Introduction
Hepatocellular carcinoma (HCC) is the principal type
of liver malignancy and the third leading cause of
cancer-associated mortality worldwide; the morbidity and mortality
rates of HCC are particularly high in China (1). The lack of reliable biomarkers for
tumorigenesis and the unclear clarification of the heterogeneous
genetic and epigenetic alterations contribute to the poor prognosis
of HCC (2).
Aerobic glycolysis, also termed the Warburg effect,
is a phenomenon by which highly proliferative malignant cells
preferentially utilize glycolysis rather than oxidative
phosphorylation, even in the presence of sufficient oxygen, to
satisfy their high nutrient requirements (3,4).
This effect is characterized by the consumption of glucose at a
higher rate and the production of more lactate compared with normal
differentiated cells. Accumulating evidence suggests that metabolic
alteration, which is one of the most consistent hallmarks of
cancer, exerts critical effects on tumor progression (5), and the aberrant expression of
glycolysis-associated molecules contributes to tumorigenesis
(6,7). Therefore, aerobic glycolysis is
pivotal to producing energy in cancer cells, indicating that the
molecules involved may be potential biomarkers and therapeutic
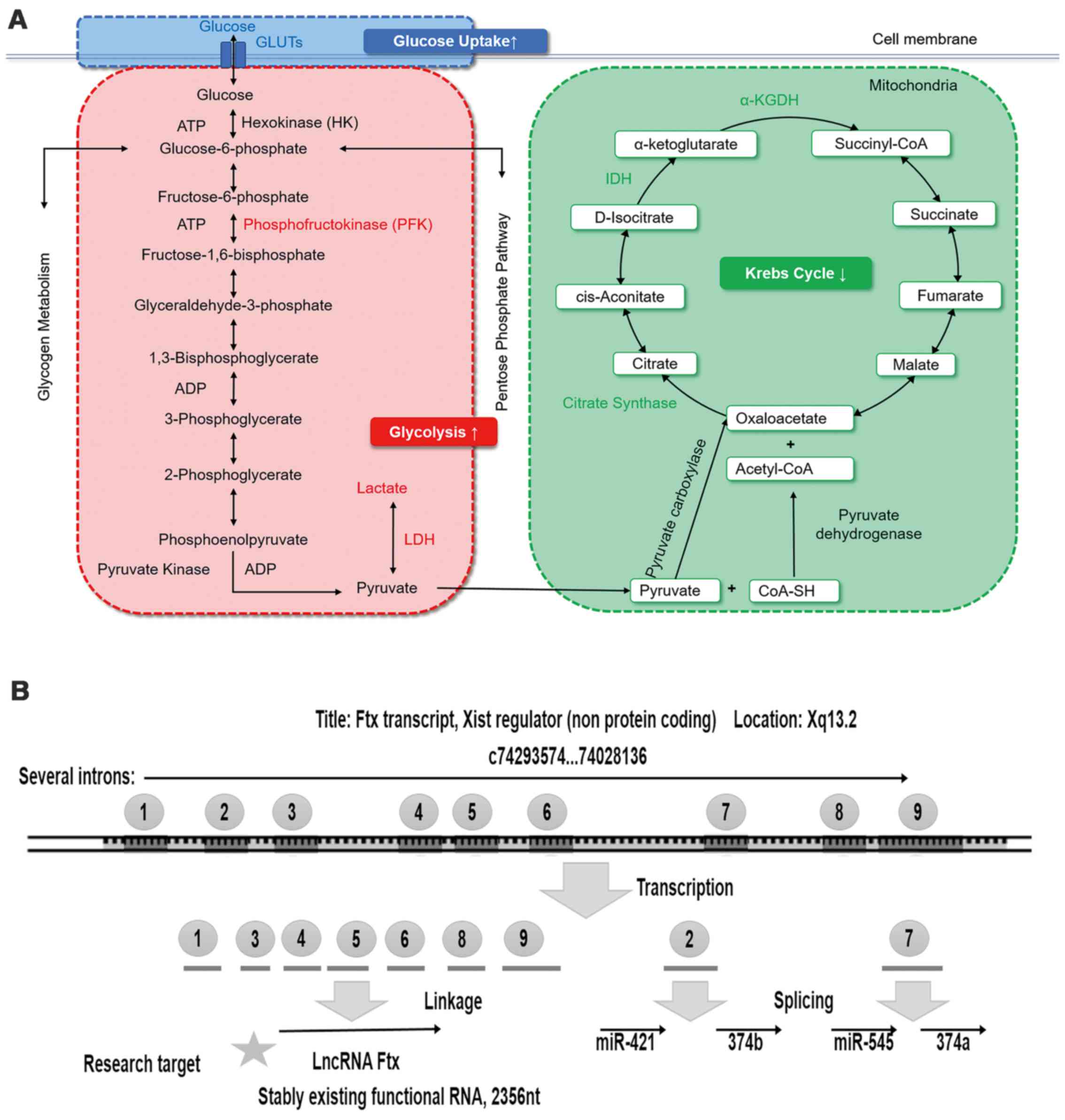
targets for HCC (8) (Fig. 1A).
Long non-coding RNAs (lncRNAs), which do not have
protein-coding ability, are a class of functional RNAs of >200
nucleotides in length and are involved in various biological
processes (9). lncRNAs modulate
target gene expression at the transcriptional and
posttranscriptional levels (10).
Recently, lncRNAs have emerged as important regulators of
carbohydrate metabolism, lipid metabolism (11,12)
and HCC development (13).
Ftx is a well-conserved noncoding gene
encoded within the X-inactivation center on the X chromosome
(14). Ftx encodes a highly
conserved transcript of 2,300 nucleotides that is termed lncRNA Ftx
(Fig. 1B). Ftx encodes nine
introns, the second and seventh of which encode two clusters of
microRNAs (miRs; miR-421/miR-374b and miR-545/miR-374a). RNA
fragments transcribed from other introns compose lncRNA Ftx. Thus,
there are no reduplicated sequences in lncRNA Ftx and the miRs. It
has been demonstrated that lncRNA Ftx/miR-545 contributes
significantly to the tumorigenesis of HCC through activation of
phosphatidylinositol 3-kinase/RAC-α serine/threonine-protein kinase
by targeting DExD/H-box helicase 58 (15). However, the specific association
between lncRNA Ftx and aerobic glycolysis, and the underlying
mechanism, remain unclear. The present study may provide a novel
insight into therapeutic interventions for HCC.
Once activated by ligands, peroxisome
proliferator-activated receptor γ (PPARγ) heterodimerizes with the
retinoid X receptor and combines with PPAR response elements to
regulate the transcription of target genes. It has been
demonstrated that PPARγ serves a vital role in steatosis-associated
hepatic tumorigenesis (16), in
addition to increasing cell sensitivity to insulin and reversing
insulin resistance (17). PPARγ
activation is additionally involved in the regulation of a number
of crucial enzymes in carbohydrate metabolism; for example, PPARγ
activation promotes insulin-responsive glucose transporter 4
(GLUT4) expression (18) and
inhibits pyruvate dehydrogenase kinase 1 (PDK1) expression
(19). Furthermore, PPARγ
activation may reduce tumor necrosis factor (TNF)α and leptin
production, thus facilitating glucose utilization and improving
insulin sensitivity in liver cells (20). However, the role of lncRNA Ftx in
PPARγ-mediated tumor metabolism remains poorly understood.
The present study investigated the aberrant status
of lncRNA Ftx and its potential target gene PPARγ to examine the
possible signaling pathway that regulates aerobic glycolysis, and
to identify a novel therapeutic target for HCC treatment.
Materials and methods
Ethics statement
Written informed consent was obtained from each
patient recruited for the present study for the use of materials.
The consent procedures and all experimental protocols were approved
by the Medical Institutional Ethical Committee of Shandong
Provincial Hospital Affiliated to Shandong University (Jinan,
China; approval no. 2017-231), according to the Declaration of
Helsinki.
Tissue specimens
A total of 73 patients with HCC were recruited
between February 2012 and January 2013 at Shandong Provincial
Hospital Affiliated to Shandong University. The inclusion criteria
were as follows: i) Patients with pathologically confirmed HCC; ii)
patients who underwent curative surgical resection; and iii)
patients >18 years old. The exclusion criteria were as follows:
i) Patients who received preoperative chemotherapy or radiotherapy;
and ii) patients with two or more primary tumors, asynchronously or
synchronously. For each patient, paired HCC tissues and adjacent
non-tumor tissues (as a control) were fresh-frozen in liquid
nitrogen immediately following surgical resection and stored at
−80°C. Patients with HCC were divided into metastasis (n=24) and
non-metastasis (n=49) groups, and complete capsule (n=45) and
incomplete capsule (n=28) groups, according to their
clinicopathological features.
Cell culture and reagents
The human immortalized normal hepatic cell line LO2
and HCC cell lines (Huh7, SMMC-7721 and Bel-7402) were obtained
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China). The Bel-7402 cell line was derived from a surgical specimen
obtained in 1974 from a 53-year-old male patient with HCC and a
positive serum α-fetoprotein status (21). The Huh7 cell line is a
well-differentiated hepatocyte-derived cellular carcinoma cell
line, originally obtained from a liver tumor from a 57-year-old
Japanese male in 1982 (22). All
cell lines were cultured in Dulbecco's modified Eagle's medium
(DMEM) supplemented with 10% fetal bovine serum (FBS) (both from
Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA),
penicillin G (100 U/ml) and streptomycin (100 µg/ml), at 37°C in a
humidified atmosphere containing 5% CO2. The PPARγ
antagonist GW9662 (10 µM) and the PPARγ agonist pioglitazone (10
μM) (MedChemExpress Co., Ltd., Monmouth Junction, NJ, USA)
were dissolved in dimethyl sulfoxide (DMSO; as a vehicle) and added
to the cell culture medium of Huh7 and Bel-7402 cells,
respectively. Following lncRNA Ftx overexpression, Huh7 cells were
treated with GW9662 or vehicle for 24 h, whereas following lncRNA
Ftx downregulation, Bel-7402 cells were treated with pioglitazone
or vehicle for 24 h.
Lentiviral transfections and construction
of stable cell lines
Transfections of lentivirus (LV)-Ftx and its
negative control LV-CON220 (Ubi-MCS-SV40-EGFP-IRES-puromycin) and
LV-Ftx-RNA interference (RNAi) and its negative control LV-CON077
(hU6-MCS-Ubiquitin-EGFP-IRES-puromycin) were performed using a
lentivirus (Shanghai GeneChem Co., Ltd., Shanghai, China),
according to the manufacturer's protocol. To obtain cell lines
stably expressing lncRNA Ftx, Huh7 cells were transfected with the
LV-Ftx, polybrene and enhanced infection solution (Shanghai
GeneChem Co., Ltd.) and selected with puromycin (2 μg/ml)
for 24 h. LV-CON220 was used as a control. To produce cell lines
with stably interfered expression of lncRNA Ftx, Bel-7402 cells
were transfected with LV-Ftx-RNAi, polybrene and enhanced infection
solution (Shanghai GeneChem Co., Ltd.) and selected with puromycin
(2 μg/ml) for 24 h. LV-CON077 was used as a control. The
stably overexpressing or interfered cell lines were validated by
RT-qPCR. Huh7 clones with ~12 times increased lncRNA Ftx expression
levels compared with the normal control were chosen as Ftx, and
Bel-7402 clones with ~74.68% decreased lncRNA Ftx expression levels
compared with the normal control were chosen as short hairpin
(sh)-Ftx. Their negative controls were termed Ftx-NC and sh-NC,
respectively.
Cell proliferation assay
The transfected Bel-7402 and Huh7 cells were seeded
onto 96-well plates (Corning Incorporated, Corning, NY, USA) at
densities of ~3,000 and 5,000 cells/well, respectively. Following
an overnight incubation, 10 μl Cell Counting Kit-8 (CCK-8;
Dojindo Molecular Technologies, Inc., Kumamoto, Japan) solution was
added to each well and incubated at 37°C for 3 h. Subsequently, the
optical density values were measured at 450 nm using a Multiskan Go
spectrophotometer (Thermo Fisher Scientific, Inc.).
Cell invasion and migration assays
Cell invasion and migration were assayed using
Transwell chambers (Corning Incorporated) with and without Matrigel
(BD Biosciences, Franklin Lakes, NJ, USA), respectively. For the
invasion assay, each Transwell chamber was coated with 60 μl
Matrigel and placed into a 24-well plate. Following lentiviral
transfection, Bel-7402 cells and Huh7 cells (1×105) were
seeded into each chamber in serum-free medium, and the lower
chambers were loaded with DMEM supplemented with 10% FBS. A total
of 48 h subsequently, non-migrated cells in the upper chambers were
removed with cotton swabs. For the migration assay, HCC cells
(5×104) were seeded in the upper chambers in serum-free
medium without a Matrigel membrane, and the lower chambers were
loaded with DMEM supplemented with 10% FBS. A total of 36 h
subsequently, HCC cells in the upper chambers that had not migrated
were removed with cotton swabs, and the migrated cells were fixed
in 100% methanol at room temperature for 30 min. The cells on the
bottom surface of the membrane were stained with hematoxylin (Wuhan
Servicebio Technology Co., Ltd., Wuhan, China) at room temperature
for 20 min. Cell images were obtained in high-power (x400
magnification) fields using a phase-contrast microscope (Leica
DM4000B; Leica Microsystems, GmbH, Wetzlar, Germany).
Measurement of aerobic glycolysis
Analysis of glucose consumption
Following transfection, cells were seeded into
6-well plates, and after 6 h, the culture medium was changed to
complete medium and incubated for a further 48 h. Subsequently, the
medium was collected to measure the glucose concentrations, and the
cells were harvested to obtain protein lysates. Glucose
concentrations were detected with a glucose assay kit (cat. no.
361500; Shanghai Rongsheng Biotech Co., Ltd., Shanghai, China),
according to the manufacturer's protocol. All values were
normalized to the total protein levels determined using a Pierce
bicinchoninic acid (BCA) protein assay kit (Thermo Fisher
Scientific, Inc.).
Measurement of lactate generation
A total of ~1×105 cells were seeded onto
6-well plates and cultured for 48 h. Subsequently, the culture
medium was used to determine the lactate concentration using a
lactate assay kit (cat. no. KGT023; Nanjing KeyGen Biotech Co.,
Ltd., Nanjing, China), and the HCC cells were harvested to
determine the protein concentration according to the manufacturer's
protocol. All lactate concentration values were normalized to the
corresponding protein concentration values.
Detection of glycolytic enzymes
The enzymic activity levels of isocitrate
dehydrogenase 1 (IDH1), α-ketoglutarate dehydrogenase (OGDH),
citrate synthase (CS), phosphofruc-tokinase, liver type (PFKL), and
lactate dehydrogenase (LDH) were analyzed using an IDH1
mitochondrial assay (cat. no. BC2160), OGDH assay (cat. no. BC0710)
(Beijing Solarbio Science & Technology Co., Ltd., Beijing,
China), CS assay (cat. no. A108), PFKL assay (cat. no. A129) and
LDH assay (cat. no. A020-1) (all from Nanjing Jiancheng
Bioengineering Institute, Nanjing, China), respectively, according
to the manufacturers' protocols. All values were normalized to the
total protein levels.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cultured Huh7 cells or
Bel-7402 cells, or frozen tissues, using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). A total of 1
μg RNA was reverse-transcribed into cDNA with an RT reagent
kit (Takara Bio, Inc., Otsu, Japan), according to the
manufacturer's protocol. The conditions were as follows: 37°C for
15 min, 85°C for 5 sec and 4°C for 10 min. The amplification was
detected using a SYBR Premix Ex Taq kit (Takara Bio, Inc.) and a
LightCycler® 480 Real-Time PCR system (Roche
Diagnostics, Indianapolis, IN, USA). The thermocycling conditions
were as follows: Pre-incubation at 95°C for 5 min; 45 cycles of
denaturation at 95°C for 10 sec, annealing at 60°C for 10 sec, and
extension at 72°C for 10 sec; a melting cycle at 95°C for 5 sec,
65°C for 1 min, and 97°C with continuous per 5°C acquisition of
fluorescence; and finally cooling at 40°C for 30 sec. The primer
sequences are listed in Table I.
The relative gene expression values are presented according to the
2−ΔΔCq method (23),
relative to β-actin.
 | Table ISequences of primers. |
Table I
Sequences of primers.
| Primer | Sequence
(5′→3′) |
|---|
| TNFα | F
CTGCCTGCTGCACTTTGGAG |
| R
ACATGGGCTACAGGCTTGTCACT |
| PDK1 | F
GTCACAGAGGAGCGTTTCTGG |
| R
TGCCGCCAGAAACATAAATGAGG |
| LEP | F
CACCAGGATCAATGACATTTCACA |
| R
AGCCCAGGAATGAAGTCCAAAC |
| GLUT1 | F
TGTGGGCATGTGCTTCCAGTA |
| R
CGGCCTTTAGTCTCAGGAACTTTG |
| GLUT4 | F
GGGCTGAGACAGGGACCATAAC |
| R
CATGAGCAATGGCATCCAGAA |
| IDH1 | F
AATCAGTGGCGGTTCTGTGGTA |
| R
ACTTGGTCGTTGGTGGCATC |
| PFKL | F
GCATTTATGTGGGTGCCAAAGTC |
| R
AGCCAGTTGGCCTGCTTGA |
| OGDH | F
GGCTACGTGTTGACGCCATA |
| R
CTCAACTTAGCAGCACAAGTCCTTA |
| CS | F
GTCTGGCTAACACAGCTGCAGA |
| R
CATGGCCATAGCCTGGAACA |
| PPARγ | F
TTGTTCCAGGGAAATTCACTGC |
| R
CGCCGTAAATTATTTCTAAACC |
| LncRNA Ftx | F
GAATGTCCTTGTGAGGCAGTTG |
| R
TGGTCACTCACATGGATGATCTG |
| ACTB | F
TGGCACCCAGCACAATGAA |
| R
CTAAGTCATAGTCCGCCTAGAAGCA |
Western blotting
Total protein was extracted from tumor cells with
Radioimmunoprecipitation Assay Lysis Buffer (Beyotime Institute of
Biotechnology, Haimen, China) and phenylmethanesulfonyl fluoride
(Beijing Solarbio Science & Technology Co., Ltd.), and the
concentrations were determined with a Pierce BCA protein assay kit
(Thermo Fisher Scientific, Inc.). A total of 40 μg protein
was separated by 10 or 12% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The membranes, following blocking with 10%
skimmed milk at room temperature for 1 h, were probed with the
following primary antibodies at 4°C overnight: Rabbit anti-PPARγ
(1:1,000; cat. no. ab191407; Abcam, Cambridge, UK), rabbit anti-CS
(1:1,000; cat. no. 16131-1-AP), rabbit anti-PFKL (1:1,000; cat. no.
15652-1-AP), mouse anti-TNFα (1:1,000; cat. no. 60291-1-lg) (Wuhan
Sanying Biotechnology, Wuhan, China), rabbit anti-OGDH (1:250; cat.
no. bs-17710R; BIOSS, Beijing, China), rabbit anti-IDH1 (1:500;
cat. no. PB0632), rabbit anti-GLUT1 (1:500; cat. no. PB0439),
rabbit anti-PDK1 (1:200; cat. no. BA4499), rabbit anti-leptin
(1:100; cat. no. BA1231) and rabbit anti-GLUT4 (1:500; cat. no.
PB0143) (Wuhan Boster Biological Technology, Ltd., Wuhan, China).
Subsequently, the membranes were incubated with horseradish
peroxidase-conjugated goat anti-rabbit or anti-mouse IgG secondary
antibodies (cat. no. ZB-2301 or ZB-2305, respectively; OriGene
Technologies, Inc., Beijing, China) at a dilution of 1:8,000 at
room temperature for 1 h, followed by enhanced chemiluminescence
reagent (EMD Millipore, Billerica, MA, USA). The protein bands were
visualized using an Amersham Imager 600 (GE Healthcare, Chicago,
IL, USA). Total protein levels were normalized to tubulin-β
(1:1,000; cat. no. BM1453; Wuhan Boster Biological Technology,
Ltd.) expression on the same membrane, and the bands were
quantified using ImageJ k 1.45 software (National Institutes of
Health, Bethesda, MD, USA).
lncRNA target prediction
Bioinformatics analysis of predicted lncRNA targets
was performed using the nucleotide BLASTn program (blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch).
Statistical analysis
The data were obtained from at least three
independent experiments and are presented as the mean ± standard
error of the mean (unless otherwise stated). Pearson's correlation
(r) was utilized to measure correlations and logarithmic regression
was used to derive the equation of the slope. Statistical analyses
were performed using GraphPad Prism 6 (GraphPad Software, Inc., La
Jolla, CA, USA) and SPSS 22.0 (IBM Corporation, Armonk, NY, USA).
The significance of differences was evaluated by Student's t-tests
(two-tailed) for two-group comparisons, and the one-way analysis of
variance and the Bonferroni post hoc test for multiple comparisons.
P<0.05 was considered to indicate a statistically significant
difference.
Results
lncRNA Ftx is upregulated in human HCC
tissues and significantly associated with poor prognosis-associated
clinicopathological features
To examine the expression of lncRNA Ftx, RT-qPCR was
performed to analyze 73 HCC and adjacent non-tumorous liver
samples. Compared with the non-tumorous control tissues, lncRNA Ftx
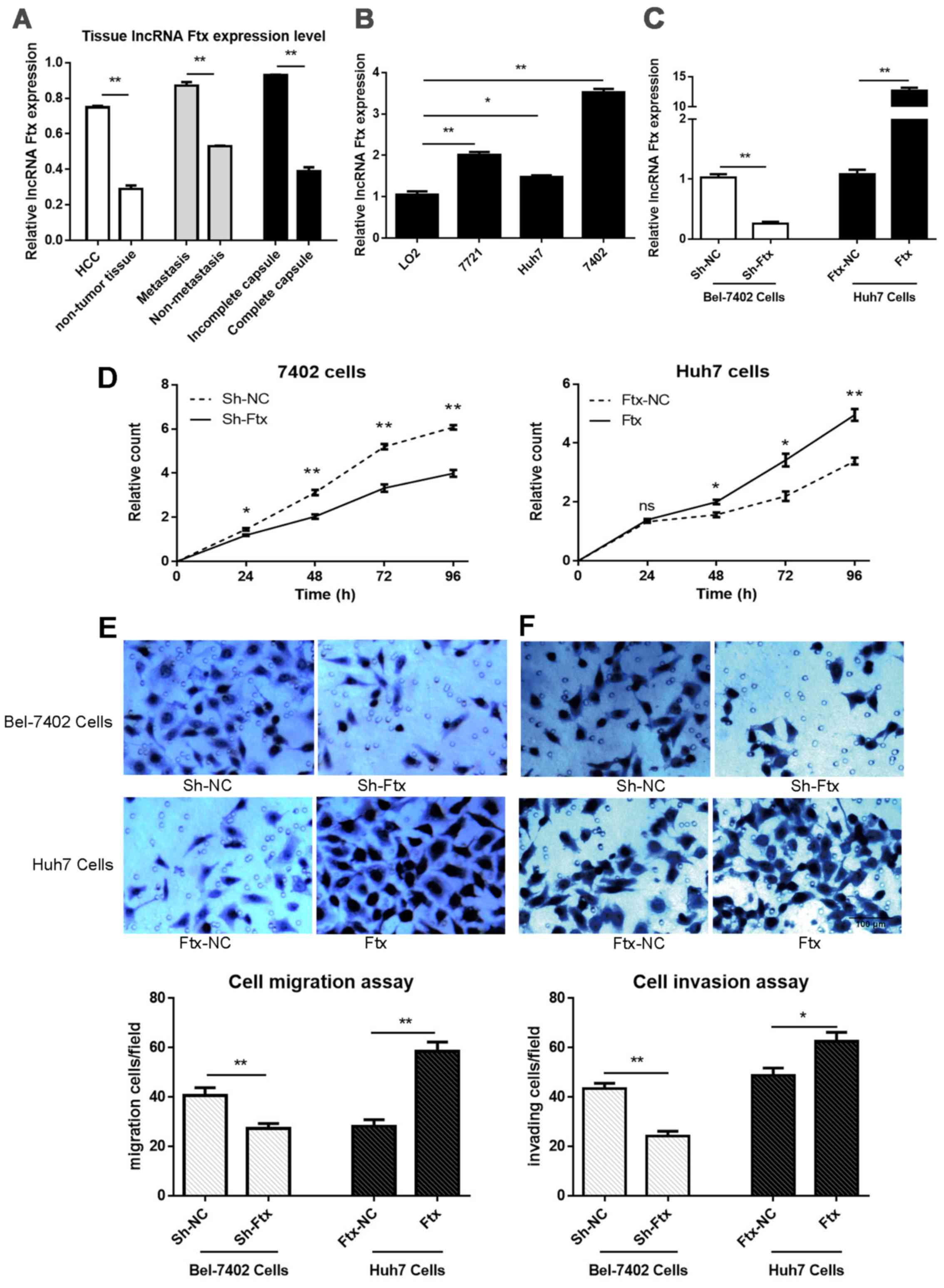
expression was markedly higher in the HCC tissues (Fig. 2A). The clinicopathological
characteristics, including metastasis, tumor capsule, histological
grade and tumor size, are summarized in Table II.
 | Figure 2lncRNA Ftx is upregulated in HCC
tissues and promotes HCC cell proliferation, invasion and migration
in vitro. (A) Expression levels of lncRNA Ftx were increased
in HCC tissues (n=73) compared with matched adjacent normal tissues
(n=73), as determined by RT-qPCR. Patients with HCC were divided
into metastasis (n=24) and non-metastasis (n=49) groups, and
complete capsule (n=45) and incomplete capsule (n=28) groups,
according to their clinicopathological features. lncRNA Ftx
expression levels were increased in the metastasis group compared
with the non-metastasis group, and decreased in the complete
capsule group compared with the incomplete capsule group. (B)
lncRNA Ftx expression levels in the three human HCC cell lines
(Bel-7402, Huh7 and SMMC7721) compared with a non-transformed liver
cell line (LO2). (C) Identification of lentiviral transfection
efficiency. Bel-7402 cells were transfected with sh-Ftx or sh-NC,
and Huh7 cells were transfected with Ftx or Ftx-NC. The
transfection efficacy was determined by RT-qPCR. (D) Cell
proliferation was markedly decreased in lncRNA Ftx-knockdown
Bel-7402 cells compared with the negative control cells, according
to the CCK-8 assay (left column). lncRNA Ftx-overexpressing Huh7
cells had significantly higher cell proliferation compared with the
negative control cells, according to the CCK-8 assay (right
column). (E) Bel-7402 cells and (F) Huh7 cells were transfected,
and cell migration and invasion assays were performed using
Transwell membranes and Matrigel-coated Transwell membranes,
respectively (×400 magnification). Representative images of the
migration and invasion chambers and the average counts from five
random microscopic fields are shown. The experiments were repeated
at least three times and yielded similar results; the error bars
represent the mean ± standard error of the mean.
*P<0.05 and **P<0.01. lncRNA, long
non-coding RNA; HCC, hepatocellular carcinoma; sh, short hairpin;
NC, negative control; CCK-8, cell counting kit-8; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. |
 | Table IIClinicopathological characteristics
of patients with hepatocellular carcinoma. |
Table II
Clinicopathological characteristics
of patients with hepatocellular carcinoma.
| Characteristic | No. of patients
(n=73) |
|---|
| Age, years | |
| <50 | 27 |
| ≥50 | 46 |
| Sex | |
| Male | 47 |
| Female | 26 |
| Tumor size | |
| <5 cm | 29 |
| ≥5 cm | 44 |
| Histological
grade | |
| Good | 14 |
| Moderate | 40 |
| Poor | 19 |
| Metastasis | |
| With | 24 |
| Without | 49 |
| Tumor capsule | |
| Complete | 45 |
| Incomplete | 28 |
In addition, to determine the potential role of
lncRNA Ftx in HCC, correlations between the expression status of
lncRNA Ftx and important clinical features associated with tumor
progression and disease prognosis were analyzed. Although there was
no significant association between lncRNA Ftx expression and age,
sex, tumor size or histological grade, high lncRNA Ftx expression
levels were positively associated with incomplete capsules and HCC
metastasis (Fig. 2A).
Taken together, the results indicated that the
upregulation of lncRNA Ftx expression is associated with poor
prognosis-associated clinicopathological features, suggesting that
lncRNA Ftx is involved in HCC tumorigenesis.
lncRNA Ftx promotes HCC cell
proliferation, invasion and migration in vitro
To choose suitable cell lines for functional
research, lncRNA Ftx expression was screened in a panel of HCC cell
lines and a non-neoplastic hepatic cell line (LO2). It was
identified that the expression levels of lncRNA Ftx were
significantly increased in the HCC cell lines compared with the LO2
cell line. Additionally, the expression level of lncRNA Ftx was the
highest in Bel-7402 cells and the lowest in Huh7 cells (Fig. 2B).
The present study aimed to overexpress and knock
down lncRNA Ftx and investigate the general role of lncRNA Ftx.
Thus, lncRNA Ftx was overexpressed in Huh7 cells as the base level
of lncRNA Ftx was low, and it was knocked down in Bel-7402 cells as
the base level of lncRNA Ftx was high. Bel-7402 cells were
transfected with LV-Ftx-RNAi (sh-Ftx) and its negative control
LV-CON077 (sh-NC), while Huh7 cells were transfected with LV-Ftx
(Ftx) and its negative control LV-CON220 (Ftx-NC). RT-qPCR was used
to confirm the transfection efficiency. lncRNA Ftx expression in
the overexpressing Huh7 cells was ~12 times higher compared with
the normal control cells, while the inhibition rate of lncRNA Ftx
in Bel-7402 cells was ~74.68% (Fig.
2C).
Tumor cell proliferation, invasion and migration are
pivotal steps in tumorigenesis. First, CCK-8 assays were performed
to measure cell viability and determine whether lncRNA Ftx has an
impact on tumor cell proliferation. Cell proliferation was
significantly increased in lncRNA Ftx-overexpressing Huh7 cells
(via LV-Ftx transfection) compared with negative control cells at
the 48, 72 and 96 h time-points, whereas decreased Bel-7402 cell
viability occurred following endogenous lncRNA Ftx knockdown (via
LV-Ftx-RNAi transfection), even at 24 h (Fig. 2D). These results suggested that
lncRNA Ftx supports HCC cell proliferation.
In addition, to assess the effect of lncRNA Ftx on
HCC cell invasion and migration ability, cell invasion and
migration assays were performed using stably transfected cells. As
presented in Fig. 2E and F,
significant migration and invasion decreases were found in lncRNA
Ftx-knockdown Bel-7402 cells compared with negative control cells,
and cell migration and invasion increases were observed in lncRNA
Ftx-overexpressing Huh7 cells; these findings suggested that lncRNA
Ftx enhances the invasion and migration ability of HCC cell lines
in vitro.
Collectively, the present data demonstrated that the
overex-pression of lncRNA Ftx exerts a promoting effect on HCC cell
proliferation, migration and invasion.
Measurement of the Warburg effect
The measurement of the Warburg effect comprises
three parts: The analysis of glucose consumption, the measurement
of the glycolytic pathway, and the detection of the Krebs cycle by
analyzing the activity and expression of the involved enzymes and
the assessment of relative products in carbohydrate metabolism
(Fig. 3). Bel-7402 cells exhibited
increased GLUT1, PFKL and PPARγ mRNA expression levels and
decreased IDH1, CS and OGDH mRNA expression levels compared with
Huh7 cells (Fig. 4A), indicating
that lncRNA Ftx may promote glycolytic metabolism in HCC cells.
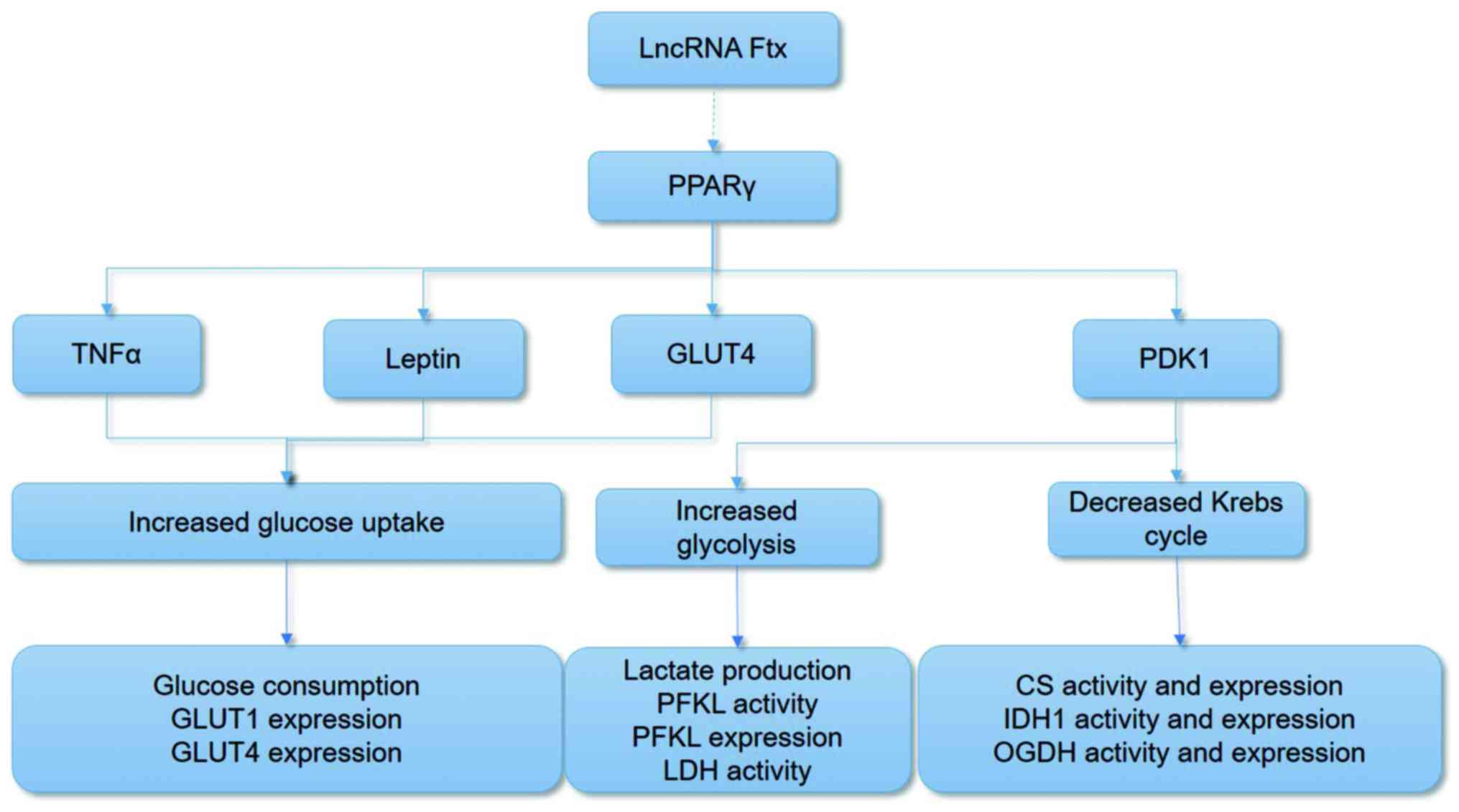 | Figure 3Illustrations of the aerobic
glycolysis measurement methods. lncRNA, long non-coding RNA; PPARγ,
peroxisome proliferator-activated receptor γ; TNFα, tumor necrosis
factor α; GLUT, glucose transporter; PFKL, phosphofructokinase,
liver type; LDH, lactate dehydrogenase; CS, citrate synthase; IDH1,
isocitrate dehydrogenase 1; OGDH, α-ketoglutarate
dehydrogenase. |
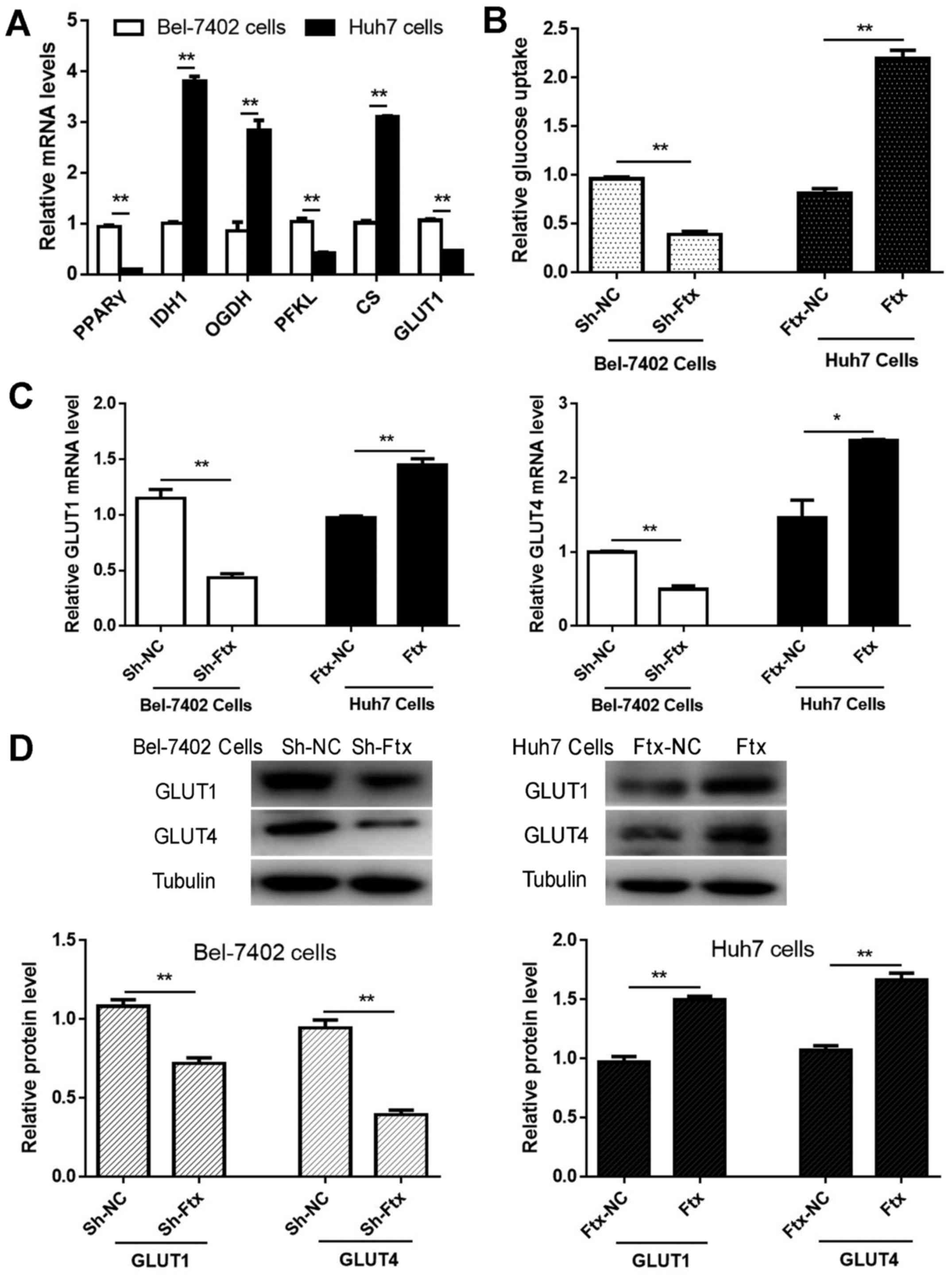 | Figure 4lncRNA Ftx facilitates glucose
consumption via GLUTs in hepatocellular carcinoma cells. (A)
Detection of GLUT1, PFKL, PPARγ, IDH1, CS and OGDH mRNA expression
levels in Bel-7402 cells and Huh7 cells. (B) Measurement of glucose
consumption levels in transfected HCC cells with a glucose assay
kit. Bel-7402 cells and Huh7 cells were transfected, and the (C)
mRNA and (D) protein expression levels of GLUT1 and GLUT4 were
assayed by reverse transcription-quantitative polymerase chain
reaction and western blotting, respectively. The mean and standard
error of the mean of three independent experiments performed in
triplicate are presented. *P<0.05 and
**P<0.01. lncRNA, long non-coding RNA; PPARγ,
peroxisome proliferator-activated receptor γ; GLUT, glucose
transporter; PFKL, phosphofructokinase, liver type; CS, citrate
synthase; OGDH, α-ketoglutarate dehydrogenase; IDH1, isocitrate
dehydrogenase 1; sh, short hairpin. |
lncRNA Ftx facilitates glucose
consumption through GLUTs in HCC cells
The majority of cancer cells generate more ATP by
increasing glucose utilization (24). To investigate whether lncRNA Ftx
may affect aerobic glycolysis in HCC, a glucose assay kit was used.
lncRNA Ftx-overexpressing Huh7 cells had significantly increased
glucose consumption compared with negative control cells, while
lncRNA Ftx-knockdown Bel-7402 cells had reduced glucose utilization
(Fig. 4B); these findings
suggested that lncRNA Ftx may be involved in aerobic glycolysis in
HCC. GLUTs, as facilitative-type glucose transporters, are highly
expressed in a number of types of cancer and are involved in
cellular glucose consumption (25). To gain an understanding of the
mechanisms through which lncRNA Ftx promotes glucose utilization in
HCC cells, the GLUT1 and GLUT4 mRNA and protein expression levels
were measured following lentiviral transfection. As presented in
Fig. 4C and D, the mRNA and
protein expression levels of GLUT1 and GLUT4 were significantly
decreased following the downregulation of lncRNA Ftx in Bel-7402
cells. Consistently, the opposite results occurred with the
upregulation of lncRNA Ftx in Huh7 cells. Taken together, the
present findings suggested that GLUT1 and GLUT4 may be involved in
lncRNA Ftx-mediated HCC glucose uptake.
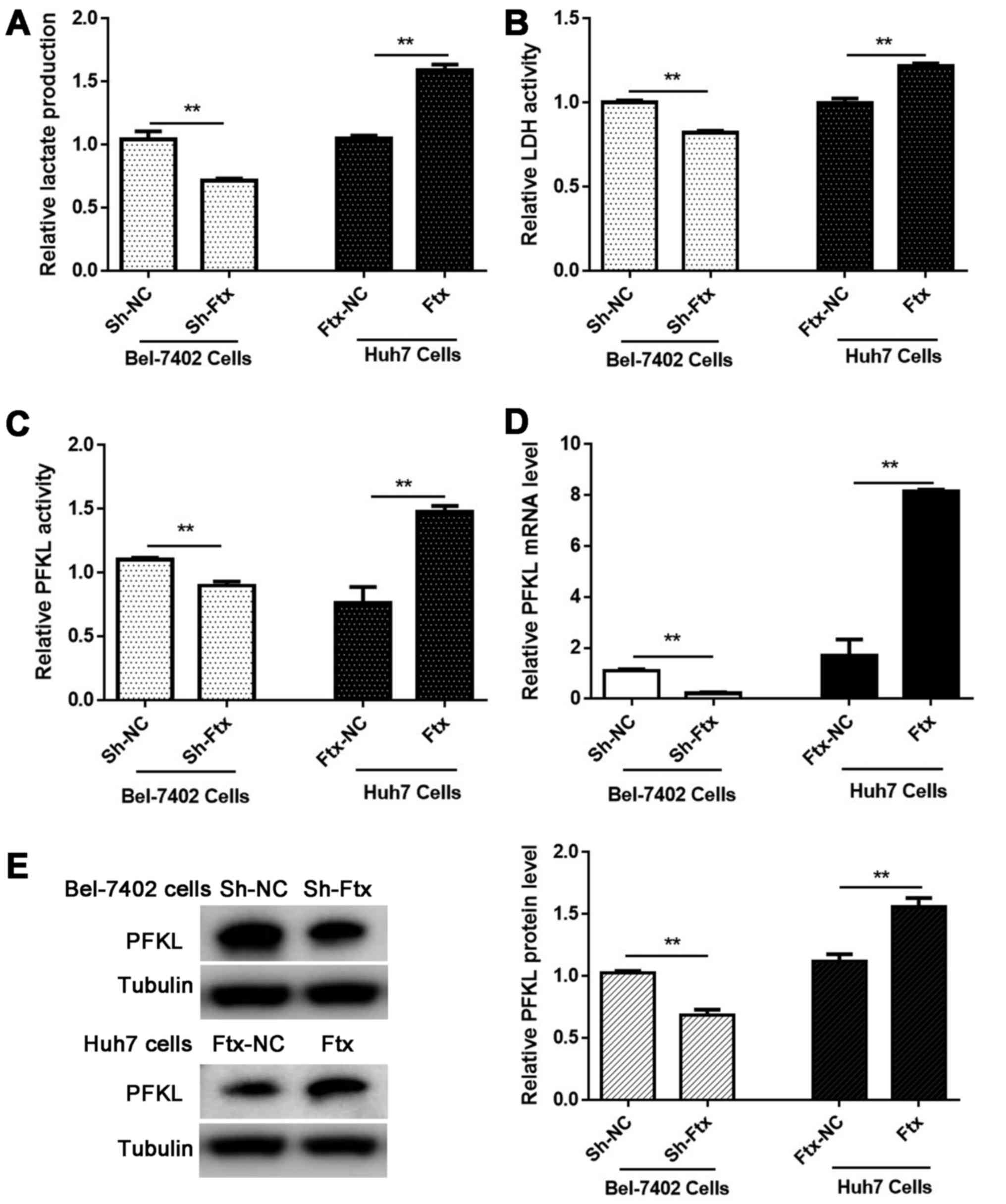
lncRNA Ftx favors lactate production by
regulating glycolytic enzymes in vitro
To evaluate glycolytic activity, a lactate assay kit
was used to measure lactate production. lncRNA Ftx knockdown
significantly decreased the lactate production in Bel-7402 cells,
and lncRNA Ftx overexpression enhanced the lactate production in
Huh7 cells (Fig. 5A). Glycolysis
is regulated by slowing down or speeding up certain steps in the
glycolytic pathway, and this regulation is accomplished by
inhibiting or activating the involved enzymes. Furthermore, PFKL, a
kinase enzyme that phosphorylates 6-phosphofructose, is a key
regulatory enzyme in glycolytic metabolism. To elucidate the
regulatory mechanism of lncRNA Ftx, the enzymic activity levels of
LDH and PFKL and the mRNA and protein expression levels of PFKL
were evaluated in lncRNA Ftx-overexpressing Huh7 cells and lncRNA
Ftx-knockdown Bel-7402 cells, respectively. A positive association
was observed between the lncRNA Ftx expression level and the
enzymic activity levels of LDH and PFKL, and the mRNA and protein
levels of PFKL (Fig. 5B–E),
indicating that lncRNA Ftx enhances the activity and expression of
glycolytic enzymes to increase lactate production in HCC cells.
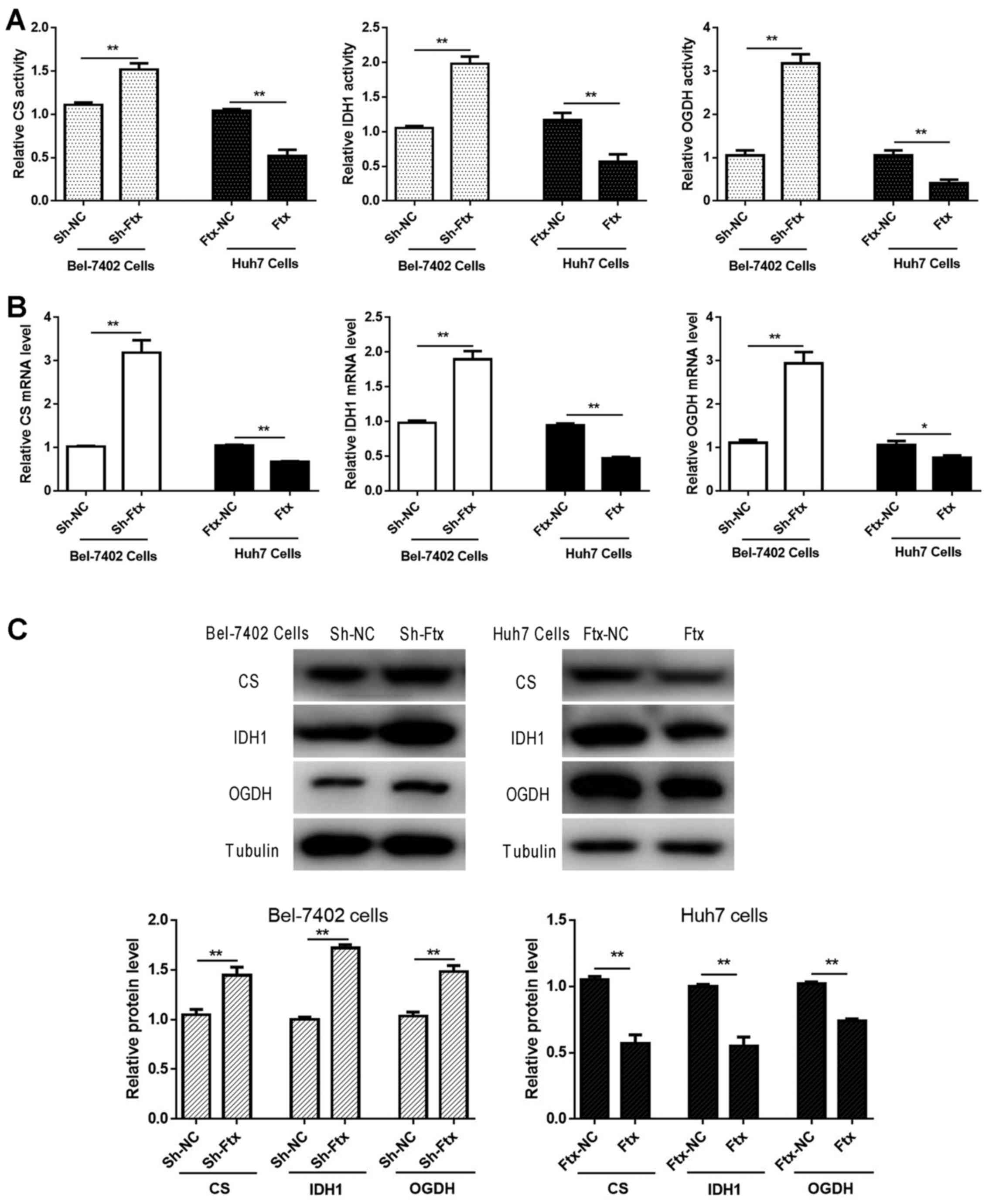
lncRNA Ftx weakens Krebs-cycle-associated
molecules in HCC cells
As the Krebs cycle is determined by the activity and
expression of relative enzymes, the enzymatic activity and mRNA and
protein expression levels of CS, IDH1 and OGDH were measured. As
presented in Fig. 6, the activity,
mRNA and protein expression levels of CS, IDH1 and OGDH were
significantly enhanced with the downregulation of lncRNA Ftx in
Bel-7402 cells, whereas the overexpression of lncRNA Ftx in Huh7
cells impaired the activity and expression levels. The results
revealed that lncRNA Ftx promotes the Warburg effect by impairing
the activity and expression of Krebs-cycle-associated molecules in
HCC cells.
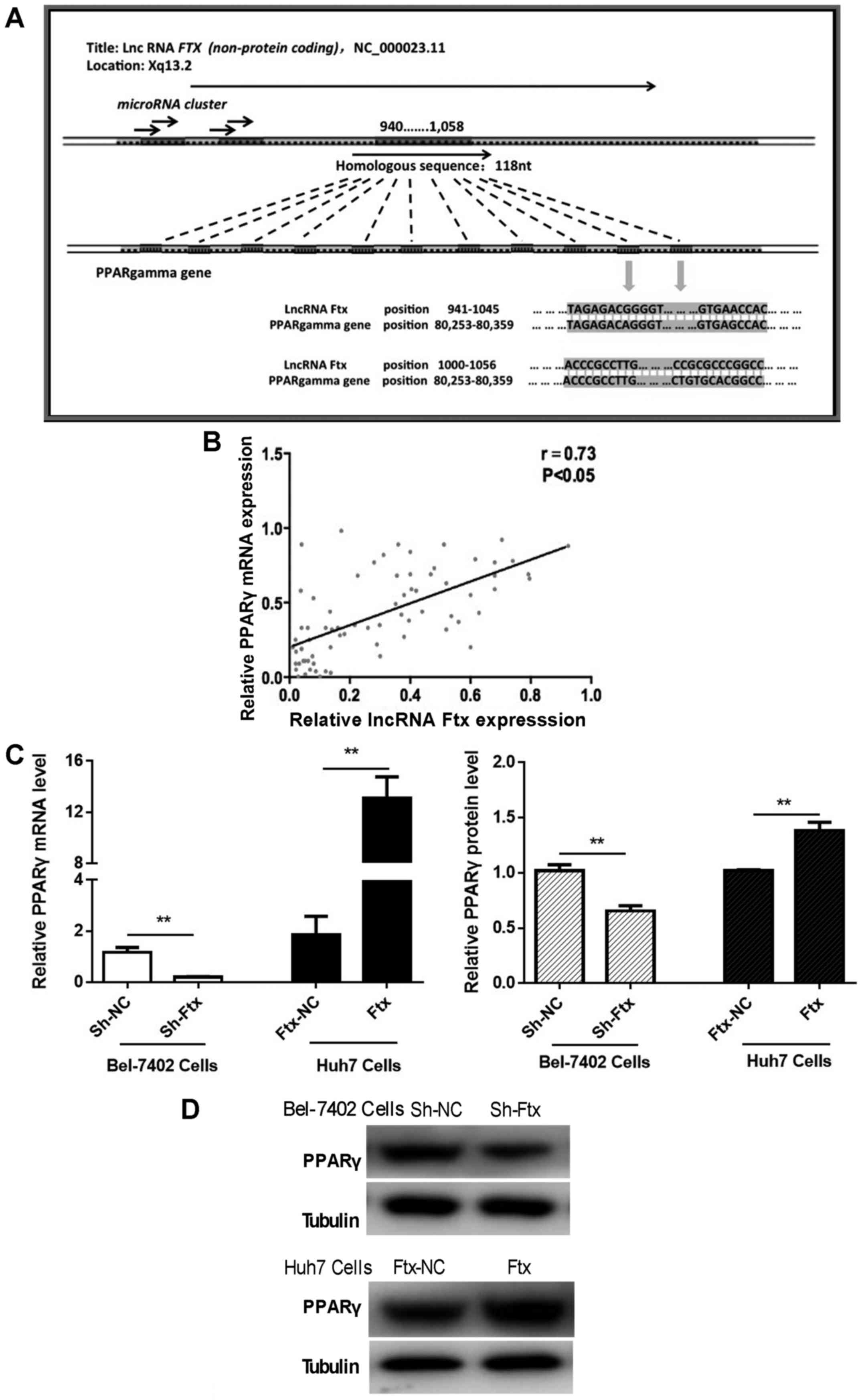
lncRNA Ftx expression is positively
correlated with PPARγ expression in HCC tissues and cells
To determine the underlying mechanism of the lncRNA
Ftx-induced promotion of HCC cell tumorigenesis and aerobic
glycolysis, bioinformatics analysis was performed to predict the
possible target genes or proteins of lncRNA Ftx. According to the
nucleotide BLASTn program, the 940-1058 nt region of Ftx (a
length of 118 nt) was highly homologous with PPARγ (Fig. 7A). The present results suggested
that lncRNA Ftx may directly target the PPARγ gene and
regulate the transcriptional and post-transcriptional expression of
PPARγ.
To assess PPARγ expression levels and their
potential association with lncRNA Ftx in HCC tissues, the mRNA
expression levels of PPARγ were compared in 73 pairs of HCC and
adjacent normal tissue samples using RT-qPCR. The results
demonstrated a positive correlation between the expression levels
of lncRNA Ftx and PPARγ in HCC tissues (Fig. 7B). To identify the PPARγ expression
status in HCC cells, mRNA and protein expression levels were
assessed by RT-qPCR and western blotting upon knocking down and
overexpressing lncRNA Ftx in Bel-7402 cells and Huh7 cells,
respectively. Consistent with the tissue results, PPARγ mRNA and
protein expression levels were positively associated with lncRNA
Ftx levels (Fig. 7C and D).
Together, these results suggested that PPARγ
is a target gene of lncRNA Ftx and functions downstream of lncRNA
Ftx in HCC cells.
Alterations in PPARγ partially abolish
lncRNA Ftx-mediated HCC aerobic glycolysis in vitro
To confirm whether the promotion of aerobic
glycolysis by lncRNA Ftx is mediated through the PPARγ pathway,
PPARγ was suppressed with its antagonist GW9662 in lncRNA
Ftx-overexpressing Huh7 cells, and activated with its agonist
pioglitazone in lncRNA Ftx-knockdown Bel-7402 cells. Subsequently,
the glucose consumption, lactate production, LDH activity and mRNA
expression levels of relative enzymes and molecules were analyzed.
As presented in Fig. 8A, PPARγ
activation enhanced glucose uptake, lactate production, LDH
activity and the mRNA expression of GLUTs and PFKL, and suppressed
the expression of CS, IDH1 and OGDH in lncRNA Ftx-knockdown
Bel-7402 cells. The opposite results were observed in Huh7 cells
(Fig. 8B). These results indicated
that PPARγ may act as an important functional downstream mediator
of lncRNA Ftx in HCC.
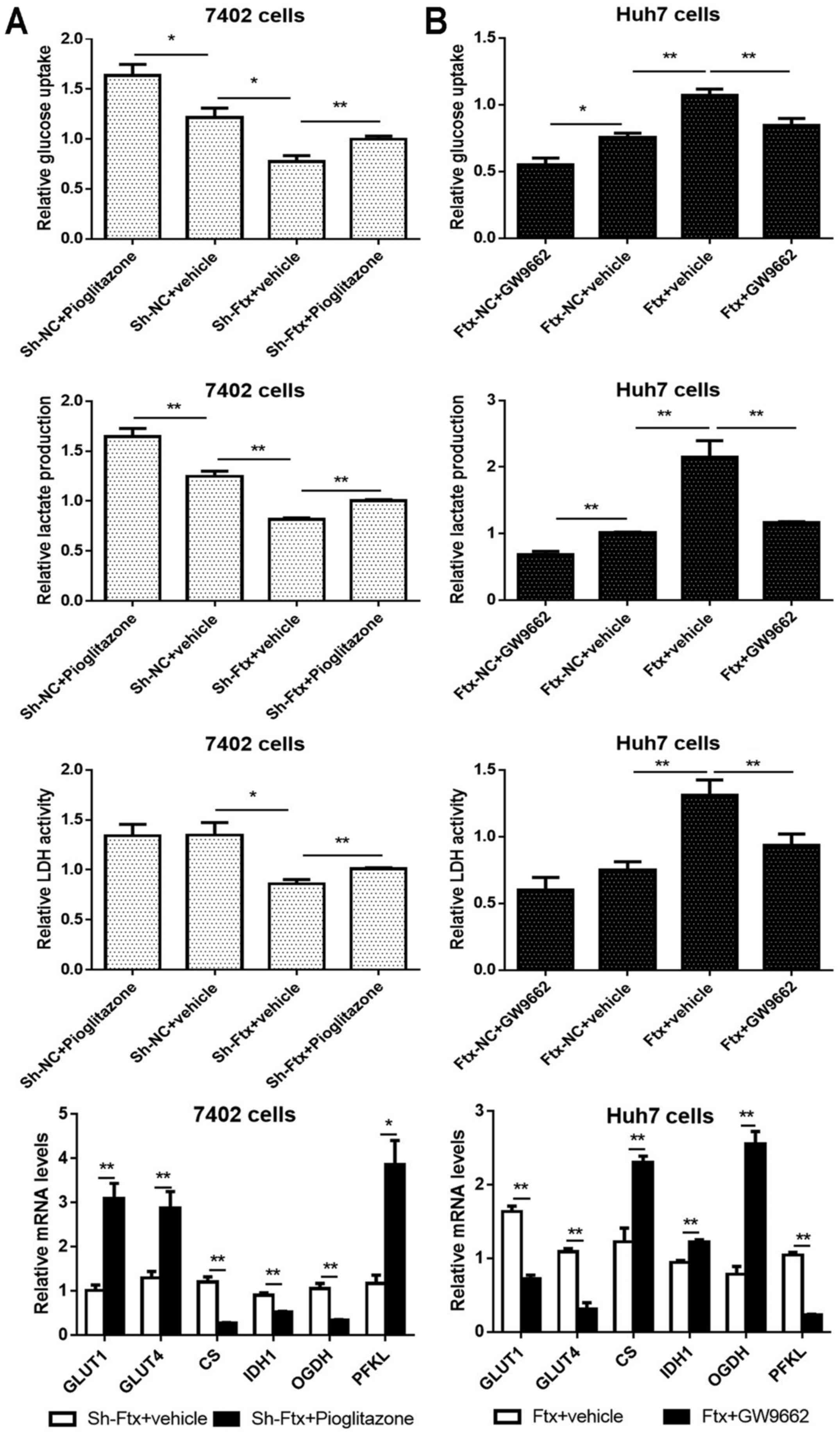 | Figure 8Alterations in PPARγ partially
abolish lncRNA Ftx-mediated HCC aerobic glycolysis in vitro.
(A) PPARγ activation induced effects opposite to those caused by
lncRNA Ftx knockdown in Bel-7402 cells (left panel). (B) PPARγ
inhibition partially abrogated the effects of lncRNA Ftx
overexpression in Huh7 cells (right panel). The experiments were
repeated at least three times and yielded similar results; the
error bars represent the mean ± standard error of the mean.
*P<0.05 and **P<0.01. PPARγ, peroxisome
proliferator-activated receptor γ; sh, short hairpin; NC, negative
control; lncRNA, long non-coding RNA; GLUT, glucose transporter;
CS, citrate synthase; IDH1, isocitrate dehydrogenase 1; OGDH,
α-ketoglutarate; PFKL, phosphofructokinase, liver type. |
lncRNA Ftx promotes the activation of the
PPARγ pathway: TNFα, leptin and PDK1
To further validate the underlying mechanisms, the
protein and mRNA expression levels of PPARγ pathway effectors
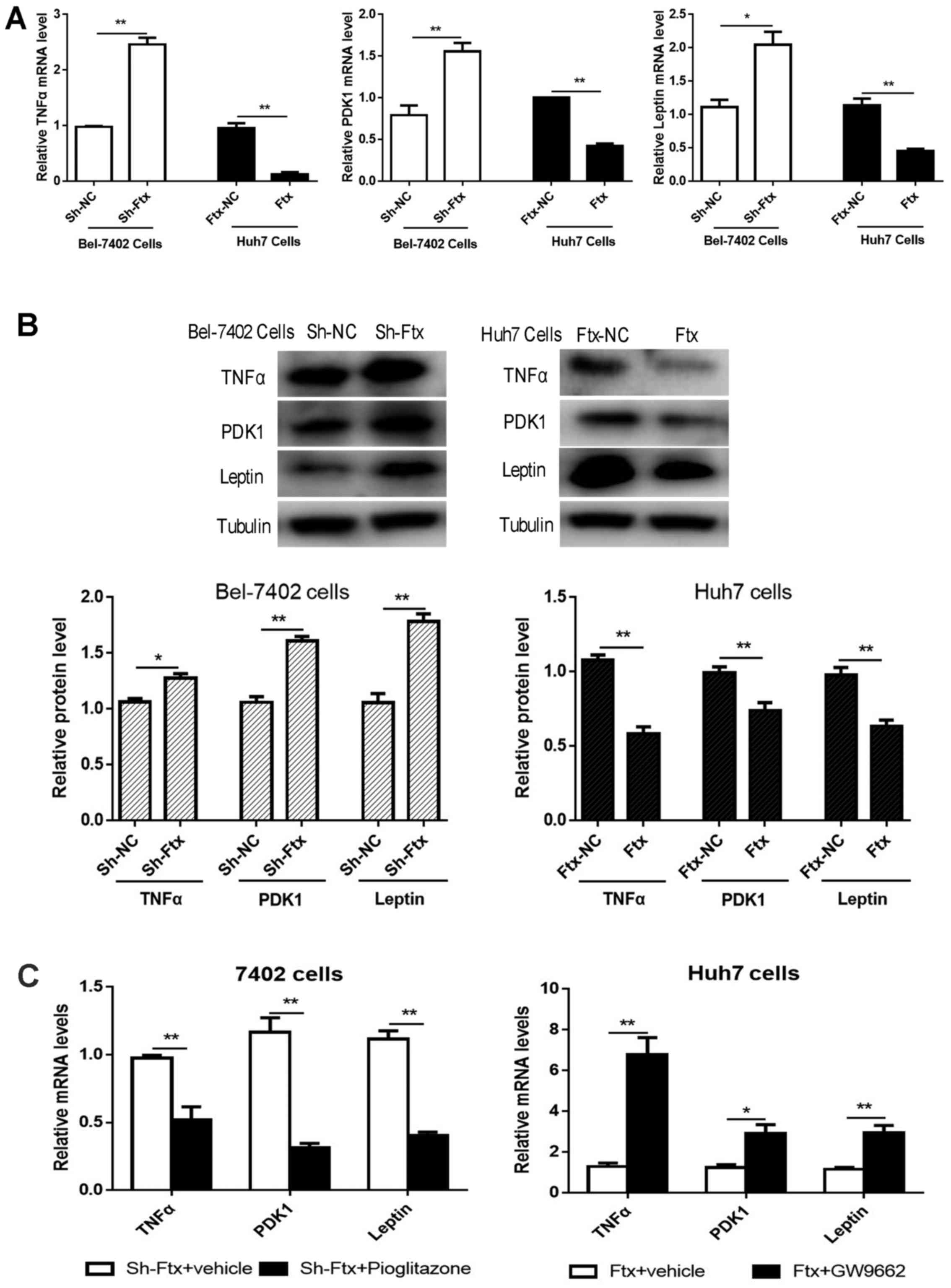
(TNFα, leptin and PDK1) were examined. It was observed that upon
lncRNA Ftx overexpression in Huh7 cells, TNFα, leptin and PDK1
expression levels were decreased significantly, whereas in Bel-7402
cells, the opposite results were observed (Fig. 9A and B). As presented in Fig. 9C, pioglitazone downregulated the
expression of TNFα, leptin and PDK1, while GW9662 upregulated their
expression. Taken together, these data further support the
hypothesis that lncRNA Ftx overexpression promotes the activation
of the PPARγ pathway, thereby attenuating TNFα, leptin and PDK1
expression to maintain increased glycolysis in HCC.
Discussion
lncRNAs have been increasingly recognized to serve
principal functions in multiple biological processes, including the
regulation of gene expression and chromatin conformation (26). Accumulating evidence has confirmed
that a number of lncRNAs are abnormally expressed, and associated
with tumor progression and prognosis in HCC (27). Therefore, the discovery of
tumorigenesis-associated lncRNAs and the corresponding mechanisms
may provide a novel insight into the diagnosis and treatment of
HCC.
Recently, lncRNA Ftx was proposed to be a novel
prognostic predictor and a prospective therapeutic target for HCC
(28). Nevertheless, the molecular
mechanisms through which lncRNA Ftx exerts its role in
tumorigenesis remain largely unclear. In the present study, lncRNA
Ftx was identified as an HCC tumor oncogene. First, lncRNA Ftx was
significantly upregulated in HCC tissues and cell lines and
associated with major clinicopathological features. Second, the
ectopic expression of lncRNA Ftx accelerated proliferation,
invasion, migration and aerobic glycolysis in vitro.
Finally, it was identified that lncRNA Ftx exerted its
tumor-promoting function, at least partially, via PPARγ. Taken
together, these results support the further investigation of lncRNA
Ftx as a therapeutic target for HCC.
The novel discovery of the present study was that
the upregulation of lncRNA Ftx may facilitate HCC tumorigenesis
through the PPARγ pathway, which is pivotal in lipid and
carbohydrate metabolism. Based on the nucleotide BLASTn program,
Ftx is highly homologous with PPARγ, including the
promoter region of PPARγ. Thus, it was hypothesized that
lncRNA Ftx may directly interact with the promoter region or
transcription factors of the PPARγ gene, thus influencing
the transcriptional levels of PPARγ. Alternatively, lncRNA
Ftx may regulate the expression of PPARγ by competitively sponging
miRs; however, this hypothesis requires further investigation. In
investigating the molecular mechanism of the promoting potential of
the lncRNA Ftx/PPARγ axis, it was observed that lncRNA Ftx induced
PPARγ overexpression to reduce the expression of downstream
signaling proteins (TNFα, leptin and PDK1) in Huh7 cells; these
proteins have been demonstrated to be downregulated by PPARγ
(18-20). Furthermore, the PPARγ agonist and
antagonist exerted similar effects on TNFα, leptin and PDK1.
Therefore, the newly identified lncRNA Ftx/PPARγ axis provides an
innovative approach to HCC tumorigenesis and indicates a potential
target for HCC therapeutics.
The significance of the present study was also
highlighted by the novel role of the lncRNA Ftx/PPARγ axis in
promoting the glycolytic phenotype, which frequently correlates
with HCC pathogenesis and worse clinical outcomes (29). Thus, future studies may focus on
uncovering the essential role and underlying mechanism of aerobic
glycolysis in HCC progression; this mechanism may be used for
possible therapeutic interventions. The present data indicated that
lncRNA Ftx facilitated glucose consumption, glucose transporter
(GLUT1 and GLUT4) expression, lactate production, and glycolytic
enzyme (LDH and PFKL) activity and expression. Conversely, lncRNA
Ftx suppressed Krebs cycle-associated enzyme (CS, IDH1 and OGDH)
activity and expression. Given that the PPARγ antagonist GW9662
partially rescued lncRNA Ftx overexpression-induced glycolysis
increases, and the PPARγ agonist pioglitazone partially abolished
lncRNA Ftx knockdown-mediated glycolysis decreases, it was
hypothesized that there may be alternative targets of lncRNA Ftx
that contribute to enhancing glycolysis.
Consistently, numerous lncRNAs have previously been
reported to be associated with malignant carbohydrate metabolism
(30,31). Therefore, targeting key metabolic
enzymes is an important method for treating cancer (32). In the present study, it was
observed that lncRNA Ftx, by activating the PPARγ pathway,
regulated key glycolytic genes (including GLUT4, PDK1
and PFKL) and promoted glucose metabolism in HCC cells,
indicating that lncRNA Ftx may serve as a novel
metabolism-targeting therapeutic agent against HCC. In addition,
recent studies have indicated that lncRNA Ftx inhibits
cardiomyocyte apoptosis (33) and
promotes glioma (34), colorectal
cancer (35) and renal cancer
(36) growth, suggesting that
lncRNA Ftx is a general target for antitumor therapy.
Taken together, the present results provide the
first evidence, to the best of our knowledge, that lncRNA Ftx
promotes aerobic glycolysis and HCC cell progression by activating
the PPARγ pathway. However, certain questions remain unclear and
require further study. First, PPARγ may be proposed as the
downstream effector of lncRNA Ftx, although direct binding was not
proven in the present study and requires investigation. In
addition, the alternative targets of lncRNA Ftx upon enhancing
glycolysis merit investigation, in addition to in vivo
experiments.
In conclusion, the present study recognized lncRNA
Ftx to be a novel promoter of HCC progression and glycolysis by
targeting the PPARγ pathway. Therefore, lncRNA Ftx has the
potential to be a promising diagnostic biomarker, a novel
prognostic predictor and a therapeutic target for HCC. Targeting
this aberrantly activated pathway may provide a novel approach for
HCC therapy and merits further investigation.
Acknowledgments
Not applicable.
Funding
The present study was funded by grants from the
National Natural Science Foundation of China (grant nos. 81472685
and 81600469), the Major Special Plan of Science and Technology of
Shandong Province (grant no. 2015ZDXX0802A01), the Natural Science
Foundation of Shandong Province (grant nos. ZR2016HB06 and
ZR2017MH035), and the Science and Technology Development Projects
of Shandong Province (grant no. 2016GSF201126).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
All authors involved helped to perform the research.
CQ and QZ designed the experiment and revised the manuscript. XL
and QZ performed the experiments. JQ and WW performed statistical
data analysis. XL, DZ and ZL wrote the manuscript. All authors read
and approved the content of the manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from each
patient recruited for the present study for the use of materials.
The consent procedures and all experimental protocols were approved
by the Medical Institutional Ethics Committee of Shandong
Provincial Hospital Affiliated to Shandong University (Jinan,
China; approval no. 2017-231), according to the Declaration of
Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar
|
|
3
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar
|
|
4
|
Phan LM, Yeung SC and Lee MH: Cancer
metabolic reprogramming: Importance, main features, and potentials
for precise targeted anti-cancer therapies. Cancer Biol Med.
11:1–19. 2014.
|
|
5
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar
|
|
6
|
Lincet H and Icard P: How do glycolytic
enzymes favour cancer cell proliferation by nonmetabolic functions.
Oncogene. 34:3751–3759. 2015. View Article : Google Scholar
|
|
7
|
Han T, Kang D, Ji D, Wang X, Zhan W, Fu M,
Xin HB and Wang JB: How does cancer cell metabolism affect tumor
migration and invasion. Cell Adhes Migr. 7:395–403. 2013.
View Article : Google Scholar
|
|
8
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar
|
|
9
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar
|
|
10
|
Taylor DH, Chu ET, Spektor R and Soloway
PD: Long non-coding RNA regulation of reproduction and development.
Mol Reprod Dev. 82:932–956. 2015. View Article : Google Scholar
|
|
11
|
Zhu X, Wu YB, Zhou J and Kang DM:
Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via
increasing FoxO1 expression. Biochem Biophys Res Commun.
469:319–325. 2016. View Article : Google Scholar
|
|
12
|
Qi M, Zhou Q, Zeng W, Shen M, Liu X, Luo
C, Long J, Chen W, Zhang J and Yan S: Analysis of long non-coding
RNA expression of lymphatic endothelial cells in response to type 2
diabetes. Cell Physiol Biochem. 41:466–474. 2017. View Article : Google Scholar
|
|
13
|
Yang Y, Chen L, Gu J, Zhang H, Yuan J,
Lian Q, Lv G, Wang S, Wu Y, Yang YT, et al: Recurrently deregulated
lncRNAs in hepatocellular carcinoma. Nat Commun. 8:144212017.
View Article : Google Scholar
|
|
14
|
Chureau C, Chantalat S, Romito A, Galvani
A, Duret L, Avner P and Rougeulle C: Ftx is a non-coding RNA which
affects Xist expression and chromatin structure within the
X-inactivation center region. Hum Mol Genet. 20:705–718. 2011.
View Article : Google Scholar
|
|
15
|
Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y,
Jia Y, Li Q, Zhang H, Tu K, et al: Ftx non coding RNA-derived
miR-545 promotes cell proliferation by targeting RIG-I in
hepatocellular carcinoma. Oncotarget. 7:25350–25365. 2016.
|
|
16
|
Patitucci C, Couchy G, Bagattin A, Cañeque
T, de Reyniès A, Scoazec JY, Rodriguez R, Pontoglio M, Zucman-Rossi
J, Pende M, et al: Hepatocyte nuclear factor 1α suppresses
steatosis-associated liver cancer by inhibiting PPARγ
transcription. J Clin Invest. 127:1873–1888. 2017. View Article : Google Scholar
|
|
17
|
Janani C and Ranjitha Kumari BD: PPAR
gamma gene - a review. Diabetes Metab Syndr. 9:46–50. 2015.
View Article : Google Scholar
|
|
18
|
Wu Z, Xie Y, Morrison RF, Bucher N and
Farmer SR: PPARgamma induces the insulin-dependent glucose
transporter GLUT4 in the absence of C/EBPalpha during the
conversion of 3T3 fibroblasts into adipocytes. J Clin Invest.
101:22–32. 1998. View
Article : Google Scholar
|
|
19
|
Lecarpentier Y, Claes V, Vallée A and
Hébert JL: Thermodynamics in cancers: Opposing interactions between
PPAR gamma and the canonical WNT/beta-catenin pathway. Clin Transl
Med. 6:142017. View Article : Google Scholar
|
|
20
|
Rangwala SM and Lazar MA: Peroxisome
proliferator-activated receptor gamma in diabetes and metabolism.
Trends Pharmacol Sci. 25:331–336. 2004. View Article : Google Scholar
|
|
21
|
Chen R, Zhu D, Ye X, Shen D and Lu R:
Establishment of three human liver carcinoma cell lines and some of
their biological characteristics in vitro. Sci Sin. 23:236–247.
1980.
|
|
22
|
Nakabayashi H, Taketa K, Miyano K, Yamane
T and Sato J: Growth of human hepatoma cells lines with
differentiated functions in chemically defined medium. Cancer Res.
42:3858–3863. 1982.
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Ferreira LM: Cancer metabolism: The
Warburg effect today. Exp Mol Pathol. 89:372–380. 2010. View Article : Google Scholar
|
|
25
|
Adekola K, Rosen ST and Shanmugam M:
Glucose transporters in cancer metabolism. Curr Opin Oncol.
24:650–654. 2012. View Article : Google Scholar
|
|
26
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar
|
|
27
|
Klingenberg M, Matsuda A, Diederichs S and
Patel T: Non-coding RNA in hepatocellular carcinoma: Mechanisms,
biomarkers and therapeutic targets. J Hepatol. 67:603–618. 2017.
View Article : Google Scholar
|
|
28
|
Zhao Q, Li T, Qi J, Liu J and Qin C: The
miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in
HBV-related hepatocellular carcinoma and promotes tumorigenesis and
tumor progression. PLoS One. 9:e1097822014. View Article : Google Scholar
|
|
29
|
Iansante V, Choy PM, Fung SW, Liu Y, Chai
JG, Dyson J, Del Rio A, D'Santos C, Williams R, Chokshi S, et al:
ARP14 promotes the Warburg effect in hepatocellular carcinoma by
inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat
Commun. 6:78822015. View Article : Google Scholar
|
|
30
|
Zhang P, Cao L, Fan P, Mei Y and Wu M:
LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses
glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep.
17:1204–1220. 2016. View Article : Google Scholar
|
|
31
|
Li Z, Li X, Wu S, Xue M and Chen W: Long
non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase
2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci.
105:951–955. 2014. View Article : Google Scholar
|
|
32
|
Hamanaka RB and Chandel NS: Targeting
glucose metabolism for cancer therapy. J Exp Med. 209:211–215.
2012. View Article : Google Scholar
|
|
33
|
Long B, Li N, Xu XX, Li XX, Xu XJ, Guo D,
Zhang D, Wu ZH and Zhang SY: Long noncoding RNA FTX regulates
cardiomyocyte apoptosis by targeting miR-29b-1-5p and Bcl2l2.
Biochem Biophys Res Commun. 495:312–318. 2018. View Article : Google Scholar
|
|
34
|
Zhang W, Bi Y, Li J, Peng F, Li H, Li C,
Wang L, Ren F, Xie C, Wang P, et al: Long noncoding RNA FTX is
upregulated in gliomas and promotes proliferation and invasion of
glioma cells by negatively regulating miR-342-3p. Lab Invest.
97:447–457. 2017. View Article : Google Scholar
|
|
35
|
Guo XB, Hua Z, Li C, Peng LP, Wang JS,
Wang B and Zhi QM: Biological significance of long non-coding RNA
FTX expression in human colorectal cancer. Int J Clin Exp Med.
8:15591–15600. 2015.
|
|
36
|
He X, Sun F, Guo F, Wang K, Gao Y, Feng Y,
Song B, Li W and Li Y: Knockdown of long noncoding RNA FTX inhibits
proliferation, migration, and invasion in renal cell carcinoma
cells. Oncol Res. 25:157–166. 2017. View Article : Google Scholar
|