Introduction
Cervical cancer affects ~500,000 individuals each
year in women worldwide. It is the major cause of cancer death in
women especially in developing countries (1). As known, persistent infection of
high-risk types of human papillomavirus (HPV) is the leading cause
of cervical cancer (2–5). Besides, genetic factors and personal
lifestyle also involve in the progression from precancerous disease
to invasive cervical cancer. However, the exact etiology of
cervical cancer remains unclear. Despite the improvement of
therapeutic methods and diagnostic tools, the overall prognosis of
patients has not been improved (2,6).
Many patients do not respond well to any currently available
treatments (7). Therefore,
exploration of pathogenesis and identification of novel biomarkers
is imperative.
The receptor for activated protein kinase C (RACK1)
is a homolog of the G protein β-subunit. It was originally
identified as an anchoring protein for activated protein kinase C
(PKC) in cytoplasm (8,9). RACK1 was reported to be involved in
tumorigenesis and tumor progression. Many researchers have
demonstrated that RACK1 plays a pivotal role in various biological
responses, including signal transduction (10–14),
cell growth and migration (15–18),
cell apoptosis and chemoresistance (19,20).
RACK1 was also used as an excellent predictor for clinical
prognosis in hepatocellular carcinoma (HCC) and oral squamous cell
carcinoma (OSCC) (21). With more
studies on RACK1, it was found that the effect of RACK1 was diverse
in different types of tumors. RACK1 was found to be upregulated and
to promote tumorigenicity in non-small cell lung cancer (12,22),
pulmonary adenocarcinoma (23,24),
hepatocellular carcinoma (25),
neuroblastoma (18), and
esophageal squamous cell carcinoma (17). On the contrary, the expression
level of RACK1 was found to be reduced in gastric cancer (13,14,26),
Besides, RACK1 decreased tumorigenicity of colon cells (27). It is speculated that RACK1 exert
diverse effects in different types of cancers via its interaction
with different partners and through various signaling pathways.
However, the exact mechanisms of the aberrant expression and
opposed roles of RACK1 are still unclear.
In this study, we found that RACK1 is highly
expressed in cervical cancer tissues and decreases cell senescence
and promotes the invasion and migration of cervical cancer cells.
Furthermore, we found that the overexpression of RACK1 upregulates
the expression of NF-κB, cyclin D1 and CDK4 and downregulates the
expression of p53, p38, p21 and STATA1 in vitro. These
results suggest that RACK1 promotes cell growth, invasion and
migration in cervical cancer cells probably by affecting the p53
signaling pathway.
Materials and methods
Cell culture and transfection
The human cervical cancer cells, CaSki, were
obtained from the ATCC (Manassas, VA, USA) and kept in our
laboratory. Cells were cultured in RPMI-1640 supplemented with 10%
fetal bovine serum (FBS) (both from Gibco Life Technologies, Grand
Island, NY, USA). Cells were grown at 37°C in the presence of 5%
CO2. Cells were transfected using Lipofectamine 2000
(Invitrogen by Life Technologies, USA) according to the
manufacturer's protocol as we described previously (28). For RACK1-overexpressed stable
cells, 1,000 µg/ml G418 (Sigma, St. Louis, MO, USA) was
added 48 h after transfection and sustained for >14 days. The
maintainance concentration of G418 was 500 µg/ml, and it was
used throughout the cell culture. We verified the transfection
effectiveness and RACK1-overexpressed stable cells by western blot
assay. We also established the transient transfected RACK1-silenced
CaSki cells by transfecting with the plasmid of
SD.U6/neo/GFP-RACK1-RNAi and SD/U6/neo/GFP-NC using Lipofectamine
2000. Briefly, cells (2×105 cells per well) were seeded
24 h prior to the transfection. For each transfection, 2 µg
of SD/U6/neo/GFP-NC or SD.U6/neo/GFP-RACK1-RNAi plasmid was
transfected into CaSki cells, respectively. The cells were digested
and resuspended for further experiments 24 h to 36 h later.
Patient samples
Twenty-five participants were recruited between 2012
and 2016 at the Third Xiangya Hospital, Central South University
(Changsha, Hunan, China). Consent forms were obtained from
individual patients, and experimental protocols were approved by
the Institutional Review Board of the Third Xiangya Hospital. The
clinical stages of cervical cancer were defined according to the
International Federation of Gynecology and Obstetrics (FIGO). All
subjects enrolled in the study were Chinese. Cervical cancer tissue
and corresponding non-tumor normal tissue were collected, and each
biopsy sample was divided into two sections, one was submitted to
routine histological diagnosis, and the remaining section was used
for western blotting and immunohistochemistry experiments.
Immunohistochemistry (IHC) and evaluation
of staining
Immunohistochemistry was done using the peroxidase
anti-peroxidase technique following a microwave antigen retrieval
procedure. Mouse anti-RACK1 was purchased from Santa Cruz
Biotechnology (cat. no. sc-17754; Dallas, TX, USA). Antibody
against RACK1 (1:200) was overlaid on cervical cancer and
corresponding non-tumor normal tissue sections and incubated
overnight at 4°C. Secondary antibody incubation (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) was performed at room
temperature for 30 min. Color reaction was developed by using 3,
3′-diaminobenzidine tetrachloride (DAB) chromogen solution. All
slides were counterstained with hematoxylin. Positive control
slides were included in every experiment in addition to the
internal positive controls. The specificity of the antibody was
determined with matched IgG isotype antibody as a negative
control.
Sections were blindly evaluated by two investigators
in an effort to provide a consensus on staining patterns by light
microscopy (Olympus, Japan). RACK1 staining was assessed according
to the methods described by Hara and Okayasu with minor
modifications (29). Each case was
rated according to a score that added a scale of intensity of
staining to the area of staining. At least 10 high-power fields
were chosen randomly, and >1,000 cells were counted for each
section. The intensity of staining was graded on the following
scale: 0, no staining; 1+, mild staining; 2+, moderate staining;
3+, intense staining. The area of staining was evaluated as
follows: 0, no staining of cells in any microscopic fields; 1+,
<30% of tissue stained positive; 2+, between 30 and 60% stained
positive; 3+, >60% stained positive. The minimum score when
summed (extension + intensity) was, therefore, 0, and the maximum,
6. A combined staining score (extension + intensity) of ≤2 was
considered to be a low staining; a score between 3 and 4 was
considered to be a moderate staining; whereas a score between 5 and
6 was considered to be a strong staining.
β-galactosidase staining
The detection of cellular senescence was performed
using a senescence-associated β-galactosidase staining kit (C0602;
Beyotime, China) according to the manufacturer's protocol. Images
were captured by a light microscope (CKX41; Olympus). The
β-galactosidase positive cells (blue) were considered
senescent.
Colony formation and CCK8 assay
Eight hundred cells were seeded per well in 6-well
plates and cultured for 14 days at 37°C. After incubation, cells
were fixed with 4% paraformaldehyde solution and stained with 0.1%
crystal violet solution. The number of colonies with >50 cells
was counted and photographed. The CCK8 assay was carried according
to the protocol (7Sea-Cell Couting kit; 7Sea Biotech, China. Cell
suspension (200 µl) was seeded in 96-well cell culture
plates at a density of 1,000 cells/well and incubated at 37°C for
six days. The density of cells were detected every 24 h. The CCK8
solution (20 µl) was added to each well and incubated at
37°C for 2 h. Then the absorbance at 450 nm was measured using a
Paradigm Detection Platform (Beckman Coulter, Brea, CA, USA).
Wound healing assay
For wound healing assay, 5×105 cells were
seeded per well in 6-well plates, when the cell density grown to
90%, a straight scratch in the cell monolayer was created by a
10-µl pipette tip. Images of the scratched area (wound) were
taken at the time point of 0, 24, 48 and 72 h under a microscope
(CKX41; Olympus).
Cell invasion and migration assay
The cell invasion experiment was performed using
Corning Matrigel Invasion Chamber in 24-well plate (8-µm
pore size; Corning Life Sciences, Lowell, MA, USA). A total of
2.5×104 cells in 500 µl of serum-free medium were
added to the top chamber. Growth medium (750 µl) with 20%
FBS was added into the lower chamber. After incubation in a
humidified incubator with 5% CO2 for 48 h at 37°C, parts
of cells were invaded to the lower surface of the upper chamber.
Fixed the cells with 4% paraformaldehyde and stained with crystal
violet. The cells on the upper surface of the upper chamber were
gently removed by a cotton swab. Images of the stained cells were
then captured under a microscope at ×100 magnification and cells
from at least five randomly selected fields were counted for each
experiment. The cell migration assay was performed similarly with
the cell invasion experiment by using Falcon Cell Culture Inserts
(8-µm pore size; Corning Life Sciences), which without
Matrigel coating on the upper surface of the Transwell filters.
After 24-h incubation in a humidified incubator, the chamber were
fixed with 4% paraformaldehyde and stained with crystal violet.
Images were then captured and counted cells from at least five
randomly selected fields for each experiment.
RNA extraction and quantitative real-time
PCR
Total RNA was extracted from cells using the TRIzol
reagent (Invitrogen, Carlsbad, CA, USA), and was converted to cDNA
using the All-in-One™ First-Strand cDNA Synthesis kit (GeneCopoeia,
Rockville, MD, USA). Quantitative real-time PCR was performed using
ChamQ™ SYBR® qPCR Master Mix (Vazyme, Nanjing, China),
according to the manufacturer's instructions. Data were normalized
to the expression level of GAPDH. The primers used in this
manuscript were as follows: MMP-2 (left-atgacagctgcaccactgag,
right-atttgttgcccaggaaagtg), MMP-3 (left-tgctttgtcctttgatgctg,
right-ggaagagatggccaaaatga), MMP-9 (left-ttgacagcgacaagaagtgg,
right-gccattcacgtcgtcc ttat) and MMP-10 (left-ggctctttcactcagccaac,
right-tcccgaaggaacagattttg).
Flow cytometry analysis of cell cycle and
cell apoptosis
Cell cycle and cell apoptosis analysis was carried
out by flow cytometry. Cells (5×105/well) were seeded in
6-well cell culture plates and incubated in a humidified atmosphere
of 5% CO2 at 37°C. Twenty-four hours later, cells were
harvested with 0.5% trypsin and centrifuged. For cell cycle
analysis, cells were fixed in ice-cold 70% ethanol overnight at
4°C. After being washed thrice with cold PBS, cells were
resuspended in 500 µl of PBS, and 10 µl RNAseA was
added for 5 min. Subsequently, 10 µl PI was added into the
cell resuspension solution, and incubated for 30 min at 4°C. The
cells were finally washed twice with PBS before analysis. Apoptotic
analysis was performed using the Hoechst 33342/PI Apoptosis assay
kit (BestBio, Shanghai, China). The samples were washed twice with
ice-cold PBS and resuspended in 500 µl staining buffer, and
then incubated with 5 µl of Hoechst 33342 and 5 µl of
PI in the dark for 20 min at 4°C. The cells were finally washed
twice with PBS before analysis. Analysis was performed on MoFlo™XDP
High-Performance Cell Sorter (Beckman Coulter) and the data were
analyzed with the summit v5.2 Software (Beckman Coulter).
Mitochondrial membrane potential
detection
The mitochondrial membrane potential (MMP) was
detected using a JC-1 fluorescent probe (MultiSciences Biotech Co.,
Ltd., Hangzhou, China), which is a cationic lipophilic dye that
exhibits potential-dependent accumulation in mitochondria. In
normal cells, the dye concentrates in the mitochondrial matrix,
where it forms red fluorescent aggregates. Any event that
dissipates the mitochondrial membrane potential prevents the
accumulation of the JC-1 dye in the mitochondria and the dye is
dispersed throughout the entire cell leading to a shift from red to
green fluorescence. Thus, a change in JC-1 fluorescence emission
from red to green indicates depolarization of the mitochondrial
membrane. In our experiments, cells were harvested and resuspended
in 1 ml warm staining buffer at ~1×106 cells/ml. One
microliter of 2 mM JC-1 was added into the cell suspension and
incubated at 37°C for 30 min. Washed cells were resuspended in 500
µl PBS. It was then detected by a flow cytometer MoFlo™ XDP
(Beckman Coulter). CCCP was used as a positive control and perform
standard compensation.
Western blot analysis
Western blot analysis was performed as we described
previously (28). In brief, cells
and tissues were lysed in RIPA buffer (CWBio, Beijing, China), then
50 µg of lysates were separated in 10% SDS-PAGE gels and
transferred onto a PVDF membrane (Hyclone Laboratories, Logan, UT,
USA) and then blocked (5% non-fat milk dissolved in TBS-Tween-20)
for 1-2 h. The membranes were then incubated overnight at 4°C with
primary antibodies. Primary antibodies used in this study include:
mouse anti-RACK1 (Santa Cruz Biotechnology, Dallas, TX, USA;
1:1,000 dilution), rabbit anti-c-rel, rabbit anti-stat1, rabbit
anti NF-κB-P65 (Immunoway Technology, Newark, DE, USA; 1:1,000
dilution), rabbit anti NF-κB-p65 (phospho-Ser536) polyclonal
antibody (Immunoway Technology, Newark, DE, USA; 1:1,000 dilution),
mouse anti-CDK4, rabbit anti-cyclin D1, mouse anti-P53, rabbit
anti-P21, rabbit anti-MAPK14/P38 (Boster Technology, Wuhan, China;
1:500 dilution), and rabbit anti-cyclin A (Affinity, Ancaster,
Canada; 1:1,000 dilution). After washing three times by TBST, the
membranes were incubated with anti-rabbit or anti-mouse
HRP-conjugated secondary antibody (Santa Cruz Biotechnology;
1:5,000 dilution). The detection was finally performed on the
ChemiDoc XRS+ Molecular Imager (Bio-Rad) using the
Luminata Forte Western HRP Substrate (Millipore Corp., Billerica,
MA, USA). Rabbit anti-GAPDH (Santa Cruz Biotechnology; 1:3,000
dilution) was used as a loading control.
Statistical analysis
Differences of non-parametric variables were
analyzed by the Mann-Whitney U test. Differences of the
quantitative variables between groups were analyzed by Student's
t-test using SPSS 11.0 program (SPSS, Chicago, IL, USA). A value of
P<0.05 was considered statistically significant.
Results
RACK1 expression is high in cervical
cancer
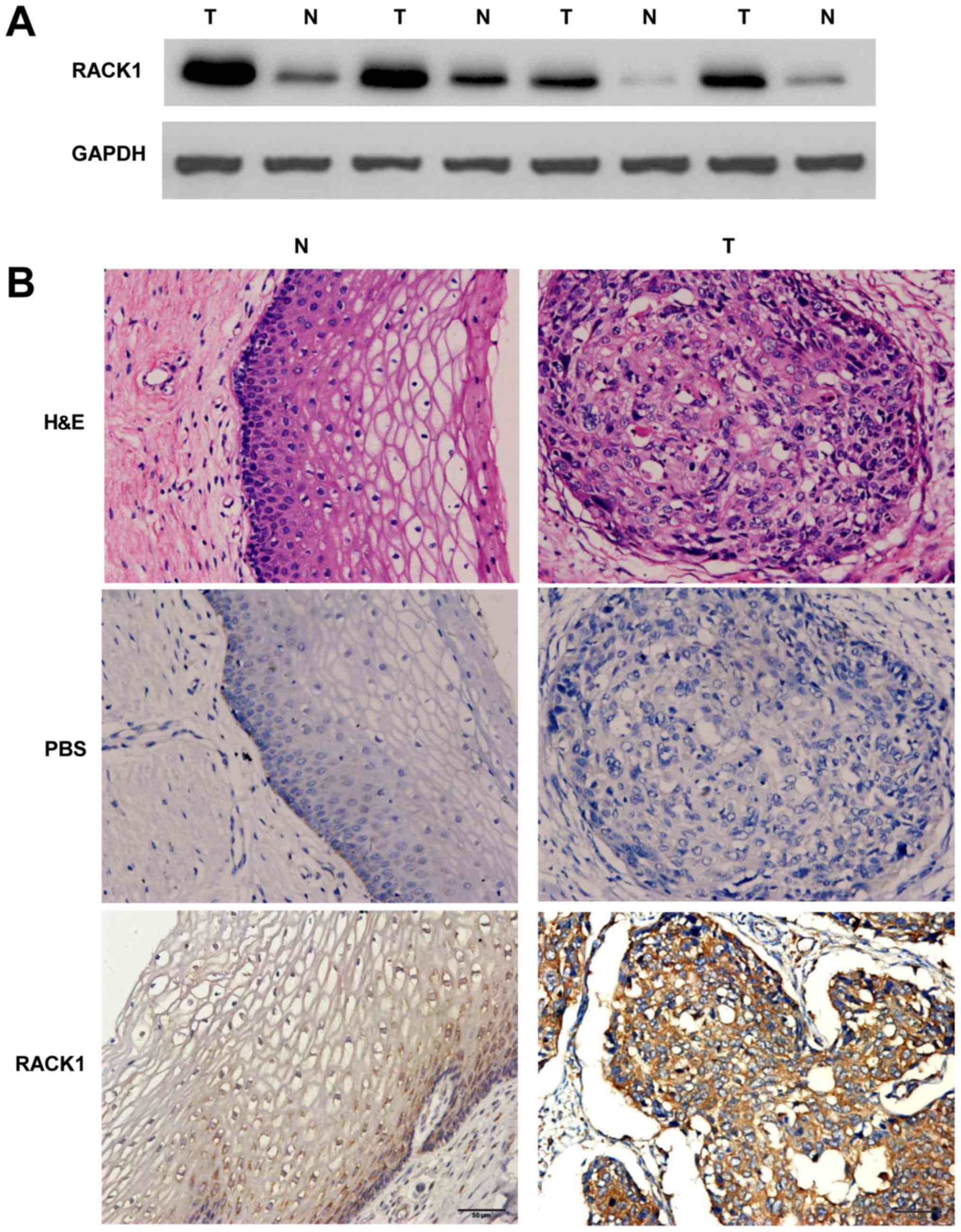
To detect the expression levels of the RACK1 in
cervical cancer and the adjacent non-cancerous tissues, western
blotting of RACK1 was performed. In comparison with the adjacent
non-cancerous tissues, the expression level was identified to be
greater in cervical cancer tissues (Fig. 1A). Meanwhile, immunohistochemistry
(IHC) was carried out with anti-bodies against RACK1 in cervical
cancer and the adjacent non-cancerous tissues. We randomly selected
25 participants who were diagnosed cervical cancer patients,
collecting cervical cancer tissues and the adjacent non-cancerous
tissues for IHC detecting (Table
I). Our study showed a similar pattern in protein expression
with western blot results. There was 68% (17/25) high score of
RACK1 in cervical cancer tissues and 28.0% (7/25) in the adjacent
non-cancerous tissues. The distribution of low score was 8.0%
(2/25) and 32.0% (8/25) in cervical cancer and the adjacent
non-cancerous tissues, respectively (P=0.012 <0.05) (Fig. 1B and Table II). These results identified that
RACK1 is highly expressed in cervical cancer.
 | Table IThe clinical information of cervical
cancer patients contributing to this study. |
Table I
The clinical information of cervical
cancer patients contributing to this study.
| Samples | Age (years) | HPV type | Cell type |
Differentiation | Stage |
|---|
| 1 | 57 | 31 | Squamous
carcinoma | Moderate | IIa |
| 2 | 45 | 16 | Squamous
carcinoma | Moderate | IIb |
| 3 | 62 | 52 | Squamous
carcinoma | Moderate | IIa |
| 4 | 51 | 16 | Squamous
carcinoma | Moderate | IIb |
| 5 | 65 | 16 | Squamous
carcinoma | Moderate | IIb |
| 6 | 48 | (−) | Squamous
carcinoma | Moderate | IIa |
| 7 | 52 | 16 | Squamous
carcinoma | Poor | IIb |
| 8 | 51 | 16 | Squamous
carcinoma | Poor | IIa |
| 9 | 39 | 16 | Squamous
carcinoma | Moderate | IIb |
| 10 | 49 | 52 | Squamous
carcinoma | Moderate | Ib1 |
| 11 | 50 | 16 | Squamous
carcinoma | Moderate | IIa |
| 12 | 47 | 16 | Squamous
carcinoma | Moderate | IIb |
| 13 | 50 | 16 | Squamous
carcinoma | Moderate | IIa |
| 14 | 52 | 45 | Squamous
carcinoma | Moderate | IIa |
| 15 | 45 | 16 | Squamous
carcinoma | Moderate | IIa |
| 16 | 52 | 33 | Squamous
carcinoma | Moderate | IIa |
| 17 | 46 | 18 | Squamous
carcinoma | Moderate | IIb |
| 18 | 40 | 16 | Squamous
carcinoma | Moderate | Ib1 |
| 19 | 54 | 16 | Squamous
carcinoma | Poor | Ib2 |
| 20 | 50 | 18 | Squamous
carcinoma | Poor | IIb |
| 21 | 47 | 16 | Squamous
carcinoma | Poor | Ib2 |
| 22 | 62 | 16 | Squamous
carcinoma | Moderate | IIb |
| 23 | 40 | 16 | Squamous
carcinoma | Moderate | Ib1 |
| 24 | 58 | 16,33 | Squamous
carcinoma | Moderate | IIb |
| 25 | 68 | 16 | Squamous
carcinoma | Moderate | IIb |
 | Table IIThe difference of RACK1 expression
between cervical cancer and the adjacent non-cancerous tissues. |
Table II
The difference of RACK1 expression
between cervical cancer and the adjacent non-cancerous tissues.
| Samples | n | Score
| P-value |
|---|
| Low (0–2) | Moderate (3–4) | High (5–6) |
|---|
| Cervical
cancer | 25 | 2 (8.0%) | 6 (24.0%) | 17 (68.0%) | 0.012a |
| Non-cancerous
tissues | 25 | 8 (32.0%) | 10 (40.0%) | 7 (28.0%) | |
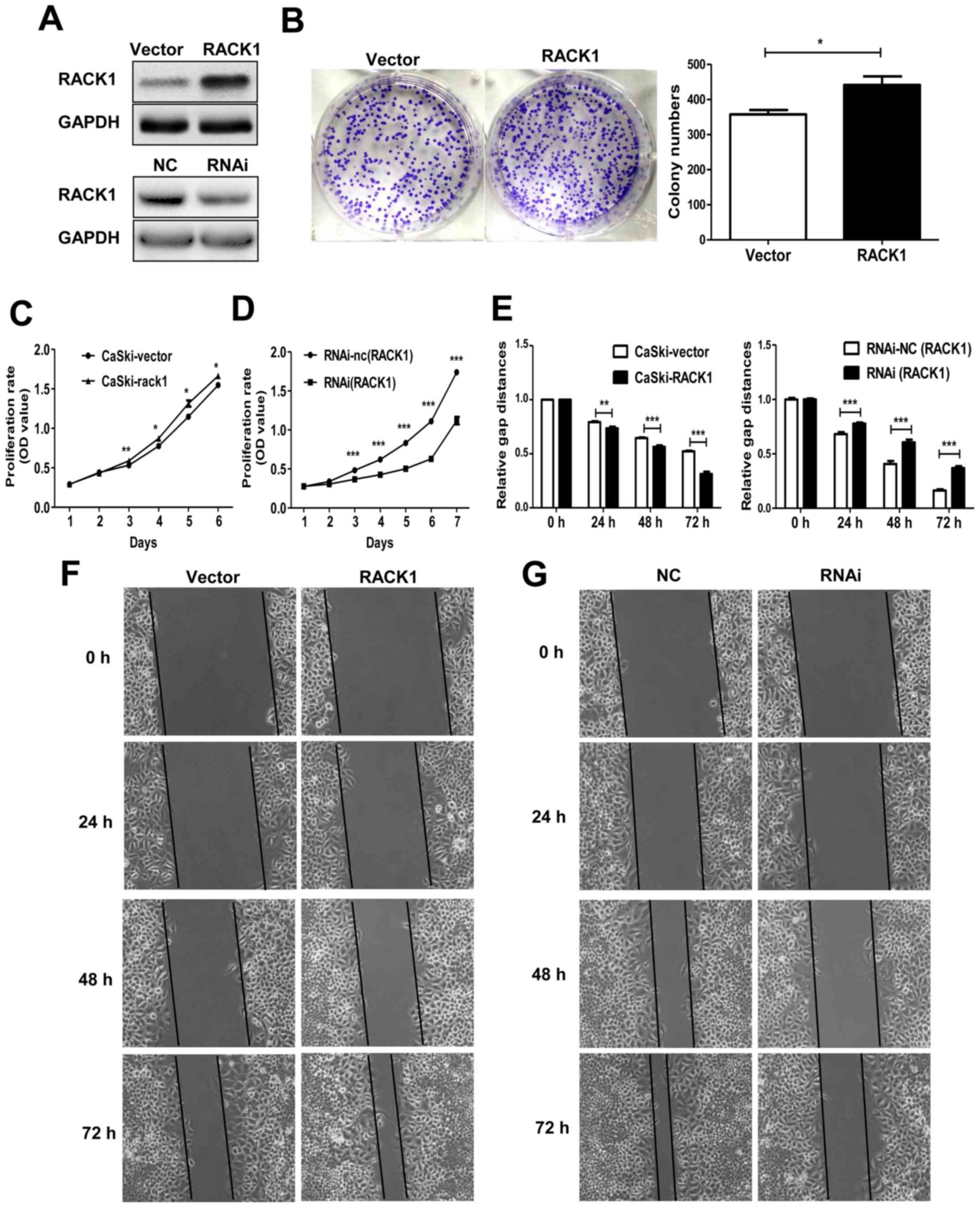
RACK1 overexpression promotes the
proliferation of cervical cancer cells
To elucidate the function of RACK1 in the biological
behavior of cervical cancer cells, the CaSki cells were transfected
with the plasmid pEGFP-N1-RACK1 or control vector pEGFP-N1-vector
to generate stable transfected CaSki-RACK1 and CaSki-vector cell
lines. After demonstrating the RACK1 expression level by western
blot analysis (Fig. 2A), the
spontaneous proliferation of CaSki-RACK1 and CaSki-vector cells
were determined by the colony formation assay and CCK8 assay,
respectively. The colony formation assay showed that RACK1
overexpression promoted the colony formation ability (Fig. 2B, P=0.023). CCK8 analysis showed
that RACK1-overexpression promoted cell proliferation in CaSki
cells (Fig. 2C). We also performed
CCK8 assay on RACK1-silenced CaSki cells. Twenty-four hours after
transfection with RACK1-RNAi or the control plasmid RNAi-NC, CaSki
cells were digested and resuspended for the CCK8 assay. It showed
that the proliferation of CaSki cells significantly decreased after
RACK1 was silenced (Fig. 2D). The
transfection effect was demonstrated by western blotting (Fig. 1A). Therefore, we considered that
RACK1 promoted the proliferation of cervical cancer cells in
vitro.
RACK1 overexpression promotes the cell
invasion and migration and inhibits the cell senescence in cervical
cancer cells
Cell invasion and migration is an important step
during tumor metastasis progress. We detected the migration ability
of RACK1-stable expressing CaSki-RACK1 cells and control
CaSki-vector cells by wound healing assay in vitro. We found
that RACK1 promoted the cell migration in cervical cancer cells
(Fig. 2E and F). We also performed
the wound healing assay in RACK1-silenced CaSki cells. Twenty-four
hours after we transfected with RACK1-RNAi or the control plasmid
RNA-NC, CaSki cells were digested and resuspended in 6-well cell
culture plate. Images of the scratched area were photographed every
24 h. It showed that the migration rate of CaSki cells decreased
after RACK1 was silenced (Fig. 2E and
G).
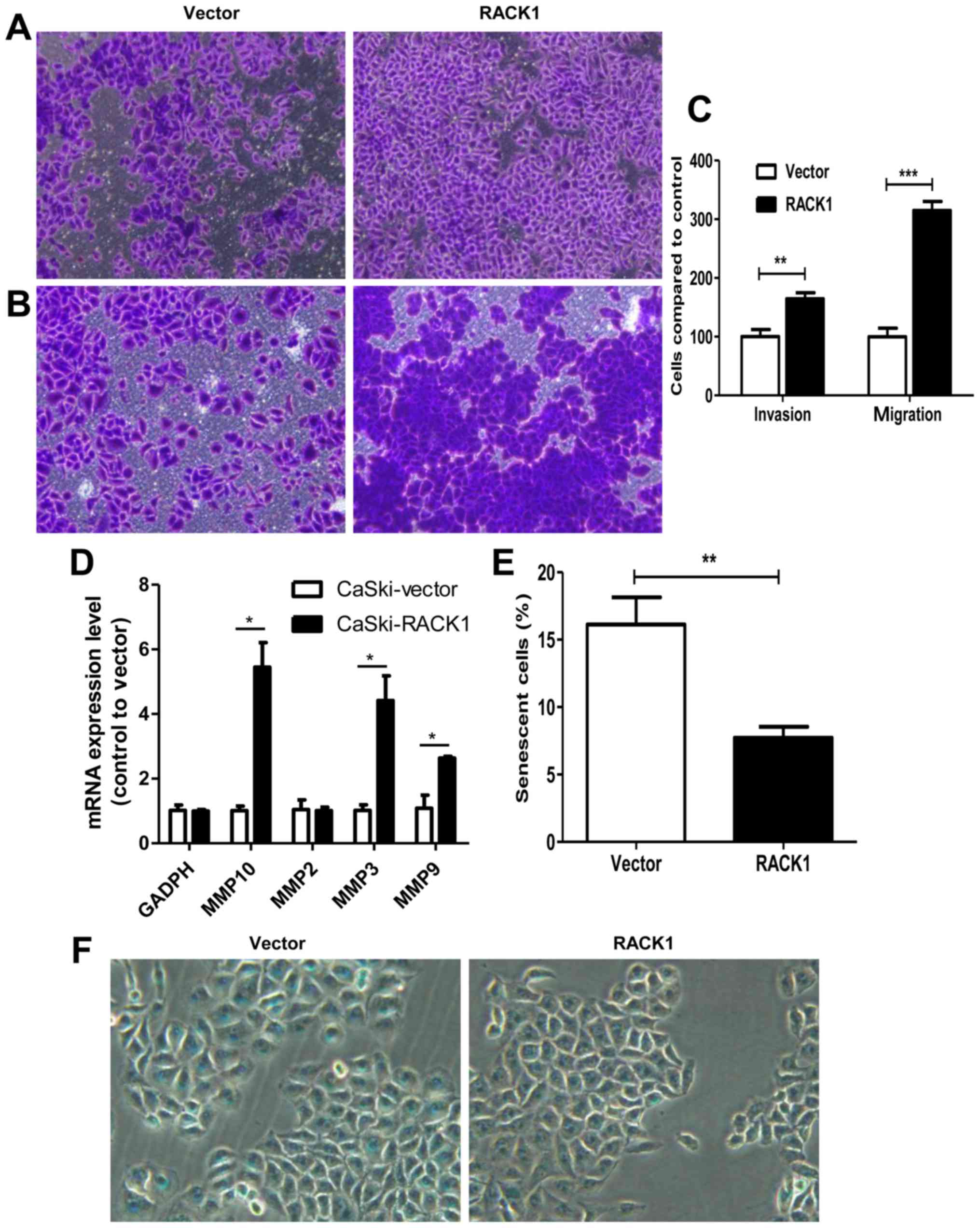
Furthermore, Transwell invasion and migration assays
demonstrated that RACK1-overexpressed CaSki cells had a higher
invasion and migration capability. For migration assay, 24-h
incubation after cell-planting, we found that cells migrated into
the downside of the chamber in RACK1-overexpressed CaSki cells were
3 times more than the control group (Fig. 3A and C, P<0.0001). For invasion
assay, 48-h incubation after cell-planting, we found that cell
invasion speed in RACK1-overexpressed CaSki cells was 1.64 times
higher than the control group (Fig. 3B
and C, P=0.0015). We further detected the mRNA expression
levels of MMP-2, MMP-3, MMP-9 and MMP-10 by qPCR technology. We
found that MMP-3, MMP-9 and MMP-10 were upregulated in
RACK1-overexpressed CaSki cells (Fig.
3D, P=0.041, 0.016 and 0.02, respectively). The results showed
that CaSki-RACK1 cells have higher invasion ability compared to
control CaSki-vector cells. This further confirmed that RACK1 may
contribute to tumor invasion and migration in cervical cancer.
Senescence is a stable cell cycle arrest that plays
an important role in tumor development or tumor suppression. We
further explored whether RACK1 has a function on cell senescence by
β-galactosidase staining. As shown in Fig. 3E and F, the number of
β-galactosidase-positive cells in control CaSki-vector cells was
much higher than RACK1 overexpression CaSki cells (16.12 vs. 7.71%,
P=0.0012). It revealed that RACK1 decreased cell senescence in
cervical cancer cells.
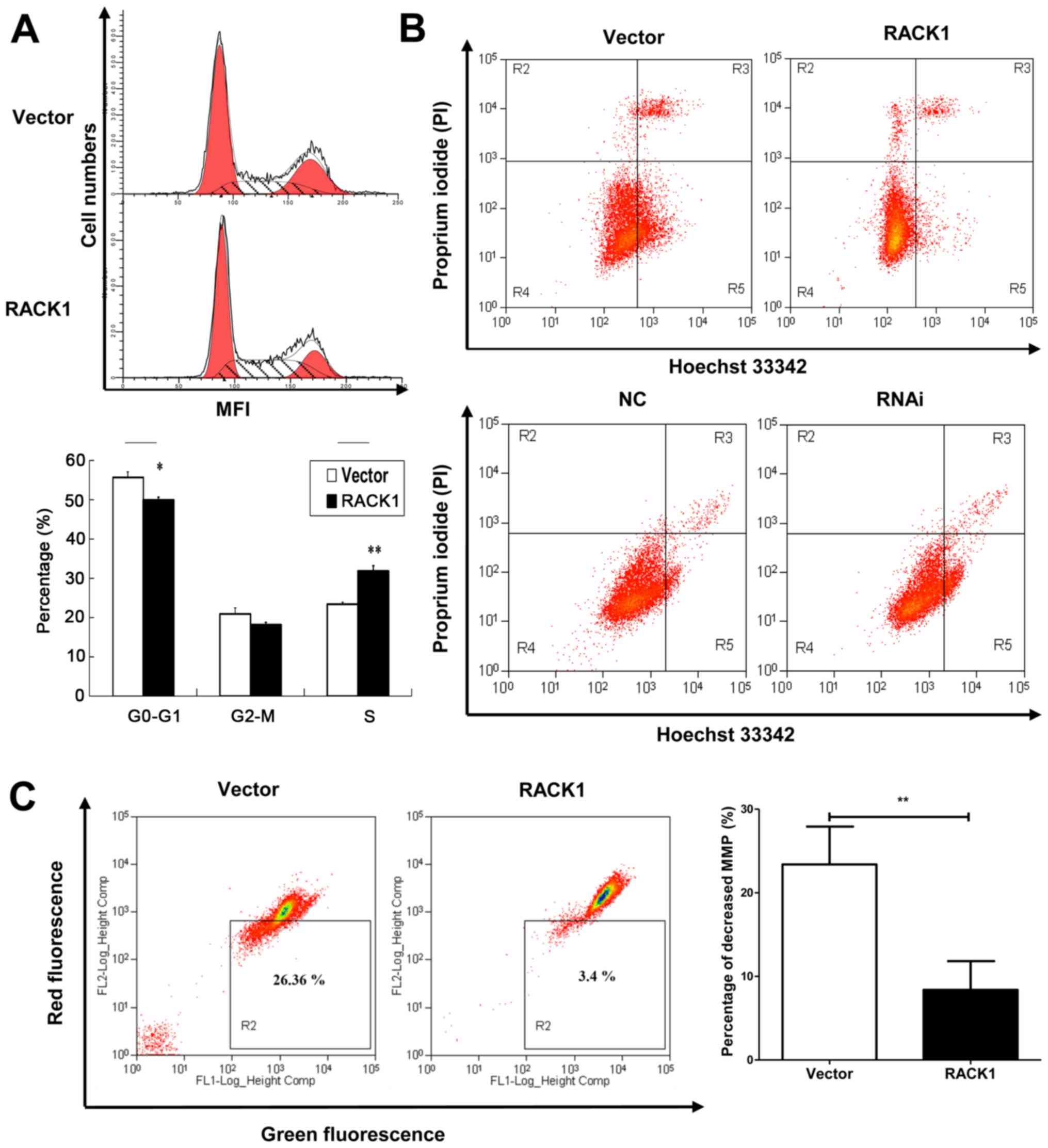
RACK1 overexpression induces S phase
accumulation in cell cycle analysis in cervical cancer cells
Cell cycle analysis was performed by flow cytometry.
Percentages of cells at different stages were statistically
analyzed. We observed that RACK1 overexpressed CaSki cells had an S
phase accumulation (31.88%), compared to empty vector transfected
CaSki cells (23.41%) (Fig. 4A).
This indicated that RACK1 may contribute to DNA synthesis in cell
cycle and promote cell proliferation.
RACK1 suppresses cell apoptosis of
cervical cancer cells
To further explore the role of RACK1 in cervical
cancer, we detected cell apoptosis using Hoechst 33342/PI double
stain assay and we analyzed by flow cytometry both the
RACK1-overexpressed and RACK1-silenced CaSki cells. Hoechst 33342
stains the condensed chromatin in apoptotic cells more brightly
than normal chromatin. Propidium iodide (PI) is only permeant to
dead cells and late apoptotic cells. The staining pattern makes it
possible to distinguish normal, apoptotic, and dead cell
populations. The cells with Hoechst (−)/PI (-) were considered
normal, the Hoechst (+)/PI (−) cells were thought to be early
apoptotic cells, the Hoechst (+)/PI (+) cells were regarded as late
apoptotic cells, and the Hoechst (−)/PI (+) cells were considered
to be necrotic cells. As shown in Fig.
4B, in CaSki stable cell lines, the proportion of apoptotic
cells in RACK1 overexpressed cells were lower than vector
transfected cells (the rate of late apoptotic was 4.53 and 5.99 for
RACK1 and vector respectively, and the rate of early apoptotic was
4.60 and 24.23 for RACK1 and vector respectively) (Fig. 4B, upper image). While in
RACK1-silenced CaSki cells, the apoptotic rate was higher than the
control group (the rate of late apoptotic was 2.27 and 2.00 for
RACK1-RNAi and RNAi-NC transfected CaSki cells respectively, and
the rate of early apoptotic was 13.60 and 5.47 for RACK1-RNAi and
RNAi-NC transfected CaSki cells respectively) (Fig. 4B, lower image). The results implied
that RACK1 could suppress cell apoptosis.
RACK1 affects the mitochondrial membrane
potential in cervical cancer cells
The dissipation of the mitochondrial membrane
potential (Δψm) is known as an early event in apoptosis. So we
examined the mitochondrial apoptosis by JC-1 staining. A decline of
mitochondrial membrane potential during apoptosis was represented
by a decrease in red fluorescence intensity and increase in green
fluorescence intensity. As demonstrated in our study, the
percentage of cells with loss of Δψm in RACK1-overexpressed CaSki
cells was less than control groups (Fig. 4C, P=0.0019). It was demonstrated
that RACK1 may inhibit cell mitochondrial apoptosis in cervical
cancer cells.
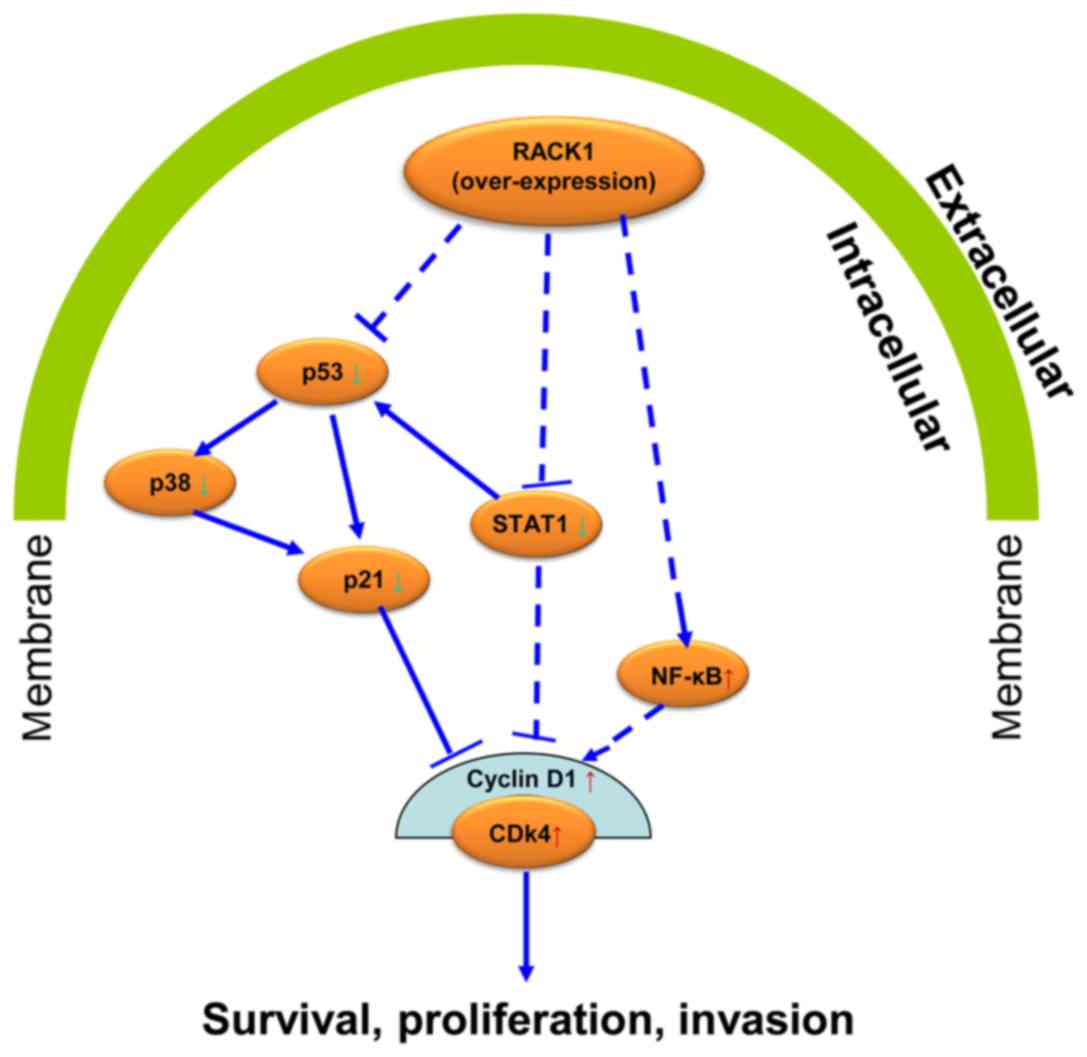
RACK1 overexpression affects the
expression of p53, p38, p21, STAT1, NF-κB, cyclin D1 and CDK4 in
vitro
To uncover the possible mechanism of RACK1 in
cervical cancer, we tested the expression levels of key molecules
in p53 signaling pathway by western blot technology. The p53, p38
and p21 were down-regulated in CaSki cells in which Rack1 was
overexpressed, whereas, the overexpression of Rack1 downregulated
the expression level of STAT1. Furthermore, NF-κB, cyclin D1 and
CDK4 was upregulated in CaSki cells in which Rack1 was
overexpressed (Fig 5). Our results
suggested that RACK1 overexpression may upregulate the expression
of NF-κB, cyclin D1 and CDK4 and downregulated the expression of
p53, p38, p21 and STAT1 in vitro.
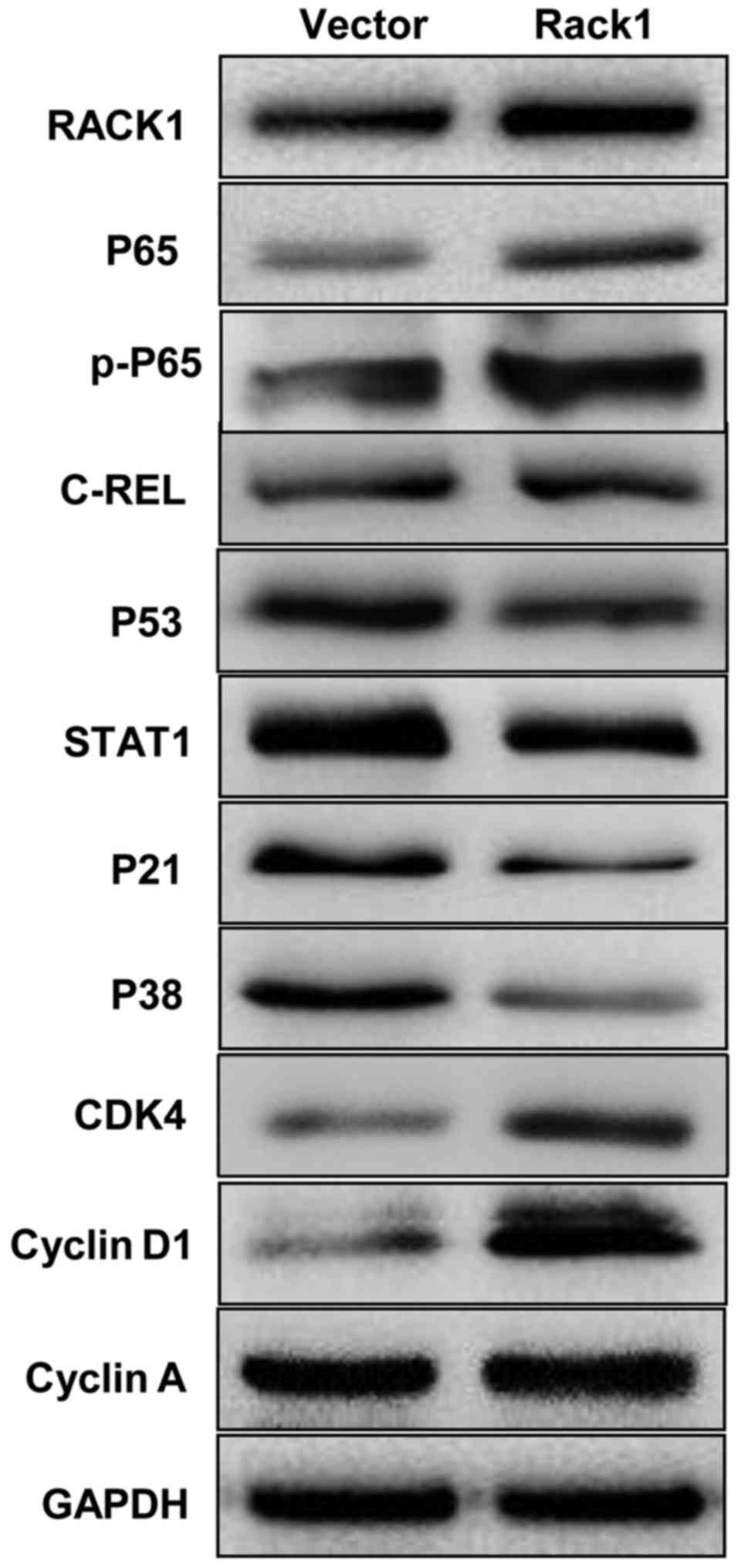 | Figure 5Western blot analysis indicates that
RACK1 overexpression affected the expression of p53, p38, p21,
STAT1, NF-κB, cyclin D1 and CDK4 of cervical cancer cell in
vitro. The protein expression levels of p53, p38, p21, STAT1,
NF-κB P65, cyclin D1 and CDK4 in RACK1-overexpressed stable
cervical cancer cell line of CaSki were analysed by western
blotting. Vector, CaSki cells transfected with the pEGFP-N1
plasmid, Rack1, CaSki cells transfected with the pEGFP-N1-RACK1
plasmid. |
Discussion
New cervical cancers (530,000) are diagnosed
annually and there are 275,000 deaths from the disease worldwide
(http://globocan.iarc.fr/factsheets/cancers/cervix.asp).
Persistent infection with high-risk types of human papillomavirus
(HPV) is known to be one of the main causes of cervical cancer.
However, the exact etiology of cervical carcinoma remains poorly
understood. In our study, we found that RACK1 expresses highly in
cervical cancer tissues compared with the adjacent non-cancerous
tissues. RACK1 may play important roles in tumorigenesis and
progression in cervical cancer.
We first detected the expression level of RACK1 in
cervical cancer tissues and the adjacent non-cancerous tissues by
western blot analysis and immunohistochemistry (IHC). The results
showed that RACK1 is higher expressed in the cancer tissues than in
the adjacent non-cancerous tissues. Further studies showed that
endogenous overexpressed RACK1 promoted the proliferation, invasion
and migration, and decreased cell senescence of cervical cancer
cells in vitro. Previous studies demonstrated that RACK1 is
expressed aberrantly in cancer cells and has diverse effects in
different types of tumors (30–34).
It can interact with multiple signaling molecules, including Akt,
Bcl-2 (35), MCM7 (36), FGFR1 and PKM2 (37). Wang et al found that RACK1
antagonized TNF-α-induced cell death by promoting p38 activation
(38). Li et al found that
RACK1 was upregulated in proliferating pancreatic ductal
adenocarcinoma (PDAC) cells, and involved in regulating cell cycle
and apoptosis of PDAC cells by interact with cyclin D1, BCL-2 and
caspase-3 (15). In our study, we
found that RACK1 induced S phase accumulation in cell cycle
analysis and suppressed cell apoptosis in cervical cancer cells.
Besides, RACK1 increased the mitochondrial membrane potential (Δψm)
levels to prevent mitochondrial apoptosis in cervical cancer cells.
As known, dysregulation of the cell cycle underlies the aberrant
cell proliferation that characterizes cancer and loss of cell cycle
checkpoint control promotes genetic instability (39–43).
To further explore the possible mechanism of RACK1 in cervical
cancer, we detected the expression levels of some key molecules in
p53 signaling pathway by western blot analysis in vitro and
found that RACK1 upregulated the expression of NF-κB, cyclin D1 and
CDK4 and downregulated the expression of p53, p38, p21 and STAT1.
It is well known that the P53-P21-CDK-cyclin D signaling pathway
plays a pivotal role in cell cycle regulation. P53 positively
regulates the expression of P21, which can bind with cyclin D1 or
cyclin A, and inhibit the activity of cyclin-CDK complex regulating
the cell cycle (44–47). STAT1 has been reported to have
tumor suppressor function and regulate cell apoptosis and cell
cycle (48,49). It is a negative regulator of MDM2
and can also act as a coactivator interacting directly with P53
(50). Besides, substantial
evidence has demonstrated that NF-κB P65 plays compelling role in
cell apoptosis (51–53). Combining with our results, it is
confirmed that RACK1 promote cell growth and inhibit cell apoptosis
in cervical cancer. We further speculated that possibly RACK1 acts
on its effect in cervical cancer by affecting the p53 signaling
pathway (Fig. 6). This conjecture
requires further experiments to show the regulatory mechanism among
these molecules in cell senescence and cell migration and invasion
of cervical cancer cells.
Abbreviations:
|
RACK1
|
receptor for activated C kinase 1
|
|
TP53
|
tumor protein p53
|
|
STAT1
|
signal transducer and activator of
transcription 1
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
IHC
|
immunohistochemistry
|
|
GADPH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
Acknowledgments
This study was supported by the National Natural
Sciences Foundation of China (81672685, 81402270, 81272975 and
81672993); Key Project of Hunan Provincial Natural Science
Foundation (12JJ2044); the Key Planned Science and Technology
Project of Hunan Province (2012FJ2014 and 2011FJ3153); the 111
Project (111-2-12); the Natural Science Foundation of Hunan
Province (2016JC2035); the Planned Project of Development and
Reform Commission of Hunan Province (2012-1493-1); the Planned
Project of Department of health of Hunan Province (B2011-030,
B2012-029); the Planned Project of Key Subject Construction of the
Third Xiangya Hospital, Central South University; the Open-End Fund
for the Valuable and Precision Instruments of Central South
University. This study was also supported by Hunan Provincial
Innovation Foundation for Postgraduate (CX2015B057).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yugawa T and Kiyono T: Molecular
mechanisms of cervical carcinogenesis by high-risk human
papillomaviruses: Novel functions of E6 and E7 oncoproteins. Rev
Med Virol. 19:97–113. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Latsuzbaia A, Tapp J, Nguyen T, Fischer M,
Arbyn M, Weyers S and Mossong J: Analytical performance evaluation
of Anyplex II HPV28 and Euroarray HPV for genotyping of cervical
samples. Diagn Microbiol Infect Dis. 85:318–322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang A, Dadaglio G, Oberkampf M, Di Carlo
S, Peduto L, Laubreton D, Desrues B, Sun CM, Montagutelli X and
Leclerc C: B cells promote tumor progression in a mouse model of
HPV-mediated cervical cancer. Int J Cancer. 139:1358–1371. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chatzistamatiou K, Moysiadis T, Moschaki
V, Panteleris N and Agorastos T: Comparison of cytology, HPV DNA
testing and HPV 16/18 genotyping alone or combined targeting to the
more balanced methodology for cervical cancer screening. Gynecol
Oncol. 142:120–127. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Scotto L, Narayan G, Nandula SV,
Subramaniyam S, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M,
Schneider A, Arias-Pulido H, et al: Integrative genomics analysis
of chromosome 5p gain in cervical cancer reveals target
over-expressed genes, including Drosha. Mol Cancer. 7:582008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dowen SE, Neutze DM, Pett MR, Cottage A,
Stern P, Coleman N and Stanley MA: Amplification of chromosome 5p
correlates with increased expression of Skp2 in HPV-immortalized
keratinocytes. Oncogene. 22:2531–2540. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ron D and Mochly-Rosen D: Agonists and
antagonists of protein kinase C function, derived from its binding
proteins. J Biol Chem. 269:21395–21398. 1994.PubMed/NCBI
|
|
9
|
McCahill A, Warwicker J, Bolger GB,
Houslay MD and Yarwood SJ: The RACK1 scaffold protein: A dynamic
cog in cell response mechanisms. Mol Pharmacol. 62:1261–1273. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng R, Jiang B, Ma J, Ma Z, Wan X, Liu H,
Chen Z, Cheng Q and Chen R: Forced downregulation of RACK1 inhibits
glioma development by suppressing Src/Akt signaling activity. Oncol
Rep. 30:2195–2202. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu J, Meng J, Du Y, Huang Y, Jin Y, Zhang
J, Wang B, Zhang Y, Sun M and Tang J: RACK1 promotes the
proliferation, migration and invasion capacity of mouse
hepatocellular carcinoma cell line in vitro probably by PI3K/Rac1
signaling pathway. Biomed Pharmacother. 67:313–319. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi S, Deng YZ, Zhao JS, Ji XD, Shi J,
Feng YX, Li G, Li JJ, Zhu D, Koeffler HP, et al: RACK1 promotes
non-small-cell lung cancer tumorigenicity through activating sonic
hedgehog signaling pathway. J Biol Chem. 287:7845–7858. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yong-Zheng X, Wan-Li M, Ji-Ming M and
Xue-Qun R: Receptor for activated protein kinase C 1 suppresses
gastric tumor progression through nuclear factor-κB pathway. Indian
J Cancer. 52(Suppl 3): E172–E175. 2015. View Article : Google Scholar
|
|
14
|
Chen L, Min L, Wang X, Zhao J, Chen H, Qin
J, Chen W, Shen Z, Tang Z, Gan Q, et al: Loss of RACK1 promotes
metastasis of gastric cancer by inducing a miR-302c/IL8 signaling
loop. Cancer Res. 75:3832–3841. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Xiao Y, Fan S, Xiao M, Wang X, Chen
X, Li C, Zong G, Zhou G and Wan C: RACK1 overexpression associates
with pancreatic ductal adenocarcinoma growth and poor prognosis.
Exp Mol Pathol. 101:176–186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng H, Gong PG, Li JB, Cai LM, Yang L,
Liu YY, Yao KT and Li X: The important role of the receptor for
activated C kinase 1 (RACK1) in nasopharyngeal carcinoma
progression. J Transl Med. 14:1312016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu F, Tao Z, Wang M, Li G, Zhang Y, Zhong
H, Xiao H, Xie X and Ju M: RACK1 promoted the growth and migration
of the cancer cells in the progression of esophageal squamous cell
carcinoma. Tumour Biol. 34:3893–3899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu F, Zhang C, Wu WJ and Wu YM: RACK1
downregulation suppresses migration and proliferation of
neuroblastoma cell lines. Oncol Rep. 27:1646–1652. 2012.PubMed/NCBI
|
|
19
|
Ruan Y, Sun L, Hao Y, Wang L, Xu J, Zhang
W, Xie J, Guo L, Zhou L, Yun X, et al: Ribosomal RACK1 promotes
chemoresistance and growth in human hepatocellular carcinoma. J
Clin Invest. 122:2554–2566. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao X, Xue A, Fang Y, Shu P, Ling J, Hou
Y, Shen K, Qin J, Sun Y and Qin X: RACK1 overexpression is linked
to acquired imatinib resistance in gastrointestinal stromal tumor.
Oncotarget. 7:14300–14309. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Z, Zhang B, Jiang L, Zeng X, Chen Y,
Feng X, Guo Y and Chen Q: RACK1, an excellent predictor for poor
clinical outcome in oral squamous carcinoma, similar to Ki67. Eur J
Cancer. 45:490–496. 2009. View Article : Google Scholar
|
|
22
|
Li JJ and Xie D: RACK1, a versatile hub in
cancer. Oncogene. 34:1890–1898. 2015. View Article : Google Scholar
|
|
23
|
Zhong X, Li M, Nie B, Wu F, Zhang L, Wang
E and Han Y: Overexpressions of RACK1 and CD147 associated with
poor prognosis in stage T1 pulmonary adenocarcinoma. Ann Surg
Oncol. 20:1044–1052. 2013. View Article : Google Scholar
|
|
24
|
Nagashio R, Sato Y, Matsumoto T, Kageyama
T, Satoh Y, Shinichiro R, Masuda N, Goshima N, Jiang SX and Okayasu
I: Expression of RACK1 is a novel biomarker in pulmonary
adenocarcinomas. Lung Cancer. 69:54–59. 2010. View Article : Google Scholar
|
|
25
|
Guo Y, Wang W, Wang J, Feng J, Wang Q, Jin
J, Lv M, Li X, Li Y, Ma Y, et al: Receptor for activated C kinase 1
promotes hepatocellular carcinoma growth by enhancing
mitogen-activated protein kinase kinase 7 activity. Hepatology.
57:140–151. 2013. View Article : Google Scholar
|
|
26
|
Deng YZ, Yao F, Li JJ, Mao ZF, Hu PT, Long
LY, Li G, Ji XD, Shi S, Guan DX, et al: RACK1 suppresses gastric
tumorigenesis by stabilizing the β-catenin destruction complex.
Gastroenterology. 142:812–823.e15. 2012. View Article : Google Scholar
|
|
27
|
Mamidipudi V, Dhillon NK, Parman T, Miller
LD, Lee KC and Cartwright CA: RACK1 inhibits colonic cell growth by
regulating Src activity at cell cycle checkpoints. Oncogene.
26:2914–2924. 2007. View Article : Google Scholar
|
|
28
|
Liao S, Xiao S, Zhu G, Zheng D, He J, Pei
Z, Li G and Zhou Y: CD38 is highly expressed and affects the
PI3K/Akt signaling pathway in cervical cancer. Oncol Rep.
32:2703–2709. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hara A and Okayasu I: Cyclooxygenase-2 and
inducible nitric oxide synthase expression in human astrocytic
gliomas: Correlation with angiogenesis and prognostic significance.
Acta Neuropathol. 108:43–48. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin Y, Cui M, Teng H, Wang F, Yu W and Xu
T: Silencing the receptor of activated C-kinase 1 (RACK1)
suppresses tumorigenicity in epithelial ovarian cancer in vitro and
in vivo. Int J Oncol. 44:1252–1258. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shen F, Yan C, Liu M, Feng Y and Chen Y:
RACK1 promotes prostate cancer cell proliferation, invasion and
metastasis. Mol Med Rep. 8:999–1004. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li J, Guo Y, Feng X, Wang Z, Wang Y, Deng
P, Zhang D, Wang R, Xie L, Xu X, et al: Receptor for activated C
kinase 1 (RACK1): A regulator for migration and invasion in oral
squamous cell carcinoma cells. J Cancer Res Clin Oncol.
138:563–571. 2012. View Article : Google Scholar
|
|
33
|
Dave JM, Kang H, Abbey CA, Maxwell SA and
Bayless KJ: Proteomic profiling of endothelial invasion revealed
receptor for activated C kinase 1 (RACK1) complexed with vimentin
to regulate focal adhesion kinase (FAK). J Biol Chem.
288:30720–30733. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gandin V, Senft D, Topisirovic I and Ronai
ZA: RACK1 function in cell motility and protein synthesis. Genes
Cancer. 4:369–377. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu B, Wang C, Chen P, Wang L and Cheng Y:
RACK1 promotes radiation resistance in esophageal cancer via
regulating AKT pathway and Bcl-2 expression. Biochem Biophys Res
Commun. 491:622–628. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fei L, Ma Y, Zhang M, Liu X, Luo Y, Wang
C, Zhang H, Zhang W and Han Y: RACK1 promotes lung cancer cell
growth via an MCM7/RACK1/Akt signaling complex. Oncotarget.
8:40501–40513. 2017.PubMed/NCBI
|
|
37
|
Zhou C, Chen T, Xie Z, Qin Y, Ou Y, Zhang
J, Li S, Chen R and Zhong N: RACK1 forms a complex with FGFR1 and
PKM2 and stimulates the growth and migration of squamous lung
cancer cells. Mol Carcinog. Apr 18–2017.Epub ahead of print.
View Article : Google Scholar
|
|
38
|
Wang Q, Zhou S, Wang JY, Cao J, Zhang X,
Wang J, Han K, Cheng Q, Qiu G, Zhao Y, et al: RACK1 antagonizes
TNF-α-induced cell death by promoting p38 activation. Sci Rep.
5:142982015. View Article : Google Scholar
|
|
39
|
Diaz-Moralli S, Tarrado-Castellarnau M,
Miranda A and Cascante M: Targeting cell cycle regulation in cancer
therapy. Pharmacol Ther. 138:255–271. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Williams GH and Stoeber K: The cell cycle
and cancer. J Pathol. 226:352–364. 2012. View Article : Google Scholar
|
|
41
|
Aarts M, Linardopoulos S and Turner NC:
Tumour selective targeting of cell cycle kinases for cancer
treatment. Curr Opin Pharmacol. 13:529–535. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vermeulen K, Van Bockstaele DR and
Berneman ZN: The cell cycle: A review of regulation, deregulation
and therapeutic targets in cancer. Cell Prolif. 36:131–149. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Schafer KA: The cell cycle: A review. Vet
Pathol. 35:461–478. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Waga S, Li R and Stillman B: p53-induced
p21 controls DNA replication. Leukemia. 11(Suppl 3): 321–323.
1997.PubMed/NCBI
|
|
45
|
Harper JW, Elledge SJ, Keyomarsi K,
Dynlacht B, Tsai LH, Zhang P, Dobrowolski S, Bai C, Connell-Crowley
L and Swindell E: Inhibition of cyclin-dependent kinases by p21.
Mol Biol Cell. 6:387–400. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Besson A, Dowdy SF and Roberts JM: CDK
inhibitors: Cell cycle regulators and beyond. Dev Cell. 14:159–169.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li D, Dai C, Yang X, Wang F, Yu X and Xiao
X: Tang S. Critical role of p21 on olaquindox-induced mitochondrial
apoptosis and S-phase arrest involves activation of PI3K/AKT and
inhibition of Nrf2/HO-1 pathway. Food Chem Toxicol. 108:148–160.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kim HS and Lee MS: STAT1 as a key
modulator of cell death. Cell Signal. 19:454–465. 2007. View Article : Google Scholar
|
|
49
|
Zhang Y, Zhang Y, Yun H, Lai R and Su M:
Correlation of STAT1 with apoptosis and cell-cycle markers in
esophageal squamous cell carcinoma. PLoS One. 9:e1139282014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Townsend PA, Scarabelli TM, Davidson SM,
Knight RA, Latchman DS and Stephanou A: STAT-1 interacts with p53
to enhance DNA damage-induced apoptosis. J Biol Chem.
279:5811–5820. 2004. View Article : Google Scholar
|
|
51
|
Aggarwal BB: Tumour necrosis factors
receptor associated signalling molecules and their role in
activation of apoptosis, JNK and NF-kappaB. Ann Rheum Dis. 59(Suppl
1): i6–i16. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kerr JF, Wyllie AH and Currie AR:
Apoptosis: A basic biological phenomenon with wide-ranging
implications in tissue kinetics. Br J Cancer. 26:239–257. 1972.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Perkins ND: The diverse and complex roles
of NF-κB subunits in cancer. Nat Rev Cancer. 12:121–132.
2012.PubMed/NCBI
|