Breast cancer is the most common and aggressive
tumor type affecting women. The typical characteristics of the
majority of patients with breast cancer are that they express
receptors for estrogen (ER) and progesterone receptor (PR) and
respond to hormonal therapy (1).
Hormonal therapy, chemical therapy and radiotherapy have led to
increased survival rates. However, the occurrence rate of breast
cancer has still increased. Triple-negative breast cancer (TNBC),
characterized by the lack of ER, PR and human epidermal growth
factor receptor (HER)-2 (ErbB-2, C-erbB2 or Her2/neu) expression,
is insensitive to hormonal therapy, chemotherapy and radiotherapy,
and is more likely to result in anticancer resistance. Therefore,
the development of novel therapeutic strategies is critical for
these patients. Recent studies have suggested that breast cancer
stem cells (CSCs), a small subpopulation of cells, have the ability
to self-renew and differentiate into the entire tumor. They are
resistant to chemotherapy and radiotherapy, and can result in tumor
recurrence, even following surgery (1). Hence, there is a need for the
development of novel strategies that target these cells. Cell death
is a complex phenomenon in multicellular organisms, which can occur
under both physiological and pathological conditions. Apoptosis,
autophagy and necrosis are three common cell death processes. It is
generally recognized that necrosis is a form of non-programmed cell
death and is independent of the caspase family. Apoptosis and
autophagy are programmed cell death pathways. The 2016 Noble Prize
for Physiology or Medicine was awarded to Dr Yoshinori Ohsumi, due
to his work on the mechanisms of autophagy. Autophagy, a conserved
catabolic pathway, can promote cell homeostasis during nutrient
deprivation (2). Recent data have
illustrated that mutations in genes involved in autophagy play a
critical role in the pathogenesis of diverse diseases, including
cancer (3–6). Specifically, autophagy has been shown
to be associated with CSC differentiation. However, the functional
importance of autophagy remains unclear in breast cancer and CSCs.
Thus, the present review aimed to elucidate the role and related
mechanisms of autophagy in breast cancer and breast CSCs. This
topic may aid in the development of a novel therapeutic strategy
for breast cancer in the future.
Three main types of autophagy are currently
recognized: Micro, macro and chaperone-dependent autophagy. These
processes are made possible due to specific enzymes and
autophagy-related genes (ATGs) (7). Within this review, the term autophagy
refers to macroautophagy, the process through which unnecessary or
dysfunctional cellular components are removed via the union of
lysosomes and autophagosomes to create autolysosomes. Autophagy
levels are usually lower under normal conditions compared with
starvation or nutrition-deficiency conditions. Autophagy can only
be induced extensively by internal or external stimulation. It can
be notably inhibited by knocking down ATGs, which leads to the
damaged proteins and cell organelles, such as the mitochondria, not
being removed, thus creating a toxic environment that can affect
the survival of normal cells. In addition, autophagy provides a
supply of metabolic precursors for macromolecular synthesis during
nutrient stress. The cytosolic targets of autophagy include
proteins, lipid droplets, glycogen, granules, nucleic acids and
whole organelles, such as the mitochondria. Autophagy also plays a
critical role in sustaining ATP production while under energy
failure.
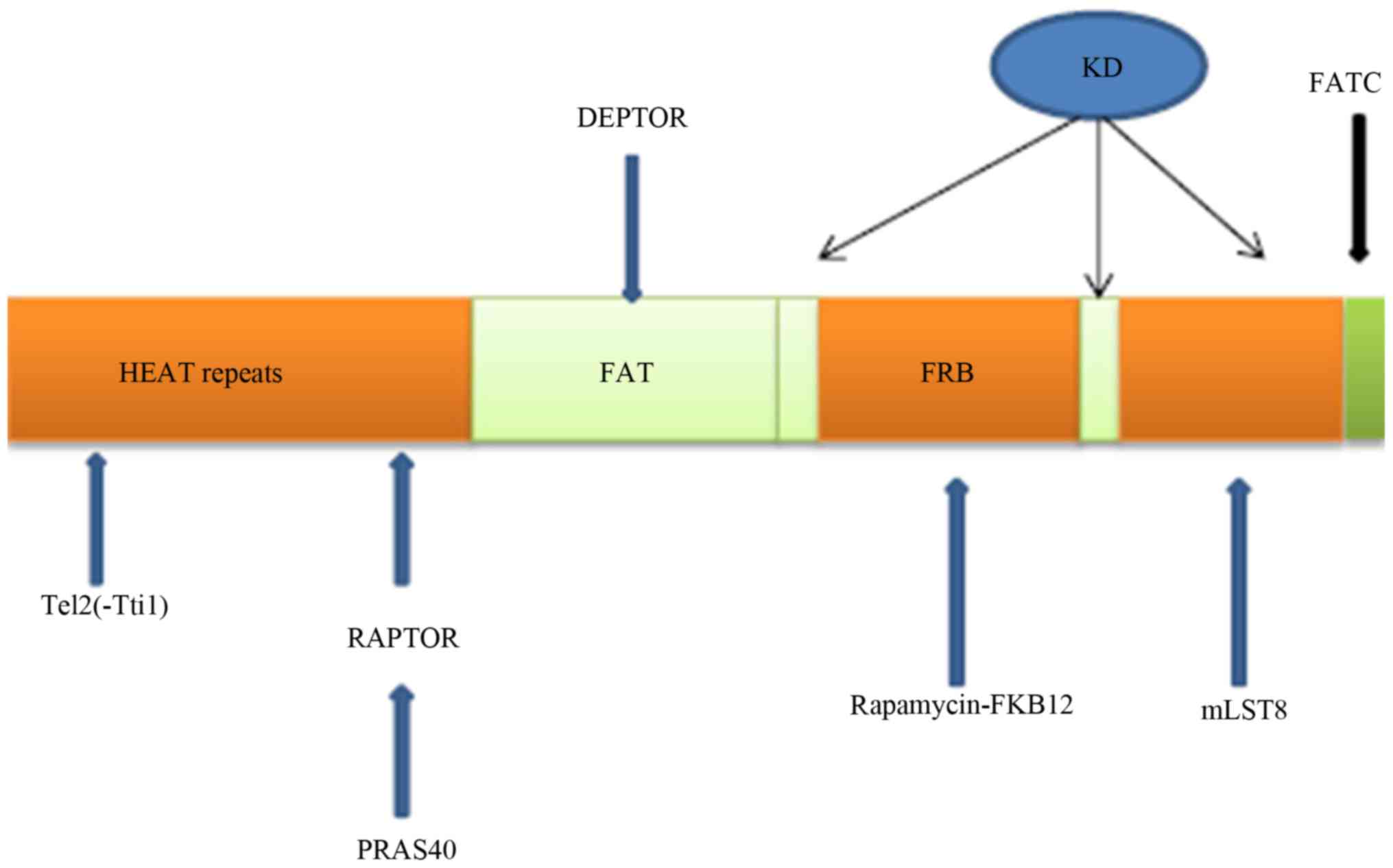
Residues 1-1,375 of the mammalian target of
rapamycin (mTOR) are relatively undefined compared with its other
sections; however, the N-terminal part is mainly formed by HEAT
repetitions (8). The well-defined
structure of mTOR consists of the FAT/REB/kinase/FATC domains
(Fig. 1). The kinase domain (KD)
binds ATP within itself, whereas the FRB domain binds
rapamycin-FKB12 within itself. The components of the well-defined
parts of mTOR are kinase (joined by ATP), FATC, REB and FAT
(9,10). The KD serves as a joint between
mTOR and mLST8, playing a considerable role in stabilizing the
reaction between mTOR and the receptor (11). Tti1 and Tel2 form a supporting
platform for mTOR function, and that of many other compounds. The
Tel2 part binds to HEAT repeat regions in mTOR (12,13).
DEPTOR binds to mTOR at the FAT domain and inhibits the mTOR
function (14). mTOR is part of
TORC1 and TORC2, which are both protein structures. Therefore,
there are many molecules that are able to affect mTOR function.
The first regulatory stage of autophagy is the
de-repression of mTOR. TORC1 (hereafter referred to as the TOR
complex) is sensitive to rapamycin. The physiological consequences
of mammalian TORC1 deregulation demonstrated that the inhibition of
mTOR may be useful in the treatment of cancer (15). mTORC2 can regulate autophagy via
Akt-FoxO3 (16,17). mTOR consists of the mTOR catalytic
subunit, including the regulatory-associated protein of mTOR
(Raptor), the proline-rich AKT substrate of 40 kDa (PRAS40) and the
G protein β-subunit-like protein (GβL), also known as mLST8
(Fig. 1).
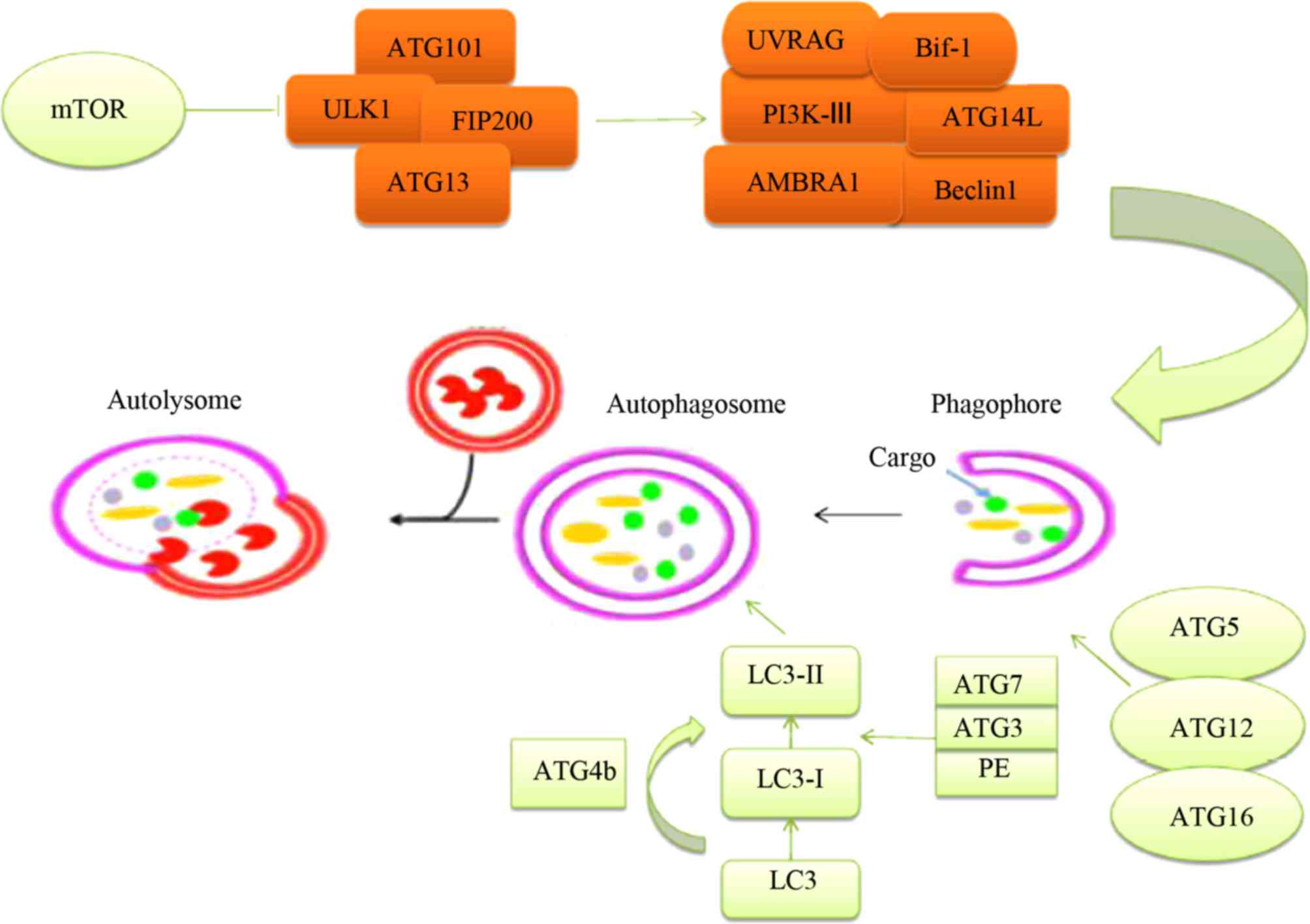
Structures, such as Unc-51-like kinase 1 (ULK1) and
FAK-family interacting protein of 200 kDa (FIP200) are formed due
to the activation of autophagy-related proteins, which occurs
following the inhibition of mTOR (Fig.
2) (18). GTPases can promote,
in the activated areas, the departure of mTOR to the external
section of the lysosome. Vacuolar ATPase enriches activities
associated with amino acids as a supportive function, binding
lysosomes and GTPases (19–21).
Inactive mTORC1 does not generate a matrix, such as the
transcription factor EB phosphorylation, being able to regulate ATG
functions when moving to the nucleus (22).
The phagophore is dependent on the combined action
of a number of factors, including Beclin1, UV radiation resistance
associated (UVRAG), Bif-1, autophagy and beclin 1 regulator 1
(AMBRA1), as well as others in its viral version (Fig. 2). The function of the class-III
PI3K is critical for vesicle nucleation. ATGs are equally important
and control the elongation of vesicles. Two protein structural
complexes are required to achieve an increase in the autophagosome
membrane: ATG5-ATG12-ATG16 and LC3-PE. These complexes can
transform LC3-I to LC3-II in the cytosol, particularly in the
autophagosome (23). LC3 can be
converted into LC3-I by joining with ATG4, and thereafter LC3-I can
be converted into LC3-II by joining ATG7, ATG3 and PE
(phosphatidylethanolamine). ATG10 can replace ATG7 in being bound
to ATG5, and this enables ATG5 to bind to ATG12. LC3-II is the
active form of LC3-I, and can attach to the phagophore
membrane.
Certain pathways can regulate mTOR, including
adenosine monophosphate-activated protein kinase (AMPK) and
PI3K/AKT. mTOR can be considered as a part of the PI3K pathway,
which is significant to autophagy. mTOR exerts a crucial effect on
autophagy as a downstream component in the signaling pathway of
PI3K. Under standard conditions, phosphorylated PI3K phosphorylates
AKT, which inhibits tuberous sclerosis complex 1/2 (TSC1/2) and
then activates mTOR. Subsequently, mTOR mediates the expression of
several autophagy proteins, ultimately suppressing the autophagy
process. AMPK plays a fundamental role as a catalyst in the
regulation of energy in a cell. It is activated when intracellular
ATP levels become lower than normal. Autophagy can be regulated
through the AMPK pathway in breast cancer, and inhibition of the
AMPK signaling pathway affects the process of autophagy in breast
cancer (24). Activated AMPK
activates autophagy by targeting the ULK1 complex.
Cells in the TME, such as endothelial cells,
fibroblasts or even mesenchymal stem cells, are thought to regulate
breast CSCs, when they secrete different signaling molecules
associated with survival, proliferation or differentiation
(30). The constant appearance of
CSCs in breast cancer has been associated with signaling pathways
associated with embryonic development, such as the Notch and
hedgehog pathways. These proteins promote cellular reproduction and
specialization through signaling routes associated with paracrine
and autocrine processes (31).
Sonic, Indian and Desert are the three types of hedgehog gene in
mammals (32). The activation of
the smoothened transmembrane protein is promoted by interactions
between hedgehog proteins and the Patched transmembrane protein
(33). CSCs appear to be sustained
due to the hedgehog pathway. Proteins of the Notch type have four
transmembrane glycoproteins and five ligands (34). This signaling route is associated
with the reproduction and specialization of cells (35). The Notch pathway is activated due
to the release of its intracellular domain through the nucleus by
proteolytic cleavage, and interaction with receptor extracellular
domains. In WNT/β-catenin signaling, the transcriptional activator,
β-catenin, is maintained at low levels through continuous
degradation. Once WNT binds to the Frizzled receptors, β-catenin is
then released from the multi-protein destruction complex (including
APC and GSK-3β) and translocates from the cytoplasm to the nucleus,
thus activating several proliferation-related target genes
(32). During development, it
controls the fate of the cells; however, in adults it supports
tissue self-renewal.
Autophagy is a key factor in cell growth; therefore,
the inhibition of autophagy can decrease tumor growth.
Cancer-associated fibroblasts (CAFs) play an essential role in
malignant cancer progression. Beclin1 and LC3-II/I protein
conversion levels in CAFs are higher than those in normal
fibroblasts (NFs). CAF autophagy can enhance triple-negative breast
cancer (TNBC) cell proliferation (36). In addition, Research has been
indicated that anticancer therapy may induce autophagy (37), affecting tumor cell proliferation.
Therefore, the combination of autophagy inhibition and chemotherapy
may become an effective therapy for breast cancer (37). Chloroquine (CQ), an autophagy
inhibitor, removes CSCs via an epigenetic mechanism by altering DNA
methylation. CQ causes mitochondrial damage, resulting in excessive
oxidative DNA damage and subsequent cell death in TNBC CSCs
(38).
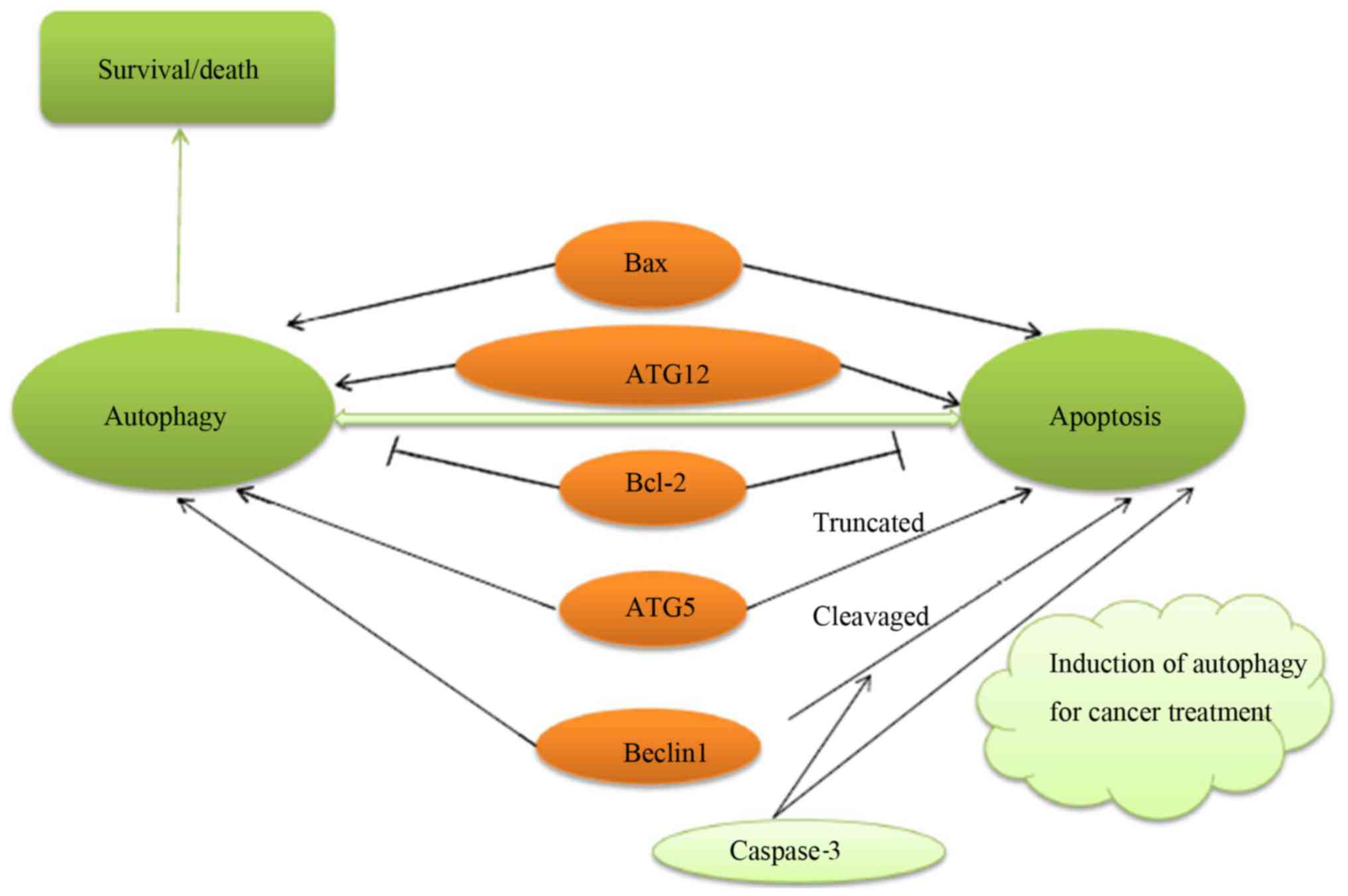
The characteristics of type I cell death include a
reduction in cell volume, the destruction of the cellular
cytoskeleton, the condensation and fragmentation of nuclear
content, and the formation of apoptotic bodies that are disposed by
phagocytes. There are two different pathways that can mediate
apoptosis: The receptor pathway and the mitochondrial pathway. It
has been indicated that many types of cancer cells may lack
death-associated protein kinase (DAPK). Phosphorylated Beclin1 can
induce the release of Beclin1 from Bcl-2-related proteins and
induce autophagy. The protein cleaved by caspases, Bcl-2, is a
negative regulator of Beclin1. This protein can inhibit autophagy
and enhance apoptosis. ATG5 is cleaved by calpains to produce an
N-terminal ATG5 cleavage product, which is believed to induce
cytochrome c release from mitochondria. Full-length Beclin1
and ATG5 oppose this process by regulating the autophagic
degradation of active caspase-8 (39). This suggests that autophagy and
apoptosis may have the same regulatory factors (Fig. 3).
Cell death and survival are notably complex
processes, and both autophagy and apoptosis play significant roles.
Based on the aforementioned analysis, we know that there are common
regulatory factors between autophagy and apoptosis. In addition,
Yousefi et al reported that knocking down ATGs decreased
sensitivity to anticancer therapy (40). Lu et al discovered that the
inhibition of autophagy enhanced the apoptosis induced by
parthenolide in breast cancer (24). Consequently, there is a close
association between autophagy and apoptosis. The investigation of
this association is critical for the understanding of the
mechanisms of autophagy in breast cancer and CSCs.
Invasion and metastasis are important
characteristics of tumors and also increase the risk of cancer.
Autophagy provides a survival mechanism that endowes cancer cells
with metabolic flexibility, allowing for their survival in
nutrient- and oxygen-poor TMEs. Cancer cells tend to metastasize
under nutrient- and oxygen-poor conditions. Studies using surrogate
markers have identified a close association between increased
autophagy and metastasis. Increased punctae staining for
microtubule-associated light chain B (LC3B) has been shown to be
associated with lymph node metastasis and decreased survival in
human breast cancer, whereas melanoma metastases demonstrated
increased LC3B staining compared with matched primary tumor samples
(4). Moreover, in response to
hypoxia and metabolic stress, necrosis frequently occurs in
carcinoma in situ, which can enhance inflammation and promote
inflammatory cell infiltration, which favors prometastatic immunity
(41,42). Autophagy effectively suppresses
necrosis and subsequent inflammatory cell infiltration from tumor
sites by providing energy and metabolic precursors, and restrains
metastasis. Based on this hypothesis, there is a complex
interaction between autophagy and metastasis.
Epithelial-to-mesenchymal transition (EMT) is important for cancer
spreading and dissemination. Two primary regulators of EMT, Snail
and Slug, which can promote EMT by inducing the loss of epithelial
(E)-cadherin-mediated cell-cell adhesion, are associated with
autophagy. Autophagy deficiency has been discovered to promote EMT
by stabilizing Twist1 (43).
Recently, some studies on autophagy and metastasis have identified
novel therapeutic methods. For example, the downregulation of
macroautophagy associated with ATG5 by chaperone-regulated
autophagy was identified to promote breast cancer cell metastasis
(44).
Autophagy plays a significant role in all stages of
cancer development. Recently, it was reported that autophagy is a
'double-edged sword'; it can inhibit cancerous growth, and can also
suppress inflammation, tissue injury and chromatin instability in
the initial stages of cancer, which is mostly relative to the
formation of cancer. During other stages, autophagy may perform a
complex function depending on the internal and external context
(39). One of the typical features
of cancer is proliferation, which can promote metastasis and lead
to nutrient deficiency. Autophagy tends to induce the metastatic
process by maintaining and spreading cell survival, and causes the
cells to enter a dormant state if they cannot establish stable
contact with the extracellular matrix in the new environment
(39). However, in fully
transformed carcinoma cells, the deficiency of autophagy can
facilitate malignant tumor metastasis and neoplasia. Moreover, it
may also facilitate breast cancer progression, independent of
genotoxic stress and genomic instability. Kongara et al
(45) reported that a low
expression of Beclin1 was linked to phosphorus (Ser73) accumulation
in human breast cancer. When cancer cells are able to be placed
under conditions with metabolic and genotoxic stress due to
progression and metastasis, and therapy is accepted, autophagy
shifts to tumor-promoting mechanisms by maintaining the survival of
the tumor cells (46). Hypoxia and
acidic material can lead to metabolic pressure, while chemotherapy
and radiotherapy lead to genotoxic pressure. Autophagy can decrease
sensitivity to anticancer treatments, eliminate organelle damage,
DNA fragmentation, and preserve the integrity of the cells. Based
on these effects, the inhibition of autophagy can enhance the
sensitivity to anticancer therapy.
In mice with autophagy dysfunction, damaged
mitochondria and complexes with p62 and ubiquitinated proteins
accumulate, which can result in the massive generation of reactive
oxygen species (ROS), and can lead to DNA mutation and chromatin
condensation (47 and refs therein). Since genetic damage can
increase the risk of tumor occurrence, the gene protection
mechanism regulated by autophagy is likely to involve the
inhibition of tumor development. Under continuous stimulation,
tumor cells with autophagy deficiency often fail to sustain their
survival processes, leading to chronic death. The chronic death of
cells causes an inflammatory response that can further promote the
development of cancer. Autophagy dysfunction may induce necrosis
and inflammation through the release of pro-inflammatory factors.
For example, high mobility group protein B1 (HMGB1), generated by
necrotic cells, binds to cell surface receptor of advanced
glycation end products (RAGE), which can activate transcription
factor nuclear factor (NF)-κB and cause inflammation (48). Similarly, nucleic acids released
from necrotic cells stimulate the inflammatory response by
activating Toll-like receptors (49). Additionally, excessive autophagy
can lead to autophagic death. Under continuous stress and
progressive autophagy, cells die due to self-consumption (50), following the high expression of
Beclin1 and massive autophagosomes.
Autophagy is considered to be a conserved cellular
process, consisting of the response of cells to various external
stimuli, such as radiation and the low concentration of oxygen or
nutrients. Morever, CSCs are considered to depend on their
surroundings to maintain development and growth. Autophagy is also
activated by environmental factors, as aforementioned and it
affects the surrounding tumor environment, regulating the activity
between tumor cells and stromal components, such as fibroblasts,
immune cells and endothelial elements. The inhibition of autophagy
has been demonstrated to enhance the sensitization of tumor cells
to treatment; however, this has not been clearly demonstrated for
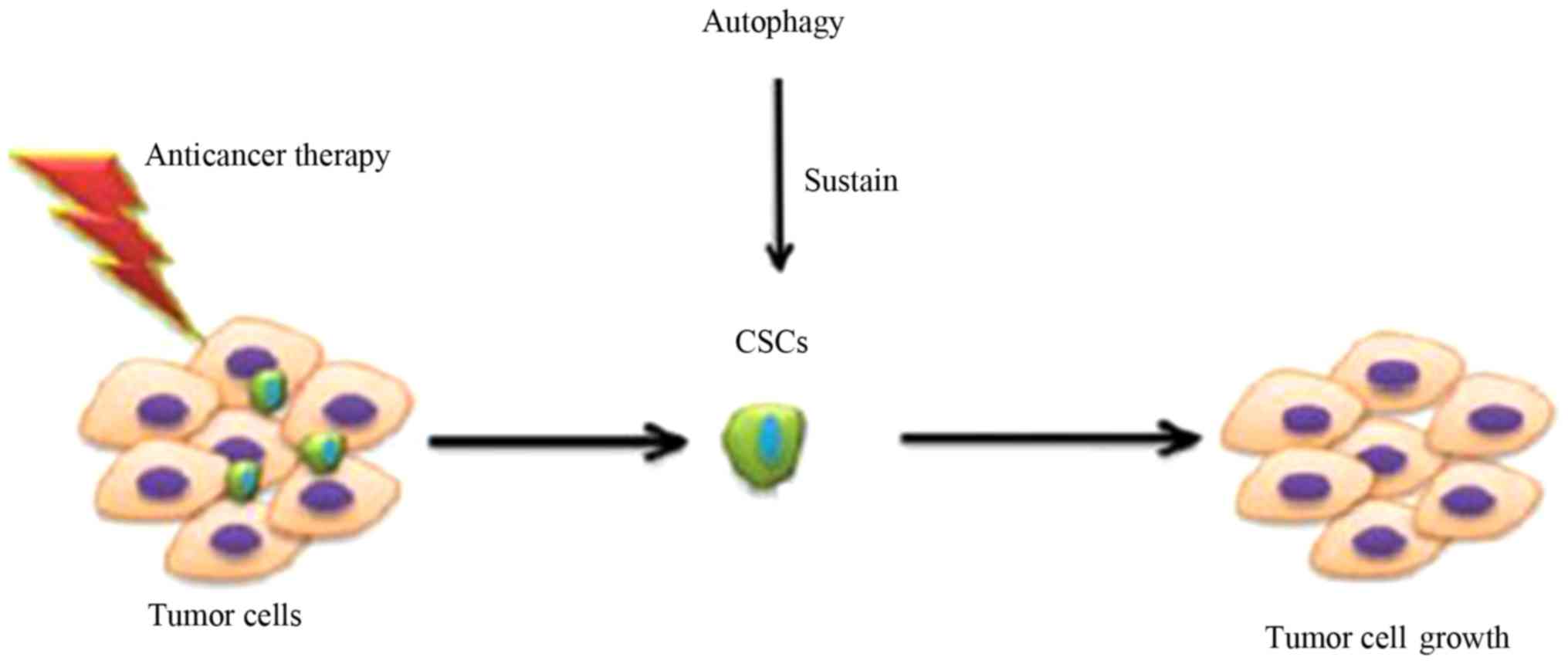
CSCs. In recent studies, the de-activation of autophagy-related
genes in order to inhibit autophagy was observed to decrease the
effects of CSCs (63,65) (Fig.
4). Combining this method with traditional cancer therapies can
improve the rate of cancer elimination and curative treatment. It
has been demonstrated that autophagy can drive CSC levels in breast
cancer (51–53), and that some ATGs, such as Beclin1
and ATG4A, serve a function in sustaining breast CSCs. This is
consistent with the theory that autophagy plays a supportive role
in CSCs (52,53).
Furthermore, certain studies have focused on breast
CSC treatment. Autophagy may play a dual role in the therapeutic
process of targeting breast CSCs. For example, Zhao et al
discovered that autophagy played a positive role, increasing the
sensitivity of glioma stem cells to X-ray radiation (54). Autophagy induced by Rottlerin can
promote breast CSC apoptosis (55,56).
However, Wei et al found that autophagy inhibited colon CSC
apoptosis induced by photodynamic therapy (57). In breast cancer, the inhibition of
autophagy can decrease the drug-resistance of breast CSCs, as well
as reduce the ability of breast CSCs to form tumors (55,58).
Therefore, further research is required to elucidate the complex
association between autophagy and breast CSCs.
Autophagy plays an important role in breast cancer
and breast CSCs, and is controlled by ATGs. Knowing more about the
roles of ATGs may be crucial to improving the effectiveness of
breast cancer therapy. There are some molecules that can regulate
breast cancer, as well as breast CSCs (Table I).
In addition, there are certain pathways that are
associated with breast CSCs, which can regulate cellular
differentiation and survival and are closely associated with tumors
(61). Recent research has
indicated that the downstream target of AKT, mTOR, is also
essential to breast CSCs (79).
Based on these findings, mTOR is a key control point for autophagy.
Thus, the PI3K/AKT signaling pathway may play an important role in
the autophagy of breast CSCs. Transforming growth factor (TGF)-β
can be mediated by SMAD2/SMAD3, resulting in the regulation of
autophagy. The Wnt signaling pathway may be linked to autophagy,
and the inhibition of the Wnt signaling pathway can induce
autophagy. Via transcription factor 4 (TCF4), β-catenin can
suppress the formation of autophagic vacuoles, and suppress the
expression of p62/SQSTM1 (80).
The overexpression of LC3-II and Beclin1 is contrary to the role of
the Notch signaling pathway. The inhibition of the hedgehog pathway
exerts the same effects as those induced by the inhibition of the
Wnt and Notch pathway (81).
Autophagy may protect the anticancer resistance
response of breast cancer cells and maintain the activity of breast
CSCs. In addition, autophagy can exert effects on the initiation,
proliferation and progression of tumors and CSCs. The inhibition of
autophagy can increase cancer cell death. Therefore, it is
important to increase our knowledge of inhibitors of autophagy.
There are some inhibitors of autophagy (Table II) that can exert an effect on
breast cancer and breast CSCs. Understanding their current stage in
research and development is essential for breast cancer therapy. In
addition, these compounds can suppress autophagy, which could be
critical to identifying new therapeutic mechanisms.
In addition, there are other medications that can
exert effects on autophagy, including eriocalyxin B (93), tetrandrine (94) and suberoylanilide hydroxamic acid,
which is also critical in breast cancer therapy (95). Based on our analysis, the majority
of inhibitors of autophagy may play an important role in both
breast cancer and breast CSCs. However, further research is
required in order to determine whether chloroquine or
3-methyladeninecan can directly inhibit breast cancer cell growth.
Therefore, more attention should be paid to the novel therapeutic
mechanisms of these autophagy-associated drugs.
Recent studies have revealed autophagy-dependent
pathways in different subtypes of breast cancer. It would thus be
helpful to discuss the role of autophagy in breast cancer
subtypes.
TNBC, a subgroup of tumors, do not express
clinically significant levels of ER, PR and HER2, and thus cannot
be treated with endocrine or anti-prognosis when compared to other
tumor subtypes. TNBC is believed to be invasive. Some studies have
reported that radiotherapy is effective for TNBC treatment,
although the treatment effects are limited (96,97).
There are a large amount of factors, including autophagy that can
lead to limited therapeutic effects. Thus, the development of novel
therapeutic strategies is essential for TNBC therapy. For example,
Liu et al demonstrated that the inhibition of autophagy led
to low levels of Chk1, which impaired the DNA repair capacity and
diminished the ability to repair DNA double-strand breaks via
homologous recombination (98).
Zhou et al demonstrated that MK-8776, an inhibitor of Chk1
increased the radiosensitivity of TNBC cell lines by inhibiting
autophagy in vitro. MK-8776 may have potential as a
radiotherapy sensitization agent (99). Insulin-like growth factor (IGF)-1
receptor (IGF-1R) activated by binding IGF-1 results in cell
proliferation, metastasis and drug resistance, and IGF-1R promotes
the survival and proliferation of TNBC cell lines (100). NVP-AEW541, an inhibitor of
IGF-1R, inhibits TNBC cell proliferation and induces autophagy. The
role of autophagy induced by NVP-AEW541 is unclear. However,
subsequent results have confirmed that the inhibition of autophagy
enhances NVP-AEW541-induced cell growth suppression and the
apoptosis of TNBC cells (101).
Recently, autophagy has been reported to function both as a tumor
suppressor mechanism and a survival mechanism, according to the
tumor cell context. Salt-inducible kinase 2 (a member of the AMPK
family; SIK2) is essential for survival, particularly in the
claudin-low subtype. It has been reported that SIK2 restrains the
autophagic flux to support TNBC survival (102). Furthermore, excessive autophagic
cell death may provide a novel therapeutic approach for cancer
therapy instead of the induction of apoptosis. Gao et al
demonstrated that small-molecule RL71 triggered excessive
autophagic cell death as a potential therapeutic strategy in TNBC
(103).
Luminal breast cancer (Luminal A, Luminal B)
accounts for 50–60% of all breast cancer cases. Although Luminal BC
is sensitive to anticancer therapy, a conspicuous proportion of
patients with breast will gradually develop resistance to
anticancer therapy and relapse. Autophagy is deemed to play a
critical role in response to resistance to therapy. For example,
adriamycin, a first-line chemotherapeutic agent, plays an essential
role in cancer. However, a high basal level of autophagy was
demonstrated in adriamycin-resistant MCF-7 cells, and the silencing
of TRPC5 (an inducer of autophagy) and the inhibition of autophagy
reversed the resistance to adriamycin (104). Resistin is a novel adipokine that
is upregulated in patients with breast cancer and promotes breast
cancer cell growth, invasion and migration. In a previous study,
resistin via an increased level of autophagy, inhibited the
doxorubicin-induced apoptosis of MCF-7 cells (105). In addition, autophagy seems to
play a dual role in luminal breast cancer, as well as TNBC.
Autophagic cell death is characterized by an extensive sequestrated
cytoplasm, resulting in cell death with the formation of
autophagosomes or autolysosomes. Autophagic cell death can inhibit
cancer development. Researches have shown that tetrandrine
increases the autophagic flux in MCF-7 cells, resulting in cell
death (94). The GABARAPL1 protein
belongs to the ATG8 family whose members are involved in autophagy.
GABARAPL1 acts as a tumor suppressor protein associated with
autophagic vesicles and regulating autophagic flux in breast cancer
(106).
HER2-positive breast cancer accounts for
approximately 20–30% of all breast cancer cases. Anti-human
epidermal growth factor 2 (HER2) therapy has been approved as a
standard practice for patients with HER2-positive breast cancer,
leading to the improvement of patient prognosis during the past
decade. However, this resistance to anti-HER2 treatment has been a
primary issue in clinical practice. Recently, autophagy has been
reported to be associated with resistance to therapy. For example,
autophagy protects from cytotoxicity induced by trastuzumab in
breast cancer with HER2 overexpression (107). Beclin1 (also known as Becn-1), an
autophagy-related gene, is important to initiate the phases of
autophagy. Researches have demonstrated that a deficiency of Becn-1
enhances the sensitivity to HER2-targeted therapy, implying that
the inhibition of autophagy in conjunction with HER2 inhibition is
critical for promoting tumor regression, and that autophagy
stimulation can transform the effectiveness of HER2 treatments
(108). In parallel with these
results, the knockdown ATG12 has been shown to suppress tumor
growth and to sensitize trastuzumab-resistant xenografts to
trastuzumab (77). The ability of
chloroquine to block autophagy by inhibiting lysosomal proteases
and preventing the fusion between autophagosomes and autolysosomes
has established chloroquine as the most widely used drug for the
inhibition of autophagy in vitro and in vivo.
Connecting chloroquine with trastuzumab-based regimens may
therefore improve outcomes among women with autophagy-addicted
HER2-positive breast cancer (109).
In addition, we summarized the inhibitors of
autophagy in different breast cancer cell lines (Table III). A recent study demonstrated
that the level of autophagy differed between different subtypes of
breast cancer. TNBC had substantially more autophagosomes than
other types of breast cancer. Basal autophagy was high and was not
influenced by chemotherapy in MDA-MB-231 cells. The expression of
LC3b was similar in the control group and chemotherapy group.
Compared with the MDA-MB-231 cells, basal autophagy was low in the
MCF-7 cells and increased with chemotherapy (110). Treatment with chloroquine may
lead to synergistic effects with chemotherapy. The combination
treatment between doxorubicin and chloroquine has exhibited
synergistic effects in TNBC compared with MCF-7 cells. TNBC cell
lines showed significantly higher levels of activated STAT3 by Tyr
phosphorylation than the luminal cell lines. These cells were
believed to be STAT3-dependent. The inhibition of autophagy
decreased STAT3 phosphorylation in TNBC cell lines, and this
reduction in STAT3 activity induced cell death. The inhibition of
autophagy was more effective in TNBC than luminal cell lines
(111).
In conclusion, autophagy is regulated by a complex
network of stress responses. The function of autophagy in breast
cancer remains unclear. Autophagy can be reduced at various
developmental and metastatic phases of breast cancer, and cn even
be a primary cell death pathway in some tumors with apoptosis
deficiency. By contrast, autophagy can effectively maintain the
existence of tumors under stimulation. Moreover, further research
is required to elucidate the association between autophagy and
breast CSCs. This review outlined the various elements and
processes that are involved in the association between breast CSCs
and autophagy. As these factors promote the development of tumors,
their comprehensive study under different circumstances and
environments is critical for the development of novel new treatment
strategies for breast cancer. Autophagy seems to play an effective
role in breast cancer and breast CSCs. However, its function is
linked to a number of other factors, including metabolic reactions,
immunoreactions and the TME. Therefore, our aim is to better
understand the autophagy-related molecular mechanisms and signaling
pathways, and to devote more attention to the association between
breast cancer, CSCs, and autophagy. It is hoped that this will
result in a meaningful strategy that could provide novel approaches
to breast cancer therapy.
The authors would like to apologize to the many
authors whose studies are important, but could not be cited due to
space limitations. We are grateful for the members of the
Department of Pathology of Dalian Medical University for their
helpful discussions and suggestions during the course of this
study.
The authors declare that they have no competing
interests.
|
1
|
Dandawate PR, Subramaniam D, Jensen RA and
Anant S: Targeting cancer stem cells and signaling pathways by
phytochemicals: Novel approach for breast cancer therapy. Semin
Cancer Biol. 40–41:192–208. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vessoni AT, Filippi-Chiela EC, Menck CF
and Lenz G: Autophagy and genomic integrity. Cell Death Differ.
20:1444–1454. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mizushima N, Levine B, Cuervo AM and
Klionsky DJ: Autophagy fights disease through cellular
self-digestion. Nature. 451:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mowers EE, Sharifi MN and Macleod KF:
Autophagy in cancer metastasis. Oncogene. 36:1619–1630. 2017.
View Article : Google Scholar :
|
|
5
|
Ruocco N, Costantini S and Costantini M:
Blue-print autophagy: Potential for cancer treatment. Mar Drugs.
14:142016. View Article : Google Scholar
|
|
6
|
Wang C, Hu Q and Shen HM: Pharmacological
inhibitors of autophagy as novel cancer therapeutic agents.
Pharmacol Res. 105:164–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee JS, Kim YJ, Kim CL and Lee GM:
Differential induction of autophagy in caspase-3/7 down-regulating
and Bcl-2 overexpressing recombinant CHO cells subjected to sodium
butyrate treatment. J Biotechnol. 161:34–41. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Knutson BA: Insights into the domain and
repeat architecture of target of rapamycin. J Struct Biol.
170:354–363. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sauer E, Imseng S, Maier T and Hall MN:
Conserved sequence motifs and the structure of the mTOR kinase
domain. Biochem Soc Trans. 41:889–895. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang H, Rudge DG, Koos JD, Vaidialingam B,
Yang HJ and Pavletich NP: mTOR kinase structure, mechanism and
regulation. Nature. 497:217–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim DH, Sarbassov DD, Ali SM, Latek RR,
Guntur KV, Erdjument-Bromage H, Tempst P and Sabatini DM: GbetaL, a
positive regulator of the rapamycin-sensitive pathway required for
the nutrient-sensitive interaction between raptor and mTOR. Mol
Cell. 11:895–904. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kaizuka T, Hara T, Oshiro N, Kikkawa U,
Yonezawa K, Takehana K, Iemura S, Natsume T and Mizushima N: Tti1
and Tel2 are critical factors in mammalian target of rapamycin
complex assembly. J Biol Chem. 285:20109–20116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takai H, Wang RC, Takai KK, Yang H and de
Lange T: Tel2 regulates the stability of PI3K-related protein
kinases. Cell. 131:1248–1259. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peterson TR, Laplante M, Thoreen CC,
Sancak Y, Kang SA, Kuehl WM, Gray NS and Sabatini DM: DEPTOR is an
mTOR inhibitor frequently overexpressed in multiple myeloma cells
and required for their survival. Cell. 137:873–886. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wullschleger S, Loewith R and Hall MN: TOR
signaling in growth and metabolism. Cell. 124:471–484. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mammucari C, Milan G, Romanello V, Masiero
E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J,
et al: FoxO3 controls autophagy in skeletal muscle in vivo. Cell
Metab. 6:458–471. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao J, Brault JJ, Schild A, Cao P, Sandri
M, Schiaffino S, Lecker SH and Goldberg AL: FoxO3 coordinately
activates protein degradation by the autophagic/lysosomal and
proteasomal pathways in atrophying muscle cells. Cell Metab.
6:472–483. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moscat J and Diaz-Meco MT: p62 at the
crossroads of autophagy, apoptosis, and cancer. Cell.
137:1001–1004. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sancak Y, Bar-Peled L, Zoncu R, Markhard
AL, Nada S and Sabatini DM: Ragulator-Rag complex targets mTORC1 to
the lysosomal surface and is necessary for its activation by amino
acids. Cell. 141:290–303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sancak Y, Peterson TR, Shaul YD, Lindquist
RA, Thoreen CC, Bar-Peled L and Sabatini DM: The Rag GTPases bind
raptor and mediate amino acid signaling to mTORC1. Science.
320:1496–1501. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zoncu R, Bar-Peled L, Efeyan A, Wang S,
Sancak Y and Sabatini DM: mTORC1 senses lysosomal amino acids
through an inside-out mechanism that requires the vacuolar
H(+)-ATPase. Science. 334:678–683. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Settembre C, Zoncu R, Medina DL, Vetrini
F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et
al: A lysosome-to-nucleus signalling mechanism senses and regulates
the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Copetti T, Bertoli C, Dalla E, Demarchi F
and Schneider C: p65/RelA modulates BECN1 transcription and
autophagy. Mol Cell Biol. 29:2594–2608. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu C, Wang W, Jia Y, Liu X, Tong Z and Li
B: Inhibition of AMPK/autophagy potentiates parthenolide-induced
apoptosis in human breast cancer cells. J Cell Biochem.
115:1458–1466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Abdal Dayem A, Choi HY, Yang GM, Kim K,
Saha SK and Cho SG: The anti-cancer effect of polyphenols against
breast cancer and cancer stem cells: Molecular mechanisms.
Nutrients. 8:581–618. 2016. View Article : Google Scholar :
|
|
27
|
Ricardo S, Vieira AF, Gerhard R, Leitão D,
Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F and Paredes
J: Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression
distribution within intrinsic molecular subtype. J Clin Pathol.
64:937–946. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Idowu MO, Kmieciak M, Dumur C, Burton RS,
Grimes MM, Powers CN and Manjili MH: CD44(+)/CD24(−/low) cancer
stem/progenitor cells are more abundant in triple-negative invasive
breast carcinoma phenotype and are associated with poor outcome.
Hum Pathol. 43:364–373. 2012. View Article : Google Scholar
|
|
29
|
Ahmed MA, Aleskandarany MA, Rakha EA,
Moustafa RZ, Benhasouna A, Nolan C, Green AR, Ilyas M and Ellis IO:
A CD44−/CD24+ phenotype is a poor prognostic
marker in early invasive breast cancer. Breast Cancer Res Treat.
133:979–995. 2012. View Article : Google Scholar
|
|
30
|
Fonseca NA, Cruz AF, Moura V, Simões S and
Moreira JN: The cancer stem cell phenotype as a determinant factor
of the heterotypic nature of breast tumors. Crit Rev Oncol Hematol.
113:111–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ingham PW and McMahon AP: Hedgehog
signaling in animal development: Paradigms and principles. Genes
Dev. 15:3059–3087. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
McMahon AP, Ingham PW and Tabin CJ:
Developmental roles and clinical significance of hedgehog
signaling. Curr Top Dev Biol. 53:1–114. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Murone M, Rosenthal A and de Sauvage FJ:
Sonic hedgehog signaling by the patched-smoothened receptor
complex. Curr Biol. 9:76–84. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bray SJ: Notch signalling: A simple
pathway becomes complex. Nat Rev Mol Cell Biol. 7:678–689. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ishii H, Iwatsuki M, Ieta K, Ohta D,
Haraguchi N, Mimori K and Mori M: Cancer stem cells and
chemoradiation resistance. Cancer Sci. 99:1871–1877. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang M, Zhang J, Huang Y, Ji S, Shao G,
Feng S, Chen D, Zhao K, Wang Z and Wu A: Cancer-associated
fibroblasts autophagy enhances progression of triple-negative
breast cancer cells. Med Sci Monit. 23:3904–3912. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun R, Shen S, Zhang YJ, Xu CF, Cao ZT,
Wen LP and Wang J: Nanoparticle-facilitated autophagy inhibition
promotes the efficacy of chemotherapeutics against breast cancer
stem cells. Biomaterials. 103:44–55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liang DH, Choi DS, Ensor JE, Kaipparettu
BA, Bass BL and Chang JC: The autophagy inhibitor chloroquine
targets cancer stem cells in triple negative breast cancer by
inducing mitochondrial damage and impairing DNA break repair.
Cancer Lett. 376:249–258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bincoletto C, Bechara A, Pereira GJS,
Santos CP, Antunes F, Peixoto da-Silva J, Muler M, Gigli RD,
Monteforte PT, Hirata H, et al: Interplay between apoptosis and
autophagy, a challenging puzzle: New perspectives on antitumor
chemotherapies. Chem Biol Interact. 206:279–288. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yousefi S, Perozzo R, Schmid I, Ziemiecki
A, Schaffner T, Scapozza L, Brunner T and Simon HU:
Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis.
Nat Cell Biol. 8:1124–1132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
DeNardo DG, Barreto JB, Andreu P, Vasquez
L, Tawfik D, Kolhatkar N and Coussens LM: CD4(+) T cells regulate
pulmonary metastasis of mammary carcinomas by enhancing protumor
properties of macrophages. Cancer Cell. 16:91–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mukhopadhyay S, Panda PK, Sinha N, Das DN
and Bhutia SK: Autophagy and apoptosis: Where do they meet?
Apoptosis. 19:555–566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yao D, Wang P, Zhang J, Fu L, Ouyang L and
Wang J: Deconvoluting the relationships between autophagy and
metastasis for potential cancer therapy. Apoptosis. 21:683–698.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Han Q, Deng Y, Chen S, Chen R, Yang M,
Zhang Z, Sun X, Wang W, He Y, Wang F, et al: Downregulation of
ATG5-dependent macroautophagy by chaperone-mediated autophagy
promotes breast cancer cell metastasis. Sci Rep. 7:47592017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kongara S, Kravchuk O, Teplova I, Lozy F,
Schulte J, Moore D, Barnard N, Neumann CA, White E and Karantza V:
Autophagy regulates keratin 8 homeostasis in mammary epithelial
cells and in breast tumors. Mol Cancer Res. 8:873–884. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kuraishy A, Karin M and Grivennikov SI:
Tumor promotion via injury- and death-induced inflammation.
Immunity. 35:467–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kung CP, Budina A, Balaburski G,
Bergenstock MK and Murphy M: Autophagy in tumor suppression and
cancer therapy. Crit Rev Eukaryot Gene Expr. 21:71–100. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
White E and DiPaola RS: The double-edged
sword of autophagy modulation in cancer. Clin Cancer Res.
15:5308–5316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Choi KS: Autophagy and cancer. Exp Mol
Med. 44:109–120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Denton D, Nicolson S and Kumar S: Cell
death by autophagy: Facts and apparent artefacts. Cell Death
Differ. 19:87–95. 2012. View Article : Google Scholar :
|
|
51
|
Maycotte P, Jones KL, Goodall ML, Thorburn
J and Thorburn A: Autophagy supports breast cancer stem cell
maintenance by regulating IL6 secretion. Mol Cancer Res.
13:651–658. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wolf J, Dewi DL, Fredebohm J,
Müller-Decker K, Flechtenmacher C, Hoheisel JD and Boettcher M: A
mammosphere formation RNAi screen reveals that ATG4A promotes a
breast cancer stem-like phenotype. Breast Cancer Res. 15:R1092013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gong C, Bauvy C, Tonelli G, Yue W,
Deloménie C, Nicolas V, Zhu Y, Domergue V, Marin-Esteban V,
Tharinger H, et al: Beclin 1 and autophagy are required for the
tumorigenicity of breast cancer stem-like/progenitor cells.
Oncogene. 32:2261–2272. 1–11. 2013. View Article : Google Scholar :
|
|
54
|
Zhao Y, Huang Q, Yang J, Lou M, Wang A,
Dong J, Qin Z and Zhang T: Autophagy impairment inhibits
differentiation of glioma stem/progenitor cells. Brain Res.
1313:250–258. 2010. View Article : Google Scholar
|
|
55
|
Singh BN, Kumar D, Shankar S and
Srivastava RK: Rottlerin induces autophagy which leads to apoptotic
cell death through inhibition of PI3K/Akt/mTOR pathway in human
pancreatic cancer stem cells. Biochem Pharmacol. 84:1154–1163.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kumar D, Shankar S and Srivastava RK:
Rottlerin-induced autophagy leads to the apoptosis in breast cancer
stem cells: Molecular mechanisms. Mol Cancer. 12:1712013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wei MF, Chen MW, Chen KC, Lou PJ, Lin SY,
Hung SC, Hsiao M, Yao CJ and Shieh MJ: Autophagy promotes
resistance to photodynamic therapy-induced apoptosis selectively in
colorectal cancer stem-like cells. Autophagy. 10:1179–1192. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yue W, Hamaï A, Tonelli G, Bauvy C,
Nicolas V, Tharinger H, Codogno P and Mehrpour M: Inhibition of the
autophagic flux by salinomycin in breast cancer
stem-like/progenitor cells interferes with their maintenance.
Autophagy. 9:714–729. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chiang GG and Abraham RT: Targeting the
mTOR signaling network in cancer. Trends Mol Med. 13:433–442. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu K, Liu P and Wei W: mTOR signaling in
tumorigenesis. Biochim Biophys Acta. 1846:638–654. 2014.PubMed/NCBI
|
|
61
|
Mateo F, Arenas EJ, Aguilar H,
Serra-Musach J, de Garibay GR, Boni J, Maicas M, Du S, Iorio F,
Herranz-Ors C, et al: Stem cell-like transcriptional reprogramming
mediates metastatic resistance to mTOR inhibition. Oncogene.
36:2737–2749. 2017. View Article : Google Scholar :
|
|
62
|
Zhang L, Fu L, Zhang S, Zhang J, Zhao Y,
Zheng Y, He G, Yang S, Ouyang L and Liu B: Discovery of a small
molecule targeting ULK1-modulated cell death of triple negative
breast cancer in vitro and in vivo. Chem Sci (Camb). 8:2687–2701.
2017. View Article : Google Scholar
|
|
63
|
Jang JE, Eom JI, Jeung HK, Cheong JW, Lee
JY, Kim JS and Min YH: Targeting AMPK-ULK1-mediated autophagy for
combating BET inhibitor resistance in acute myeloid leukemia stem
cells. Autophagy. 13:761–762. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhou Y, Rucker EB III and Zhou BP:
Autophagy regulation in the development and treatment of breast
cancer. Acta Biochim Biophys Sin (Shanghai). 48:60–74. 2016.
|
|
65
|
Yeo SK, Wen J, Chen S and Guan JL:
Autophagy differentially regulates distinct breast cancer stem-like
cells in murine models via EGFR/Stat3 and Tgfβ/Smad signaling.
Cancer Res. 76:3397–3410. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Nagy P, Kovács L, Sándor GO and Juhász G:
Stem-cell-specific endocytic degradation defects lead to intestinal
dysplasia in Drosophila. Dis Model Mech. 9:501–512. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu K, Zhao Q, Liu P, Cao J, Gong J, Wang
C, Wang W, Li X, Sun H, Zhang C, et al: ATG3-dependent autophagy
mediates mitochondrial homeostasis in pluripotency acquirement and
maintenance. Autophagy. 12:2000–2008. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang L, Li J, Ouyang L, Liu B and Cheng
Y: Unraveling the roles of Atg4 proteases from autophagy modulation
to targeted cancer therapy. Cancer Lett. 373:19–26. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Antonelli M, Strappazzon F, Arisi I,
Brandi R, D'Onofrio M, Sambucci M, Manic G, Vitale I, Barilà D and
Stagni V: ATM kinase sustains breast cancer stem-like cells by
promoting ATG4C expression and autophagy. Oncotarget.
8:21692–21709. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu H, He Z, von Rütte T, Yousefi S,
Hunger RE and Simon HU: Down-regulation of autophagy-related
protein 5 (ATG5) contributes to the pathogenesis of early-stage
cutaneous melanoma. Sci Transl Med. 5:202ra1232013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Debnath J: The multifaceted roles of
autophagy in tumors-implications for breast cancer. J Mammary Gland
Biol Neoplasia. 16:173–187. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chaterjee M and van Golen KL: Breast
cancer stem cells survive periods of farnesyl-transferase
inhibitor-induced dormancy by undergoing autophagy. Bone Marrow
Res. 2011:3629382011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Memni H, Macherki Y, Klayech Z,
Ben-Haj-Ayed A, Farhat K, Remadi Y, Gabbouj S, Mahfoudh W, Bouzid
N, Bouaouina N, et al: E-cadherin genetic variants predict survival
outcome in breast cancer patients. J Transl Med. 14:3202016.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhuang W, Li B, Long L, Chen L, Huang Q
and Liang Z: Induction of autophagy promotes differentiation of
glioma-initiating cells and their radiosensitivity. Int J Cancer.
129:2720–2731. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Qin Z, Xue J, He Y, Ma H, Jin G, Chen J,
Hu Z, Liu X and Shen H: Potentially functional polymorphisms in
ATG10 are associated with risk of breast cancer in a Chinese
population. Gene. 527:491–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Sanchez CG, Penfornis P, Oskowitz AZ,
Boonjindasup AG, Cai DZ, Dhule SS, Rowan BG, Kelekar A, Krause DS
and Pochampally RR: Activation of autophagy in mesenchymal stem
cells provides tumor stromal support. Carcinogenesis. 32:964–972.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Cufí S, Vazquez-Martin A,
Oliveras-Ferraros C, Corominas-Faja B, Urruticoechea A,
Martin-Castillo B and Menendez JA: Autophagy-related gene 12
(ATG12) is a novel determinant of primary resistance to
HER2-targeted therapies: Utility of transcriptome analysis of the
autophagy interactome to guide breast cancer treatment. Oncotarget.
3:1600–1614. 2012. View Article : Google Scholar
|
|
78
|
Chang SJ, Ou-Yang F, Tu HP, Lin CH, Huang
SH, Kostoro J, Hou MF, Chai CY and Kwan AL: Decreased expression of
autophagy protein LC3 and stemness
(CD44+/CD24−/low) indicate poor prognosis in
triple-negative breast cancer. Hum Pathol. 48:48–55. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Carpenter RL, Sirkisoon S, Zhu D, Rimkus
T, Harrison A, Anderson A, Paw I, Qasem S, Xing F, Liu Y, et al:
Combined inhibition of AKT and HSF1 suppresses breast cancer stem
cells and tumor growth. Oncotarget. 8:73947–73963. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Petherick KJ, Williams AC, Lane JD,
Ordóñez-Morán P, Huelsken J, Collard TJ, Smartt HJ, Batson J, Malik
K, Paraskeva C, et al: Autolysosomal β-catenin degradation
regulates Wnt-autophagy-p62 crosstalk. EMBO J. 32:1903–1916. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang Y, Han C, Lu L, Magliato S and Wu T:
Hedgehog signaling pathway regulates autophagy in human
hepatocellular carcinoma cells. Hepatology. 58:995–1010. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Espina V and Liotta LA: What is the
malignant nature of human ductal carcinoma in situ? Nat Rev Cancer.
11:68–75. 2011. View Article : Google Scholar
|
|
83
|
Yang H, Zheng Y, Zhang Y, Cao Z and Jiang
Y: Mesenchymal stem cells derived from multiple myeloma patients
protect against chemotherapy through autophagy-dependent activation
of NF-κB signaling. Leuk Res. 60:82–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Huang S, Wang D, Zhang S, Huang X, Wang D,
Ijaz M and Shi Y: Tunicamycin potentiates paclitaxel-induced
apoptosis through inhibition of PI3K/AKT and MAPK pathways in
breast cancer. Cancer Chemother Pharmacol. 80:685–696. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sharma N, Thomas S, Golden EB, Hofman FM,
Chen TC, Petasis NA, Schönthal AH and Louie SG: Inhibition of
autophagy and induction of breast cancer cell death by mefloquine,
an antimalarial agent. Cancer Lett. 326:143–154. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ma YW, Liu YZ and Pan JX: Verteporfin
induces apoptosis and eliminates cancer stem-like cells in uveal
melanoma in the absence of light activation. Am J Cancer Res.
6:2816–2830. 2016.
|
|
87
|
Shi TT, Yu XX, Yan LJ and Xiao HT:
Research progress of hydroxychloroquine and autophagy inhibitors on
cancer. Cancer Chemother Pharmacol. 79:287–294. 2017. View Article : Google Scholar
|
|
88
|
Solomon VR, Almnayan D and Lee H: Design,
synthesis and characterization of novel quinacrine analogs that
preferentially kill cancer over non-cancer cells through the
down-regulation of Bcl-2 and up-regulation of Bax and Bad. Eur J
Med Chem. 137:156–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Siddharth S, Nayak D, Nayak A, Das S and
Kundu CN: ABT-888 and quinacrine induced apoptosis in metastatic
breast cancer stem cells by inhibiting base excision repair via
adenomatous polyposis coli. DNA Repair (Amst). 45:44–55. 2016.
View Article : Google Scholar
|
|
90
|
Mishra P, Dauphinee AN, Ward C, Sarkar S,
Gunawardena AHLAN and Manjithaya R: Discovery of pan autophagy
inhibitors through a high-throughput screen highlights
macro-autophagy as an evolutionarily conserved process across 3
eukaryotic kingdoms. Autophagy. 13:1556–1572. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Liang S, Chen Z, Jiang G, Zhou Y, Liu Q,
Su Q, Wei W, Du J and Wang H: Activation of GPER suppresses
migration and angiogenesis of triple negative breast cancer via
inhibition of NF-κB/IL-6 signals. Cancer Lett. 386:12–23. 2017.
View Article : Google Scholar
|
|
92
|
Torrente E, Parodi C, Ercolani L, De Mei
C, Ferrari A, Scarpelli R and Grimaldi B: Synthesis and in vitro
anticancer activity of the first cass of dual inhibitors of
REV-ERBβ and autophagy. J Med Chem. 58:5900–5915. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhou X, Yue GG, Chan AM, Tsui SK, Fung KP,
Sun H, Pu J and Lau CB: Eriocalyxin B, a novel autophagy inducer,
exerts anti-tumor activity through the suppression of
Akt/mTOR/p70S6K signaling pathway in breast cancer. Biochem
Pharmacol. 142:58–70. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wong VKW, Zeng W, Chen J, Yao XJ, Leung
ELH, Wang QQ, Chiu P, Ko BCB and Law BYK: Tetrandrine, an activator
of autophagy, induces autophagic cell death via PKC-α inhibition
and mTOR-dependent mechanisms. Front Pharmacol. 8:3512017.
View Article : Google Scholar
|
|
95
|
Han H, Li J, Feng X, Zhou H, Guo S and
Zhou W: Autophagy-related genes are induced by histone deacetylase
inhibitor suberoylanilide hydroxamic acid via the activation of
cathepsin B in human breast cancer cells. Oncotarget.
8:53352–53365. 2017.PubMed/NCBI
|
|
96
|
Chen X, Yu X, Chen J, Yang Z, Shao Z,
Zhang Z, Guo X and Feng Y: Radiotherapy can improve the
disease-free survival rate in triple-negative breast cancer
patients with T1-T2 disease and one to three positive lymph nodes
after mastectomy. Oncologist. 18:141–147. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chen X, Yu X, Chen J, Zhang Z, Tuan J,
Shao Z, Guo X and Feng Y: Analysis in early stage triple-negative
breast cancer treated with mastectomy without adjuvant
radiotherapy: Patterns of failure and prognostic factors. Cancer.
119:2366–2374. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Liu EY, Xu N, O'Prey J, Lao LY, Joshi S,
Long JS, O'Prey M, Croft DR, Beaumatin F, Baudot AD, et al: Loss of
autophagy causes a synthetic lethal deficiency in DNA repair. Proc
Natl Acad Sci USA. 112:773–778. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhou ZR, Yang ZZ, Wang SJ, Zhang L, Luo
JR, Feng Y, Yu XL, Chen XX and Guo XM: The Chk1 inhibitor MK-8776
increases the radiosensitivity of human triple-negative breast
cancer by inhibiting autophagy. Acta Pharmacol Sin. 38:513–523.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Davison Z, de Blacquière GE, Westley BR
and May FEB: Insulin-like growth factor-dependent proliferation and
survival of triple-negative breast cancer cells: Implications for
therapy. Neoplasia. 13:504–515. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wu W, Ma J, Shao N, Shi Y, Liu R, Li W,
Lin Y and Wang S: Co-targeting IGF-1R and autophagy enhances the
effects of cell growth suppression and apoptosis induced by the
IGF-1R inhibitor NVP-AEW541 in triple-negative breast cancer cells.
PLoS One. 12:e01692292017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Maxfield KE, Macion J, Vankayalapati H and
Whitehurst AW: SIK2 restricts autophagic flux to support
triple-negative breast cancer survival. Mol Cell Biol.
36:3048–3057. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Gao J, Fan M, Peng S, Zhang M, Xiang G, Li
X, Guo W, Sun Y, Wu X, Wu X, et al: Small-molecule RL71-triggered
excessive autophagic cell death as a potential therapeutic strategy
in triple-negative breast cancer. Cell Death Dis. 8:e30492017.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhang P, Liu X, Li H, Chen Z, Yao X, Jin J
and Ma X: TRPC5-induced autophagy promotes drug resistance in
breast carcinoma via CaMKKβ/AMPKα/mTOR pathway. Sci Rep.
7:31582017. View Article : Google Scholar
|
|
105
|
Liu Z, Shi A, Song D, Han B, Zhang Z, Ma
L, Liu D and Fan Z: Resistin confers resistance to
doxorubicin-induced apoptosis in human breast cancer cells through
autophagy induction. Am J Cancer Res. 7:574–583. 2017.PubMed/NCBI
|
|
106
|
Poillet-Perez L, Jacquet M, Hervouet E,
Gauthier T, Fraichard A, Borg C, Pallandre JR, Gonzalez BJ, Ramdani
Y, Boyer-Guittaut M, et al: GABARAPL1 tumor suppressive function is
independent of its conjugation to autophagosomes in MCF-7 breast
cancer cells. Oncotarget. 8:55998–56020. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Rodríguez CE, Reidel SI, Bal de Kier Joffé
ED, Jasnis MA and Fiszman GL: Autophagy protects from
trastuzumab-induced cytotoxicity in HER2 overexpressing breast
tumor spheroids. PLoS One. 10:e01379202015. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zambrano J, Yeh ES and Zambrano J:
Autophagy and apoptotic crosstalk: Mechanism of therapeutic
resistance in HER2-positive breast cncer. Breast Cancer (Auckl).
10:13–23. 2016.
|
|
109
|
Cufí S, Vazquez-Martin A,
Oliveras-Ferraros C, Corominas-Faja B, Cuyàs E, López-Bonet E,
Martin-Castillo B, Joven J and Menendez JA: The anti-malarial
chloroquine overcomes primary resistance and restores sensitivity
to trastuzumab in HER2-positive breast cancer. Sci Rep. 3:24692013.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Garbar C, Mascaux C, Giustiniani J,
Merrouche Y and Bensussan A: Chemotherapy treatment induces an
increase of autophagy in the luminal breast cancer cell MCF7, but
not in the triple-negative MDA-MB231. Sci Rep. 7:72012017.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Maycotte P, Gearheart CM, Barnard R, Aryal
S, Mulcahy Levy JM, Fosmire SP, Hansen RJ, Morgan MJ, Porter CC,
Gustafson DL, et al: STAT3-mediated autophagy dependence identifies
subtypes of breast cancer where autophagy inhibition can be
efficacious. Cancer Res. 74:2579–2590. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Wang S, Wang K, Wang H, Han J and Sun H:
Autophagy is essential for flavopiridol-induced cytotoxicity
against MCF-7 breast cancer cells. Mol Med Rep. 16:9715–9720. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Chang CT, Korivi M, Huang HC, Thiyagarajan
V, Lin KY, Huang PJ, Liu JY, Hseu YC and Yang HL: Inhibition of ROS
production, autophagy or apoptosis signaling reversed the
anticancer properties of Antrodia salmonea in triple-negative
breast cancer (MDA-MB-231) cells. Food Chem Toxicol. 103:1–17.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zheng N, Liu L, Liu WW, Li F, Hayashi T,
Tashiro SI, Onodera S and Ikejima T: Crosstalk of ROS/RNS and
autophagy in silibinin-induced apoptosis of MCF-7 human breast
cancer cells in vitro. Acta Pharmacol Sin. 38:277–289. 2017.
View Article : Google Scholar :
|
|
115
|
Liu ZY, He KW, Song XG, Wang XZ, Zhuo PY,
Wang XW, Ma QH, Huo ZJ and Yu ZY: Effect of autophagy inhibitor
combined with EGFR inhibitor on triple-negative breast cancer
MDA-MB-468 and MDA-MB-231 cells. Zhonghua Zhong Liu Za Zhi.
38:417–424. 2016.In Chinese. PubMed/NCBI
|
|
116
|
Tran AT, Ramalinga M, Kedir H, Clarke R
and Kumar D: Autophagy inhibitor 3-methyladenine potentiates
apoptosis induced by dietary tocotrienols in breast cancer cells.
Eur J Nutr. 54:265–272. 2015. View Article : Google Scholar
|
|
117
|
Liu Z, He K, Ma Q, Yu Q, Liu C, Ndege I,
Wang X and Yu Z: Autophagy inhibitor facilitates gefitinib
sensitivity in vitro and in vivo by activating mitochondrial
apoptosis in triple negative breast cancer. PLoS One.
12:e01776942017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Wang H, Wang W, Xu Y, Yang Y, Chen X, Quan
H and Lou L: Aberrant intracellular metabolism of T-DM1 confers
T-DM1 resistance in human epidermal growth factor receptor
2-positive gastric cancer cells. Cancer Sci. 108:1458–1468. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Gong C, Hu C, Gu F, Xia Q, Yao C, Zhang L,
Qiang L, Gao S and Gao Y: Co-delivery of autophagy inhibitor ATG7
siRNA and docetaxel for breast cancer treatment. J Control Release.
266:272–286. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Shen P, Chen M, He M, Chen L, Song Y, Xiao
P, Wan X, Dai F, Pan T and Wang Q: Inhibition of ERα/ERK/P62
cascades induces 'autophagic switch' in the estrogen
receptor-positive breast cancer cells exposed to gemcitabine.
Oncotarget. 7:48501–48516. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Li HC, Xia ZH, Chen YF, Yang F, Feng W,
Cai H, Mei Y, Jiang YM, Xu K and Feng DX: Cantharidin inhibits the
growth of triple-negative breast cancer cells by suppressing
autophagy and inducing apoptosis in vitro and in vivo. Cell Physiol
Biochem. 43:1829–1840. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Gu Y, Chen T, Li G, Xu C, Xu Z, Zhang J,
He K, Zheng L, Guan Z, Su X, et al: Lower Beclin 1 downregulates
HER2 expression to enhance tamoxifen sensitivity and predicts a
favorable outcome for ER positive breast cancer. Oncotarget.
8:52156–52177. 2016.
|