Introduction
Basaloid squamous cell carcinoma (BSC) was first
reported by Wain et al (1)
to occur in the head and neck region. BSC may occur in various
other sites, including esophagus (2), lung (3), anus (4), uterine cervix (5), penis (6), and urinary bladder (7). BSC of the esophagus (BSCE) is a rare
and uncommon variant of squamous cell carcinoma (SCC), with a
reported incidence ranging from 1.0 to 8.7% (8–16) in
Japan and 0.4 to 11.3% (2,17–24)
in other countries.
BSCE has six typical components on histology: solid
nest with central necrosis, cribriform pattern, ductal
differentiation, microcyst and/or trabecular nests, hyaline-like
material deposition, and coexistence of SCC components (16). Because of these histological
varieties, BSCE is difficult to distinguish from adenoid cystic
carcinoma, small cell carcinoma, poorly differentiated SCC, or
adenosquamous carcinoma (2,23,25,26).
Furthermore, it is even more difficult to diagnose BSCE based on
the histological examination of endoscopic biopsy specimens, with a
low diagnostic accuracy of only 0–10% (23,24,27).
BSCE has been frequently diagnosed as SCC on endoscopic biopsy
specimens, probably because of the fact that BSCE frequently
presents as a submucosal tumor-like structure covered with normal
epithelium or SCC (14,15). Therefore, sampling multiple and
deeper sites is recommended for diagnosis by endoscopic biopsy
(24,27). Some diagnostic approaches using
immunoreactivity (2,9,12,16,18,19)
or polymerase chain reaction (PCR) (28,29)
have been reported, but none of these showed high specificity for
BSCE.
The prognosis of BSCE is still controversial. Some
studies stated no significant difference between BSCE and SCC
(2,20), whereas others stated poorer
prognosis of BSCE than that of SCC (16,17,23).
Some authors specified that BSCE shows a poor degree of
differentiation, high proliferative activity (2), aggressive biological behavior
(16), high telomerase activity
(20), and a worse prognosis than
that in SCC in advanced cases (30). Meanwhile, other authors mentioned
that the treatment for BSCE is similar to that for SCC of the
esophagus (15,23). The rarity of and difficulty in the
proper diagnosis of BSCE (15) may
be responsible for this diversity. Therefore, proper diagnosis is
mandatory for analyzing the outcome and determining the suitable
treatment for this disease entity.
We have previously reported the use of comprehensive
gene expression analysis (CGEA) to identify some disease-specific
genes (31,32). The present study aimed to improve
the diagnostic accuracy for BSCE by attempting to extract the genes
expressed in it. From CGEA of esophagectomy specimens, we
constructed, verified, and evaluated a gene expression scoring
system for the proper diagnosis of BSCE.
Materials and methods
Patient selection
We initially enrolled all 113 esophageal cancer
patients who underwent esophagectomy and/or endoscopic biopsy at
Fukushima Medical University Hospital from January 2008 to July
2015. Among these patients, 14 patients (1 in stage 0, 4 in stage
II, 4 in stage III, 4 in stage IVa, 1 in stage IVb and 1 in unknown
stage) were not followed in our department, and one surviving
patient (stage II) denied to participate in research. These cases
were excluded.
Ethics statement
This study was approved by the ethics committee of
Fukushima Medical University (approval no. 1953). Written informed
consent was obtained from 98 patients.
Commercially available esophageal
specimens
From US Biomax Inc. (Rockville, MD, USA), we
purchased 20 formalin-fixed paraffin-embedded (FFPE) specimens that
were diagnosed to have BSCE components. These FFPE specimens were
reviewed by three pathologists before inclusion.
Specimen sampling
Small fractions (7×7 mm) of the cancerous site and
normal mucosa were removed from each surgical specimen and were
immediately frozen in liquid nitrogen before performing CGEA.
Residual tissue specimens were fixed in formalin and then embedded
in paraffin before pathological examination.
For the biopsy specimens, tiny fractions (3×3 mm) of
the esophageal epithelium, including cancerous and normal sites,
were obtained endoscopically; they were immediately frozen
separately in liquid nitrogen before performing CGEA. Another
specimen from near the first biopsy site was obtained
endoscopically; it was fixed in formalin and embedded in paraffin
before pathological examination. We made an effort to sample
multiple and deeper sites by endoscopic biopsy. We ascertained that
the frozen specimens for CGEA and the FFPE specimens for
pathological examination had identifiable pathological
features.
Pathological review
The surgical, endoscopic biopsy and commercially
available FFPE specimens were stained with hematoxylin and eosin
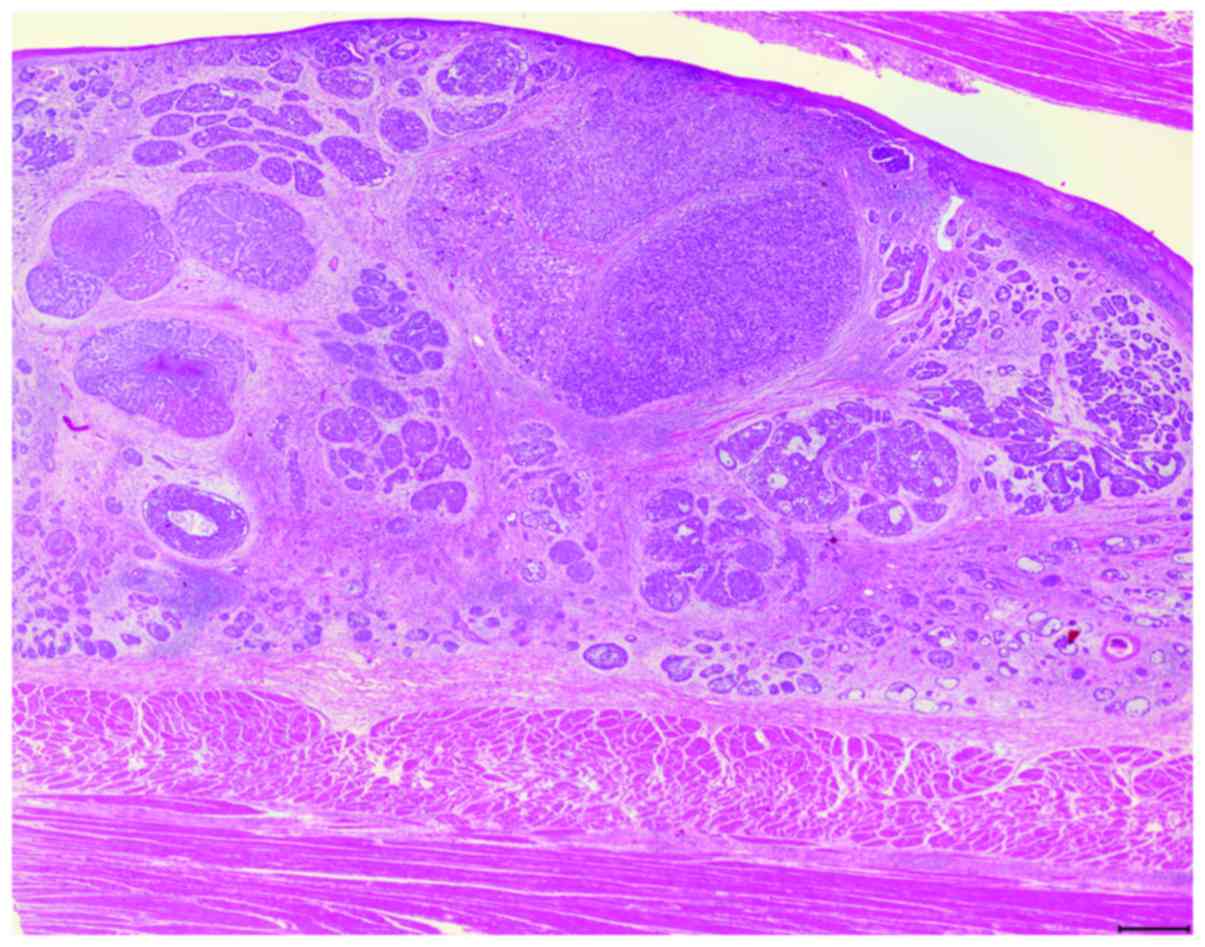
and were reviewed by three pathologists (Fig. 1). BSCE was defined using the
criteria described by Wain et al (1); the six component histological
features reported by Imamhasan et al (16) were also evaluated. In this study,
tumors that contained some BSC components within SCC were
categorized as BSCE.
Comprehensive gene expression
analysis
Frozen specimens were processed for total RNA
extraction using Isogen (Nippon Gene Co., Ltd., Tokyo, Japan) and
for poly(A)+RNA purification using MicroPoly(A) Purist kit (Ambion,
Austin, TX, USA). Commercially available FFPE specimens were
processed for total RNA extraction using ISOGEN PB kit (Nippon Gene
Co., Ltd.). The human common reference RNA was prepared by mixing
equal amounts of total RNA and poly(A)+RNA, which were extracted
from 22 human cancer cell lines (A431, A549, AKI, HBL-100, HeLa,
HepG2, HL60, IMR-32, Jurket, K562, KP4, MKN7, NK-92, Raji, RD,
Saos-2, SK-N-MC, SW-13, T24, U251, U937, and Y79).
The DNA microarray that used poly(A)+RNA was named
system 1; a set of synthetic polynucleotides (80-mers) representing
31,797 species of human transcript sequences was printed on a glass
slide using a custom arrayer. The DNA microarray that used total
RNA was named system 2; a set of synthetic polynucleotides
(80-mers) representing 14,400 species of human transcript sequences
was printed on a glass slide using a custom arrayer. For RNA of the
samples, SuperScript II (Invitrogen Life Technologies, Carlsbad,
CA, USA) and Cyanine 5-dUTP (Perkin-Elmer Inc., Boston, MA, USA)
were used to synthesize labeled cDNA from 2 µg of poly(A)+RNA in
system 1 and 5 µg of total RNA in system 2. Using the same method
for the reference RNA, Cyanine 3-dUTP (Perkin-Elmer Inc.) was used
to synthesize labeled cDNA from 2 µg of poly(A)+RNA in system 1 and
5 µg of total RNA in system 2.
Hybridization was performed with a Labeling and
Hybridization kit (MicroDiagnostic, Tokyo, Japan). Signals were
measured using a GenePix 4000B Scanner (Axon Instruments, Inc.,
Union City, CA, USA) and then processed into the primary expression
ratios of the cyanine 5 intensity of each specimen to the cyanine 3
intensity of the human common reference RNA. Each ratio was
normalized using GenePix Pro 3.0 software (Axon Instruments, Inc.).
The primary expression ratios were converted into log2 values,
which were designated as log ratios or converted value. Data were
processed using Microsoft Excel software (Microsoft, Bellevue, WA,
USA) and MDI gene expression analysis software package
(MicroDiagnostic) (33).
Statistical analysis
Clustering analysis was performed using group
average method with an Expression View Pro (MicroDiagnostic).
The cut-off score was determined by receiver
operating characteristic (ROC) curve analysis with the aim of
validating the gene scoring system. The optimal cut-off score for
the definitive diagnosis of BSCE was assessed and determined by
area under the ROC curve (AUC) analysis of the maximum values of
sensitivity and specificity. ROC curve analysis was performed using
the software program SPSS version 23 (SPSS, Inc., Chicago, IL,
USA).
Refinement steps to identify candidate
genes from CGEA of surgical specimens
Step 1: Genes with fluorescence intensity below the
detection limit in two or more of the seven BSCE specimens were
excluded. Step 2: The genes with a converted value of ≥1 in at
least one of the 57 surgical specimens were selected. Step 3: The
mean or average of the converted values of the chosen genes were
calculated, and the genes that met the following requirement were
selected: average value - converted value ≥1. Step 4: Clustering
analysis was performed on the chosen genes.
Construction of gene expression scoring
system for BSCE
Step 5: The mean of the converted values of the
genes that were expressed in six specimens of the BSCE cluster was
calculated; genes with an average value of ≥1 were selected. Step
6: Genes with fluorescence intensity below the detection limit were
excluded in more than half of the non-BSCE specimens. Step 7: The
standard deviation (SD) of the converted values of the genes that
were expressed in the non-BSCE specimens were calculated; genes
with an SD of <0.5 were selected. Step 8: Genes that met the
following requirement were selected: average value of six specimens
in the BSCE cluster - average value of non-BSCE specimens of ≥1.
Step 9: A t-test was used to compare the average value of six
specimens between the BSCE cluster and the non-BSCE specimens;
genes with a P-value of <0.01 were selected. Step 10: The
converted values of the selected genes from all specimens were
added as gene expression scores, which were arranged in ascending
order.
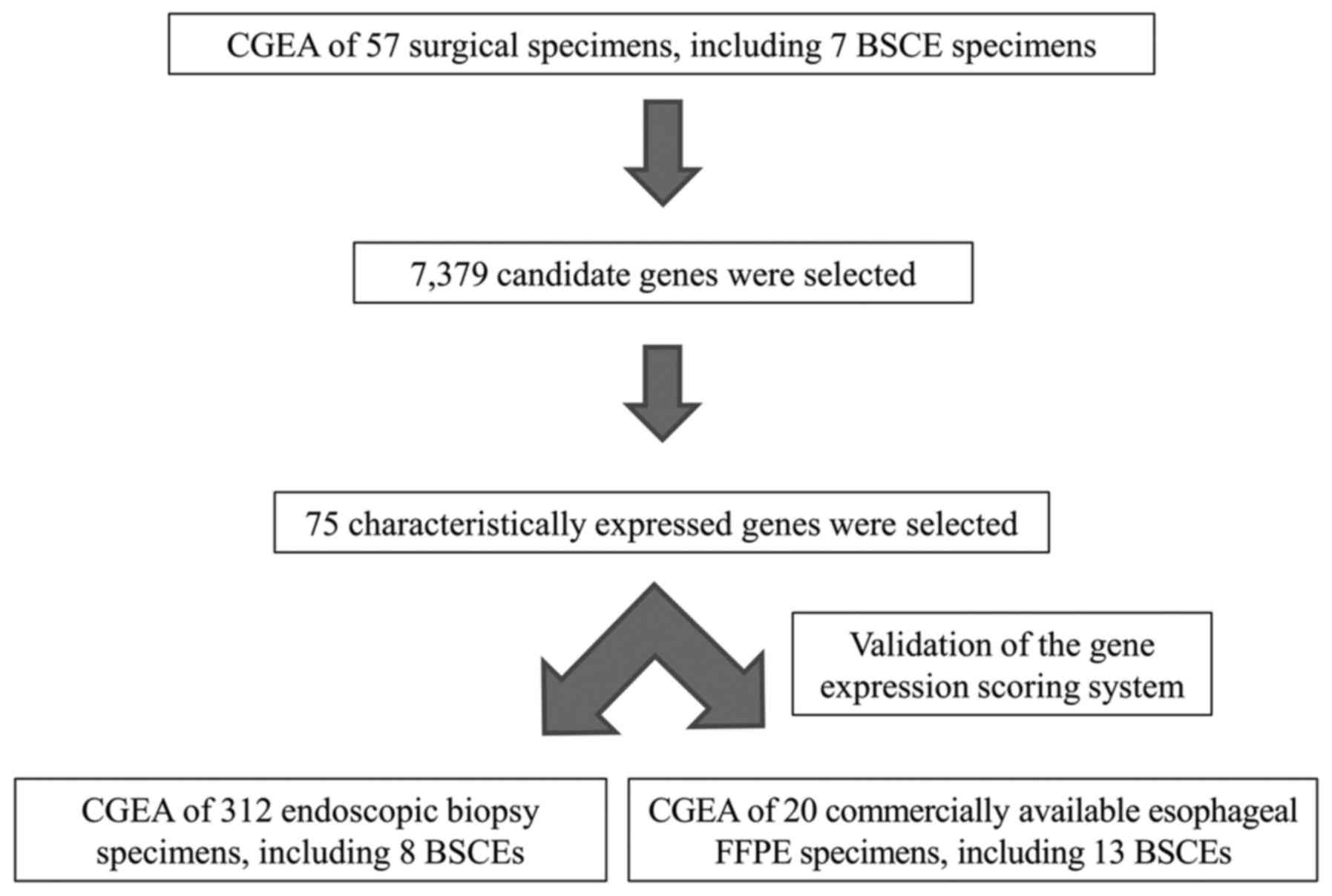
Study design
First, using CGEA of the surgical specimens, genes
that were characteristically expressed in BSCE were identified;
subsequently, a gene expression scoring system was constructed to
more accurately diagnose BSCE. Second, the accuracy of our scoring
system was validated using biopsy and commercially available FFPE
specimens. The study design is shown in Fig. 2.
Results
Pathological diagnosis of the specimens
for CGEA
Of the 98 patients, surgical specimens were obtained
from 30, endoscopic biopsy specimens from 80, and both from 12.
Seven cases of BSCE were included in this study, six cases
underwent esophagectomy and one case underwent only endoscopic
biopsy. We obtained more than one specimen from each individual,
and all specimens were subjected to gene expression analysis. The
number of specimens that contained enough amount of RNA for CGEA
was 369 (57 surgical and 312 endoscopic) (Table I). The surgical specimens comprised
26 normal esophageal tissues, 23 SCCs, seven BSCEs, and one
neuro-endocrine carcinoma (NEC). Biopsy specimens comprised 229
normal esophageal tissues, 51 SCCs, eight BSCEs, one NEC, 21
adenocarcinomas, and two intraepithelial neoplasias. Commercially
available FFPE specimens comprised two SCCs, 13 BSCEs, and five
NECs. All commercially available FFPE specimens were also subjected
to CGEA.
 | Table ISources of specimens for CGEA. |
Table I
Sources of specimens for CGEA.
| Histology | Surgical
specimens
(N=57) | Biopsy
specimens
(N=312) | Commercially
available FFPE specimens
(N=20) |
|---|
| Normal esophageal
tissue | 26 | 229 | 0 |
| SCC | 23 | 51 | 2 |
| BSCE | 7 | 8 | 13 |
| NEC | 1 | 1 | 5 |
| Adenocarcinoma | 0 | 21 | 0 |
| Intraepithelial
neoplasia | 0 | 2 | 0 |
Clinicopathological characteristics
Patients with BSCE, including five men and one
woman, with a mean age of 63 (range, 55–68) years, underwent
esophagectomy; their clinicopathological characteristics are listed
in Table II. Only three of six
patients (50%) were diagnosed as having BSCE by preoperative
endoscopic biopsy. The mean tumor size was 28.6 (range, 12–45) mm.
Based on the seventh Union for International Cancer Control
tumor-node-metastasis classification of malignant tumors, the
pathological stage was stage I in four patients, stage II in one,
and stage III in one. Within a mean follow-up period of 35 (range,
7–60) months, two patients died because of BSCE, one remained alive
with recurrence, and three remained alive without recurrence.
 | Table IIClinicopathologic features of six
esophagectomy cases. |
Table II
Clinicopathologic features of six
esophagectomy cases.
| Age, sex | Biopsy
diagnosis | Pathologic
diagnosis size | Tumor (mm) | Tumor type | Depth of invasion
(pT) | Lymph node
metastasis (pN) | Lymphatic invasion
(ly) | Venous invasion
(v) | UICC stage | Prognosis |
|---|
| 1 | 68, M | ASC | BSCE | 42 | Type 2 | T1 | N0 | 0 | 1 | IA | 58 months-died
(lung metastasis) |
| 2 | 55, M | SCC | BSCE | 30 | Type 5 | T1 | N0 | 0 | 2 | IA | 60
months-alive |
| 3 | 56, M | SCC | BSCE | 25 | Type 3 | T4 | N3 | 2 | 2 | IIIC | 19 months-died
(lung metastasis) |
| 4 | 68, F | SCC (first), BSCE
(second) | BSCE | 18 | Type 2 | T1 | N0 | 0 | 1 | IA | 37
months-alive |
| 5 | 64, M | BSCE | BSCE | 12 | Type 0-IIa | T1 | N1 | 0 | 0 | IIB | 29 months-alive (LN
metastasis) |
| 6 | 65, M | BSCE | BSCE, SCC | 5, 45 | Type 0-IIc | T1 | N0 | 0 | 0 | IA | 7 months-alive |
Comprehensive gene expression analysis of
the surgical specimens
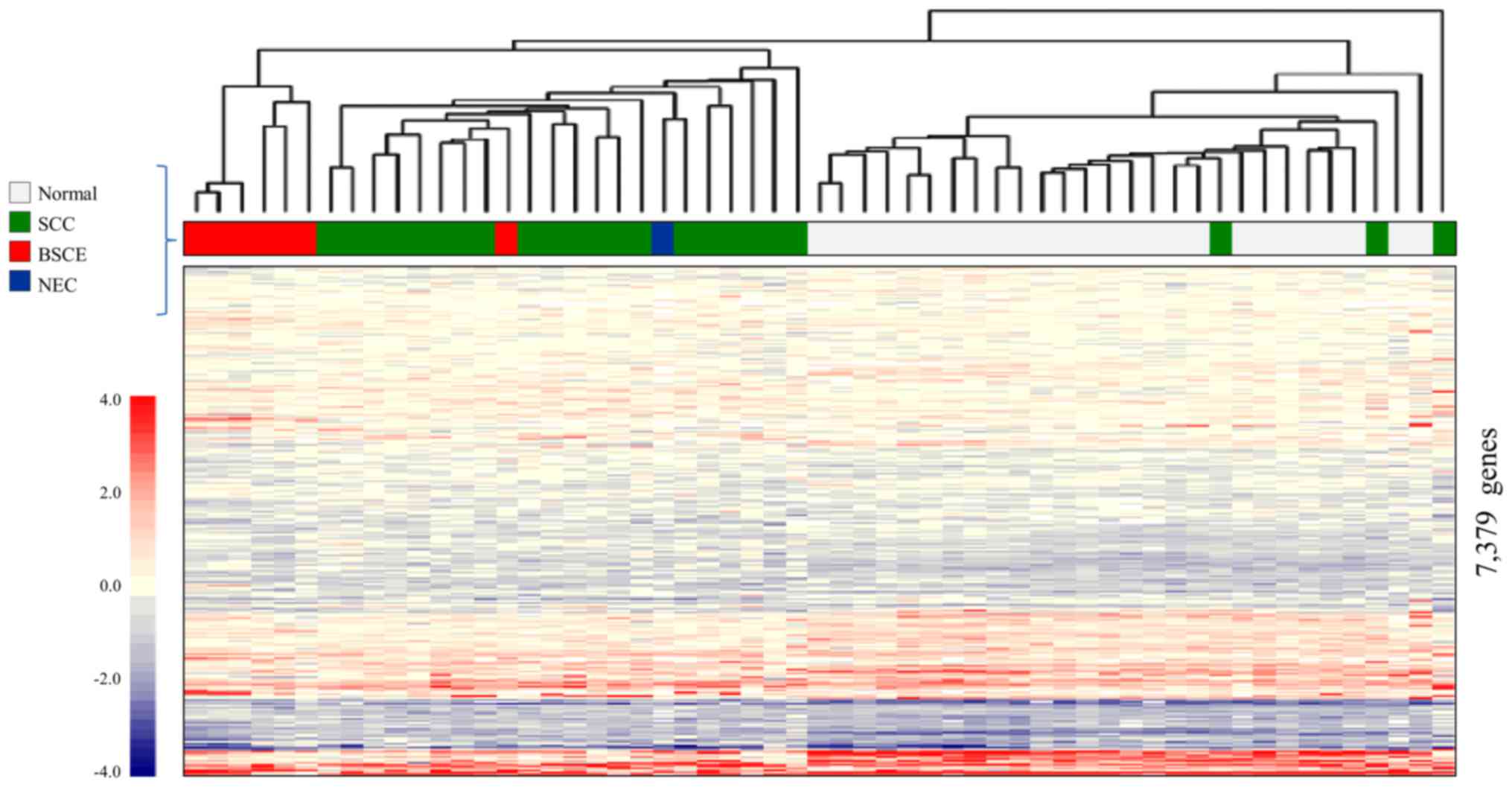
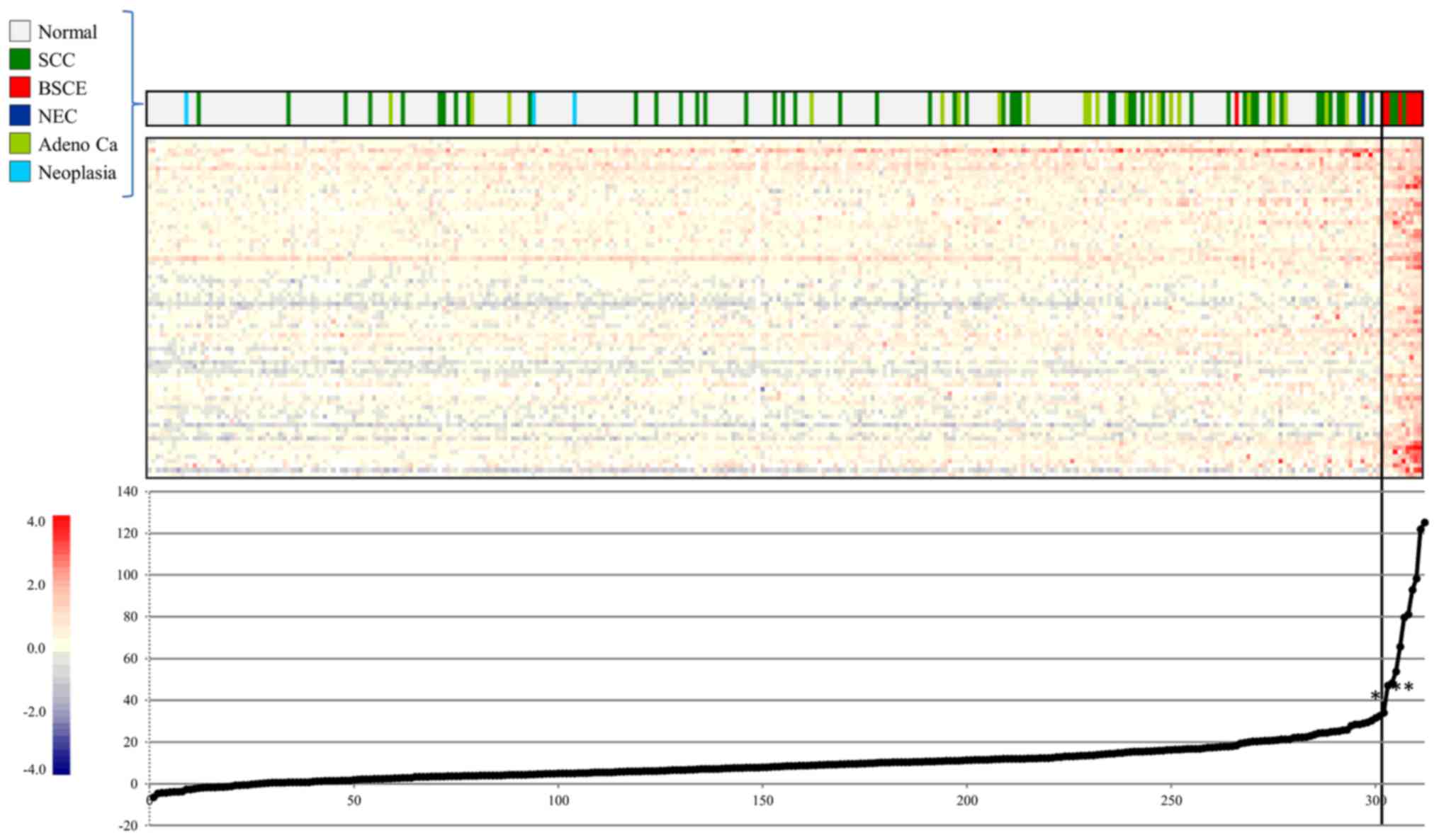
Fig. 3 shows the
result of CGEA of 57 surgical specimens, including seven BSCE
specimens (one specimen from cases 1, 2, 3, 5 and three specimens
from case 4); 10,027 genes were selected in step 1; 9,004 in step
2; and 7,379 in step 3.
A two-dimensional hierarchical clustering analysis
of 7,379 genes yielded three different clusters: the 1) BSCE
cluster, which comprised six of the seven BSCE specimens; 2) SCC
cluster, which mainly comprised SCC; and 3) normal cluster, which
mainly comprised normal esophageal tissue. It was possible to
distinguish BSCE specimens from the others using this analysis.
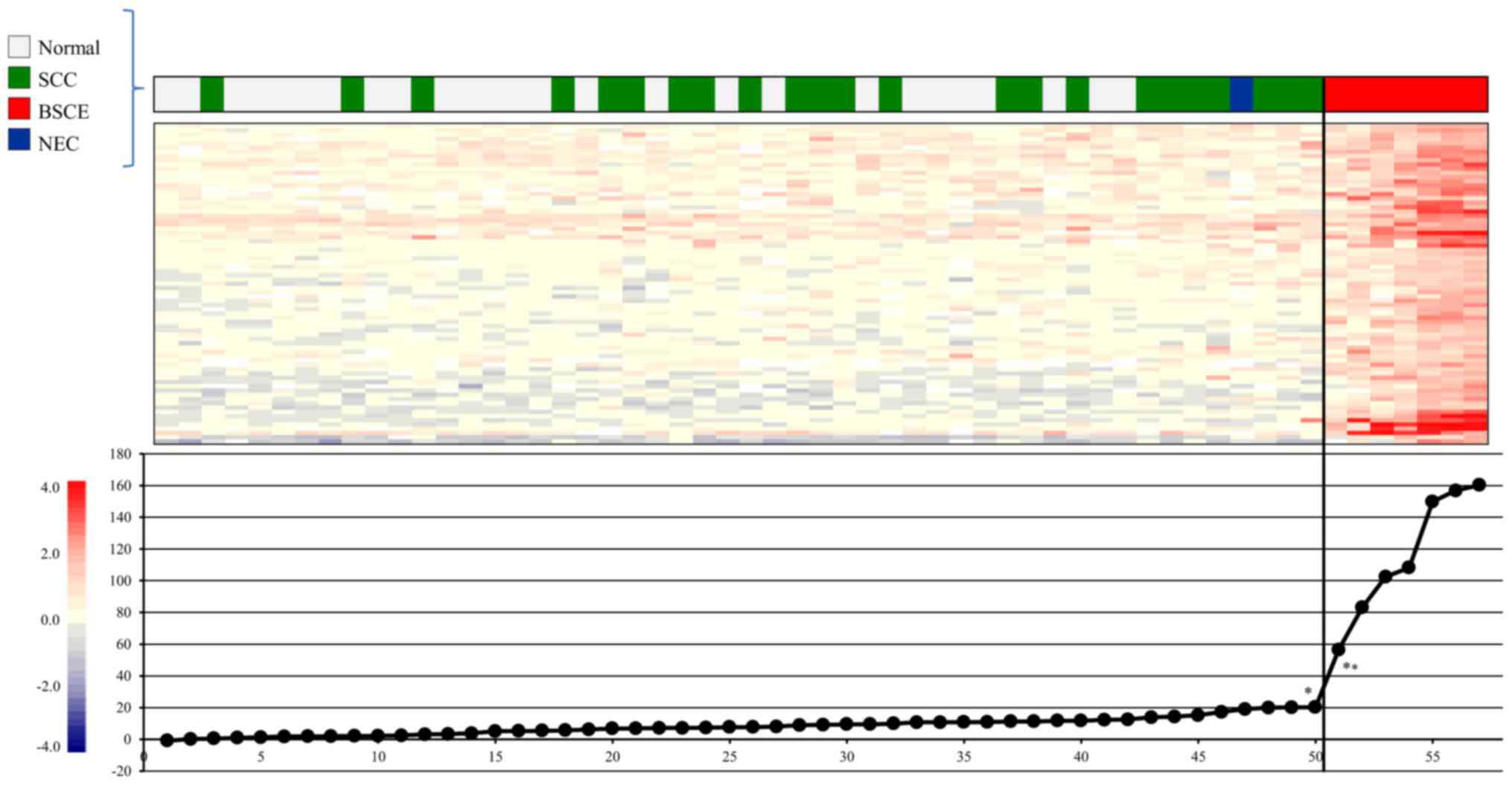
Gene expression scoring system for
BSCE
We selected BSCE-specific candidate maker genes and
attempted to construct a gene expression scoring system for more
accurate diagnosis of BSCE, and 986, 972, 243, and 100 genes were
sequentially selected from steps 5, 6, 7, and 8, respectively.
Finally, 75 genes were selected in step 9 (Table III) and subjected to
extrapolation of the gene expression score, as described in step
10. Our gene expression scoring system, which set the cut-off score
at 56.5, very clearly distinguished the seven BSCE specimens from
the non-BSCE specimens (Fig.
4).
 | Table IIIGenes characteristically expressed in
BSCE based on CGEA of surgical specimens. |
Table III
Genes characteristically expressed in
BSCE based on CGEA of surgical specimens.
| No. | ID | Symbol | Name |
|---|
| 1 | NM_001033568.2 | RHOT1 | Ras homolog family
member T1 (RHOT1), transcript variant 1 |
| 2 | NM_015690.4 | STK36 | Serine/threonine
kinase 36 (STK36), transcript variant 1 |
| 3 | NM_000915.3 | OXT |
Oxytocin/neurophysin I prepropeptide
(OXT) |
| 4 | NM_001409.3 | MEGF6 | Multiple
EGF-like-domains 6 (MEGF6) |
| 5 | NM_021197.3 | WFDC1 | WAP four-disulfide
core domain 1 (WFDC1) |
| 6 | NM_020796.4 | SEMA6A | Sema domain,
transmembrane domain (TM), cytoplasmic domain, (semaphorin) 6A
(SEMA6A) |
| 7 | NM_153213.3 | ARHGEF19 | Rho guanine
nucleotide exchange factor (GEF) 19 (ARHGEF19) |
| 8 | XM_005261771.3 | PLA2G6 | Phospholipase A2,
group VI (cytosolic, calcium-independent) (PLA2G6), transcript
variant X18 |
| 9 | NM_000933.3 | PLCB4 | Phospholipase C, β4
(PLCB4), transcript variant 1 |
| 10 | NM_023110.2 | FGFR1 | Fibroblast growth
factor receptor 1 (FGFR1), transcript variant 1 |
| 11 | AK055081.1 | | cDNA FLJ30519 fis,
clone BRAWH2000859 |
| 12 | NM_000346.3 | SOX9 | SRY (sex
determining region Y)-box 9 (SOX9) |
| 13 | NM_025176.4 | NINL | Ninein-like
(NINL) |
| 14 | NM_014698.2 | TMEM63A | Transmembrane
protein 63A (TMEM63A) |
| 15 | NM_020870.3 | SH3RF1 | SH3 domain
containing ring finger 1 (SH3RF1) |
| 16 | NM_001110514.1 | EBF4 | Early B-cell factor
4 (EBF4) |
| 17 | NR_036481.2 | FGD5P1 | FYVE, RhoGEF and PH
domain containing 5 pseudogene 1 (FGD5P1), non-coding RNA |
| 18 | NM_005117.2 | FGF19 | Fibroblast growth
factor 19 (FGF19) |
| 19 | NM_032192.3 | PPP1R1B | Protein phosphatase
1, regulatory (inhibitor) subunit 1B (PPP1R1B) |
| 20 | NM_020659.3 | TTYH1 | Tweety family
member 1 (TTYH1), transcript variant 1 |
| 21 | NM_145804.2 | ABTB2 | Ankyrin repeat and
BTB (POZ) domain containing 2 (ABTB2) |
| 22 | NM_194302.3 | CCDC108 | Coiled-coil domain
containing 108 (CCDC108), transcript variant 1 |
| 23 | NM_002995.2 | XCL1 | Chemokine (C motif)
ligand 1 (XCL1) |
| 24 | NM_001940.3 | ATN1 | Atrophin 1 (ATN1),
transcript variant 2 |
| 25 | AK021565.1 | | cDNA FLJ11503 fis,
clone HEMBA1002113 |
| 26 | NM_006941.3 | SOX10 | SRY (sex
determining region Y)-box 10 (SOX10) |
| 27 | NM_003222.3 | TFAP2C | Transcription
factor AP-2γ (activating enhancer-binding protein 2γ) (TFAP2C) |
| 28 | NM_003963.2 | TM4SF5 | Transmembrane 4 L
six family member 5 (TM4SF5) |
| 29 | NM_002180.2 | IGHMBP2 | Immunoglobulin
mu-binding protein 2 (IGHMBP2) |
| 30 | NM_015696.4 | GPX7 | Glutathione
peroxidase 7 (GPX7) |
| 31 | NM_017789.4 | SEMA4C | Sema domain,
immunoglobulin domain (Ig), transmembrane domain (TM) and short
cytoplasmic domain, (semaphorin) 4C (SEMA4C) |
| 32 | NM_178502.3 | DTX3 | Deltex 3, E3
ubiquitin ligase (DTX3), transcript variant 1 |
| 33 | NM_014937.3 | INPP5F | Inositol
polyphosphate-5-phosphatase F (INPP5F), transcript variant 1 |
| 34 | NM_001380.4 | DOCK1 | Dedicator of
cytokinesis 1 (DOCK1), transcript variant 2 |
| 35 | NM_007081.2 | RABL2B | RAB, member of RAS
oncogene family-like 2B (RABL2B), transcript variant 2 |
| 36 | AK055044.1 | TARBP1 | TAR (HIV-1)
RNA-binding protein 1 (TARBP1) |
| 37 | NM_006312.5 | NCOR2 | Nuclear receptor
corepressor 2 (NCOR2), transcript variant 1 |
| 38 | NM_007270.4 | FKBP9 | FK506-binding
protein 9, 63 kDa (FKBP9), transcript variant 1 |
| 39 | NM_016162.3 | ING4 | Inhibitor of growth
family, member 4 (ING4), transcript variant 1 |
| 40 | NM_005937.3 | MLLT6 | Myeloid/lymphoid or
mixed-lineage leukemia; translocated to 6 (MLLT6) |
| 41 | AK021700.1 | | cDNA FLJ11638 fis,
clone HEMBA1004323 |
| 42 | NM_015662.2 | IFT172 | Intraflagellar
transport 172 (IFT172) |
| 43 | NM_032501.3 | ACSS1 | Acyl-CoA synthetase
short-chain family member 1 (ACSS1), transcript variant 1 |
| 44 | NM_016102.3 | TRIM17 | Tripartite motif
containing 17 (TRIM17), transcript variant 1 |
| 45 | NM_152753.3 | SCUBE3 | Signal peptide, CUB
domain, EGF-like 3 (SCUBE3), transcript variant 1 |
| 46 | NM_133455.3 | EMID1 | EMI domain
containing 1 (EMID1), transcript variant 1 |
| 47 | NM_014640.4 | TTLL4 | Tubulin tyrosine
ligase-like family member 4 (TTLL4) |
| 48 | NM_001161616.2 | RGL3 | Ral guanine
nucleotide dissociation stimulator-like 3 (RGL3), transcript
variant 1 |
| 49 | NM_024798.2 | SNX22 | Sorting nexin 22
(SNX22), transcript variant 1 |
| 50 | NM_032781.3 | PTPN5 | Protein tyrosine
phosphatase, non-receptor type 5 (striatum-enriched) (PTPN5),
transcript variant 2 |
| 51 | NM_005996.3 | TBX3 | T-box 3 (TBX3),
transcript variant 1 |
| 52 | NM_000875.4 | IGF1R | Insulin-like growth
factor 1 receptor (IGF1R), transcript variant 1 |
| 53 | NM_178238.3 | PILRB | Paired
immunoglobin-like type 2 receptor β (PILRB) |
| 54 | NM_152748.3 | KIAA1324L | KIAA1324-like
(KIAA1324L), transcript variant 1 |
| 55 | NM_003505.1 | FZD1 | Frizzled class
receptor 1 (FZD1) |
| 56 | NM_173812.4 | DPY19L2 | Dpy-19-like 2
(C. elegans) (DPY19L2) |
| 57 | NM_032447.3 | FBN3 | Fibrillin 3
(FBN3) |
| 58 | NM_001987.4 | ETV6 | Ets variant 6
(ETV6) |
| 59 | NM_017563.3 | IL17RD | Interleukin 17
receptor D (IL17RD) |
| 60 | NM_032040.4 | CCDC8 | Coiled-coil domain
containing 8 (CCDC8) |
| 61 | NM_018257.2 | PCMTD2 |
Protein-L-isoaspartate (D-aspartate)
O-methyltransferase domain containing 2 (PCMTD2), transcript
variant 1 |
| 62 | NM_152730.5 | TBC1D32 | TBC1 domain family,
member 32 (TBC1D32), transcript variant 1 |
| 63 | NM_152739.3 | HOXA9 | Homeobox A9
(HOXA9) |
| 64 | NM_021156.3 | TMX4 | Thioredoxin-related
transmembrane protein 4 (TMX4) |
| 65 | NM_002507.3 | NGFR | Nerve growth factor
receptor (NGFR) |
| 66 | NM_004776.3 | B4GALT5 | UDP-Gal:betaGlcNAc
β 1,4-galactosyltransferase, polypeptide 5 (B4GALT5) |
| 67 | NM_015544.2 | TMEM98 | Transmembrane
protein 98 (TMEM98), transcript variant 1 |
| 68 | NM_001852.3 | COL9A2 | Collagen, type IX,
α2 (COL9A2) |
| 69 | NM_005247.2 | FGF3 | Fibroblast growth
factor 3 (FGF3) |
| 70 | NM_002523.2 | NPTX2 | Neuronal pentraxin
II (NPTX2) |
| 71 | NM_001853.3 | COL9A3 | Collagen, type IX,
α3 (COL9A3) |
| 72 | NM_001851.4 | COL9A1 | Collagen, type IX,
α1 (COL9A1), transcript variant 1 |
| 73 | NM_014289.3 | CAPN6 | Calpain 6
(CAPN6), |
| 74 | NM_002336.2 | LRP6 | Low-density
lipoprotein receptor-related protein 6 (LRP6) |
| 75 | NM_001692.3 | ATP6V1B1 | ATPase,
H+ transporting, lysosomal 56/58 kDa, V1 subunit B1
(ATP6V1B1) |
Validation of the gene expression scoring
system
Using CGEA, we calculated 75 gene expression scores,
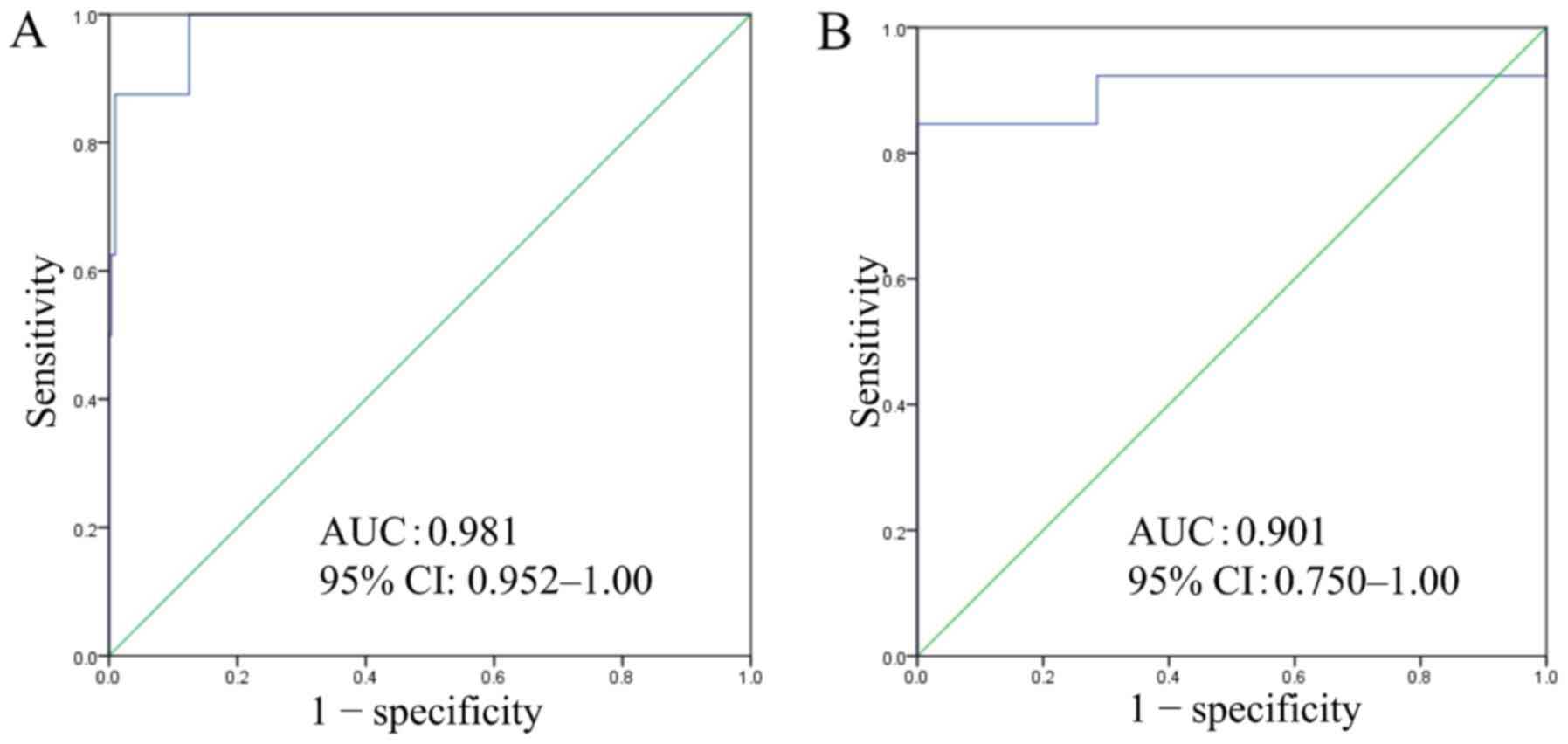
which were arranged in ascending order (Fig. 5). ROC curve analysis of the gene
expression scoring system using biopsy specimens yielded an optimal
cut-off score of 40.5, with an AUC of 0.981, sensitivity of 87.5%,
and specificity of 99.0% (Fig.
6A).
By the same procedure, ROC curve analysis of the
gene expression scoring system using commercially available FFPE
specimens, including 13 BSCE, yielded an optimal cut-off score of
34.9, with AUC of 0.901, sensitivity of 92.3%, and specificity of
71.4% (Fig. 6B).
Discussion
Using CGEA of esophagectomy specimens, we identified
the 75 genes that were characteristically expressed in BSCE to
construct a gene expression scoring system, which made it possible
to distinguish BSCE from non-BSCE in biopsy and commercially
available FFPE specimens with high sensitivity and specificity. To
our knowledge, this is the first report to show an accurate
diagnostic modality that could significantly contribute in
improving the diagnosis and treatment of BSCE.
Diagnosing BSCE using endoscopic biopsy specimens is
difficult, with a reported diagnostic accuracy of only 0% to 10%
(23,24,27).
We too were unable to accurately diagnose BSCE using preoperative
endoscopic biopsy in three of six surgical patients. To overcome
this difficulty, immunohistochemical staining was performed.
Cytokeratin (CK) subtypes, including CK13 (12), CK14 (16), and CK19 (9,12,19),
were attempted, but they failed to show specific properties, as did
p53 and Rb protein (12,34,35).
On the other hand, a combination of immunohistochemical staining
and PCR analysis was previously tested for differential diagnosis:
Bcl-2 expression together with c-myc amplification was demonstrated
to be more frequent in BSCE than in SCC (28), but the specificity of this test was
low (43.5%). In this study, we identified the 75 genes that were
characteristically expressed in BSCE. None of these genes were
mentioned in previous reports. Of 75 genes, collagen related genes
(COL9A2,COL9A3,COL9A1) and fibroblast growth factor related genes
(FGF19, FGF3) might be concerning the characteristics of BSCE based
on the association with genes identified in this study.
In one study, comprehensive gene expression
profiling was performed for endoscopic biopsy specimens of
esophageal SCC (36). However,
CGEA in our institution is different from that in the other
institution. We had previously extracted some disease-specific
genes through the CGEA system at our institution (31,32).
Our CGEA has three features: 1) it can analyze small samples, like
endoscopic biopsy specimens; 2) it can be performed without RNA
amplification; and 3) the gene expression ratio of all types of
samples can be compared with human common reference RNA.
Comprehensive gene expression analysis is done to compare the
expression levels of the human common reference RNA which was
prepared from 22 human cancer cell lines. Profiles were obtained
even from the BSC samples comprising SCC components. A group of
genes specific for BSC were selected by comparing BSC containing
SCC component with non-BSC (SCC). A group of genes that were
expressed in SCC components were eliminated by selection process
for the genes specific for BSC. Therefore, the group of genes
specific for BSC can distinguish between the BSC and non-BSC even
though BSC samples included SCC components. Introducing of the
scoring system enabled us to differentiate BSC from non-BSC if the
sample contains a limited portion of BSC with SCC components. In
this study, we selected 75 genes for constructing a gene expression
scoring system for the proper diagnosis of BSCE. There have been no
reports on studies in which a gene expression scoring system was
constructed to diagnose cancers. This scoring system is a novel,
precise, and powerful tool for diagnosing BSCE in both endoscopic
biopsy and FFPE specimens.
There are several limitations in this study. First,
the biopsy specimens obtained for histology and CGEA were not
identical, although we tried as much as possible to choose
specimens that were adjacent to each other. In addition, biopsy
samples might not hit the component of BSC for accurate diagnosis
by pathological and genetical analysis. Thus, biopsy samples should
be obtained from multiple sites in deep portion of tumor to obtain
histological characteristics of BSC. In contrast, with this method,
we are able to diagnose BSC from very small amount of specimens as
long as it contains BSC component. Second, the parameters of
processing the commercially available FFPE specimens (i.e.,
interval between resection and fixation, duration of fixation) were
not controlled. Third, the cut-off scores varied among the sources
(surgical, endoscopic biopsy, and FFPE specimens); this may have
affected the attainment of reasonable specificity and sensitivity.
In this study, the number of patients was too small to enable
comparison of the prognoses between BSCE and SCC. These limitations
should be addressed using a larger number of cases in the future.
Nevertheless, we believe that this method elucidates the proper
diagnosis of very rare cases of BSCE. Furthermore, it may be able
to clarify whether the prognosis of patients with BSCE is similar
to or poorer than that of patients with SCC. Lastly, recently
(December 1, 2016) International Cell Line Authentication Committee
released Version 8.0 of database of cross-contaminated or
misidentified cell lines (37), in
which we found five cell lines (AKI human melanoma, HBL-100 human
breast carcinoma, human gastric carcinoma MKN-7, SK-N-MC human
neuroblastoma and U937 lymphoma histiocytic cells) among our 22
reference cell lines have been contaminated with HeLa cervical
adenocarcinoma cells, human cells of unknown origin, a cell line of
unknown origin, human Sarcoma (Ewing's) cells and a cell line of
unknown origin, respectively. Even with this condition of reference
cell lines it is obvious that our results would not be affected
since we only used a relative ratio of BSCE against SCC, but not an
absolute ratio to reference cell lines in order to select the
responsible genes for the scoring system.
In conclusion, using CGEA of esophagectomy
specimens, we identified 75 genes that are characteristically
expressed in BSCE; a gene expression scoring system constructed
from these data enabled us to distinguish BSCE from non-BSCE with
high sensitivity and specificity, even on endoscopic biopsy
specimens. We believe that this scoring system can be a novel
method that may significantly contribute to improving the
diagnostic accuracy for BSCE.
Acknowledgments
This work was partially supported by grants for
translational research programs from the New Energy and Industrial
Technology Development Organization (Tokyo, Japan) and Fukushima
Prefecture. The authors would like to thank Enago (www.enago.jp) for the English language review.
Glossary
Abbreviations
Abbreviations:
|
BSC
|
basaloid squamous cell carcinoma
|
|
BSCE
|
BSC of the esophagus
|
|
CGEA
|
comprehensive gene expression
analysis
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
AUC
|
area under the curve
|
|
CI
|
confidence interval
|
|
SCC
|
squamous cell carcinoma
|
|
PCR
|
polymerase chain reaction
|
|
NEC
|
neuroendocrine carcinoma
|
|
CK
|
cytokeratin
|
|
SD
|
standard deviation
|
|
ROC
|
receiver operating characteristic
|
References
|
1
|
Wain SL, Kier R, Vollmer RT and Bossen EH:
Basaloid-squamous carcinoma of the tongue, hypopharynx, and larynx:
Report of 10 cases. Hum Pathol. 17:1158–1166. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sarbia M, Verreet P, Bittinger F,
Dutkowski P, Heep H, Willers R and Gabbert HE: Basaloid squamous
cell carcinoma of the esophagus: Diagnosis and prognosis. Cancer.
79:1871–1878. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brambilla E, Moro D, Veale D, Brichon PY,
Stoebner P, Paramelle B and Brambilla C: Basal cell (basaloid)
carcinoma of the lung: A new morphologic and phenotypic entity with
separate prognostic significance. Hum Pathol. 23:993–1003. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chetty R, Serra S and Hsieh E: Basaloid
squamous carcinoma of the anal canal with an adenoid cystic
pattern: Histologic and immunohistochemical reappraisal of an
unusual variant. Am J Surg Pathol. 29:1668–1672. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brainard JA and Hart WR: Adenoid basal
epitheliomas of the uterine cervix: A reevaluation of distinctive
cervical basaloid lesions currently classified as adenoid basal
carcinoma and adenoid basal hyperplasia. Am J Surg Pathol.
22:965–975. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cubilla AL, Reuter VE, Gregoire L, Ayala
G, Ocampos S, Lancaster WD and Fair W: Basaloid squamous cell
carcinoma: a distinctive human papilloma virus-related penile
neoplasm: a report of 20 cases. Am J Surg Pathol. 22:755–761. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vakar-López F and Abrams J: Basaloid
squamous cell carcinoma occurring in the urinary bladder. Arch
Pathol Lab Med. 124:455–459. 2000.PubMed/NCBI
|
|
8
|
Takubo K, Mafune K, Tanaka Y, Miyama T and
Fujita K: Basaloid-squamous carcinoma of the esophagus with marked
deposition of basement membrane substance. Acta Pathol Jpn.
41:59–64. 1991.PubMed/NCBI
|
|
9
|
Abe K, Sasano H, Itakura Y, Nishihira T,
Mori S and Nagura H: Basaloid-squamous carcinoma of the esophagus.
A clinicopathologic, DNA ploidy, and immunohistochemical study of
seven cases. Am J Surg Pathol. 20:453–461. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koide N, Koike S, Adachi W, Amano J, Usuda
N and Nagata T: Immunohistochemical expression of bcl-2 protein in
squamous cell carcinoma and basaloid carcinoma of the esophagus.
Surg Today. 27:685–691. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kawahara K, Makimoto K, Maekawa T,
Yamamoto S, Shiraishi T, Takahashi S, Shirakusa T, Nakayama Y and
Kikuchi M: An immunohistochemical examination of basaloid squamous
cell carcinoma of the esophagus: Report of a case. Surg Today.
31:655–659. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ohashi K, Horiguchi S, Moriyama S, Hishima
T, Hayashi Y, Momma K, Hanashi T, Izumi Y, Yoshida M and Funata N:
Superficial basaloid squamous carcinoma of the esophagus. A
clinicopathological and immunohistochemical study of 12 cases.
Pathol Res Pract. 199:713–721. 2003.
|
|
13
|
Yoshioka S, Tsujinaka T, Fujitani K and
Kawahara K: Prognostic analysis of four cases of basaloid cell
carcinoma of the esophagus and 60 reported cases in Japan. Jpn J
Gastroenterol Surg. 37:290–295. 2004. View Article : Google Scholar
|
|
14
|
Kobayashi Y, Nakanishi Y, Taniguchi H,
Sekine S, Igaki H, Tachimori Y, Kato H, Matsubara H, Okazumi S and
Shimoda T: Histological diversity in basaloid squamous cell
carcinoma of the esophagus. Dis Esophagus. 22:231–238. 2009.
View Article : Google Scholar
|
|
15
|
Saito S, Hosoya Y, Zuiki T, Hyodo M, Lefor
A, Sata N, Nagase M, Nakazawa M, Matsubara D, Niki T, et al: A
clinicopathological study of basaloid squamous carcinoma of the
esophagus. Esophagus. 6:177–181. 2009. View Article : Google Scholar
|
|
16
|
Imamhasan A, Mitomi H, Saito T, Hayashi T,
Takahashi M, Kajiyama Y and Yao T: Immunohistochemical and
oncogenetic analyses of the esophageal basaloid squamous cell
carcinoma in comparison with conventional squamous cell carcinomas.
Hum Pathol. 43:2012–2023. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang XH, Sun GQ, Zhou XJ, Guo HF and
Zhang TH: Basaloid squamous carcinoma of esophagus:a
clinicopathological, immunohistochemical and electron microscopic
study of sixteen cases. World J Gastroenterol. 4:397–403. 1998.
View Article : Google Scholar
|
|
18
|
Cho KJ, Jang JJ, Lee SS and Zo JI:
Basaloid squamous carcinoma of the oesophagus: A distinct neoplasm
with multipotential differentiation. Histopathology. 36:331–340.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang Z, Shen Y, Liang Y and Wu X:
Basaloid squamous cell carcinoma of the esophagus: An
immunohistochemical study of 8 cases. Chin Med J (Engl).
114:1084–1088. 2001.
|
|
20
|
Lam KY, Law S, Luk JM and Wong J:
Oesophageal basaloid squamous cell carcinoma: A unique
clinicopathological entity with telomerase activity as a prognostic
indicator. J Pathol. 195:435–442. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Klaase JM, Hulscher JBF, Offerhaus GJA,
ten Kate FJ, Obertop H and van Lanschot JJ: Surgery for unusual
histopathologic variants of esophageal neoplasms: A report of 23
cases with emphasis on histopathologic characteristics. Ann Surg
Oncol. 10:261–267. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li TJ, Zhang YX, Wen J, Cowan DF, Hart J
and Xiao SY: Basaloid squamous cell carcinoma of the esophagus with
or without adenoid cystic features. Arch Pathol Lab Med.
128:1124–1130. 2004.PubMed/NCBI
|
|
23
|
Chen SB, Weng HR, Wang G, Yang JS, Yang
WP, Li H, Liu DT and Chen YP: Basaloid squamous cell carcinoma of
the esophagus. J Cancer Res Clin Oncol. 138:1165–1171. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang BH, Cheng GY, Xue Q, Gao SG, Sun KL,
Wang YG, Mu JW and He J: Clinical outcomes of basaloid squamous
cell carcinoma of the esophagus: A retrospective analysis of 142
cases. Asian Pac J Cancer Prev. 14:1889–1894. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Akagi I, Miyashita M, Makino H, Nomura T,
Ohkawa K and Tajiri T: Basaloid squamous cell carcinoma of the
esophagus: Report of two cases. J Nippon Med Sch. 75:354–360. 2008.
View Article : Google Scholar
|
|
26
|
Nishimura W, Naomoto Y, Hamaya K, Toda S,
Miyagi K and Tanaka N: Basaloid-squamous cell carcinoma of the
esophagus: Diagnosis based on immunohistochemical analysis. J
Gastroenterol Hepatol. 16:586–590. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kato T, Morita T, Fujita M, Miyasaka Y,
Horita S, Watanabe Y and Kato H: Basaloid-squamous carcinoma of the
esophagus: Report of a case. Surg Today. 30:163–167. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sarbia M, Loberg C, Wolter M, Arjumand J,
Heep H, Reifenberger G and Gabbert HE: Expression of Bcl-2 and
amplification of c-myc are frequent in basaloid squamous cell
carcinomas of the esophagus. Am J Pathol. 155:1027–1032. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bellizzi AM, Woodford RL, Moskaluk CA,
Jones DR, Kozower BD and Stelow EB: Basaloid squamous cell
carcinoma of the esophagus: Assessment for high-risk human
papillomavirus and related molecular markers. Am J Surg Pathol.
33:1608–1614. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Arai T, Aida J, Nakamura KI, Ushio Y and
Takubo K: Clinicopathologic characteristics of basaloid squamous
carcinoma of the esophagus. Esophagus. 8:169–177. 2011. View Article : Google Scholar
|
|
31
|
Miyamoto K, Iwadate M, Yanagisawa Y, Ito
E, Imai J, Yamamoto M, Sawada N, Saito M, Suzuki S, Nakamura I, et
al: Cathepsin L is highly expressed in gastrointestinal stromal
tumors. Int J Oncol. 39:1109–1115. 2011.PubMed/NCBI
|
|
32
|
Okabe N, Ezaki J, Yamaura T, Muto S, Osugi
J, Tamura H, Imai J, Ito E, Yanagisawa Y, Honma R, et al: FAM83B is
a novel biomarker for diagnosis and prognosis of lung squamous cell
carcinoma. Int J Oncol. 46:999–1006. 2015.PubMed/NCBI
|
|
33
|
Miura A, Honma R, Togashi T, Yanagisawa Y,
Ito E, Imai J, Isogai T, Goshima N, Watanabe S and Nomura N:
Differential responses of normal human coronary artery endothelial
cells against multiple cytokines comparatively assessed by gene
expression profiles. FEBS Lett. 580:6871–6879. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Owonikoko T, Loberg C, Gabbert HE and
Sarbia M: Comparative analysis of basaloid and typical squamous
cell carcinoma of the oesophagus: A molecular biological and
immunohistochemical study. J Pathol. 193:155–161. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Baba Y, Ishimoto T, Harada K, Kosumi K,
Murata A, Miyake K, Hiyoshi Y, Kurashige J, Iwatsuki M, Iwagami S,
et al: Molecular characteristics of basaloid squamous cell
carcinoma of the esophagus: analysis of KRAS, BRAF, and PIK3CA
mutations and LINE-1 methylation. Ann Surg Oncol. 22:3659–3665.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Motoori M, Takemasa I, Yamasaki M, Komori
T, Takeno A, Miyata H, Takiguchi S, Fujiwara Y, Yasuda T, Yano M,
et al: Prediction of the response to chemotherapy in advanced
esophageal cancer by gene expression profiling of biopsy samples.
Int J Oncol. 37:1113–1120. 2010.PubMed/NCBI
|
|
37
|
International Cell Line Authentication
Committee: Register of Misidentified Cell Lines, version 8.0.
December 1–2016, http://iclac.org/databases/cross-contaminations/.
|