Introduction
Lung cancer is the leading cause of
cancer-associated mortality worldwide; in 2012, 1.8 million new
cases of lung cancer were diagnosed and it was responsible for 1.6
million deaths (1). In the past
decade, epidermal growth factor receptor-tyrosine kinase inhibitors
(EGFR-TKIs), including gefitinib, erlotinib and afatinib, have been
widely used in the treatment of patients with non-small cell lung
cancer (NSCLC). The EGFR gene encodes a pharmacologically
targetable tyrosine kinase, and patients with NSCLC harboring
EGFR mutations, such as deletions in exon 19 and L858R in
exon 21, exhibit notable responses to EGFR-TKIs, resulting in
improved prognoses compared to those achieved with standard
chemotherapies (2–7). In addition, osimertinib, which is a
third-generation TKI that specifically targets the T790M secondary
EGFR mutation, has been demonstrated to exhibit clinical
efficacy (8). Since the
therapeutic effect of EGFR-TKIs is strongly dependent upon the
EGFR mutation status of patients (9), reliable mutation detection methods
are required to facilitate personalized lung cancer treatments.
At present, in clinical practice, the molecular
testing guidelines (10) for the
selection of patients with lung cancer for EGFR-TKI administration
recommend using validated polymerase chain reaction (PCR)-based
assays and specimens containing sufficient cancer cells to provide
accurate results. Although tissue specimens, such as transbronchial
biopsy or surgically resected specimens should be prioritized, the
guidelines also recommend using cytological specimens, as it is
often difficult to obtain sufficient tissue specimens for mutation
analysis. Numerous studies have reported the sensitivity and
reliability of using cytology specimens for EGFR testing (11–13).
Furthermore, recent advances in highly sensitive genotyping have
allowed the development of liquid biopsies, which examine
circulating tumor cells or freely circulating cell-free DNA (cfDNA)
isolated from the serum or plasma (14–16).
A liquid biopsy is a useful and minimally invasive method,
particularly for patients who require rebiopsy to confirm acquired
EGFR T790M mutation (16).
However, the amount of cfDNA in the bloodstream is extremely low;
therefore, liquid biopsies require highly sensitive assay
platforms, which are often slow to perform and yield limited
detection rates (17–19).
Cytology specimens, including bronchial lavage fluid
(BLF), are usually obtained directly from the tumor location, and
their supernatants are expected to have a higher amount of cfDNA
derived from tumor cells than the blood. If cfDNA supernatants from
cytology specimens are available for EGFR mutation
detection, cell pellets can be reserved for additional
morphological and molecular analyses. Our previous study revealed a
novel rapid point-of-care system for the detection of EGFR
mutations using droplet-PCR (d-PCR) to assess cell pellets of
cytology specimens (EGFR d-PCR assay) (20). This EGFR d-PCR assay reduced
the reaction time to <10 min and exhibited sensitivity as high
as that achieved using conventional PCR assays. The purpose of the
present study was to validate the performance of the EGFR
d-PCR assay in assessing cfDNA from supernatants obtained from
cytology specimens. Briefly, the results of the assay were compared
to those achieved via the EGFR d-PCR assay of
cytology-specimen cell pellets, as well as those obtained via a
cytological diagnosis of the corresponding cell pellets. In
addition, conventional EGFR assays were conducted using tissue or
cytology specimens in order to confirm the accuracy of the
EGFR d-PCR assay using cytology specimens.
Materials and methods
Patients
The present study enrolled 90 patients who had been
diagnosed with or were radiologically suspected to have lung cancer
(including benign disease), and whose cytological specimens were
submitted to the Department of Laboratory Medicine at the Shinshu
University Hospital (Matsumoto, Japan) between July and November
2016. All patients received medical treatments or follow-up
examinations in Shinshu University Hospital following specimen
collection. The 90 cytological specimens comprised the following:
80 samples, BLF; nine samples, pleural effusion (PE); and one
sample, spinal fluid (SP). BLF specimens were obtained following
transbronchial lung biopsies (TBLB), which were performed to either
assess an undiagnosed lung mass, or perform T790M screening in
patients with diagnosed lung cancer. PE and SP specimens were
obtained via fine-needle aspiration from patients with advanced
lung cancer. Of the 15 patients that were revealed to be positive
for EGFR mutations, as determined by the EGFR d-PCR
assay using cytology specimens, 14 formalin-fixed paraffin-embedded
(FFPE) tissue specimens (11 samples, TBLB; three samples, surgical
resection) and one FFPE cell block from a cytology specimen (PE)
were collected to confirm the mutation status using conventional
assays. All patients provided written informed consent for their
participation in the present study, which was reviewed and approved
by the medical ethics committee of the Shinshu University School of
Medicine.
Processing and pathological evaluation of
specimens
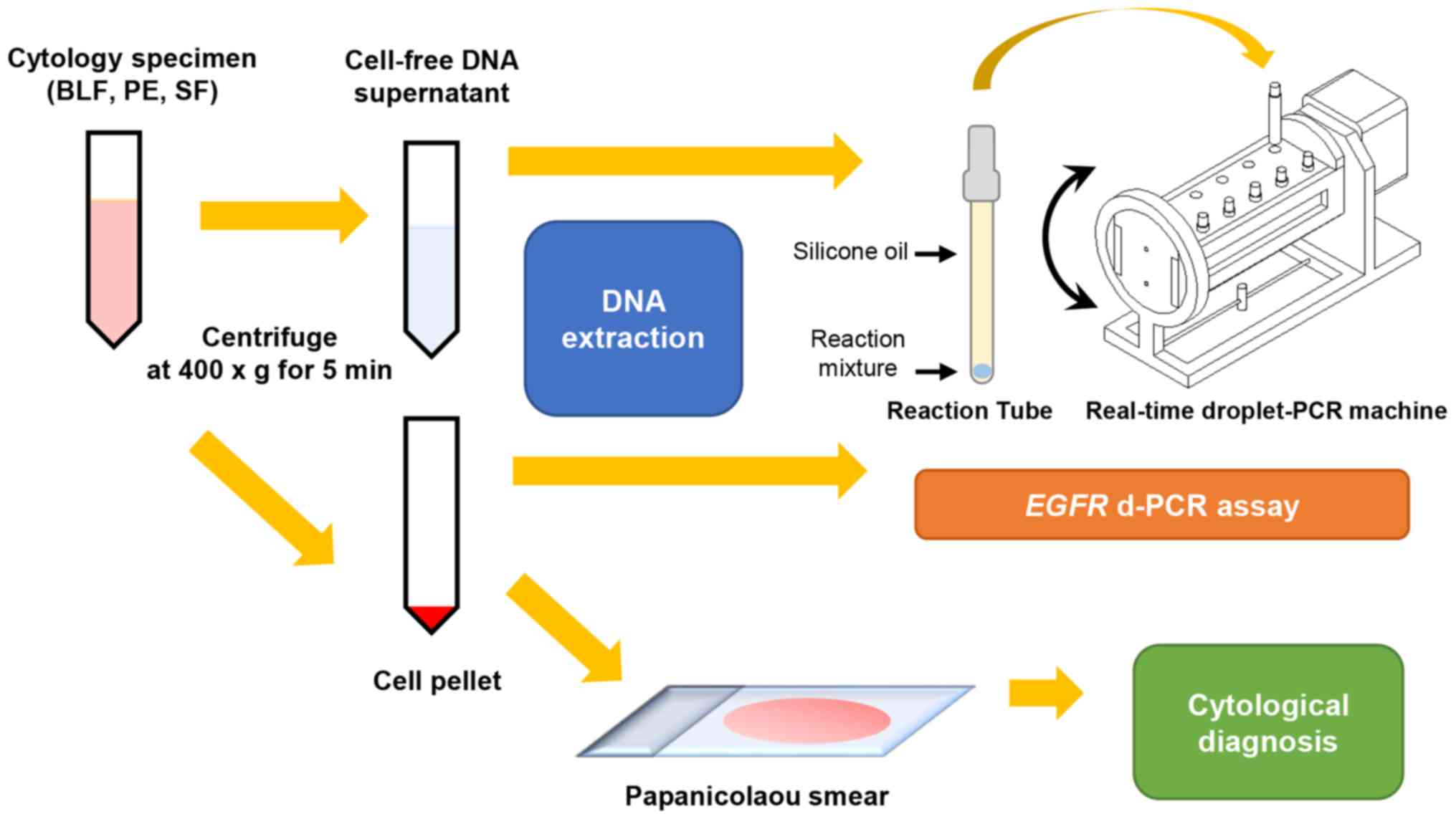
Processing of the materials is shown in Fig. 1. Briefly, each cytological specimen
was immediately centrifuged (400 × g, 5 min, room temperature) upon
reception by the department, and was then divided into a
cell-pellet and a cfDNA supernatant fraction. The cell pellet was
further divided into two portions, one of which was used to prepare
the Papanicolaou smear for cytological analysis, whereas the other
was used for DNA extraction. Cytological diagnosis was performed by
one cytotechnologist, and reviewed by two pathologists. Specimens
were classified according to the standardized terminology proposed
by the Papanicolaou Society of Cytopathology, as either
‘malignant’, ‘suspicious of malignancy’, ‘neoplastic, benign
neoplasm, low-grade carcinoma’, ‘atypical’, ‘negative for
malignancy’ or ‘non-diagnostic’ (21). Both fresh cell pellets and cfDNA
supernatants were stored at −20°C.
DNA extraction
DNA was extracted from cell-pellet portions and 1.5
ml cell-free supernatant aliquots using the QIAamp DNA Blood Mini
kit (Qiagen, Inc., Valencia, CA, USA) according to the
manufacturer’s protocol. For FFPE tissue and cell block specimens,
the QIAamp DNA FFPE Tissue kit (Qiagen, Inc.) was used to extraxt
DNA according to the manufacturer’s protocol.
For the EGFR d-PCR assay, the concentration
of extracted DNA from each specimen was quantified via
spectrophotometry using a NanoDrop ND100 instrument (NanoDrop
Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA),
and was adjusted to a concentration of <10 ng/μl with AE
Buffer from the QIAamp DNA extraction kit.
EGFR d-PCR assay
The EGFR d-PCR assay was performed using a
d-PCR machine (Seiko Epson Corporation, Suwa, Japan) as previously
described (20). Briefly, the
EGFR d-PCR assay is designed to detect three major
EGFR mutations: L858R in exon 21, E746_A750del in exon 19
and T790M in exon 20, in 8 min and 10 sec. The primers and probes
used in the present study were as follows: L858R, forward
5′-GCTTGGTGCACCGCGACCTG-3′, reverse 5′-CGCACCCAGCAGTTTGGCAC-3′,
probe 5′-6FAM-AGCCAGGAACGTACTG GTGAAAACACCGCA-BHQ-1-3′;
E746_A750del, forward 5′-GGCAGCATGTGGCACCATC-3′, reverse
5′-GTTGGCTTTCGGAGATGTAT-3′, probe
5′′-6FAM-TCTCACCTTCTGGGATCCAGAGTCCCT-BHQ-1-3′; T790M, forward
5′-CCCCACGTGTGCCGCCTG-3′, reverse 5′-GCCGAAGGGCATGAGCTGTA-3′, and
probe 5′-6FAM-TGGGCATCTGCCTCACCTCCACCGTGCA-BHQ-1-3′. Each reaction
mixture contained genomic DNA (<10 ng/μl), Platinum Taq
DNA polymerase (Thermo Fisher Scientific, Inc., Waltham, MA, USA),
appropriate primers (800 nmol/l), TaqMan probe (300 nmol/l) and
sufficient reaction buffer [comprised of Tris-HCl (pH 9.0), KCl and
MgCl2] to reach a total volume of 10 μl. A
1.6-μl aliquot of each reaction mixture was placed in each
reaction tube filled with silicone oil, and subjected to the
following reaction conditions: 98°C for 10 sec, followed by 40
cycles at 98°C for 5 sec, and either 60°C for 6 sec (L858R) or 55°C
for 6 sec (E746_A750del and T790M). PCR results were determined to
be either EGFR-mutation positive or negative according to
threshold fluorescence level values of 4.7, 4.7 and 6.8 for L858R,
E746_A750del and T790M, respectively. The EGFR d-PCR assay
was previously shown to exhibit high sensitivity, and concordance
with the commercial PCR therascreen assay, which uses the
Scorpions-amplification-refractory mutation system method, as
performed using the therascreen® EGFR RGQ PCR kit
(Qiagen, Inc.) (20).
Conventional EGFR mutation PCR
assays
Conventional assays were performed using FFPE tissue
or cell block specimens using either a Rotor-Gene Q 5plex HRM
instrument with the therascreen® EGFR RGQ PCR kit
(Qiagen, Inc.) or a Roche Cobas® EGFR mutation test v2
(Roche Molecular Diagnostics, Branchburg, NJ, USA) according to
manufacturer’s protocols. Both assays are approved as companion
diagnostics in the United States, Europe and Japan (20,22).
According to the manufacturer’s protocols, the therascreen assay
detects 29 types of somatic mutation in EGFR in 1 h and 45
min, whereas the Cobas v2 assay detects 42 types in 1.5–2 h. The
detection limits for L858R, E756_A750del and T790M are 1.26, 1.64
and 7.02% of mutant DNA in the therascreen assay, and 3.96–5.32,
1.39–2.53 and 2.04–3.03% of mutant DNA in the Cobas v2 assay,
respectively.
Statistical analysis
Concordance rates and Cohen’s kappa coefficients
were used to examine the agreement of the assay results achieved
via an assessment of the two different specimen types. A kappa
coefficient (K) value of zero was considered to indicate
that there was no agreement beyond that which occurred by chance,
whereas a K value of 1.00 was considered to indicate perfect
agreement. The following K value ranges: 0–0.20, 0.21–0.40,
0.41–0.60, 0.61–0.80, and 0.81–1 were considered to represent
slight, fair, moderate, substantial and almost perfect agreement
between the compared results, respectively (23). P<0.05 was considered to indicate
a statistically significant difference. All data were statistically
analyzed using JMP® 13 software (SAS Institute Inc.,
Cary, NC, USA).
Results
Characteristics of patients and
specimens
Patient characteristics are shown in Table I. The mean age of the 90 patients
was 71.1 years (range, 42–86 years), all patients were Japanese,
and of East Asian ethnicity, and 55 (61.1%) and 35 (38.9%) of the
patients were male and female, respectively. The final diagnoses by
comprehensive clinical, radiological and pathological analysis were
lung cancer for 74 (82.2%), metastases from other organs for three
(3.3%), malignant lymphoma for one (1.1%), cancer of unknown
primary origin for two (2.2%), benign disease for three (3.3%), and
unknown for seven (7.8%) patients. At the time of diagnosis, 27
(36.5%), five (6.6%), 15 (20.3%) and 25 (33.8%) of the 74 lung
cancer cases were clinically staged (TNM classification 7th
edition) (24) as stage I, II, III
and IV, respectively. The stage of the remaining two (2.7%) lung
cancer cases was unknown.
 | Table IClinical characteristics of the 90
patients. |
Table I
Clinical characteristics of the 90
patients.
| Characteristic | Values |
|---|
| Age, years | |
| Mean | 71.1 |
| Range | 42-86 |
| Male/female, n
(%) | 55 (61.1)/35
(38.9) |
| Final diagnosis, n.
(%) | |
| Lung cancer | 74 (82.2) |
| Metastasis from
other organs |
3 (3.3) |
| Malignant
lymphoma |
1 (1.1) |
| Cancer of unknown
primary origin |
2 (2.2) |
| Benign
disease |
3 (3.3) |
| Unknown |
7 (7.8) |
| Stage of lung
cancer, n (%) | |
| I | 27 (36.5%) |
| II |
5 (6.6%) |
| III | 15 (20.3%) |
| IV | 25 (33.8%) |
| Unknown |
2 (2.7%) |
Cytological diagnosis of cell pellets classified the
90 specimens as follows: ‘malignant’, 28 (31.1%); ‘suspicious for
malignancy’, nine (10.0%); ‘neoplastic, benign neoplasm, low-grade
carcinoma’, none (0%); ‘atypical’, 12 (13.3%); ‘negative for
malignancy’, 40 (44.4%); and ‘non-diagnostic’, one (1.1%).
The mean concentration of DNA extracted from the
cfDNA supernatants and cell pellets, prior to adjustment, was 7.4
(range 1.2–152.8 ng/μl) and 47.2 ng/μl (1.3–314.1
ng/μl), respectively.
Comparison of cfDNA-supernatant and
cell-pellet assay results
The results of the EGFR d-PCR assays performed using
cfDNA supernatants and cell pellets were compared (Tables II and III), and concordance rates were
determined (Table IV). The total
number of patients with EGFR mutations detected using cfDNA
supernatants was 12 (13.3%), compared to 15 (16.7%) using cell
pellets. L858R, E746_ A750del and T790M mutations were identified
in nine, four and two cfDNA supernatant samples respectively, and
in 10, six and three cell pellet samples, respectively. The number
of patients negative for EGFR mutations was 78 (86.7%) using
cfDNA supernatants, and 75 (83.3%) using cell pellets. Three
patients had more than one EGFR mutation, one had a triple
mutation (L858R, E746_A750del and T790M), and two had double
mutations (L858R and T790M). The concordance rates between the
EGFR d-PCR assay results obtained using cfDNA supernatants
and cell pellet samples were 96.7% [K=0.87, 95% confidence
interval (CI) 0.73–1.00, P<0.0001] for all three EGFR mutations
in combination, 98.9% (K=0.94, 95% CI 0.83–1.00,
P<0.0001) for L858R alone, 98.9% (K=0.79, 95% CI
0.51–1.00, P<0.0001) for E746_A750del alone, and 98.9%
(K=0.79, 95% CI 0.40–1.00, P<0.0001) for T790M alone.
 | Table IIResults of the EGFR d-PCR
assay in 90 patients using cell-free DNA supernatants and cell
pellets. |
Table II
Results of the EGFR d-PCR
assay in 90 patients using cell-free DNA supernatants and cell
pellets.
| EGFR
mutations | Cell-free DNA
supernatants | Cell pellets |
|---|
| Positive | 12 (13.3%) | 15 (16.7%) |
| Exon 21 L858R | 9a,b | 10a,b |
| Exon 19
E746_A750del | 4a | 6a |
| Exon 20 T790M | 2a,b | 3a,b |
| Negative | 78 (86.7%) | 75 (83.3%) |
| Total | 90 | 90 |
 | Table IIIComparison of assay results obtained
for each epidermal growth factor receptor mutation using cell-free
DNA supernatants and cell pellets. |
Table III
Comparison of assay results obtained
for each epidermal growth factor receptor mutation using cell-free
DNA supernatants and cell pellets.
| Cell-free DNA
supernatants | Cell pellets
|
|---|
| Positive | Negative | Total |
|---|
| Exon 21 L858R | | | |
| Positive | 9 | 0 | 9 |
| Negative | 1 | 80 | 81 |
| Total | 10 | 80 | 90 |
| Exon 19
E746_A750del | | | |
| Positive | 4 | 0 | 4 |
| Negative | 2 | 84 | 86 |
| Total | 6 | 84 | 90 |
| Exon 20 T790M | | | |
| Positive | 2 | 0 | 2 |
| Negative | 1 | 87 | 88 |
| Total | 3 | 87 | 90 |
 | Table IVConcordance rates between the results
obtained using cell-free DNA supernatants and cell pellets. |
Table IV
Concordance rates between the results
obtained using cell-free DNA supernatants and cell pellets.
| Mutation | Concordance rate
(%) | K
coefficient | 95% CI | P-value |
|---|
| EGFR
mutation | 96.7 | 0.87 | 0.73-1.00 | <0.0001 |
| L858R | 98.9 | 0.94 | 0.83-1.00 | <0.0001 |
| E746_A750del | 98.9 | 0.79 | 0.51-1.00 | <0.0001 |
| T790M | 98.9 | 0.79 | 0.40-1.00 | <0.0001 |
Comparison of the EGFR assay results
obtained using cfDNA supernatants and the corresponding cytological
diagnosis obtained from cell pellets
The EGFR d-PCR assay results achieved using
cfDNA supernatant were then compared with the cytological diagnosis
of the corresponding cell pellets (Table V). Using cfDNA supernatants as
templates, the EGFR d-PCR assay detected EGFR
mutations in 12 samples. The corresponding cytological diagnosis
was ‘malignant’ in seven patients (53.8%), ‘suspicious for
malignancy’ in one patient (7.7%), ‘atypical cells’ in one patient
(7.7%), and ‘negative for malignancy’ in three patients
(30.8%).
 | Table VComparison of the cell-free
DNA-supernatant assay and corresponding cell-pellet cytological
diagnosis results. |
Table V
Comparison of the cell-free
DNA-supernatant assay and corresponding cell-pellet cytological
diagnosis results.
| Cytological
diagnosis of cell pellets | EGFR d-PCR
assay results of cell-free DNA supernatants
|
|---|
| Positive | Negative | Total |
|---|
| Malignant | 7 | 21 | 28 |
| Suspicious for
malignancy | 1 | 8 | 9 |
| Neoplastic, benign
neoplasm, low-grade carcinoma | 0 | 0 | 0 |
| Atypical | 1 | 11 | 12 |
| Negative for
malignancy | 3 | 37 | 40 |
| Non-diagnostic | 0 | 1 | 1 |
| Total | 12 | 78 | 90 |
EGFR status analyzed by conventional EGFR
assays using FFPE tissue or cell block specimens
The cytological diagnoses of cell pellets and the
results of a conventional EGFR assay using FFPE tumor tissue
or cell block specimens for the patients whose cytology specimens
(cfDNA supernatants or cell pellets) were positive for EGFR
mutations, as determined using the EGFR d-PCR assay, are shown in
Table VI. For the 15 EGFR
mutation-positive patients, FFPE tumor tissue specimens were
available for 14 patients (11 TBLBs and three surgical resections)
and a PE FFPE cellblock specimen was available for one patient.
EGFR mutations were detected in all of the specimens using
conventional assays. Only two patients exhibited different results
between the EGFR d-PCR assay using cell pellets and
conventional assays using FFPE tumor tissues or cell block
specimens: L858R and T790M vs. L858R in patient #16, and L858R,
E746_A750del and T790M vs. exon 19 deletion in patient #29.
 | Table VIEGFR status in patients with
EGFR mutation-positive results according to the EGFR
d-PCR assay using tumor tissue or cytology specimens and
conventional assays. |
Table VI
EGFR status in patients with
EGFR mutation-positive results according to the EGFR
d-PCR assay using tumor tissue or cytology specimens and
conventional assays.
| Patient #
(specimen) | Cytological
diagnosis of cell pellets | EGFR d-PCR
assay results
| Conventional
EGFR assay results
|
|---|
| Cell-free DNA
supernatants | Cell pellets | FFPE tumor tissue
or cellblock |
|---|
| #4 (BLF) | Malignant | Negative | E746_A750del | Exon 19
Deletiona (TBLB, Cobas v2) |
| #7 (PE) | Malignant | L858R, T790M | L858R, T790M | L858R, T790M (PE
cellblock, Cobas v2) |
| #16 (BLF) | Malignant | L858R | L858R, T790M | L858R (TBLB,
therascreen) |
| #18 (BLF) | Malignant | E746_A750del | E746_A750del | Deletionsa (TBLB, therascreen) |
| #24 (BLF) | Negative for
malignancy | Negative | L858R | L858R (TBLB,
therascreen) |
| #27 (BLF) | Negative for
malignancy | E746_A750del | E746_A750del | Deletionsa (TBLB, therascreen) |
| #29 (BLF) | Malignant | L858R,
E746_A750del, T790M | L858R,
E746_A750del, T790M | Deletionsa (TBLB, therascreen) |
| #39 (BLF) | Malignant | L858R | L858R | L858R (TBLB, Cobas
v2) |
| #53 (BLF) | Negative for
malignancy | L858R | L858R | L858R (TBLB, Cobas
v2) |
| #58 (BLF) | Negative for
malignancy | L858R | L858R | L858R (SR,
therascreen) |
| #67 (BLF) | Malignant | E746_A750del | E746_A750del | Deletionsa (SR, therascreen) |
| #74 (BLF) | Malignant | Negative | E746_A750del | Deletionsa (SR, therascreen) |
| #75 (BLF) | Atypical | L858R | L858R | L858R (TBLB,
therascreen) |
| #83 (BLF) | Malignant | L858R | L858R, T790M | L858R, T790M (TBLB,
Cobas v2) |
| #87 (BLF) | Suspicious for
malignancy | L858R | L858R | L858R (TBLB,
therascreen) |
Effectiveness of EGFR-TKIs
Of the 12 patients with cfDNA supernatants found to
be positive for EGFR mutations, eight received EGFR-TKI
therapy using gefitinib, erlotinib or osimertinib, based on the
conventional EGFR assay results achieved using FFPE tumor
tissue or cell block specimens: four TBLBs and one PE cell block
were analyzed with the Cobas v2 assay, and three TBLBs with the
therascreen assay. Six of these eight patients (75%) exhibited a
positive response to EGFR-TKI therapy and four patients (50%) were
continuing this therapy at the data cut-off point in September
2017.
Discussion
Due to recent advances in molecular targeted
therapies and genomic technologies, such as next-generation
sequencing (NGS), multiple and parallel molecular testing is now
available and is recommended to facilitate improved treatment
strategies for patients with lung cancer (25,26).
However, these processes often consume extensive resources, and
integrating their results into standard clinical care regimes is
complex and sometimes challenging (27,28).
Furthermore, although obtaining appropriate tumor specimens with a
sufficient number of cancer cells from patients is crucial for
accurate molecular testing, doing so is often problematic or
impossible (26). Therefore,
simple, rapid and cost-effective methods of molecular testing,
which take full advantage of the limited materials available in the
clinical setting, such as cytology specimens, are urgently
required. This is particularly important in cases of frequent
oncogenic genomic alteration, including EGFR mutation. In
the present study, the suitability of an EGFR d-PCR assay,
which has a reaction time of <10 min, for analyzing cfDNA from
cytology specimen supernatants was determined. The results of this
assay were compared with those of an EGFR d-PCR assay using
cell pellets, cytological diagnoses and conventional EGFR
assays using FFPE tumor tissue or cell block specimens. In
addition, the results were compared with the observed clinical
effectiveness of EGFR-TKI therapy.
The EGFR d-PCR assay results produced using
cfDNA supernatants exhibited substantial agreement with those
obtained using cell pellets, and detected EGFR mutations
even in specimens cytologically diagnosed as ‘suspicious for
malignancy’, ‘atypical’ and ‘negative for malignant cells.’ One
reason for this is the difference in detection sensitivity between
cytological analysis with the Papanicolaou smear for the cancer
cells and the EGFR d-PCR assay for target mutations. Since
the Papanicolaou smear uses only a few drops from cell pellets
derived from specimens such as BLF, which usually do not contain
thousands of cells, the detection sensitivity of cytological
analyses is limited. Conversely, the detection limits of the
EGFR d-PCR assay were previously determined using
mutation-positive cancer cell line mixtures; the results
demonstrated that the detection limit of each mutation was 0.5,
0.05 and 0.5% of mutation-positive cancer cells, for L858R,
E756_A750del and T790M, respectively (20). Furthermore, the PCR assay can
detect tumor DNA in the absence of cancer cells, whereas
cytological analysis requires cancer cells by definition. These
findings indicated that the detection sensitivity of the
EGFR d-PCR assay is superior to cytological analysis. In
addition, cfDNA supernatants from cytology specimens contain a
relatively high amount of cancer DNA. While cfDNA from normal cells
is derived mainly from apoptotic processes (29), that generated by cancer cells is
derived from apoptotic and necrotic processes associated with high
cellular turnover (30,31). Therefore, more cfDNA should be
isolated from cancer cells than from normal cells in cytology
supernatants. Although current guidelines for EGFR assays
recommend making cell blocks from cell pellets of cytology
specimens (10), our previous
study demonstrated that cfDNA supernatants had significantly lower
quantification cycle values for EGFR-mutation detection than
cell blocks in conventional PCR assays (32).
Previous studies have demonstrated the use of cfDNA
supernatants obtained from cytology specimens for EGFR
mutation assays in patients with NSCLC, using either PCR or direct
sequencing methods (33–36). However, the majority of these
studies used PE cytology specimens, which usually contain greater
number of cancer cells compared to BLF specimens. Kawahara et
al (37) and Park et al
(38) previously compared the
EGFR mutation status determined by subjecting cfDNA
supernatants from bronchial cytology samples (such as BLF,
bronchoalveolar washing and bronchial brushing), to that determined
by subjecting the corresponding tumor tissue samples to
conventional PCR-based assays, for 51 and 20 patients,
respectively. The concordance rate between the two types of results
was 94.1 (48/51) and 75.0% (9/12) for each study, respectively. To
the best of our knowledge, the present study has analyzed the
largest number of bronchial cytology-specimen cfDNA using the
EGFR d-PCR assay of any study conducted to date. A rate of
96.7% concordance was achieved between the results that were
obtained using cfDNA supernatants, and those that were obtained
using cell pellets, and furthermore, this was achieved using a much
shorter reaction time than necessary for conventional PCR-based
assays.
In addition to the marked concordance of assay
results between cfDNA supernatants and cell pellets, the
EGFR mutations detected by the EGFR d-PCR assay using
cytology specimens (cfDNA supernatants or cell pellets) were highly
consistent with those detected by conventional assays using tumor
tissues or cytology specimens. Our previous study reported complete
concordance between BLF cell pellet and FFPE tumor tissue assay
results in 49 patients with NSCLC using the current companion
diagnostic of EGFR-TKI therapy, the therascreen assay (32). In another study, we also detected
complete concordance between the results achieved using the
EGFR d-PCR assay and the therascreen assay to assess cell
pellets of cytology specimens collected from 80 patients with NSCLC
(20). Therefore, with respect to
L858R, E746_A750del and T790M, the EGFR d-PCR assay results
appear to be as reliable as current companion diagnostics using
FFPE tumor tissues.
The objective response rate for EGFR-TKI therapy was
75.0% (6/8) in patients with NSCLC whose cfDNA super-natants were
shown to be positive for mutations using the EGFR d-PCR
assay. This finding suggested that the results of the EGFR
d-PCR assay obtained using cfDNA supernatants correlate with the
clinical effectiveness of EGFR-TKIs. At present, EGFR
mutations detectable by the EGFR d-PCR assay are limited to
three mutations: L858R, E746_A750del and T790M. However, L858R and
E746_A750del represent ~90% of oncogenic EGFR mutations
(39). Furthermore, patients with
NSCLC with these mutations have been reported to exhibit better
responses to EGFR-TKI therapy than those that harbor more minor
EGFR mutations, including insertions in exon 20, L861Q in
exon 21 or exon 19 deletions starting at codon L747 (40,41).
Furthermore, application of the EGFR d-PCR assay using cfDNA
supernatants reserves the cell pellets of cytology specimens, which
can thus be subjected to additional comprehensive and detailed
genotyping using multiplex PCR assays and/or NGS. Since the
EGFR d-PCR assay using cfDNA supernatants detects mutations
more quickly and with greater sensitively than the cytological
diagnosis of cell pellets, it may also be useful as a screening and
confirmation method for lung cancer diagnosis.
In two patients, the EGFR d-PCR assay using
cytology specimens detected more mutations than the conventional
EGFR assays using FFPE tissue specimens. It is assumed that
DNA in FFPE specimens is negatively affected by the
formalin-fixation process, including DNA fragmentation and
cross-linking formation (42).
Notably, several studies (42,43),
including our previous study (32), have demonstrated the strong
negative impact of fixation on EGFR mutation detection
efficiency in patients with lung cancer. Furthermore, the
EGFR d-PCR assay has a notable detection limit compared to
conventional assays (20),
indicating that mutations detected only by the EGFR d-PCR
assay may indeed be real mutations. In the present study, there was
a rare case of a triple mutation detected only by the EGFR
d-PCR assay; however, triple EGFR mutations have previously
been reported in lung cancer (44). Furthermore, since the patient had
already received EGFR-TKI treatment for >1 year when the
cytology specimen was obtained, it is not surprising that the
patient had acquired secondary mutations such as T790M.
A limitation of the present study was that the
optimal amount of supernatant that should be used for cfDNA
extraction is unknown. While extracting DNA from a greater volume
of supernatant would increase the detection sensitivity of the
assay, it would also incrementally increase the time and effort
required for DNA extraction. Therefore, further study is required
to analyze the association between the amount of utilized
supernatant and extracted DNA, and to develop more efficient DNA
extraction methods for liquid specimens. This may also enable the
current EGFR d-PCR assay to be further developed into a
liquid biopsy using cfDNA from blood serum or plasma. EGFR
d-PCR liquid biopsy would provide a novel, fast and minimally
invasive screening method for detecting EGFR mutations in
patients with lung cancer.
In conclusion, the present study demonstrated that
using the EGFR d-PCR assay to analyze cfDNA from cytology
specimen supernatants is a rapid, sensitive and reliable method for
the detection of EGFR mutations, which furthermore reserves
cell pellets for use in other morphological and molecular analyses.
This assay may therefore be considered a promising novel
point-of-care testing method that may enable patients with NSCLC to
receive EGFR-TKI therapy as soon as possible.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
EGFR
|
epidermal growth factor receptor
|
|
EGFR-TKIs
|
EGFR tyrosine kinase inhibitors
|
|
EGFR d-PCR assay
|
EGFR droplet-polymerase chain
reaction
|
|
cfDNA
|
cell-free DNA
|
|
BLF
|
bronchial lavage fluid
|
|
PE
|
pleural effusion
|
|
SF
|
spinal fluid
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
Acknowledgments
Not applicable.
Funding
The present study was supported by the Japan Society
for the Promotion of Science (JSPS) Grants-in-Aid for Scientific
Research (KAKENHI; grant no. 25460434). This work was also
supported, in-kind, by the Seiko Epson Corporation (Suwa, Nagano,
Japan).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
SA designed the study and wrote the initial draft of
the manuscript. AY assisted in study design, data analysis,
interpretation and preparation of the manuscript. KS, SA and TN
performed DNA extractions and PCR assays using patients’
cytological and tissue specimens. YK and RN are cytotechnologists
that performed cytological analysis of patients’ specimens. SA and
AY are pathologists that reviewed the patients’ cytological
diagnosis. HY contributed to patients’ clinical data collection,
specimen collection and critically reviewed the manuscript. KM and
AY assisted in developing the EGFR d-PCR assay and optimized
the assay conditions. TH contributed to the acquisition of funding,
general supervision, data interpretation and critically reviewed
the manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent for
their participation in the present study, which was reviewed and
approved by the medical ethics committee of the Shinshu University
School of Medicine (Matsumoto, Japan).
Consent for publication
All patients provided written informed consent.
Competing interests
Akemi Yamaguchi, one of the authors of this study,
was an employee of Seiko Epson Corporation. The remaining authors
have no conflicts of interest to disclose.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar
|
|
2
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al North-East Japan Study Group: Gefitinib or chemotherapy for
non-small-cell lung cancer with mutated EGFR. N Engl J Med.
362:2380–2388. 2010. View Article : Google Scholar
|
|
3
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et
al West Japan Oncology Group: Gefitinib versus cisplatin plus
docetaxel in patients with non-small-cell lung cancer harbouring
mutations of the epidermal growth factor receptor (WJTOG3405): An
open label, randomised phase 3 trial. Lancet Oncol. 11:121–128.
2010. View Article : Google Scholar
|
|
4
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar
|
|
5
|
Rosell R, Carcereny E, Gervais R,
Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R,
Pallares C, Sanchez JM, et al Spanish Lung Cancer Group in
collaboration with Groupe Français de Pneumo-Cancérologie and
Associazione Italiana Oncologia Toracica: Erlotinib versus standard
chemotherapy as first-line treatment for European patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(EURTAC): A multicentre, open-label, randomised phase 3 trial.
Lancet Oncol. 13:239–246. 2012. View Article : Google Scholar
|
|
6
|
Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang
Y, Li W, Hou M, Shi JH, Lee KY, et al: Afatinib versus cisplatin
plus gemcitabine for first-line treatment of Asian patients with
advanced non-small-cell lung cancer harbouring EGFR mutations
(LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet
Oncol. 15:213–222. 2014. View Article : Google Scholar
|
|
7
|
Yang JC, Wu YL, Schuler M, Sebastian M,
Popat S, Yamamoto N, Zhou C, Hu CP, O’Byrne K, Feng J, et al:
Afatinib versus cisplatin-based chemotherapy for EGFR
mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6):
Analysis of overall survival data from two randomised, phase 3
trials. Lancet Oncol. 16:141–151. 2015. View Article : Google Scholar
|
|
8
|
Wang S, Cang S and Liu D: Third-generation
inhibitors targeting EGFR T790M mutation in advanced non-small cell
lung cancer. J Hematol Oncol. 9:342016. View Article : Google Scholar
|
|
9
|
Fukuoka M, Wu YL, Thongprasert S,
Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu
DT, Saijo N, et al: Biomarker analyses and final overall survival
results from a phase III, randomized, open-label, first-line study
of gefitinib versus carboplatin/paclitaxel in clinically selected
patients with advanced non-small-cell lung cancer in Asia (IPASS).
J Clin Oncol. 29:2866–2874. 2011. View Article : Google Scholar
|
|
10
|
Leighl NB, Rekhtman N, Biermann WA, Huang
J, Mino-Kenudson M, Ramalingam SS, West H, Whitlock S and
Somerfield MR: Molecular testing for selection of patients with
lung cancer for epidermal growth factor receptor and anaplastic
lymphoma kinase tyrosine kinase inhibitors: American Society of
Clinical Oncology endorsement of the College of American
Pathologists/International Association for the study of lung
cancer/association for molecular pathology guideline. J Clin Oncol.
32:3673–3679. 2014. View Article : Google Scholar
|
|
11
|
Smouse JH, Cibas ES, Jänne PA, Joshi VA,
Zou KH and Lindeman NI: EGFR mutations are detected comparably in
cytologic and surgical pathology specimens of nonsmall cell lung
cancer. Cancer. 117:67–72. 2009.
|
|
12
|
Reynolds JP, Tubbs RR, Minca EC, MacNamara
S, Almeida FA, Ma PC, Pennell NA and Cicenia JC: EGFR mutational
genotyping of liquid based cytology samples obtained via fine
needle aspiration (FNA) at endobronchial ultrasound of non-small
cell lung cancer (NSCLC). Lung Cancer. 86:158–163. 2014. View Article : Google Scholar
|
|
13
|
Lozano MD, Labiano T, Echeveste J, Gurpide
A, Martín-Algarra S, Zhang G, Sharma A and Palma JF: Assessment of
EGFR and KRAS mutation status from FNAs and core-needle biopsies of
non-small cell lung cancer. Cancer Cytopathol. 123:230–236. 2015.
View Article : Google Scholar
|
|
14
|
Malapelle U, Sirera R, Jantus-Lewintre E,
Reclusa P, Calabuig-Fariñas S, Blasco A, Pisapia P, Rolfo C and
Camps C: Profile of the Roche cobas® EGFR mutation test
v2 for non-small cell lung cancer. Expert Rev Mol Diagn.
17:209–215. 2017. View Article : Google Scholar
|
|
15
|
Marchetti A, Del Grammastro M, Felicioni
L, Malatesta S, Filice G, Centi I, De Pas T, Santoro A, Chella A,
Brandes AA, et al: Assessment of EGFR mutations in circulating
tumor cell preparations from NSCLC patients by next generation
sequencing: Toward a real-time liquid biopsy for treatment. PLoS
One. 9:e1038832014. View Article : Google Scholar
|
|
16
|
Remon J, Caramella C, Jovelet C, Lacroix
L, Lawson A, Smalley S, Howarth K, Gale D, Green E, Plagnol V, et
al: Osimertinib benefit in EGFR-mutant NSCLC patients with
T790M-mutation detected by circulating tumour DNA. Ann Oncol.
28:784–790. 2017.
|
|
17
|
Sacher AG, Paweletz C, Dahlberg SE, Alden
RS, O’Connell A, Feeney N, Mach SL, Jänne PA and Oxnard GR:
Prospective validation of rapid plasma genotyping for the detection
of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol.
2:1014–1022. 2016. View Article : Google Scholar
|
|
18
|
Sacher AG, Komatsubara KM and Oxnard GR:
Application of plasma genotyping technologies in non-small cell
lung cancer: A practical review. J Thorac Oncol. 12:1344–1356.
2017. View Article : Google Scholar
|
|
19
|
Huang WL, Chen YL, Yang SC, Ho CL, Wei F,
Wong DT, Su WC and Lin CC: Liquid biopsy genotyping in lung cancer:
Ready for clinical utility? Oncotarget. 8:18590–18608. 2017.
|
|
20
|
Asaka S, Yoshizawa A, Matsuda K, Yamaguchi
A, Yamamoto H, Shiina T, Nakata R, Ogawa K, Zhang M and Honda T: A
novel, rapid point-of-care test for lung cancer patients to detect
epidermal growth factor receptor gene mutations by using real-time
droplet-PCR and fresh liquid cytology specimens. Oncol Rep.
37:1020–1026. 2017. View Article : Google Scholar
|
|
21
|
Layfield LJ, Baloch Z, Elsheikh T, Litzky
L, Rekhtman N, Travis WD, Zakowski M, Zarka M and Geisinger K:
Standardized terminology and nomenclature for respiratory cytology:
The Papanicolaou Society of Cytopathology guidelines. Diagn
Cytopathol. 44:399–409. 2016. View Article : Google Scholar
|
|
22
|
Vallée A, Le Loupp AG and Denis MG:
Efficiency of the Therascreen® RGQ PCR kit for the
detection of EGFR mutations in non-small cell lung carcinomas. Clin
Chim Acta. 429:8–11. 2014. View Article : Google Scholar
|
|
23
|
Landis JR and Koch GG: The measurement of
observer agreement for categorical data. Biometrics. 33:159–174.
1977. View Article : Google Scholar
|
|
24
|
Sobin LH, Gospodarowicz MK and Wittekind
Ch: International Union Against Cancer (UICC) TNM Classification of
Malignant Tumours. 7th edition. Wiley-Blackwell; Oxford: 2009
|
|
25
|
Gravitz L: Therapy: This time it’s
personal. Nature. 509:S52–S54. 2014. View Article : Google Scholar
|
|
26
|
Soo RA, Stone ECA, Cummings KM, Jett JR,
Field JK, Groen HJM, Mulshine JL, Yatabe Y, Bubendorf L, Dacic S,
et al: Scientific advances in thoracic oncology 2016. J Thorac
Oncol. 12:1183–1209. 2017. View Article : Google Scholar
|
|
27
|
Doble B, John T, Thomas D, Fellowes A, Fox
S and Lorgelly P: Cost-effectiveness of precision medicine in the
fourth-line treatment of metastatic lung adenocarcinoma: An early
decision analytic model of multiplex targeted sequencing. Lung
Cancer. 107:22–35. 2017. View Article : Google Scholar
|
|
28
|
Harada S, Arend R, Dai Q, Levesque JA,
Winokur TS, Guo R, Heslin MJ, Nabell L, Nabors LB, Limdi NA, et al:
Implementation and utilization of the molecular tumor board to
guide precision medicine. Oncotarget. 8:57845–57854. 2017.
View Article : Google Scholar
|
|
29
|
Suzuki N, Kamataki A, Yamaki J and Homma
Y: Characterization of circulating DNA in healthy human plasma.
Clin Chim Acta. 387:55–58. 2008. View Article : Google Scholar
|
|
30
|
Li CN, Hsu HL, Wu TL, Tsao KC, Sun CF and
Wu JT: Cell-free DNA is released from tumor cells upon cell death:
A study of tissue cultures of tumor cell lines. J Clin Lab Anal.
17:103–107. 2003. View Article : Google Scholar
|
|
31
|
Jahr S, Hentze H, Englisch S, Hardt D,
Fackelmayer FO, Hesch RD and Knippers R: DNA fragments in the blood
plasma of cancer patients: Quantitations and evidence for their
origin from apoptotic and necrotic cells. Cancer Res. 61:1659–1665.
2001.
|
|
32
|
Asaka S, Yoshizawa A, Nakata R, Negishi T,
Yamamoto H, Shiina T, Shigeto S, Matsuda K, Kobayashi Y and Honda
T: Utility of bronchial lavage fluids for epithelial growth factor
receptor mutation assay in lung cancer patients: Comparison between
cell pellets, cell blocks and matching tissue specimens. Oncol
Lett. 15:1469–1474. 2018.
|
|
33
|
Asano H, Toyooka S, Tokumo M, Ichimura K,
Aoe K, Ito S, Tsukuda K, Ouchida M, Aoe M, Katayama H, et al:
Detection of EGFR gene mutation in lung cancer by mutant-enriched
polymerase chain reaction assay. Clin Cancer Res. 12:43–48. 2006.
View Article : Google Scholar
|
|
34
|
Soh J, Toyooka S, Aoe K, Asano H, Ichihara
S, Katayama H, Hiraki A, Kiura K, Aoe M, Sano Y, et al: Usefulness
of EGFR mutation screening in pleural fluid to predict the clinical
outcome of gefitinib treated patients with lung cancer. Int J
Cancer. 119:2353–2358. 2006. View Article : Google Scholar
|
|
35
|
Kimura H, Fujiwara Y, Sone T, Kunitoh H,
Tamura T, Kasahara K and Nishio K: EGFR mutation status in
tumour-derived DNA from pleural effusion fluid is a practical basis
for predicting the response to gefitinib. Br J Cancer.
95:1390–1395. 2006. View Article : Google Scholar
|
|
36
|
Lin J, Gu Y, Du R, Deng M, Lu Y and Ding
Y: Detection of EGFR mutation in supernatant, cell pellets of
pleural effusion and tumor tissues from non-small cell lung cancer
patients by high resolution melting analysis and sequencing. Int J
Clin Exp Pathol. 7:8813–8822. 2014.
|
|
37
|
Kawahara A, Fukumitsu C, Taira T, Abe H,
Takase Y, Murata K, Yamaguchi T, Azuma K, Ishii H, Takamori S, et
al: Epidermal growth factor receptor mutation status in cell-free
DNA supernatant of bronchial washings and brushings. Cancer
Cytopathol. 123:620–628. 2015. View Article : Google Scholar
|
|
38
|
Park S, Hur JY, Lee KY, Lee JC, Rho JK,
Shin SH and Choi CM: Assessment of EGFR mutation status using
cell-free DNA from bronchoalveolar lavage fluid. Clin Chem Lab Med.
55:1489–1495. 2017. View Article : Google Scholar
|
|
39
|
Mitsudomi T, Kosaka T and Yatabe Y:
Biological and clinical implications of EGFR mutations in lung
cancer. Int J Clin Oncol. 11:190–198. 2006. View Article : Google Scholar
|
|
40
|
Naidoo J, Sima CS, Rodriguez K, Busby N,
Nafa K, Ladanyi M, Riely GJ, Kris MG, Arcila ME and Yu HA:
Epidermal growth factor receptor exon 20 insertions in advanced
lung adenocarcinomas: Clinical outcomes and response to erlotinib.
Cancer. 121:3212–3220. 2015. View Article : Google Scholar
|
|
41
|
Lee VH, Tin VP, Choy TS, Lam KO, Choi CW,
Chung LP, Tsang JW, Ho PP, Leung DK, Ma ES, et al: Association of
exon 19 and 21 EGFR mutation patterns with treatment outcome after
first-line tyrosine kinase inhibitor in metastatic non-small-cell
lung cancer. J Thorac Oncol. 8:1148–1155. 2013. View Article : Google Scholar
|
|
42
|
Dedhia P, Tarale S, Dhongde G, Khadapkar R
and Das B: Evaluation of DNA extraction methods and real time PCR
optimization on formalin-fixed paraffin-embedded tissues. Asian Pac
J Cancer Prev. 8:55–59. 2007.
|
|
43
|
Harada S, Agosto-Arroyo E, Levesque JA,
Alston E, Janowski KM, Coshatt GM and Eltoum IA: Poor cell block
adequacy rate for molecular testing improved with the addition of
Diff-Quik-stained smears: Need for better cell block processing.
Cancer Cytopathol. 123:480–487. 2015. View Article : Google Scholar
|
|
44
|
Uchibori K, Inase N, Araki M, Kamada M,
Sato S, Okuno Y, Fujita N and Katayama R: Brigatinib combined with
anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated
non-small-cell lung cancer. Nat Commun. 8:147682017. View Article : Google Scholar
|