Introduction
Hepatocellular carcinoma (HCC) is known as the most
common liver malignancy worldwide, with extremely high incidence
and mortality rate (1–5). In China, HCC frequently occurs owing
to chronic infection of hepatitis B virus (HBV) (6–9).
Other conditions, such as alcoholic hepatitis, non-alcoholic fatty
liver disease, hemochromatosis and diabetes, also contribute to the
development of HCC (10–12). Up to now, liver transplantation and
tumor resection still have been the most effective treatments for
HCC, whereas the high metastasis and postoperative recurrence rates
barricade the prognosis of HCC patients, especially HCC patients in
advanced stage (13,14). As the current treatment options are
limited, it is of great significance to investigate the underlying
mechanism of HCC, which might provide novel insights into the
diagnosis and treatment of HCC patients.
Long non-coding RNAs (lncRNAs) represent the
non-protein coding RNAs with the length from 200 nucleotides to 100
kb (15–17). Recent evidence has clarified that
various lncRNAs could act as key regulators in disease development,
epigenetic gene regulation or transcriptional regulation (18–23).
Importantly, lncRNAs can regulate the proliferation, invasion,
metastasis and apoptosis of malignancies (24–29).
In HCC, growing evidence has demonstrated that the differential
expression of lncRNAs could affect the development and progression
of HCC by regulating the self-renewal ability of liver cancer stem
cells and other biological functions (30–34).
Moreover, it has been reported that the lncRNA expression might be
also associated with chemotherapy resistance of HCC patients
(35–37).
lncRNA HOTTIP, also identified as HOXA-AS6,
HOXA13-AS1 and NCRNA00213, is located on 7p15.2, with the NCBI Gene
ID: 100316868 (38). Moreover,
HOTTIP functions as an independent biomarker in multiple cancers,
such as breast cancer and esophageal squamous cell carcinoma
(39–42). The high HOTTIP expression could
inhibit the growth of glioma, increase the chemoresistance of
osteosarcoma and promote tumor invasion of gastric cancer (33,43,44).
Although it has been shown that HOTTIP was upregulated in HCC
compared with normal tissues and could be related to disease
progression and predict the outcome of HCC; nonetheless, the
detailed functions and mechanism of HOTTIP in HCC remain elusive
(45).
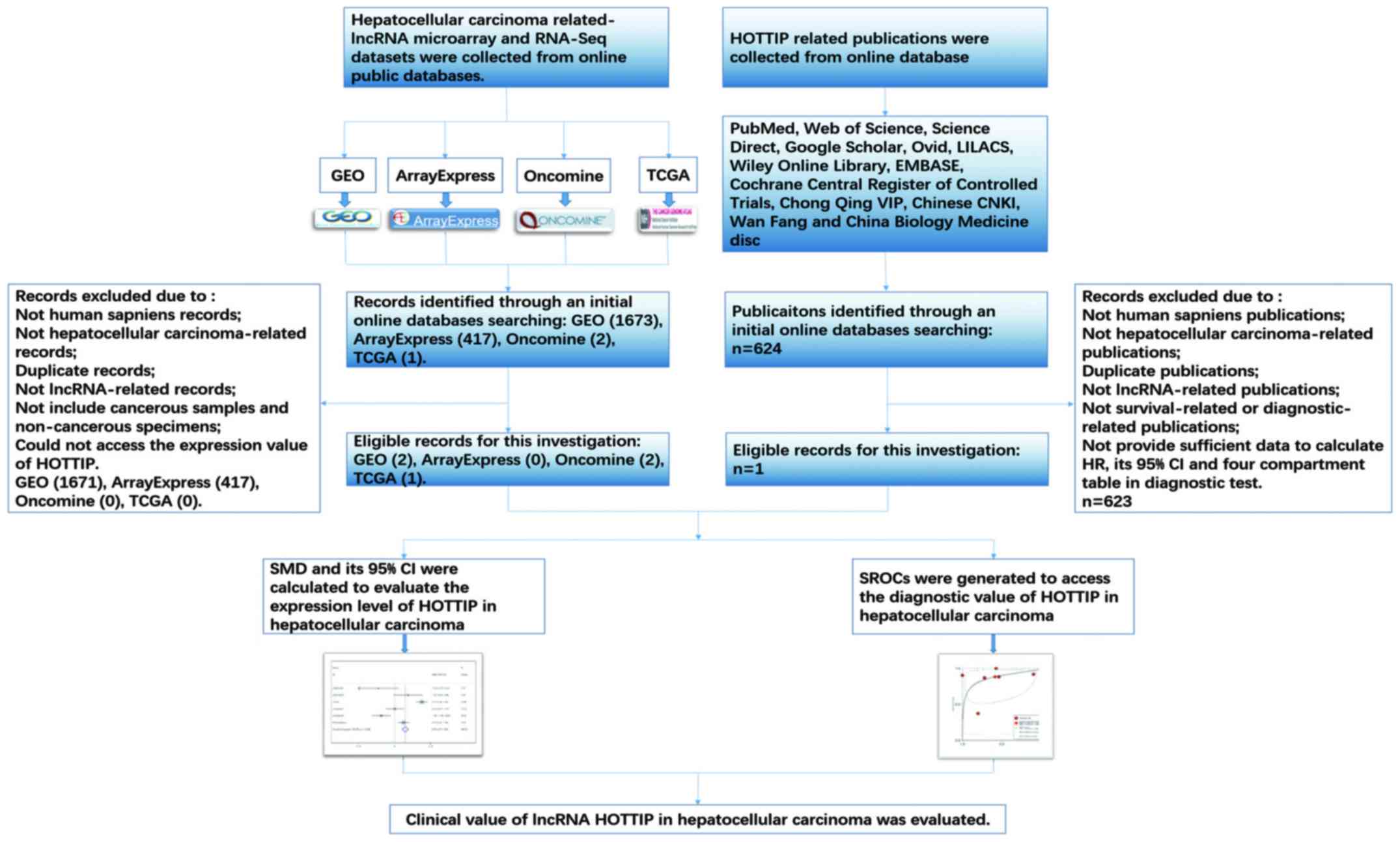
In the present study, we investigated the expression
of HOTTIP in HCC and normal liver. Furthermore, we combined Gene
Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA), Multi
Experiment Matrix (MEM) and Oncomine database, quantitative reverse
transcription-polymerase chain reactions (qRT-PCR), and
meta-analysis to assess the clinical role and the potential
molecular mechanism of HOTTIP in HCC. Additionally, bioinformatics
analysis, which contain Gene Ontology (GO), Kyoto Encyclopedia of
Genes and Genomes (KEGG) and network analysis, were applied to
explore the underlying functions, pathways and networks of the
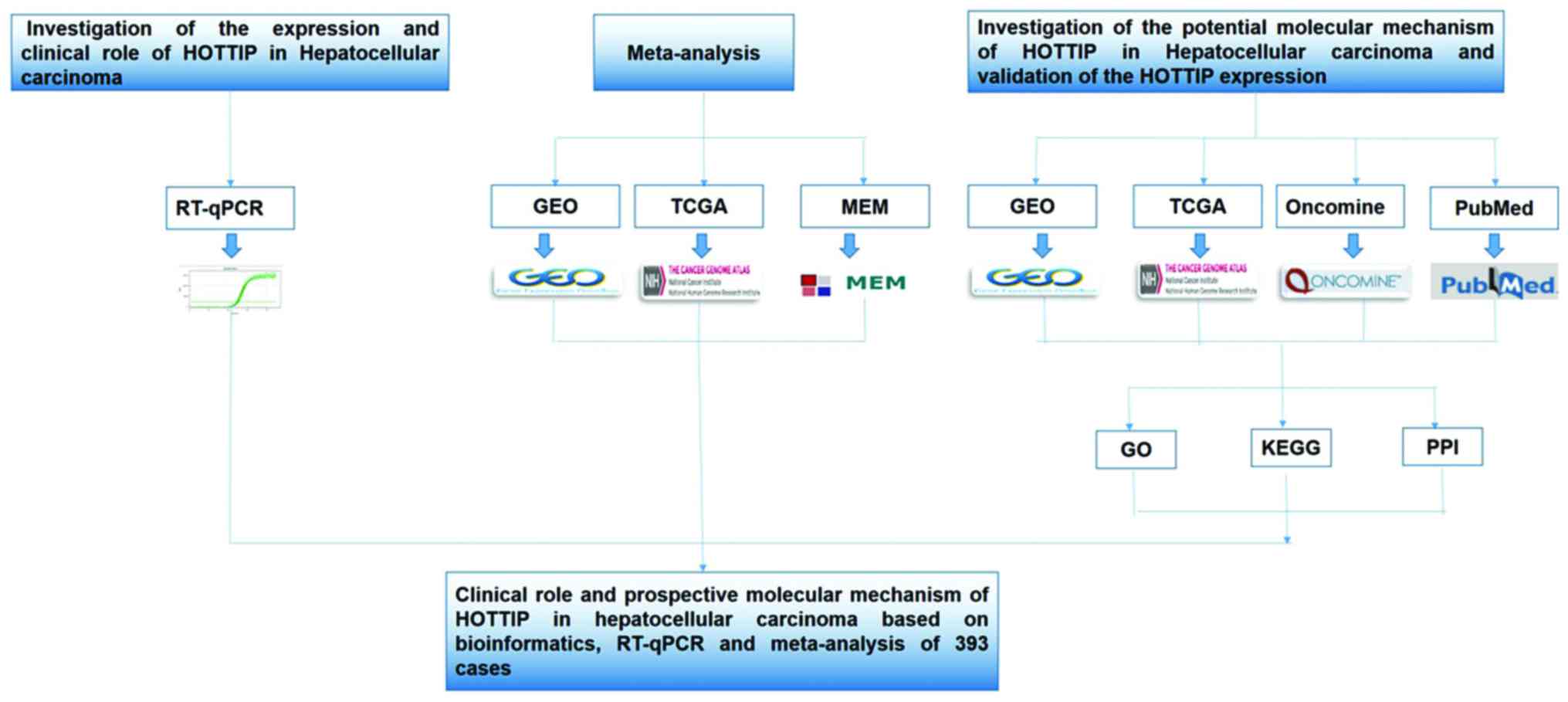
potential genes (46–48). A flow chart of this study is shown
in Fig. 1.
Materials and methods
Quantitative real-time PCR
A total of 41 HCC cases, between January 2012 and
March 2013, were collected from the Department of Pathology, First
Affiliated Hospital of the Guangxi Medical University (Nanning,
Guangxi, China). The samples were collected randomly from patients
undergoing surgical resection without treatment. All methods were
performed based on the relevant regulations and guidelines. Also,
all experimental protocols have been approved by the Ethical
Committee of the First Affiliated Hospital of Guangxi Medical
University, and the clinicians and patients have signed the consent
forms for the use of their tissues in the study. In this study, the
total RNA was extracted via a Takara PrimeScript RT reagent kit
based on the manufacturer's instructions. Then, the total RNA was
used for cDNA synthesis through the Takara PrimeScript RT reagent
kit according to the instructions. Then, qRT-PCR was operated using
a LightCycler 480 Real-time PCR system (Roche, Shanghai, China).
The specific primers were employed as follows: HOTTIP forward,
5′-CACACTCACATTCGCACACT-3′; reverse, 5′-TCCAGAACTAAGCCAGCCATA-3′.
GAPDH (internal control) forward, 5′-AGTGGCAA AGTGGAGATT-3′;
reverse, 5′-GTGGAGTCATACTGGAACA-3′ (49). Results were normalized to the GAPDH
expression and calculated based on the ∆Ct method (50,51).
HOTTIP and HCC: a meta-analysis
HCC-related HOTTIP microarray and RNA-seq datasets
were downloaded from TCGA, the National Center of Biotechnology
Information (NCBI) GEO (http://www.ncbi.nlm.nih.gov/geo/), ArrayExpress
(http://www.ebi.ac.uk/arrayexpress/)
and Oncomine (https://www.oncomine.org/resource/main.html). In
addition, publications associated with the diagnostic value of
HOTTIP in HCC were also selected from 12 online databases: PubMed,
Web of Science, Science Direct, Google Scholar, Ovid, LILACS, Wiley
Online Library, EMBASE, Cochrane Central Register of Controlled
Trials, Chong Qing VIP, Chinese CNKI, Wan Fang and China Biology
Medicine disc. The retrieval date was up to April 20, 2017 with the
following keywords: (HOTTIP or HOXA-AS6 or HOXA13-AS1 or
NCRNA00213) and (malignan* OR cancer OR tumor OR tumor OR neoplas*
OR carcinoma) and (hepatocellular OR liver OR hepatic OR HCC). The
literature retrieval was assessed and cross-checked by two
independent investigators (Jia-Cheng Huang and Wen-Ya Pan). Group
discussion was carried out if there was disagreement. The number of
true-positives (tp), true-negatives (tn), false-positives (fp) and
false-negatives (fn) was extracted. When no direct data was found
from a study, a basic formula (such as 'sensitivity' = tp/(tp+fn),
'specificity' = tn/(tn+fp)) would be used to calculate the
incidence.
Validation of the expression of HOTTIP in
HCC
TCGA (http://cancergenome.nih.gov/) is a collection of SNP
arrays, DNA methylation, miRNA-seq, exome sequencing, RNA-seq, and
more (52,53). TCGA can be also applied to
investigate the complicated cancer genomics expression and clinical
parameters. In this study, RNA-Seq data from individuals with HCC,
which were calculated on IlluminaHiSeq RNASeq platform, were
achieved from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/), containing 171
HCC tissues and 28 adjacent normal liver tissue samples up to April
1, 2017. The expression data of HOTTIP were displayed as reads per
million (RPM) and the HOTTIP expression level was normalized by
Deseq package of R language for further analysis. Student's t-test
(SPSS Inc., Chicago, IL, USA) was performed for the statistical
analysis of the differential expression of HOTTIP between HCC and
non-cancerous liver tissues. Also, the relationship between HOTTIP
and the clinicopathological parameters in HCC was identified based
on the original data from TCGA database. Then, the clinical
diagnostic value of HOTTIP was analyzed by a receiver operating
characteristic (ROC) curve. Furthermore, the original data in
Oncomine database was also applied to verify the HOTTIP expression
in HCC (54).
The potential functions and pathways
associated with HOTTIP
To further investigate the co-expressed genes
associated with HOTTIP, MEM (http://biit.cs.ut.ee/mem/index.cgi), an open-access
resource, was utilized to explore the co-expressed genes of HOTTIP
based on Affymetrix Gene Chip Human Genome U133 Plus 2.0 Array
platform (55). Differentially
expressed genes in TCGA were extracted via R language package DESeq
based on the following criteria: adjusted P<0.05 and the
absolute log2 fold change >1 (56,57).
Also, the genes differentially expressed in GEO (http://www.ncbi.nlm.nih.gov/geo/) database were
selected using the GEO2R online tool (58–61).
The overlapping genes were identified and compared using Venn
diagrams (available online: http://bioinformatics.psb.ugent.be/webtools/Venn/).
Then, bioinformatics analyses including GO, KEGG and network
analysis were applied to explore the potential functions, pathways
and networks of the overlapping genes as described (62,63).
In this process, Database for Annotation, Visualization and
Integrated Discovery (DAVID: available online: http://david.abcc.ncifcrf.gov/) was applied to
perform GO and KEGG analyses. In addition, three independent
categories [biological process (BP), cellular component (CC) and
molecular function (MF)] were derived from GO analysis. Besides, a
functional network was constructed via Cytoscape (version 2.8,
http://cytoscape.org).
Construction of protein-protein
interaction (PPI) network
The interaction pairs of these co-expressed genes
were explored with Search Tool for the Retrieval of Interacting
Genes (STRING; version 9.0, http://string-db.org) (64). The STRING database supplies a
global perspective for as many organisms as feasible. The known and
predicted interactions are integrated and scored. The interaction
pairs in PPI network were selected with the combined score
>0.4.
Statistical analysis
The high-throughput expression data were
log2-transformed. The mean ± standard deviation (mean ± SD) was
calculated through SPSS 22.0 (IBM, NY, USA) to estimate the HOTTIP
expression level in each dataset. The HOTTIP expression between
normal liver tissue and HCC was evaluated by Student's t-test.
Student's t-test was also used to evaluate the relationships
between HOTTIP expression and the clinicopathological parameters.
The comparison between subgroups was performed via one-way analysis
of variance (ANOVA). A Mann-Whitney U test or Kruskal-Wallis H test
was conducted for non-normally distributed variables. Mining for
co-expression genes across hundreds of datasets was carried out via
novel rank aggregation and visualization methods. A P-value of
<0.05 was identified to be statistically significant (two sides)
using SPSS 22.0.
In the diagnostic meta-analysis, all the statistical
analysis was performed by STATA 14.0 (STATA Corp., College, TX,
USA). Heterogeneity between the included studies was assessed by
Cochrane's Q test and I2 statistic, and I2
>50% represented significant heterogeneity. Publication bias was
evaluated by Deek's funnel plot asymmetry test. A P-value of
<0.05 was regarded significant publication bias. To investigate
the underlying diagnostic performance of HOTTIP in HCC, we
performed summary receiver operating characteristic (SROC) curves
and calculated area under the curve (AUC) with 95% CIs and the
corresponding sensitivity and specificity by using Meta-DISc
software (65). An AUC value of
0.5–0.7 was regarded as low diagnostic capability; an AUC of
0.7–0.9 represented a moderate diagnostic ability; an AUC of
>0.9 indicated a high diagnostic accuracy. We also applied STATA
14.0 (STATA Corp.) to conduct continuous variable meta-analysis.
Fixed model (Mantel-Haenszel method) was applied at first and
random model (Der Simonian and Laird method) was used when there
existed significant heterogeneity. Funnel plot was described to
test publication bias.
Results
The clinical value of HOTTIP expression
in HCC
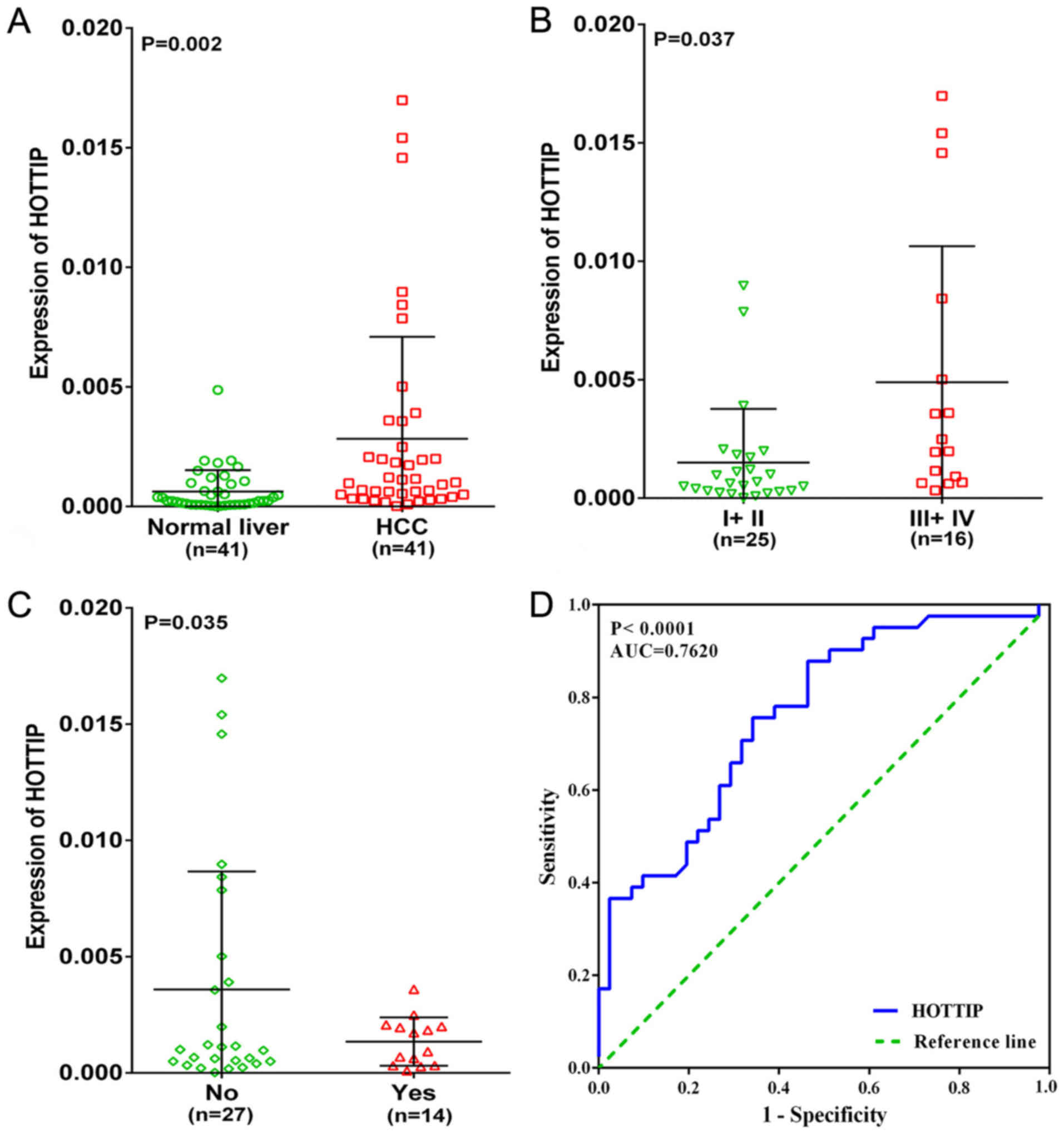
In the present study, an upregulated trend in HOTTIP
level in HCC tissues (4.67-fold) was found when compared to normal
liver tissues (P=0.002, Fig. 2A)
based on qRT-PCR. We also investigated the relationship between
different expression of HOTTIP and clinicopathological parameters.
As a result, HOTTIP expression was highly expressed in III +IV and
no cirrhosis groups (P<0.05, Table
I and Fig. 2B and C) in HCC,
but no statistical significance was found in other
clinicopathological parameters. In addition, the diagnostic value
of the HOTTIP level in HCC was assessed by a ROC curve and the AUC
of HOTTIP was 0.762 (95% CI, 0.660–0.864, P-value of <0.001,
Fig. 2D), indicating a moderate
diagnostic value of the HOTTIP level in HCC.
 | Table IDifferential expression of HOTTIP of
other clinicopathological parameters in HCC tissue based on
qRT-PCR. |
Table I
Differential expression of HOTTIP of
other clinicopathological parameters in HCC tissue based on
qRT-PCR.
| Clinicopathological
features | N | HOTTIP expression
|
|---|
| Fold change | T | P-value |
|---|
| Tissues | | | | |
| Normal liver | 41 | 1.00 | 3.226 | 0.002 |
| HCC | 41 | 4.67 | | |
| Age | | | | |
| <50 | 19 | 3.33 | 1.233 | 0.226 |
| ≥50 | 22 | 6.00 | | |
| Sex | | | | |
| Male | 31 | 5.17 | 0.775 | 0.443 |
| Female | 10 | 3.17 | | |
| Cirrhosis | | | | |
| No | 27 | 6.00 | −2.212 | 0.035 |
| Yes | 14 | 2.17 | | |
| TNM | | | | |
| I+ II | 25 | 2.50 | −2.255 | 0.037 |
| III +IV | 16 | 8.17 | | |
| Metastasis | | | | |
| No | 38 | 4.17 | −0.754 | 0.528 |
| Yes | 3 | 10.83 | | |
| Tumor diameter
(cm) | | | | |
| <7 | 11 | 2.67 | −1.126 | 0.267 |
| ≥7 | 30 | 5.50 | | |
| Vascular
invasion | | | | |
| No | 35 | 5.00 | 0.772 | 0.445 |
| Yes | 6 | 2.67 | | |
| Complete
capsule | | | | |
| No | 16 | 4.67 | 0.050 | 0.961 |
| Yes | 25 | 4.83 | | |
|
Differentiation | | | | |
| Low | 10 | 3.17 | F=2.194 | 0.125 |
| Moderate | 27 | 4.67 | | |
| High | 4 | 8.33 | | |
| AFP | | | | |
| Negative | 22 | 5.50 | −1.409 | 0.169 |
| Positive | 13 | 2.67 | | |
| Embolus | | | | |
| No | 39 | 4.83 | 0.791 | 0.433 |
| Yes | 2 | 0.83 | | |
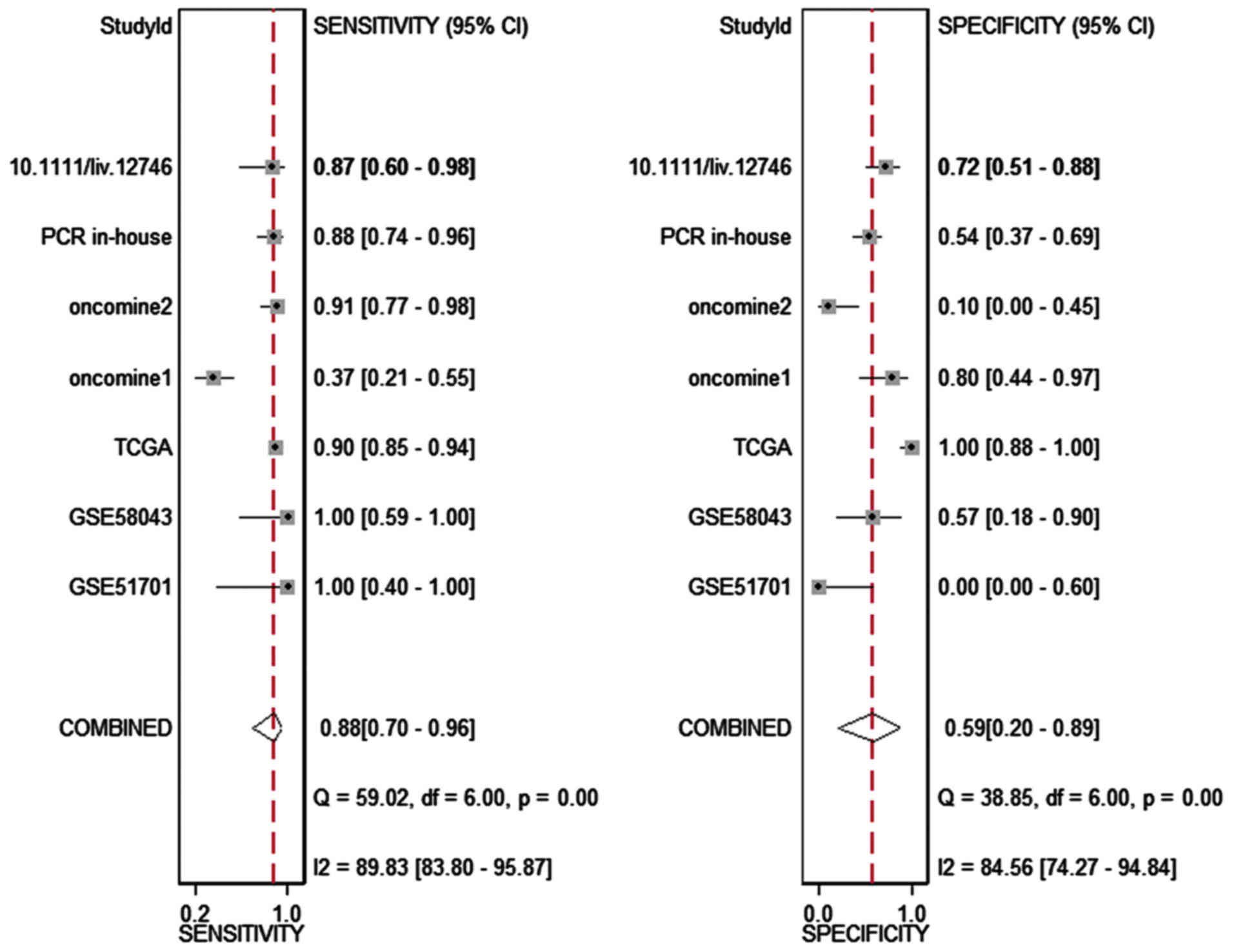
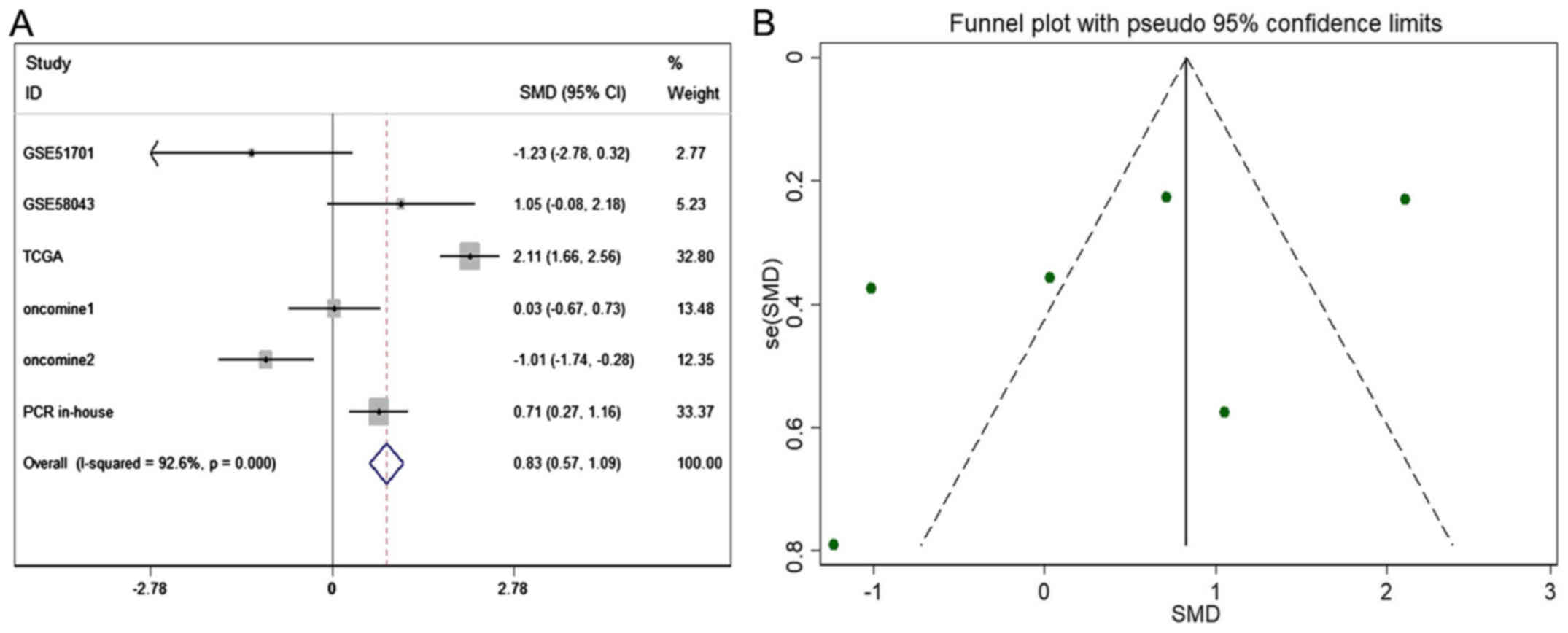
HOTTIP and HCC: a meta-analysis
The meta-analysis included 393 cases from multiple
centers [two datasets in GEO (GSE58043 and GSE51701), the original
data in TCGA, two probe sets (1564069_at and 1564070_s_at) of
Wurmbach Liver in Oncomine and one publication (45)]. Thus, the pooled sensitivity and
specificity of HOTTIP was 0.88 (0.70–0.96) and 0.59 (0.20–0.89,
Fig. 3), respectively. In
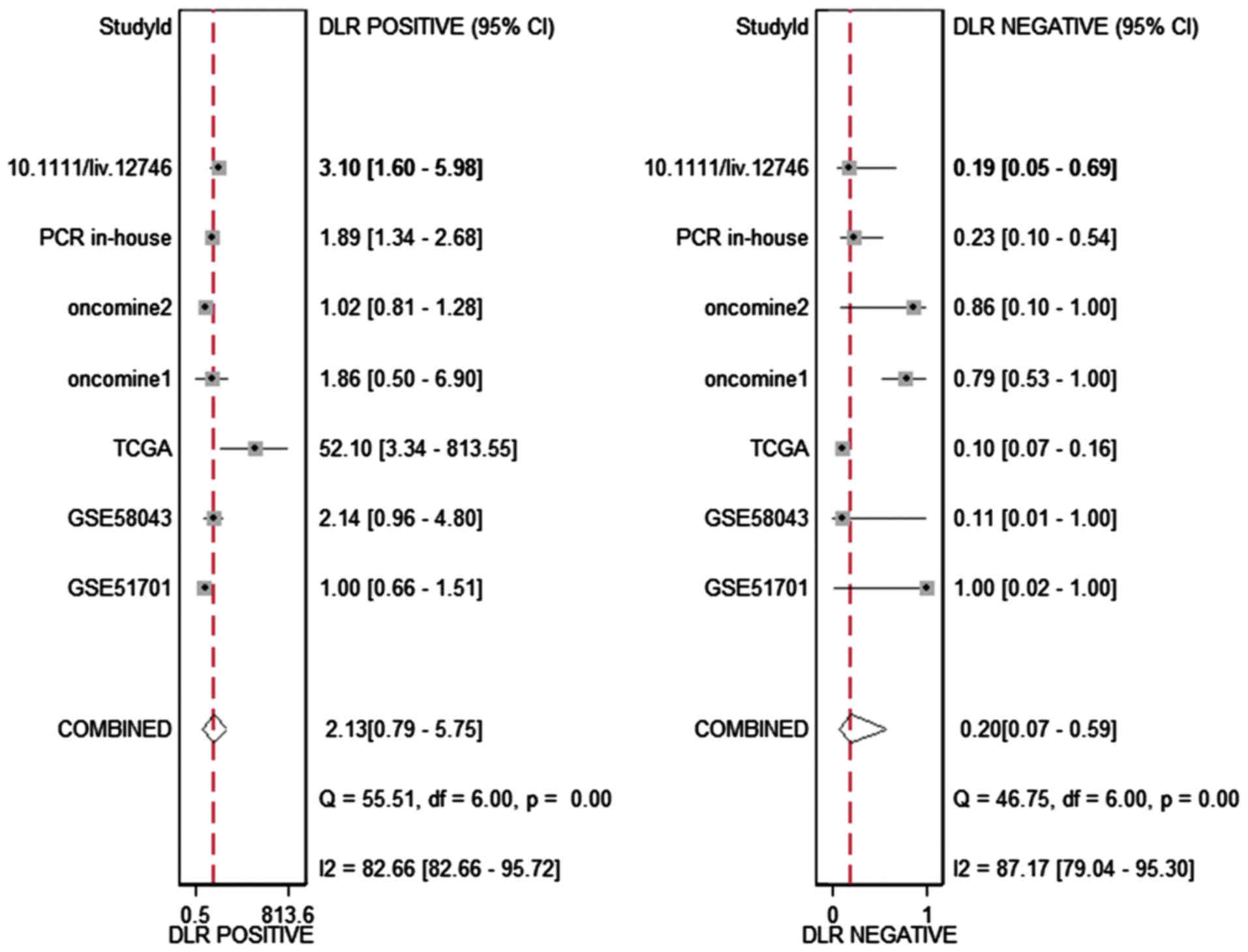
addition, the DLR-positive and DLR-negative were 2.13 (0.79–5.75)
and 0.20 (0.07–0.59, Fig. 4),
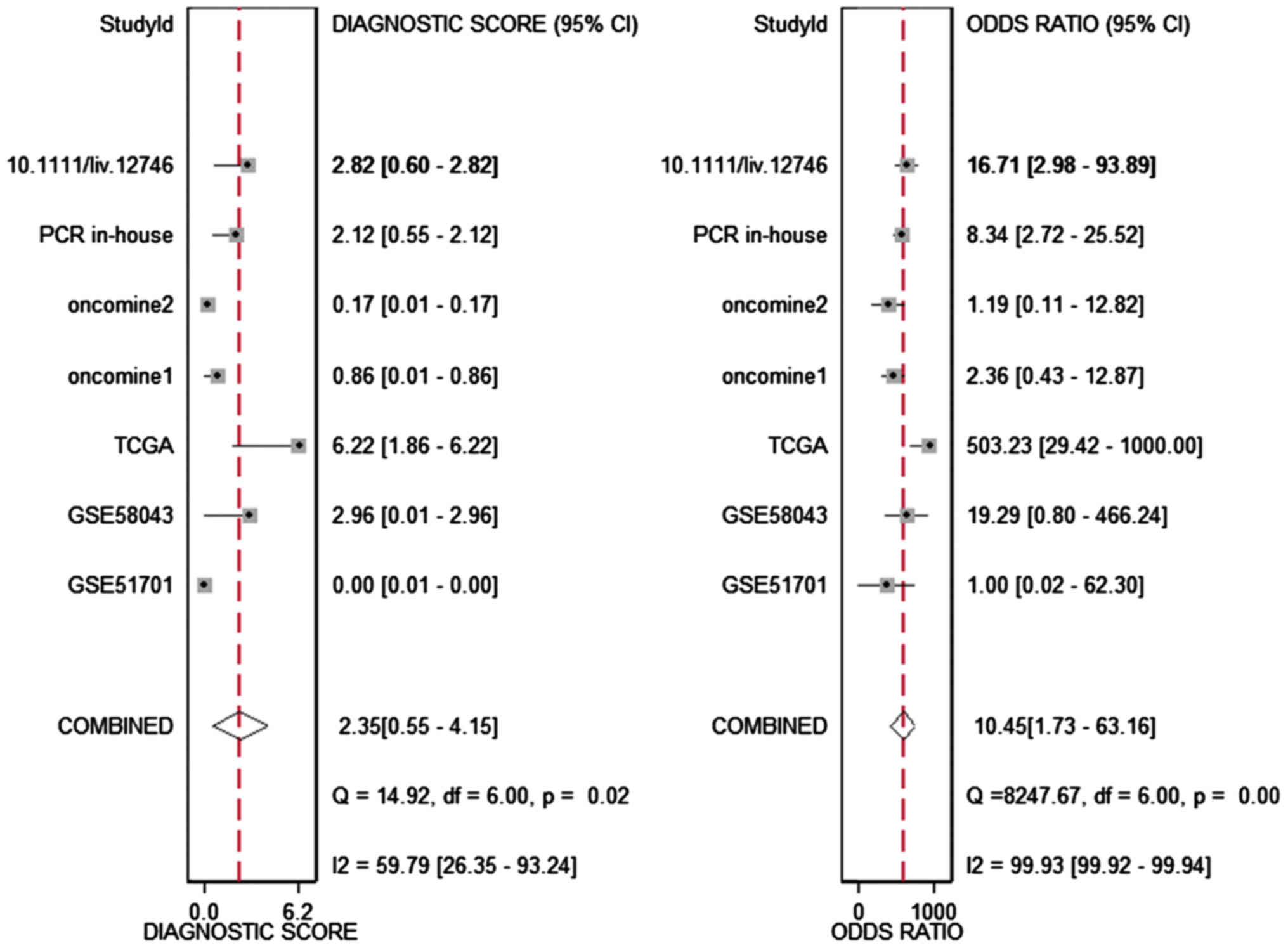
respectively. The diagnostic score and odds ratio were 2.35
(0.55–4.15) and 10.45 (1.73–63.16, Fig. 5), respectively. The AUC of SROC was
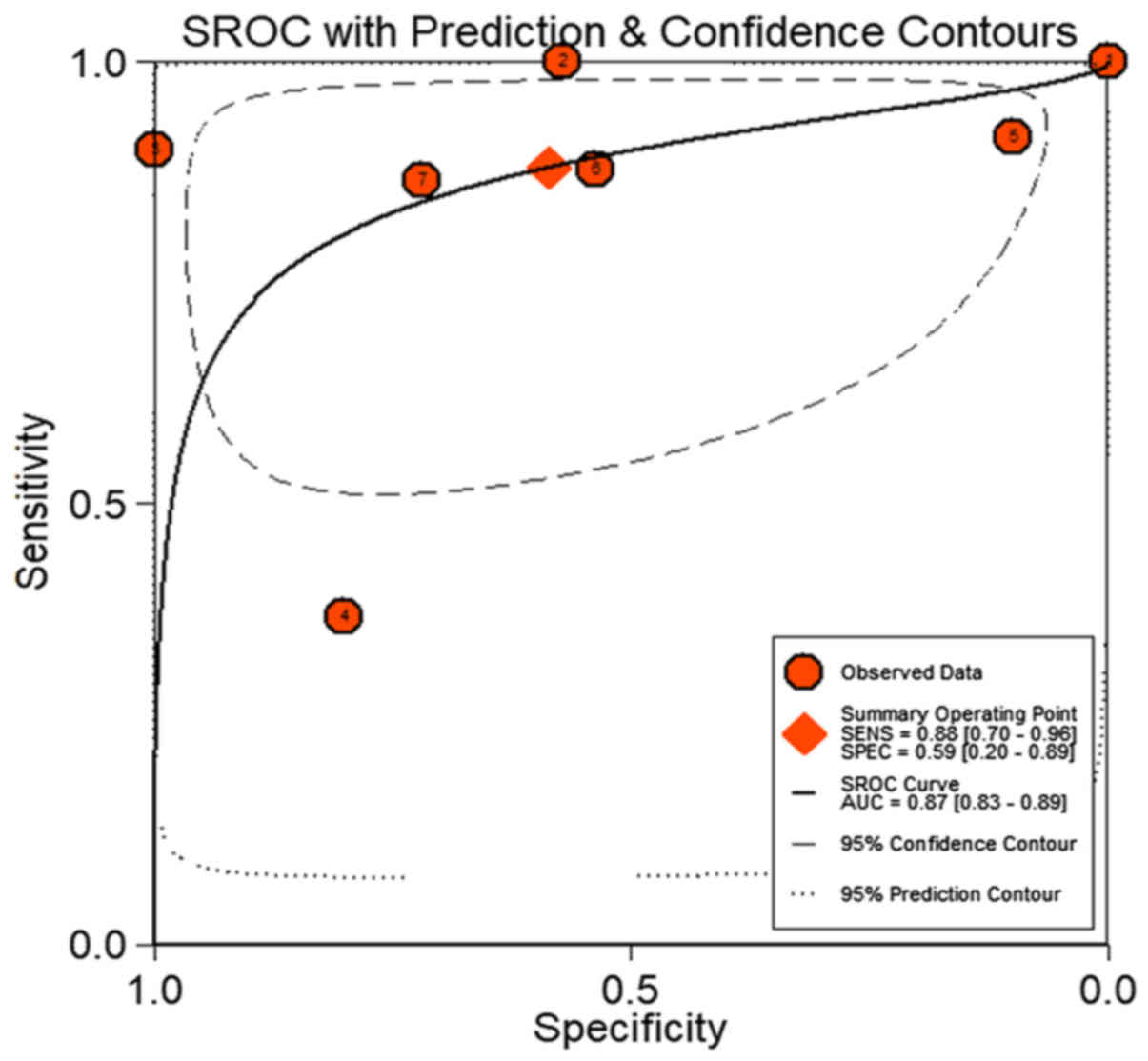
0.87 (0.83–0.89, Fig. 6), which
indicated a moderate diagnostic value of HOTTIP in HCC. The
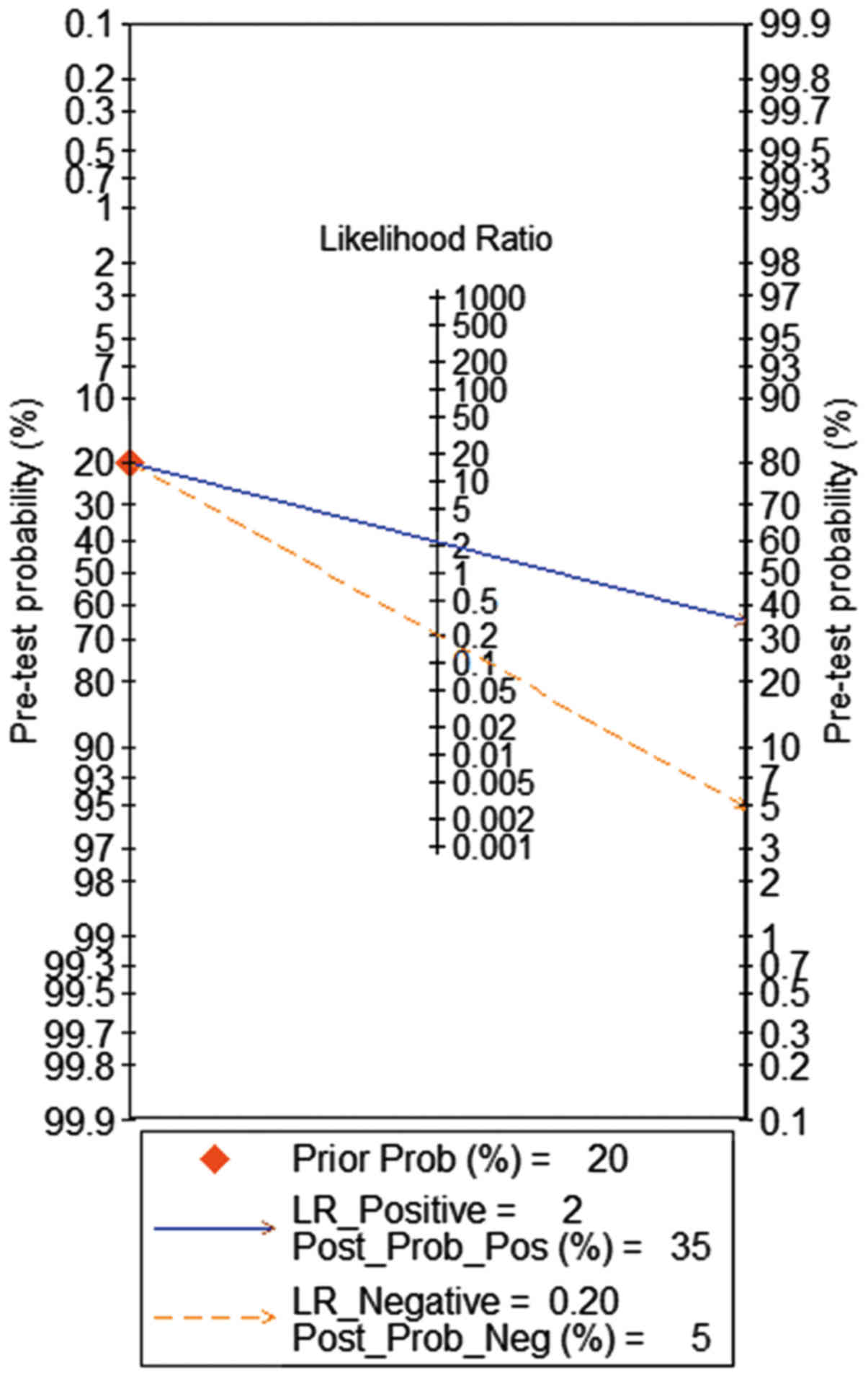
pre-test probability was 20% when the positive and negative
pre-test probability was 35 and 5% (Fig. 7), respectively. As for the
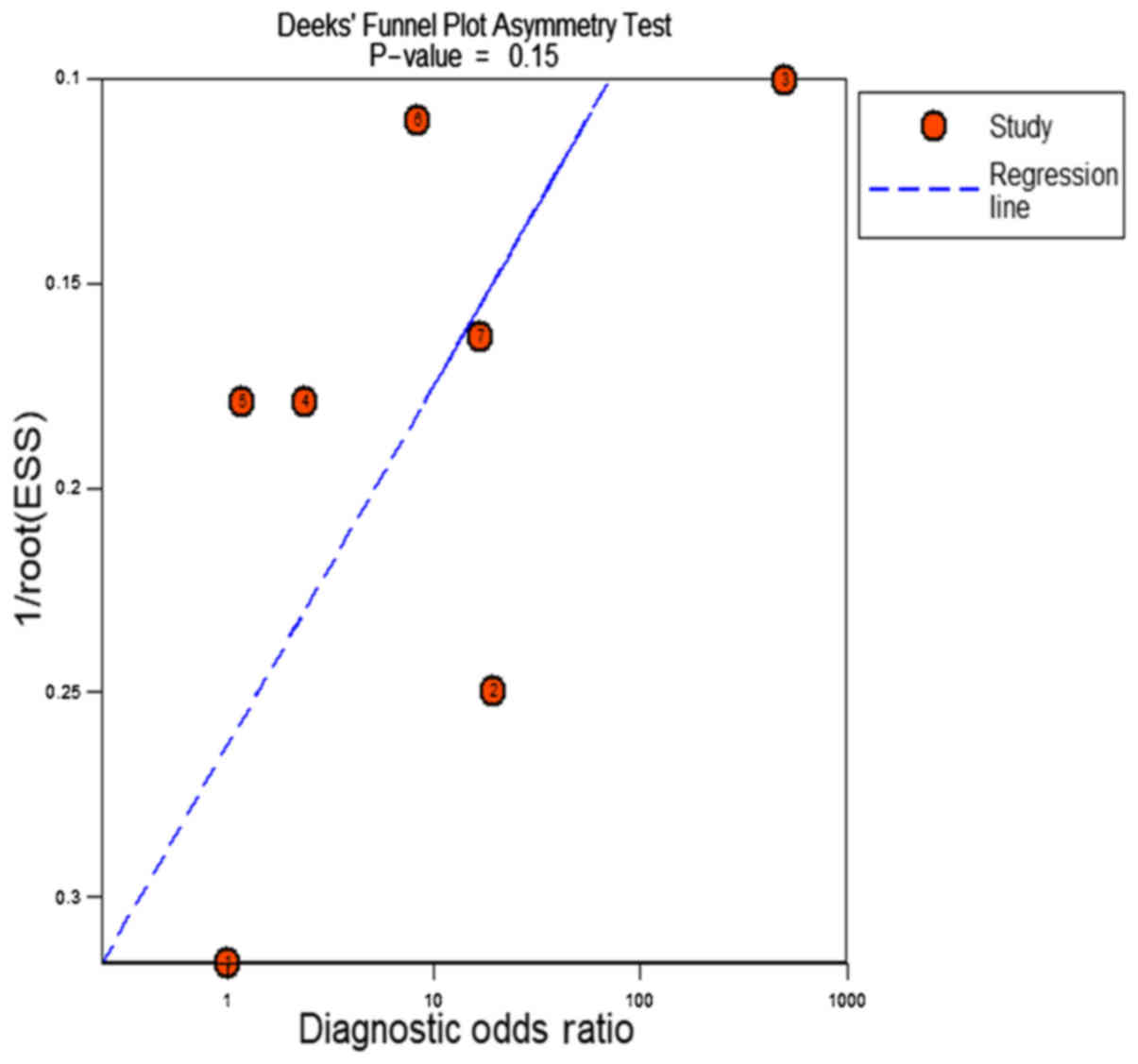
publication bias, no significant publication bias was found
(P=0.15, Fig. 8).
As for the expression of HOTTIP in HCC compared to
non-cancerous group, a fix-effect model was selected to calculate
the pooled standard mean deviation (SMD) and 95% CI and the
combined SMD reached 0.83 (0.57, 1.09), indicating a statistically
significant higher expression of HOTTIP could be found in HCC than
in normal control groups (P<0.001, Fig. 9A). In addition, no report bias was
found in our study (P>0.05, Fig.
9B). Above all, a flow chart of this meta-analysis is shown in
Fig. 10.
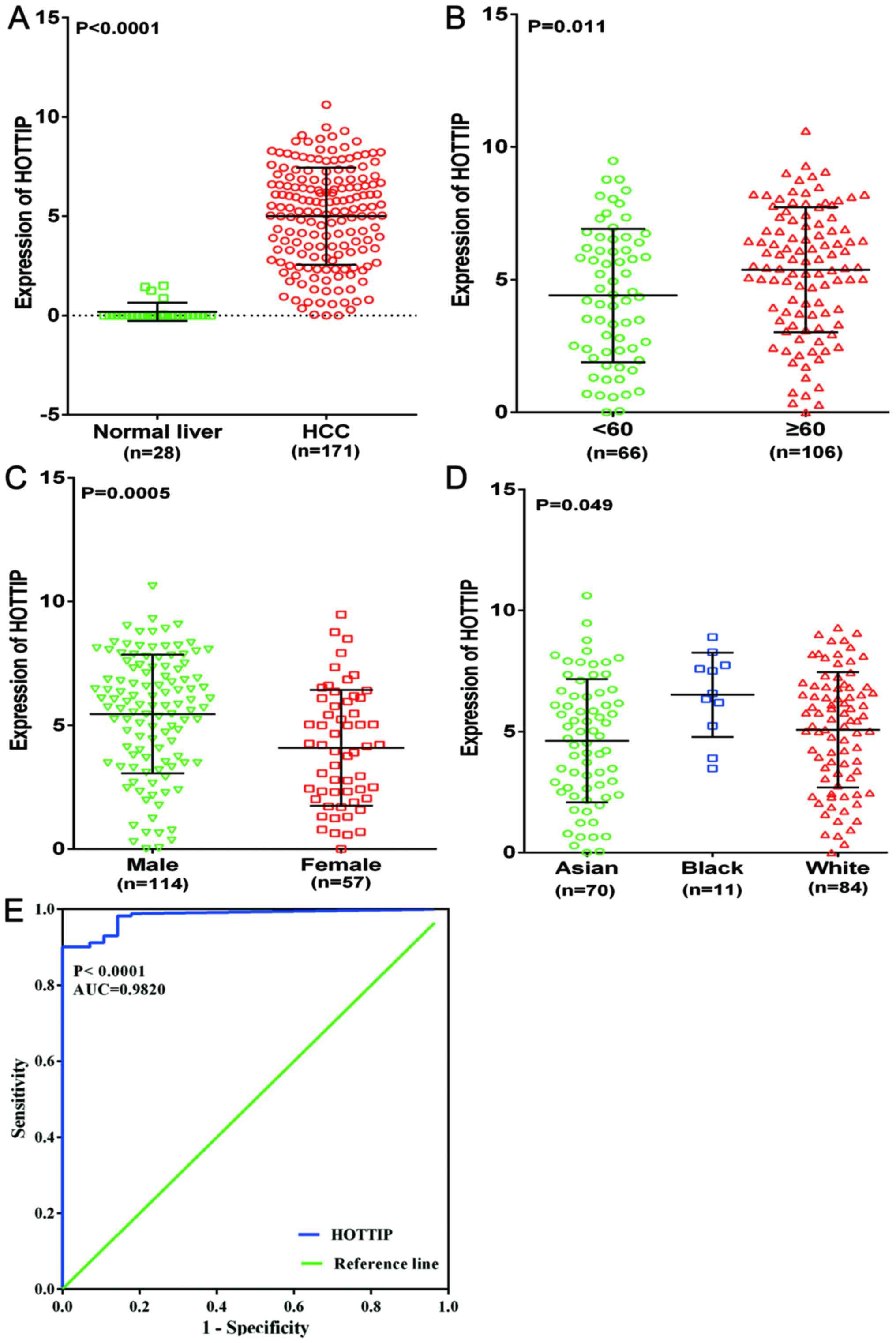
Validation of the expression of HOTTIP in
HCC
To further explore the differential expression of
HOTTIP between HCC and non-cancerous liver tissues, we performed a
clinical research based on the original data in TCGA. One HCC
cohort, which was comprised of 171 HCC cases and 28 non-cancerous
liver cases, was extracted. Increased expression of HOTTIP was
observed in HCC tissues (5.001±2.453) compared with the
non-cancerous tissues (0.182±0.459, P<0.001, Fig. 11A). With regard to the
clinicopathological parameters, we found that HOTTIP expression was
highly expressed in age (≥60), sex (male), race (white) compared to
that in the control group (P-value of <0.05, Table II and Fig. 11B–D). For the other
clinicopathological characteristics, no statistical significance
was found based on TCGA database. Moreover, the AUC of HOTTIP
reached 0.982 (95% CI, 0.966–0.998, P<0.0001, Fig. 11E), which indicated a high
diagnostic value of the HOTTIP level in HCC.
 | Table IIDifferential expression of HOTTIP of
clinicopathological parameters in HCC tissue based on TCGA
database. |
Table II
Differential expression of HOTTIP of
clinicopathological parameters in HCC tissue based on TCGA
database.
| Clinicopathological
features | N | HOTTIP expression
|
|---|
| Mean ± SD | T | P-value |
|---|
| Tissues | | | | |
| Normal liver | 28 | 0.182±0.459 | 8.16 | <0.0001 |
| HCC | 171 | 5.001±2.453 | | |
| Age | | | | |
| <60 | 66 | 4.405±2.513 | −2.563 | 0.011 |
| ≥60 | 106 | 5.376±2.349 | | |
| Sex | | | | |
| Male | 114 | 5.457±2.393 | 3.556 | 0.0005 |
| Female | 57 | 4.089±2.332 | | |
| Race | | | | |
| White | 84 | 5.077±2.379 | F=3.078 | 0.049 |
| Black | 11 | 6.522±1.740 | | |
| Yellow | 70 | 4.622±2.540 | | |
| T (tumor) | | | | |
| T1+ T2 | 133 | 5.099±2.372 | 1.236 | 0.218 |
| T3+ T4 | 37 | 4.539±2.657 | | |
| Vascular
invasion | | | | |
| No | 95 | 4.898±2.405 | 0.521 | 0.603 |
| Yes | 51 | 5.118±2.509 | | |
| Patological
grade | | | | |
| g1+g2 | 107 | 5.068±.2.434 | 0.569 | 0.570 |
| g3+g4 | 61 | 4.847±2.382 | | |
| Stage | | | | |
| I+ II | 126 | 5.065±2.333 | 1.544 | 0.125 |
| III +IV | 32 | 4.341±2.520 | | |
| Recurrence | | | | |
| No | 151 | 4.942±2.468 | 1.334 | 0.184 |
| Yes | 10 | 6.008±2.029 | | |
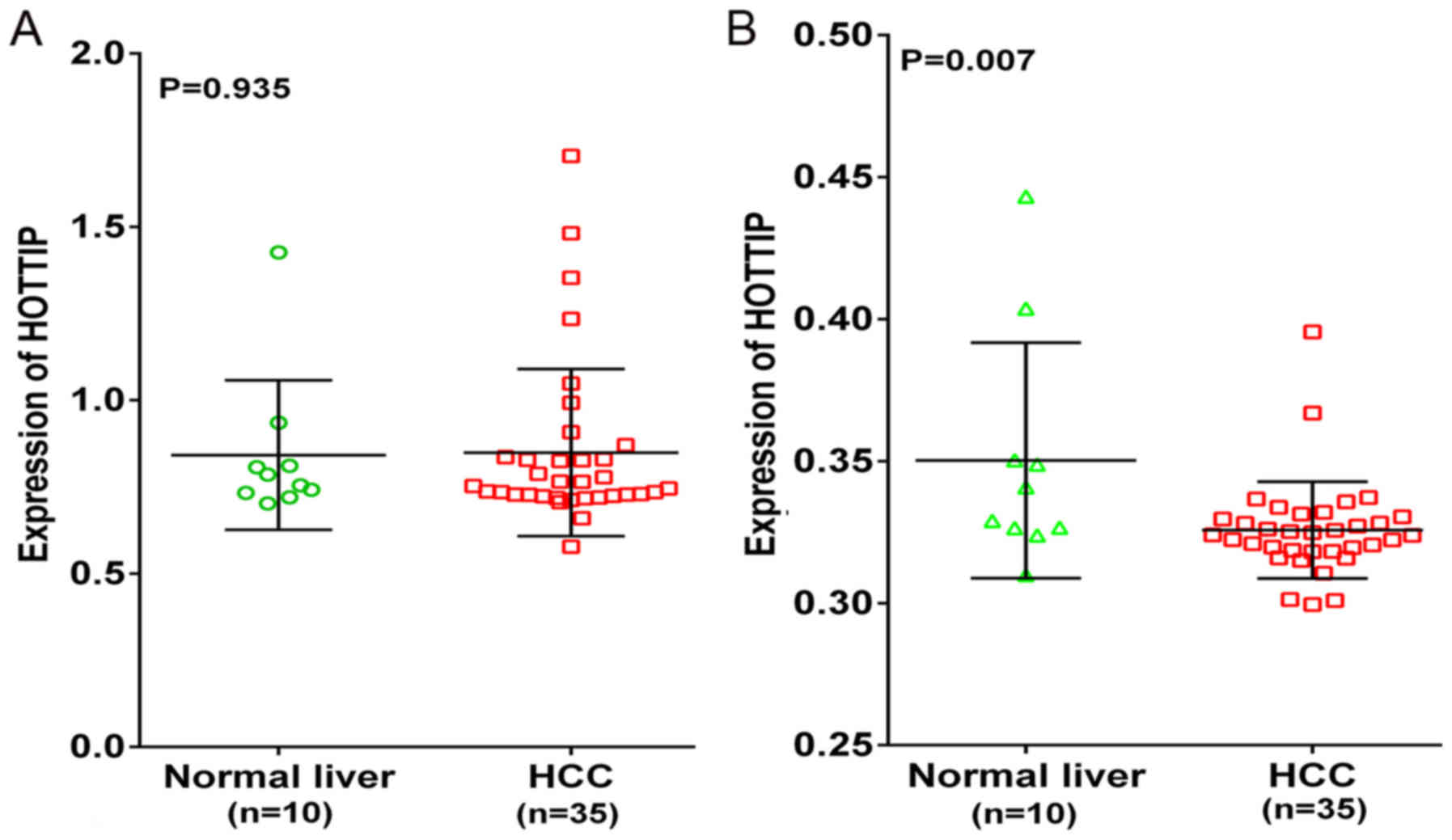
Additionally, two probe sets (1564069_at and
1564070_s_at) of Wurmbach Liver in Oncomine were used to validate
the HOTTIP expression. However, an opposite trend was found in the
two probe sets (Fig. 12). HOTTIP
was downregulated in 1564070_s_at probe set compared to the normal
liver, which was inconsistent with the results of qRT-PCR and TCGA.
Also, the opposite trend of HOTTIP expression might be a source of
heterogeneity in meta-analysis.
The potential pathways associated with
HOTTIP
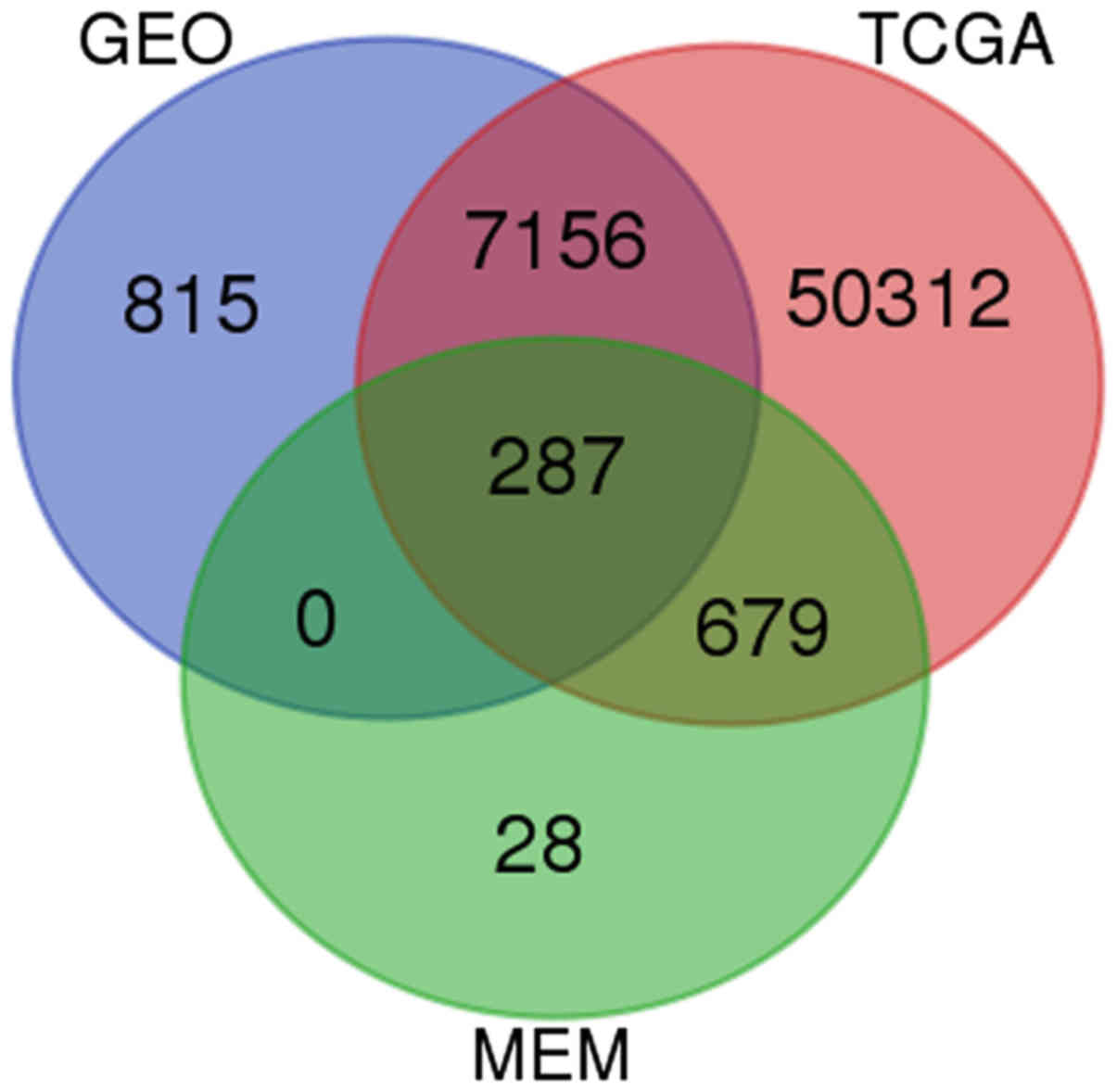
Based on GEO, TCGA and MEM database, 287 overlapped
genes were selected (Fig. 13). In
addition, GO and pathway analyses were performed using these 287
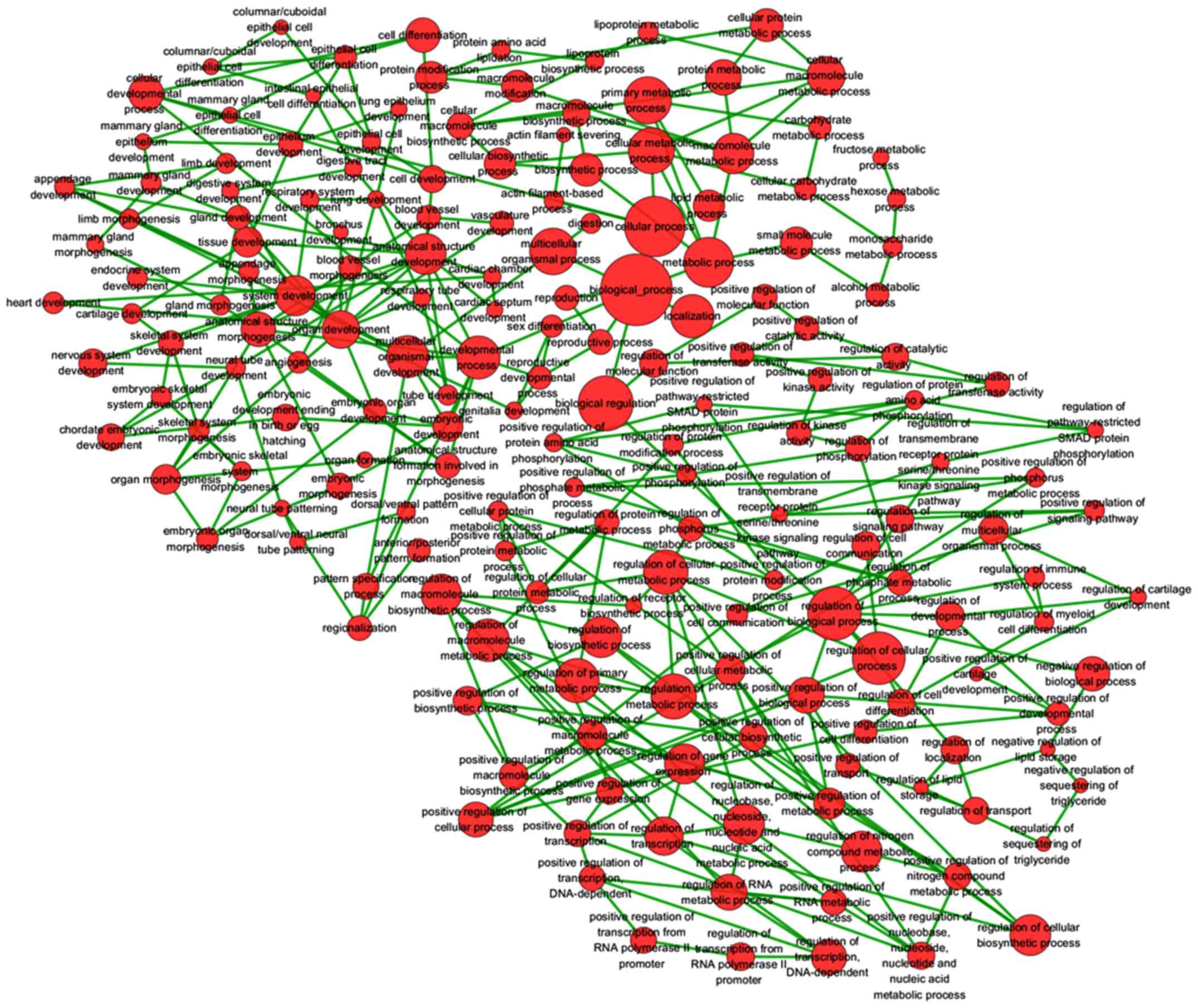
genes. The most strongly enriched GO terms were identified as
follows: embryonic morphogenesis, gland development, transcription
factor activity and extrinsic to membrane (Table III). To better affect the
functions of these genes, a function network was constructed
according to GO analysis (Fig.
14). Besides, the KEGG pathway analysis confirmed that these
genes were significantly involved in PPAR signaling pathway, Fc γ
R-mediated phagocytosis and endocytosis (Table IV). Altogether, the GO and KEGG
pathway analysis revealed that HOTTIP might participate in the
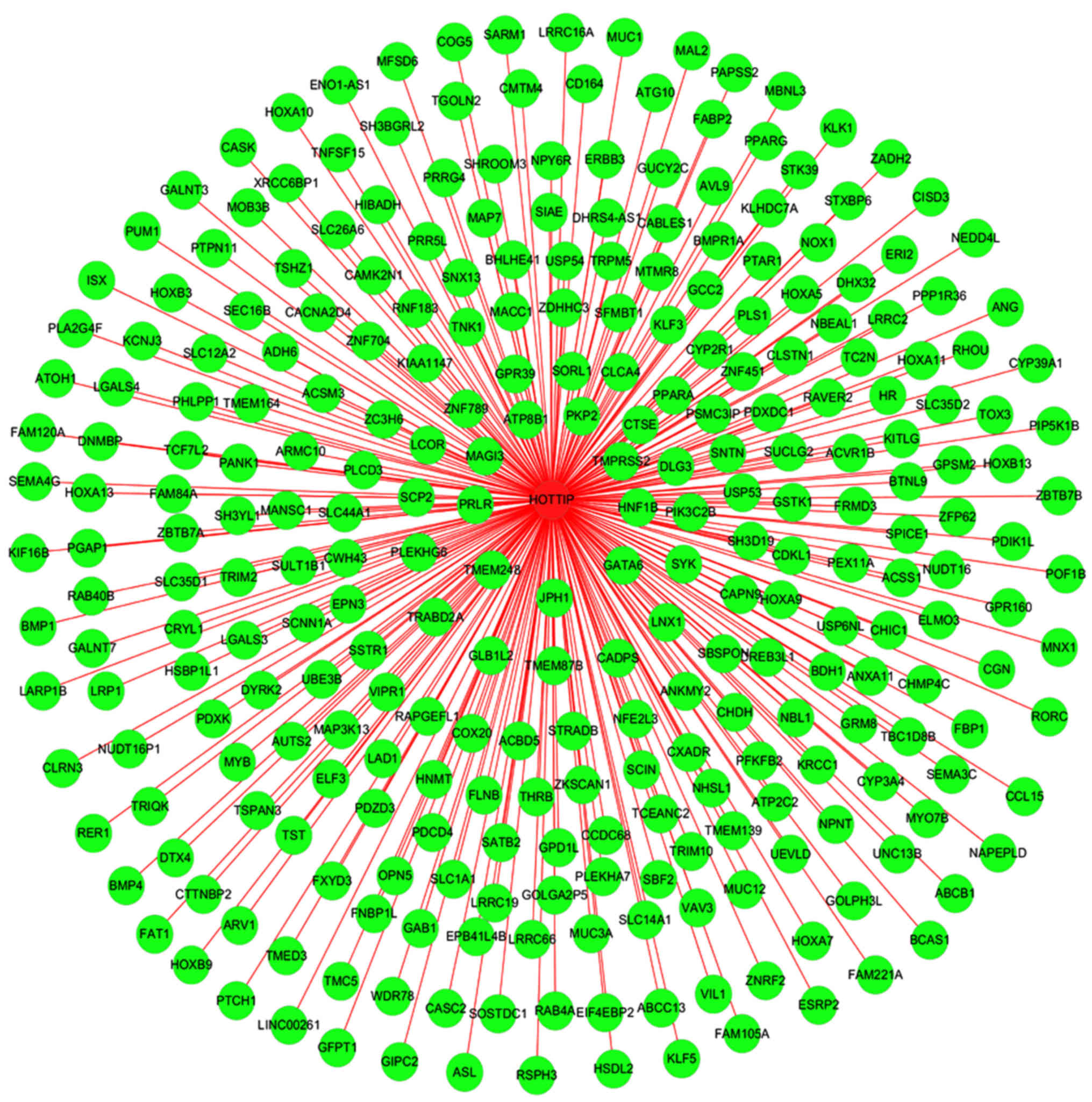
biological mechanism of HCC. In addition, a gene network of the 287
genes was constructed in the present study (Fig. 15), from which we could easily
observe relationships between HOTTIP and these potential genes.
 | Table IIITop 5 enrichment GO terms (BP, CC and
MF) of the potential genes of HOTTIP. |
Table III
Top 5 enrichment GO terms (BP, CC and
MF) of the potential genes of HOTTIP.
| GO ID | Term | Ontology | Count | P-value |
|---|
| GO:0048732 | Gland
development | BP | 12 | 5.32E-06 |
| GO:0007389 | Pattern
specification process | BP | 16 | 1.08E-05 |
| GO:0003002 |
Regionalization | BP | 13 | 3.84E-05 |
| GO:0048598 | Embryonic
morphogenesis | BP | 16 | 5.50E-05 |
| GO:0051216 | Cartilage
development | BP | 8 | 1.12E-04 |
| GO:0043565 | Sequence-specific
DNA binding | MF | 23 | 3.61E-04 |
| GO:0003700 | Transcription
factor activity | MF | 31 | 5.20E-04 |
| GO:0008289 | Lipid binding | MF | 17 | 2.81E-03 |
| GO:0004385 | Guanylate kinase
activity | MF | 3 | 1.31E-02 |
| GO:0042803 | Protein
homodimerization activity | MF | 12 | 2.08E-02 |
| GO:0019898 | Extrinsic to
membrane | CC | 23 | 3.54E-06 |
| GO:0045177 | Apical part of
cell | CC | 13 | 1.32E-05 |
| GO:0016324 | Apical plasma
membrane | CC | 11 | 2.63E-05 |
| GO:0044459 | Plasma membrane
part | CC | 52 | 4.28E-04 |
| GO:0005624 | Membrane
fraction | CC | 22 | 7.97E-03 |
 | Table IVKEGG pathway enrichment analysis of
the potential genes of HOTTIP. |
Table IV
KEGG pathway enrichment analysis of
the potential genes of HOTTIP.
| KEGG ID | KEGG term | Count | P-value | Gene symbol |
|---|
| hsa04666 | Fc γ R-mediated
phagocytosis | 5 | 5.94E-02 | VAV3, SCIN,
PIP5K1B, PLA2G4F, SYK |
| hsa04144 | Endocytosis | 7 | 6.52E-02 | ACVR1B, EPN3,
CHMP4C, ERBB3, RAB4A, PIP5K1B, NEDD4L |
| hsa03320 | PPAR signaling
pathway | 4 | 9.13E-02 | PPARA, PPARG,
FABP2, SCP2 |
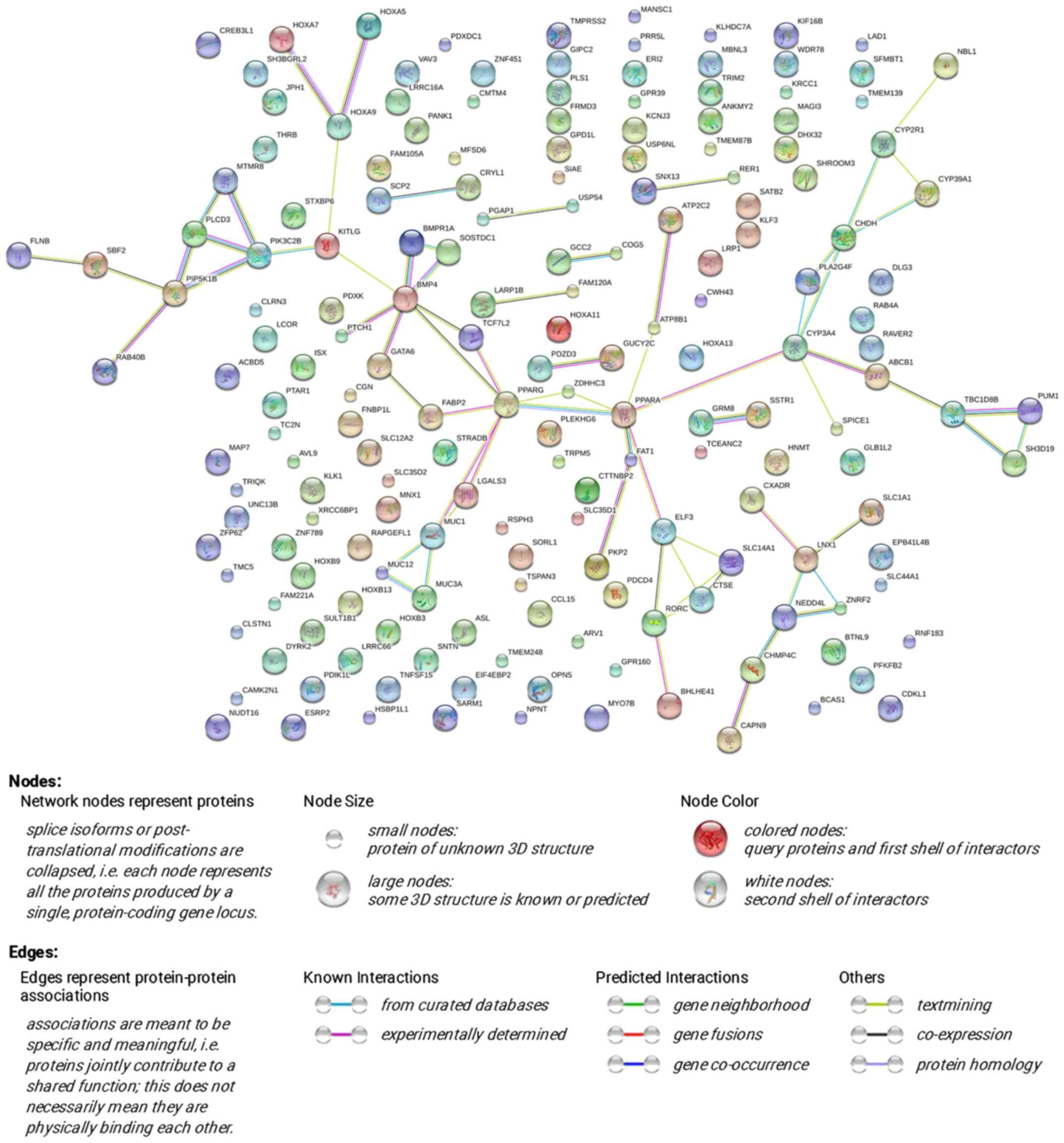
The PPI network was constructed through STRING
online and a total of 72 PPI pairs with combined score >0.4 were
noted (Fig. 16). Also, PPARA had
the highest degree (degree 6) according to the PPI network.
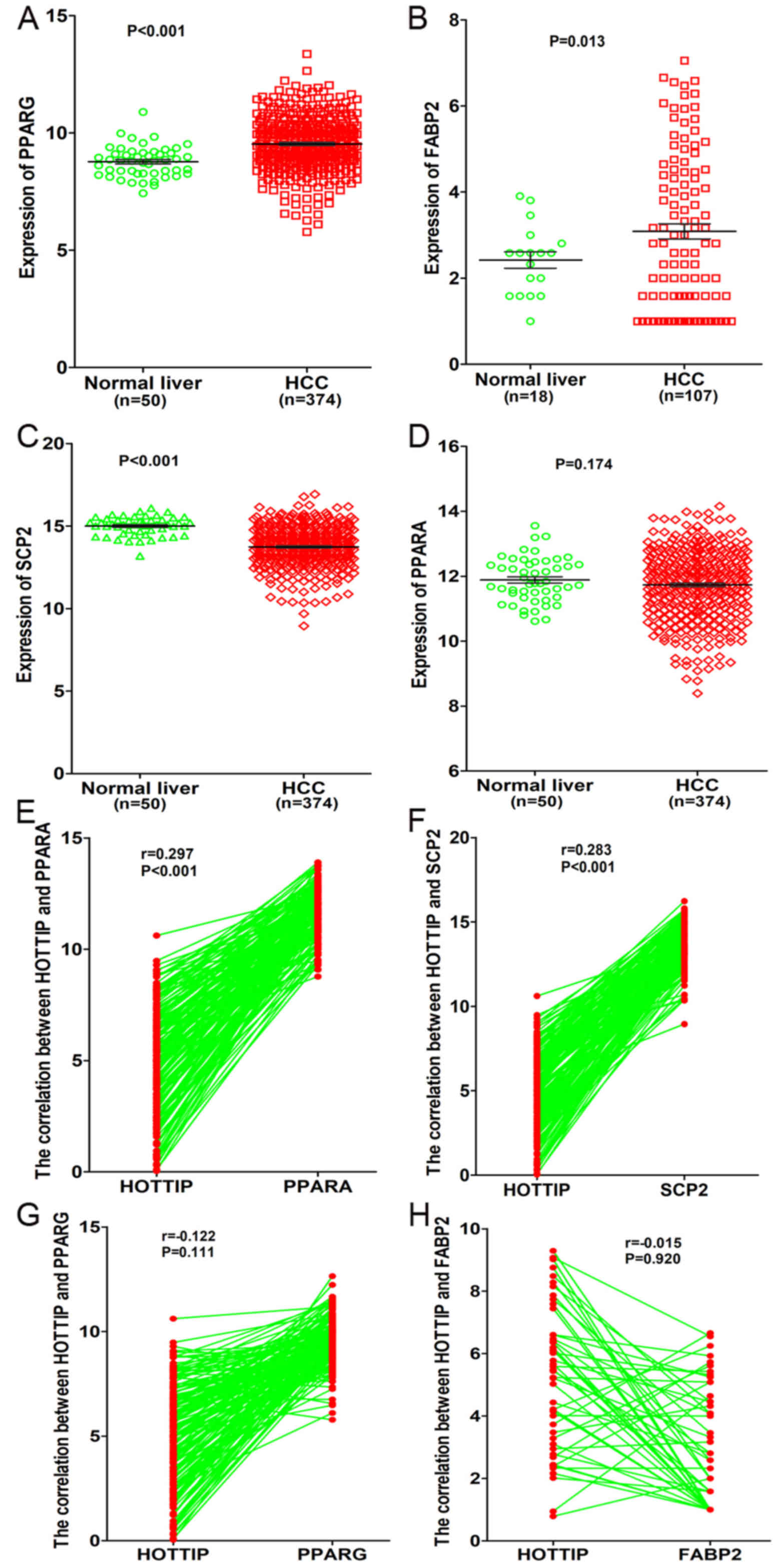
In addition, a total of four genes (PPARA, PPARG,
FABP2, SCP2) related to PPAR signaling pathway were detected based
on KEGG pathway analysis. Moreover, we investigated the expression
of these four genes and their correlations with HOTTIP based on the
original data in TCGA. We found that both PPARG and FABP2 were
highly expressed in HCC compared to normal liver (P<0.05,
Fig. 17A and B), whereas SCP2 was
highly expressed in normal liver (14.99±0.082 vs. 13.74±0.064,
P<0.001, Fig. 17C). However,
only a minor difference was found in PPARA expression between
normal liver and HCC (11.88±0.098 vs. 11.73±0.054, P=0.174,
Fig. 17D). As for the correlation
between HOTTIP and these genes, we discovered that HOTTIP had a
positive correlation with PPARA or SCP2 (P<0.001, Fig. 17E and F), whereas a negative
correlation was found between HOTTIP and PPARG and FABP2
(P>0.05, Fig. 17G and H).
Based on the aforementioned results, we hypothesized that HOTTIP
may influence the SCP2 expression of PPAR signaling pathway to
participate in different biological processes of HCC.
Discussion
Up to now, countless studies have confirmed that
lncRNAs could participate in various chemical and biological
processes, such as chromosome remodeling, transcription, cancer
metastasis and posttranscriptional processing (66,67).
Many studies have demonstrated that lncRNAs were related to
tumorigenesis and development of HCC through various pathways,
including regulation of apoptosis, cell cycle and chemotherapy
resistance in HCC tissue or cell lines (68–70).
lncRNAs have opened an avenue of cancer genomics.
To date, several studies have investigated the
effect and potential mechanism of HOTTIP on HCC. Quagliata et
al (71) found that the HOTTIP
expression was associated with HCC progression and HOTTIP could be
a predictive biomarker in HCC based on the snap-frozen needle HCC
biopsies and their matched non-neoplastic counterparts. They also
clarified that HOTTIP could directly control the expression of HOXA
locus gene by interacting with the WDR5/MLL complex, but the
specific relationship between HOTTIP and HOX genes is still vague.
Tsang et al (45) found
that HOTTIP was an oncogenic lncRNA and highly expressed in HCC
tissues based on qRT-PCR, and HOTTIP could contribute to
hepatocarcinogenesis via targeting tumor suppressive miR-125b,
which was verified by luciferase reporter assay and functional
analysis. Also, the migratory ability of HCC cells could be
inhibited after silencing HOTTIP expression. Ge et al
(72), using dual luciferase
reporter assays, confirmed that HOTTIP was a significant oncogene
in HCC and miR-192/-204-HOTTIP axis was a significant molecular
pathway during tumorigenesis of HCC. They also demonstrated the
prognostic and potential therapeutic roles of HOTTIP. In
comparison, we designed this study using RT-qPCR, meta-analysis and
bioinformatics to further investigate the effect of HOTTIP in HCC.
Interestingly, we discovered that HOTTIP was a tumorigenic gene and
HOTTIP expression was highly expressed in TNM (III +IV), age (≥60),
sex (male), race (white) and cirrhosis (no) compared to that in the
control groups. In the present study, ROC curve was applied to
evaluate the association between HOTTIP expression and the
diagnostic value, and the AUC of HOTTIP indicated the potential
diagnostic value of HOTTIP level in HCC. Moreover, this is the
first meta-analysis to investigate the expression and diagnostic
value of HOTTIP in HCC. As a result, the SMD of the meta-analysis
validated the higher expression of HOTTIP in HCC. Furthermore, in
the diagnostic meta-analysis, 393 cases from GEO, TCGA, Oncomine
and publications were included. The meta-analysis was performed to
evaluate the validity of HOTTIP for the detection of HCC. The
sensitivity of the HOTTIP assay in the included parts ranged from
70 to 96%, and the specificity of HOTTIP range from 20 to 89%. The
combined values of sensitivity (0.88) and specificity (0.59)
demonstrated the accuracy of HOTTIP for the detection of HCC. Also,
our results clarified that the SROC curve was located near the
upper left corner. The AUC was 0.87, which indicated a moderate
diagnostic accuracy (73). The PLR
and NLR were presented to measure the diagnostic accuracy of
HOTTIP. Likelihood ratios >10 or <0.1 was identified as high
accuracy. A PLR value of 2.13 suggested that patients with HCC had
an ~2.13-fold higher chance of being HOTTIP assay-positive. In
addition, the NLR (0.20) showed that if the HOTTIP result was
negative, the chance that this patient has HCC was ~20%. Hence, the
high diagnostic accuracy in meta-analysis confirmed our results.
However, there were some limitations in our meta-analysis. The
heterogeneity (high I-square values) is unavoidable, partly because
of the opposite results in one probe set of Oncomine. Furthermore,
blinding in 4 included databases was not certain, which also
contributed to the heterogeneity. In addition, only two
publications concerned the prognosis of HOTTIP. In this case, the
number of studies restricted the prognostic meta-analysis.
As reported, HOX gene encoded-transcription could
regulate cell fate and embryonic development (71). According to GO and KEGG analyses,
we found that the most strongly enriched functional terms were
embryonic morphogenesis, gland development, transcription factor
activity and extrinsic to membrane. Also, the HOTTIP co-expressed
genes were significantly related to PPAR signaling pathway. Several
studies have demonstrated that PPAR signaling pathway played a
vital role in HCC, but no studies were found on HOTTIP and PPAR
signaling pathway (74,75). In the present study, we
hypothesized that HOTTIP could play a significant role in HCC via
PPAR signaling pathway, but the specific underlying mechanism of
HOTTIP in HCC still needs to be investigated with further research.
We also investigated the genes from PPAR signaling pathway and the
hub genes from PPI. In addition, we hypothesized that HOTTIP may
influence the SCP2 expression of PPAR signaling pathway to
participate in different biological processes of HCC. To verify our
hypothesis, we intend to perform various experiments, including
proliferation, invasion and metastasis assays, chicken
embryochorioallantoic membrane and nude mouse models. The clinical
significance and the molecular mechanism of HOTTIP on the
biological function of HCC will be investigated from the
perspectives of molecule, cell, tissue and animal. However, the
circulating HOTTIP in HCC patients has higher clinical value in
evaluating the real diagnostic potential of HOTTIP than HOTTIP in
tissues. We also intend to perform in-depth exploration on the
circulating HOTTIP in HCC patients in the future. Focusing on the
new insight of HOTTIP, this study aimed to provide a new biomarker
or therapeutic target for HCC.
Acknowledgments
This study was supported by the Fund of National
Natural Science Foundation of China (NSFC81560386) and the Fund of
Guangxi Medical University Training Program for Distinguished Young
Scholars (2017). The authors acknowledge the data provided by the
TCGA database.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
qRT-PCR
|
quantitative reverse
transcription-polymerase chain reactions
|
|
GEO
|
Gene Expression Omnibus
|
|
TCGA
|
The Cancer Genome Atlas
|
|
MEM
|
Multi Experiment Matrix
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
lncRNAs
|
long non-coding RNAs
|
|
ROC
|
receiver operating characteristic
|
|
BP
|
biological process
|
|
CC
|
cellular component
|
|
MF
|
molecular function
|
|
DAVID
|
Database for Annotation, Visualization
and Integrated Discovery
|
|
NCBI
|
National Center of Biotechnology
Information
|
|
mean ± SD
|
mean ± standard deviation
|
|
SROC
|
summary receiver operating
characteristic
|
References
|
1
|
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng
Y, Bi J, Qu K, Zhang J and Liu C: Down-regulation of miR-146b-5p by
long noncoding RNA MALAT1 in hepatocellular carcinoma promotes
cancer growth and metastasis. Oncotarget. 8:28683–28695.
2017.PubMed/NCBI
|
|
2
|
Lu PH, Chen MB, Liu YY, Wu MH, Li WT, Wei
MX, Liu CY and Qin SK: Identification of sphingosine kinase 1
(SphK1) as a primary target of icaritin in hepatocellular carcinoma
cells. Oncotarget. 8:22800–22810. 2017.PubMed/NCBI
|
|
3
|
He R, Gao L, Ma J, Peng Z, Zhou S, Yang L,
Feng Z, Dang Y and Chen G: The essential role of MTDH in the
progression of HCC: A study with immunohistochemistry, TCGA,
meta-analysis and in vitro investigation. Am J Transl Res.
9:1561–1579. 2017.
|
|
4
|
Mo Z, Zheng S, Lv Z, Zhuang Y, Lan X, Wang
F, Lu X, Zhao Y and Zhou S: Senescence marker protein 30 (SMP30)
serves as a potential prognostic indicator in hepatocellular
carcinoma. Sci Rep. 6:393762016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang WT, Wang HL, Yang H, Ren FH, Luo YH,
Huang CQ, Liang YY, Liang HW, Chen G and Dang YW: Lower expressed
miR-198 and its potential targets in hepatocellular carcinoma: A
clinicopathological and in silico study. Onco Targets Ther.
9:5163–5180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu Y, Qi Y, Luo J, Yang J, Xie Q, Deng C,
Su N, Wei W, Shi D, Xu F, et al: Hepatitis B Virus X protein
stimulates proliferation, wound closure and inhibits apoptosis of
HuH-7 cells via CDC42. Int J Mol Sci. 18:182017. View Article : Google Scholar
|
|
8
|
Yu C, Cao Q, Chen P, Yang S, Gong X, Deng
M, Ruan B and Li L: Tissue transglutaminase 2 exerts a
tumor-promoting role in hepatitis B virus-related hepatocellular
carcinoma. Tumour Biol. 37:16269–16274. 2016. View Article : Google Scholar
|
|
9
|
Gong X, Wei W, Chen L, Xia Z and Yu C:
Comprehensive analysis of long non-coding RNA expression profiles
in hepatitis B virus-related hepatocellular carcinoma. Oncotarget.
7:42422–42430. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dengler M, Staufer K, Huber H, Stauber R,
Bantel H, Weiss KH, Starlinger P, Pock H, Klöters-Plachky P,
Gotthardt DN, et al: Soluble Axl is an accurate biomarker of
cirrhosis and hepatocellular carcinoma development: Results from a
large scale multicenter analysis. Oncotarget. 8:46234–46248.
2017.PubMed/NCBI
|
|
11
|
Ye Q, Qian BX, Yin WL, Wang FM and Han T:
Association between the HFE C282Y, H63D polymorphisms and the risks
of non-alcoholic fatty liver disease, liver cirrhosis and
hepatocellular carcinoma: An updated systematic review and
meta-analysis of 5,758 cases and 14,741 controls. PLoS One.
11:e01634232016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Joshi K, Kohli A, Manch R and Gish R:
Alcoholic liver disease: High risk or low risk for developing
hepatocellular carcinoma? Clin Liver Dis. 20:563–580. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fairman J, Liu KH and Menne S: Prevention
of liver tumor formation in woodchucks with established
hepatocellular carcinoma by treatment with cationic liposome-DNA
complexes. BMC Cancer. 17:1722017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hectors SJ, Wagner M, Bane O, Besa C,
Lewis S, Remark R, Chen N, Fiel MI, Zhu H, Gnjatic S, et al:
Quantification of hepatocellular carcinoma heterogeneity with
multiparametric magnetic resonance imaging. Sci Rep. 7:24522017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie X, Pan J, Wei L, Wu S, Hou H, Li X and
Chen W: Gene expression profiling of microRNAs associated with UCA1
in bladder cancer cells. Int J Oncol. 48:1617–1627. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun XJ, Wang Q, Guo B, Liu XY and Wang B:
Identification of skin-related lncRNAs as potential biomarkers that
involved in Wnt pathways in keloids. Oncotarget. 8:34236–34244.
2017.PubMed/NCBI
|
|
17
|
Ma G, Tang M, Wu Y, Xu X, Pan F and Xu R:
LncRNAs and miRNAs: Potential biomarkers and therapeutic targets
for prostate cancer. Am J Transl Res. 8:5141–5150. 2016.
|
|
18
|
Wilusz JE: Long noncoding RNAs: Re-writing
dogmas of RNA processing and stability. Biochim Biophys Acta.
1859:128–138. 2016. View Article : Google Scholar
|
|
19
|
Yuan X, Wang J, Tang X, Li Y, Xia P and
Gao X: Berberine ameliorates nonalcoholic fatty liver disease by a
global modulation of hepatic mRNA and lncRNA expression profiles. J
Transl Med. 13:242015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei Y and Niu B: Role of MALAT1 as a
prognostic factor for survival in various cancers: A systematic
review of the literature with meta-analysis. Dis Markers.
2015:1646352015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei Y and Zhang X: Transcriptome analysis
of distinct long non-coding RNA transcriptional fingerprints in
lung adenocarcinoma and squamous cell carcinoma. Tumour Biol.
37:16275–16285. 2016. View Article : Google Scholar
|
|
22
|
Li Y, Li W, Liang B, Li L, Wang L, Huang
H, Guo S, Wang Y, He Y, Chen L, et al: Identification of cancer
risk lncRNAs and cancer risk pathways regulated by cancer risk
lncRNAs based on genome sequencing data in human cancers. Sci Rep.
6:392942016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ,
Wang CY, Zhang HM, Zhang RX, Zhang JJ, et al: LncRNA HULC enhances
epithelial-mesenchymal transition to promote tumorigenesis and
metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1
signaling pathway. Oncotarget. 7:42431–42446. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang ZL, Chen RP, Zhou XT, Zhan HL, Hu
MM, Liu B, Wu GD and Wu LF: Long non-coding RNA MEG3 induces cell
apoptosis in esophageal cancer through endoplasmic reticulum
stress. Oncol Rep. 37:3093–3099. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Zhang Q, Zhang M, Yuan M, Wang Z,
Zhang J, Zhou X, Zhang Y, Lin F, Na H, et al: DC - SIGNR by
influencing the lncRNA HNRNPKP2 upregulates the expression of CXCR4
in gastric cancer liver metastasis. Mol Cancer. 16:782017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Zhang Z, Xiong L, Guo C, Jiang T,
Zeng L, Li G and Wang J: SNHG1 lncRNA negatively regulates
miR-199a-3p to enhance CDK7 expression and promote cell
proliferation in prostate cancer. Biochem Biophys Res Commun.
487:146–152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xue F, Liu Y, Chu H, Wen Y, Yan L, Tang Q,
Xiao E, Zhang D and Zhang H: eIF5A2 is an alternative pathway for
cell proliferation in cetuximab-treated epithelial hepatocellular
carcinoma. Am J Transl Res. 8:4670–4681. 2016.PubMed/NCBI
|
|
28
|
Deng LL, Chi YY, Liu L, Huang NS, Wang L
and Wu J: LINC00978 predicts poor prognosis in breast cancer
patients. Sci Rep. 6:379362016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong
M, Dang Y, Feng Z and Chen G: Clinical implication of long
non-coding RNA NEAT1 expression in hepatocellular carcinoma
patients. Int J Clin Exp Pathol. 8:5395–5402. 2015.PubMed/NCBI
|
|
30
|
Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B,
Du Y, Gao G, Tian Y, He L, et al: LncBRM initiates YAP1 signalling
activation to drive self-renewal of liver cancer stem cells. Nat
Commun. 7:136082016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cao C, Sun J, Zhang D, Guo X, Xie L, Li X,
Wu D and Liu L: The long intergenic noncoding RNA UFC1, a target of
MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to
increase levels of beta-catenin in HCC cells. Gastroenterology.
148:415–426. e4182015. View Article : Google Scholar
|
|
32
|
Zhou M, Zhang XY and Yu X: Overexpression
of the long non-coding RNA SPRY4-IT1 promotes tumor cell
proliferation and invasion by activating EZH2 in hepatocellular
carcinoma. Biomed Pharmacother. 85:348–354. 2017. View Article : Google Scholar
|
|
33
|
Xu LM, Chen L, Li F, Zhang R, Li Z-Y, Chen
F-F and Jiang X-D: Over-expression of the long non-coding RNA
HOTTIP inhibits glioma cell growth by BRE. J Exp Clin Cancer Res.
35:1622016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang X, Tang W, Li R, He R, Gan T, Luo Y,
Chen G and Rong M: Downregulation of microRNA-132 indicates
progression in hepatocellular carcinoma. Exp Ther Med.
12:2095–2101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jin W, Chen L, Cai X, Zhang Y, Zhang J, Ma
D, Cai X, Fu T, Yu Z, Yu F, et al: Long non-coding RNA TUC338 is
functionally involved in sorafenib-sensitized hepatocarcinoma cells
by targeting RASAL1. Oncol Rep. 37:273–280. 2017. View Article : Google Scholar
|
|
36
|
Yin X, Zheng SS, Zhang L, Xie XY, Wang Y,
Zhang BH, Wu W, Qiu S and Ren ZG: Identification of long noncoding
RNA expression profile in oxaliplatin-resistant hepatocellular
carcinoma cells. Gene. 596:53–88. 2017. View Article : Google Scholar
|
|
37
|
Lin L, Zhang YD, Chen ZY, Chen Y and Ren
CP: The clinicopathological significance of miR-149 and PARP-2 in
hepatocellular carcinoma and their roles in chemo/radiotherapy.
Tumour Biol. 37:12339–12346. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Colombo T, Farina L, Macino G and Paci P:
VT1: A rising star among oncogenic long noncoding RNAs. BioMed Res
Int. 2015:3042082015. View Article : Google Scholar
|
|
39
|
Liu FT, Xue QZ, Zhang Y, Hao TF, Luo HL
and Zhu PQ: Long non-coding RNA HOXA transcript at the distal tip
as a putative biomarker of metastasis and prognosis: A
meta-analysis. Clin Lab. 62:2091–2098. 2016. View Article : Google Scholar
|
|
40
|
Yang Y, Qian J, Xiang Y, Chen Y and Qu J:
The prognostic value of long noncoding RNA HOTTIP on clinical
outcomes in breast cancer. Oncotarget. 8:6833–6844. 2017.
|
|
41
|
Chen Z, He A, Wang D, Liu Y and Huang W:
Long noncoding RNA HOTTIP as a novel predictor of lymph node
metastasis and survival in human cancer: A systematic review and
meta-analysis. Oncotarget. 8:14126–14132. 2017.
|
|
42
|
Chen X, Han H, Li Y, Zhang Q, Mo K and
Chen S: Upregulation of long noncoding RNA HOTTIP promotes
metastasis of esophageal squamous cell carcinoma via induction of
EMT. Oncotarget. 7:84480–84485. 2016.PubMed/NCBI
|
|
43
|
Li Z, Zhao L and Wang Q: Overexpression of
long non-coding RNA HOTTIP increases chemoresistance of
osteosarcoma cell by activating the Wnt/β-catenin pathway. Am J
Transl Res. 8:2385–2393. 2016.
|
|
44
|
Ye H, Liu K and Qian K: Overexpression of
long noncoding RNA HOTTIP promotes tumor invasion and predicts poor
prognosis in gastric cancer. Onco Targets Ther. 9:2081–2088.
2016.PubMed/NCBI
|
|
45
|
Tsang FH, Au SL, Wei L, Fan DN, Lee JM,
Wong CC, Ng IO and Wong CM: Long non-coding RNA HOTTIP is
frequently up-regulated in hepatocellular carcinoma and is targeted
by tumour suppressive miR-125b. Liver Int. 35:1597–1606. 2015.
View Article : Google Scholar
|
|
46
|
Xu X, Wang X, Fu B, Meng L and Lang B:
Differentially expressed genes and microRNAs in bladder carcinoma
cell line 5637 and T24 detected by RNA sequencing. Int J Clin Exp
Pathol. 8:12678–12687. 2015.
|
|
47
|
Subramanian Y, Kaliyappan K and
Ramakrishnan KS: Facile hydrothermal synthesis and characterization
of Co2GeO4/r-GO@C ternary nanocomposite as
negative electrode for Li-ion batteries. J Colloid Interface Sci.
498:76–84. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fu L, Xu Y, Hou Y, Qi X, Zhou L, Liu H,
Luan Y, Jing L, Miao Y, Zhao S, et al: Proteomic analysis indicates
that mitochondrial energy metabolism in skeletal muscle tissue is
negatively correlated with feed efficiency in pigs. Sci Rep.
7:452912017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sang Y, Zhou F, Wang D, Bi X, Liu X, Hao
Z, Li Q and Zhang W: Up-regulation of long non-coding HOTTIP
functions as an oncogene by regulating HOXA13 in non-small cell
lung cancer. Am J Transl Res. 8:2022–2032. 2016.PubMed/NCBI
|
|
50
|
Dai J, Wu H, Zhang Y, Gao K, Hu G, Guo Y,
Lin C and Li X: Negative feedback between TAp63 and Mir-133b
mediates colorectal cancer suppression. Oncotarget. 7:87147–87160.
2016.PubMed/NCBI
|
|
51
|
Wu H, Zhou J, Zeng C, Wu D, Mu Z, Chen B,
Xie Y, Ye Y and Liu J: Curcumin increases exosomal TCF21 thus
suppressing exosome-induced lung cancer. Oncotarget. 7:87081–87090.
2016.PubMed/NCBI
|
|
52
|
Bornstein S, Schmidt M, Choonoo G, Levin
T, Gray J, Thomas CR Jr, Wong M and McWeeney S: IL-10 and integrin
signaling pathways are associated with head and neck cancer
progression. BMC Genomics. 17:382016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zeng JH, Xiong DD, Pang YY, Zhang Y, Tang
RX, Luo DZ and Chen G: Identification of molecular targets for
esophageal carcinoma diagnosis using miRNA-seq and RNA-seq data
from The Cancer Genome Atlas: A study of 187 cases. Oncotarget.
8:35681–35699. 2017.PubMed/NCBI
|
|
54
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Adler P, Kolde R, Kull M, Tkachenko A,
Peterson H, Reimand J and Vilo J: Mining for coexpression across
hundreds of datasets using novel rank aggregation and visualization
methods. Genome Biol. 10:R1392009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tang W, Liao Z and Zou Q: Which
statistical significance test best detects oncomiRNAs in cancer
tissues? An exploratory analysis. Oncotarget. 7:85613–85623.
2016.PubMed/NCBI
|
|
57
|
Li CQ, Huang GW, Wu ZY, Xu YJ, Li XC, Xue
YJ, Zhu Y, Zhao JM, Li M, Zhang J, et al: Integrative analyses of
transcriptome sequencing identify novel functional lncRNAs in
esophageal squamous cell carcinoma. Oncogenesis. 6:e2972017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dinh TA, Vitucci EC, Wauthier E, Graham
RP, Pitman WA, Oikawa T, Chen M, Silva GO, Greene KG, Torbenson MS,
et al: Comprehensive analysis of The Cancer Genome Atlas reveals a
unique gene and non-coding RNA signature of fibrolamellar
carcinoma. Sci Rep. 7:446532017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Seiler R, Black PC, Thalmann G, Stenzl A
and Todenhöfer T: Is The Cancer Genome Atlas (TCGA) bladder cancer
cohort representative of invasive bladder cancer? Urol Oncol.
35:458.e1–458.e7. 2017. View Article : Google Scholar
|
|
60
|
Gao H, Wang H and Yang W: Identification
of key genes and construction of microRNA-mRNA regulatory networks
in multiple myeloma by integrated multiple GEO datasets using
bioinformatics analysis. Int J Hematol. 106:99–107. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Tan W, Song Y, Mo C, Jiang S and Wang Z:
Analysis of gene expression profile microarray data in complex
regional pain syndrome. Mol Med Rep. 16:3371–3378. 2017.PubMed/NCBI
|
|
62
|
Mirsafian H, Ripen AM, Leong WM, Chear CT,
Bin Mohamad S and Merican AF: Transcriptome profiling of monocytes
from XLA patients revealed the innate immune function dysregulation
due to the BTK gene expression deficiency. Sci Rep. 7:68362017.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhao Z, Bai J, Wu A, Wang Y, Zhang J, Wang
Z, Li Y, Xu J and Li X: Co-LncRNA: Investigating the lncRNA
combinatorial effects in GO annotations and KEGG pathways based on
human RNA-Seq data. Database (Oxford). Sep 10–2015.Epub ahead of
print. View Article : Google Scholar
|
|
64
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering
C, et al: STRING v9.1: Protein-protein interaction networks, with
increased coverage and integration. Nucleic Acids Res.
41D:D808–D815. 2013.
|
|
65
|
Zamora J, Abraira V, Muriel A, Khan K and
Coomarasamy A: Meta-DiSc: A software for meta-analysis of test
accuracy data. BMC Med Res Methodol. 6:312006. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xiong DD, Feng ZB, Cen WL, Zeng JJ, Liang
L, Tang RX, Gan XN, Liang HW, Li ZY, Chen G, et al: The clinical
value of lncRNA NEAT1 in digestive system malignancies: A
comprehensive investigation based on 57 microarray and RNA-seq
datasets. Oncotarget. 8:17665–17683. 2017.PubMed/NCBI
|
|
68
|
Lu S, Zhou J, Sun Y, Li N, Miao M, Jiao B
and Chen H: The noncoding RNA HOXD-AS1 is a critical regulator of
the metastasis and apoptosis phenotype in human hepatocellular
carcinoma. Mol Cancer. 16:1252017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Xu LC, Chen QN, Liu XQ, Wang XM, Chang QM,
Pan Q, Wang L and Wang YL: Up-regulation of LINC00161 correlates
with tumor migration and invasion and poor prognosis of patients
with hepatocellular carcinoma. Oncotarget. 8:56168–56173.
2017.PubMed/NCBI
|
|
70
|
Wang Y, Hu Y, Wu G, Yang Y, Tang Y, Zhang
W, Wang K, Liu Y, Wang X and Li T: Long noncoding RNA PCAT-14
induces proliferation and invasion by hepatocellular carcinoma
cells by inducing methylation of miR-372. Oncotarget.
8:34429–34441. 2017.PubMed/NCBI
|
|
71
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long noncoding RNA HOTTIP/HOXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar :
|
|
72
|
Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L,
Han S, Yuan Q and Yang M: MiRNA-192 [corrected] and miRNA-204
directly suppress lncRNA HOTTIP and interrupt GLS1-mediated
glutaminolysis in hepatocellular carcinoma. PLoS Genet.
11:e10057262015. View Article : Google Scholar
|
|
73
|
Swets JA: Measuring the accuracy of
diagnostic systems. Science. 240:1285–1293. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Lu G, Zhang G, Zheng X, Zeng Y, Xu Z, Zeng
W and Wang K: c9, t11- conjugated linoleic acid induces HCC cell
apoptosis and correlation with PPAR-γ signaling pathway. Am J
Transl Res. 7:2752–2763. 2015.
|
|
75
|
Li J, Huang Q, Long X, Zhang J, Huang X,
Aa J, Yang H, Chen Z and Xing J: CD147 reprograms fatty acid
metabolism in hepatocellular carcinoma cells through
Akt/mTOR/SREBP1c and P38/PPARα pathways. J Hepatol. 63:1378–1389.
2015. View Article : Google Scholar : PubMed/NCBI
|