Introduction
Renal cell carcinoma (RCC) is the most common
malignancy of the adult kidney, which accounts for ~3% of adult
malignant tumors (1,2). The most common subtype of RCC is
clear cell RCC (ccRCC), which accounts for ~80% of all diagnosed
cases (3). Progress has been made
with regards to the diagnosis and treatment of ccRCC; however, its
incidence continues to rise and ~30% of newly diagnosed patients,
and 20–40% of postoperative patients, will experience metastasis or
local recurrence (4–6). Recent advances have been made in
ccRCC treatment strategies, including targeted therapies, which
have resulted in therapeutic improvements (7); however, the majority of treated
patients eventually develop progressive disease due to acquired
resistance (8,9). Therefore, it is essential to
elucidate the molecular mechanisms underlying ccRCC progression and
metastasis, which may contribute to the development of novel
strategies for the treatment of ccRCC.
The perilipin (PLIN) family consists of five
members, including PLIN1, PLIN2, PLIN3, PLIN4 and PLIN5, which are
all characterized as lipid droplet (LD) proteins in adipocytes
(10). These proteins are
expressed in numerous species, including mammals, Drosophila
and Dictyostelium (11–16),
in which they are involved in various intracellular activities,
such as lipid metabolism and transport, intracellular trafficking
and signaling, and cytoskeletal organization (13,17,18).
It is well known that ccRCC is characterized by the presence of
intracellular LDs, which consist of a neutral lipid core surrounded
by a phospholipid monolayer and chimeric LD surface proteins
(19). PLIN family members
regulate lipid metabolism and are involved in the tumorigenesis and
development of various types of cancer. For example, PLIN1, which
is primarily expressed in adipose and steroidogenic cells, has been
reported to be involved in breast cancer progression and may act as
a tumor suppressor gene (20). In
cervical cancer, high PLIN3 expression is correlated with advanced
tumor stages and poor patient prognosis (21).
PLIN2 was originally considered an RNA transcript,
and is markedly involved in adipocyte differentiation (22,23).
It is commonly expressed in numerous types of tissue, including the
mammary gland, liver and skeletal muscle (24–26).
It has previously been reported that PLIN2 abundance is directly
associated with the levels of intracellular lipids, and increased
expression levels of PLIN2 have been observed in several specific
diseases involving fat accumulation (27). Recent studies have demonstrated
that numerous types of tumor overexpress PLIN2 (28,29).
Notably, the majority of these tumors have clear cell histology
(30,31). Two major studies revealed the
prognostic value of the transcript and protein levels of PLIN2 in
ccRCC (32,33). Furthermore, a recent study provided
evidence to suggest that PLIN2 is upregulated in ccRCC and directly
proportional to hypoxia-inducible factor (HIF)2α activation
(34); however, to the best of our
knowledge, a functional analysis of PLIN2 in ccRCC has not yet been
performed. Therefore, the present study aimed to investigate
whether PLIN2 expression is associated with clinicopathological
features of ccRCC and patient survival. In addition, the function
of PLIN2 in ccRCC was examined in vitro.
Materials and methods
Renal cancer tissue samples
Between January 2014 and December 2016, 80 pairs of
ccRCC and adjacent normal renal tissues and 7 benign renal
angiomyolipoma tissues were collected from patients who received
partial or radical nephrectomy at the Wuhan Union Hospital (Wuhan,
China). Among these, 40 pairs of resected tissues underwent protein
expression analysis by western blotting. The remaining tissues were
fixed in 10% formalin for 24 h at room temperature and embedded in
paraffin for immunohistochemistry (IHC). None of the patients
received preoperative or postoperative adjuvant anticancer therapy.
The present study and experimental procedures were approved by the
Human Research Ethics Committee of Huazhong University of Science
and Technology (Wuhan, China). Written informed consent was
obtained from the patients/patients' families.
Cell lines and cell culture
The ACHN, 786-O, A-498, OS-RC-2, Caki-1 and HK-2
cell lines were purchased from the American Type Culture Collection
(Manassas, VA, USA). Cells were cultured in Dulbecco's modified
Eagle's medium (HyClone; GE Healthcare, Logan, UT, USA) containing
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and 1% penicillin-streptomycin solution. Cells
were maintained at 37°C in an incubator containing 5%
CO2.
Transient transfection assay
Small interfering (si)RNA sequences specifically
targeting PLIN2 (si-PLIN2) and negative control siRNA (si-NC) were
chemically synthesized by Shanghai GenePharma Co., Ltd. (Shanghai,
China). The off-target effects of all siRNA sequences were detected
using the Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). For
transfection, ccRCC cells were seeded in 6-well plates at 50–70%
confluence. Cells (3×105) were transfected with 100 pmol
siRNA sequences using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to our previous study (35). A total of 48 h post-transfection,
cells were used for subsequent assays. The si-PLIN2 sequence was as
follows: 5′-CAGCCAUCAACUCAGAUUGUU-3′.
IHC
ccRCC tissues and adjacent normal tissues were
sequentially fixed in formalin, dehydrated and embedded in
paraffin. Subsequently, IHC was conducted by incubating tissue
sections (4 µm) with a primary rabbit PLIN2 polyclonal
antibody (1:100; A6276; ABclonal Biotech Co., Ltd., Wuhan, China)
overnight at 4°C. Subsequently, after washing three times with PBS,
the sections were incubated with goat anti-rabbit secondary
antibody (1:200; GB23303; Servicebio, Inc., Woburn, MA, USA) at
room temperature for 2 h.
Cell proliferation analysis
A-498 cells were transfected with si-PLIN2 or si-NC.
Subsequently, the cells were added to each well of 96-well plates
at a density of 3×103/well. Cell proliferation rate was
determined using the Cell Counting kit-8 (CCK-8) (Dojindo Molecular
Technologies, Inc, Rockville, MD, USA) every 24 h, according to the
manufacturer's protocol. Briefly, 10 µl CCK-8 solution was
added to each well. After 4 h, the optical density of each well was
measured at 450 nm. Each experiment was conducted in triplicate in
three independent experiments.
Migration assay
Boyden Transwell chambers (Corning Incorporated,
Corning, NY, USA) containing 8-µm membrane filters were
applied to 24-well plates. Cells (1×104) were seeded
into the upper chamber in serum-free medium, whereas the bottom
chamber was filled with complete medium supplemented with 10% FBS.
After 24 h at 37°C, the cells on the upper surface were washed with
PBS and cells on the lower surface were fixed with 100% methanol
for 10 min and stained with 0.05% crystal violet for 10 min at room
temperature. The average number of migrated cells was counted in
five randomly selected fields under a microscope (Olympus
CX41-32C02; Olympus Corporation, Tokyo, Japan). Experiments were
independently repeated in triplicate.
Invasion assays
Invasion assays were performed as previously
described (36). Briefly, 24-well
Transwell chambers with 8-µm membrane filters (Corning
Incorporated) were precoated with Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA). Cells (2×104) were seeded into
the upper chamber in serum-free medium, whereas the lower chamber
was filled with complete medium containing 10% FBS. Following
incubation for 24 h at 37°C, cells on the lower surface were fixed
with 100% methanol for 10 min and stained with 0.05% crystal violet
for 10 min at room temperature. Five random fields were captured
using an optical microscope at 100× magnification (Olympus
CX41-32C02; Olympus Corporation). Experiments were independently
repeated in triplicate.
Western blotting
Tissues and cells were lysed in
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China) containing a protease inhibitor
cocktail tablet (Roche Diagnostics, Indianapolis, IN, USA) and 1 mM
phenylmethylsulfonyl fluoride. Subsequently, the protein
concentrations were measured using a bicinchoninic acid kit
(Beyotime Institute of Biotechnology), according to the
manufacturer's protocol. Total proteins (30 µg) were
separated by 10% SDS-PAGE and were then transferred to
polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Bedford,
MA, USA) at 90 V for 90 min. The PVDF membranes were blocked in PBS
containing 5% nonfat milk for 1 h at room temperature, and were
then incubated with primary antibodies against PLIN2 (1:1,000;
A6276; ABclonal Biotech Co., Ltd.) and GAPDH (1:3,000; BM3876;
Wuhan Boster Biological Technology, Ltd., Wuhan, China) overnight
at 4°C. Subsequently, the membranes were incubated with secondary
antibodies (1:3,000; GB23303; Servicebio, Inc.) for 2 h at room
temperature. Finally, the proteins were visualized using
ChemiDoc-XRS+ (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Bioinformatics analysis
PLIN2 mRNA expression and clinical data of The
Cancer Genome Atlas (TCGA) ccRCC dataset (TCGA_KIRC) were
downloaded from the Xena Functional Genomics Explorer of University
of California Santa Cruz (https://xenabrowser.net/heatmap/) (37). To provide understanding of the
biological pathways involved in ccRCC pathogenesis via the PLIN2
pathway, a gene set enrichment analysis (GSEA) was conducted to
analyze the enrichment of given gene sets (c2.cgp.v6.0.symbols.gmt)
in TCGA ccRCC dataset using GSEA software (http://www.broadinstitute.org/gsea) (38). For enriched gene sets, those with a
false discovery rate (FDR) <25% and nominal P<0.05 following
the performance of 1,000 permutations were considered significantly
enriched.
Statistical analysis
All data were analyzed using GraphPad Prism 5.0
(GraphPad Software, Inc., La Jolla, CA, USA) and SPSS 22.0 (IBM
Corporation, Armonk, NY, USA). Numerical data are presented as the
means ± standard deviation. Student's t-test was used to assess
differences in PLIN2 expression between each ccRCC subgroup.
Survival information was evaluated via Kaplan-Meier analysis and
was compared using the log-rank test. The correlation between PLIN2
mRNA expression and clinicopathological parameters of patients with
ccRCC was evaluated using χ2 test. The significant
differences in PLIN2 expression among various T stage groups and
grade groups were estimated using one-way analysis of variance
followed by Tukey's post hoc test to assess multiple comparisons.
Receiver operator characteristic (ROC) curve analysis was used to
assess the prognostic value of PLIN2 with regards to various ccRCC
clinicopathological factors. Univariate and multivariate analyses
were performed using a Cox proportional hazard regression model.
P<0.05 was considered to indicate a statistically significant
difference.
Results
PLIN2 is strongly upregulated and
associated with various types of clinicopathological factors in
ccRCC tissues
To investigate the role of PLIN2 in ccRCC
development, the present study examined the mRNA expression levels
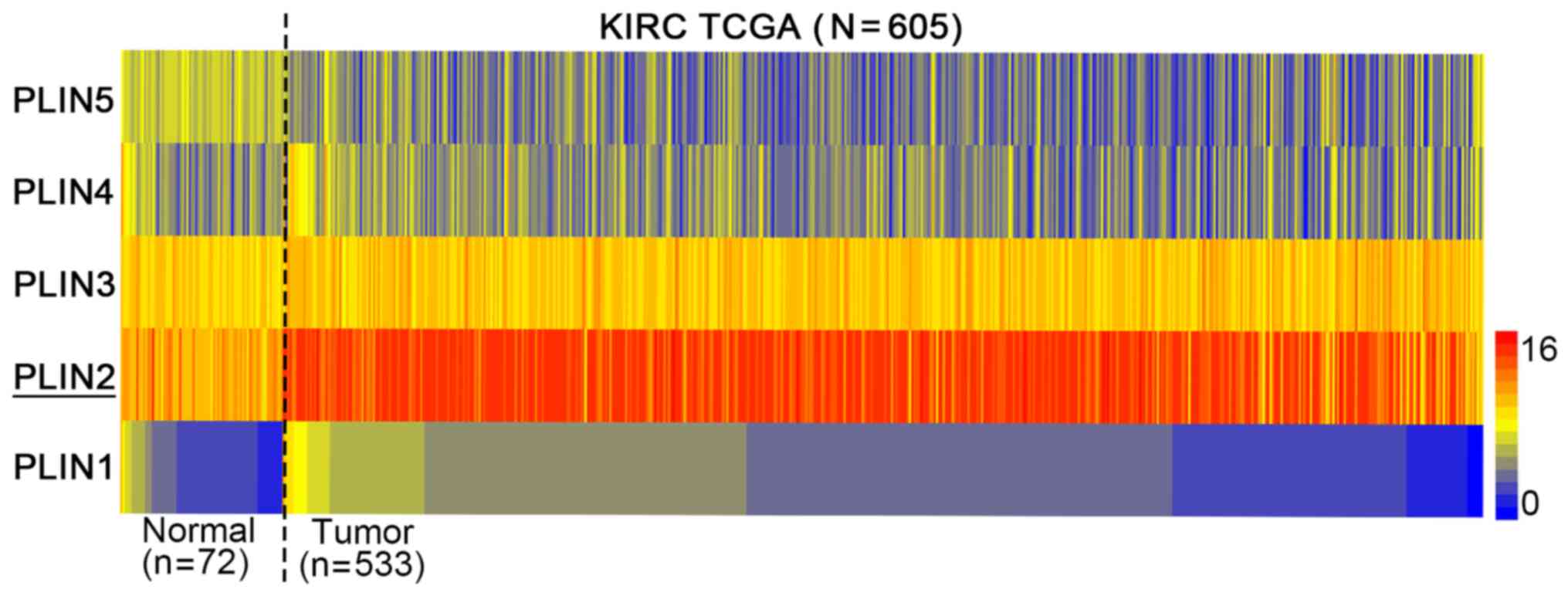
of the five PLIN family members (PLIN1-5) in TCGA database. The
heat map indicated that among the five PLIN members, PLIN2
exhibited the most obvious difference in expression between normal
tissues and ccRCC (Fig. 1).
Therefore, PLIN2 was chosen for subsequent investigation.
The present study explored the mRNA expression
levels of PLIN2 in ccRCC cancer tissues and adjacent normal tissues
from TCGA; PLIN2 expression was significantly elevated in tumor
tissues compared with in normal tissues (Fig. 2A). Subsequent validation was
conducted in 72 pairs of matched ccRCC tissues and adjacent normal
tissues. As expected, PLIN2 expression was significantly increased
in tumor tissues compared with in paired normal tissues (Fig. 2B). Furthermore, TCGA dataset
revealed that PLIN2 expression was increased in patients ≥60 years
old compared with in those <60 years old (Fig. 2C). Elevated PLIN2 expression was
also significantly associated with sex, T stage and grade in ccRCC
(Fig. 2D–F). PLIN2 expression
levels tended to decrease with increasing tumor T stage and G
grade. Markedly, PLIN2 expression was clearly increased in patients
with VHL alteration-positive ccRCC compared with in those with VHL
alteration-negative ccRCC (Fig.
2G). Clinicopathological information for the 526 ccRCC tissues
in TCGA dataset is listed in Table
I. There was a significant association between high PLIN2
expression and age or sex, which is consistent with the
aforementioned results. In addition, the χ2 test
performed for VHL alteration and PLIN2 expression demonstrated that
PLIN2 expression levels were significantly associated with VHL
alteration status (P=0.007).
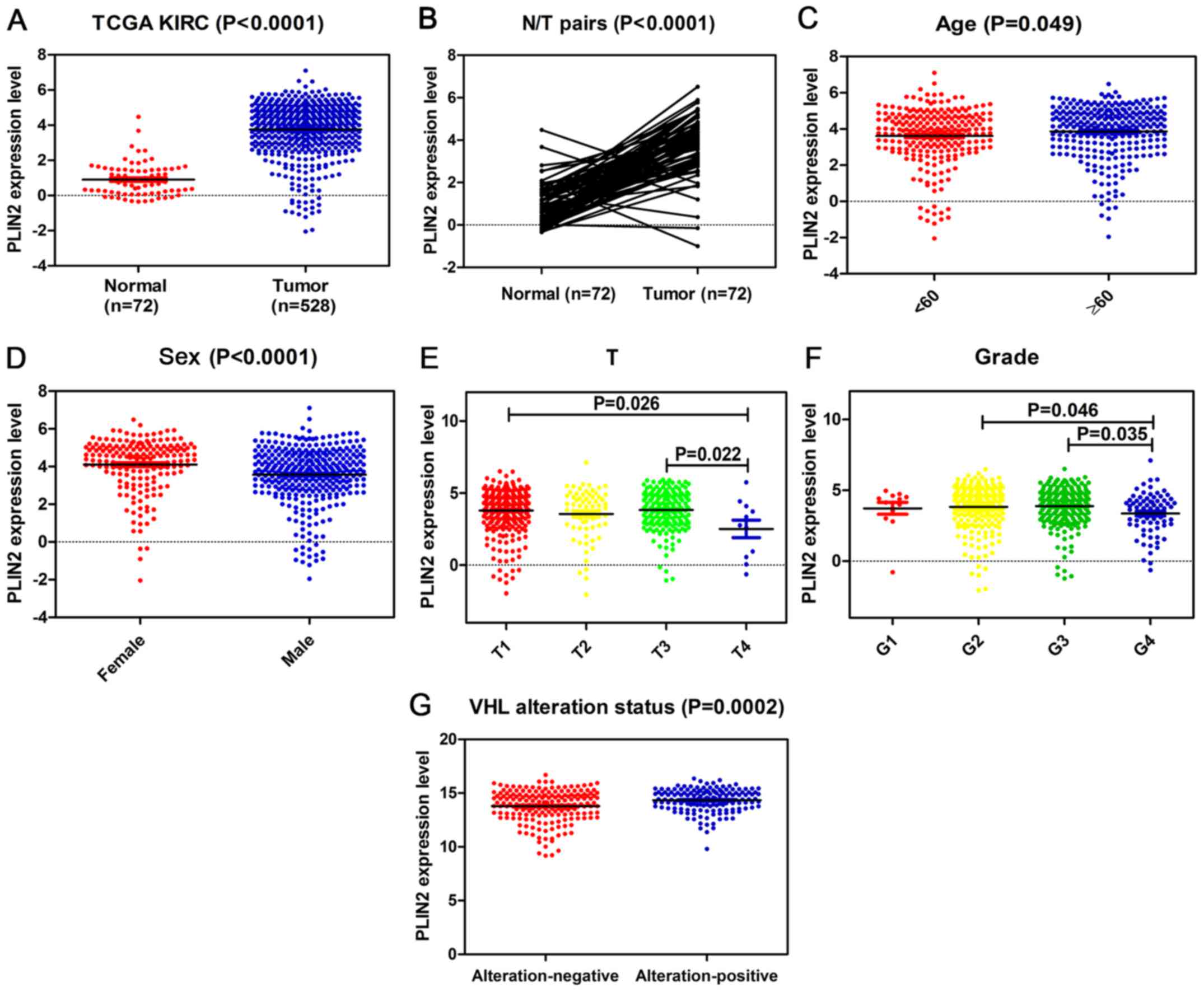 | Figure 2PLIN2 expression is upregulated in
ccRCC, and is associated with various clinicopathological factors
in ccRCC tissues. The mRNA expression levels of PLIN2 were obtained
from TCGA dataset, which contained 72 adjacent normal tissues and
528 ccRCC tissues. The mRNA expression levels of PLIN2 were
compared with respect to various clinicopathological factors: (A)
Cancer vs. normal tissues, (B) tumor vs. adjacent normal tissues,
(C) age, (D) sex, (E) T stage, (F) grade and (G) VHL alteration
status. ccRCC, clear cell renal cell carcinoma; PLIN, perilipin;
TCGA, The Cancer Genome Atlas; VHL, von Hippel-Lindau. |
 | Table IAssociation between PLIN2 mRNA
expression and clinicopathological parameters of patients with
clear cell renal cell carcinoma. |
Table I
Association between PLIN2 mRNA
expression and clinicopathological parameters of patients with
clear cell renal cell carcinoma.
A, Patient
characteristics
|
|---|
| Parameter | No. | PLIN2 mRNA
expression
| P-value |
|---|
| Low (n=263) | High) (n=263 |
|---|
| Age (years) | | | | |
| <60 | 243 | 140 | 103 | |
| ≥60 | 283 | 123 | 160 | 0.002 |
| Sex | | | | |
| Female | 185 | 64 | 119 | |
| Male | 341 | 199 | 144 | 0.000 |
| T stage | | | | |
| T1 or T2 | 337 | 161 | 176 | |
| T3 or T4 | 189 | 102 | 87 | 0.203 |
| N stage | | | | |
| N0 or NX | 510 | 252 | 258 | |
| N1 | 16 | 11 | 5 | 0.203 |
| M stage | | | | |
| M0 or MX | 448 | 216 | 232 | |
| M1 | 78 | 47 | 31 | 0.065 |
| G grade | | | | |
| G1 or G2 | 246 | 119 | 126 | |
| G3 or G4 | 280 | 144 | 137 | 0.541 |
| TNM stage | | | | |
| I + II | 319 | 152 | 167 | |
| III + IV | 207 | 111 | 96 | 0.211 |
|
| B, VHL status |
|
| Parameter | No. | PLIN2 mRNA
expression
| P-value |
| Low (n=172) | High (n=171) |
|
| VHL alteration | | | | |
| Negative | 190 | 108 | 82 | |
| Positive | 153 | 64 | 89 | 0.007 |
Association between high PLIN2 expression
and good clinical outcome in patients with ccRCC
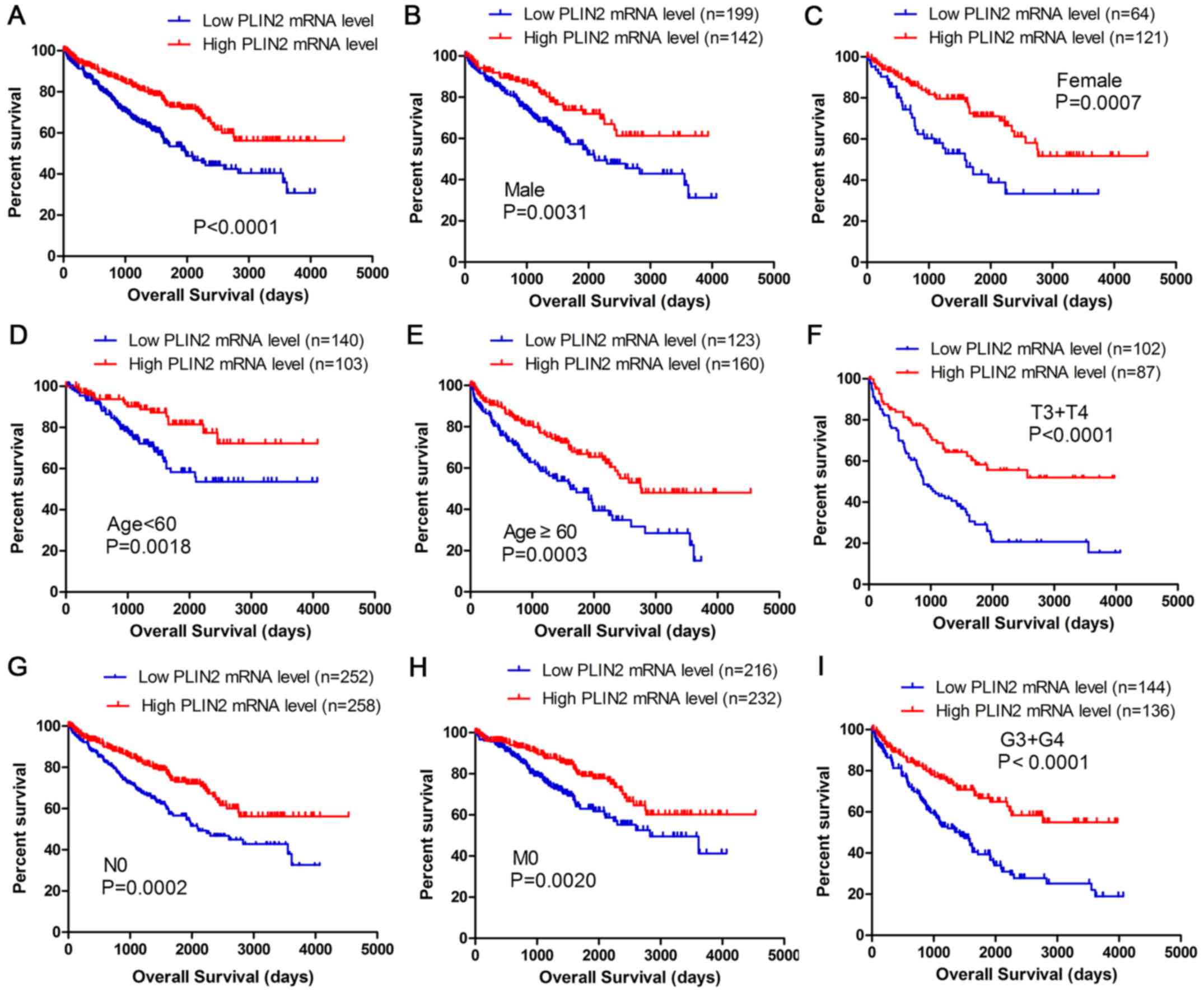
Kaplan-Meier survival analysis was used to determine
overall survival (OS) according to PLIN2 expression. Patients with
high PLIN2 expression exhibited better OS (P<0.0001) (Fig. 3A). Furthermore, OS analysis with
regards to PLIN2 expression was conducted in various subgroups of
patients with ccRCC. The results demonstrated that high PLIN2
expression may be considered a useful prognostic indicator for
patients with ccRCC with the following characteristics: Male
(Fig. 3B) or female (Fig. 3C), aged <60 (Fig. 3D) or ≥60 years old (Fig. 3E), T3 + T4 stage (Fig. 3F), N0 stage (Fig. 3G), M0 stage (Fig. 3H) and G3 + G4 grade (Fig. 3I). The prognostic value of each
clinicopathological factor, including PLIN2 expression status, was
evaluated for OS (Table II).
Univariate Cox proportion hazard ratio (HR) analysis indicated that
age (HR, 1.803; P<0.001), T stage (HR, 3.120; P<0.001), N
stage (HR, 3.823; P<0.001), M stage (HR, 4.346; P<0.001), G
grade (HR, 2.639; P<0.001) and PLIN2 expression status (HR,
0.524; P<0.001) were associated with OS. Further multivariate
analysis demonstrated that age (HR, 1.652; P=0.002), T stage (HR,
1.619; P=0.010), N stage (HR, 2.219; P=0.013), M stage (HR, 2.448;
P<0.001), G grade (HR, 1.677; P=0.006) and PLIN2 expression (HR,
0.586; P=0.001) could be considered independent prognostic
indicators of OS.
 | Table IIUnivariate and multivariate analyses
of PLIN2 mRNA expression and patient survival. |
Table II
Univariate and multivariate analyses
of PLIN2 mRNA expression and patient survival.
| Variable | Univariate analysis
| Multivariate
analysisc
|
|---|
| HRa | 95% CIb | P-value | HR | 95% CI | P-value |
|---|
| Overall survival
(n=526) | | | | | | |
| Age (years) | 1.803 | 1.318–2.468 | <0.001 | 1.652 | 1.199–2.276 | 0.002 |
| Sex | 0.948 | 0.697–1.290 | 0.736 | | | |
| T stage | 3.120 | 2.306–4.220 | <0.001 | 1.619 | 1.122–2.337 | 0.010 |
| N stage | 3.823 | 2.070–7.061 | <0.001 | 2.219 | 1.182–4.163 | 0.013 |
| M stage | 4.346 | 3.192–5.918 | <0.001 | 2.448 | 1.702–3.522 | <0.001 |
| G grade | 2.639 | 1.885–3.697 | <0.001 | 1.677 | 1.163–2.420 | 0.006 |
| PLIN2 | 0.524 | 0.386–0.711 | <0.001 | 0.586 | 0.429–0.803 | 0.001 |
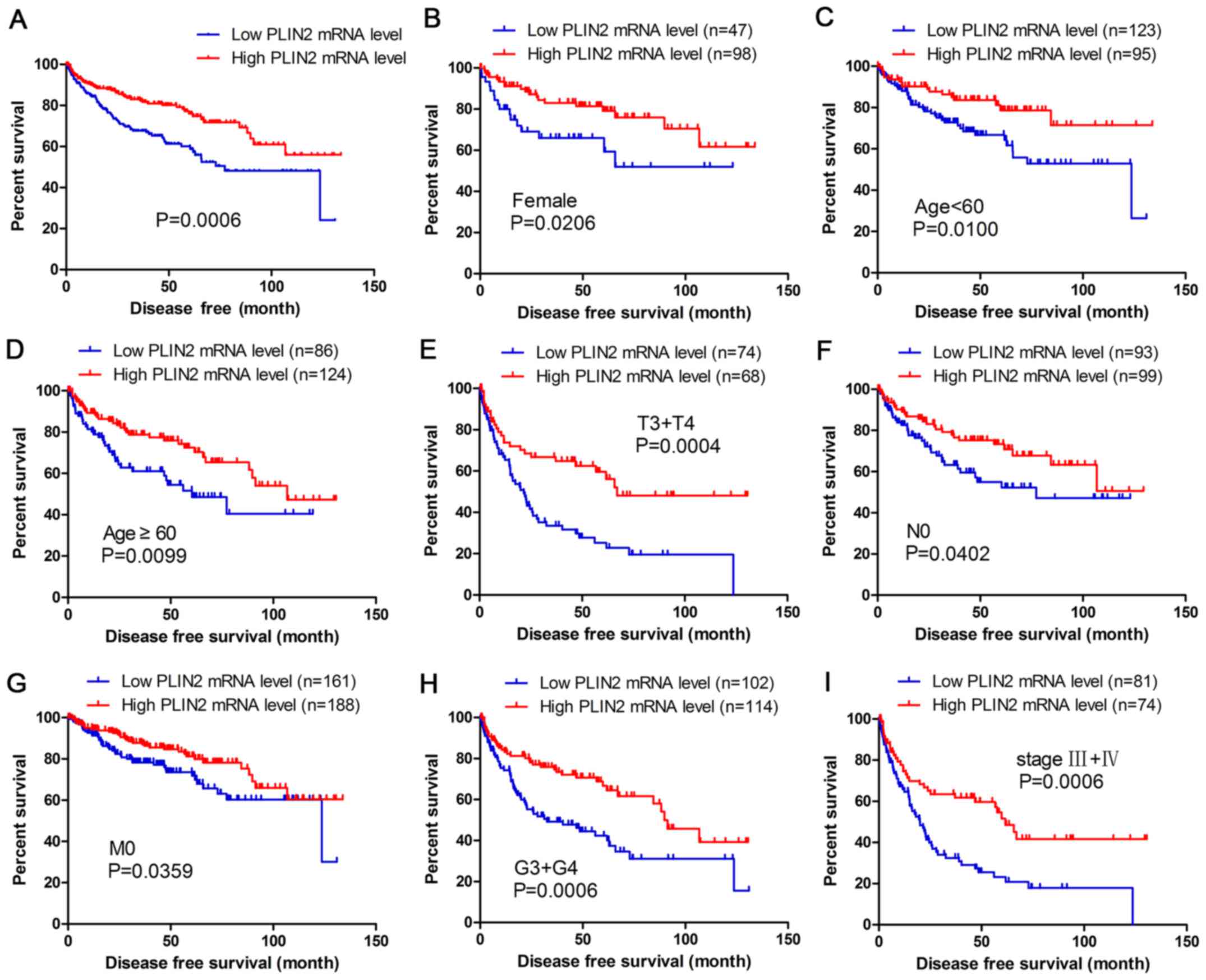
The present study analyzed the association between
PLIN2 expression and disease-free survival (DFS) of patients with
ccRCC by Kaplan-Meier analysis. The results demonstrated that
patients with high PLIN2 expression exhibited better DFS compared
with those with low PLIN2 expression (Fig. 4A; P<0.001). Furthermore, DFS
analysis with regards to PLIN2 expression was performed in
subgroups of patients with ccRCC. As expected, the results
demonstrated that high PLIN2 expression was a potential prognostic
indicator for patients with ccRCC with the following
characteristics: Female (Fig. 4B),
aged <60 (Fig. 4C) or ≥60 years
(Fig. 4D), T3 + T4 stage (Fig. 4E), N0 stage (Fig. 4F), M0 stage (Fig. 4G), G3 + G4 grade (Fig. 4H) and stage III + IV (Fig. 4I).
Association between high PLIN2 expression
and diagnostic role in patients with ccRCC
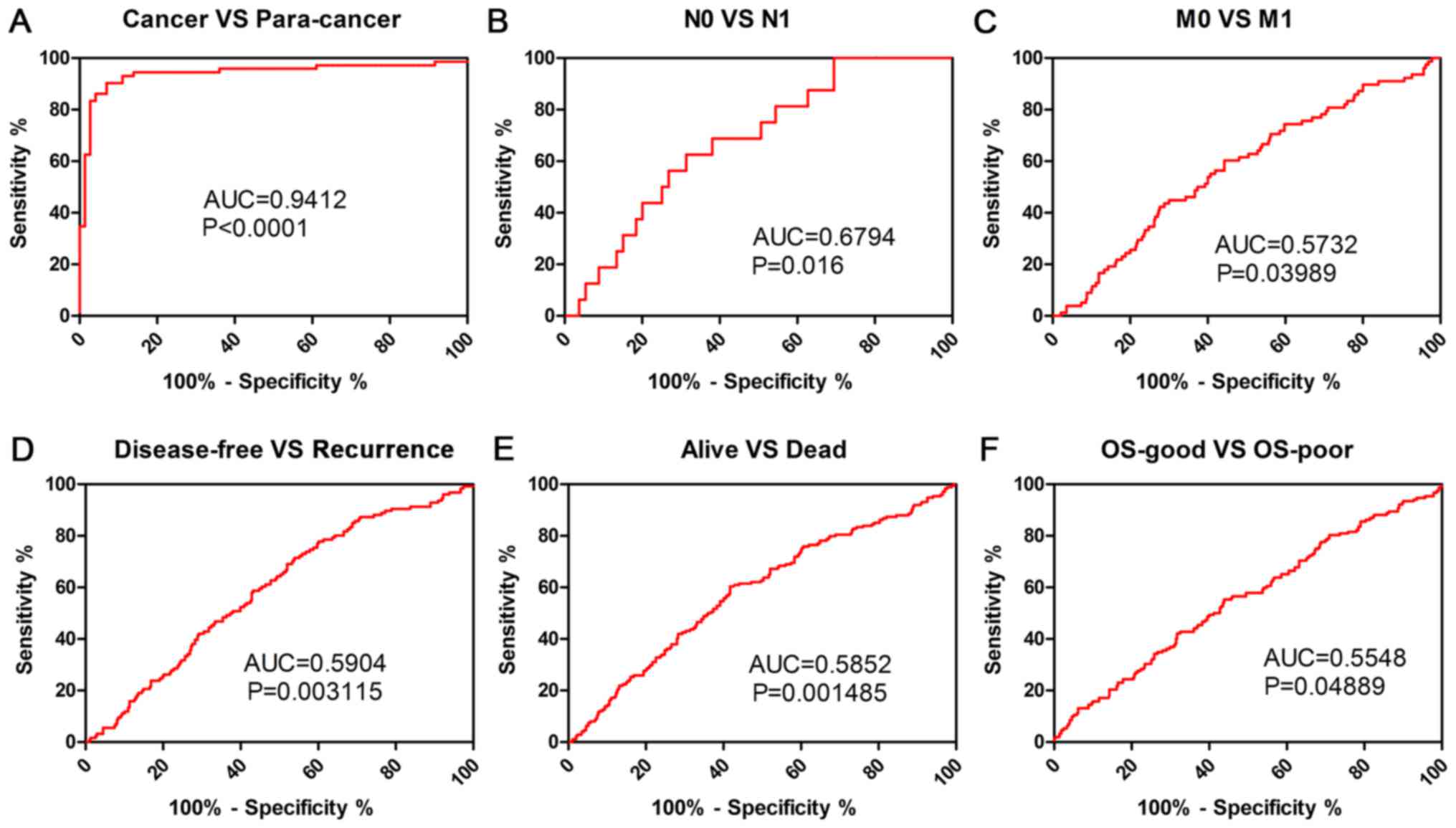
Clinicopathological factors were analyzed using ROC
curve to investigate the diagnostic role of PLIN2 in patients with
ccRCC. The results demonstrated that PLIN2 could sufficiently
discriminate ccRCC from paired normal tissues with an area under
the curve (AUC) of 0.9412 (Fig.
5A; P<0.0001). In addition, ROC curve analysis was conducted
with regards to PLIN2 expression in various subgroups of patients
with ccRCC. The results demonstrated that PLIN2 expression may be
an effective diagnostic indicator for patients with ccRCC with the
following characteristics: N0 vs. N1 stage (Fig. 5B; AUC=0.6794, P=0.016), M0 vs. M1
stage (Fig. 5C; AUC=0.5732,
P=0.03989), disease-free vs. recurrence (Fig. 5D; AUC=0.5904, P=0.003115), alive
vs. dead (Fig. 5E; AUC=0.5852,
P=0.001485), OS-good vs. OS-poor (Fig.
5F; AUC=0.5548, P=0.04889).
PLIN2 upregulation is verified in ccRCC
cells and tissues, and regulates biological pathways in ccRCC
pathogenesis
To further validate the results of TCGA dataset,
western blotting was conducted to detect the protein expression
levels of PLIN2 in ccRCC cells and tissues (Fig. 6A and B). In addition, IHC was
conducted in 40 paired ccRCC tumor tissues and adjacent normal
tissues (Fig. 6C). The results
demonstrated that PLIN2 protein expression was significantly
elevated in tumor cells and tissues compared with in immortalized
renal epithelial cells and adjacent normal tissues.
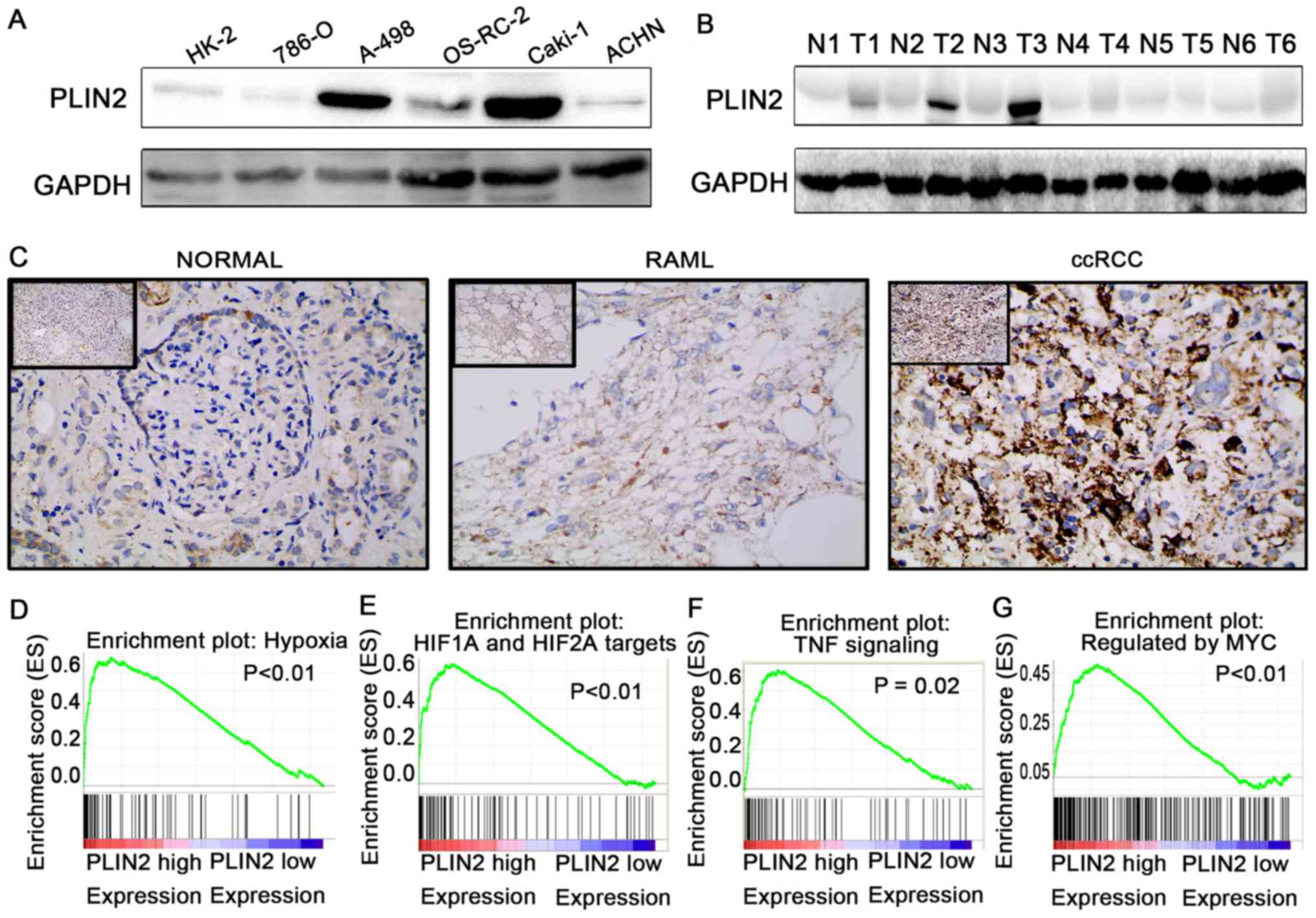 | Figure 6PLIN2 is upregulated in ccRCC cells
and tissues, and regulates biological pathways. Western blotting of
PLIN2 expression in (A) renal cancer cell lines and (B) ccRCC
tissues. (C) Immunohistochemistry of PLIN2 expression in ccRCC
tissues, benign RAML tissues and paired normal tissues.
Representative images are shown (magnification, ×200 in main
images, ×40 in upper left images). Gene set enrichment analysis
compared low PLIN2 and high PLIN2 expression groups in The Cancer
Genome Atlas database. Enrichment curves are shown for activated
gene sets related to (D) hypoxia, (E) HIF1α and HIF2α targets, (F)
TNF signaling and (G) regulated by Myc. ccRCC, clear cell renal
cell carcinoma; HIF, hypoxia-inducible factor; N, normal; PLIN,
perilipin; RAML, renal angiomyolipomas; T, tumor; TNF, tumor
necrosis factor. |
To elucidate how PLIN2 is involved in ccRCC
pathogenesis, GSEA was performed to gain further insight into the
biological pathways in TCGA database. GSEA is a computational tool
that determines whether a predefined set of genes shows
statistically significant, concordant differences between two
biological statuses. The GSEA results demonstrated that the gene
signatures of hypoxia pathway, HIF1α and HIF2α signaling, tumor
necrosis factor signaling and Myc signaling were associated with
patients with higher PLIN2 expression compared with those with
lower PLIN2 expression (Fig. 6D–G;
P<0.05).
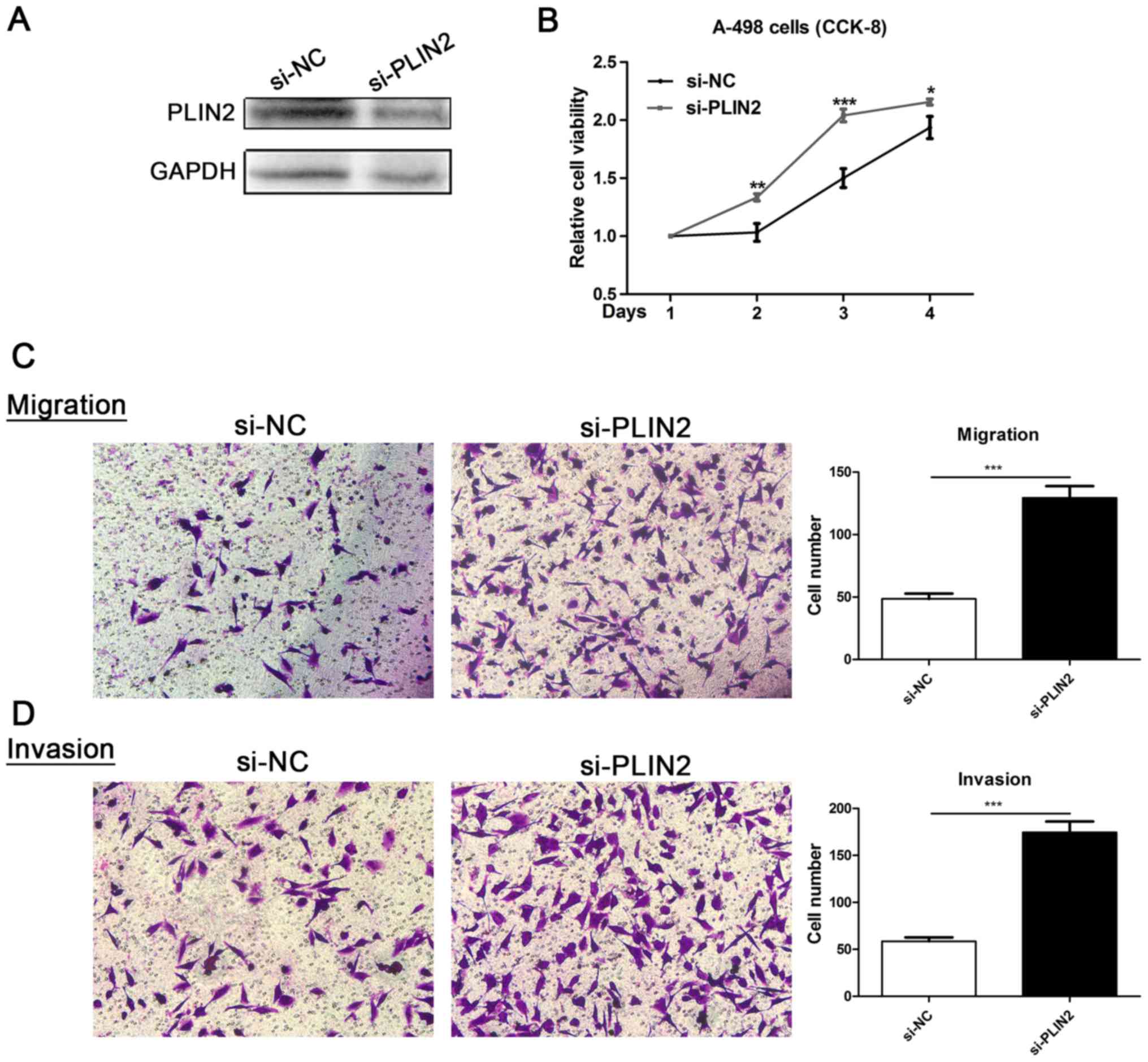
Roles of PLIN2 in ccRCC cell lines
To evaluate the functional role of PLIN2 in ccRCC,
ccRCC cell lines with PLIN2 knockdown were generated. Decreased
PLIN2 protein expression was observed in A-498 cells
post-transfection with si-PLIN2 oligonucleotide sequences (Fig. 7A). Proliferation of A-498 cells
transfected with si-PLIN2 was markedly enhanced compared with in
cells transfected with si-NC (Fig.
7B). Furthermore, Transwell assays were conducted to assess the
migration and invasion of ccRCC cells; downregulation of PLIN2
significantly promoted migration and invasion ability compared with
in the si-NC group (Fig. 7C and
D). These data revealed that PLIN2 may attenuate the migration
and invasion of ccRCC cells.
Discussion
The present study investigated the expression
pattern, clinical significance and biological functions of PLIN2 in
ccRCC, including its effects on cell proliferation, migration and
invasion. PLIN2 expression was revealed to be elevated in ccRCC
tissues compared with in adjacent normal tissues, and was
associated with good prognosis of patients with ccRCC. Furthermore,
the results demonstrated that knockdown of PLIN2 enhanced
proliferation, migration and invasion of ccRCC cells.
PLIN2 is a member of the perilipin protein family,
which consists of five members, namely PLIN1-5. PLIN2 serves a
vital role in fatty acid uptake and LD formation, and is believed
to function in intracellular lipid metabolism (26,28,39).
Investigations into PLIN2 in several types of cancer have indicated
that it has an important role in tumorigenesis. Numerous studies
have reported that PLIN2 levels are markedly increased in specific
types of cancer in plasma (29),
urine (40) and tumor samples.
Several types of cancer, such as Burkitt lymphoma (41), colorectal cancer (29), lung adenocarcinoma (28) and ccRCC (40), exhibit high levels of PLIN2. In
addition, PLIN2 is relevant to tumor maintenance in numerous
malignancies; high PLIN2 expression has been observed in the
majority of malignant melanomas (42), and overexpression of PLIN2 is
correlated with a markedly worse prognosis in breast cancer
(43). This area of investigation
remains superficial, at least for some types of cancer; however,
these findings indicated that PLIN2 may serve an important role in
tumorigenesis or tumor progression.
Previous studies have provided information regarding
the association between PLIN2 expression and ccRCC. A previous
study reported that PLIN2 expression was markedly increased in
cases of low-stage, low-grade or VHL alteration-positive ccRCC
(32), which is consistent with
the present findings. These results suggested that PLIN2 levels may
represent the tumor differentiation status in ccRCC. By conducting
high-density oligonucleotide microarrays on 33 ccRCC tissues and
nine normal kidney samples, Yao et al (44) identified 149 significantly
differentially expressed genes. When matching them with other
microarray data, it was demonstrated that PLIN2 was overexpressed
in both datasets. The present study observed increased levels of
PLIN2 in clinical ccRCC tissues compared with in paired normal
tissues. The mRNA expression levels of PLIN2 were further validated
by TCGA database, which revealed significantly increased PLIN2
expression in 72 ccRCC tumor tissues compared with in paired normal
tissues. In addition to validation at the protein level, IHC
results demonstrated that the expression levels of PLIN2 were
significantly higher in ccRCC tissues compared with in normal
kidney tissues and benign renal angiomyolipoma tissues. These
results demonstrated that PLIN2 may function as an oncogene,
serving an important role in the tumorigenesis of ccRCC.
Age is a risk factor for numerous types of cancer,
including ccRCC; however, the present study demonstrated that PLIN2
expression was increased in patients ≥60 years group compared with
in those <60 years old. These findings indicated that high PLIN2
expression is associated with poor prognosis, which is in
contradiction with the follow-up results. Conversely, multivariate
regression analysis demonstrated that age and PLIN2 expression
levels were independent prognostic factors for ccRCC; age was a
risk factor (HR, 1.652; P=0.002) and PLIN2 was a protective factor
(HR, 0.586; P=0.001). PLIN2 may therefore be considered a
prognostic factor that is not affected by age. Although PLIN2
expression levels may decrease as the tumor progresses, the role of
age in tumor progression remains unclear; therefore, there is
statistical significance between the two factors, but they do not
necessarily have clinical significance.
The present results revealed that high PLIN2
expression was associated with good OS (P<0.001) and DFS
(P=0.0006), and may be considered a diagnostic biomarker in
patients with ccRCC with various clinicopathological
characteristics. Three previous studies also concluded that
increased PLIN2 mRNA expression is associated with a satisfactory
cancer-specific survival in patients with ccRCC (32,33,44)
and metastatic lesions exhibited low expression (32,44).
Notably, the present study demonstrated that knockdown of PLIN2
promoted ccRCC cell invasion and migration in vitro.
Furthermore, PLIN2 inhibited ccRCC cell proliferation; 4 days
following PLIN2 knockdown, the proliferative capacity of A-498
cells was significantly elevated compared with in cells in the
control group. In addition, the present study demonstrated that
PLIN2 expression levels tended to decrease with increasing tumor
grade and T stage. These findings suggested that after PLIN2 is
transcriptionally activated, the expression levels of PLIN2 may be
gradually downregulated alongside the dedifferentiation processes
in the carcinogenic progression of ccRCC. These results indicated
that PLIN2 may function as an anti-oncogene, serving an important
role in the progression of ccRCC.
Previous studies have elucidated the potential
mechanism underlying PLIN2 overexpression. Yao et al
(32) revealed that overexpression
of PLIN2 may be induced by disruption of the VHL/HIF pathway in
ccRCC. Furthermore, it has been reported that HIF2α may enhance
PLIN2 expression, thus promoting lipid storage, endoplasmic
reticulum homeostasis and cell viability in ccRCC (34). The present study demonstrated that
VHL alteration-positive ccRCC was associated with increased PLIN2
expression. Furthermore, GSEA demonstrated that the HIF1α and HIF2α
signaling pathways were significantly enriched in response to high
PLIN2 expression in patients with ccRCC. It may be hypothesized
that inactivation of VHL, which activates downstream HIF
expression, contributes to increased expression of PLIN2 in ccRCC.
It is well known that PLIN2 is transcriptionally activated by the
peroxisome proliferator-activated receptor (PPAR)-mediated pathway
(45). Conversely, Qiu et
al (34) reported that
constitutive HIF2α activity, rather than PPARγ, PPARα or HIF1α,
regulates PLIN2 in ccRCC cell lines and primary patient samples. We
aim to further explore the molecular mechanisms underlying PLIN2
overexpression and its associated signaling pathways involved in
renal cancer in future studies.
To the best of our knowledge, the present study is
the first comprehensive study establishing the functional role of
PLIN2 in ccRCC tumorigenesis and progression. The results also
indicated that PLIN2 may be considered a potential novel biomarker
for predicting prognosis of patients with ccRCC. However, one of
the potential limitations of our study should be addressed; the
exact mechanism by which PLIN2 inhibits ccRCC progression was not
investigated. Therefore, further explorations to elucidate the
underlying molecular mechanisms are required.
In conclusion, the present results demonstrated that
PLIN2 expression was significantly elevated in ccRCC tissues and
cells, and was correlated with numerous important clinical factors
in patients with ccRCC. Upregulation of PLIN2 expression was
associated with good prognosis of ccRCC. In vitro
experiments also indicated that PLIN2 knockdown may enhance
proliferation, migration and invasion of RCC cells. These findings
suggested that PLIN2 may be considered a novel prognostic biomarker
in ccRCC and a specific diagnostic indicator for patients with
ccRCC. In addition, it could be a potential novel target for the
clinical treatment of ccRCC.
Acknowledgments
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81672524 and
81672528), the Clinical Research Physician Program of Tongji
Medical College, Huazhong University of Science and Technology
(grant no. 5001530015) and the Independent innovation foundation of
Huazhong University of Science and Technology (grant no.
118530309).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ and HR designed the study. QC, KC, KW, ZS and LB
carried out data acquisition and analysis. QC, HR, DL and KC
performed the majority of the experiments. QC, TX and HX wrote the
manuscript and conducted immunohistochemistry analyses. CW and XM
collected the clinical samples and managed the clinical data. GC
and JT contributed to bioinformatics analysis. HY and KC were
involved in project management, and contributed to preparing and
making figures and tables. HY and XZ supervised the study. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study and experimental procedures were
approved by the Human Research Ethics Committee of Huazhong
University of Science and Technology (Wuhan, China). Written
informed consent was obtained from the patients/patients'
families.
Consent for publication
Written informed consent was obtained from the
patients/patients' families.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moch H, Cubilla AL, Humphrey PA, Reuter VE
and Ulbright TM: The 2016 WHO Classification of Tumours of the
Urinary System and Male Genital Organs-Part A: Renal, penile, and
testicular Tumours. Eur Urol. 70:93–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Novara G, Ficarra V, Antonelli A, Artibani
W, Bertini R, Carini M, Cosciani Cunico S, Imbimbo C, Longo N,
Martignoni G, et al: SATURN Project-LUNA Foundation: Validation of
the 2009 TNM version in a large multi-institutional cohort of
patients treated for renal cell carcinoma: Are further improvements
needed? Eur Urol. 58:588–595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gong J, Maia MC, Dizman N, Govindarajan A
and Pal SK: Metastasis in renal cell carcinoma: Biology and
implications for therapy. Asian J Urol. 3:286–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pichler M, Hutterer GC, Chromecki TF,
Jesche J, Kampel-Kettner K, Rehak P, Pummer K and Zigeuner R:
External validation of the Leibovich prognosis score for
nonmetastatic clear cell renal cell carcinoma at a single European
center applying routine pathology. J Urol. 186:1773–1777. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Albiges L, Fay AP, Xie W, Krajewski K,
McDermott DF, Heng DY, Dariane C, DeVelasco G, Lester R, Escudier
B, et al: Efficacy of targeted therapies after PD-1/PD-L1 blockade
in metastatic renal cell carcinoma. Eur J Cancer. 51:2580–2586.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oudard S and Vano Y: The role of
rechallenge with targeted therapies in metastatic renal-cell
carcinoma. Curr Opin Urol. 25:402–410. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zanardi E, Verzoni E, Grassi P, Necchi A,
Giannatempo P, Raggi D, De Braud F and Procopio G: Clinical
experience with temsirolimus in the treatment of advanced renal
cell carcinoma. Ther Adv Urol. 7:152–161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bickel PE, Tansey JT and Welte MA: PAT
proteins, an ancient family of lipid droplet proteins that regulate
cellular lipid stores. Biochim Biophys Acta. 1791:419–440. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kimmel AR, Brasaemle DL, McAndrews-Hill M,
Sztalryd C and Londos C: Adoption of PERILIPIN as a unifying
nomenclature for the mammalian PAT-family of intracellular lipid
storage droplet proteins. J Lipid Res. 51:468–471. 2010. View Article : Google Scholar :
|
|
12
|
Brasaemle DL: Thematic review series:
adipocyte biology. The perilipin family of structural lipid droplet
proteins: stabilization of lipid droplets and control of lipolysis.
J Lipid Res. 48:2547–2559. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ducharme NA and Bickel PE: Lipid droplets
in lipogenesis and lipolysis. Endocrinology. 149:942–949. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Londos C, Sztalryd C, Tansey JT and Kimmel
AR: Role of PAT proteins in lipid metabolism. Biochimie. 87:45–49.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu X, Gruia-Gray J, Copeland NG, Gilbert
DJ, Jenkins NA, Londos C and Kimmel AR: The murine perilipin gene:
The lipid droplet-associated perilipins derive from
tissue-specific, mRNA splice variants and define a gene family of
ancient origin. Mamm Genome. 12:741–749. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miura S, Gan JW, Brzostowski J, Parisi MJ,
Schultz CJ, Londos C, Oliver B and Kimmel AR: Functional
conservation for lipid storage droplet association among Perilipin,
ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and
Dictyostelium. J Biol Chem. 277:32253–32257. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thiele C and Spandl J: Cell biology of
lipid droplets. Curr Opin Cell Biol. 20:378–385. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zehmer JK, Huang Y, Peng G, Pu J, Anderson
RG and Liu P: A role for lipid droplets in inter-membrane lipid
traffic. Proteomics. 9:914–921. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Walther TC and Farese RV Jr: Lipid
droplets and cellular lipid metabolism. Annu Rev Biochem.
81:687–714. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou C, Wang M, Zhou L, Zhang Y, Liu W,
Qin W, He R, Lu Y, Wang Y, Chen XZ, et al: Prognostic significance
of PLIN1 expression in human breast cancer. Oncotarget.
7:54488–54502. 2016.PubMed/NCBI
|
|
21
|
Szigeti A, Minik O, Hocsak E, Pozsgai E,
Boronkai A, Farkas R, Balint A, Bodis J, Sumegi B and Bellyei S:
Preliminary study of TIP47 as a possible new biomarker of cervical
dysplasia and invasive carcinoma. Anticancer Res. 29:717–724.
2009.PubMed/NCBI
|
|
22
|
Jiang HP, Harris SE and Serrero G:
Molecular cloning of a differentiation-related mRNA in the
adipogenic cell line 1246. Cell Growth Differ. 3:21–30.
1992.PubMed/NCBI
|
|
23
|
Jiang HP and Serrero G: Isolation and
characterization of a full-length cDNA coding for an adipose
differentiation-related protein. Proc Natl Acad Sci USA.
89:7856–7860. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chong BM, Reigan P, Mayle-Combs KD,
Orlicky DJ and McManaman JL: Determinants of adipophilin function
in milk lipid formation and secretion. Trends Endocrinol Metab.
22:211–217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brasaemle DL, Barber T, Wolins NE, Serrero
G, Blanchette-Mackie EJ and Londos C: Adipose
differentiation-related protein is an ubiquitously expressed lipid
storage droplet-associated protein. J Lipid Res. 38:2249–2263.
1997.PubMed/NCBI
|
|
26
|
Phillips SA, Choe CC, Ciaraldi TP,
Greenberg AS, Kong AP, Baxi SC, Christiansen L, Mudaliar SR and
Henry RR: Adipocyte differentiation-related protein in human
skeletal muscle: Relationship to insulin sensitivity. Obes Res.
13:1321–1329. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Heid HW, Moll R, Schwetlick I, Rackwitz HR
and Keenan TW: Adipophilin is a specific marker of lipid
accumulation in diverse cell types and diseases. Cell Tissue Res.
294:309–321. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang XD, Li W, Zhang N, Hou YL, Niu ZQ,
Zhong YJ, Zhang YP and Yang SY: Identification of adipophilin as a
potential diagnostic tumor marker for lung adenocarcinoma. Int J
Clin Exp Med. 7:1190–1196. 2014.PubMed/NCBI
|
|
29
|
Matsubara J, Honda K, Ono M, Sekine S,
Tanaka Y, Kobayashi M, Jung G, Sakuma T, Nakamori S, Sata N, et al:
Identification of adipophilin as a potential plasma biomarker for
colorectal cancer using label-free quantitative mass spectrometry
and protein microarray. Cancer Epidemiol Biomarkers Prev.
20:2195–2203. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mentrikoski MJ, Wendroth SM and Wick MR:
Immunohistochemical distinction of renal cell carcinoma from other
carcinomas with clear-cell histomorphology: Utility of CD10 and
CA-125 in addition to PAX-2, PAX-8, RCCma, and adipophilin. Appl
Immunohistochem Mol Morphol. 22:635–641. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Straub BK, Herpel E, Singer S, Zimbelmann
R, Breuhahn K, Macher-Goeppinger S, Warth A, Lehmann-Koch J,
Longerich T, Heid H, et al: Lipid droplet-associated PAT-proteins
show frequent and differential expression in neoplastic
steatogenesis. Mod Pathol. 23:480–492. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yao M, Huang Y, Shioi K, Hattori K,
Murakami T, Nakaigawa N, Kishida T, Nagashima Y and Kubota Y:
Expression of adipose differentiation-related protein: A predictor
of cancer-specific survival in clear cell renal carcinoma. Clin
Cancer Res. 13:152–160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tolkach Y, Lüders C, Meller S, Jung K,
Stephan C and Kristiansen G: Adipophilin as prognostic biomarker in
clear cell renal cell carcinoma. Oncotarget. 8:28672–28682. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qiu B, Ackerman D, Sanchez DJ, Li B,
Ochocki JD, Grazioli A, Bobrovnikova-Marjon E, Diehl JA, Keith B
and Simon MC: HIF2α-dependent lipid storage promotes endoplasmic
reticulum homeostasis in clear-cell renal cell carcinoma. Cancer
Discov. 5:652–667. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ruan H, Li X, Yang H, Song Z, Tong J, Cao
Q, Wang K, Xiao W, Xiao H, Chen X, et al: Enhanced expression of
caveolin-1 possesses diagnostic and prognostic value and promotes
cell migration, invasion and sunitinib resistance in the clear cell
renal cell carcinoma. Exp Cell Res. 358:269–278. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu R, Qin X, Ji C, Zeng W, Yang Y and Tan
W: Pygopus 2 promotes kidney cancer OS-RC-2 cells proliferation and
inva-sionin vitroandin vivo. Asian J Urol. 2:151–157. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cline MS, Craft B, Swatloski T, Goldman M,
Ma S, Haussler D and Zhu J: Exploring TCGA pan-cancer data at the
UCSC cancer genomics browser. Sci Rep. 3:26522013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Targett-Adams P, Chambers D, Gledhill S,
Hope RG, Coy JF, Girod A and McLauchlan J: Live cell analysis and
targeting of the lipid droplet-binding adipocyte
differentiation-related protein. J Biol Chem. 278:15998–16007.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Morrissey JJ, Mobley J, Figenshau RS,
Vetter J, Bhayani S and Kharasch ED: Urine aquaporin 1 and
perilipin 2 differentiate renal carcinomas from other imaged renal
masses and bladder and prostate cancer. Mayo Clin Proc. 90:35–42.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ambrosio MR, Piccaluga PP, Ponzoni M,
Rocca BJ, Malagnino V, Onorati M, De Falco G, Calbi V, Ogwang M,
Naresh KN, et al: The alteration of lipid metabolism in Burkitt
lymphoma identifies a novel marker: Adipophilin. PLoS One.
7:e443152012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fujimoto M, Matsuzaki I, Yamamoto Y,
Yoshizawa A, Warigaya K, Iwahashi Y, Kojima F, Furukawa F and
Murata SI: Adipophilin expression in cutaneous malignant melanoma.
J Cutan Pathol. 44:228–236. 2017. View Article : Google Scholar
|
|
43
|
Lucenay KS, Doostan I, Karakas C, Bui T,
Ding Z, Mills GB, Hunt KK and Keyomarsi K: Cyclin E associates with
the lipogenic enzyme ATP-citrate lyase to enable malignant growth
of breast cancer cells. Cancer Res. 76:2406–2418. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yao M, Tabuchi H, Nagashima Y, Baba M,
Nakaigawa N, Ishiguro H, Hamada K, Inayama Y, Kishida T, Hattori K,
et al: Gene expression analysis of renal carcinoma: Adipose
differentiation-related protein as a potential diagnostic and
prognostic biomarker for clear-cell renal carcinoma. J Pathol.
205:377–387. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Targett-Adams P, McElwee MJ, Ehrenborg E,
Gustafsson MC, Palmer CN and McLauchlan J: A PPAR response element
regulates transcription of the gene for human adipose
differentiation-related protein. Biochim Biophys Acta. 1728:95–104.
2005. View Article : Google Scholar : PubMed/NCBI
|