Introduction
Keratocystic odontogenic tumor (KCOT), previously
known as odontogenic keratocyst, is a benign cystic lesion often
manifesting locally in an aggressive fashion and recurring at a
high rate (1). According to WHO
classification, KCOT is defined as a benign neoplasm of odontogenic
origin (2). Most KCOTs arise in
the mandible, and KCOT is identified according to a characteristic
histological appearance in which the cystic space is lined with a
uniform parakeratinized squamous epithelium. The basal cells are
aligned, with vertically elongated nuclei, and mitotic activity is
higher than that of other odontogenic cysts (3). As described in detail previously
(4), ~5% of KCOTs occur as
multiple lesions, and some of those cases occur as a manifestation
of basal cell nevus syndrome (BCNS). BCNS, also known as Gorlin
syndrome, is a rare autosomal dominant disorder (5), characterized by developmental
abnormalities, such as calcification of the falx cerebri, multiple
nevi, palmar and plantar pits, and skeletal deformity and
predisposition to malignant cancers, including basal cell carcinoma
(BCC), medulloblastoma, rhabdomyosarcoma, as well as benign tumors,
such as KCOTs, ovarian and cardiac fibromas (6). Among these tumors, >80% of BCNS
patients from all populations suffer KCOTs. The gene encoding the
Hedgehog (Hh) receptor, Patched1 (PTCH1) has been identified
to be responsible for BCNS (7).
PTCH1 is a transmembrane co-receptor for Hh family proteins, which
are secreted signaling molecules required for embryonic development
and adult tissue homeostasis (8).
Various types of PTCH1 mutations were found in most BCNS patients;
however, a genotype-phenotype relationship was not observed
(9,10). After the discovery of the link
between BCNS and Hh signaling, mutation analyses of sporadic
BCNS-related neoplasms were intensively performed. Recent analysis
showed that >80% of sporadic KCOTs had a mutated PTCH1
gene (11). In addition,
heterozygous Ptch1 knockout mice (Ptch1+/−), an
animal model of BCNS (12), and
Sufu+/− mice (13) also develop jaw keratocysts.
Furthermore, overexpression of Gli2 under the control of the
keratin 5 promoter induced keratocyst formation (14). The animal model data strongly
support the idea that activation of Hh signaling is an important
factor in the induction of KCOTs. However, the study of KCOT has
been hindered because a suitable in vitro biological system
has not been available. To date, establishment of only one KCOT
cell line has been reported (15).
In this study, we successfully established immortalized KCOT cell
lines from a patient with BCNS (iKCOT1) and from a sporadic case
(sKCOT1) and found that these cells showed stem cell and
mesenchymal cell properties. By exposing cells to a high calcium
concentration, these cells showed epithelial keratinocyte
properties. A possible mechanism of the onset of KCOTs will be
discussed.
Materials and methods
Patients
KCOT tissues derived from BCNS patient NS11 or from
a sporadic KCOT from a 25-year-old Japanese woman were kindly
donated to our department. Patient NS11 was diagnosed as BCNS and a
point mutation in intron 3 (c.584+2T>G) of PTCH1 was
detected in this patient (10).
The parents of the patients gave their written informed consent for
participation in this study. This study was approved by the Medical
Ethics Board of Hyogo College of Medicine (No. 108), and is in
accordance with the principles expressed in the Declaration of
Helsinki.
Cell culture and establishment of
immortalized cells
KCOT cells (iKCOT1 and sKCOT1) were maintained in
Epilife medium supplemented with HKGS (both from Cascade Biologics,
Portland, OR, USA) or F-medium (16). Lentiviral vectors, CSII-CMV-cyclin
D1, CSII-CMV-CDK4R24C and CSII-CMV-TERT were used to immortalize
KCOT cells. Production and infection of recombinant lentiviruses
with the vesicular stomatitis virus G glycoprotein (VSV-G) were
described previously (17). The
lentivirus backbone vector, CSII-MV-RfA, was a gift from Dr Miyoshi
(Riken, BRC). After several passages, established cell lines were
designated as iKCOT1 and sKCOT1.
Cell culture in 3-D life hydrogels
To analyze the ability to form 3-D structures, 3-D
life hydrogel (Cellendes GmbH, Reutlingen, Germany) was used. Cells
were incubated in the Hydrogel in the presence of RGD-peptides for
12 days according to the manufacturer's MDCK cyst formation
protocol. Cells were fixed in 4% formaldehyde in phosphate-buffered
saline [PBS(−)]. After permeabilization, cells were stained with
rhodamine phalloidin (Cytoskeleton, Denver, CO, USA) and mounted
using Vectashield containing DAPI (Vector Laboratories, Burlingame,
CA, USA). Imaging was carried out using a Zeiss LS780 confocal
microscope.
DNA and RNA isolation and sequencing
analysis
Genomic DNAs from KCOT cell lines were extracted
using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany).
All coding exons and intron-exon boundaries were amplified by PCR
and sequenced as described previously (10).
For analysis of mRNA, total RNA was extracted from a
sKCOT1 cell line using the QIAamp RNA Blood mini kit (Qiagen) and
subjected to RT-PCR using a Cells-to-cDNA® II kit
(Applied Biosystems, Foster City, CA, USA) with random primers. The
forward primer for exon 7 was 5′-GAATGGTGGATGTCATGGCTTATCC-3′, and
the reverse primer for exon 19 was 5′-ACGGCACTGAGCTTGATTCCGATGA-3′.
Amplified products were purified and cloned into pCRII (Thermo
Fisher Scientific, Waltham, MA, USA). Eleven randomly selected
clones were sequenced using M13 forward or reverse primers.
Antibodies, immunostaining and western
blotting
For immunofluorescence staining, cells were plated
in 8-well culture slides (BD Bioscience, Bedford, MA, USA) and half
of the medium in each well was changed to a high calcium medium
(1.23 mM; 1:1 mix of F medium and DMEM supplemented with 5% FCS).
Medium was changed every 3 days. Ten to twelve days later, cells
were fixed in 4% paraformaldehyde in PBS(−) and permeabilized with
0.1% Tween-20 in PBS(−). After blocking in PBS(−) with 2% BSA,
cells were subjected to immunofluorescence staining. Antibodies
were visualized with Alexa-488 or Alexa-546 secondary antibodies
(Thermo Fisher Scientific). Rhodamine phalloidin (Cytoskeleton) was
used to detect actin filaments. DNA was stained with DAPI (Vector
Laboratories). Imaging was carried out using a Zeiss LS780 confocal
microscope.
For western blotting, iKCOT1 and sKCOT1 were
harvested or incubated with low (0.65 mM) or high (1.23 mM) calcium
medium for 10–12 days. Gel electrophoresis and western blotting
were performed as described previously (18). Signals were detected by
chemiluminescence using a Pierce SuperSignal western blotting kit
(Thermo Fisher Scientific). All antibodies used are listed in
Table I.
 | Table IThe antibodies used in this
study. |
Table I
The antibodies used in this
study.
| Name | Company | Cat no. |
|---|
| Vimentin
(D21H3)XP®, rabbit mAb | Cell Signaling
Technology | 5741 |
| E-Cadherin (24E10)
rabbit mAb | Cell Signaling
Technology | 3195 |
| Slug (C19G7) rabbit
mAb | Cell Signaling
Technology | 9585 |
| Claudin-1
antibody | Cell Signaling
Technology | 4933 |
| β-Catenin (D10A8)
XP®, rabbit mAb | Cell Signaling
Technology | 8480 |
| Anti-cytokeratin 10
antibody | Abcam | Ab76318 |
| N-Cadherin (D4R1H)
XP®, Rabbit mAb | Cell Signaling
Technology | 13116 |
| Bmi1 (D20B7)
XP®, rabbit mAb | Cell Signaling
Technology | 6964 |
| Neurofilament-L
(C28E10) Rabbit mAb | Cell Signaling
Technology | 2837 |
| Monoclonal
anti-involucrin antibody produced in mouse | Sigma-Aldrich | I9018 |
| Sox2 (D6D9)
XP®, rabbit mAb | Cell Signaling
Technology | 3579 |
| ZO-1 (D7D12) rabbit
mAb | Cell Signaling
Technology | 8193 |
| CD44 (156-3C11)
mouse mAb | Cell Signaling
Technology | 3570 |
| β-actin (D6A8)
rabbit mAb | Cell Signaling
Technology | 8457 |
| Anti-mouse IgG (H+L
chain) HRP | MBL | PM009-7 |
| Anti-rabbit IgG
(H+L chain) HRP | MBL | 458 |
| Alexa Fluor 488
goat anti-mouse IgG (H+L) | Invitrogen | A11001 |
| Alexa Fluor 546
goat anti-rabbit IgG (H+L) | Invitrogen | A11035 |
Results
Establishment of immortalized KCOT cell
lines from patients with and without BCNS
We initially tried to culture cells from the KCOT of
the patient with BCNS using standard culture methods for normal
keratinocytes of the oral mucosa. In primary culture, KCOT cell
colonies grew slowly, and predominantly consisted of closely packed
polygonal cells with epithelial morphology. KCOT cells could be
subcultured 3 to 4 times, but the polygonal cells were replaced by
larger flattened cells and subsequently became senescent and could
no longer be cultured. Therefore, we attempted to immortalize
primary human KCOT cells by introducing CDK4R24C
(inhibitor-resistant form of CDK4), cyclinD1 (which can inactivate
Rb by phosphorylation) and hTERT genes using lentiviral vectors.
The combination of CDK4R24C, cyclin D1 and hTERT resulted in
efficient prolonged proliferation of the cells and they were
designated as iKCOT1, and they were cultured for >50 passages.
To our knowledge, this is the first study of the successful
establishment of immortalized KCOT cells derived from a patient
with BCNS. In addition, we also immortalized sporadic KCOT cells by
the same method and designated this line as sKCOT1.
Identification of PTCH1 mutations in KCOT
cell lines
We first analyzed the status of PTCH1 in both
cell lines. Sequence analysis of iKCOT1 cells showed a germline
c.584+2T>G mutation (10) and a
wild-type PTCH1 sequence indicating no evidence of loss of
heterozygosity (LOH) at least in the coding region. RT-PCR analysis
indicated that mutated and wild-type alleles were equally expressed
in this cell line (10). These
data suggested that the wild-type allele is expressed in iKCOT1
cells. sKCOT1 cells showed two mutations in the PTCH1 gene:
the c1120G>T mutation in exon 8 is a nonsense mutation and the
c3064_3065delAT mutation in exon 18 is a frameshift mutation
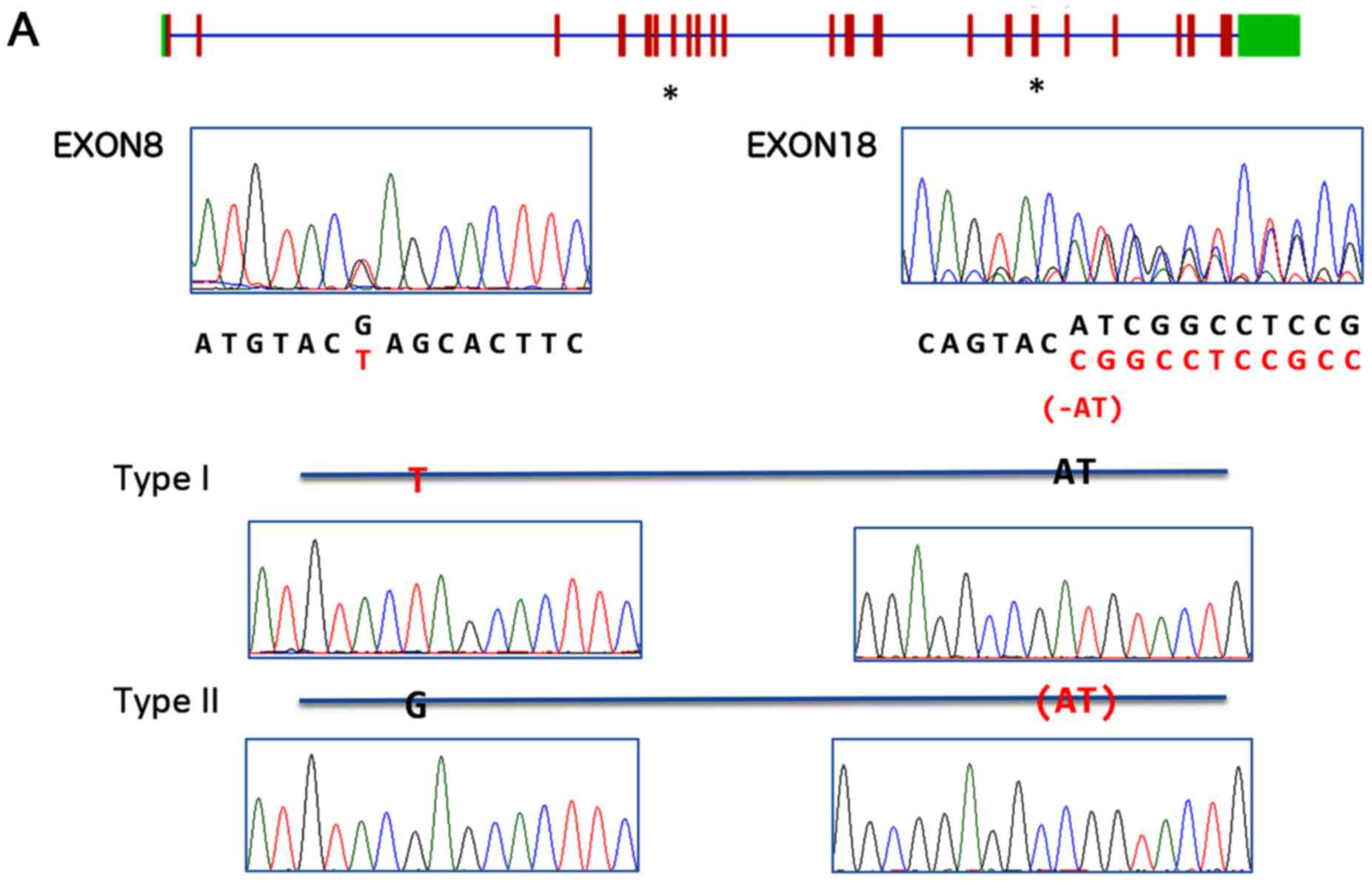
(Fig. 1A). Both mutations were
designated as loss of function mutations; therefore, we analyzed
mRNA from sKCOT1 cells to address whether these mutations were on
the same allele. After purification of mRNA from sKCOT1 cells,
RT-PCR was performed using a primer set that covers exon 8 to 18 of
the PTCH1 gene. Obtained fragments were cloned into
pCRII-TOPO (Thermo Fisher Scientific) and 11 randomly selected
independent clones were sequenced. Three out of 11 clones contained
only the exon 8 c1120G>T mutation and eight out of 11 clones had
exon 18 c3064_3065delAT mutation only (Fig. 1A). These results indicated that
each PTCH1 allele carried a single, different lethal
mutation in sKCOT1 cells. Sequence analysis of the blood sample
from the patient from which sKCOT1 cells were derived showed no
mutations in the PTCH1 gene indicating that these mutations
were sporadic and that the patient was not BCNS. In addition, we
sequenced the coding regions of the PTCH2, SMO and
SUFU genes of both KCOT cells, but did not detect any
mutation (data not shown).
Characterization in KCOT cell lines
To characterize the nature of the KCOT cell lines,
immunostaining was performed. In both cell lines in F medium, the
stem cell markers BMI1, SOX2 and CD44 and the mesenchymal cell
markers SNAI2, CDH2 and VIM, were observed. Furthermore, both cell
lines expressed NEFL, which is characteristic of neuronal cells
(Fig. 1B).
Calcium treatment induced morphological
changes and expression of epithelial or keratinocyte markers in
KCOT cells
KCOT cells retain several characteristics of
keratinocytes; therefore, we tested the response of the cell lines
to calcium treatment by increasing calcium concentration of the
culture media from 0.65 to 1.23 mM. Calcium-treated sKCOT1 cells
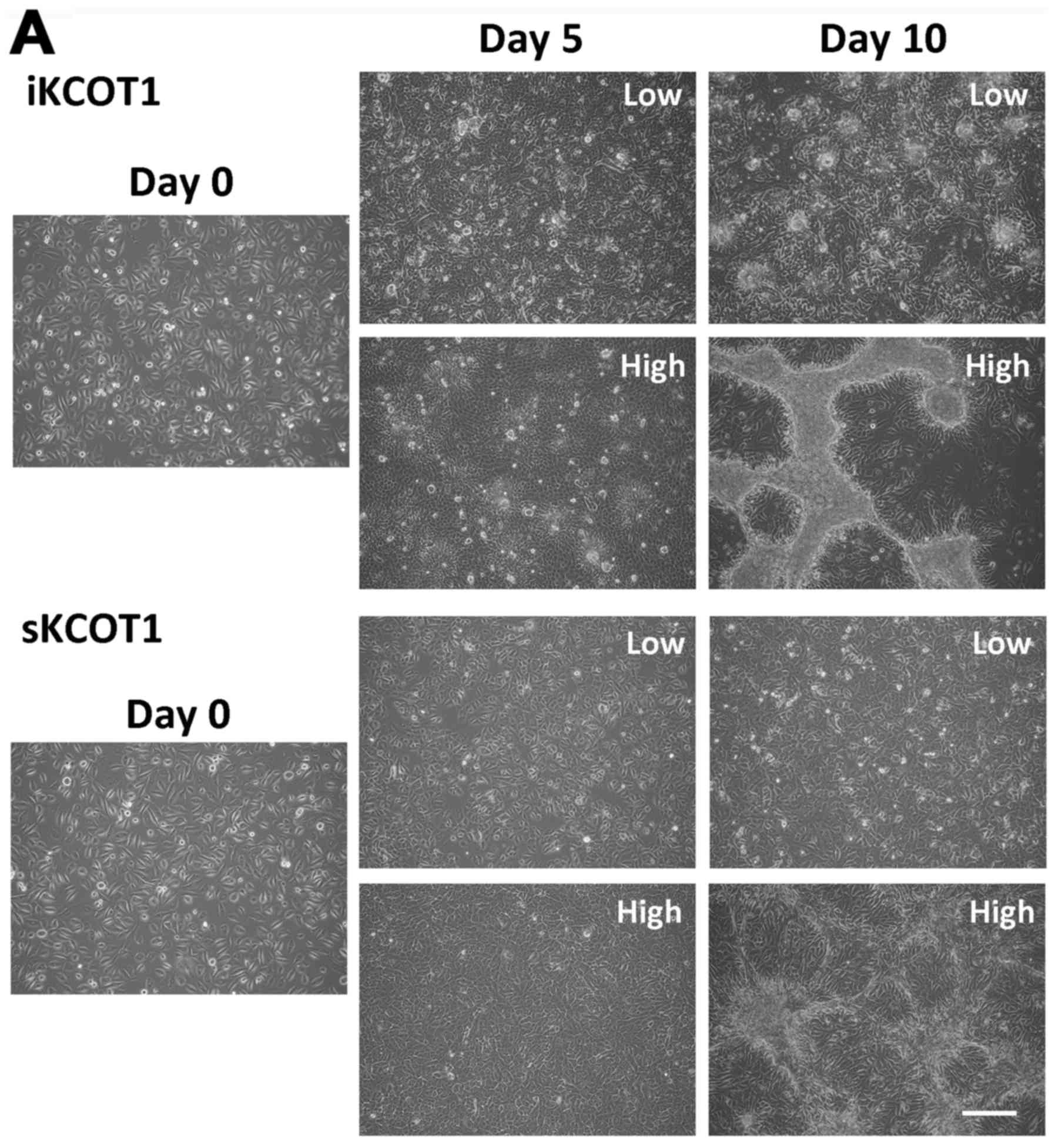
stratified and parakeratinized, and iKCOT1 cells piled up and
formed nodule-like structures by day 10 (Fig. 2A). Fig. 2B shows CDH1 and actin staining at
10 days of KCOT cells in low or high calcium medium. High
expression of CDH1 was observed especially in nodule-like structure
of iKCOT1 cells (Fig. 2B). When
cells were cultured in dishes with high calcium medium for 12 days,
sKCOT1 cells were stratified with the surface cells being of bigger
cell size and with bigger nuclei compared with the bottom cells.
Fig. 2C shows sequential images of
CDH1 (green) and DAPI (magenta) stained sKCOT1 cells. The upper
cells, predicted to be differentiated cells, expressed CDH1 and
parakeratinized. Fig. 3 shows
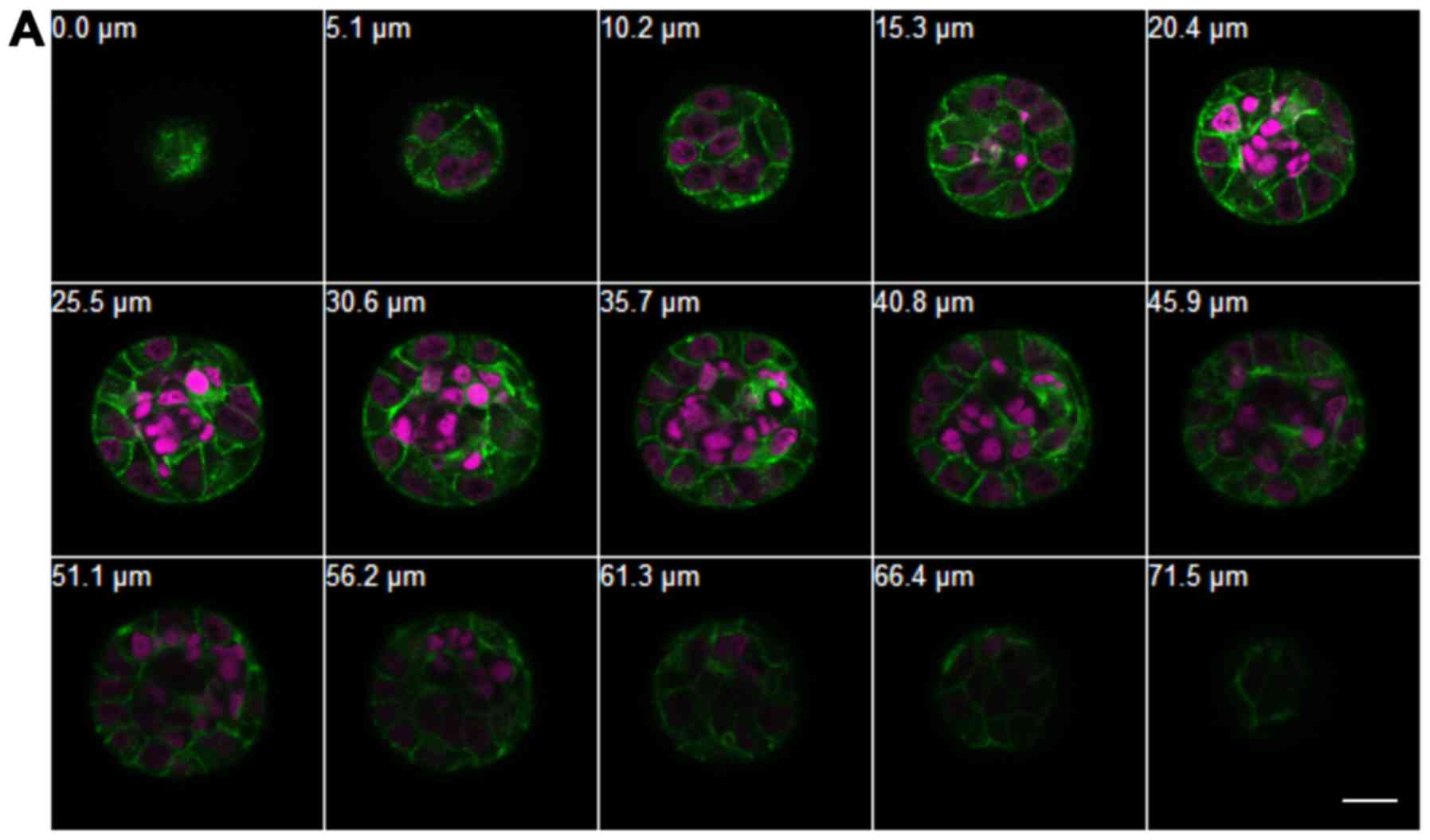
sequential images of sKCOT1 and iKCOT1 cells in 3-D hydrogels in
low calcium concentration. Under low calcium conditions, sKCOT1
cells formed spheroids with nuclei at the center that were small
and condensed, and lacked cell membranes (Fig. 3A). Conversely, iKCOT1 cells formed
spheroids with normal nuclei and cell membranes (Fig. 3B). However, under high calcium
conditions, some nuclei were condensed and lacked cell membranes,
and were located at the center of the iKCOT1 spheroids similar to
sKCOT1. These data indicated that both cell lines showed cyst-like
structures in 3-D hydrogels with different calcium
concentrations.
Because calcium treatment induced dramatic
morphological changes to the KCOT cells, we examined protein levels
by western blotting (Fig. 4). In
both cell lines, CDH1, CLDN1, KRT10 and IVL were induced by
calcium. However, levels of CDH2, VIM, TJP1, SNAI2, CD44 and BMI1
were reduced. CTNNB1 expression was reduced in sKCOT1 cells but was
not changed in iKCOT1 cells. Although an increase of calcium
concentration induced these changes in protein expression, the
expression of some proteins changed with respect to time.
Therefore, other factors, such as cell-to-cell contact or cell
proliferation, may also be involved in changes in protein
expression. NEFL expression was not influenced by calcium.
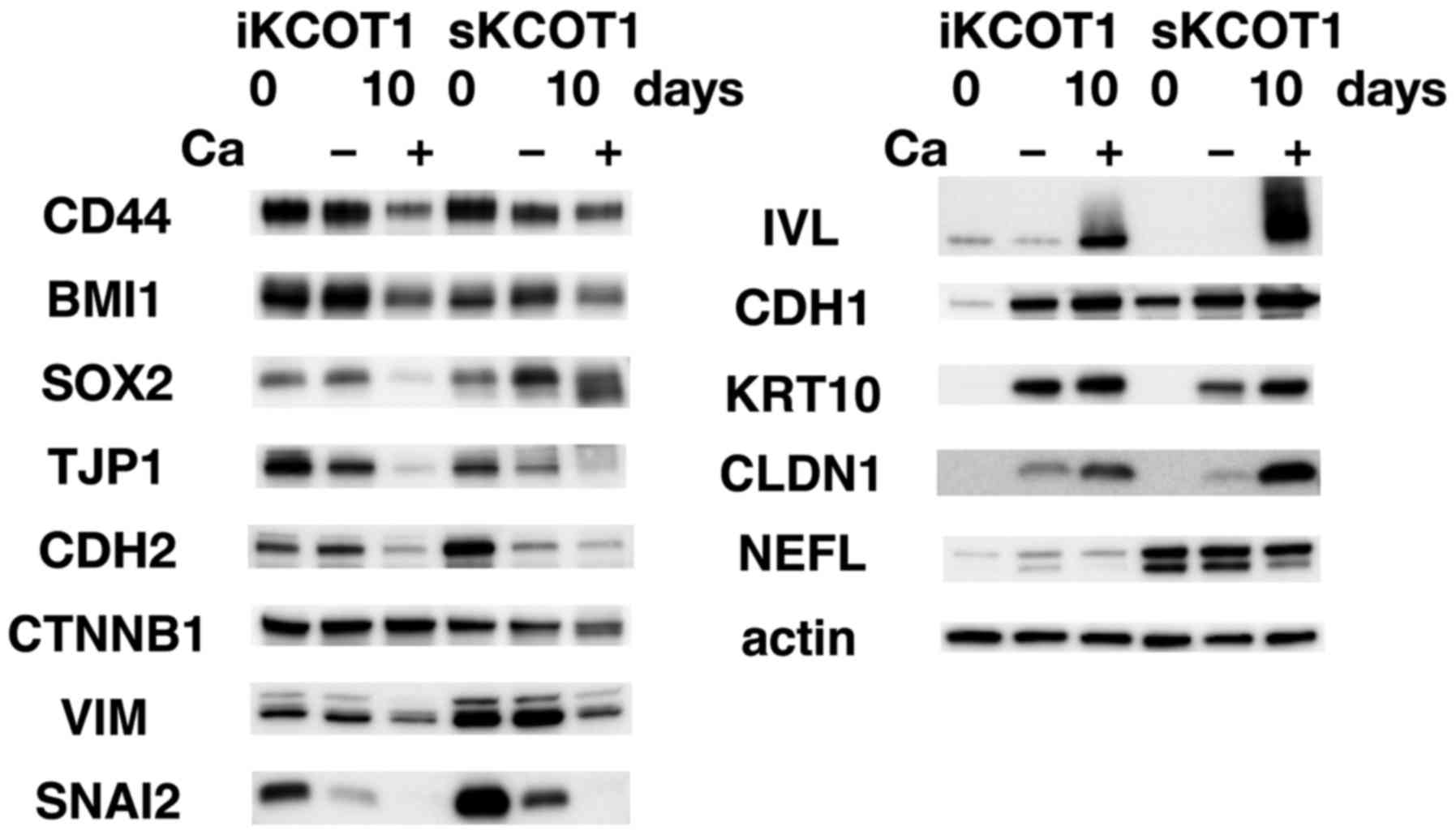 | Figure 4Protein expression in KCOT cell
lines. Western blot analysis of KCOT cells grown in low (0.65 mM)
or high (1.23 mM) calcium medium (indicating − or + respectively)
for 10 days. Stem cell markers, CD44, SOX2 and BMI1, were
expressed; however, their levels were downregulated by the presence
of calcium. Protein levels of mesenchymal-associated proteins, such
as CDH2, VIM, CTNNB1 and SNAI2 were downregulated, whereas
epithelial-associated proteins, such as CDH1 and CLDN1, were
upregulated by the high calcium medium in both KCOT cell lines.
Keratinocyte differentiation markers IVL and KRT10 were
dramatically induced by the high calcium medium. |
Discussion
Recent studies have demonstrated that mutations in
Hh signaling components induce tumors in various types of cells.
However, Hh signaling, especially during tumor formation, is not
fully understood. BCNS is a very rare inherited disease caused by
the misregulation of Hh signaling (19). Activation of Hh signaling, mainly
due to the loss of PTCH1 function, causes benign and malignant
tumors in BCNS patients. Therefore, study of BCNS derived tumors
will provide clues to comprehend Hh-related tumors, especially
tissue and ethnic specificity. Our group has intensively analyzed
BCNS and its tumors (10,20,21).
Among BCNS-associated tumors, the study of KCOTs has been hindered
because of the lack of a suitable model system. We therefore
established sporadic and BCNS-derived KCOT cell lines and analyzed
their characteristics.
PTCH1 mutations and tumor formation
iKCOT1 from a BCNS patient and sKCOT1 from a
non-BCNS sporadic patient have mutations in PTCH1. iKCOT1
cells have a germline mutation of c.584+2T>G (10) in one allele but we found no
additional mutations on the other allele. RT-PCR analysis showed
the existence of normal PTCH1 transcripts indicating that one of
the PTCH1 alleles appears to be wild-type.
Tumors occurring in Japanese BCNS patients are
usually KCOTs, and BCCs are sometimes observed in older patients
(10). BCC occurs at a low rate in
African-American patients, but at a high rate (up to 97%) in
Caucasian patients with BCNS (6).
Differences in type and onset of symptoms in BCNS patients from
different ethnic populations clearly demonstrate the participation
of other factor(s) besides PTCH1 in BCNS phenotypes. Data from BCNS
patients with BCC support Knudson's two-hit hypothesis (22,23),
a theory of oncogenesis stipulating that normal cells require two
mutagenic hits (two distinct episodes of DNA damage) to generate
cancer. In BCNS, various benign and malignant tumors, such as BCC,
KCOT, medulloblastoma and ovarian fibromas, exhibit loss of
heterozygosity (LOH) (3,24). Pan et al reported that not
all KCOTs in BCNS-derived as well as sporadic KCOT had two
PTCH1 mutations (25). Some
KCOTs, as well as the sKCOT1 cell line, carry loss of function
mutations in both PTCH1 alleles. Previously, we reported
PTCH1 mutations in both alleles of a BCNS-derived ovarian
fibroma case (20). Therefore,
loss of PTCH1 functions itself does not confirm tumor
malignancy. Further genome-wide analysis of KCOTs will elucidate
additional mutations or alterations that are involved in the
malignancy of the tumor. Retinoblastoma is a malignant eye cancer
that is common in childhood. It is known that loss of function of
both copies of the tumor suppressor RB1 gene induces
retinoblastoma. However, why depletion of the RB1 gene does
not induce other types of cancers is not well understood. Xu et
al showed that only cone precursor cells are sensitive to the
Rb defect in mice and that a cell type-specific gene network
is critical for the formation of a tumor (26). In BCNS patients or
Ptch+/− mice, PTCH1 function is halved
throughout the body; however, only specific cells or tissues show
tumor formation. These data indicated that in addition to the
depletion of PTCH1 function, cell type-specific gene
networks may specify the sensitivity of cells and the development
of tumors. Next-generation sequencing will elucidate the genes
involved in tumor formation and gene expression networks that are
essential for cell type restricted tumor formation.
KCOT cells differentiate in response to
high calcium conditions
KCOT cell characteristics were examined by
immunofluorescence and western blot analyses (Figs. 1B and 4). Both cell lines expressed stem cell
markers, such as CD44, BMI1 and SOX2, and mesenchymal cell markers,
including CDH2, VIM and SNAI2. However, levels of these proteins
were decreased under high calcium conditions. Conversely, levels of
epithelial cell markers, CDH1 and CLDN1, and also the keratinocyte
markers, KRT10 and IVL were dramatically increased (Fig. 4). These data suggested that both
cell lines have a mesenchymal stem cell-like character, and high
calcium conditions induced differentiation of epithelial
characteristics. Grachtchouk et al showed that disrupting
normal Hh signaling at the epithelial cell rests of Malassez (ERM)
induced KCOTs (14). Gao et
al induced KCOTs from ERM by disrupting TGF-β signaling
(27). ERM are odontogenic
epithelial cells in the periodontal ligaments and are normally
quiescent and predicted to work during periodontal regeneration
(28,29). ERM are known to express several
proteins that are usually associated with mesenchymal cells
(28). Therefore, ERM are a
candidate for the origin of KCOTs. Recent analysis reveals that ERM
have stem cell-like properties with capacities to undergo
epithelial-mesenchymal transition (EMT) and to differentiate into
mesenchymal lineages (29). Cells
in which proliferation or differentiation activity is activated or
repressed by abnormal Hh or TGF-β signaling in ERM may be an origin
of KCOTs. Therefore, ERM-derived KCOT cells may also have stem cell
and mesenchymal properties. Alternatively, KCOT may be a result of
EMT of ERM cells and a high calcium concentration induces reverse
transition of KCOT cells from mesenchymal cells to epithelial-like
cells. One interesting feature of these cell lines was the
expression of NEFL, which is characteristic for neuronal cells
(Fig. 4). This may be due to the
mesenchymal stem cell-like characteristics of these cell lines.
Recently, genome-wide expression analysis of fresh Ameloblastoma
and KCOT samples were carried out, the KCOTs are distinguished by
elevated levels of expression of epithelial differentiation
markers, and stem cell marker SOX2 (30). These expression profiles partially
support our KCOT cell lines as retaining original KCOT
characteristics.
In conclusion, we have established immortalized KCOT
cell lines, and have developed an in vitro culture model of
KCOT. Our experimental models may facilitate further studies to
understand the genesis of syndromic and sporadic KCOTs and provide
a unique resource to elucidate unsolved questions.
Acknowledgments
We thank all patients, their families and
collaborating doctors for participating in this study. We also
thank the Joint-Use Research Facilities at Hyogo College of
Medicine. This study was partly supported by Grants-in-Aid for
Research on Intractable Disease from the Ministry of Health, Labour
and Welfare, Japan (no. H22-120 to K.N.), and by JSPS KAKENHI
(grant nos. 24592854 and 15K11098 to Y.N. and nos. 25463123 and
16K11737 to K.N.), also supported by Grant-in Aid for Researchers,
Hyogo College of Medicine (to Y.N.) 2015.
References
|
1
|
Morgan TA, Burton CC and Qian F: A
retrospective review of treatment of the odontogenic keratocyst. J
Oral Maxillofac Surg. 63:635–639. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Philipsen HP and Reichart PA:
Classification of odontogenic tumours. A historical review J Oral
Pathol Med. 35:525–529. 2006. View Article : Google Scholar
|
|
3
|
Shear M: The aggressive nature of the
odontogenic keratocyst: Is it a benign cystic neoplasm? Part 1.
Clinical and early experimental evidence of aggressive behaviour.
Oral Oncol. 38:219–226. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
González-Alva P, Tanaka A, Oku Y,
Yoshizawa D, Itoh S, Sakashita H, Ide F, Tajima Y and Kusama K:
Keratocystic odontogenic tumor: A retrospective study of 183 cases.
J Oral Sci. 50:205–212. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cohen MM Jr: Nevoid basal cell carcinoma
syndrome: Molecular biology and new hypotheses. Int J Oral
Maxillofac Surg. 28:216–223. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kimonis VE, Goldstein AM, Pastakia B, Yang
ML, Kase R, DiGiovanna JJ, Bale AE and Bale SJ: Clinical
manifestations in 105 persons with nevoid basal cell carcinoma
syndrome. Am J Med Genet. 69:299–308. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Johnson RL, Rothman AL, Xie J, Goodrich
LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH Jr,
et al: Human homolog of patched, a candidate gene for the basal
cell nevus syndrome. Science. 272:1668–1671. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ingham PW, Nakano Y and Seger C:
Mechanisms and functions of Hedgehog signalling across the metazoa.
Nat Rev Genet. 12:393–406. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wicking C, Shanley S, Smyth I, Gillies S,
Negus K, Graham S, Suthers G, Haites N, Edwards M, Wainwright B, et
al: Most germ-line mutations in the nevoid basal cell carcinoma
syndrome lead to a premature termination of the PATCHED protein,
and no genotype-phenotype correlations are evident. Am J Hum Genet.
60:21–26. 1997.PubMed/NCBI
|
|
10
|
Fujii M, Noguchi K, Urade M, Muraki Y,
Moridera K, Kishimoto H, Hashimoto-Tamaoki T and Nakano Y: Novel
PTCH1 mutations in Japanese Nevoid basal cell carcinoma syndrome
patients: Two familial and three sporadic cases including the first
Japanese patient with medulloblastoma. J Hum Genet. 56:277–283.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qu J, Yu F, Hong Y, Guo Y, Sun L, Li X,
Zhang J, Zhang H, Shi R, Chen F, et al: Underestimated PTCH1
mutation rate in sporadic keratocystic odontogenic tumors. Oral
Oncol. 51:40–45. 2015. View Article : Google Scholar
|
|
12
|
Kimi K, Ohki K, Kumamoto H, Kondo M,
Taniguchi Y, Tanigami A and Ooya K: Immunohistochemical and genetic
analysis of mandibular cysts in heterozygous ptc knockout mice. J
Oral Pathol Med. 32:108–113. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Svärd J, Heby-Henricson K, Persson-Lek M,
Rozell B, Lauth M, Bergström A, Ericson J, Toftgård R and Teglund
S: Genetic elimination of suppressor of fused reveals an essential
repressor function in the mammalian hedgehog signaling pathway. Dev
Cell. 10:187–197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grachtchouk M, Liu J, Wang A, Wei L,
Bichakjian CK, Garlick J, Paulino AF, Giordano T and Dlugosz AA:
Odontogenic keratocysts arise from quiescent epithelial rests and
are associated with deregulated hedgehog signaling in mice and
humans. Am J Pathol. 169:806–814. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ren C, Amm HM, DeVilliers P, Wu Y,
Deatherage JR, Liu Z and MacDougall M: Targeting the sonic hedgehog
pathway in keratocystic odontogenic tumor. J Biol Chem.
287:27117–27125. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu X, Ory V, Chapman S, Yuan H, Albanese
C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, et al:
ROCK inhibitor and feeder cells induce the conditional
reprogramming of epithelial cells. Am J Pathol. 180:599–607. 2012.
View Article : Google Scholar :
|
|
17
|
Sasaki R, Narisawa-Saito M, Yugawa T,
Fujita M, Tashiro H, Katabuchi H and Kiyono T: Oncogenic
transformation of human ovarian surface epithelial cells with
defined cellular oncogenes. Carcinogenesis. 30:423–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamamura M, Noguchi K, Nakano Y, Segawa E,
Zushi Y, Takaoka K, Kishimoto H, Hashimoto-Tamaoki T and Urade M:
Functional analysis of Zyxin in cell migration and invasive
potential of oral squamous cell carcinoma cells. Int J Oncol.
42:873–880. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Athar M, Li C, Kim AL, Spiegelman VS and
Bickers DR: Sonic hedgehog signaling in Basal cell nevus syndrome.
Cancer Res. 74:4967–4975. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jimbo T, Masumoto K, Urita Y, Takayasu H,
Shinkai T, Uesugi T, Gotoh C, Sakamoto N, Sasaki T, Oto T, et al:
Nevoid basal cell carcinoma syndrome with a unilateral giant
ovarian fibroma in a Japanese 6-year-old girl. Eur J Pediatr.
173:667–670. 2014. View Article : Google Scholar
|
|
21
|
Takahashi C, Kanazawa N, Yoshikawa Y,
Yoshikawa R, Saitoh Y, Chiyo H, Tanizawa T, Hashimoto-Tamaoki T and
Nakano Y: Germline PTCH1 mutations in Japanese basal cell nevus
syndrome patients. J Hum Genet. 54:403–408. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Knudson AG Jr: Mutation and cancer:
Statistical study of retinoblastoma. Proc Natl Acad Sci USA.
68:820–823. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Levanat S, Gorlin RJ, Fallet S, Johnson
DR, Fantasia JE and Bale AE: A two-hit model for developmental
defects in Gorlin syndrome. Nat Genet. 12:85–87. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barreto DC, Gomez RS, Bale AE, Boson WL
and De Marco L: PTCH gene mutations in odontogenic keratocysts. J
Dent Res. 79:1418–1422. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pan S, Dong Q, Sun LS and Li TJ:
Mechanisms of inactivation of PTCH1 gene in nevoid basal cell
carcinoma syndrome: modification of the two-hit hypothesis. Clin
Cancer Res. 16:442–450. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu XL, Singh HP, Wang L, Qi DL, Poulos BK,
Abramson DH, Jhanwar SC and Cobrinik D: Rb suppresses human
coneprecursor-derived retinoblastoma tumours. Nature. 514:385–388.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao Y, Yang G, Weng T, Du J, Wang X, Zhou
J, Wang S and Yang X: Disruption of Smad4 in odontoblasts causes
multiple keratocystic odontogenic tumors and tooth malformation in
mice. Mol Cell Biol. 29:5941–5951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rincon JC, Young WG and Bartold PM: The
epithelial cell rests of Malassez - a role in periodontal
regeneration? J Periodontal Res. 41:245–252. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiong J, Mrozik K, Gronthos S and Bartold
PM: Epithelial cell rests of Malassez contain unique stem cell
populations capable of undergoing epithelial-mesenchymal
transition. Stem Cells Dev. 21:2012–2025. 2012. View Article : Google Scholar
|
|
30
|
Heikinheimo K, Kurppa KJ, Laiho A,
Peltonen S, Berdal A, Bouattour A, Ruhin B, Catón J, Thesleff I,
Leivo I, et al: Early dental epithelial transcription factors
distinguish ameloblastoma from keratocystic odontogenic tumor. J
Dent Res. 94:101–111. 2015. View Article : Google Scholar
|