Introduction
Lung cancer is one of the most malignant cancer
types worldwide (1), with a low
5-year survival rate of 16.6% (2).
Non-small cell lung cancer (NSCLC) is the predominant form of lung
cancer and accounts for the majority of cancer-related mortality
worldwide (3). Conventional
therapeutic strategies of chemotherapy following surgery exert
limited effects for patients with advanced NSCLC (4). A better understanding of the
molecular mechanisms underlying NSCLC resistance and the
development of personalized therapeutic strategies is urgently
required in order to improve the prognosis of patients with
NSCLC.
Recently, an improved understanding of NSCLC
pathogenesis has led to the development of multiple kinase
inhibitors, such as erlotinib, one of the known tyrosine kinase
inhibitors (TKIs). The targeting the ATP binding site of the
intracellular domain of epidermal growth factor receptor (EGFR) by
erlotinib has revolutionized the treatment of NSCLC (5). Patients with NSCLC whose tumors
harbor sensitizing and driving mutations in EGFR, can benefit from
erlotinib treatments. Moreover, there are also 3–15% of patients
with NSCLC with wild-type EGFR which respond to erlotinib with a
disease control rate (DCR) of 40–60% (6,7).
Although erlotinib has been shown to improve the prognosis of
patients with NSCLC in large randomized phase III studies, the
majority of these patients have erlotinib-refractory disease, and
thus acquire chemoresistance and suffer from cancer progression
within 6–15 months of therapy (8).
Therefore, there is an urgent need to elucidate the mechanisms of
erlotinib resistance and to discover reliable biological targets
that play important role in erlotinib resistance.
With the advanced development of whole genome and
transcriptome sequencing technologies and the ENCODE project
(9), it is widely accepted that
the majority of genomic DNA is represented in processed transcripts
lacking protein coding ability (10). Long non-coding RNAs (lncRNAs) are a
group of non-coding RNAs (ncRNAs) containing >200 nucleotides,
which have recently been found to play important regulatory role in
various diseases (11). In recent
years, emerging evidence indicates that they play important roles
in regulating cellular and biological functions, for example, by
controlling gene expression at the post-transcriptional level via
sponging microRNAs (miRNAs or miRs) (12) and modulating transcriptional gene
activation or silencing via epigenetic regulation (13). However, lncRNA RP11-838N2.4
(ENST00000581442), which is located on chromosome 18:
3466247-3478925, has been rarely reported.
Recently, increasing evidence suggests that cells
may also communicate via other mechanisms in addition to these
known methods, including the exchange of cellular fragments,
membranes or specialized organelles, such as microvesicles, which
until recently were regarded as cellular debris (14). Exosomes, which are membrane-derived
vesicles that originate from endosomal multivesicular bodies, have
a size range of 20–150 nm when released into the interstitial
fluid. These vesicles contain protein, lipids, coding or ncRNAs
derived from their donor cell cytoplasm (15) and can be taken up by other cells.
Recently, some studies have suggested that exosomes from stromal
cells can potentially affect the therapeutic response though the
transfer of proteins and lncRNAs (16). However, whether exosomes derived
from resistant cancer cells can confer drug resistance to sensitive
cells still needs to be elucidated.
In this study, we investigated the contributions of
exosome-transmitted lncRNA to erlotinib resistance and explored the
therapeutic implications for patients with NSCLC. Our results
identified the involvement of lncRNA RP11-838N2.4 in the modulation
of chemotherapeutic responses by tumor cell extracellular exosome.
These results suggest that exosomal lncRNA RP11-838N2.4 may be a
novel target for the treatment of NSCLC.
Materials and methods
Clinical samples
In total, 78 serum samples were collected from
patients with NSCLC [male/female ratio, 53/25; range of age, 41–70
(median age, 56)] who received erlotinib treatment at the
Affiliated Hospital of Shandong University of Traditional Chinese
Medicine (Jinan, China) between January, 2011 and June, 2014. In
brief, 5 ml of venous blood from each participant was collected via
venous puncture prior to the commencement of chemotherapy. Serum
was segregated via a centrifugation at 1,600 × g for 10 min at room
temperature within 2 h after collection, followed by a second
centrifugation at 12,000 × g for 10 min at 4°C to remove the
residual cellular debris. Each serum supernatant was transferred
into RNase-free tubes and stored at −80°C until use. The patients
were divided into the responding (CR + PR, 43 patients) and
non-responding (SD + PD, 35 patients) groups according to the
Response Evaluation Criteria In Solid Tumors (RECIST) (version 1.1)
(17). Written informed consent
was obtained from each participant prior to blood collection. The
study protocol was approved by the Clinical Research Ethics
Committee of the Affiliated Hospital of Shandong University of
Traditional Chinese Medicine.
Stability testing of exosomal lncRNAs in serum was
performed by exposing the serum to different conditions, including
incubation at room temperature for 0, 3, 6, 12 and 24 h, RNase A
digestion and low (pH 1.0) or high (pH 13.0) pH solution for 3 h at
room temperature followed by RT-qPCR determination of RP11-838N2.4
expression.
Cells and cell culture
The human NSCLC cell lines, HCC827 and HCC4006,
which harbor EGFR-activating mutations (18,19),
were purchased from the Chinese Academy of Sciences (Shanghai,
China). Both cell lines were cultured in RPMI-1640 medium
(BioWhittaker; Lonza, Walkersville, MD, USA) supplemented with 10
mM HEPES, 1 mM L-glutamine, 100 U/ml penicillin/streptomycin
(BioWittaker; Lonza) and heat-inactivated 10% fetal bovine serum
(FBS; Gibco/Invitrogen Inc., Carlsbad, CA, USA) at 37°C in a
humidified incubator with 5% CO2.
Reagents and treatments
Trichostatin A (TsA), RNase A and Triton X-100 were
purchased from Qiagen (Waltham, MA, USA). The cells were treated
with TsA for 24 h at the concentration of 2.5 nM followed by the
detection of RP11-838N2.4. Rnase A were used at a working
concentration of 20 µg/ml for 1 h followed by the
determination of changes in the expression of lncRNA RP11-838N2.4.
Triton X-100 was used at a volume ratio of 0.1% for 20 min to test
the existence of vesicles.
Establishment of NSCLC
erlotinib-resistant cell lines
The HCC827 and HCC4006 cells were used for the
construction of erlotinib-resistant cells. Erlotinib (s1023) was
purchased from Selleckchem (Houston, TX, USA). A total of 32 male
BALB/C nude mice (6 weeks of age with a median weight of 17.8 g)
were purchased from the Shanghai SIPPR-BK Laboratory Animal Co.
Ltd. (Shanghai, China) and maintained in microisolator cages.
Briefly, 32 mice were used for 4 passages of cell injection, and 8
mice were used for each passage of cells. For each passage, 4 mice
were used for the construction of erlotinib-resistant HCC827 cells
(HCC827/R), including 2 mice treated free of erlotinib (control)
and 2 mice treated with erlotinib; the other 4 mice were used for
the construction of erlotinib-resistant HCC4006 cells (HCC4006/R),
including 2 mice treated free of erlotinib (control) and 2 mice
treated with erlotinib. The mice were housed in a facility under
controlled pathogen-free conditions at a temperature of 28°C, 50%
humidity and were fed ad libitum with sterile chow food and
water. All surgeries were performed under sodium pentobarbital
anesthesia via intraperitoneal injection (75 mg/kg) and all efforts
were made to minimize suffering. The research protocol was approved
by the Shandong University of Traditional Chinese Medicine
Committee on Ethics regarding the Care and Use of Laboratory
Animals. Xenograft tumor volumes were evaluated by caliper
measurements of two perpendicular diameters and calculated using
the following formula: Volume = a x b2/2 ('a' represents
length and 'b' represents width). In order to obtain
erlotinib-resistant NSCLC cells, 5×106 HCC827 or HCC4006
cells were injected subcutaneously into the flanks of nude mice.
When the volume of the xenografts reached 200 mm3, the
mice were orally treated with erlotinib (40 mg/kg/day) following a
standard schedule of 4 weeks on and 2 weeks off treatment. After
one treatment course, the xenografted NSCLC cells were isolated and
transplanted into nude mice again followed by erlotinib treatment.
NSCLC cells from the 4th generation xenografts were isolated and
confirmed to be erlotinib-resistant NSCLC cells. The volume of the
4th generation xenografts following erlotinib treatment was ~150
mm3 and ~500 mm3 for the control treatment.
The established erlotinib-resistant cells were named HCC827/R and
HCC4006/R respectively, while the original HCC827 and HCC4006 cells
were parental cells.
Exosome isolation, labeling and RNA
extraction
Exosomes were extracted from the NSCLC cell culture
medium or serum samples using the ExoQuick precipitation kit (SBI;
System Biosciences, Mountain View, CA, USA) according to the
manufacturer's instructions. Briefly, the culture medium or serum
was thawed on ice and centrifuged at 3,000 × g for 15 min to remove
cells and cell debris. Subsequently, 250 µl of the
supernatant was mixed with 63 µl of the ExoQuick
precipitation kit and incubated at 4°C for 30 min following a brief
up- and -down mix, followed by centrifugation at 1,500 × g for 30
min. The supernatant was then removed by careful aspiration,
followed by another 5 min of centrifugation at 1,500 × g to remove
the residual liquid. The exosome-containing pellet was subsequently
re-suspended in 250 µl phosphate-buffered saline (PBS). The
final pellets, containing exosomes, were collected for RNA
isolation. Size distribution of exosomes was analyzed by Zetasizer
(Malvern Panalytical Ltd., Malvern, UK). Purified exosomes were
labeled with PKH26 Red Fluorescent Cell Linker kit for General Cell
Membrane Labeling (Sigma-Aldrich, St. Louis, MO, USA) as per the
manufacturer's instructions.
The extraction of RNA from the exosomes was
performed using the commercial miRNeasy Serum/Plasma kit (Qiagen),
and RNA extraction from the cell fraction was performed using
TRIzol reagent (Invitrogen) according to the manufacturer's
instructions. All RNA elution steps were carried out at 12,000 × g
for 15 sec, and the RNA was finally eluted in 15 µl
RNase-free ultra-pure water.
Transmission electron microscopy
(TEM)
The exosome pellets were re-suspended in 50
µl PBS and a drop of the suspension was placed on a sheet of
parafilm. A carbon-coated copper grid was floated on the drop for 5
min at room temperature. The grid was then removed and excess
liquid was drained by touching the grid edge against a piece of
clean filter paper. The grid was then placed onto a drop of 2%
phosphotungstic acid with pH 7.0 for ~5 sec, and the excess liquid
was drained off. The grid was allowed to dry for several minutes
and then examined using a JEM-1200 EX microscope (Jeol, Akishima,
Japan) at 80 kilo electron volts.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the serum samples or
cell lines using TRIzol reagent (Invitrogen). The RNA was then
reverse transcribed into cDNA using the SuperScript III®
(Invitrogen) and then amplified by RT-qPCR based on the TaqMan
method on a Bio-Rad CFX96 Sequence Detection system (Bio-Rad,
Berkeley, CA, USA) (20). The
thermocy-cling condition was 95°C for 10 min, followed by 40 cycles
of 95°C for 15 sec and 60°C for 1 min. The gene expression levels
were normalized to GAPDH expression. The RT-qPCR results were
analyzed and expressed as ΔΔCq (21). All the primer sequences were
synthesized by RiboBio (Guangzhou, China) and their sequences are
listed in Table I.
 | Table IThe sequences of the primers used in
RT-qPCR and the siRNA sequences. |
Table I
The sequences of the primers used in
RT-qPCR and the siRNA sequences.
| RT-qPCR primer
name | Primer sequence
(5′-3′) |
|---|
| RP11-838N2.4
(Forward) |
GTTTCCTGGAAGGGCATTTT |
| RP11-838N2.4
(Reverse) |
TCCAGCTTCTCCTTTTGCA |
| FOXO1
(Forward) |
GTGAAAACTGCGGGGAAAA |
| FOXO1
(Reverse) |
CCCCTGGACATCAGCACA |
| GAPDH
(Forward) |
GCACCGTCAAGGCTGAGAAC |
| GAPDH
(Reverse) |
ATGGTGGTGAAGACGCCAGT |
|
| siRNA name | siRNA sequence
(5′-3′) |
|
| RP11-838N2.4
sense |
GCAAAUGAAAGCUACCAAU |
| RP11-838N2.4
antisense |
AUUGGUAGCUUUCAUUUGC |
| ENST00000424980
sense |
GCACAAUAUCUUUGAACUA |
| ENST00000424980
antisense |
UAGUUCAAAGAUAUUGUGC |
| ENST00000430635
sense |
CUAGAAUCCUAAAGGCAAA |
| ENST00000430635
antisense |
UUUGCCUUUAGGAUUCUAG |
| ENST00000412816
sense |
GCTGCTTTCTCGCTTGCT |
| ENST00000412816
antisense |
CCAGGGTCCTTGGTCTCA |
| ENST00000548172
sense |
CCCATGTCGAGCAGGAAG |
| ENST00000548172
antisense |
TGGTGGTTTAGCCAAAGAAT |
| ENST00000413504
sense |
GCTGCCTTCCTTTACCTTC |
| ENST00000413504
antisense |
GCATGGGAGACAGAGTTCTT |
| NC sense |
UUCUCCGAACGUGUCACGUTT |
| NC antisense |
ACGUGACACGUUCGGAGAATT |
RNA oligoribonucleotides and cell
transfection
Small interfering RNA against lncRNAs and negative
control siRNA were synthesized by GenePharma (Shanghai, China).
Constitutive active FOXO1-A3 reagent was purchased from Invitrogen.
The NSCLC cells were plated at 5×104 cells/well in
24-well plates ~24 h prior to transfection. After the cells reached
30–50% confluence, transfection was carried out using Lipofectamine
3000 (Invitrogen) following the manufacturer's instructions.
Transfection efficiency was evaluated in each experiment by RT-qPCR
24 h later to ensure that the cells were actually transfected.
Functional experiments were then performed after sufficient
transfection for 48 h. The siRNA sequences are presented in
Table I.
Microarray analysis
RNA expression profiling was performed using the
Agilent human lncRNA microarray V.2.0 platform (GPL18109; Agilent
Technologies, Santa Clara, CA, USA). Quantile normalization and
subsequent data processing were performed using Agilent Gene Spring
Software 11.5. Heatmaps representing differentially regulated genes
were generated using Cluster software (version 3.0) (22). The microarray analysis was
performed by Beijing Genomics Institute/HuaDa-Shenzhen (Shenzhen,
China).
Bioinformatics analysis
The online database of transcription factor binding
profiles JASPAR (http://jaspar.genereg.net/) was used for prediction of
potential transcription factor binding to the promoter region of
RP11-838N2.4.
Cell viability assay
Alterations in cell viability following transfection
or erlotinib treatment was assayed using the CCK8 kit (Dojindo,
Rockville, MD, USA). In brief, the NSCLC cell lines were seeded
into a 96-well plate in triplicate and then treated with
si-RP11-838N2.4 or si-NC for different periods of time. The cell
cultures were then treated with CCK8 reagent and further cultured
for 2 h. The optical density at 450 nm was measured using a
spectrophotometer (Thermo Electron Corp., Beverly, MA, USA). The
percentage of the control samples of each cell line was calculated
thereafter.
Fluorescence in situ hybridization
analysis (FISH)
Nuclear and cytosolic fraction separation was
performed using a PARIS kit (Life Technologies/Thermo Fisher
Scientific, Waltham, MA, USA), and RNA FISH probes were designed
and synthesized by RiboBio (Guangzhou, China) according to the
manufacturer's instructions. Briefly, the cells were fixed in 4%
formaldehyde for 15 min and then washed with PBS. The fixed cells
were treated with pepsin and dehydrated through ethanol. The
air-dried cells were incubated further with 40 nM of the FISH probe
in hybridization buffer. Following hybridization, the slide was
washed, dehydrated and mounted with Prolong Gold Antifade Reagent
with DAPI for detection. The slides were visualized for
immunofluorescence with an Olympus fluorescence microscope (IX73;
Olympus Corp., Tokyo, Japan).
Chromatin immunoprecipitation (ChIP)
ChIP was performed using the EZ ChIPTM Chromatin
Immunoprecipitation kit (Millipore, Burlington, MA, USA) according
to the manufacturer's instructions. Briefly, cross-linked chromatin
was sonicated into 200–1,000 bp fragments. The chromatin was
immunoprecipitated using anti-FOXO1 antibodies (#2880, 1:1,000;
Cell Signaling Technology, Beverly, MA, USA). RT-qPCR was conducted
to detect the relative enrichment according to the method described
above.
Western blot analysis
Cell lysates were prepared with RIPA buffer
containing protease inhibitors (Sigma, St. Louis, MO, USA). Protein
concentrations were measured with the BCA Protein Assay kit
according to the manufacturer's instructions (Beyotime Institute of
Biotechnology, Shanghai, China). Equal amounts of protein (25
µg) were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and transferred onto
polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA).
The membranes were then blocked with 5% (5 g/100 ml) non-fat dry
milk in Tris-buffered saline plus Tween (TBS-T) buffer for 2 h at
room temperature. The membranes were incubated overnight at 4°C
with a 1:1,000 solution of antibodies: Anti-FOXO1 (#2880, 1:1,000,
Cell Signaling Technology), anti-cleaved PARP (#5625, 1:1,000, Cell
Signaling Technology), anti-cleaved caspase3 (#9664, 1:1,000, Cell
Signaling Technology), anti-CD9 (#13403, 1:1,000, Cell Signaling
Technology) and anti-β-actin (#4970, 1:1,000, Cell Signaling
Technology). The horseradish peroxidase-conjugated (HRP)
anti-rabbit antibody (#7074, 1:5,000, Cell Signaling Technology)
was used as secondary antibody for immunostaining for 1 h at room
temperature.
Statistical analysis
The Mann-Whitney U test, or Kruskal-Wallis test
(post hoc Mann-Whitney U test with Bonferroni's correction) was
used for evaluating the differences among clinical cohort groups or
cell groups. Receiver operating characteristic (ROC) curves and the
area under the curve (AUC) were established to discriminate
patients with NSCLC responding to treatment from those not
responding using MedCalc (MedCalc Software bvba, Ostend, Belgium).
Count dates were described as frequency and examined using Fisher's
exact test. All statistical analyses were performed with SPSS
software (version 17.0, SPSS Incorporation, Chicago, IL, USA). The
package plots and function heatmap in R software were used for
mapping. Error bars in figures represent the means ± standard
deviation (SD). The results were considered statistically
significant at P<0.05.
Results
lncRNA RP11-838N2.4 is upregulated in
erlotinib-resistant NSCLC cells
To identify the potential lncRNAs that regulate
erlotinib resistance in NSCLC, we established erlotinib-resistant
cell lines. HCC827 and HCC4006 cells are known to be sensitive to
erlotinib treatment, as they harbor EGFR activating mutations in
the tyrosine kinase domain, precisely in exon 19. To obtain
erlotinib-resistant cells, we grafted HCC827 and HCC4006 cells into
nude mice and performed cycles of erlotinib treatment along with
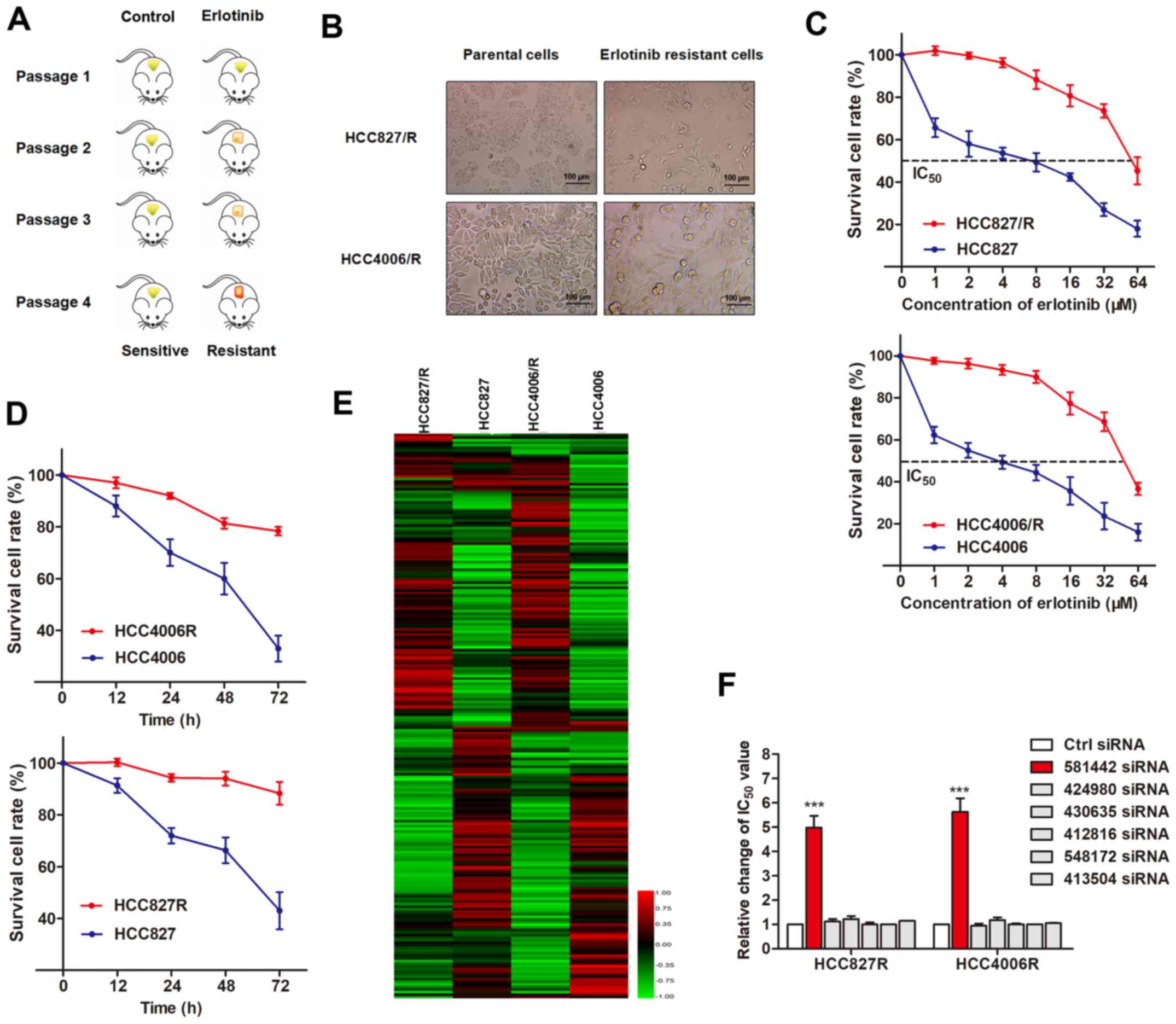
serial passaging in vivo (Fig.
1A). NSCLC xenografts from the 4th passage exhibited a poor
response to erlotinib treatment. Resistant NSCLC cells were
isolated from these xenografts and termed HCC827/R and HCC4006/R
cells, respectively. As shown in Fig.
1B, both erlotinib-resistant cells exhibited specific
morphological changes, including loss of cell polarity causing a
spindle-cell morphology, increased intercellular separation
signifying the loss of intercellular adhesion and the increased
formation of pseudopodia. Compared with the parental cells, these
established resistant cells were less responsive to erlotinib
treatment, as evidenced by increased IC50 values and an
enhanced cell viability (Fig. 1C and
D).
By using the parental and erlotinib-resistant cell
lines, we performed an lncRNA microarray assay to identify the
dysregulated lncRNAs between them. The heatmap created revealed
significant differentially expressed lncRNAs between the NSCLC
parental and resistant cell lines (Fig. 1E), which were then subjected to
validation by RT-qPCR using sensitive and resistant NSCLC cells.
From the 6 upregulated lncRNAs validated in the first round of
experiments (Table II), we
validated that the interference of lncRNA RP11-838N2.4
(ENST00000581442) reversed erlotinib resistance in
erlotinib-resistant cells, while the other 5 lncRNAs exerted
minimal effects (Fig. 1F).
Therefore, we focused on the functional role of lncRP11-838N2.4 in
this study.
 | Table IIFold change of deregulated lncRNAs
between erlotinib-resistant cells and parental cells. |
Table II
Fold change of deregulated lncRNAs
between erlotinib-resistant cells and parental cells.
| lncRNA | Average fold change
in HCC827R/HCC827 cells | Average fold change
in HCC4006R/HCC4006 cells |
|---|
|
ENST00000581442 | 6.08 | 1.69 |
|
ENST00000424980 | 5.88 | 3.02 |
|
ENST00000430635 | 4.76 | 3.35 |
|
ENST00000412816 | 2.58 | 3.05 |
|
ENST00000548172 | 3.59 | 2.39 |
|
ENST00000413504 | 3.77 | 2.43 |
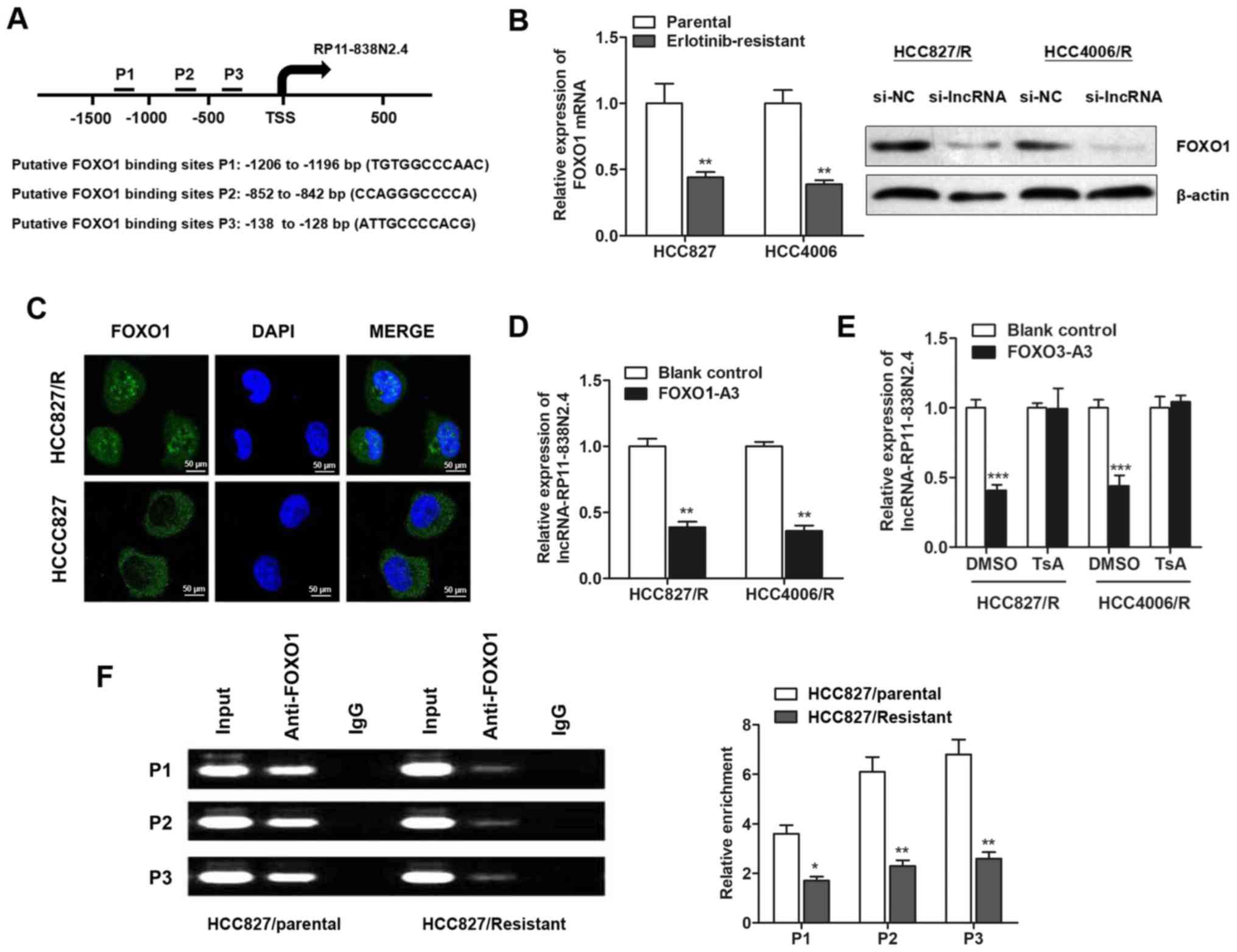
lncRP11-838N2.4 is regulated by FOXO1 in
erlotinib-resistant cells
Increasing evidence has revealed that several key
transcription factors contribute to lncRNA dysregulation in the
human cancer cells (23,24). We then sought to determine whether
the dysregulation of lncRNA RP11-838N2.4 in erlotinib-resistant
cells is due to the activation or silencing of upstream factors.
Three DNA binding elements at the promoter region of lncRNA
RP11-838N2.4 were predicted for FOXO1 by JASPAR (Fig. 2A). RT-qPCR and western blot
analysis revealed that FOXO1 expression was downregulated in
erlotinib-resistant cells when compared with the parental cells at
both the transcript and protein levels (Fig. 2B). Consistently, immunofluorescence
assay revealed that FOXO1 was less enriched in the nucleus of
HCC827 erlotinib-resistant NSCLC cells in contrast to the parental
cells (Fig. 2C). Moreover,
constitutively active FOXO1-A3 markedly inhibited the lncRNA
RP11-838N2.4 expression levels in both erlotinib-resistant cells
(Fig. 2D). Previously, FOXO1 has
been reported to act as a transcription inhibitor by recruiting
histone deacetylase (25). Thus,
we then investigated whether FOXO1 functions in a similar manner.
As expected, treatment with TsA, a well-known histone deacetylase
inhibitor, abrogated the effects of FOXO1 (Fig. 2E). We also performed ChIP assay to
further verify the enrichment of FOXO1 at the promoter region of
lncRNA RP11-838N24. As expected, FOXO1 was enriched and the
enrichment was significantly depressed in the HCC827/R cells
(Fig. 2F). Taken together, these
results identified that lncRNA RP11-838N2.4 was inactivated by
FOXO1 in erlotinib-resistant cell lines.
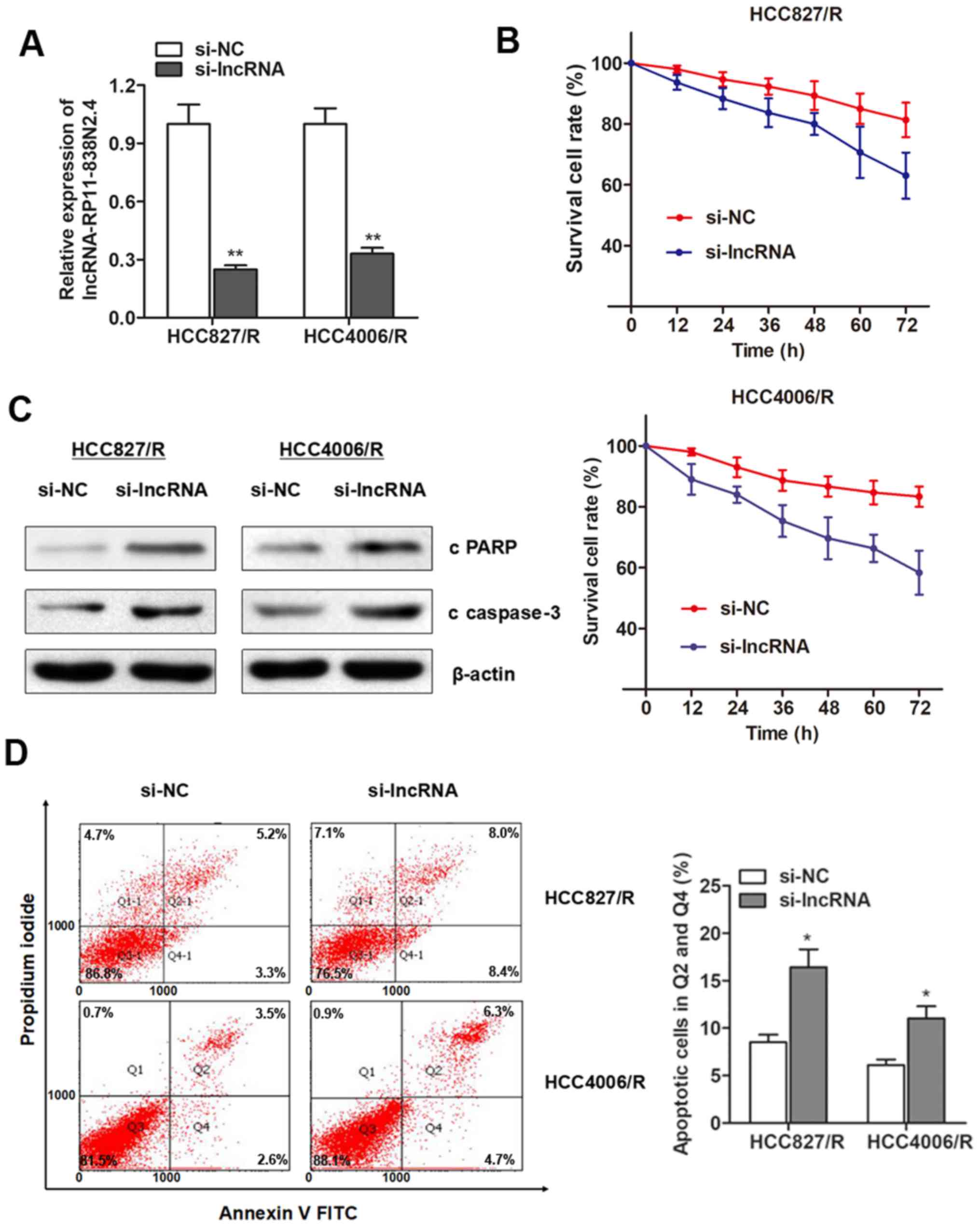
lncRNA RP11-838N2.4 is required for the
erlotinib resistance of NSCLC cells
To investigate whether lncRP11-838N2.4 is essential
for the erlotinib resistance of NSCLC cells, we performed
loss-of-function assay. siRNAs against lncRNA cRP11-838N2.4 was
synthesized and incorporated into erlotinib-resistant cells. As
shown in Fig. 3A, lncRNA
RP11-838N2.4 was markedly silenced in both cell lines. Compared
with the response of the control group, the silencing of lncRNA
RP11-838N2.4 promoted cellular cytotoxicity induced by erlotinib
treatment (Fig. 3B). Moreover, the
knockdown of lncRNA RP11-838N2.4 induced an increase in the protein
expression levels of cleaved PARP (c PARP) and cleaved caspase3 (c
caspase-3) following treatment with erlotinib (Fig. 3C). In addition, flow cytometry
indicated that lncRNA RP11-838N2.4 silencing significantly promoted
the proportion of apoptotic cells following treatment with
erlotinib in the two resistant cell lines (Fig. 3D). Collectively, these results
suggest that lncRNA RP11-838N2.4 is essential for the development
of erlotinib resistance.
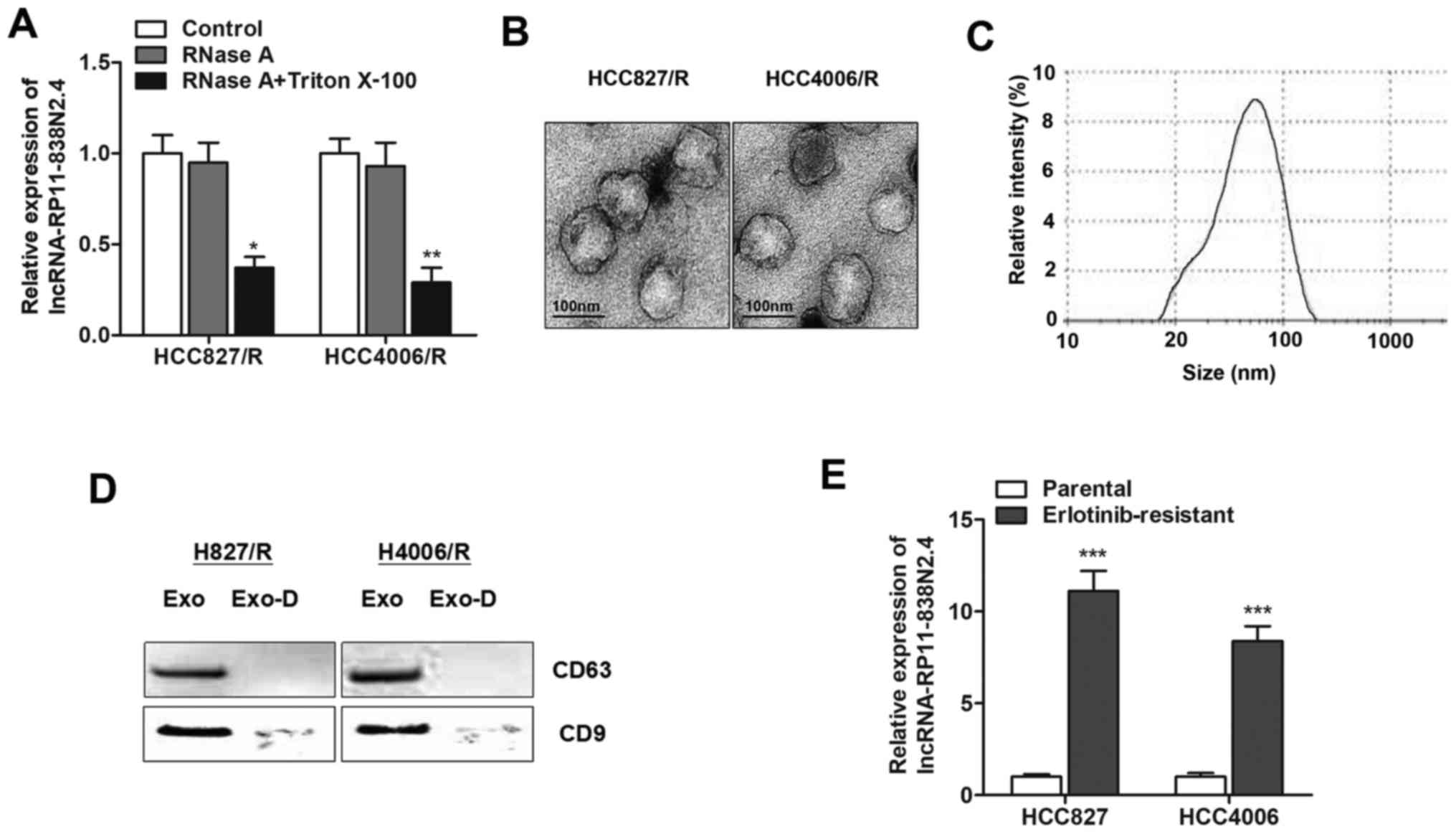
Extracellular lncRNA RP11-838N2.4 is
transferred through incorporation into exosomes
Exosomes can be actively secreted from a variety of
cell types, and lncRNAs contained in exosomes can be secreted into
the culture medium (26). In order
to investigate whether lncRNA RP11-838N2.4 is secreted through
packaging into exosomes, we detected changes in the expression
level of lncRNA RP11-838N2.4 following treatment with RNase A. As
shown in Fig. 4A, the expression
level of lncRNA RP11-838N2.4 in the culture medium was minimally
influenced upon RNase treatment; however it was significantly
decreased following treatment with RNase and Triton X-100
simultaneously, suggesting that this lncRNA was protected by the
membrane instead of being directly secreted. To validate these
results, we purified and extracted exosomes from the culture
medium. The representative micrograph obtained by TEM revealed
vesicles with a round or oval membrane, and a diameter of 20–200 nm
by TEM (Fig. 4B). Exosomes with a
size ranging from 30–150 nm in diameter accounted for 75%, with the
median value was 57.89 nm (Fig.
4C). Western blot analysis further confirmed their identity by
enriched exosome proteins, such as CD63 and CD9 (Fig. 4D). Subsequently, we determined
whether lncRNA RP11-838N2.4 was incorporated into exosomes.
Exosomes were isolated from the culture medium with the ExoQuick
purification kit followed by RT-qPCR. As expected, lncRNA
RP11-838N2.4 was detectable, and its expression level was
significantly higher in the culture medium collected from
erlotinib-resistant cells than in the culture medium from the
parental cells (Fig. 4E). These
findings strongly indicated that extracellular lncRNA RP11-838N2.4
was released by packaging into exosomes.
Exosome-mediated transfer of lncRNA
RP11-838N2.4 induces erlotinib resistance
To identify whether lncRP11-838N2.4 regulates
erlotinib resistance through the delivery of exosomes, we proved
that lncRNA RP11-838N2.4 contained in exosomes can be taken up by
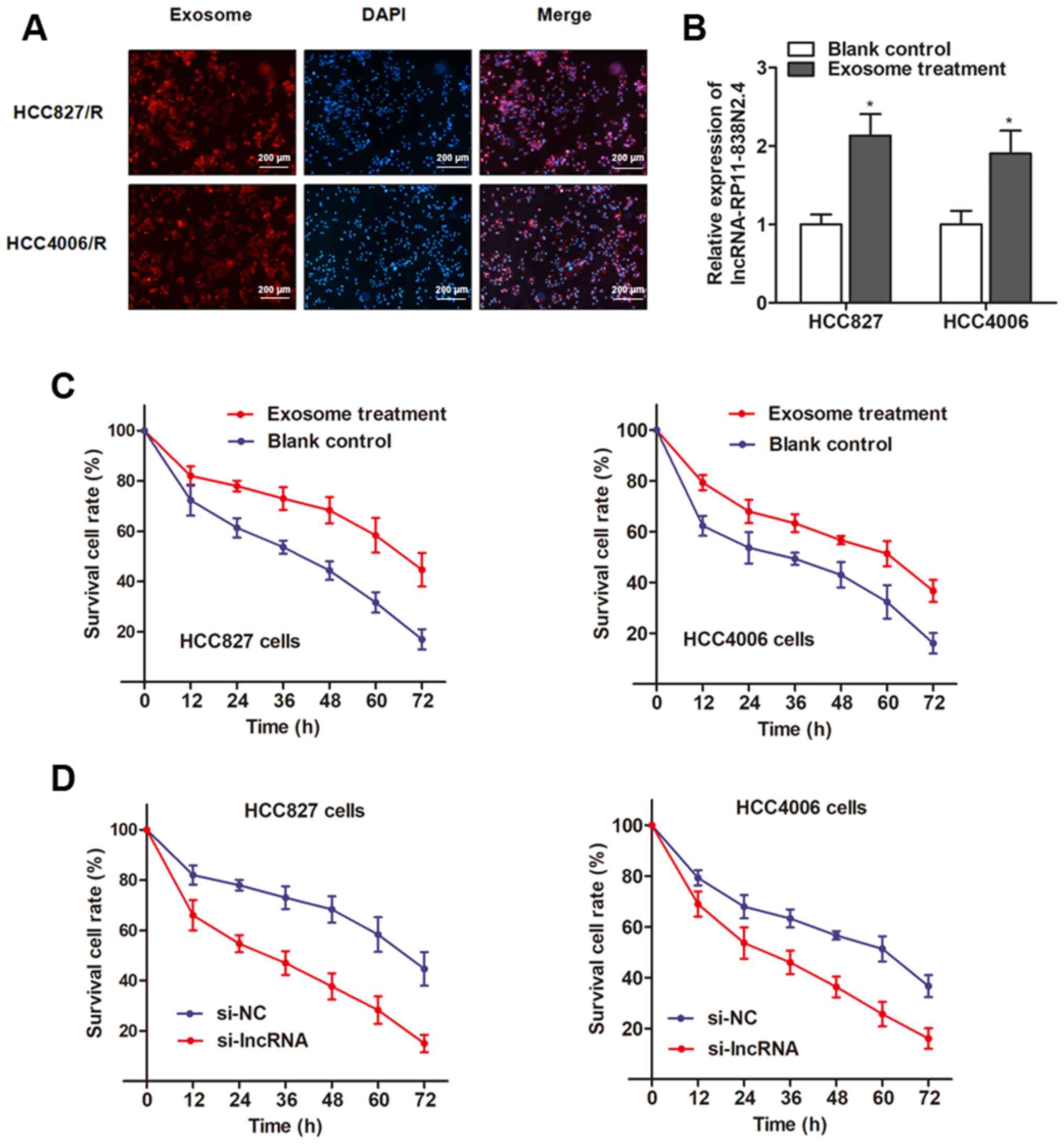
recipient cells using two approaches. Firstly, we examined whether
the secreted exosomes could be taken up by recipient cells by
labeling isolated exosomes with PKH26 dye from HCC827/R cells. The
labeled exosomes were then added and incubated with the HCC827 and
HCC4006 parental cells. As shown in Fig. 5A, the majority of the recipient
cells exhibited a red signal under a confocal microscope. Secondly,
we examined whether these exosomes could deliver lncRNA
RP11-838N2.4 to recipient cells similar to the intercellular
transfer of other ncRNAs as previously reported (27). RT-qPCR revealed an increased
expression of lncRNA RP11-838N2.4 in both recipient cells incubated
with exosomes from the HCC827/R cells (Fig. 5B). Thus, we verified that lncRNA
RP11-838N2.4 contained in exosomes could be taken by recipient
cells.
Subsequently, we determined whether the HCC827 and
HCC4006 cells with elevated exosomal lncRNA RP11-838N2.4 expression
levels exhibit an increased resistance to erlotinib treatment
compared with the control cells. As expected, both recipient cell
lines exhibited decreased cell death compared with the control
cells when treated with erlotinib (Fig. 5C). However, the knockdown of lncRNA
RP11-838N2.4 in the parental cells abrogated this effect (Fig. 5D), indicating that the increased
erlotinib-resistant potency was induced by treatment with exosomal
lncRNA RP11-838N2.4.
Serum exosomal lncRNA RP11-838N2.4 level
is upregulated in erlotinib-resistant patients
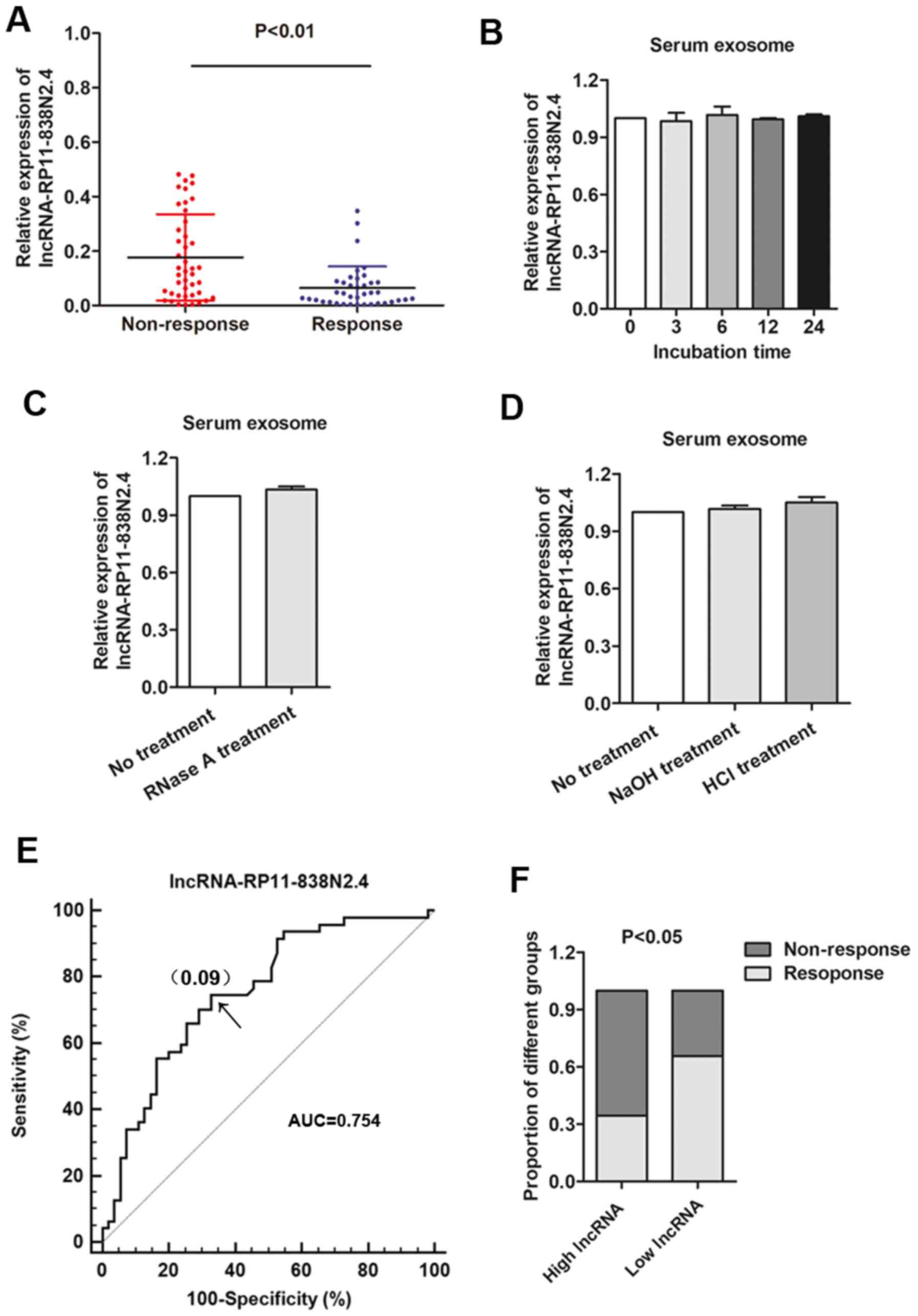
We attempted to determine the expression level of
exosomal lncRNA RP11-838N2.4 in 78 serum samples from patients with
advanced NSCLC receiving erlotinib treatment. Patients were divided
into the responding (CR + PR, 43 patients) and non-responding (SD +
PD, 35 patients) groups according to the Response Evaluation
Criteria In Solid Tumors (RECIST) (version 1.1) (17). We found that the serum exosomal
lncRNA RP11-838N2.4 level was significantly higher in patients who
did not respond to treatment than in those who responded to
erlotinib (Fig. 6A). Since a
better stability was a critical prerequisite for tumor markers, we
then examined the stability of lncRNA RP11-838N2.4 in serum
exosomes by exposing exosomes to different conditions, including
incubation at room temperature for 0, 3, 6, 12 and 24 h, RNase A
digestion, and low (pH 1.0) or high (pH 13.0) pH solution for 3 h
at room temperature. Our results revealed that the exosomal lncRNA
RP11-838N2.4 expression level was not significantly affected by any
of the experimental conditions (Fig.
6B–D), indicating that exosomal lncRNAs were stable in serum
exosomes. We then investigated the diagnostic potential of lncRNA
RP11-838N2.4 by performing ROC analysis. As shown in Fig. 6E, the AUC, diagnostic sensitivity
and specificity reached 0.754, 74.5 and 67.3% with the established
cut-offs (0.09), respectively. Under this stratification criteria
(0.09), patients were divided into the low and a high lncRNA
RP11-838N2.4 expression groups, and the proportion of patients not
responding to chemotherapy was significantly higher in the high
exosomal lncRNA RP11-838N2.4 expression group than in the low
expression group (Fig. 6F). Taken
together, serum exosomal lncRNA RP11-838N2.4 was also dysregulated
in patients with NSCLC in addition to the in vitro
observations. However, further investigations of clinical value are
warranted.
Discussion
Extensive efforts in the past have contributed to
the understanding of both the molecular and cellular mechanisms of
chemoresistance, one of the major causes for the failure of
treatment of advanced cancer types. However, little progress has
been made ever since (28). In
this study, we established erlotinib-resistant cell lines, and
further investigated the functional association between erlotinib
resistance and specific lncRNAs. Our data revealed that lncRNA
RP11-838N2.4 was inactivated by FOXO1, and was upregulated in
erlotinib-resistant cells. More importantly, we revealed that
extracellular lncRNA RP11-838N2.4 was transferred through
incorporation into exosomes, and exosomal lncRNA RP11-838N2.4
incubation promoted the erlotinib resistance of parental cells.
Clinically, the serum exosomal lncRNA RP11-838N2.4 level was
associated with erlotinib response in patients with NSCLC.
EGFR is critical in proliferation and survival
pathways, and activating mutations are often observed in NSCLC
(29). EGFR mutations occur more
frequently in Asian patients compared with Caucasian patients
(30,31). Erlotinib, the second EGFR TKI
evaluated in NSCLC, was approved by the FDA in November, 2004 based
on the results of the BR.21 trial conducted by the National Cancer
Institute of Canada Clinical Trials Group. Erlotinib has exhibited
efficacy as a second-/third-line treatment for advanced NSCLC, and
superior first-line efficacy versus chemotherapy in EGFR
mutation-positive disease (32–34).
However, acquired resistance, defined as progression after initial
benefits are observed, to targeted therapies inevitably occurs
(35). Therefore, breakthroughs
are required in the understanding and treatment of acquired
erlotinib resistance in NSCLC, particularly for patients with EGFR
mutant and ALK rearrangement-positive sites. In this study, we
established two erlotinib-resistant NSCLC cell lines, which
exhibited specific morphological changes, including the loss of
cell polarity, increased intercellular separation, and increased
formation of pseudopodia. These changes strongly indicate that
chemoresistant cells undergo an epithelial-mesenchymal transition
(EMT) process. Previous studies have also demonstrated that cancer
cells with aquired chronic chemoresistance undergo EMT in various
cancer types (36–38). Thus, the role of lncRNA
RP11-838N2.4 in erlotinib-induced EMT warrants further
investigation in our subsequent studies.
Exosomes are nano-sized vesicles released upon the
fusion of multivesicular bodies with plasma membranes in a variety
of mammalian cells (39). In fact,
exosomes exist extensively in body fluids, such as blood, urine,
ascites and amnionic fluid. Exosomes consist of lipid bilayer
membranes and numerous molecular constituents of their original
cells, including proteins and nucleic acids (40). They have been described as a means
of communication between tumor cells and other cell types, through
the transfer of constituents, such as lncRNAs (41). lncRNAs have gained marked attention
in recent years due to their aberrant expression in a wide range of
cancers and their multiple roles in cancer progression, invasion
and resistance. lncRNAs can act as activators or inhibitors to
participate in a variety of biological processes via interacting
with DNAs, mRNAs, miRNAs or proteins (42). lncRNAs can be protected by exosomes
from degradation in the circulation and can be useful for
monitoring cancer at the early stage (43,44).
Therefore, in this study, we performed microarray analysis to
identify the potential dysregulated lncRNAs in erlotinib-resistant
cells in contrast to the parental NSCLC cell lines. By using
two-step approaches, we finally identified lncRNA RP11-838N2.4 as a
potential lncRNA participating in erlotinib resistance. Moreover,
our integrated research also revealed that this lncRNA can be
packaged into exosomes when released into the extracellular culture
medium. A previous study by Liu et al demonstrated that
lncRNA RP11-838N2.4 enhanced the cytotoxic effects of temozolomide
and reversed temozolomide resistance in glioma cells (45). This suggests that lncRNA
RP11-838N2.4 may be an active regulator during the formation of
chemoresistance, which is consistent with our conclusion. Other 5
potential preliminary identified lncRNAs, which have not been
previously reported, were excluded as they exhibited minimal
effects on erlotinib resistance.
We also verified whether lncRNA RP11-838N2.4 was
incorporated into exosomes, and whether these exosomes containing
lncRNA RP11-838N2.4 could mediate erlotinib resistance. Its
expression in the culture medium was minimally influenced by the
treatment with RNase alone, but was markedly suppressed following
treatment with RNase and Triton X-100 simultaneously, suggesting
that this lncRNA was contained in vesicles, such as exosomes.
Moreover, cDNA detection with RT-qPCR from the extracted exosomes
in the culture medium revealed that the epxression lncRNA
RP11-838N2.4 was detectable and markedly increased in the culture
medium of erlotinib-resistant cells. Treatment of the parental
cells with exosomes containing lncRNA RP11-838N2.4 induced an
enhanced erlotinib resistance. To conclude, lncRNA RP11-838N2.4
participated in the development of resistance to erlotinib through
exosome-mediated transfer. Moreover, we should state that the two
NSCLC cell lines, HCC827 and HCC4006, were inconsistent used for
some experimental validation, which is a limitation of our
work.
Based on the functional observations, we then
determined the exosomal lncRNA RP11-838N2.4 level in clinical serum
samples. Several attempts have been made to use lncRNAs in serum or
plasma as useful predictors in NSCLC (46,47).
Nevertheless, these potential tumor biomarkers are often found in
relatively low abundance and are impeded by the complexity of
bodily fluids. The release of exosomes into the extracellular space
affords an opportunity to examine exosomes in body fluids, such as
blood and urine. Most importantly, exosomes contain nucleic acids
and proteins, reflecting the characteristics of cancer cells, which
provides us with the development of highly sensitive diagnostic
strategies in a non-invasive manner (48). Emerging evidence has uncovered the
unique properties of exosomes, including their ability to embed
specific lncRNAs, their stability and easy detection in the
circulatory system (49,50). As expected, our data clearly
indicated that the exosomal lncRNA RP11-838N2.4 level was
upregulated in erlotinib-resistant patients, and was associated
with erlotinib response.
In conclusion, the findings of this study suggested
that the exosome-mediated transfer of lncRNA RP11-838N2.4 induced
erlotinib resistance in NSCLC cells. Therefore, lncRNA RP11-838N2.4
can act as a potential therapeutic target which may be used to
overcome erlotinib resistance, thus enhancing the clinical benefits
of erlotinib therapy in patients with NSCLC.
Acknowledgments
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81704071, 81273704 and
81273702), the Taishan Scholar Program of Shandong Province in
China of Pulmonary disease of traditional Chinese Medicine (grant
no. ts201712096), the Natural Science Foundation of Shandong
Province (grant nos. ZR2017BH027, ZR2016HB19 and ZR2012HM093), the
Innovation Project of Shandong Academy of Medical Sciences, the
Project of Scientific and Technological Development Program of
Shandong Province (grant no. 2010GSF10242), the Project of
Scientific and Technological Development Program of Traditional
Chinese Medicine of Shandong Province (2017-180, 2011-038,
2009Z004-1, 2007-037 and 2003-43) and the Technology Program of
Shandong Academy of Medical Sciences (grant no. 2015-31).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
WZ, XC, and JY collected clinical samples and
performed the in vitro and in vivo assays; WZ and XC
created a draft of the manuscript; XL and QQ analyzed and
interpreted the data and performed statistical analysis; WZ and WQ
conceived and designed the study. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
For the use of patient samples, the study protocol
was approved by the Clinical Research Ethics Committee of the
Affiliated Hospital of Shandong University of Traditional Chinese
Medicine. Written informed consent was obtained from each
participant prior to tissue collection. For the use of animals, the
research protocol was approved by the Shandong University of
Traditional Chinese Medicine Committee on Ethics regarding the Care
and Use of Laboratory Animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar
|
|
2
|
Ettinger DS, Akerley W, Bepler G, Blum MG,
Chang A, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, Ganti AK, et
al: NCCN Non-Small Cell Lung Cancer Panel Members: Non-small cell
lung cancer. J Natl Compr Canc Netw. 8:740–801. 2010. View Article : Google Scholar
|
|
3
|
Pastorino U: Lung cancer screening. Br J
Cancer. 102:1681–1686. 2010. View Article : Google Scholar
|
|
4
|
Gettinger S and Lynch T: A decade of
advances in treatment for advanced non-small cell lung cancer. Clin
Chest Med. 32:839–851. 2011. View Article : Google Scholar
|
|
5
|
Ciardiello F and Tortora G: EGFR
antagonists in cancer treatment. N Engl J Med. 358:1160–1174. 2008.
View Article : Google Scholar
|
|
6
|
Zhu CQ, da Cunha Santos G, Ding K,
Sakurada A, Cutz JC, Liu N, Zhang T, Marrano P, Whitehead M, Squire
JA, et al: National Cancer Institute of Canada Clinical Trials
Group Study BR.21: Role of KRAS and EGFR as biomarkers of response
to erlotinib in National Cancer Institute of Canada Clinical Trials
Group Study BR.21. J Clin Oncol. 26:4268–4275. 2008. View Article : Google Scholar
|
|
7
|
Horiike A, Yamamoto N, Tanaka H,
Yanagitani N, Kudo K, Ohyanagi F, Ono A, Naito T, Murakami H, Horai
T, et al: Phase II study of erlotinib for acquired resistance to
gefitinib in patients with advanced non-small cell lung cancer.
Anticancer Res. 34:1975–1981. 2014.
|
|
8
|
Lee JY, Sun JM, Lim SH, Kim HS, Yoo KH,
Jung KS, Song HN, Ku BM, Koh J, Bae YH, et al: A phase Ib/II study
of afatinib in combination with nimotuzumab in non-small cell lung
cancer patients with acquired resistance to gefitinib or erlotinib.
Clin Cancer Res. 22:2139–2145. 2016. View Article : Google Scholar
|
|
9
|
ENCODE Project Consortium: An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar
|
|
10
|
Zhuo W, Zhang L, Zhu Y, Xie Q, Zhu B and
Chen Z: Valproic acid, an inhibitor of class I histone
deacetylases, reverses acquired Erlotinib-resistance of lung
adenocarcinoma cells: A Connectivity Mapping analysis and an
experimental study. Am J Cancer Res. 5:2202–2211. 2015.
|
|
11
|
Zhang S, Qin C, Cao G, Xin W, Feng C and
Zhang W: Systematic analysis of long noncoding RNAs in the
senescence-accelerated mouse prone 8 brain using RNA sequencing.
Mol Ther Nucleic Acids. 5:e3432016. View Article : Google Scholar
|
|
12
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar
|
|
13
|
Li P, Zhang X, Wang H, Wang L, Liu T, Du
L, Yang Y and Wang C: MALAT1 is associated with poor response to
oxaliplatin-based chemotherapy in colorectal cancer patients and
promotes chemo-resistance through EZH2. Mol Cancer Ther.
16:739–751. 2017. View Article : Google Scholar
|
|
14
|
Lee TH, D'Asti E, Magnus N, Al-Nedawi K,
Meehan B and Rak J: Microvesicles as mediators of intercellular
communication in cancer - the emerging science of cellular
'debris'. Semin Immunopathol. 33:455–467. 2011. View Article : Google Scholar
|
|
15
|
Théry C, Ostrowski M and Segura E:
Membrane vesicles as conveyors of immune responses. Nat Rev
Immunol. 9:581–593. 2009. View
Article : Google Scholar
|
|
16
|
Santos JC, Ribeiro ML, Sarian LO, Ortega
MM and Derchain SF: Exosomes-mediate microRNAs transfer in breast
cancer chemo-resistance regulation. Am J Cancer Res. 6:2129–2139.
2016.
|
|
17
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar
|
|
18
|
Fustaino V, Presutti D, Colombo T,
Cardinali B, Papoff G, Brandi R, Bertolazzi P, Felici G and Ruberti
G: Characterization of epithelial-mesenchymal transition
intermediate/hybrid phenotypes associated to resistance to EGFR
inhibitors in non-small cell lung cancer cell lines. Oncotarget.
8:103340–103363. 2017. View Article : Google Scholar
|
|
19
|
Presutti D, Santini S, Cardinali B, Papoff
G, Lalli C, Samperna S, Fustaino V, Giannini G and Ruberti G: MET
gene amplification and MET receptor activation are not sufficient
to predict efficacy of combined MET and EGFR inhibitors in EGFR
TKI-resistant NSCLC cells. PLoS One. 10:e01433332015. View Article : Google Scholar
|
|
20
|
Xu SY, Huang X and Cheong KL: Recent
advances in marine algae polysaccharides: Isolation, structure, and
activities. Mar Drugs. 15:152017. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
de Hoon MJ, Imoto S, Nolan J and Miyano S:
Open source clustering software. Bioinformatics. 20:1453–1454.
2004. View Article : Google Scholar
|
|
23
|
Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen
WM, Huang MD and Shu YQ: SP1-induced upregulation of the long
noncoding RNA TINCR regulates cell proliferation and apoptosis by
affecting KLF2 mRNA stability in gastric cancer. Oncogene.
34:5648–5661. 2015. View Article : Google Scholar
|
|
24
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar
|
|
25
|
Schmidt M, Fernandez de Mattos S, van der
Horst A, Klompmaker R, Kops GJ, Lam EW, Burgering BM and Medema RH:
Cell cycle inhibition by FoxO forkhead transcription factors
involves downregulation of cyclin D. Mol Cell Biol. 22:7842–7852.
2002. View Article : Google Scholar
|
|
26
|
Pefanis E, Wang J, Rothschild G, Lim J,
Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, et al:
RNA exosome-regulated long non-coding RNA transcription controls
super-enhancer activity. Cell. 161:774–789. 2015. View Article : Google Scholar
|
|
27
|
Kogure T, Lin WL, Yan IK, Braconi C and
Patel T: Intercellular nanovesicle-mediated microRNA transfer: A
mechanism of environmental modulation of hepatocellular cancer cell
growth. Hepatology. 54:1237–1248. 2011. View Article : Google Scholar
|
|
28
|
Wang Z, Wang N, Li W, Liu P, Chen Q, Situ
H, Zhong S, Guo L, Lin Y, Shen J, et al: Caveolin-1 mediates
chemoresistance in breast cancer stem cells via β-catenin/ABCG2
signaling pathway. Carcinogenesis. 35:2346–2356. 2014. View Article : Google Scholar
|
|
29
|
Ripamonti F, Albano L, Rossini A, Borrelli
S, Fabris S, Mantovani R, Neri A, Balsari A, Magnifico A and
Tagliabue E: EGFR through STAT3 modulates ΔN63α expression to
sustain tumor-initiating cell proliferation in squamous cell
carcinomas. J Cell Physiol. 228:871–878. 2013. View Article : Google Scholar
|
|
30
|
Rosell R, Moran T, Queralt C, Porta R,
Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M,
et al: Spanish Lung Cancer Group: Screening for epidermal growth
factor receptor mutations in lung cancer. N Engl J Med.
361:958–967. 2009. View Article : Google Scholar
|
|
31
|
Shigematsu H, Lin L, Takahashi T, Nomura
M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, et
al: Clinical and biological features associated with epidermal
growth factor receptor gene mutations in lung cancers. J Natl
Cancer Inst. 97:339–346. 2005. View Article : Google Scholar
|
|
32
|
Shepherd FA, Rodrigues Pereira J, Ciuleanu
T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S,
Smylie M, Martins R, et al National Cancer Institute of Canada
Clinical Trials Group: Erlotinib in previously treated
non-small-cell lung cancer. N Engl J Med. 353:123–132. 2005.
View Article : Google Scholar
|
|
33
|
Costa C, Molina MA, Drozdowskyj A,
Giménez-Capitán A, Bertran-Alamillo J, Karachaliou N, Gervais R,
Massuti B, Wei J, Moran T, et al: The impact of EGFR T790M
mutations and BIM mRNA expression on outcome in patients with
EGFR-mutant NSCLC treated with erlotinib or chemotherapy in the
randomized phase III EURTAC trial. Clin Cancer Res. 20:2001–2010.
2014. View Article : Google Scholar
|
|
34
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar
|
|
35
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar
|
|
36
|
Yang AD, Fan F, Camp ER, van Buren G, Liu
W, Somcio R, Gray MJ, Cheng H, Hoff PM and Ellis LM: Chronic
oxaliplatin resistance induces epithelial-to-mesenchymal transition
in colorectal cancer cell lines. Clin Cancer Res. 12:4147–4153.
2006. View Article : Google Scholar
|
|
37
|
Li P, Zhang X, Wang L, Du L, Yang Y, Liu
T, Li C and Wang C: lncRNA HOTAIR contributes to 5FU resistance
through suppressing miR-218 and activating NF-κB/TS signaling in
colorectal cancer. Mol Ther Nucleic Acids. 8:356–369. 2017.
View Article : Google Scholar
|
|
38
|
Wu W, Wang Q, Yin F, Yang Z, Zhang W,
Gabra H and Li L: Identification of proteomic and metabolic
signatures associated with chemoresistance of human epithelial
ovarian cancer. Int J Oncol. 49:1651–1665. 2016. View Article : Google Scholar
|
|
39
|
Denzer K, Kleijmeer MJ, Heijnen HF,
Stoorvogel W and Geuze HJ: Exosome: From internal vesicle of the
multivesicular body to intercellular signaling device. J Cell Sci.
113:3365–3374. 2000.
|
|
40
|
van den Boorn JG, Dassler J, Coch C,
Schlee M and Hartmann G: Exosomes as nucleic acid nanocarriers. Adv
Drug Deliv Rev. 65:331–335. 2013. View Article : Google Scholar
|
|
41
|
Kucharzewska P and Belting M: Emerging
roles of extracellular vesicles in the adaptive response of tumour
cells to microenvironmental stress. J Extracell Vesicles. 2:22013.
View Article : Google Scholar
|
|
42
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar
|
|
43
|
Yang H, Fu H, Xu W and Zhang X: Exosomal
non-coding RNAs: A promising cancer biomarker. Clin Chem Lab Med.
54:1871–1879. 2016. View Article : Google Scholar
|
|
44
|
Li Q, Shao Y, Zhang X, Zheng T, Miao M,
Qin L, Wang B, Ye G, Xiao B and Guo J: Plasma long noncoding RNA
protected by exosomes as a potential stable biomarker for gastric
cancer. Tumour Biol. 36:2007–2012. 2015. View Article : Google Scholar
|
|
45
|
Liu Y, Xu N, Liu B, Huang Y, Zeng H, Yang
Z, He Z and Guo H: Long noncoding RNA RP11-838N2.4 enhances the
cytotoxic effects of temozolomide by inhibiting the functions of
miR-10a in glioblastoma cell lines. Oncotarget. 7:43835–43851.
2016.
|
|
46
|
Tan Q, Yu Y, Li N, Jing W, Zhou H, Qiu S,
Liang C, Yu M and Tu J: Identification of long non-coding RNA 00312
and 00673 in human NSCLC tissues. Mol Med Rep. 16:4721–4729. 2017.
View Article : Google Scholar
|
|
47
|
Tan Q, Zuo J, Qiu S, Yu Y, Zhou H, Li N,
Wang H, Liang C, Yu M and Tu J: Identification of circulating long
non-coding RNA GAS5 as a potential biomarker for non-small cell
lung cancer diagnosisnon-small cell lung cancer, long non-coding
RNA, plasma, GAS5, biomarker. Int J Oncol. 50:1729–1738. 2017.
View Article : Google Scholar
|
|
48
|
Kim KM, Abdelmohsen K, Mustapic M,
Kapogiannis D and Gorospe M: RNA in extracellular vesicles. Wiley
Interdiscip Rev RNA. 8:82017. View Article : Google Scholar
|
|
49
|
Boukouris S and Mathivanan S: Exosomes in
bodily fluids are a highly stable resource of disease biomarkers.
Proteomics Clin Appl. 9:358–367. 2015. View Article : Google Scholar
|
|
50
|
Keller S, Ridinger J, Rupp AK, Janssen JW
and Altevogt P: Body fluid derived exosomes as a novel template for
clinical diagnostics. J Transl Med. 9:862011. View Article : Google Scholar
|