Introduction
Hematopoietic differentiation occurs as a result of
a distinct gene expression program, whereby the self-renewal of
pluripotent hematopoietic stem cells and sequential commitment of
intermediate progenitors, with a decreased capacity to proliferate,
is governed by specific combinations of lineage-specific
transcription factors (1).
Transcription factors (TFs), such as those in the ETS gene family,
play an integral role in hematopoiesis by coordinating the balance
between proliferation and differentiation and influencing
properties of self-renewal (2).
Therefore, unsurprisingly the dysregulation of normal ETS
transcriptional machinery plays a causal role in several human and
murine hematological malignancies associated with chromosomal
translocations or viral insertions.
Multi-stage erythroleukemia induced by Friend virus
has served as an excellent mouse model to study the effects of
specific ETS transcription factors associated with hematological
pathogenesis. Friend virus-induced erythroleukemia is characterized
by a marked expansion of erythroid progenitors. The two strains of
the Friend virus complex, polycythemia-and anemia-inducing
isolates, consist of a replication defective spleen-focus forming
virus (SFFV) and a replication-competent Friend murine leukemia
virus (F-MuLV) (3–5). The emergence of clonal tumorigenic
erythroblasts is dependent upon retroviral insertional activation
of the ETS transcription factors, spi-1/PU.1 in SFFV-induced
erythroleukemia (6), and
fli-1, in F-MuLV-induced erythroleukemia (7,8). In
both cases, tumorigenic erythroid progenitor cells are blocked in
differentiation at the proerythroblast stage, with self-renewal
capacities. SFFV induces enhancement of proerythroblasts exhibiting
properties of erythroid colony-forming (CFU-E) cells, whereas
F-MuLV-induced erythroleukemic cells exhibit properties of
erythroid burst forming (BFU-E) cells (9). Leukemic cells grown in
methylcellulose have given rise to several established
erythropoietin (Epo)-independent cell lines. Recent evidence
suggests that the maintenance of the malignant phenotype in these
cell lines is dependent upon the aberrant regulation of
fli-1 (10–12).
Fli-1, in addition to involvement in
erythroleukemia, is also overexpressed in almost all hematological
malignancies and activated as a result of translocation in Ewing's
sarcoma resulting in generation of fusion protein EWS-Fli-1 with
strong oncogenic activity (13).
In human, Fli-1 deficiency was associated with both erythroid and
megakaryocytic development (14,15).
Studies of Friend virus-induced erythroleukemia have implied that
activation of Fli-1 inhibits the commitment of erythroid
progenitors to differentiate through disruption of critical
erythroid signaling pathways, such as that of Epo and stem cell
factor (SCF). Indeed, Fli-1 has been shown to alter the expression
of erythroid lineage-associated genes, such as Rb (15), bcl-2 (16) GATA1 (17) and SHIP-1 (18).
To directly assess the role of ETS genes in
erythroid transformation, an SFFV-induced erythroleukemia cell line
was generated to ectopically express Fli-1 along with green
fluorescent protein (GFP) reporter. Using this erythroleukemic cell
line, we show that Fli-1 overexpression de-differentiates these
cells to earlier progenitor status. However, contrary to Fli-1,
when Spi-1/PU.1 is overexpressed in an F-MuLV-induced
erythroleukemia cell line, these cells differentiate to a more
mature erythroid progenitor. These data suggest that Fli-1 and
Spi-1/PU.1 function differently and target distinct erythroid
progenitors during erythroleukemogenesis.
Materials and methods
Cell culture and treatments
Erythroleukemia cell lines DP-17-17 and CB3 were
maintained in alpha-minimum essential medium (α-MEM) (Gibco, Grand
Island, NY, USA) supplemented with 10% fetal bovine serum (FBS)
(Gibco). HEK293T cells were maintained in Dulbecco's modified
Eagle's medium (DMEM) (Gibco) supplemented with 10% fetal bovine
serum (FBS) (Gibco). To induce erythroid differentiation, FACS
sorted DP17-17 cells were treated for two days with 2% dimethyl
sulfoxide (DMSO) (Sigma-Aldrich, Oakville, ON, Canada).
Differentiation assays were performed in triplicate by seeding
(1×105) cells/well in 3 ml of a 6-well plate. After 48 h
of induction with DMSO, adherent cells were removed from the
culture dish using a cell scraper for cytospin preparation and
histological analysis.
Enforced expression of Fli-1 and
Spi-1
The MigR1-Fli-1, or empty vector control plasmid,
MigR1, was triple-transfected with Lipofectamine 2000 (Invitrogen,
Burlington, Canada) into HEK293T cells, following the
manufacturer's protocol. In this transfection we included the
vesicular stomatitis virus G glycoprotein (VSVG)-expressing vector,
as well as the gag and pol virus packaging signals
were provided by Dr D. Barber, University of Toronto. Viral
supernatant was collected 48 h post-transfection. DP17-17
(2.5×106) were infected with virus, and incubated 16 h
with polybrene (8 μg/ml final concentration), as previously
described (10). Two days
post-infection, cells were sorted by flow cytometry based on the
intensity of green fluorescence. Sorted cell populations expanded
in culture, and were sorted a second time based on high intensities
of green fluorescence.
The MSCV-Spi-1/PU.1 or empty vector MSCV DNA was
transfected into CB3 cells and after 3 days selected with neomycin
(800 mg/ml) for two weeks. The G418 resistant cells were pooled and
used in this study.
qRT-PCR analysis
RNA levels were determined by quantitative real-time
PCR in a StepOne Plus thermal cycler (Applied Biosystems, Forest
City, CA, USA) using specific primers and SYBR® select
Master Mix (Life Technologies, Carlsbad, CA, USA), as described
(19). The β-actin gene was used
as control, the list of primers used for RT-PCR and qRT-PCR is as
follows: all experiments were performed in triplicates and repeated
at least two times.
The murine β-actin primers: 5′ forward
(5′-GTGACGTTGACATCCGTAAAGA-3′) and 3′ reverse
(5′-GCCGGACTCATCGTACTCC-3′). The murine GATA-1: 5′ forward
(5′-CACCCTGAACTCGTCATACC-3′) and 3′ reverse
(5′-ACCAGGGCAGAATCCACAAA-3′). The murine TAL-1: 5′ forward
(5′-CGGCAGCAGAATGTGAATGG-3′) and 3′ reverse
(5′-CTCCTGGTCATTGAGTAACTTGG-3′). The murine RUNX1: 5′ forward
(5′-TGGTGGAGGTACTAGCTGACC-3′) and 3′ reverse
(5′-CGAGTAGTTTTCATCGTTGCCT-3′).
Immunoblotting
Cells were lysed with lysis buffer (0.5% Nonidet
P-40, 50 mM Tris-HCl (pH 8.0), 120 mM NaCl, 50 mM NaF, plus 1 mM
Na3VO4, 10 g/ml aprotinin, 100 g/ml leupeptin
and 10 mM phenylmethylsulfonyl fluoride). Lysates (40 μg)
were fractionated by SDS/PAGE and transferred to a polyvinylidene
fluoride membrane (Immobilon-P, Millipore, Billerica, MA, USA). The
following antibodies were used: SHIP-1, Fli-1 (Santa Cruz
Biotechnology, Santa Cruz, CA, USA); β-actin (Sigma-Aldrich);
goat-anti-mouse, and goat anti-rabbit HRP-conjugated (Promega,
Madison, WI, USA), ERK, phospho-AKT, AKT, MYC and JAK2 antibodies
from Cell Signaling Technology (CST, Danvers, MA, USA).
Cellular proliferation assay
Transduced DP17-17 and CB3 cells, 1×104,
were plated in triplicates, removed at 24-h intervals, and cellular
proliferation was measured by performing trypan-blue exclusion
assay and counting using a hemocytometer.
Cytospin preparation and histochemical
staining
Cells (2×104 per slide) were cytospun
onto glass slides for 15 min at 1,000 rpm (Cytospin; Thermo
Shandon, USA). Cells were fixed at room temperature in methanol for
5 min and air-dried. Fixed cells were stained with May-Grunwald
stain followed by Giemsa stain per the manufacturer's protocol
(Sigma). Light microscopy images were obtained using a Leica DM LB2
microscope, Leica DFC 300FX camera, and Leica Application Suite
3.1.0 software (Leica Microsystems, Switzerland). Blinded erythroid
differential counts were performed by a hematopathologist, at
Sunnybrook Health Sciences Centre, University of Toronto. A total
of 6 cytospin slides with May-Grunwald Giemsa stains were prepared
during two separate experiments for each non-transduced,
double-sorted MigR1, and MigR1-Fli-1 DP17-vector cell groups.
Approximately 100–200 cells were counted on each slide and
categorized into one of the three defined stages; R1,
proerythroblast; R2, early basophilic erythroblast; R3, late
basophilic erythroblast. Data are presented as the percentage of
total cells analyzed.
Immunostaining and flow cytometric
analysis
Freshly isolated cells were washed twice in PBS
(Gibco) and immunostained for 15 min with the appropriate antibody;
phycoerythrin (PE)-conjugated anti-TER119 (erythroid marker),
PE-conjugated Anti-cKIT (SCF receptor), PE-conjugated anti-CD41
(glycoprotein IIb), PE-conjugated anti-CD61 (glycoprotein IIIa),
PE-conjugated anti-Gr-1 (granulocytic marker), PE-conjugated
anti-MAC-1 (monocytic marker), APC-conjugated anti-CD71
(transferrin receptor), APC-conjugated anti-SCA-1 (primitive
hematopoietic cell marker) (1:200) (eBioscience, San Diego, CA,
USA). Following antibody incubation, cells were washed once in PBS
and resuspended in 500 μl PBS. Cell sorting and analysis of
stained cells were performed using the Becton-Dickinson FACSCalibur
(BD Biosciences, San Jose, CA, USA), and the FlowJo flow cytometry
analysis software (FlowJo TreeStar Inc., Ashland, OR, USA).
Relative mean fluorescence intensity (MFI) values were based on the
unstained population controls and calculated using the Geometric
Mean statistic (average of log fluorescence). Statistical analyses
were performed using the two-tailed Student's t-test, where
P<0.05 was considered to indicate a statistically significant
difference.
Colony-forming cell assay
DP17-17 cells, transduced with the MigR1 empty
vector control or MigR1 Fli-1 expressing vector and double sorted,
were suspended in Iscove's modified Dulbecco's medium (IMDM)
(Gibco) supplemented with 2% FBS (Gibco) and added to
methylcellulose Medium (M334, or M3434, Stem Cell Technologies,
Vancouver, BC, Canada) to assay for the presence of erythroid
colony-forming units (CFU-E) and mature erythroid burst-forming
units (BFU-E) (M3334 formulation consists of 15% FBS, 1% BSA, 10
μg/ml insulin, 200 μg/ml transferrin, 3 U/ml rh EPO,
and M3434 formulation consists of 15% FBS, 1% BSA, 10 μg/ml
insulin, 200 μg/ml transferrin, 50 ng/ml rm SCF, 10 ng/ml rm
IL-3, 10 ng/ml IL-6, 3 U/ml rh EPO), according to the
manufacturer's protocol. Colonies were counted after 12 days of
culture. CFU-E and BFU-E colonies were detected by staining with
benzidine solution, 0.4% benzidine (Sigma-Aldrich) in 12% acetic
acid, with the addition of 0.3% hydrogen peroxide (Sigma).
Individual colonies were isolated from methylcellulose cultures,
cells were resuspended, and prepared for cytospins and
histochemical staining, as described above.
Transplantation assay
DP17-17 cells, transduced with the MigR1 empty
vector control or MigR1-Fli-1 expressing vector, were sorted by
flow cytometry based on high intensities of green fluorescence, as
described above. Double-sorted DP17-17 were suspended in
phosphate-buffered saline (PBS) (Gibco), in a total volume of 200
μl, and administered intravenously into the tail veins of
groups of female recipient eight-week-old DBA/2J mice, at
concentrations of 1.0×106, 1.0×105 or
1.0×104 cells. Injected mice were sacrificed if they
presented with symptoms of disease progression, such as paleness,
hunched posture, enlarged abdomen, and paralysis or difficulty
breathing. Spleen and liver samples were isolated from recipient
mice, cultured for two days, and subjected to flow cytometric
analyses, as described above. The contribution and presence of the
transduced DP17-17 cells injected was evaluated based on the
detection of green fluorescence.
Survival and statistical analysis
The mouse survival rates were computed and plotted
according to the non-parametric Kaplan-Meier analysis. Statistical
analysis was performed using the two-tailed Student's t-test with
significance considered at P<0.05, and by analysis of variance
using Origin 3.5 software (Microcal Software, Northampton, MA,
USA).
Animal care
Animal care was in accordance with the guidelines of
the institution, animal care committee and University of
Toronto.
Results
Exogenous expression of Fli-1 alters the
state of erythroid differentiation
The spleen focus forming virus (SFFV) and the Friend
murine leukemia virus (F-MuLV) induces erythroleukemia associated
with insertional activation of spi-1 (PU.1) and
fli-1, respectively (3). As
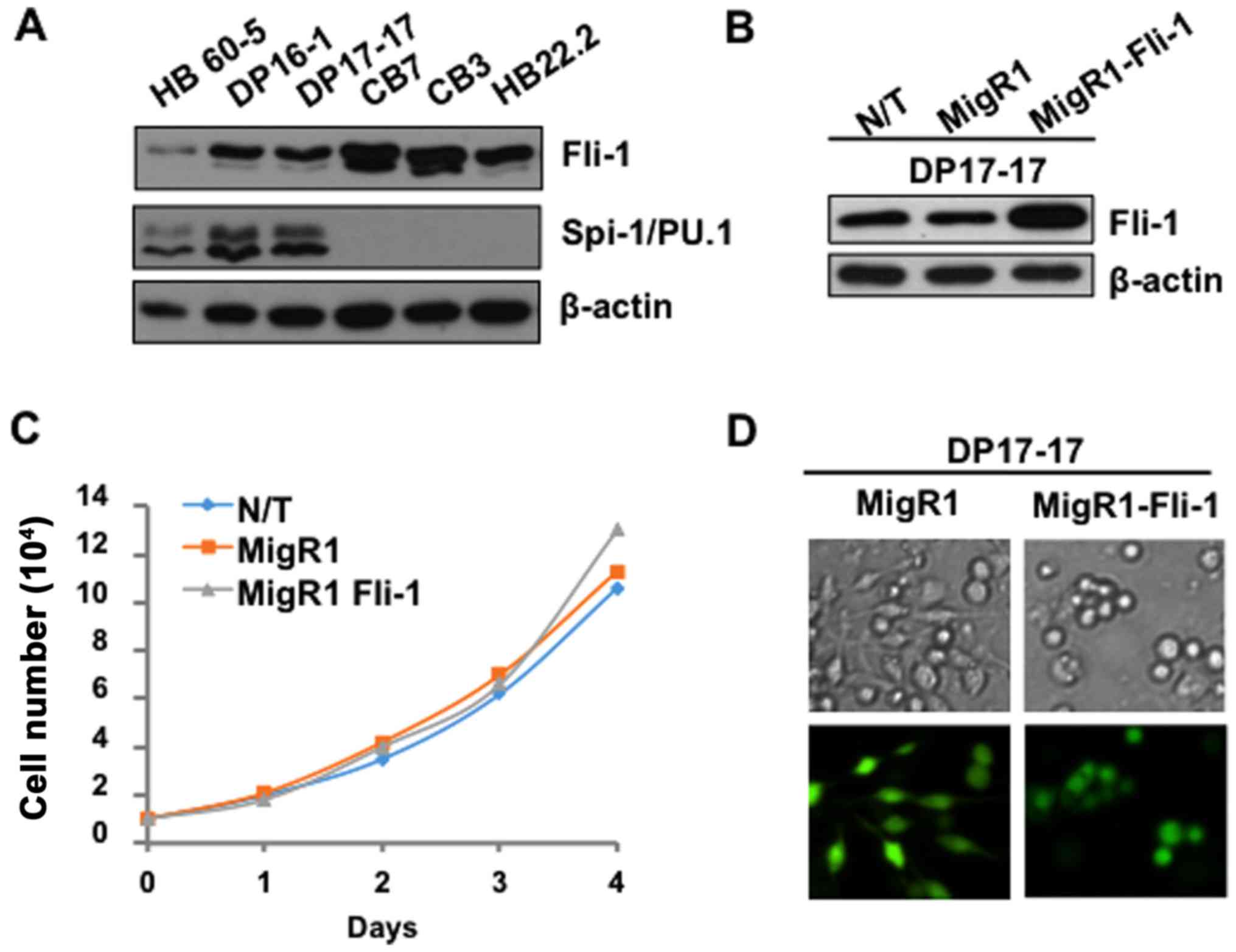
shown in Fig. 1A, Spi-1/PU.1 is
overexpressed in SFFV-induced erythroleukemia cell lines HB60-5,
DP16-1, and DP17-17, however, its expression is absent in the
F-MuLV-induced erythroleukemia cells CB7, CB3 and HB22.2,
overexpressing Fli-1. The SFFV-induced erythroleukemic cell lines
express Fli-1, albeit at significantly lower levels, compared to
F-MuLV-induced erythroleukemia cell lines (Fig. 1A).
To decipher the role of Fli-1 in erythroid
proliferation and differentiation, exogenous expression of Fli-1
was introduced into the SFFV-induced erythroleukemia cell line
DP17-17. DP17-17 cells, transduced with either the MigR1 empty
vector control, or MigR1-Fli-1 expressing retrovirus, were sorted
by flow cytometry based on the intensity of green fluorescence two
days post-infection. Sorted cell populations were isolated, and
subjected to western blot analysis to confirm enforced expression
of Fli-1 (Fig. 1B). Trypan blue
exclusion assay was performed to reveal the effects of Fli-1
overexpression on the proliferation of the SFFV-induced
erythroleukemic cells. The non-transduced (N/T), MigR1 and
MigR1-Fli-1 transduced cell populations retained a comparable
proliferation rate (Fig. 1C),
indicating that Fli-1 overexpression has no additional effect on
proliferation in these cells in culture. However, upon microscopic
examination of transduced DP17-17 cultures, a clear distinction was
observed in the gross morphology of the transduced populations.
Typically, DP17-17 cells growing in culture have both an adherent
and suspension population, and upon cell differentiation, using a
chemical inducer, these cells become mainly adherent. Under normal
culture conditions, exogenous Fli-1 expression in DP17-17 cells
resulted in a remarkable shift in the proportion of these
populations, increasing the numbers of suspension cells, compared
to the MigR1 empty vector control (Fig. 1D). This observation suggested that
Fli-1 overexpression might influence the differentiation of this
erythroleukemia cell line.
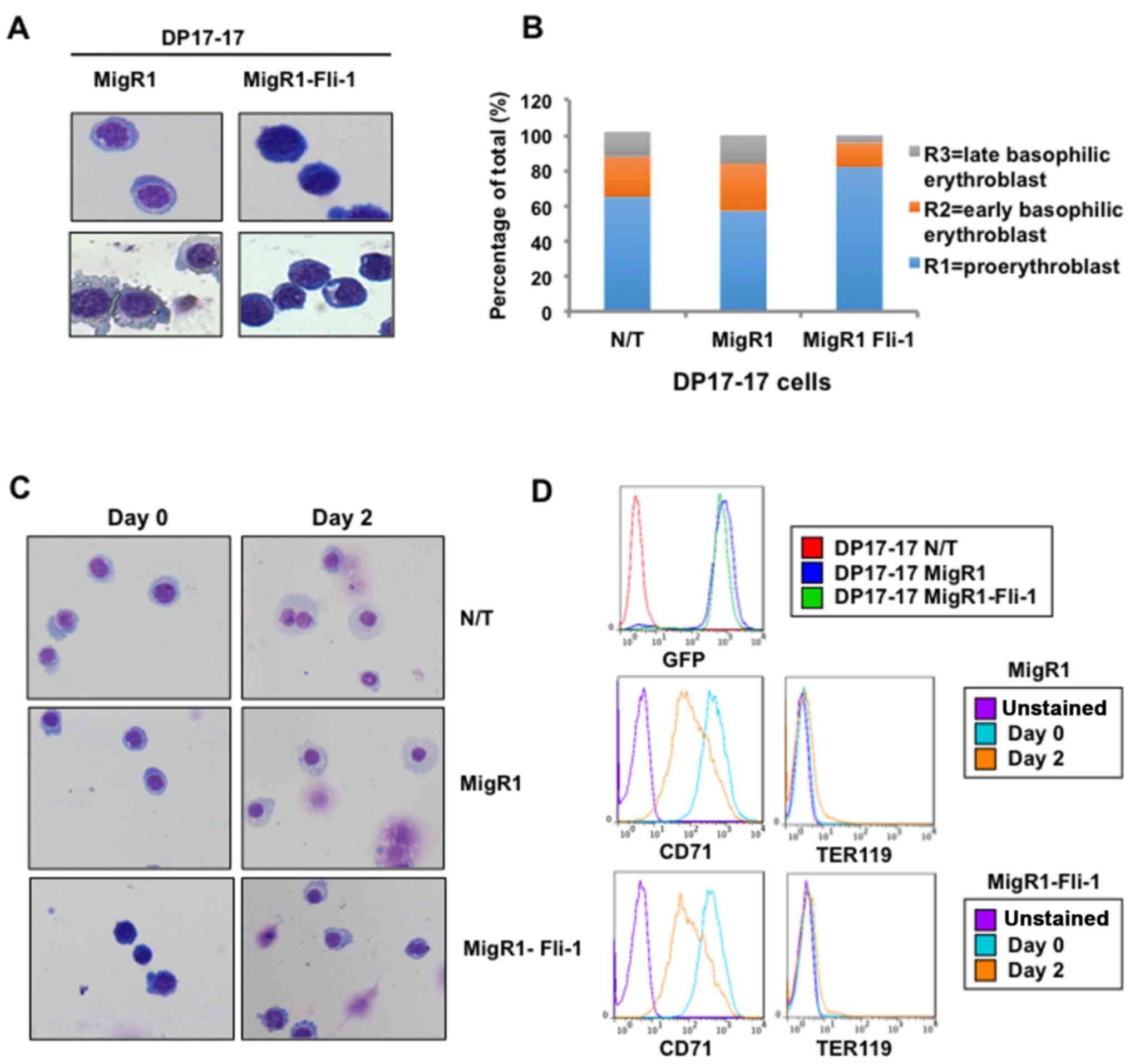
To confirm the above hypothesis, histochemical
staining of transduced DP17-17 cells was performed to distinguish
between different stages of erythroid development. Histology can
detect the morphologically defined stages of erythropoiesis
including, proerythroblasts, basophilic erythroblasts,
polychromatophilic erythroblasts, orthochromatophilic
erythroblasts, reticulocytes and mature erythrocytes. Successive
erythroid differentiation is characterized by a decrease in cell
size, increase in condensation of nuclear chromatin, more abundant
cytoplasm and increase in hemoglobinization as indicated by paler
or less saturated dye. In two separate experiments, May-Grunwald
Giemsa stains revealed that Fli-1 overexpressing DP17-17 cells
exhibit darker staining of the nuclei, denser appearance of the
nuclei chromatin (indicating immature chromatin), and deeply
basophilic cytoplasm, compared to control cells (Fig. 2A). Overall, the morphological and
expression characteristics of Fli-1 overexpressing DP17-17 cells
was indicative of a more immature cell type compared to the control
cells. To confirm our findings, a clinical hematopathologist
performed blinded erythroid differential counts of transduced
DP17-17 cell cytospin preparations stained with May-Grunwald/Giemsa
and the result is depicted in Fig.
2B.
Exogenous Fli-1 expression does not
affect the ability of Friend erythroleukemic cells to undergo
erythroid differentiation in culture
The above-mentioned data suggest the ability of
Fli-1 expression to induce features of a less-differentiated
erythroid phenotype. To determine if exogenous Fli-1 expression
also affects the potential or capacity of Friend erythroleukemic
cells to differentiate along the erythroid lineage, DP17-17-MigR1
or MigR1-Fli-1 cells were grown in the presence of DMSO. Double
sorted cell populations were seeded in triplicate and grown in
culture medium supplemented with 2% DMSO. Following 48 h of
induction, most of the cells became adherent to the culture dish
(data not shown). To examine changes in the morphology of the
DMSO-treated cells, suggestive of erythroid differentiation,
DP17-17 cells were removed from the culture dish for cytospin
preparation and histochemical staining. DMSO-induced cultures of
non-transduced and MigR1 empty control DP17-17 cells consisted
predominantly of cells resembling the orthochromatophilic stage,
with small, condensed nuclei, and paler blue, more abundant
cytoplasm (Fig. 2C), resembling
late stage erythroid differentiation. DMSO-induced cultures of
DP17-17 Fli-1 population consisted predominantly of cells
resembling the polychromatophilic stage, with slightly larger
nuclei and more deeply stained, less abundant cytoplasm compared to
the control DP17-17 cells treated with DMSO (Fig. 2C). Flow cytometry analysis revealed
that erythroid maturation by DMSO stimulation in DP17-17 Fli-1 and
control cells coincides with a decrease in the expression of the
transferrin receptor (CD71) and a slight increase in the erythroid
lineage cell surface marker TER119 (Fig. 2D). This result is consistent with
lower CD71 and higher TER119 expression in differentiated erythroid
cells beyond CFU-E stage (20).
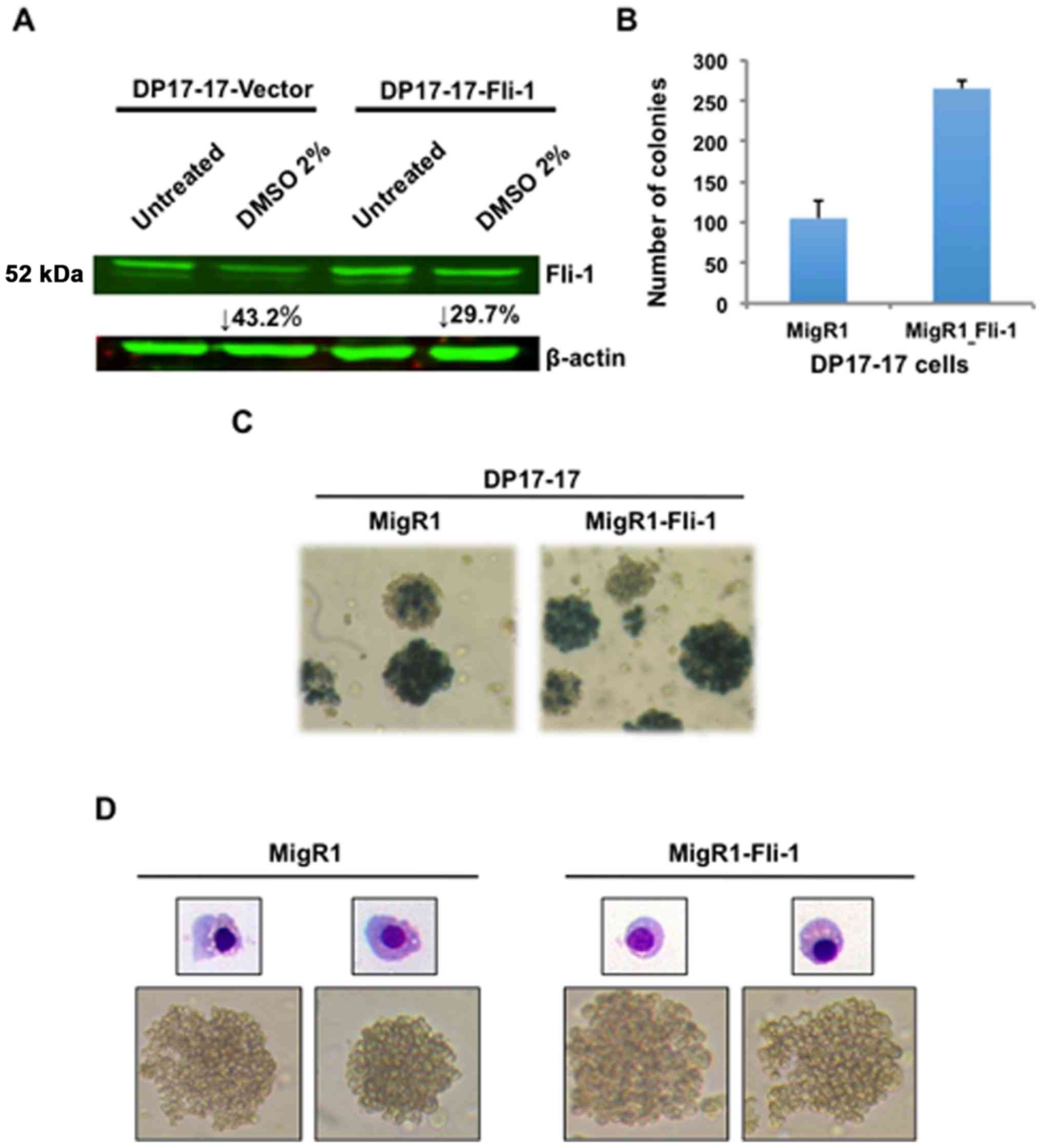
Since Fli-1 is known to be downregulated by DMSO or during
differentiation of erythroleukemia cells (15,21,22),
we examined the level of this TF before and after chemical inducer
treatment. Indeed, Fli-1 protein expression is significantly
downregulated in DP17-17-vector and to a lesser extent in DP17-17
Fli-1 cells (Fig. 3A). Therefore,
while exogenous expression of Fli-1 induces a less differentiated
phenotype, it does not affect the potential of these leukemic cells
to undergo chemically-induced erythroid differentiation.
Fli-1 overexpression increases the number
of colony-forming cells and expression of stem cell genes
To further investigate the role of exogenous Fli-1
overexpression in erythroid differentiation, a colony-forming cell
(CFC) assay was performed to quantify changes in the number of
lineage-restricted progenitors of the erythroid or megakaryocytic
lineage. Double-sorted DP17-17 cells transduced with the MigR1 or
MigR1-Fli-1 were suspended in semi-solid methylcellulose medium
supplemented with Epo, or Epo plus other additional cytokines.
CFU-E and BFU-E colonies were enumerated by staining with benzidine
solution. Individual colonies were quantified and the lineage
composition was classified based on morphological recognition by
light microscopy, and cytochemical staining of cellular cytospins
with May-Grunwald/Giemsa staining. DP17-17 Fli-1 cells produced
~2.5× more erythroid colonies compared to the appropriate controls
(Fig. 3B). Typical morphologies of
benzidine-stained cells for MigR1 or MigR1 Fli-1 expressing
colonies are presented in Fig. 3C.
Benzidine positive staining revealed that the clear majority of
colonies generated by both groups of DP17-17 cells are of erythroid
origin. Gross morphological examination by light microscopy also
revealed that most Fli-1 overexpressing colonies grew larger in
size, compared to controls (data not shown), suggesting that Fli-1
may enhance the proliferation of erythroid progenitors. Isolation
of individual colonies, and subsequent cytospin preparation for
May-Grunwald Giemsa staining confirmed the erythroid composition,
as stained cells displayed characteristic morphologies of mid to
late stage erythroblasts (Fig.
3D). The data further suggest that Fli-1 promotes hematopoietic
progenitor de-differentiation program along erythroid pathway, and
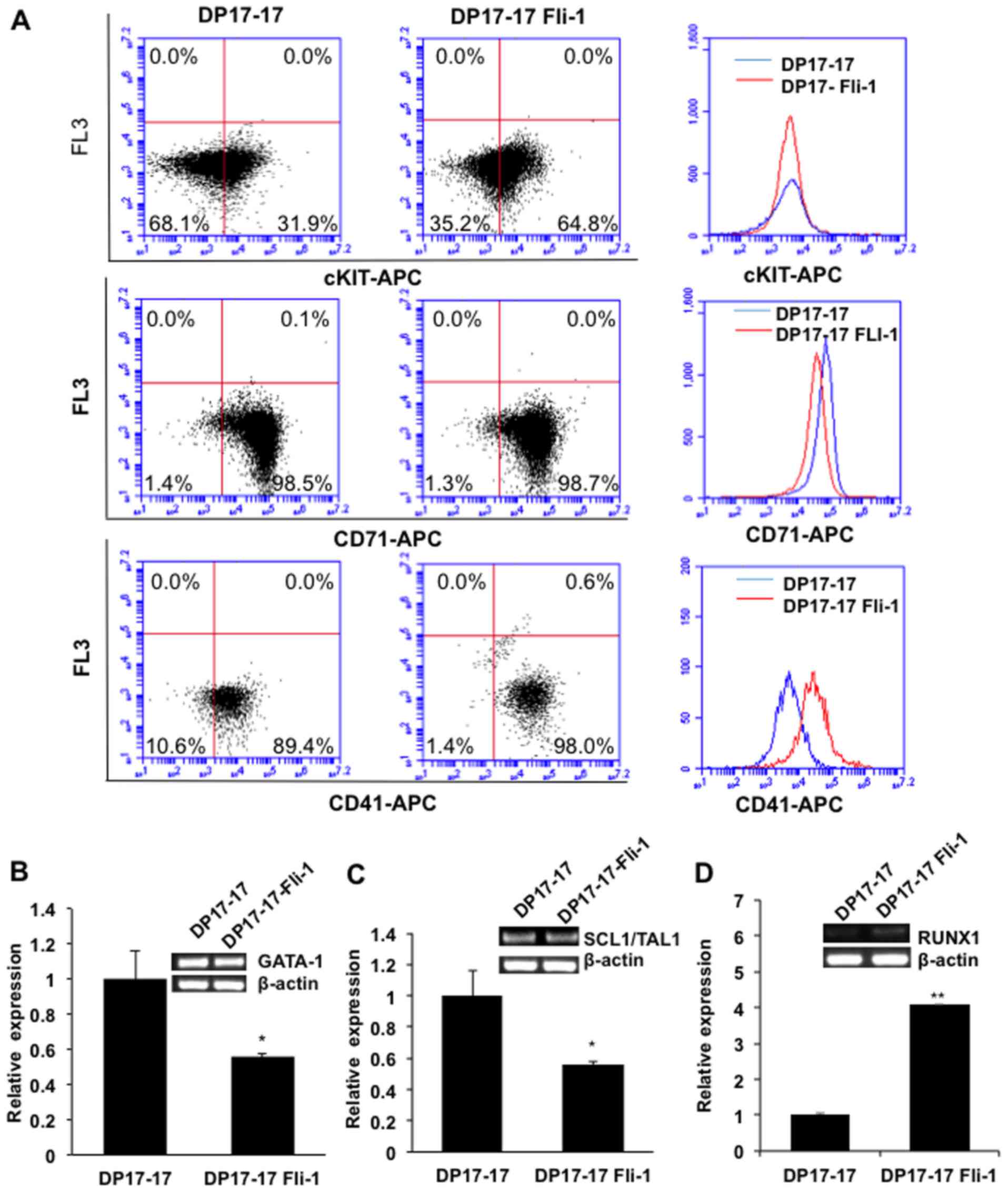
was further supported by flow cytometry, in which a higher number
of DP17-17 Fli-1 cells with higher intensity express the
stem/progenitor cell markers cKIT and CD41, when compared to
DP17-17 control cells (Fig. 4A).
No significance difference in expression of CD71 was observed
between DP17-17 Fli-1 and control cells (Fig. 4A).
Fli-1 binds and forms heptad complexes with other
members of master hematopoietic TFs GATA1/GATA2, RUNX1 and SCL/TAL1
(23). The composition of this
heptad TFs is critical for differentiation of progenitors to
erythroid and megakaryocytic lineage (22). We then determined the expression of
these genes in DP17-17 cells. Indeed, a lower expression of GATA1
was detected in DP17-17 Fli-1 than DP17-17 control cells (Fig. 4B). This result is consistent with
negative regulation of GATA-1 by Fli-1 (17). Fli-1 expression in DP17-17 Fli-1
cells increased the expression of RUNX1, but significantly reduced
SCL/TAL1 transcription when compared to the control cells (Fig. 4C and D), as previously observed
during erythroid differentiation (2).
Fli-1 overexpression in SFFV-induced
erythroleukemia increases the latency of disease progression and
alters the hematopoietic phenotype
To determine whether Fli-1 overexpression, and the
concurrent alteration of differentiation status, affects the
progression of FV-P-induced erythroleukemia, DP17-17-vector and
DP17-17 Fli-1 cells were transplanted into syngeneic DBA/2J mice.
Transduced and double-sorted DP17-17 cells (1×106,
1×105, 1×104) were intravenously injected
into the tail veins of eight-week-old DBA/2J mice, and monitored
for physiological signs of FV-P-induced erythroleukemia progression
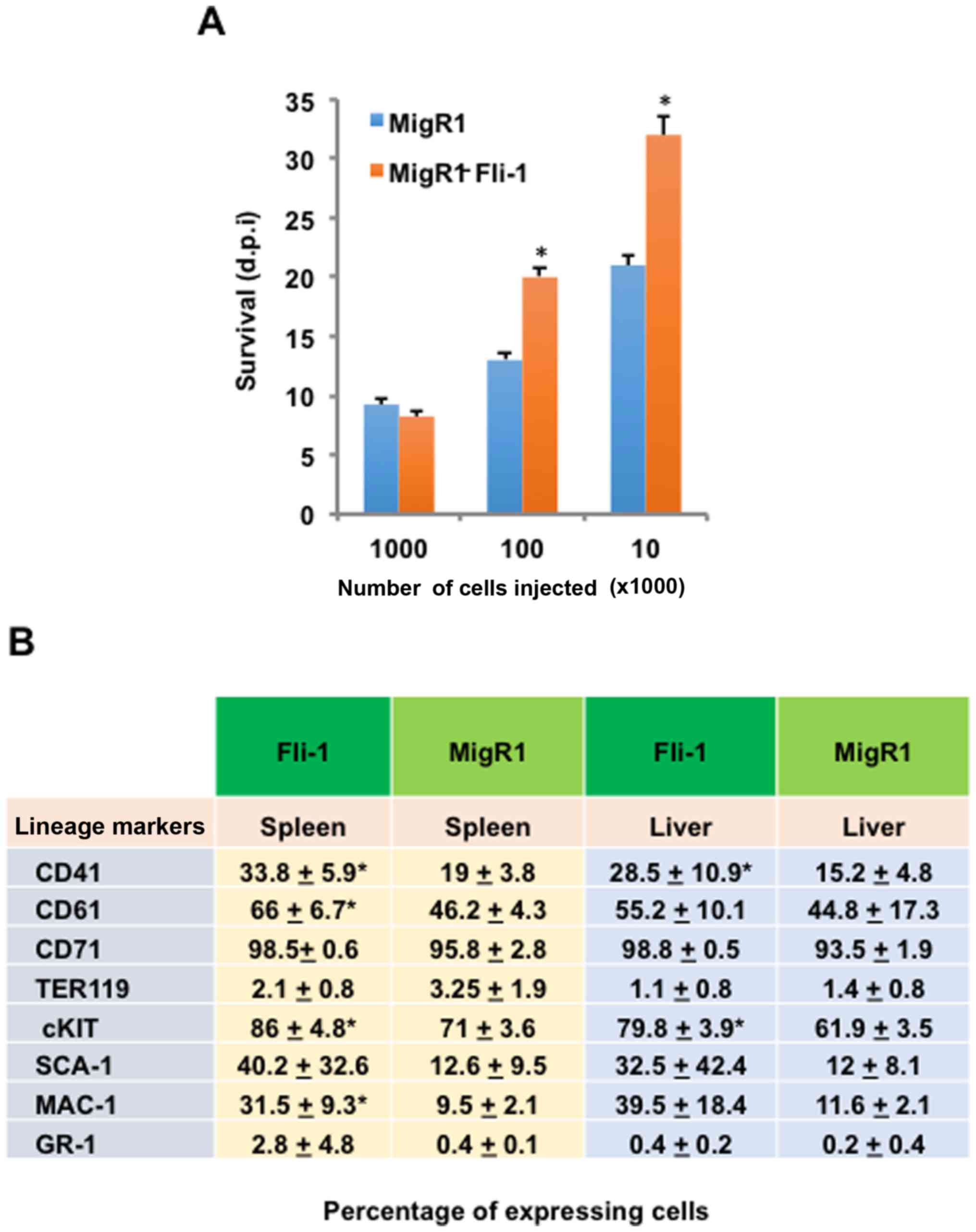
and survival. No appreciable difference in survival rate was
observed when mice received 1×106 cells (Fig. 5A). However, mice injected with
1×105 and 1×104 Fli-1 overexpressing cells
displayed a statistically significant increase in survival,
indicating growth suppression (Fig.
5A).
Friend virus-induced erythroleukemia is marked by
the emergence and expansion of transformed erythroblasts in the
spleen and liver (5). Indeed, the
size of both spleens and liver increased in DP17-17 vector and
DP17-17-Fli-1 injected mice, although no significance difference
was observed (data not shown). To further characterize the
erythroid malignancy generated in the recipient mice, tumor spleen
and liver cultures isolated from these tumorigenic mice were
subjected to flow cytometric immunophenotyping. The contribution
and presence of intravenously injected DP17-17 vector (n=9) or
DP17-17 Fli-1 (n=7) cells (1×104) was detected through
green fluorescence. Thus, the frequencies of several hematopoietic
cell surface markers were determined as a percentage of the total
GFP-positive cell population in diseased recipient mice. The
enlarged spleens and livers of both vector and Fli-1 injected mice
contained similar percentages of cells positive for CD71 and TER119
(Fig. 5B). Although the
percentages of cells positive for CD41, CD61, cKIT, and MAC-1 in
the spleen, as well as CD41 and cKIT in the liver, were
significantly higher in Fli-1 cells injected mice compared to
vector cells injected mice (Fig.
5B). Taken together, the data suggested that increased latency
of FV disease progression caused by Fli-1 overexpression might
result from an inherent change in the hematopoietic phenotype of
erythroid progenitors.
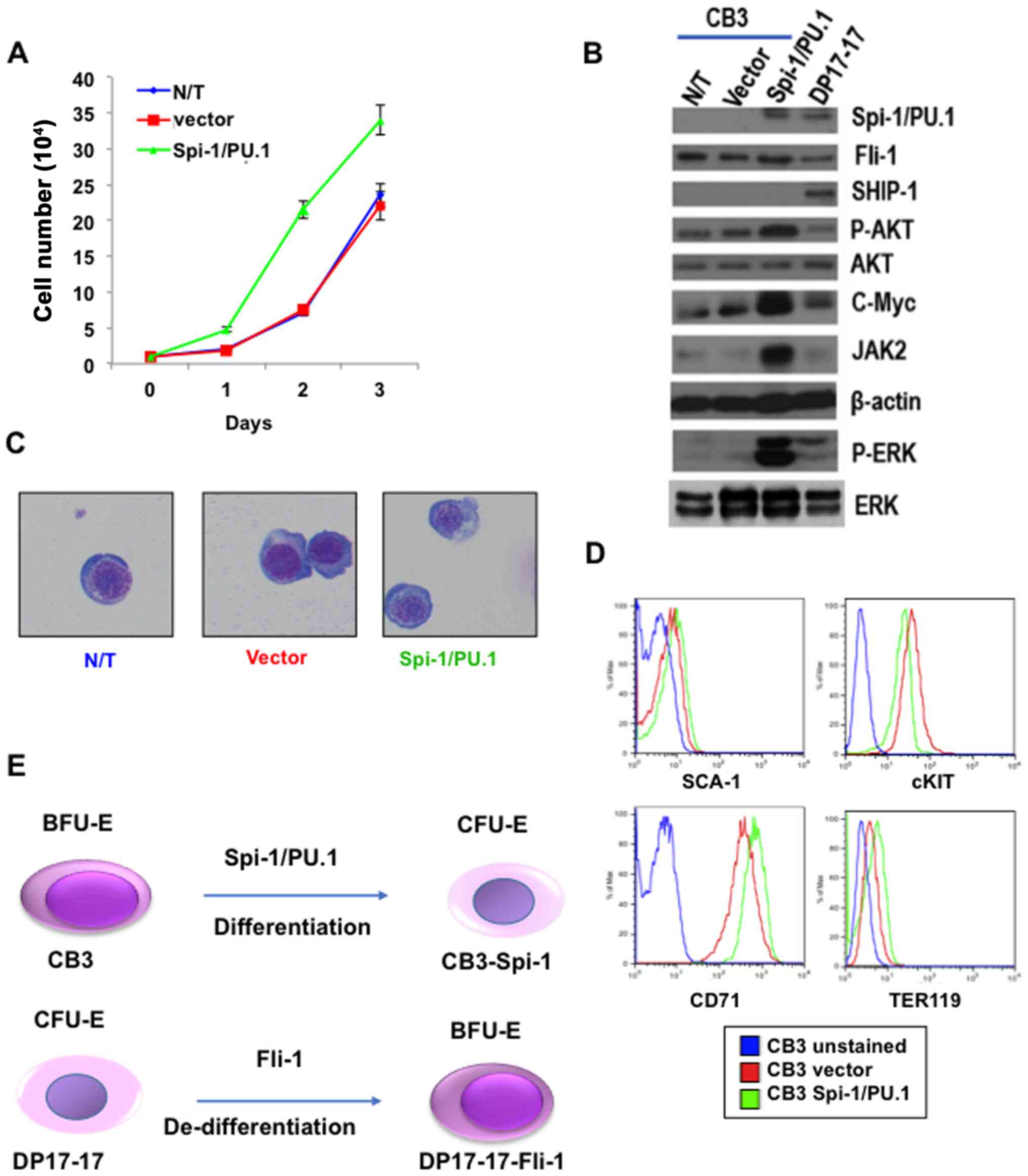
Spi-1/PU.1 transduction in the negative
producing erythroleukemia cell line CB3 promotes further erythroid
differentiation
In F-MuLV-induced erythroleukemias cell line CB3,
insertional activation of fli-1 increases the expression of
this TF, while negligible level of Spi-1 was detected in these
cells (8). We next examined if
expression of Spi-1/PU.1 in CB3 cells can alter the phenotype of
these cells through erythroid differentiation pathway. CB3-Spi-1
cells proliferate at a higher rate that CB3-vector cells in culture
(Fig. 6A). Accordingly, these
cells express a higher level of growth promoting genes, including
phospho-MAPK/ERK, phospho-AKT, cMYC and JAK2 (Fig. 6B). The Spi-1 overexpressing CB3
cells exhibit lighter staining of the nuclei with less density of
the nuclei chromatin (indicating mature chromatin), and weaker
basophilic cytoplasm, compared to control CB3-vector cells
(Fig. 6C). Moreover, while
Spi-1/PU.1 expression in CB3 cells did not affect the level of
SCA-1 on cells, it significantly increased CD71 and moderately
decreased cKIT expression (Fig.
6D). TER119 is only slightly increased in Spi-1/PU.1 expressing
CB3 cells (Fig. 6D). Higher CD71
expression is consistent with highest level of this cell surface
protein detected in CFU-E progenitors (20). Thus, while Spi-1/PU.1 expression in
erythroid progenitors transform erythroblasts at CFU-E stage of
erythroid differentiation, Fli-1 overexpression target progenitors
at BFU-E stage during erythroleukemogenesis (Fig. 6E).
Discussion
The ETS family member, fli-1, is aberrantly
expressed in several cancers, including erythroleukemia and Ewing's
sarcoma (7,8,13).
Mice homozygous for a targeted deletion of fli-1 revealed an
indispensable function for Fli-1 during embryonic development and
hematopoiesis, specifically proper megakaryocytic development
(24). Moreover, fetal livers of
homozygous Fli-1 mutants contain a significant reduction in the
number of multilineage, erythroid, and myeloid progenitors compared
to those of the wild-type and heterozygous embryos (25). Loss of function studies performed
in the Xenopus and zebrafish embryos have provided
conclusive evidence implicating Fli-1 at the top of the
transcriptional network regulating blood and endothelial cell
development within the mesoderm (26). To explore a more specific
contribution of Fli-1 aberrant regulation in malignant
transformation, we compared the phenotypes of an SFFV-induced
erythroleukemia cell line, with and without exogenous Fli-1
expression. This study demonstrated that the enforced expression of
Fli-1 in erythroleukemia cells induces de-differentiation of
progenitors toward more immature state, an observation consistent
with the role of this TF in regulation of hematopoiesis. In
contrast, when Spi-1/PU.1 was introduced into a Fli-1
overexpressing erythroleukemia cell line, these leukemic cells
expressed markers of more mature erythroid progenitors. These
results indicate that both Fli-1 and Spi-1 block erythroid
differentiation in distinct progenitors leading to the development
of erythroleukemias with distinct maturation phenotype.
TFs play a critical role in controlling
hematopoietic precursor cell fate decision, function and behavior
(1). Indeed, while exogenous
expression of Fli-1 into DP17-17 cells did not affect the
proliferation rate of these cells in culture, it resulted in
morphological characteristics, indicative of more primitive
erythroid progenitors (27). Fli-1
overexpression in these cells induces changes in the expression of
key hematopoietic TFs such as SCL/TAL1, GATA-1, RUNX1 that may
promote properties of self-renewal and favor the transition to a
more immature erythroid phenotype. Among these, GATA1 is well known
to be negatively regulated by Fli-1 (17). Although Fli-1 overexpression leads
to de-differentiation, it does not inhibit the ability of this
erythroleukemia cell line to differentiate along the erythroid
lineage in response to DMSO. Both Fli-1 and Spi-1 is known to be
downregulated during normal erythropoiesis and by DMSO (15,21,22),
as observed for Fli-1 in this study. This result then further
confirms the role of these ETS genes in hematopoiesis.
The cell surface antigens CD41 (integrin
αIIb) and CD61 (integrin β3) are expressed in
cells of the megakaryocytic lineage, and increased in DP17-17 Fli-1
cells. Megakaryocytes and erythroblasts originate from a common
myeloid progenitor (28,29), and accordingly cells derived from
megakaryocytic leukemia or erythroleukemia often display traits of
both erythroid and megakaryocytic progenitors (30–32).
The molecular mechanisms regulating differentiation of either
lineage remain unclear, however, it is likely that activation of
signaling pathways regulating self-renewal, survival, and
proliferation of the erythroid/myeloid progenitor are governed
through alterations of gene expression profiles. The expression of
Fli-1, along with the RUNX-1 transcription factor, is associated
with megakaryocytic differentiation (33,34).
The differentiation of megakaryocytes is primarily driven by
thrombopoietin (TPO), and several genes known to play an important
role in megakaryopoiesis, including the TPO receptor (TPOR) or
c-MPL (35), glycoprotein (GP) IX
and IIb, and cyclin D1 (36), all
of which contain ETS binding sites and are regulated by Fli-1
(24,37). Indeed, Fli-1 overexpressing DP17-17
cells express higher CD41. Moreover, leukemic cells isolated from
the organs of mice injected with DP17-17 Fli-1 cells revealed a
statistically significant increase in the percentage of cells
positive for CD41 and CD61 expression. The data indicate that Fli-1
overexpression enhanced acquisition of megakaryocytic features
while maintaining erythroid features, indicative of a
megakaryocytic/erythroid progenitor phenotype, and are consistent
with our recent observation that Fli-1 agonist compounds can induce
the expression of megakaryocytic-specific markers and promote
megakaryocytic differentiation in erythroleukemic cells (38).
The ectopic expression of Fli-1 also stimulated an
increase in the percentage and intensity of cKIT expressing cells
in culture and in transplanted mice. Interestingly, both MYB and
ETS proteins are candidate regulators of cKIT expression since its
promoter contains potential binding sites for these TFs (39). The positive relationship between
cKIT and Fli-1 expression may partially explain the ability of this
ETS transcriptional regulator to increase progenitor cell activity
and induce changes that resemble an earlier stage of erythroid
development. Future studies involving the regulation of cKIT by
Fli-1 should uncover the role of this TF in malignant
transformation, and its ability to alter the balance between
hematopoietic differentiation and proliferation.
When Spi-1/PU.1 was expressed in F-MuLV-induced
erythroleukemia cell line CB3, these cells exhibited characteristic
opposite to Fli-1 overexpressing DP17-17 cells. Spi1/PU.1
expressing CB3 cells grow faster in culture associated with an
increase in expression of growth promoting genes including JAK-2,
cMYC and activation of PI3K and Ras pathway. Since JAK-2 activation
regulates all these growth promoting genes and pathways, this
kinase is likely a direct or indirect target of Spi-1/PU.1
(40), which should be
investigated in an independent study. The Spi-1.PU.1 cells also
exhibited morphology of more differentiated CFU-E progenitors
associated with higher expression of CD71 and TER119, as previously
described (20). In addition, a
lower expression of cKIT and SCA-1, associated with more primitive
erythroid progenitors, detected in Spi-1/PU.1 transduced cells.
These results suggested that while Fli-1 and Spi-1/PU.1 play a
critical role in the induction of different erythroleukemias, their
oncogenic function can only be exerted in distinct erythroid
progenitors. As summarized in Fig.
6E, we propose that Fli-1 overexpression in CFU-E progenitors
can initiate a de-differentiation phenotype like BFU.E. This BFU-E
phenotype can be induced to become CFU-E through Spi-1/PU.1
overexpression.
The overexpression of Fli-1 led to increased latency
of disease progression, upon transplantation of transduced DP17-17
cells in DBA/2J syngeneic mice. This increase in disease latency of
SFFV-induced erythroleukemia seems to support the notion that Fli-1
overexpression imparts the expansion of a more immature erythroid
phenotype. This is consistent with lower proliferation ability of
cancer stem cells versus more differentiated cells arisen from
these cells within the tumors (41).
In conclusion, we have shown that Fli-1
overexpression in erythroid progenitors induces de-differentiation
program, while Spi-1/PU.1 reverses this process. This phenomenon
may be partially mediated through direct activation of distinct
Fli-1 target genes, including cKIT, CD41, GATA1, CD71 and
Spi-1/PU.1 target genes JAK-2. These results for the first time
demonstrate the role for these ETS genes in progenitor
proliferation/self-renewal and differentiation. Most importantly,
the discovery of a role for Fli-1 in self-renewal may also shed
light on the pathogenesis of diseases associated with Fli-1
aberrant regulation including erythroleukemia and Ewing's
sarcoma.
Acknowledgments
This study was supported by research grants from the
Science and Technology Department of Guizhou Province Innovation
and Project Grant (2013-6012), Thousand Talent Program of China
(WQ20135200171) and The National Natural Science Foundation of
China (81472609) to Y.B.D.
References
|
1
|
Göttgens B: Regulatory network control of
blood stem cells. Blood. 125:2614–2620. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li Y, Luo H, Liu T, Zacksenhaus E and
Ben-David Y: The ets transcription factor Fli-1 in development,
cancer and disease. Oncogene. 34:2022–2031. 2015. View Article : Google Scholar
|
|
3
|
Ben-David Y and Bernstein A: Friend
virus-induced erythroleukemia and the multistage nature of cancer.
Cell. 66:831–834. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mager D, MacDonald ME, Robson IB, Mak TW
and Bernstein A: Clonal analysis of the late stages of
erythroleukemia induced by two distinct strains of Friend leukemia
virus. Mol Cell Biol. 1:721–730. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moreau-Gachelin F: Multi-stage Friend
murine erythroleukemia: Molecular insights into oncogenic
cooperation. Retrovirology. 5:992008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moreau-Gachelin F, Tavitian A and
Tambourin P: Spi-1 is a putative oncogene in virally induced murine
erythroleukaemias. Nature. 331:277–280. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ben-David Y, Giddens EB and Bernstein A:
Identification and mapping of a common proviral integration site
Fli-1 in erythroleukemia cells induced by Friend murine leukemia
virus. Proc Natl Acad Sci USA. 87:1332–1336. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ben-David Y, Giddens EB, Letwin K and
Bernstein A: Erythroleukemia induction by Friend murine leukemia
virus: Insertional activation of a new member of the ets gene
family, Fli-1, closely linked to c-ets-1. Genes Dev. 5:908–918.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shibuya T and Mak TW: Isolation and
induction of erythroleukemic cell lines with properties of
erythroid progenitor burst-forming cell (BFU-E) and erythroid
precursor cell (CFU-E). Proc Natl Acad Sci USA. 80:3721–3725. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cui JW, Vecchiarelli-Federico LM, Li YJ,
Wang GJ and Ben-David Y: Continuous Fli-1 expression plays an
essential role in the proliferation and survival of F-MuLV-induced
erythroleukemia and human erythroleukemia. Leukemia. 23:1311–1319.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Juban G, Giraud G, Guyot B, Belin S, Diaz
JJ, Starck J, Guillouf C, Moreau-Gachelin F and Morlé F: Spi-1 and
Fli-1 directly activate common target genes involved in ribosome
biogenesis in Friend erythroleukemic cells. Mol Cell Biol.
29:2852–2864. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Papetti M and Skoultchi AI: Reprogramming
leukemia cells to terminal differentiation and growth arrest by RNA
interference of PU.1. Mol Cancer Res. 5:1053–1062. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Delattre O, Zucman J, Plougastel B,
Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau
G, et al: Gene fusion with an ETS DNA-binding domain caused by
chromosome translocation in human tumours. Nature. 359:162–165.
1992. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stockley J, Morgan NV, Bem D, Lowe GC,
Lordkipanidzé M, Dawood B, Simpson MA, Macfarlane K, Horner K, Leo
VC, et al: UK Genotyping and Phenotyping of Platelets Study Group:
Enrichment of FLI1 and RUNX1 mutations in families with excessive
bleeding and platelet dense granule secretion defects. Blood.
122:4090–4093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tamir A, Howard J, Higgins RR, Li YJ,
Berger L, Zacksenhaus E, Reis M and Ben-David Y: Fli-1, an
Ets-related transcription factor, regulates erythropoietin-induced
erythroid proliferation and differentiation: Evidence for direct
transcriptional repression of the Rb gene during differentiation.
Mol Cell Biol. 19:4452–4464. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lesault I, Quang CT, Frampton J and
Ghysdael J: Direct regulation of BCL-2 by Fli-1 is involved in the
survival of FLI-1-transformed erythroblasts. EMBO J. 21:694–703.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Athanasiou M, Mavrothalassitis G,
Sun-Hoffman L and Blair DG: FLI-1 is a suppressor of erythroid
differentiation in human hematopoietic cells. Leukemia. 14:439–445.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lakhanpal GK, Vecchiarelli-Federico LM, Li
YJ, Cui JW, Bailey ML, Spaner DE, Dumont DJ, Barber DL and
Ben-David Y: The inositol phosphatase SHIP-1 is negatively
regulated by Fli-1 and its loss accelerates leukemogenesis. Blood.
116:428–436. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li YJ, Liu G, Xia L, Xiao X, Liu JC,
Menezes ME, Das SK, Emdad L, Sarkar D, Fisher PB, et al:
Suppression of Her2/Neu mammary tumor development in mda-7/IL-24
transgenic mice. Oncotarget. 6:36943–36954. 2015.PubMed/NCBI
|
|
20
|
Shimizu R, Engel JD and Yamamoto M:
GATA1-related leukaemias. Nat Rev Cancer. 8:279–287. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Blaybel R, Théoleyre O, Douablin A and
Baklouti F: Down-regulation of the Spi-1/PU.1 oncogene induces the
expression of TRIM10/HERF1, a key factor required for terminal
erythroid cell differentiation and survival. Cell Res. 18:834–845.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pereira R, Quang CT, Lesault I, Dolznig H,
Beug H and Ghysdael J: FLI-1 inhibits differentiation and induces
proliferation of primary erythroblasts. Oncogene. 18:1597–1608.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tijssen MR, Cvejic A, Joshi A, Hannah RL,
Ferreira R, Forrai A, Bellissimo DC, Oram SH, Smethurst PA, Wilson
NK, et al: Genome-wide analysis of simultaneous GATA1/2, RUNX1,
FLI1, and SCL binding in megakaryocytes identifies hematopoietic
regulators. Dev Cell. 20:597–609. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hart A, Melet F, Grossfeld P, Chien K,
Jones C, Tunnacliffe A, Favier R and Bernstein A: Fli-1 is required
for murine vascular and megakaryocytic development and is
hemizygously deleted in patients with thrombocytopenia. Immunity.
13:167–177. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Spyropoulos DD, Pharr PN, Lavenburg KR,
Jackers P, Papas TS, Ogawa M and Watson DK: Hemorrhage, impaired
hematopoiesis, and lethality in mouse embryos carrying a targeted
disruption of the Fli1 transcription factor. Mol Cell Biol.
20:5643–5652. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu F, Walmsley M, Rodaway A and Patient
R: Fli1 acts at the top of the transcriptional network driving
blood and endothelial development. Curr Biol. 18:1234–1240. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Göttgens B, Nastos A, Kinston S, Piltz S,
Delabesse EC, Stanley M, Sanchez MJ, Ciau-Uitz A, Patient R and
Green AR: Establishing the transcriptional programme for blood: The
SCL stem cell enhancer is regulated by a multiprotein complex
containing Ets and GATA factors. EMBO J. 21:3039–3050. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Akashi K, Traver D, Miyamoto T and
Weissman IL: A clonogenic common myeloid progenitor that gives rise
to all myeloid lineages. Nature. 404:193–197. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ceredig R, Rolink AG and Brown G: Models
of haematopoiesis: Seeing the wood for the trees. Nat Rev Immunol.
9:293–300. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hassan HT and Freund M: Characteristic
biological features of human megakaryoblastic leukaemia cell lines.
Leuk Res. 19:589–594. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Drexler HG, Matsuo Y and MacLeod RA:
Malignant hematopoietic cell lines: In vitro models for the study
of erythroleukemia. Leuk Res. 28:1243–1251. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tallack MR and Perkins AC:
Megakaryocyte-erythroid lineage promiscuity in EKLF null mouse
blood. Haematologica. 95:144–147. 2010. View Article : Google Scholar :
|
|
33
|
Yamada H, Sekikawa T, Iwase S, Arakawa Y,
Suzuki H, Agawa M, Akiyama M, Takeda N and Horiguchi-Yamada J:
Segregation of megakaryocytic or erythroid cells from a
megakaryocytic leukemia cell line (JAS-R) by adhesion during
culture. Leuk Res. 31:1537–1543. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jackers P, Szalai G, Moussa O and Watson
DK: Ets-dependent regulation of target gene expression during
megakaryopoiesis. J Biol Chem. 279:52183–52190. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Edvardsson L, Dykes J and Olofsson T:
Isolation and characterization of human myeloid progenitor
populations - TpoR as discriminator between common myeloid and
megakaryocyte/erythroid progenitors. Exp Hematol. 34:599–609. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun S, Zimmet JM, Toselli P, Thompson A,
Jackson CW and Ravid K: Overexpression of cyclin D1 moderately
increases ploidy in megakaryocytes. Haematologica. 86:17–23.
2001.PubMed/NCBI
|
|
37
|
Pang L, Xue HH, Szalai G, Wang X, Wang Y,
Watson DK, Leonard WJ, Blobel GA and Poncz M: Maturation
stage-specific regulation of megakaryopoiesis by pointed-domain Ets
proteins. Blood. 108:2198–2206. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu T, Yao Y, Zhang G, Wang Y, Deng B,
Song J, Li X, Han F, Xiao X, Yang J, et al: A screen for Fli-1
transcriptional modulators identifies PKC agonists that induce
erythroid to megakaryocytic differentiation and suppress
leukemogenesis. Oncotarget. 8:16728–16743. 2017.PubMed/NCBI
|
|
39
|
Ratajczak MZ, Perrotti D, Melotti P,
Powzaniuk M, Calabretta B, Onodera K, Kregenow DA, Machalinski B
and Gewirtz AM: Myb and ets proteins are candidate regulators of
c-kit expression in human hematopoietic cells. Blood. 91:1934–1946.
1998.PubMed/NCBI
|
|
40
|
Akada H, Akada S, Gajra A, Bair A,
Graziano S, Hutchison RE and Mohi G: Efficacy of vorinostat in a
murine model of polycythemia vera. Blood. 119:3779–3789. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Signer RA, Magee JA, Salic A and Morrison
SJ: Haematopoietic stem cells require a highly regulated protein
synthesis rate. Nature. 509:49–54. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang J, Socolovsky M, Gross AW and Lodish
HF: Role of Ras signaling in erythroid differentiation of mouse
fetal liver cells: Functional analysis by a flow cytometry-based
novel culture system. Blood. 102:3938–3946. 2003. View Article : Google Scholar : PubMed/NCBI
|