Introduction
Osteosarcoma is the most common primary malignant
bone tumor, which originates from mesenchymal tissue. It is
characterized by spindle-shaped stromal cells that produce
bone-like tissue (1,2). It usually occurs in adolescents with
metaphysis, mainly around the knee joint (3). According to statistical data, the
incidence of osteosarcoma is ranked first among all primary
malignant bone tumors (4). The
malignant degree of osteosarcoma is extremely high. The prognosis
is poor, and pulmonary metastasis can occur within several months,
with the survival rate being only approximately 60% following
amputation (5–7).
At present, radical surgery is still the first
choice of treatment for osteosarcoma, and is accompanied by
adjuvant chemotherapy at high doses (8–10).
With the promotion of radical surgery and high-dose chemotherapy,
the 5-year survival rate of patients with osteosarcoma has
increased to approximately 70% (11). However, there a considerable number
of patients with this disease still suffer from tumor recurrence
and distant metastasis following complex treatment (12). Distant metastasis is one of the
most important reasons for the failure of treatment in the majority
of patients with osteosarcoma (13–15).
The main reason for the distant metastasis of osteosarcoma is the
abnormal expression of tumor metastasis-related genes and
metastasis suppressor genes (16,17).
In recent years, some progress has been made in the
etiology, development, diagnosis and treatment of osteosarcoma;
however, this progress has been very gradual, and the specific
pathogenesis of the disease is not yet clear (17,18).
In addition, the lack of an effective target for the diagnosis and
treatment of osteosarcoma has made prognosis and treatment
difficult. Therefore, the identification of biological targets for
osteosarcoma may have important theoretical and clinical
implications (19).
MicroRNAs (miRNAs or miRs) are single-stranded
non-coding RNAs of 21–23 nucleotides in length (20–22).
miRNAs regulate gene expression via the specific binding to target
genes (23). Over the past
decades, a large number of miRNAs have been identified and are
believed to play an important role in the occurrence,
differentiation, proliferation and apoptosis of cells (24,25).
Recent studies have indicated that alterations in miRNA expression
are associated with the occurrence, development, diagnosis and
prognosis of cancers (26–28).
It has been previously demonstrated that miR-27 can
regulate the expression of genes involved in cell metabolism,
differentiation, apoptosis, migration and immunity (29,30);
On the other hand, an abnormal expression of miR-27 has been found
in a variety of tumors and diseases (31–33).
miR-27 regulates cell apoptosis, cell cycle progression and cell
proliferation, as well as processes via the post-transcriptional
regulation of different molecules, which indicates that miR-27 is a
key molecule in the development of tumors (30,34).
Pan et al demonsrated that miR-27a promoted osteosarcoma
cell proliferation, migration and invasion (33). However, the molecular mechanisms
underlying the role of miR-27a in the development and progression
of osteosarcoma remain largely unexplored. In this study, revere
transcription-quantitative PCR (RT-qPCR) was used to detect the
expression of miR-27a-3p in human osteosarcoma cell lines, and Cell
Counting kit-8 (CCK-8) assay, flow cytometry and Transwell assay
were used to examine the effects of miR-27a-3p on the
proliferation, apoptosis and invasion of osteosarcoma cells.
It has been previously demonstrated that epigenetic
modification plays a very important role in the occurrence and
development of tumors. The degree of DNA methylation in prostate
cancer and breast cancer is closely related to the progression of
the tumor (35). The expression of
ten-eleven translocation 1 (TET1) in prostate cancer and breast
cancer tissues is downregulated, and it has been found that the
deletion of TET1 promotes the invasion and growth of tumor cells,
and promotes the metastasis of tumor cells in a xenograft tumor
model; TET1 overexpression can reduce cell invasion and inhibit the
formation of xenograft tumors (36,37).
Online software (http://www.targetscan.org) analysis was used to
predict the possible miR-27a-3p target genes, whereby TET1 was
identified as a miR-27a-3p target gene. In addition, the luciferase
reporter gene system was used to determine whether TET1 is a
regulatory target gene of miR-27a-3p. Western blot analysis was
used to examine the effects of miR-27a-3p on the expression of
TET1, and the mRNA expression of TET1 in osteosarcoma cell lines as
detected by RT-qPCR. Further validation of the cell biological
functions of miR-27a-3p in MG-63 cells through the regulation of
TET1 was achieved by assessing the cell biological functions of
miR-27a-3p in MG-63 cells through the regulation of TET1 via the
overexpression of TET1.
Therefore, the findings of this study may provide
the experimental basis for the application of miR-27a-3p and TET1
in the treatment of osteosarcoma.
Materials and methods
Cell lines and cell culture
Normal human osteoblasts (hFOB 1.19 cells), were
cultured in mixed medium containing 0.5 mM sodium pyruvate, 2.5 mM
L-glutamine (Gln), 15 mM HEPES and 10% fetal bovine serum (FBS)
(DMEM and F12 medium were mixed 1:1). The human osteosarcoma MG-63
and MNNG/HOS Cl #5 cells were cultured in EMEM containing 2.0 mM
L-glutamine (Gln), 1.0 mM sodium pyruvate, 0.1 mM non-essential
amino acid, 1.5 g/ml sodium bicarbonate and 10% FBS Eagle's BSS.
Saos-2 cells were cultured in McCoy's 5A medium with 1.5 g/ml
sodium bicarbonate, 1.5 mM L-glutamine (Gln) 15% FBS. The U-2OS
cells were cultured in McCoy's 5A medium containing 1.5 g/ml sodium
bicarbonate, 1.5 mM L-glutamine (Gln) and 10% FBS.
All cell lines are purchased from the Cell Bank of
the Chinese Academy of Sciences (Shanghai, China). FBS, sodium
pyruvate, L-glutamine, non-essential amino acid and basic culture
medium for cell culture were all purchased from Invitrogen
(Carlsbad, CA, USA). The above-mentioned cells were cultured under
conditions of 37°C, 5% CO2 and saturated humidity. The
growth state of the cells was observed under an inverted microscope
(Leica DM IL LED; Leica, Wetzlar, Germany). When the cells reached
a 70–80% fusion state, they were digested with 0.25% trypsin
(Invitrogen). Cells at the logarithmic growth phase were collected
for use in the experiments.
RT-qPCR
RT-qPCR was used to detect the expression of
miR-27a-3p in normal human osteoblasts (hFOB 1.19) and osteosarcoma
cell lines (MG-63, MNNG/HOS Cl #5, Saos-2 and U-2OS). All cell
lines were collected in vitro and the TaqMan miRNA isolation
kit was used to extract the RNA. TaqMan microRNA Assay and TaqMan
Universal PCR Master Mix (Applied Biosystems, Foster city, CA, USA)
were used to detect the expression of mature miR-27a-3p, using U6
as an internal reference. The primers used were as follows:
miR-27a-3p forward, 5′-ACA CTC CAG CTG GGT TCA CAG TGG CTA AG-3′
and reverse, 5′-TGG TGT CGT GGA GTC G-3′; and U6 forward, 5′-CTC
GCT TCG GCA GCA CA-3′ and reverse, 5′-AAC GCT TCA CGA ATT TGC
GT-3′. The thermocycling conditions used for PCR were as follows:
Firstly pre-heating at 95°C for 10 min, followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing at 60°C for 60 sec, and
extension at 72°C 90 sec. All experiments were conducted in
triplicate. The RT-qPCR experimental results were analyzed by using
the relative quantification method (38).
Following transfection of the MG-63 cells with
miR-27a-3p inhibitor and inhibitor negative control for 48 h, the
TaqMan miRNA isolation kit was used to respectively extract the RNA
from the cells in each group, and miR-27a-3p expression in the
MG-63 cells of each group was detected by RT-qPCR as described
above.
The mRNA expression of TET1 in the osteosarcoma
cells was also detected by RT-qPCR. Total RNA was extracted from
the osteosarcoma cells using TRIzol reagent (Invitrogen), and the
first strand cDNA was synthesized using the First Strand cDNA
Synthesis kit (Qiagen, Hilden, Germany). GAPDH was used as an
internal reference gene to detect the mRNA expression of mRNA TET1.
The primers used were as follows: TET1 forward,
5′-GCCAGCAGAAGACCAACT-3′ and reverse, 5′-TCCAGAGGCACAACAACA-3′;
GAPDH forward, 5′-GACCTGACCTGCCGTCTA-3′ and reverse,
5′-AGGAGTGGGTGTCGCTGT-3′.
The thermocycling conditions were as follows:
Firstly pre-heating at 95°C for 10 min, followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing at 60°C for 60 sec, and
extension at 72°C 90 sec. All experiments were conducted in
triplicate. The RT-qPCR experimental results are analyzed using the
relative quantification method (38).
Cell treatment
Normal cultured MG-63 cells were inoculated into
6-well plates at a density of 3×105 cells/well. After
cell adherence, according to the instructions of the provider of
Lipofectamine 2000 transfection reagent (Invitrogen), trans-fection
with miR-27a-3p inhibitor, inhibitor negative control (provided by
GenePharma, Shanghai, China), TET1 siRNA or siRNA control
(scramble) (provided by Guangzhou RiboBio Co., Ltd., Guangzhou,
China) was carried out. At the same time, the normal control group
consisted of untransfected cells. MEM culture medium without serum
was used to dilute the miR-27a-3p inhibitor, inhibitor negative
control, TET1 siRNA and siRNA control, respectively. The diluted
Lipofectamine 2000 was mixed with miR-27a-3p inhibitor or inhibitor
negative control or TET1 siRNA or siRNA control. After gentle
mixing, incubation was carried out at room temperature for 20 min
to form the complex. The compound was added to the culture plate of
MG63 cells, which was placed in an incubator at 37°C and a 5%
CO2 volume fraction. After 5 h, the culture medium was
replaced with MEM medium containing 10% fetal bovine serum and
culture was continued for 48 h.
CCK-8 assay
The normal cultured MG-63 cells were inoculated into
96-well plates with 1×106/ml, and the volume of each
well was 100 µl. Following cell adherence, and following the
instructions of the provider of Lipofectamine 2000 (Invitrogen),
transfection with miR-27a-3p inhibitor, or inhibitor negative
control was carried out, and the normal control group consisted of
untransfected cells. Following transfection for 24, 48 and 72 h, 10
µl CCK-8 solution (Sigma-Aldrich, St. Louis, MO, USA) were
added to each well of a 96-well plate and the plates were then
placed in a CO2 incubator for continuous cultivation for
48 h. A microplate reader (BioTek Instruments, Inc., Winooski, VT,
USA) is used to detect the OD value at the 450-nm position.
Analysis of cell apoptosis
The normal cultured MG-63 cells were inoculated into
96-well plates. Following transfection, the MG-63 cells were
digested by trypsin, and washed with PBS. Annexin V-FITC and PI
staining solution (Beyotime Institute of Biotechnology, Haimen,
China) were then added. The 96-well plates were placed at room
temperature for 15 min. After screen filtering, flow cytometric
analysis was performed (flow cytometer; BD Biosciences, Palo Alto,
CA, USA) and FCM cellQuest software (BD Biosciences) was used for
cell counting. FACsuite software (BD Biosciences) was adopted to
analyze the data.
Transwell invasion assay
The cell invasive ability was examined by Transwell
invasion assay. The MG-63 cells transfected miR-27a-3p inhibitor
and inhibitor negative control were added to the upper section of a
Transwell chamber (Corning Inc., Corning, NY, USA). The bottom of
the Transwell chamber was filled with 500 µl of medium
containing 10% FBS. Following 48 h of incubation, the MG-63 cells
on the top surface of the chamber were removed using a cotton swab.
The MG-63 cells which had passed through the Transwell chamber were
fixed with 4% paraformaldehyde for 15 min at room temperature and
stained with a crystal violet solution (Beyotime Institute of
Biotechnology) for 10 min. A total of 8 visual fields were observed
at random using a Leica microscope (Leica Microsystems, Bensheim,
Germany). Image pro plus 6.0 software (Media Cybernetics Inc.,
Silver Spring, MD, USA) was used to count the cells passing through
the Transwell polycarbonate film. The cells passing through the
Transwell polycarbonate film were the invading cells.
Luciferase reporter gene system
assay
TargetScan (http://www.targetscan.org/) was used to predict the
gene targeted by miR-27a-3p, and TET1 was predicted as a potential
target. The luciferase reporter plasmid was constructed, which
included the miR-27a-3p complementary binding site wild-type 3′-UTR
sequence segments of TET1 (TET1 3′-UTR WT) and mutations of 3′-UTR
sequence segments of TET1 (TET1 3′-UTR MUT). The normal cultured
293 cells (the Cell Bank of the Chinese Academy of Sciences,
Shanghai, China) was inoculated into the wells of a 6-well plate at
a density of 3×105/ml cells/well, and the volume of each pore was
1,000 µl. The luciferase plasmid containing TET1 3′-UTR WT
or TET1 3′-UTR MUT and miR-27a-3p mimic or mimic negative-control
(GenePharma) were co-transfected into the 293 cells. Following
transfection for 48 h, the 293 cells were collected and the
luciferase activity was determined using a microplate reader
(BioTek Instruments, Inc.).
Western blot analysis
Total protein was extracted from the osteosarcoma
cells using RIPA buffer (Beyotime Institute of Biotechnology) and
the quantitative analysis of the protein concentration was
performed using a BCA assay kit (Beyotime Institute of
Biotechnology). A total of 50 µg of total protein was
separated for each lane, and then was transferred onto PVDF
membranes (Bio-Rad, Hercules, CA, USA). Subsequently, the PVDF
membranes were blocked with blocking buffer containing 5% skim milk
powder, 10 mM Tris-HCl, 150 mM NaCl and 0.1% Tween-20, pH 7.5, at
room temperature for 1 h. This was followed by the addition of the
primary antibodies [mouse anti-human vimentin monoclonal antibody
(1:2,000 dilution; ab8979), rabbit anti-human E-cadherin polyclonal
antibody (1:1,000 dilution; ab15148), rabbit anti human TET1
(ab105475) and α-catenin monoclonal (ab51032) antibodies (1:2,000
dilution) (all from Abcam, Cambridge, MA, USA) and rabbit
anti-human β-actin monoclonal antibody (1:1,000 dilution; A5441;
Sigma-Aldrich)] and incubation of the PVDF membranes at 4°C
overnight, followed by continuous incubation with secondary
antibodies [HRP-labeled goat anti-rabbit IgG (A6154) or goat
anti-mouse IgG (A4416); Sigma-Aldrich; 1:2,000 dilution] at 37°C
for 1 h. Finally, the PVDF membranes were treated with enhanced
chemiluminescence reagent (Pierce, Rockford, IL, USA), and exposed
to medical X-ray film. The relative content of vimentin,
E-cadherin, α-catenin and TET1 was determined using PDQuest
software (Bio-Rad).
Statistical analysis
SPSS17.0 statistical analysis software was used for
the statistical analysis of the experimental data. The t-test was
used for comparisons between 2 groups. One-way ANOVA followed by
Tukey's post hoc test was used for comparisons of 3 or more groups
(groups or variables) to determine statistical significance. The
results were considered to be statistically significant at a value
of P<0.05.
Results
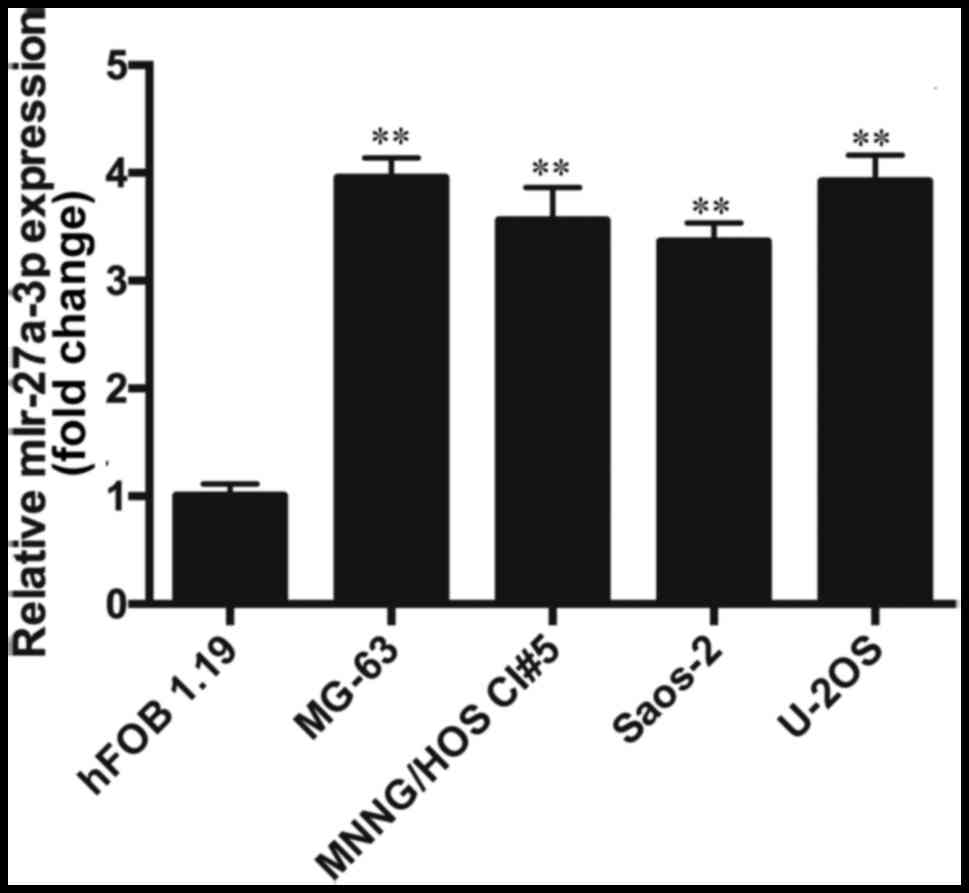
miR-27a-3p is upregulated in human
osteosarcoma cells
The results from RT-qPCR revealed that the
expression of miR-27a-3p in the osteosarcoma cell lines (MNNG/HOS
Cl #5, MG-63, U-2OS and Saos-2) was significantly higher than that
in the normal human osteoblasts (hFOB 1.19) (P<0.01). The
expression of miR-27a-3p in the osteosarcoma cell lines was
approximately 3.5-fold higher than that in the hFOB 1.19 cells
(Fig. 1), which indicated that the
upregulation of miR-27a-3p may play an important role in the
development of osteosarcoma. As the expression of miR-27a-3p was
highest in the MG-63 cells, these cells were selected for use in
transfection experiments. In the future, we aim to perform further
studies using multiple cell lines.
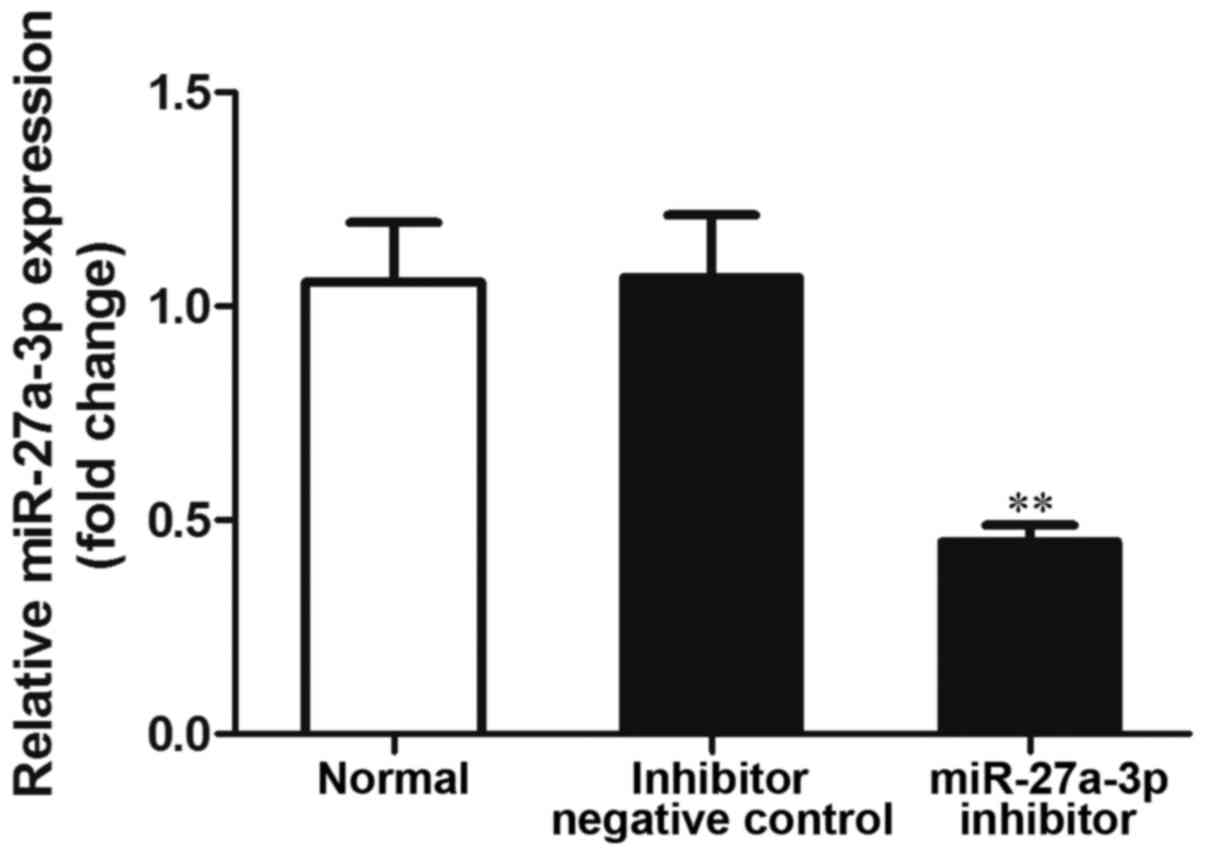
miR-27a-3p inhibition suppresses cell
proliferation and induces cell apoptosis
In order to examine the effects of miR-27a-3p on
osteosarcoma cells, miR-27a-3p inhibitor was transfected into the
MG-63 cells, and the expression of miR-27a-3p was detected by
RT-qPCR. The results revealed that the expression of miR-27a-3p was
significantly lower in the group transfected with the miR-27a-3p
inhibitor than in the normal control group (untransfected cells;
P<0.01), and the negative control group (P<0.01) (Fig. 2). These data indicated that the use
of miR-27a-3p inhibitor effectively inhibited its expression.
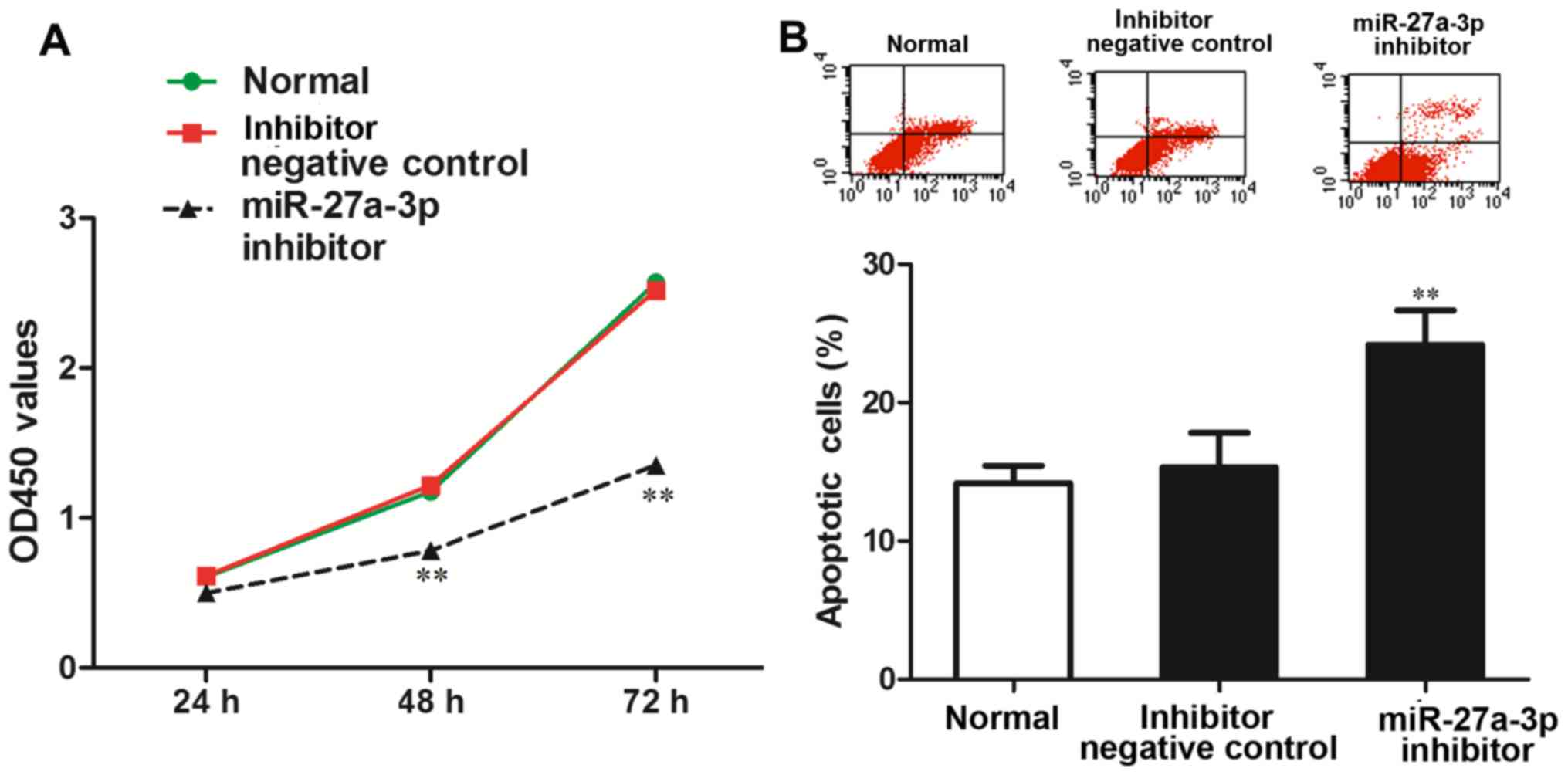
The effect of miR-27a-3p on MG-63 cell viability was
determined by CCK-8 assay. The results revealed that the OD450
value was significantly lower in the group transfected with the
miR-27a-3p inhibitor than in the normal control group and negative
control group (P<0.01) (Fig.
3A), which demonstrated that the inhibition of miR-27a-3p
expression reduced the ability of the MG-63 cells to proliferate,
suggesting that miR-27a-3p promotes the proliferation of MG-63
cells.
The effect of miR-27a-3p on the apoptosis of the
MG-63 cells was detected by flow cytometric analysis. The results
revealed that the proportion of apoptotic cells in the group
transfected with the miR-27a-3p inhibitor was significantly higher
than that in the normal control group and negative control group
(P<0.01) (Fig. 3B). These data
demonstrated that the inhibition of miR-27a-3p promoted MG-63 cell
apoptosis.
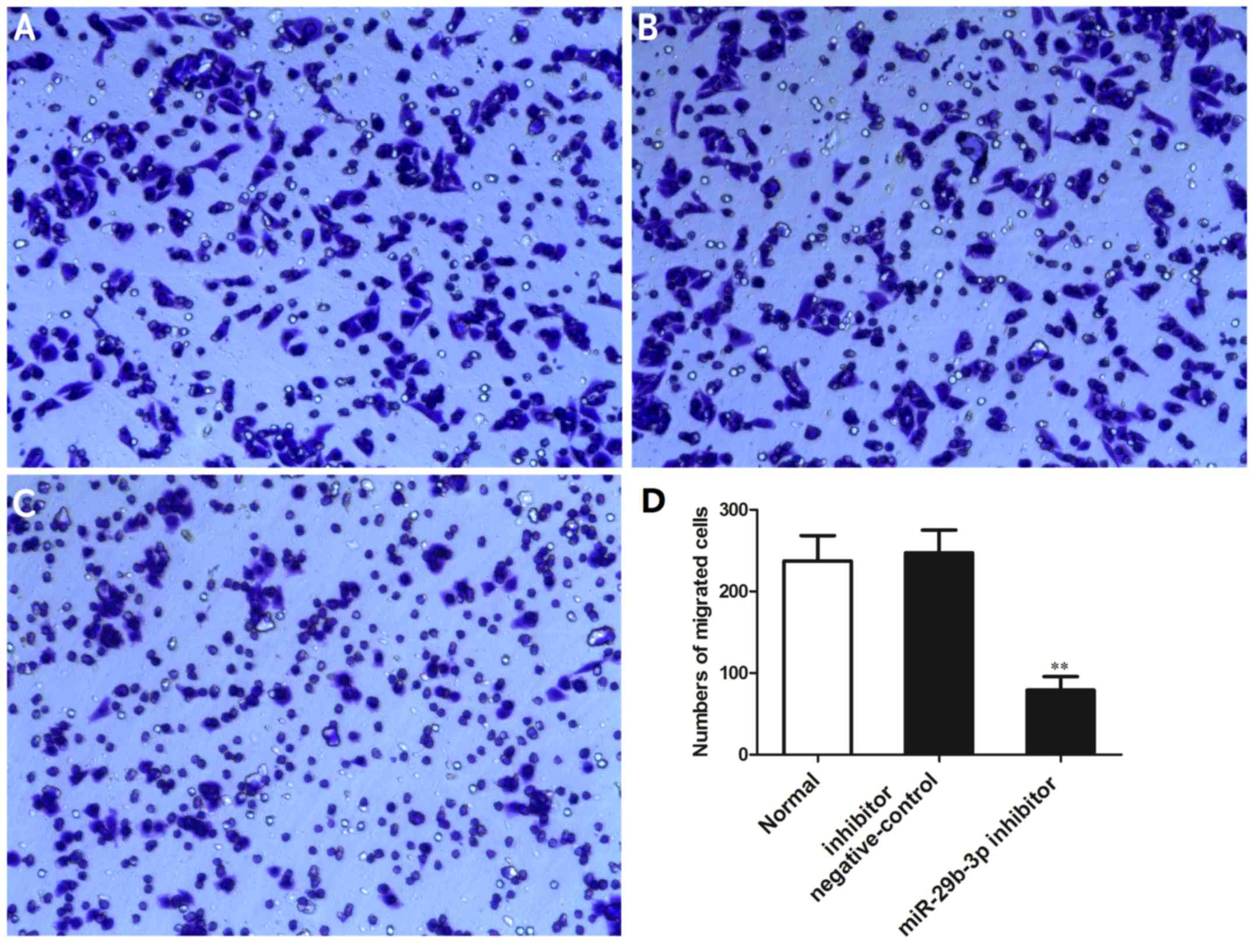
Downregulation of miR-27a-3p suppresses
the invasive ability of the MG-63 cells
The effects of miR-27a-3p on the invasive ability of
the MG-63 cells were examined by Transwell invasion assay. The
results indicated that the number of cells that passed through the
Transwell chamber was significantly reduced in the group
transfected with the miR-27a-3p inhibitor compared with the normal
control group and negative control group (P<0.01) (Fig. 4). These data demonstrated that the
downregulation of miR-27a-3p suppressed the invasive ability of the
osteosarcoma cells.
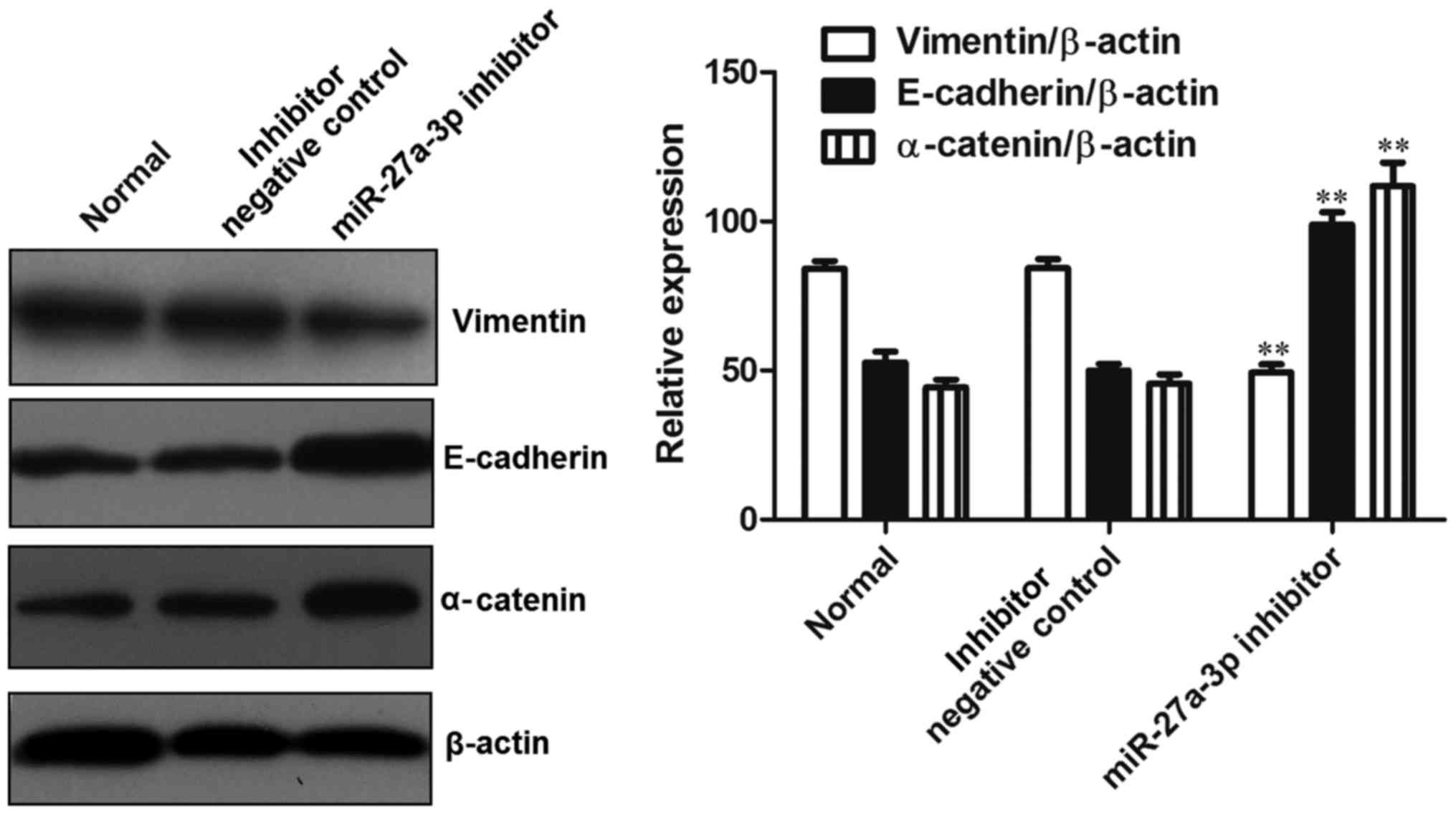
In order to explore the mechanisms responsible for
the promoting effects of miR-27a-3p on the invasion of osteosarcoma
cells, the expression levels of epithelial-mesenchymal transition
(EMT)-associated proteins (vimentin, E-cadherin and α-catenin) were
detected by western blot analysis. The results revealed that,
compared with the control group, the expression of vimentin was
significantly decreased (P<0.01), while the expression of
E-cadherin and α-catenin was significantly increased (P<0.01) in
the group transfected with the miR-27a-3p inhibitor (Fig. 5). These data indicated that the
downregulation of miR-27a-3p suppressed the expression of vimentin
and induced the expression of E-cadherin and α-catenin, thus
preventing EMT.
TET1 is the target gene of
miR-27a-3p
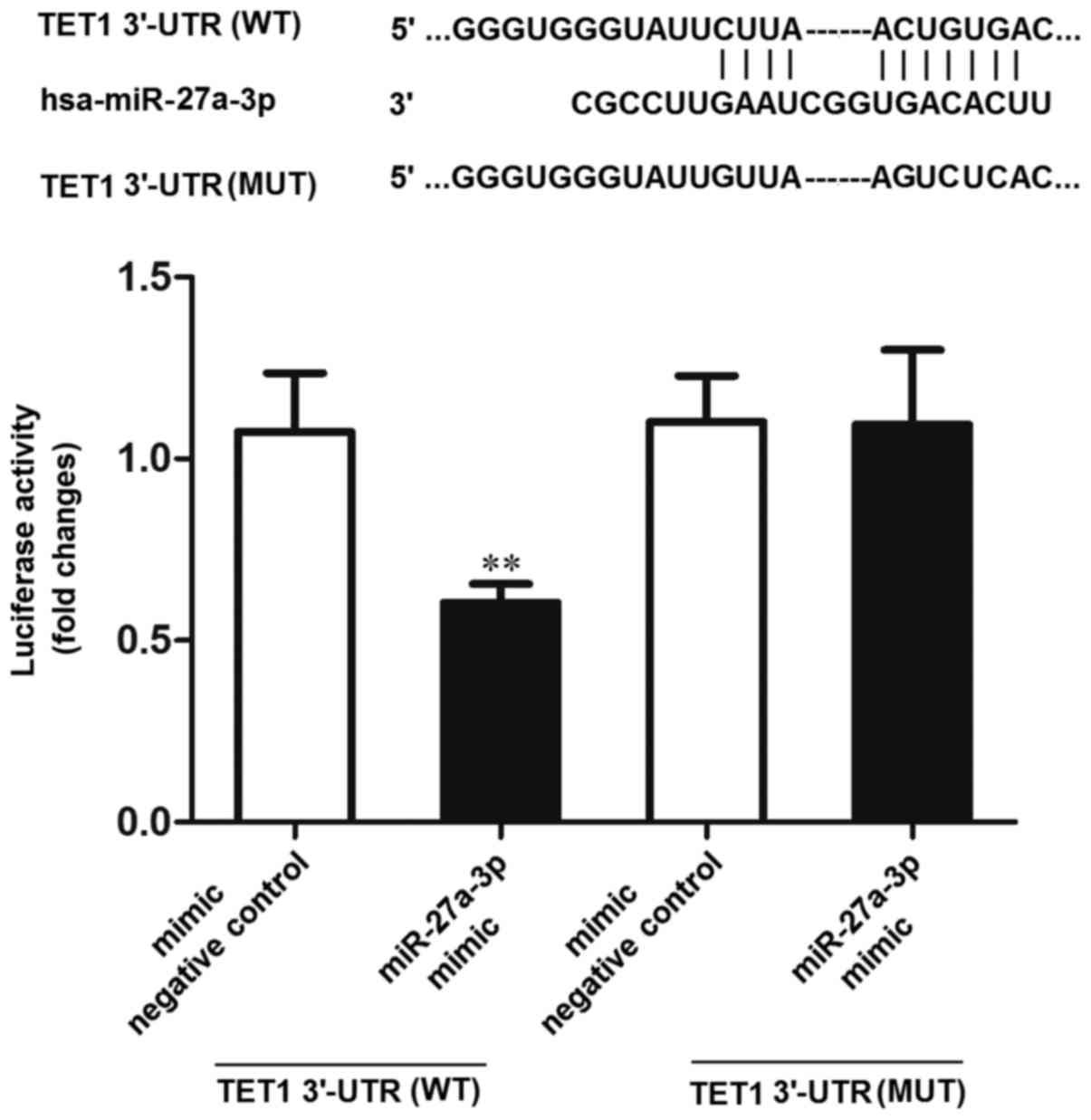
The TargetScan tools (http://www.targetscan.org/) were used to predict the
potential targets of miR-27a-3p. The results indicated that TET1
was one of the potential targets of miR-27a-3p. Subsequently, the
dual-luciferase reporter gene system was used to verify whether
TET1 was the target of miR-27a-3p. The results revealed that the
luciferase activity was significantly decreased in the 293 cells
co-transfected with miR-27a-3p mimic and TET1 3′-UTR (WT), compared
to the cells co-transfected with the negative control (mimic
negative control) and TET1 3′-UTR (WT) or miR-27a-3p mimic and TET1
3′-UTR (MUT) (P<0.01) (Fig. 6).
These results indicated that miR-27a-3p may directly interact with
the target site of the TET1 3′-UTR.
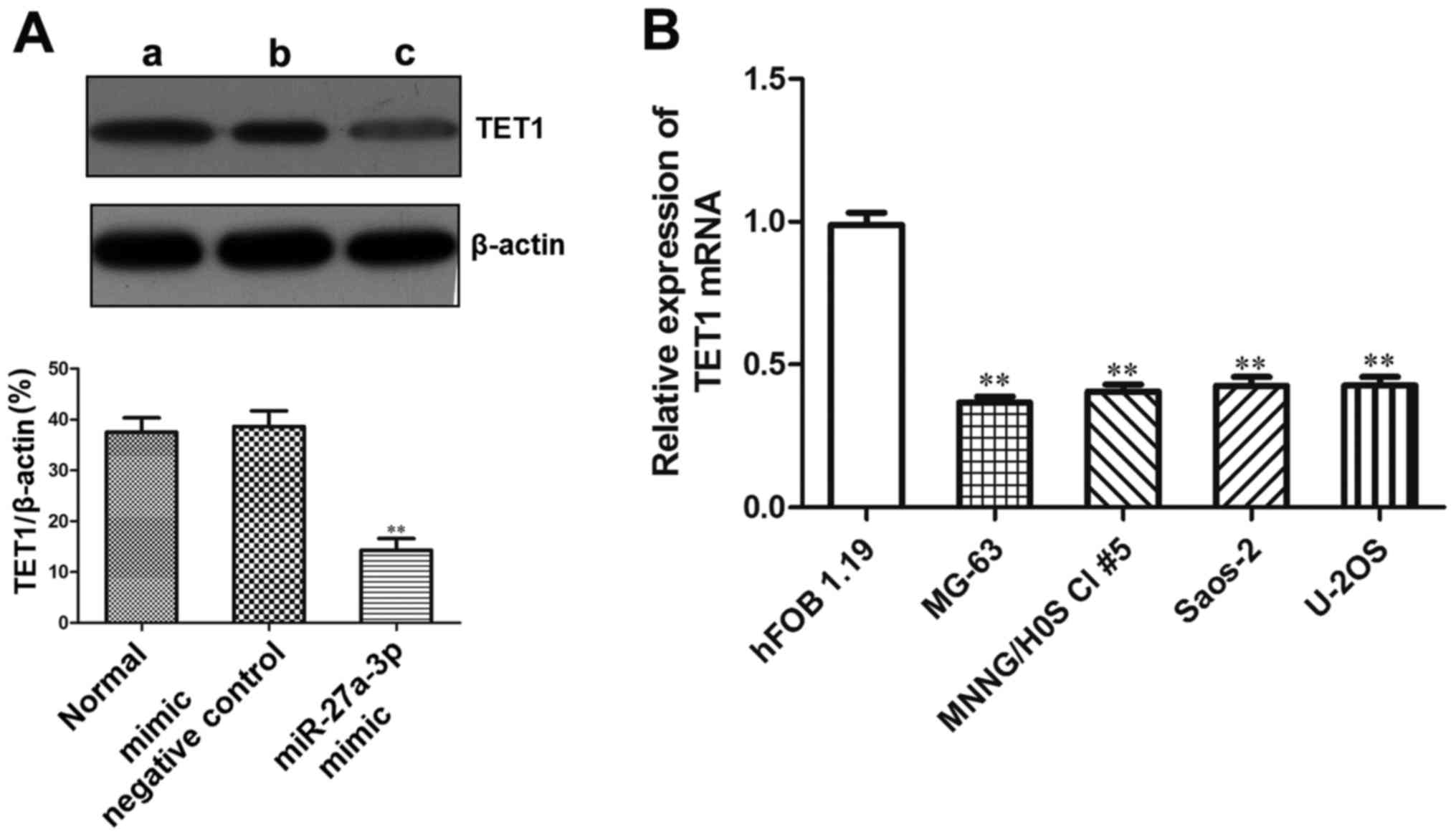
In order to explore the role of miR-27a-3p in
regulating the expression of TET1, western blot analysis was used
to detect the protein expression of TET1 in the MG-63 cells
transfected with miR-27a-3p. The results revealed that the protein
expression of TET1 was significantly lower in the group transfected
with the miR-27a-3p mimic (P<0.01) (Fig. 7A), which demonstrated that
miR-27a-3p inhibited the protein expression of TET1 in the MG-63
cells.
Since TET1 was found to be a target gene of
miR-27a-3p, and miR-27a-3p expression was upregulated in human
osteosarcoma cells, we then wished to examine TET1 expression in
osteosarcoma cells. RT-qPCR was used to detect the mRNA expression
of TET1 in the human osteosarcoma cell lines. The results revealed
that the mRNA expression of TET1 in the osteosarcoma cell lines
(MNNG/HOS Cl #5, MG-63, U-2OS and Saos-2) was significantly lower
than that in the normal human osteoblasts (hFOB 1.19) (P<0.01).
Its expression in the osteosarcoma cells was approximately 1/3
lower than that in the hFOB1.19 cells (Fig. 7B). These data demonstrated that
TET1 was the target gene of miR-27a-3p in osteosarcoma cells.
miR-27a-3p affects cellular biological
functions by regulating TET1
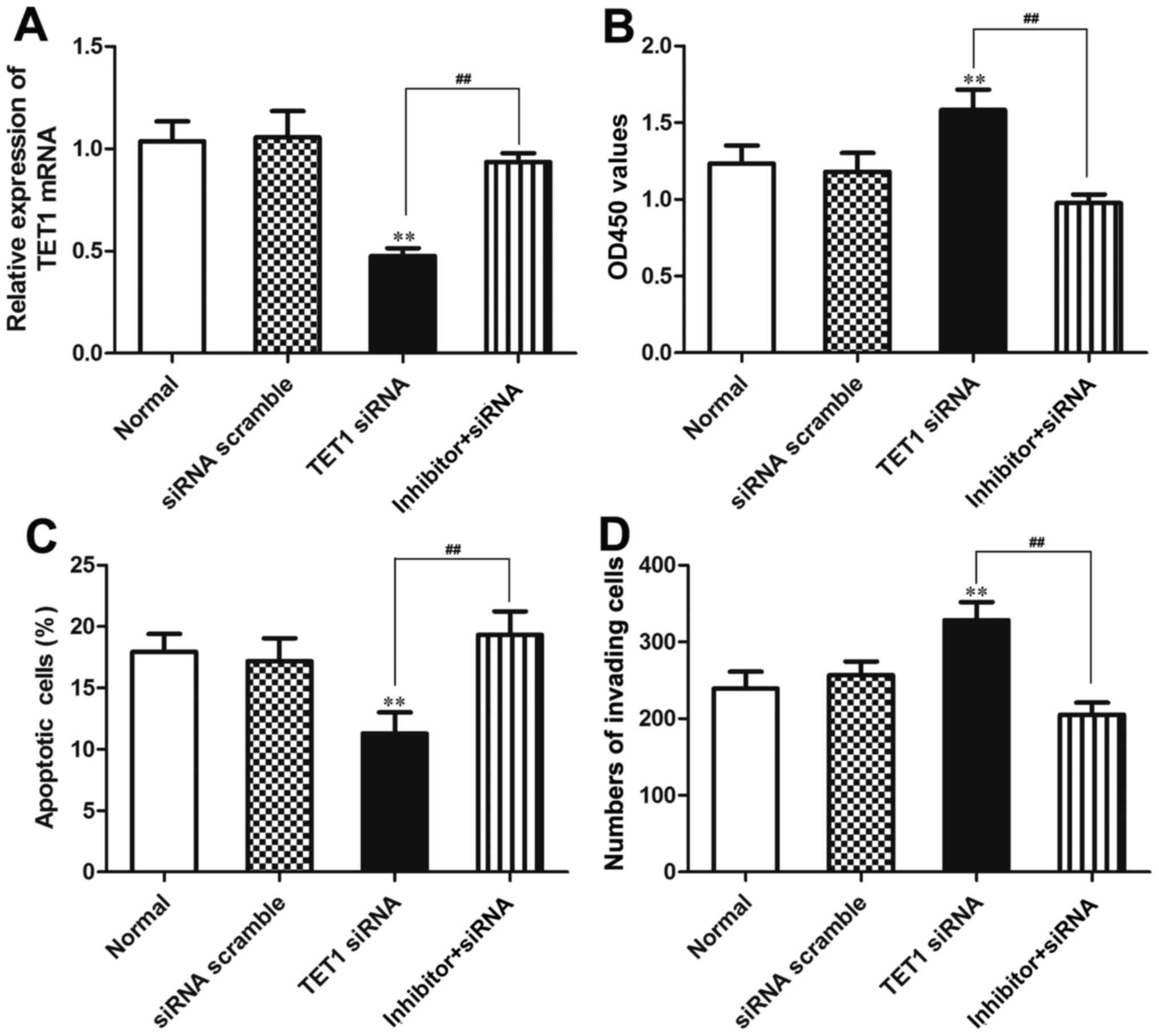
Our data indicated that TET1 was a target gene of
miR-27a-3p. In order to verify that miR-27a-3p plays a biological
role in osteosarcoma cells by regulating TET1, miR-27a-3p inhibitor
and TET1 siRNA were co-transfected into the MG-63 cells using
Lipofectamine 2000. We further verified that the expression of TET1
was regulated by miR-27a-3p in the MG-63 cells. The results
revealed that the mRNA expression of TET1 was significantly
decreased (P<0.01) in the group transfected with TET1 siRNA, and
was upregulated in the group co-transfected with miR-27a-3p
inhibitor and TET1 siRNA compared to the group transfected with
TET1 siRNA (P<0.01). The mRNA expression of TET1 did not differ
significantly between the normal control group (Normal) and the
siRNA negative control group (siRNA scramble) (Fig. 8A).
CCK-8 assay, flow cytometry and Transwell assay were
used to further verify that miR-27a-3p plays a biological role by
regulating the expression of TET1. The results from CCK-8 assay
revealed that the OD450 value was significantly increased in the
group transfected with TET1 siRNA compared to that in the group
co-transfected with miR-27a-3p inhibitor and TET1 siRNA
(P<0.01), which indicated that the viability of the MG-63 cells
was enhanced in the group transfected with TET1 siRNA compared to
that in the group co-transfected with miR-27a-3p inhibitor and TET1
siRNA. Thus, the inhibition of TET1 expression may promote the
proliferation of MG-63 osteosarcoma cells. The OD450 value of the
MG-63 cells in the group co-transfected with miR-27a-3p inhibitor
and TET1 siRNA did not differ significantly compared with that in
the normal control group and siRNA negative control group (siRNA
scramble) (Fig. 8B). The results
from flow cytometry demonstrated that the number of apoptotic cells
in the group co-transfected with miR-27a-3p inhibitor and TET1
siRNA was significantly higher than that in the group transfected
with TET1 siRNA (P<0.01); no significant differences were
observed between the normal control group and the siRNA negative
control group (Fig. 8C). The
results from Transwell assay revealed that the number of cells
invading through the Transwell chamber in the group co-transfected
with the miR-27a-3p inhibitor and TET1 siRNA was significantly
lower than that in the group transfected with TET1 siRNA
(P<0.01); no significant differences were found between the
normal control group and the siRNA negative control group (Fig. 8D). These data demonstrated that
TET1 inhibition attenuated the biological effects induced by
miR-27a-3p inhibition in the MG-63 cells, also suggesting that
miR-27a-3p affects cellular biological functions by regulating TET1
in osteosarcoma.
Discussion
In different tumor types, miRNAs play two different
roles: As tumor suppressor genes and as oncogenes. Their main
mechanisms of action include the loss of loci, amplification,
mutations, alterations at the epigenetic level and the abnormal
expression of transcription factors (39,40).
miR-27a is an evolutionarily conserved family, and has been found
to have an abnormal expression in certain types of tumors, such as
in breast cancer, gastric cancer and esophageal cancer (35,41,42).
However, the role of miR-27a in osteosarcoma has not yet been fully
elucidated. Therefore, it is necessary to further examine the role
and mechanisms of action of miR-27a in osteosarcoma.
There are a number of studies available on miR-27
and tumors. Zhou et al reported that miR-27a-3p plays the
role of a proto-oncogene in gastric cancer via the
BAK-SMAC/DIABLO-XIAP axis (44).
miR-27a acts as a proto-oncogene by regulating the expression of
MHC class I in colorectal cancer (43). In addition, a number of studies
have confirmed that miR-27a-3p is related to the sensitivity of
cells to chemotherapy and radiotherapy (44). Therefore, in this study, RT-qPCR
was used to detect the expression of miR-27a-3p in human
osteosarcoma cell lines. The results revealed that the expression
of miR-27a-3p was markedly upregulated in the osteosarcoma cell
lines compared with the normal osteoblasts, hFOB 1.19 cells, which
suggested that miR-27a-3p plays an important role in the
development of osteosarcoma.
In order to explore the role of miR-27a-3p in
osteosarcoma, the effects of miR-27a-3p on the viability, apoptosis
and invasive ability of osteosarcoma cells were determined, and the
results revealed that miR-27a-3p inhibition decreased the
proliferative ability, contributed to cell apoptosis, and reduced
the invasive ability of the osteosarcoma cells. Based on these
findings, it can be speculated that miR-27a-3p suppresses the
apoptosis of osteosarcoma cells, and promotes the growth and
invasion of osteosarcoma cells. Metastasis is the leading cause of
mortality in patients with osteosarcoma (45).
The main reason for the distant metastasis of
osteosarcoma is the abnormal expression of tumor metastasis-related
genes and metastasis suppressor genes (16,17).
Increasing evidence has indicated that EMT is associated with the
invasive and metastatic behavior of cells during cancer progression
(46,47). In the process of EMT in tumors, the
expression of epithelial markers (E-cadherin and α-catenin)
undergoes a downregulation and the expression of mesenchymal
markers (vimentin) undergoes an upregulation, which indicates that
epithelium-derived tumor cells are losing cell polarity, the
connections between cells become loose and intracellular
cytoskeletal proteins undergo recombination (48). Therefore, in this study, the
expression of EMT-related proteins (E-cadherin, α-catenin and
vimentin) was detected in the MG-63 cells. The results revealed
that miR-27a-3p inhibition decreased the expression of vimentin,
and induced the expression of E-cadherin and α-catenin, which
indicated that the downregulation of miR-27a-3p suppressed EMT in
osteosarcoma. On the other hand, it also indicated that miR-27a-3p
promoted the expression of vimentin, inhibited by the expression of
E-cadherin and α-catenin, so as to promote the invasion and
migration of osteosarcoma cells.
miRNAs are small (approximately 21 nucleotides in
length) non-coding RNAs that play pivotal roles in cellular and
developmental processes by regulating gene expression at the
post-transcriptional level (49).
Our data demonstrated that miR-27a-3p interacted with TET1, and
that miR-27a-3p inhibited the expression of TET1 protein in
osteosarcoma cells. TET1 inhibits the invasion of cancer cells by
inhibiting the methylation of matrix metalloproteinase 2 and 3, and
maintaining the expression of tissue inhibitor of matrix
metalloproteinases to achieve the inhibition of cancer cell
invasion (50). In order to
explore whether miR-27a-3p plays a biological role by regulating
TET1, miR-27a-3p inhibitor and TET1 siRNA were co-transfected into
the MG-63 cells. Our data demonstrated that TET1 inhibition
attenuated the biological effects induced by miR-27a-3p inhibition
in osteosarcoma cells, also suggesting that miR-27a-3p promoted the
malignant phenotypes of osteosarcoma by targeting TET1.
In conclusion, our data demonstrated that miR-27a-3p
is upregulated in human osteosarcoma cells. miR-27a-3p inhibition
suppressed the viability and invasive ability of the osteosarcoma
cells, and promoted cell apoptosis. At the same time, miR-27a-3p
inhibition increased the expression of E-cadherin and α-catenin,
and decreased the expression of vimentin in osteosarcoma cells. Our
data also demonstrated that miR-27a-3p interacted with TET1, and
TET1 knockdown attenuated the biological effects induced by
miR-27a-3p in osteosarcoma cells. on the whole, this study
demonstrates that miR-27a-3p promotes the malignant phenotypes of
osteosarcoma by targeting TET1; thus, miR-27a-3p may prove to be a
potential prognostic marker and therapeutic target for patients
with osteosarcoma.
Acknowledgments
Not applicable.
Notes
[1]
Funding
This study was funded by the Natural Science
Foundation of Jiangsu Province (grant no. BK20131199), by the
fifty-fifth batch of China Postdoctoral Science Foundation (grant
no. 2014M551640) and by the Project of Jiangsu provincial Health
and Family Planning Commission (grant no. H201524).
[2] Availability
of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
[3] Authors'
contributions
JZ and JL designed the research and wrote the
manuscript. JL and ML prepared and completed most of the
experiments. JL, ML, XL and FL collected the data, and analyzed and
interpreted the data. All authors have read and approved the final
manuscript.
[4] Ethics
approval and consent to participate
Not applicable.
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Bilbao-Aldaiturriaga N, Askaiturrieta Z,
Granado-Tajada I, Goričar K, Dolžan V, Garcia-Miguel P, Garcia de
Andoin N, Martin-Guerrero I and Garcia-Orad A; For The Slovenian
Osteosarcoma Study Group: A systematic review and meta-analysis of
MDM2 polymorphisms in osteosarcoma susceptibility. Pediatr Res.
80:472–479. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anderson PM, Bielack SS, Gorlick RG,
Skubitz K, Daw NC, Herzog CE, Monge OR, Lassaletta A, Boldrini E,
Pápai Z, et al: A phase II study of clinical activity of SCH 717454
(robatumumab) in patients with relapsed osteosarcoma and Ewing
sarcoma. Pediatr Blood Cancer. 63:1761–1770. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nikitovic D, Kavasi RM, Berdiaki A,
Papachristou DJ, Tsiaoussis J, Spandidos DA, Tsatsakis AM and
Tzanakakis GN: Parathyroid hormone/parathyroid hormone-related
peptide regulate osteosarcoma cell functions: Focus on the
extracellular matrix (Review). Oncol Rep. 36:1787–1792. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liao LQ, Yan HH, Mai JH, Liu WW, Li H, Guo
ZM, Zeng ZY and Liu XK: Radiation-induced osteosarcoma of the
maxilla and mandible after radiotherapy for nasopharyngeal
carcinoma. Chin J Cancer. 35:892016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bami M, Mavrogenis AF, Angelini A,
Milonaki M, Mitsiokapa E, Stamoulis D and Soucacos PN: Bone
morphogenetic protein signaling in musculoskeletal cancer. J Cancer
Res Clin Oncol. 142:2061–2072. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Angelini A, Mavrogenis AF, Trovarelli G,
Ferrari S, Picci P and Ruggieri P: Telangiectatic osteosarcoma: A
review of 87 cases. J Cancer Res Clin Oncol. 142:2197–2207. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Z, Li B, Ren Y and Ye Z: T-cell-based
immunotherapy for osteosarcoma: Challenges and opportunities. Front
Immunol. 7:3532016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Osasan S, Zhang M, Shen F, Paul PJ, Persad
S and Sergi C: Osteogenic sarcoma: A 21st century review.
Anticancer Res. 36:4391–4398. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nataraj V, Rastogi S, Khan SA, Sharma MC,
Agarwala S, Vishnubhatla S and Bakhshi S: Prognosticating
metastatic osteosarcoma treated with uniform chemotherapy protocol
without high dose methotrexate and delayed metastasectomy: A single
center experience of 102 patients. Clin Transl Oncol. 18:937–944.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu K, Dai HB and Qiu ZL: mTOR signaling in
osteosarcoma: Oncogenesis and therapeutic aspects (Review). Oncol
Rep. 36:1219–1225. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee JA, Jeon DG, Cho WH, Song WS, Yoon HS,
Park HJ, Park BK, Choi HS, Ahn HS, Lee JW, et al: Higher
gemcitabine dose was associated with better outcome of osteosarcoma
patients receiving gemcitabine-docetaxel chemotherapy. Pediatr
Blood Cancer. 63:1552–1556. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kebudi R, Ozger H, Kızılocak H, Bay SB and
Bilgiç B: Osteosarcoma after hematopoietic stem cell
transplantation in children and adolescents: Case report and review
of the literature. Pediatr Blood Cancer. 63:1664–1666. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Adamopoulos C, Gargalionis AN, Piperi C
and Papavassiliou AG: Recent advances in mechanobiology of
osteosarcoma. J Cell Biochem. 118:232–236. 2017. View Article : Google Scholar
|
|
14
|
Li CJ, Liu XZ, Zhang L, Chen LB, Shi X, Wu
SJ and Zhao JN: Advances in bone-targeted drug delivery systems for
neoadjuvant chemotherapy for osteosarcoma. Orthop Surg. 8:105–110.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Abarrategi A, Tornin J, Martinez-Cruzado
L, Hamilton A, Martinez-Campos E, Rodrigo JP, González MV, Baldini
N, Garcia-Castro J and Rodriguez R: Osteosarcoma: Cells-of-origin,
cancer stem cells, and targeted therapies. Stem Cells Int.
2016:36317642016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang SD, Li HY, Li BH, Xie T, Zhu T, Sun
LL, Ren HY and Ye ZM: The role of CTLA-4 and PD-1 in anti-tumor
immune response and their potential efficacy against osteosarcoma.
Int Immunopharmacol. 38:81–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ding L, Congwei L, Bei Q, Tao Y, Ruiguo W,
Heze Y, Bo D and Zhihong L: mTOR: An attractive therapeutic target
for osteosarcoma? Oncotarget. 7:50805–50813. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ravindra VM, Eli IM, Schmidt MH and
Brockmeyer DL: Primary osseous tumors of the pediatric spinal
column: Review of pathology and surgical decision making. Neurosurg
Focus. 41:E32016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hurley C, McCarville MB, Shulkin BL, Mao
S, Wu J, Navid F, Daw NC, Pappo AS and Bishop MW: Comparison of
(18) F-FDG-PET-CT and bone scintigraphy for evaluation of osseous
metastases in newly diagnosed and recurrent osteosarcoma. Pediatr
Blood Cancer. 63:1381–1386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stope MB, Koensgen D, Weimer J, Paditz M,
Burchardt M, Bauerschlag D and Mustea A: The future therapy of
endometrial cancer: microRNA's functionality, capability, and
putative clinical application. Arch Gynecol Obstet. 294:889–895.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fang Q, Xu T, Wu C, Zhou S and Sun H:
Biotargets in neural regeneration. Biotarget. 1:62017. View Article : Google Scholar
|
|
23
|
Wang DD, Chen X, Yu DD, Yang SJ, Shen HY,
Sha HH, Zhong SL, Zhao JH and Tang JH: miR-197: A novel biomarker
for cancers. Gene. 591:313–319. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang C, Zheng SD, Wu HJ and Chen SJ:
Regulatory mechanisms of the molecular pathways in fibrosis induced
by microRNAs. Chin Med J (Engl). 129:2365–2372. 2016. View Article : Google Scholar
|
|
25
|
Lee K and Ferguson LR: MicroRNA biomarkers
predicting risk, initiation and progression of colorectal cancer.
World J Gastroenterol. 22:7389–7401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matin F, Jeet V, Clements JA, Yousef GM
and Batra J: MicroRNA theranostics in prostate cancer precision
medicine. Clin Chem. 62:1318–1333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rajasekaran S, Pattarayan D, Rajaguru P,
Sudhakar Gandhi PS and Thimmulappa RK: MicroRNA regulation of acute
lung injury and acute respiratory distress syndrome. J Cell
Physiol. 231:2097–2106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Connelly CM, Moon MH and Schneekloth JS
Jr: The emerging role of RNA as a therapeutic target for small
molecules. Cell Chem Biol. 23:1077–1090. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Li J, Sun X, Chen J, Sun X, Zheng J
and Chen R: MicroRNA-27a functions as a tumor suppressor in renal
cell carcinoma by targeting epidermal growth factor receptor. Oncol
Lett. 11:4217–4223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen S, Sun YY, Zhang ZX, Li YH, Xu ZM and
Fu WN: Transcriptional suppression of microRNA-27a contributes to
laryngeal cancer differentiation via GSK-3β-involved Wnt/β-catenin
pathway. Oncotarget. 8:14708–14718. 2017.PubMed/NCBI
|
|
31
|
Jiang H, Zhang G, Wu JH and Jiang CP:
Diverse roles of miR-29 in cancer (review). Oncol Rep.
31:1509–1516. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kollinerova S, Vassanelli S and Modriansky
M: The role of miR-29 family members in malignant hematopoiesis.
Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 158:489–501.
2014.PubMed/NCBI
|
|
33
|
Pan W, Wang H, Jianwei R and Ye Z:
MicroRNA-27a promotes proliferation, migration and invasion by
targeting MAP2K4 in human osteosarcoma cells. Cell Physiol Biochem.
33:402–412. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou L, Liang X, Zhang L, Yang L, Nagao N,
Wu H, Liu C, Lin S, Cai G and Liu J: miR-27a-3p functions as an
oncogene in gastric cancer by targeting BTG2. Oncotarget.
7:51943–51954. 2016.PubMed/NCBI
|
|
35
|
Hu D and Shilatifard A: Epigenetics of
hematopoiesis and hematological malignancies. Genes Dev.
30:2021–2041. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Forloni M, Gupta R, Nagarajan A, Sun LS,
Dong Y, Pirazzoli V, Toki M, Wurtz A, Melnick MA, Kobayashi S, et
al: Oncogenic EGFR represses the TET1 DNA demethylase to induce
silencing of tumor suppressors in cancer cells. Cell Reports.
16:457–471. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li L, Li C, Mao H, Du Z, Chan WY, Murray
P, Luo B, Chan AT, Mok TS, Chan FK, et al: Epigenetic inactivation
of the CpG demethylase TET1 as a DNA methylation feedback loop in
human cancers. Sci Rep. 6:265912016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
39
|
Zhou K, Liu M and Cao Y: New insight into
microRNA functions in cancer: Oncogene-microRNA-tumor suppressor
gene network. Front Mol Biosci. 4:462017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kaur S, Lotsari-Salomaa JE,
Seppänen-Kaijansinkko R and Peltomäki P: MicroRNA methylation in
colorectal cancer. Adv Exp Med Biol. 937:109–122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang W, Zhu J, Su S, Wu W, Liu Q, Su F and
Yu F: miR-27 as a prognostic marker for breast cancer progression
and patient survival. PLoS One. 7:e517022012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tanaka K, Miyata H, Sugimura K, Fukuda S,
Kanemura T, Yamashita K, Miyazaki Y, Takahashi T, Kurokawa Y,
Yamasaki M, et al: miR-27 is associated with chemoresistance in
esophageal cancer through transformation of normal fibroblasts to
cancer-associated fibroblasts. Carcinogenesis. 36:894–903. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Colangelo T, Polcaro G, Ziccardi P, Pucci
B, Muccillo L, Galgani M, Fucci A, Milone MR, Budillon A,
Santopaolo M, et al: Proteomic screening identifies calreticulin as
a miR-27a direct target repressing MHC class I cell surface
exposure in colorectal cancer. Cell Death Dis. 7:e21202016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhou S, Huang Q, Zheng S, Lin K, You J and
Zhang X: miR-27a regulates the sensitivity of breast cancer cells
to cisplatin treatment via BAK-SMAC/DIABLO-XIAP axis. Tumour Biol.
37:6837–6845. 2016. View Article : Google Scholar
|
|
45
|
Mirabello L, Koster R, Moriarity BS,
Spector LG, Meltzer PS, Gary J, Machiela MJ, Pankratz N, Panagiotou
OA, Largaespada D, et al: A genome-wide scan identifies variants in
NFIB associated with metastasis in patients with osteosarcoma.
Cancer Discov. 5:920–931. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lv YF, Dai H, Yan GN, Meng G, Zhang X and
Guo QN: Downregulation of tumor suppressing STF cDNA 3 promotes
epithelial-mesenchymal transition and tumor metastasis of
osteosarcoma by the Wnt/GSK-3β/β-catenin/Snail signaling pathway.
Cancer Lett. 373:164–173. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu Y and Jiang M: The revolution of lung
cancer treatment: From vaccines, to immune checkpoint inhibitors,
to chimeric antigen receptor T therapy. Biotarget. 1:72017.
View Article : Google Scholar
|
|
48
|
Sun L and Fang J: Epigenetic regulation of
epithelial-mesenchymal transition. Cell Mol Life Sci. 73:4493–4515.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tufekci KU, Oner MG, Genc S and Genc K:
MicroRNAs and multiple sclerosis. Autoimmune Dis.
2011:8074262010.PubMed/NCBI
|
|
50
|
Lu HG, Zhan W, Yan L, Qin RY, Yan YP, Yang
ZJ, Liu GC, Li GQ, Wang HF, Li XL, et al: TET1 partially mediates
HDAC inhibitor-induced suppression of breast cancer invasion. Mol
Med Rep. 10:2595–2600. 2014. View Article : Google Scholar : PubMed/NCBI
|