |
|
Differerntiation | |
| Well | 3 |
| Moderate | 4 |
| Poor | 13 |
| Histology | |
|
Non-adenocarcinoma | 9 |
|
Adenocarcinoma | 11 |
| Lymph node
metastasis | |
| Yes | 11 |
| No | 9 |
| TNM stage | |
| I | 7 |
| II | 8 |
| III | 4 |
| IV | 1 |
Discussion
EMT is characterized by the loss of cellular
polarity and adhesion and by enhanced invasive and migratory
properties. Tumor cells treated with TGF-β1 have been shown to
display decreased expression of E-cadherin and cytokeratins while
overexpressing vimentin and N-cadherin, which is a hallmark of EMT
(21,25). Studies of circulating tumor cells
(CTCs) have shown that a subpopulation of CTCs shows
characteristics of EMT, such as the overexpression of Twist, Snail,
Slug, vimentin and FOXC1, and suggested that EMT induced by TGF-β1
is linked to the metastatic spread of cancer cells (26,27).
Tumor cells that acquire a mesenchymal phenotype are released into
peripheral blood; however, not all of these can form metastases.
The cancer stem cell hypothesis suggests that the rare population
of cancer cells that establishes metastases has characteristics of
CSCs. However, there is heterogeneity among CSCs. Brabletz et
al (28) proposed the concept
of migratory CSCs. Compared to stationary CSCs, migratory CSCs gain
migratory and metastatic properties by undergoing EMT in the
primary tumor. In squamous cell carcinoma (SCC), Biddle et
al (29) found that
CD44highESAhigh cells retained epithelial
characteristics (non-EMT CSCs) and that
CD44highESAlow cells were migratory and had
mesenchymal traits characteristic of EMT CSCs. In this study, we
investigated whether TGF-β1 can induce EMT in both CSCs and
non-CSCs during the initial stage of tumor metastasis in NSCLC.
For this purpose, we sorted a
CD133+/CD326+ cell subpop-ulation from A549
cells cultured in serum-free medium. These cells overexpressed
stemness-associated genes, show increased tumorigenicity, and could
be regarded as CSLCs. After treatment with TGF-β1, the expression
of vimentin, Snail and Slug was upregulated both in CSLCs and in
unsorted A549 cells. In contrast, the expression of E-cadherin was
decreased by TGF-β1. Consistent with apparent EMT, CSLCs treated
with TGF-β1 had the strongest capacity to invade in Transwell
invasion assay. Thus, we concluded that both CSLCs and non-CSLCs
can undergo EMT. The goal of this study was to assess the
contribution of microRNAs in CSLCs undergoing EMT.
To establish the effect of TGF-β1 treatment on the
expression of microRNAs in non-stem cells and CSLCs, we compared
the expression of microRNAs between two pairs of treatment groups:
A549 vs A549/TGF-β1 and CD133+/CD326+ vs
CD133+/CD326+/TGF-β1. Four microRNA
expression profiles, A549, A549/TGF-β1,
CD133+/CD326+ and
CD133+/CD326+/TGF-β1, were analyzed using
bioinformatics approaches, including GO analysis, pathway analysis
and graph theory (30–33) (Table
II). Several miRNAs have an established role in EMT; however,
the function of miR-181b-5p in promoting EMT in CSCs has not been
reported. We therefore focused on the role of miR-181b-5p in
EMT.
Previous studies showed that miR-181b functions as a
tumor suppressor or a tumor promoter in different human
malignancies (34–36). However, miR-181b-5p was rarely
reported in lung cancer. Liu et al (37) reported that miR-181b is
downregulated in NSCLC tissues compared with the normal adjacent
tissues. In contrast, Cinegaglia et al (38) found that miR-181b was overexpressed
in lung adenocarcinoma compared to normal lung tissue. Similar
results have been recently reported. Tian et al (39) characterized the expression profiles
of miRNAs in sera and in tissues collected from NSCLC patients and
found that miR-181b-5p was upregulated in serum and tissue of SCC
patients. In our study, we modulated the expression of miR-181b-5p
in A549 cells and CD133+/CD326+ cells and
found that miR-181b-5p enhanced invasion and metastasis in
vitro and in vivo. E-cadherin expression was negatively
correlated with miR-181b-5p expression, suggesting that E-cadherin
may be a target gene of miR-181b-5p. However, we observed that not
all of normal A549 cells treated with agomiR-181b-5p lost their
epithelial morphology and A549/TGF-β1 cells treated with
antagomiR-181b-5p did not lose their mesenchymal morphology
observably (Fig. 5D and E),
implying a lack of direct correlation between E-cadherin expression
and morphology. Additionally, CDH1 was not identified as a
potential target gene of miR-181b-5p, suggesting that miR-181b-5p
does not directly bind to E-cadherin mRNA. Transfection of
TGF-β1-treated A549 cells or CSLCs with antagomiR-181b-5p did not
reduce their invasive capacity to the levels of untreated cells
(Fig. 3C–E), demonstrating that
miR-181b-5p is one of several regulators involved in
TGF-β1-dependent EMT.
Finally, we compared the levels of miR-181b-5p in
peripheral blood of healthy humans and in patients with NSCLC. We
found that the levels of miR-181b-5p were increased in peripheral
blood of NSCLC patients and specifically in patients with stage
II/III/IV disease. These data and the results discussed above
suggest that miR-181b-5p may be a diagnostic biomarker of
NSCLCs.
In conclusion, this study provides evidence that
miR-181b-5p mediates TGF-β1-induced EMT by suppressing E-cadherin
expression both in normal A549 cells and
CD133+/CD326+ cells which have
characteristics of CSLCs. miR-181b-5p may be a diagnostic marker
and a new therapeutic target in NSCLC. Further studies are required
to reveal the mechanism of miR-181b-5p suppression of the
E-cadherin expression and to analyze the association between
miR-181b-5p and NSCLC.
Acknowledgments
This study was supported by grants in aid (CSTC,
2011AB5032) from Chongqing Science and Technology Commission,
China. We are grateful to Dr Sheng Lin from the Oncology
Department, The Affiliated Hospital of Southwest Medical University
for helpful discussion and Dr Xinwei Diao from the Pathology
Department of Xinqiao Hospital for pathologic technical
support.
References
|
1
|
Gupta G, Singh R, Kotasthane DS and
Kotasthane VD: Myelodysplastic syndromes/neoplasms: Recent
classification system based on World Health Organization
Classification of Tumors - International Agency for Research on
Cancer for Hematopoietic and Lymphoid Tissues. J Blood Med.
1:171–182. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ghotra VP, Puigvert JC and Danen EH: The
cancer stem cell microenvironment and anti-cancer therapy. Int J
Radiat Biol. 85:955–962. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kreso A and Dick JE: Evolution of the
cancer stem cell model. Cell Stem Cell. 14:275–291. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Florian S, Sonneck K, Hauswirth AW, Krauth
MT, Schernthaner GH, Sperr WR and Valent P: Detection of molecular
targets on the surface of CD34+/CD38− stem
cells in various myeloid malignancies. Leuk Lymphoma. 47:207–222.
2006. View Article : Google Scholar
|
|
6
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sarvi S, Mackinnon AC, Avlonitis N,
Bradley M, Rintoul RC, Rassl DM, Wang W, Forbes SJ, Gregory CD and
Sethi T: CD133+ cancer stem-like cells in small cell
lung cancer are highly tumorigenic and chemoresistant but sensitive
to a novel neuropeptide antagonist. Cancer Res. 74:1554–1565. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pak MG, Shin DH, Lee CH and Lee MK:
Significance of EpCAM and TROP2 expression in non-small cell lung
cancer. World J Surg Oncol. 10:532012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zakaria N, Yusoff NM, Zakaria Z, Lim MN,
Baharuddin PJ, Fakiruddin KS and Yahaya B: Human non-small cell
lung cancer expresses putative cancer stem cell markers and
exhibits the transcriptomic profile of multipotent cells. BMC
Cancer. 15:842015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Leung EL, Fiscus RR, Tung JW, Tin VP,
Cheng LC, Sihoe AD, Fink LM, Ma Y and Wong MP: Non-small cell lung
cancer cells expressing CD44 are enriched for stem cell-like
properties. PLoS One. 5:e140622010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang F, Qiu Q, Khanna A, Todd NW, Deepak
J, Xing L, Wang H, Liu Z, Su Y, Stass SA, et al: Aldehyde
dehydrogenase 1 is a tumor stem cell-associated marker in lung
cancer. Mol Cancer Res. 7:330–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen YC, Hsu HS, Chen YW, Tsai TH, How CK,
Wang CY, Hung SC, Chang YL, Tsai ML, Lee YY, et al: Oct-4
expression maintained cancer stem-like properties in lung
cancer-derived CD133-positive cells. PLoS One. 3:e26372008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin S, Sun JG, Wu JB, Long HX, Zhu CH,
Xiang T, Ma H, Zhao ZQ, Yao Q, Zhang AM, et al: Aberrant microRNAs
expression in CD133+/CD326+ human lung
adenocarcinoma initiating cells from A549. Mol Cell. 33:277–283.
2012. View Article : Google Scholar
|
|
14
|
Brabletz T: EMT and MET in metastasis:
Where are the cancer stem cells? Cancer Cell. 22:699–701. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thiery JP and Lim CT: Tumor dissemination:
An EMT affair. Cancer Cell. 23:272–273. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tu Z, Xie S, Xiong M, Liu Y, Yang X, Tembo
KM, Huang J, Hu W, Huang X, Pan S, et al: CXCR4 is involved in
CD133-induced EMT in non-small cell lung cancer. Int J Oncol.
50:505–514. 2017.
|
|
17
|
Abba ML, Patil N, Leupold JH and Allgayer
H: MicroRNA regulation of epithelial to mesenchymal transition. J
Clin Med. 5:52016. View Article : Google Scholar
|
|
18
|
Gibbons DL, Lin W, Creighton C, Zhang S,
Lozano G and Kurie J: Use of a murine model of NSCLC to evaluate
the role of the microRNA-200 family in regulating EMT and
metastasis. J Clin Oncol. 27:110062009.
|
|
19
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nishijima N, Seike M, Soeno C, Chiba M,
Miyanaga A, Noro R, Sugano T, Matsumoto M, Kubota K and Gemma A:
miR-200/ZEB axis regulates sensitivity to nintedanib in non-small
cell lung cancer cells. Int J Oncol. 48:937–944. 2016.PubMed/NCBI
|
|
21
|
Cao M, Seike M, Soeno C, Mizutani H,
Kitamura K, Minegishi Y, Noro R, Yoshimura A, Cai L and Gemma A:
miR-23a regulates TGF-β-induced epithelial-mesenchymal transition
by targeting E-cadherin in lung cancer cells. Int J Oncol.
41:869–875. 2012.PubMed/NCBI
|
|
22
|
Hahn S, Jackstadt R, Siemens H, Hünten S
and Hermeking H: SNAIL and miR-34a feed-forward regulation of
ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J.
32:3079–3095. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Díaz-López A, Moreno-Bueno G and Cano A:
Role of microRNA in epithelial to mesenchymal transition and
metastasis and clinical perspectives. Cancer Manag Res. 6:205–216.
2014.PubMed/NCBI
|
|
24
|
Derynck R, Muthusamy BP and Saeteurn KY:
Signaling pathway cooperation in TGF-β-induced
epithelial-mesenchymal transition. Curr Opin Cell Biol. 31:56–66.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma M, He M, Jiang Q, Yan Y, Guan S, Zhang
J, Yu Z, Chen Q, Sun M, Yao W, et al: miR-487a promotes
TGF-β1-induced EMT, the migration and invasion of breast cancer
cells by directly targeting MAGI2. Int J Biol Sci. 12:397–408.
2016. View Article : Google Scholar :
|
|
26
|
Yu M, Bardia A, Wittner BS, Stott SL, Smas
ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al:
Circulating breast tumor cells exhibit dynamic changes in
epithelial and mesenchymal composition. Science. 339:580–584. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kallergi G, Papadaki MA, Politaki E,
Mavroudis D, Georgoulias V and Agelaki S: Epithelial to mesenchymal
transition markers expressed in circulating tumour cells of early
and metastatic breast cancer patients. Breast Cancer Res.
13:R592011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brabletz T, Jung A, Spaderna S, Hlubek F
and Kirchner T: Opinion: Migrating cancer stem cells - an
integrated concept of malignant tumour progression. Nat Rev Cancer.
5:744–749. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Biddle A, Liang X, Gammon L, Fazil B,
Harper LJ, Emich H, Costea DE and Mackenzie IC: Cancer stem cells
in squamous cell carcinoma switch between two distinct phenotypes
that are preferentially migratory or proliferative. Cancer Res.
71:5317–5326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gene Ontology C; Gene Ontology Consortium:
The Gene Ontology (GO) project in 2006. Nucleic Acids Res.
34:D322–D326. 2006. View Article : Google Scholar :
|
|
31
|
Draghici S, Khatri P, Tarca AL, Amin K,
Done A, Voichita C, Georgescu C and Romero R: A systems biology
approach for pathway level analysis. Genome Res. 17:1537–1545.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guo CJ, Pan Q, Li DG, Sun H and Liu BW:
miR-15b and miR-16 are implicated in activation of the rat hepatic
stellate cell: An essential role for apoptosis. J Hepatol.
50:766–778. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Joung JG, Hwang KB, Nam JW, Kim SJ and
Zhang BT: Discovery of microRNA-mRNA modules via population-based
probabilistic learning. Bioinformatics. 23:1141–1147. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhi F, Shao N, Wang R, Deng D, Xue L, Wang
Q, Zhang Y, Shi Y, Xia X, Wang S, et al: Identification of 9 serum
microRNAs as potential noninvasive biomarkers of human astrocytoma.
Neuro Oncol. 17:383–391. 2015. View Article : Google Scholar :
|
|
35
|
Zheng Y, Lv X, Wang X, Wang B, Shao X,
Huang Y, Shi L, Chen Z, Huang J and Huang P: miR-181b promotes
chemoresistance in breast cancer by regulating Bim expression.
Oncol Rep. 35:683–690. 2016.
|
|
36
|
Wang J, Sai K, Chen FR and Chen ZP:
miR-181b modulates glioma cell sensitivity to temozolomide by
targeting MEK1. Cancer Chemother Pharmacol. 72:147–158. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu Y, Hu X, Xia D and Zhang S:
MicroRNA-181b is down-regulated in non-small cell lung cancer and
inhibits cell motility by directly targeting HMGB1. Oncol Lett.
12:4181–4186. 2016.PubMed/NCBI
|
|
38
|
Cinegaglia NC, Andrade SC, Tokar T,
Pinheiro M, Severino FE, Oliveira RA, Hasimoto EN, Cataneo DC,
Cataneo AJ, Defaveri J, et al: Integrative transcriptome analysis
identifies deregulated microRNA-transcription factor networks in
lung adenocarcinoma. Oncotarget. 7:28920–28934. 2016.PubMed/NCBI
|
|
39
|
Tian F, Shen Y, Chen Z, Li R, Lu J and Ge
Q: Aberrant miR-181b-5p and miR-486-5p expression in serum and
tissue of non-small cell lung cancer. Gene. 591:338–343. 2016.
View Article : Google Scholar : PubMed/NCBI
|