Introduction
Hepatocellular carcinoma (HCC) is one of the most
aggressive malignant tumors with respect to the disease course and
mortality rate (1) Although
numerous therapeutic strategies have been introduced, the prognosis
of patients with HCC remains poor due to the frequent metastasis
and recurrence (2,3). Therefore, it is crucial to identify
specific molecular targets and provide the basis for early
diagnosis of HCC and targeted therapy, in order to achieve optimal
outcomes and improve the quality of life and life expectancy of HCC
patients.
MicroRNAs (miRNAs or miRs) are small, non-coding,
single-stranded RNAs that regulate key cell processes, such as
proliferation, differentiation and apoptosis (4–9), and
exert major functional effects on various disorders, including
numerous, if not all, cancers (10–13).
Abnormalities in miRNA expression profiles may be caused by
different mechanisms, such as amplifications, deletions, or
sequence changes in the miRNA loci, epigenetic silencing and the
dysregulation of transcription factors targeting specific miRNAs
(14). Over the past decades,
miRNAs have provided a powerful tool for health care intervention
in a variety of tumor types (10–13).
Increasing interest in the development of miRNAs as novel
therapeutic biomarkers stems from the ability to interfere with
miRNA levels through selective miRNA mimics or anti-miRs (15).
Recently, a number of miRNAs have been implicated as
modulators of cell necroptosis (16,17),
a newly discovered pathway of regulated cell necrosis governed by
receptor-interacting protein (RIP)1, RIP3 and mixed lineage kinase
domain-like (MLKL) protein, which constitutes a critical
cell-killing mechanism in response to severe stress, chemotherapy
or inflammatory factors (18).
Previous studies have demonstrated that necroptosis is associated
with cancer initiation and progression, playing a dual and opposing
role. Accordingly, while enhanced necroptosis concurs with cancer
cell death, extensive cell death also increases the risk of
proliferation and the metastasis of surviving cells by promoting
the formation of reactive oxygen species, triggering the
inflammatory process and inhibiting the immune response (19). In this regard, emphasis has been
placed on the crucial role of certain miRNAs in the transcriptional
regulation of the nucleotide-binding oligomerization domain-like
receptor family, pyrin domain-containing−3 (NRLP3) inflammasome, an
intracellular multiprotein complex that contributes to necroptosis
upon the production of pro-inflammatory cytokines (20), with activity involved in the
pathogenesis and progression of several cancers, including HCC
(21,22). In this context, recent
investigations revealed that the caspase-8 gene (Casp-8) is
a key controller of NLRP3 activation (23–26);
however, the mechanisms underlying this process have not yet been
fully elucidated.
The aim of the present study was to identify
potential interactions between a panel of miRNAs predicted by
computational scoring as necroptotic pathway regulators and
Casp-8 in HCC specimens and cell lines. We identified three
miRNAs, namely miR-371-5p, miR-373 and miR-543, which were
upregulated in HCC tissues compared with adjacent non-cancerous
tissues. These miRNAs have the ability to bind to their target
Casp-8 mRNA transcript in order to facilitate the
necroptosis of HCC cells. Additionally, the data presented herein
confirm the well-known association between miR-223 and NLRP3, as
already reported in different disease contexts (27–31).
The findings of this study identified a key piece in
the complex molecular puzzle of HCC, thereby providing new insight
for a better understanding of the pathogenic mechanisms of this
type of tumor. The assessment of the necroptotic pathway regulation
by miRNAs depicts the starting point for a novel miRNA-based
diagnostic, prognostic and therapeutic approach in HCC.
Materials and methods
Patient selection
This a retrospective study of tumor tissues from 38
patients with HCC treated by surgical resection at the University
Hospital of Messina between 2011 and 2015. This study was approved
by the local Institutional Review Board (IRB) of the University
Hospital of Messina, Messina, Italy. The study protocol conforms to
the ethical guidelines of the 1975 Declaration of Helsinki. Written
informed consent was obtained from all subjects at the moment of
the surgical procedures. For research purposes, no consent for
retrospective de-identified data analysis was required as deemed by
the IRB.
No patients received neoadjuvant therapy
(radiotherapy and/or chemotherapy). The diagnosis of HCC was based
on explant evaluation according to the revised WHO classification
tumors (32). The clinical
characteristics of the patients are presented in Table I.
 | Table IClinical characteristics of the
patients with HCC. |
Table I
Clinical characteristics of the
patients with HCC.
| Sex
(male/female) | 27/11 |
| Mean age (range,
years) | 68±11 |
|
HBsAg/Anti-HCV/NBNC | 8/13/17 |
| Child-Pugh grading
(A/B/C) | 21/17/0 |
| Stage
(I/II/IIIA/IIIB/IIIC/IV) | |
| 0/26/12/0/0/0 | |
| Total bilirubin
(mg/dl) | 44±10 |
| Albumin (g/dl) | 3.5± 0.4 |
| PT (INR) | 4±0.8 |
| Ascites (−/+) | 20/18 |
Tissue samples
HCC tissues and their normal liver counterparts were
frozen immediately after surgery and stored at −80°C for
biomolecular analyses.
Cells and cell culture
The Hep3B-hCG cells were purchased from
Sigma-Aldrich Italy (Milan, Italy) and maintained in Dulbecco's
modified Eagle's medium (DMEM) with high glucose (HyClone, Logan,
UT, USA) supplemented with 10% FBS. The hNHeps™ (human normal
hepatocytes) cells were purchased from Lonza (Lonza, Walkersville,
MD, USA) and maintained in hepatocyte culture medium (Lonza).
RNA extraction, cDNA synthesis and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was extracted from the HCC tissues and
cell cultures using TRIzol reagent (Invitrogen/Thermo Fisher
Scientific, Inc., Waltham, MA USA), according to the manufacturer's
instructions. Total RNA concentration and integrity were examined
using an Agilent Bioanalyzer (Agilent Technologies, Inc., Santa
Clara, CA, USA). Subsequently, 300 ng of total mRNA per sample was
reverse transcribed into cDNA using the High Capacity cDNA Reverse
Transcription kit (Applied Biosystems/Thermo Fisher Scientific,
Inc.).
RT-qPCR for Casp-8 and NLRP3 expression was
performed using a standard TaqMan PCR kit procedure on an AB-7300
RT-PCR system (Applied Biosystems/Thermo Fisher Scientific, Inc.)
The reaction was performed with one cycle of 95°C for 5 min
followed by 40 cycles at 95°C for 15 sec, 52°C for 20 sec and 72°C
for 30 sec. The mRNA levels of Casp-8 and NLRP3 were
normalized to endogenous β-actin (Applied Biosystems).
Primers and probes were as previously reported (33,34).
The control was represented by hNHeps™
cells (described above) whose expression was considered as equal to
1
Relative fold expression and changes were calculated
using the 2−∆∆Cq method (35).
miRNA isolation, cDNA synthesis and qPCR
analysis
miRNAs were extracted from the HCC tissues and cells
using the miRVana Isolation kit (Ambion/Thermo Fisher Scientific,
Inc.), following the manufacturer's instructions. The enriched
miRNA fraction was converted in cDNA using the TaqMan MicroRNA
Reverse Transcriptase kit (Applied Biosystems/Thermo Fisher
Scientific, Inc.). hsa-miR-137, hsa-miR-223, hsa-miR-324,
hsa-miR-371-5p, hsa-miR-372, hsa-miR-373, hsa-miR-455, hsa-miR-543,
hsa-miR-595, and hsa-miR-661 were predicted by computational
scoring as necroptotic pathway regulators (36,37).
For miRNA quantification, 2 µl of cDNA was
used for each specific miRNA TaqMan Small RNA assay (Applied
Biosystems/Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. All reverse transcription reactions,
including no-template controls and reverse transcription controls,
were performed in triplicate. RNU6 small nuclear RNA was used to
normalize miRNA expression levels owing to its claimed expression
stability and its wide use as loading control in several published
miRNA expression studies (38,39).
Relative fold expression and changes were calculated using the
2−ΔΔCq method (35).
Target prediction tools
miRNA/Casp-8 mRNA interactions were identified by
means of bioinformatics algorithms that predict miRNA target sites
(40). Specifically, three online
databases, microRNA.org (www.microrna.org), miRDB (http://mirdb.org/miRDB/), and TargetScan (www.targetscan.org), were used.
Oligonucleotide transfection
hsa-miR-371-5p, hsa-miR-373 and hsa-miR-543
mimics/inhibitors (Qiagen, Milan, Italy) were transfected into the
Hep3B-hCG cells using HiPerFect according to the
manufacturer's instructions (Qiagen). The cells were transfected
twice with 100 pmol of oligonucleotide per well (0.5×106
cells) at 24-h intervals. The transfected cells were assayed 48 h
after the second transfection.
Plasmid constructs and transient
transfection
The plasmid containing the full-length Casp-8
cDNA was obtained as previously described (33). The 3′-UTR of Casp-8 and the
mutation sequences were amplified by PCR using the primers with a
BglII restriction site on each 5′ or 3′ strand. The PCR
products were inserted into the BglII sites of the
pGL3-control vector (Promega, Madison, WI, USA) and identified by
DNA sequencing.
The wild-type plasmids were created containing the
3′UTR of Casp-8 with the complementary sequence of miR-371-5p
(pGL3-Casp-8-3′UTR wild 1), miR-373 (pGL3-Casp-8-3′UTR wild 2) and
miR-543 (pGL3-Casp-8-3′UTR wild 3). Three mutant plasmids were
generated without the complementary sequence of miR-371-5p
(pGL3-Casp-8-3′UTR mut 1), miR-373 (pGL3-Casp-8-3′UTR mut 2) and
miR-543 (pGL3-Casp-8-3′UTR mut 3).
For the luciferase reporter assay, the cells were
seeded on 24-well plates and co-transfected using Lipofectamine
2000 (Invitrogen/Thermo Fisher Scientific, Inc.) with 100 ng per
well of the resulting luciferase UTR-report vector, 2 ng per well
of pRLCMV vector (internal control, Promega), and 20 ng per well of
miR-371-5p, miR-373 and miR-543 mimics or inhibitors following the
manufacturer's instructions (Qiagen). After 24 h, the cells were
lysed, and the relative luciferase activity was assessed with the
Dual-Luciferase Assay Reporter System (Promega).
Necroptosis assay
The Hep3B-hCG cells (2×103
cells/well) were pre-treated with the pan-caspase inhibitor, Z-VAD
(10 µM), for 1 h in order to induce RIPK3-dependent
necroptosis, in the presence or absence of miRNA mimics and
inhibitors. Tumor necrosis factor (TNF)-α (50 ng/ml) was then added
and the cells were incubated for an additional 72 h. Cell viability
was assessed by MTT assay. In brief, 20 µl of 5 mg/ml stock
solution of 3-(4,5-dimetylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (Sigma-Aldrich) was added to each well, and the cells were
incubated for 1 h at 37°C in a humidified atmosphere of 5%
CO2. The supernatants were carefully removed, and 100
µl of DMSO (vehicle) was added to each well. The plates were
subsequently shaken for 20 min at room temperature, and the
absorbance of the solution was measured at 570 nm. The absorbance
of the untreated cells was set as 100% and cell survival was
expressed as a percentage of this value.
Statistical analysis
Statistical analysis was performed using INSTAT,
version 3.0 and PRISM version 4.0 software (GraphPad, San Diego,
CA, USA). One-way analysis of variance (ANOVA), followed by a
pair-wise multiple comparison test (Bonferroni correction), was
performed to identify the differences among the groups. The
association between Casp-8 mRNA expression levels and
clinicopathological characteristics was evaluated using Pearson's
Chi-square test. Pearson's correlation analysis was used to
estimate the correlation between the Casp-8 mRNA levels and
the miR-371-5p, miR-373 and miR-543 expression profiles, as well as
between NLRP3 abundance and miR-223 expression. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of Casp-8 in HCC samples
Casp-8 is a cysteine protease that is a key
regulator of cell apoptosis; however, its functional significance
as a negative regulator of programmed cell necrosis has attracted
growing attention over the years (41–43).
Since different programmed cell death pathways, including
necroptosis, have been recently implicated in hepatocarcinogenesis
(44), in this study, we addressed
the question of whether Casp-8 plays a role in the
programmed necrotic death of HCC cells. The values for
Casp-8 mRNA levels were first determined in the normal liver
(NL) samples in order to determine the cut-off point for
Casp-8 expression levels in the HCC samples. Casp-8
expression levels (as defined by the ratio between the values
measured in NL samples over those of hNHeps™ cells) were between
0.95 and 1.6 (mean, 1.29±0.16). Values ≤0.81 (determined as the
mean minus 3 s.d.) were considered to represent the underexpression
of Casp-8. As shown in Fig.
1A, a marked decrease in the Casp-8 mRNA level was found
in 34/38 (89.5%) HCC samples. Box plots showing the individual
means and scores of Casp-8 mRNA expression in the NL and HCC
specimens are presented in Fig.
2B. Likewise, a marked decrease in Casp-8 expression
levels was observed in the HCC cell line, Hep3B-hCG,
compared with the normal hepatocyte cell line, hNHeps™ (Fig. 1C). In addition, in order to gain
insight into the role of Casp-8 in HCC, we analyzed the
association between Casp-8 expression and various patient
characteristics, including gender, age, Child-Pugh grade and stage
(Table II). No significant
association was observed between Casp-8 expression and the
clinical variables considered (P>0.05).
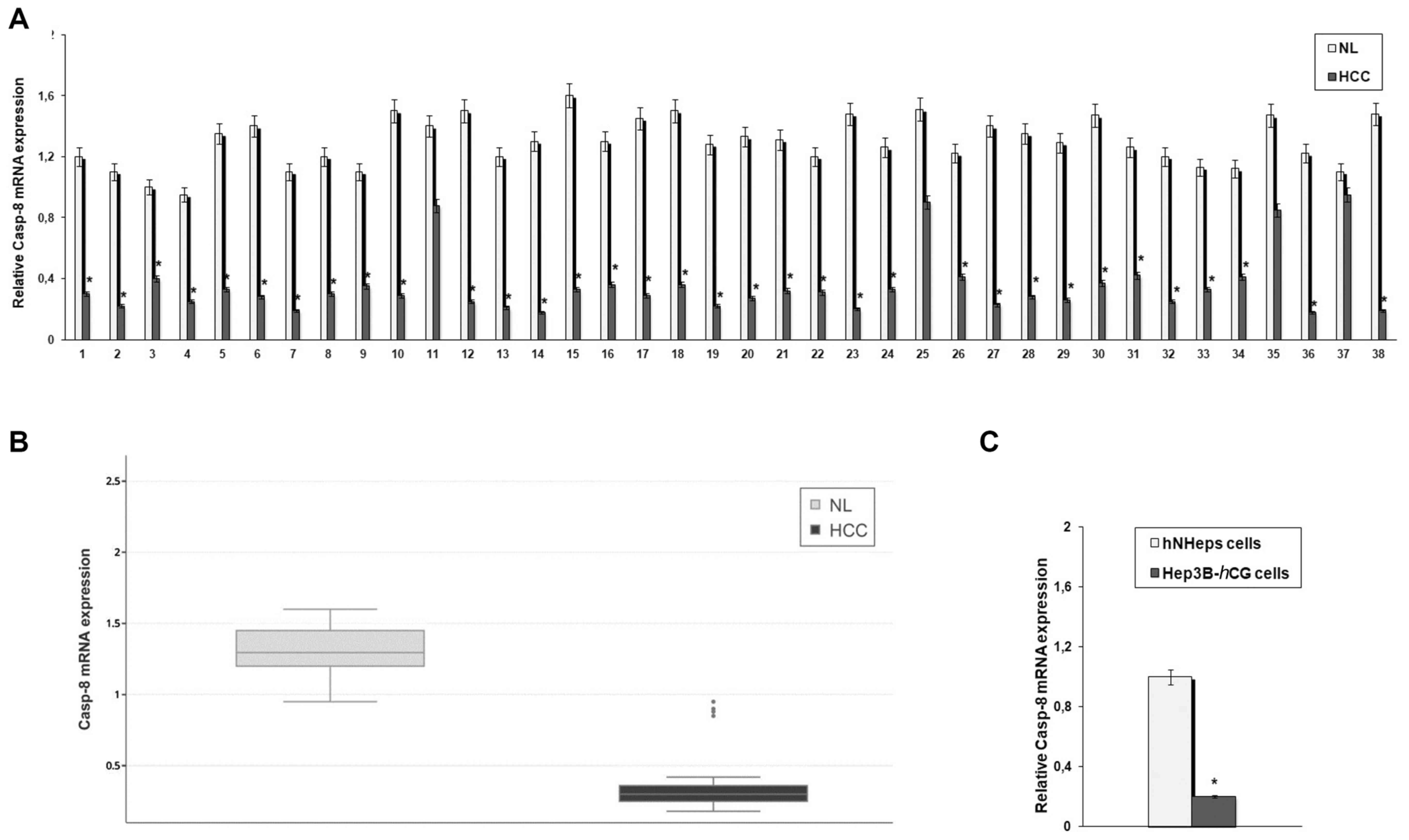 | Figure 1Expression levels of caspase-8
(Casp-8) in hepatocellular carcinoma (HCC) tissues and
cells. (A) Total RNA, extracted from 38 HCC and paired normal liver
(NL) tissues, was reverse-transcribed and analyzed by quantitative
polymerase chain reaction (qPCR). The mRNA levels of Casp-8
were normalized to the housekeeping gene, β-actin, as an internal
control. Data are presented as the means ± standard deviation of 3
independent experiments. *P<0.001, compared to NL
specimens. (B) Box plots relative to Casp-8 mRNA expression
in HCC and NL tissues. (C) Total RNA, extracted from the human HCC
cell line, Hep3B-hCG, and human normal hepatocytes
(hNHeps™), was reverse transcribed and analyzed by qPCR. The mRNA
levels of Casp-8 were normalized by using the housekeeping
gene β-actin as internal control. Data are presented as the means ±
standard deviation of 3 independent experiments.
*P<0.001, compared to the hNHeps™ cells. |
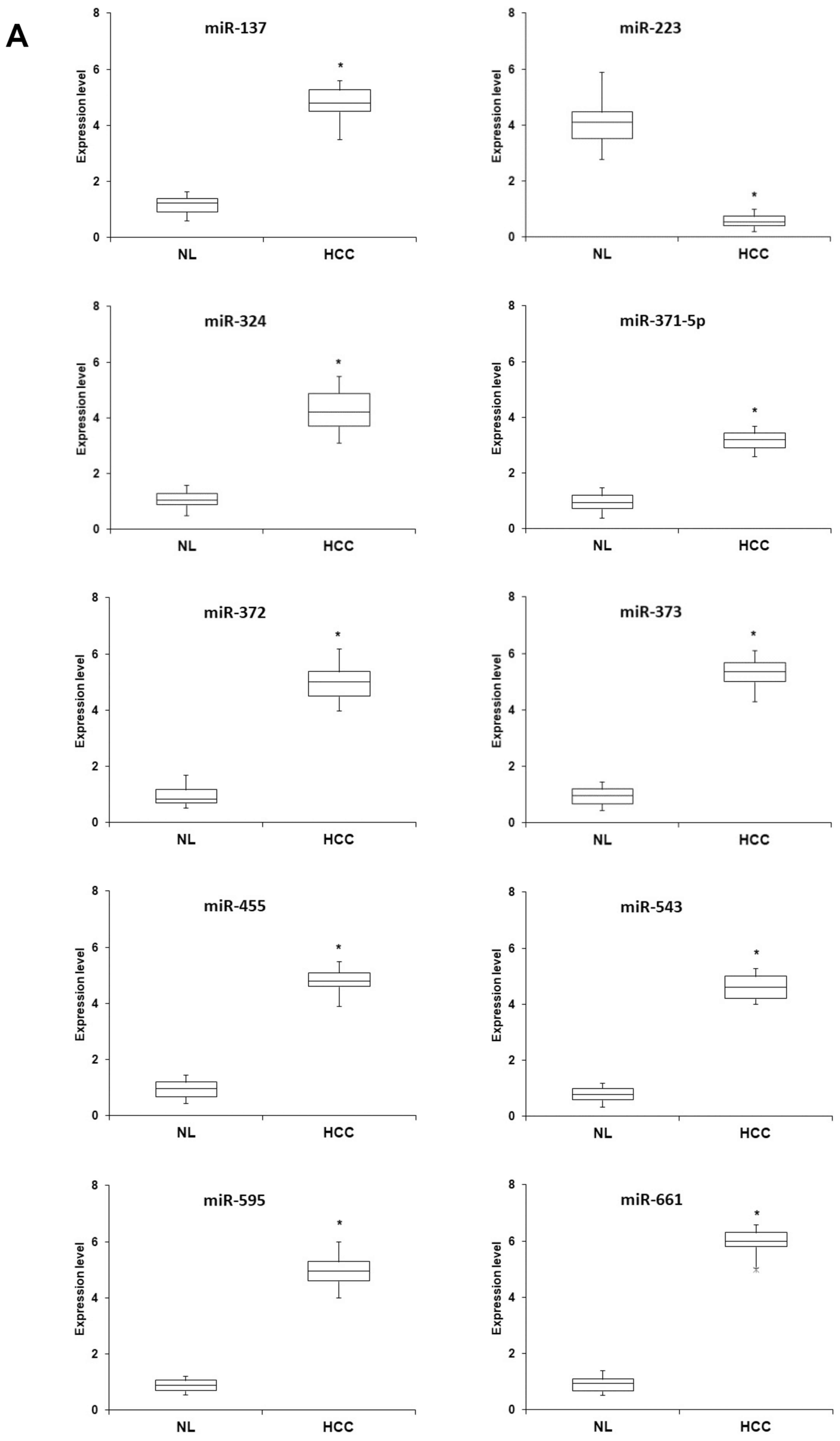 | Figure 2Expression profiles of a panel of
miRNAs implicated in programmed cell necrosis in hepatocellular
carcinoma (HCC) tissues and cells. (A) Total RNA was extracted from
38 HCC and the paired normal liver (NL) tissues, reverse
transcribed and analyzed by quantitative polymerase chain reaction
(qPCR) to determine the amounts of miR-137, miR-223, miR-324,
miR-371-5p, miR-372, miR-373, miR-455, miR-543, miR-595 and
miR-661. Box plots showing means and scores of miRNA expression in
NL and HCC specimens are shown. Data are representative of three
independent experiments. (B) Total RNA, extracted from the human
HCC cell line Hep3B-hCG and human normal hepatocytes
(hNHeps™), was reverse transcribed and analyzed by qPCR to
determine the relative amounts of miR-137, miR-223, miR-324,
miR-371-5p, miR-372, miR-373, miR-455, miR-543, miR-595 and
miR-661. Data are presented as the means ± standard deviation of 3
independent experiments. *P<0.001, compared to the
hNHeps™ cells. |
 | Table IIAssociation between Casp-8
expression and clinicopathological characteristics. |
Table II
Association between Casp-8
expression and clinicopathological characteristics.
| Variables | Casp-8 mRNA
expression
| P-value |
|---|
| Low (n-34) | High (n=4) |
|---|
| Sex | | | 0.854 |
| Male | 24 | 3 | |
| Female | 10 | 1 | |
| Age (years) | | | 0.911 |
| ≤60 | 16 | 2 | |
| ≥60 | 18 | 2 | |
| Child-Pugh
grading | | | 0.401 |
| A | 18 | 3 | |
| B | 16 | 1 | |
| Stage | | | 0.765 |
| II | 23 | 3 | |
| IIIA | 11 | 1 | |
Prediction of miRNAs regulating
necroptotic cell death that target Casp-8
To identify the potential upstream executioners of
Casp-8 signaling in necrotic cascades, we first explored the
expression profiles of a panel of miRNAs predicted by computational
scoring as necroptotic pathway regulators (36,37).
Significantly higher levels of hsa-miR-137, hsa-miR-324,
hsa-miR-371-5p, hsa-miR-372, hsa-miR-373, hsa-miR-455, hsa-miR-543,
hsa-miR-595 and hsa-miR-661, and lower levels of miR-223, were
observed in the HCC tissues compared with the NL specimens
(Fig. 2A). A similar expression
pattern of this miRNA panel was noted in the HCC cell line,
Hep3B-hCG, compared with the normal hepatocyte cell line,
hNHeps™ (Fig. 2B). Subsequently,
among these miRNAs, we searched for those that may have seed
regions able to bind to the 3′UTR of Casp-8 by using
predictive bioinformatics programs (microRNA.org,
miRDB and TargetScan). We identified miR-371-5p, miR-373 and
miR-543 as potential putative Casp-8-targeting miRNAs. Of
note, Pearson's correlation analysis revealed a strong negative
correlation between the miR-371-5p, miR-373 and miR-543 expression
profiles and Casp-8 mRNA levels in the paired HCC tumor
tissues (r=−0.689 for miR-371-5p; r=−0.611 for miR-373; and
r=−0.606 for miR-543).
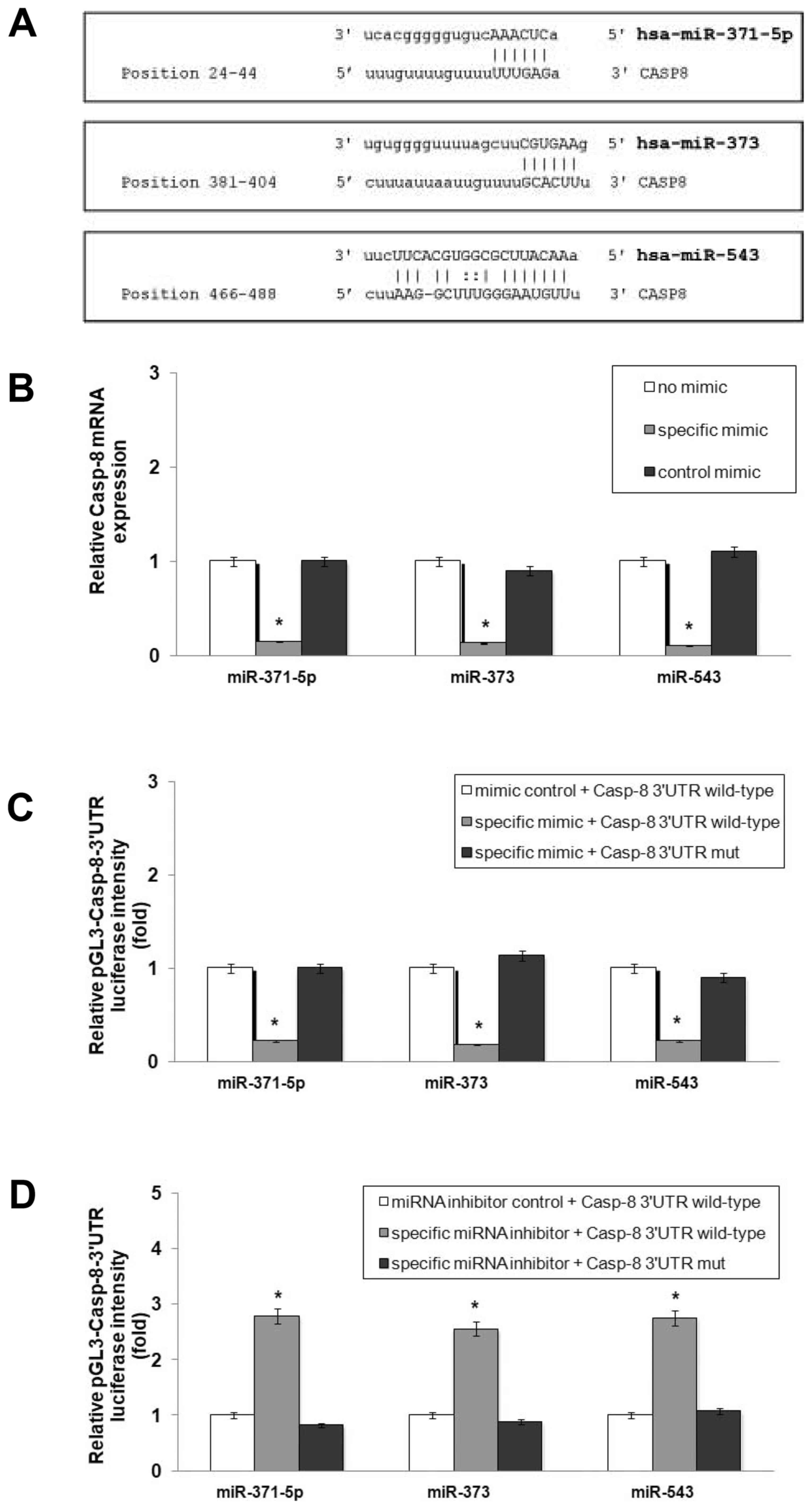
miR-371-5p, miR-373 and miR-543
participate in the regulation of Casp-8 expression
To validate the direct interaction of miR-371-5p,
miR-373 and miR-543 with Casp-8 mRNA (Fig. 3A), wild-type plasmids containing
the 3′UTR of Casp-8 with complementary sequence of
miR-371-5p (pGL3-Casp-8-3′UTR wild 1), miR-373 (pGL3-Casp-8-3′UTR
wild 2) and miR-543 (pGL3-Casp-8-3′UTR wild 3) and the mutant
plasmids without complementary sequence of miR-371-5p
(pGL3-Casp-8-3′UTR mut 1), miR-373 (pGL3-Casp-8-3′UTR mut 2) and
miR-543 (pGL3-Casp-8-3′UTR mut 3) were co-transfected into the
Hep3B-hCG cells together with specific mimics or inhibitors.
We found that the enforced expression of miR-371-5p, miR-373 and
miR-543 induced a marked decrease in Casp-8 expression
(Fig. 3B), accompanied by a
significant decrease in the luciferase activity of
pGL3-Casp-8-3′UTR wild-type plasmids (Fig. 3C; P<0.001), whereas the
transcription of the pGL3-Casp-8-3′UTR mut vectors was not affected
(Fig. 3C). Upon blocking the
expression of the three miRNAs with specific inhibitors, luciferase
intensity markedly increased in cells transfected with the
pGL3-Casp-8-3′UTR wild-type vectors (P<0.001). No change was
observed in the cells transfected with the pGL3-Casp-8-3′UTR mut
plasmids (Fig. 3D). These results
provide evidence of Casp-8 as a target gene of miR-371-5p,
miR-373 and miR-543 in our setting.
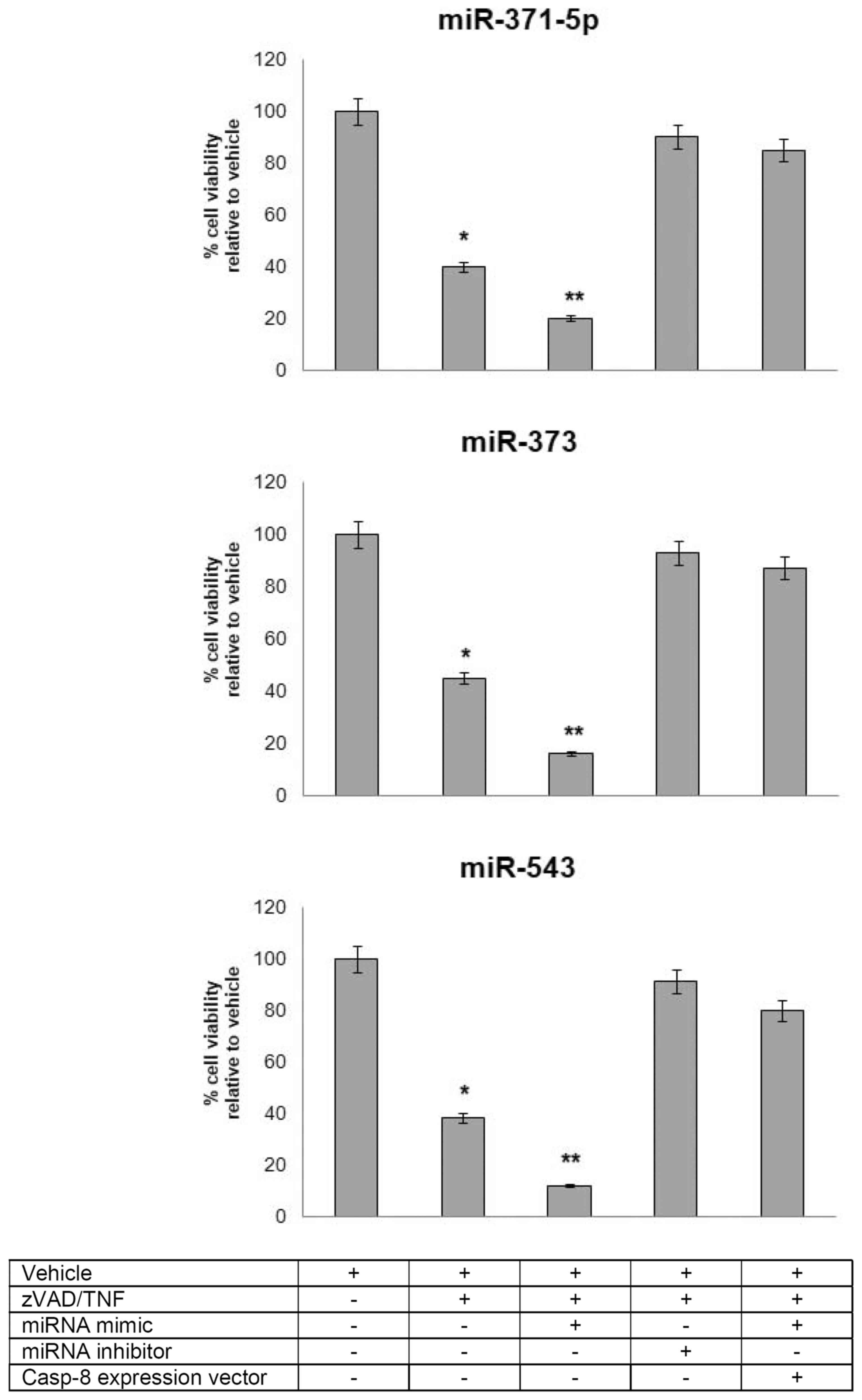
miR-371-5p, miR-373 and miR-543 regulated
necroptosis of HCC cells by targeting Casp-8
Only a limited number of studies to date have
reported the mechanisms through which miRNAs regulate necroptotic
cell death (16,17). Of note, miR-874 has been shown to
enhance necroptosis by targeting Casp-8 (45). Hence, given the functional
interaction of miR-371-5p, miR-373 and miR-543 with Casp-8,
in this study, we attempted to elucidate whether these miRNAs play
a role in the necrotic cascade orchestrated by Casp-8.
Hep3B-hCG cells were first pre-treated with the pan-caspase
inhibitor, Z-VAD (10 µM), in the presence or absence of
miRNA mimics and inhibitors, and then exposed to TNF-α. The
enforced expression of miR-371-5p, miR-373 and miR-543 markedly
increased Z-VAD/TNF-α-induced cell necroptosis, whereas the
knockdown of miR-371-5p, miR-373 and miR-543 led to a marked
reduction of programmed cell necrosis. Of note, the pro-necroptotic
effects exerted by the miRNA mimics were abolished when Casp-8 was
ectopically overexpressed, thus indicating that this three-miRNA
signature was able to regulate the necroptosis of Hep3B-hCG
cells via targeting Casp-8 (Fig. 4).
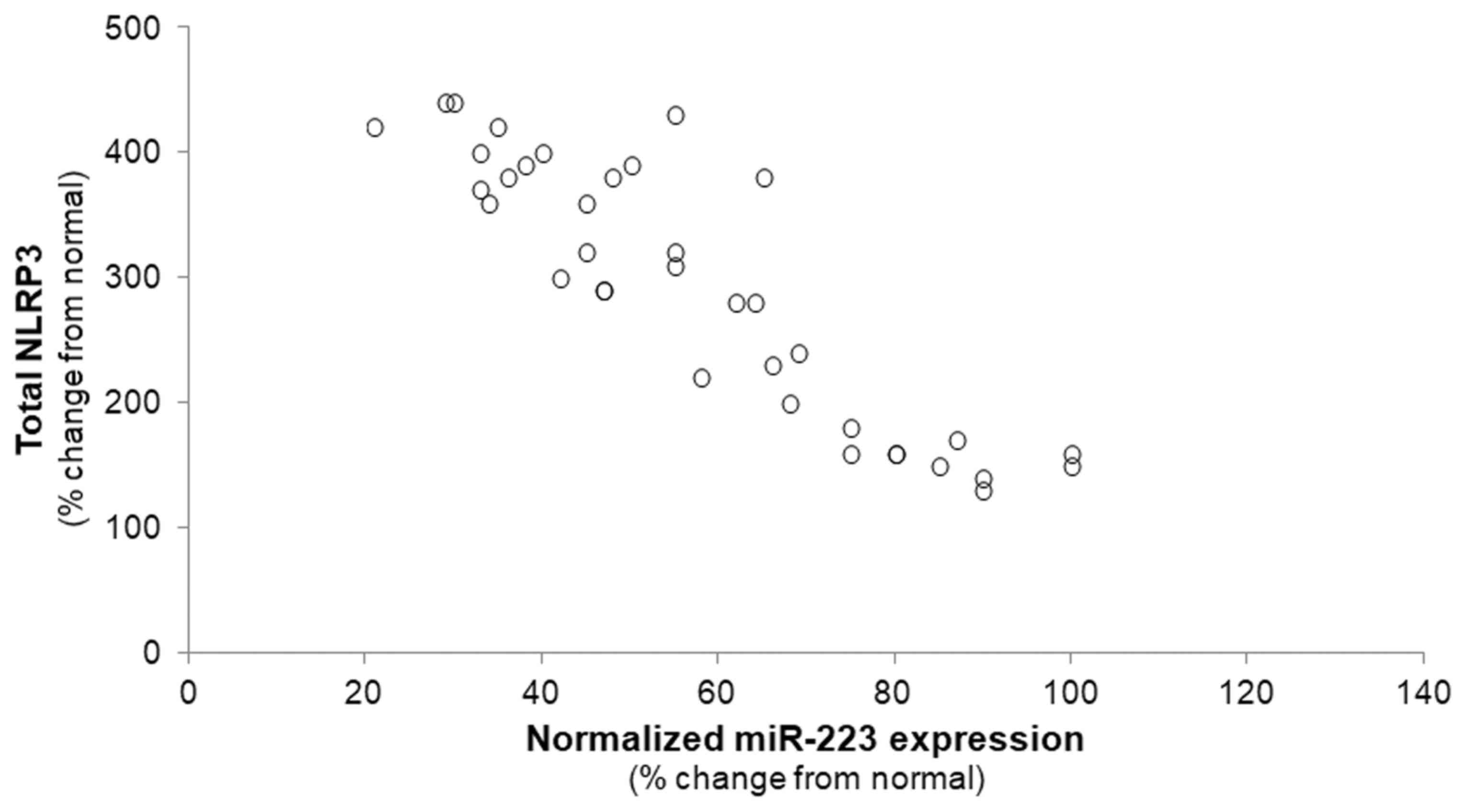
Inverse association between miR-223 and
NLRP3 in HCC samples
The miR-223-dependent post-transcriptional
regulation of the NLRP3 inflammasome has been well validated in a
variety of cell types (27,30);
however, this functional link has not been established yet in HCC.
Therefore, in this study, we proceeded to investigate whether an
inverse correlation between miR-223 abundance and NLRP3 mRNA
expression also exists in HCC. A significant decrease in the
miR-223 level was accompanied by a corresponding increase in
NLRP3 in the HCC samples compared with the NL specimens
(r=−0.3455, P<0.01), supporting the hypothesis that miR-223
likely also targets NLRP3 in HCC (Fig. 5).
Discussion
Necroptosis has recently attracted increasing
attention as a newly identified type of programmed cell death that
occurs when cells are subjected to severe stress or are exposed to
chemotherapeutic agents or inflammatory factors (46–49).
This type of regulated necrosis has been implicated in the
pathogenesis of a variety of human diseases, such as
neurodegeneration, ischemic reperfusion injury, Gaucher's disease,
progressive atherosclerotic lesions, TNF-mediated hypothermia and
systemic inflammation (50).
However, the role of necroptosis in cancer remains controversial,
as some authors consider this phenomenon a double-edged sword
(19). Indeed, although a number
of studies have reported that necroptosis prevents the onset and
progression of cancer and promotes its therapy by proving a
plethora of cancer cell lines highly sensitive and responsive to
necroptosis inducers (18),
emerging data have revealed that necroptosis may in fact contribute
to cancer development (51–53).
The rapid and extensive release of damage-associated molecules
[e.g., interleukin (IL)-1 family cytokines] from necroptotic cancer
cells may act as a potent inducer of inflammation (54), which in turn leads to tumorigenesis
through the stimulation of angiogenesis and metastasis, as well as
to the mitigation of the adaptive immune responses (55,56).
miRNAs were recently identified as new participants in regulated
necrosis (57); however, the
underlying mechanisms have not yet been fully elucidated. The
findings of the present study indicated that a novel three-miRNA
signature, including miR-371-5p, miR-373 and miR-543, is critically
involved in programmed necroptotic death of HCC cells. This
assertion was strongly supported by functional experiments in which
enforced expression of these miRNAs markedly increased
Z-VAD/TNF-α-induced necroptosis in Hep3B-hCG cells, whereas
their specific knockdown was able to significantly attenuate
programmed necrosis (Fig. 4).
Indeed, among all the miRNAs initially predicted by
computational scoring as necroptotic pathway regulators, we
restricted our investigations to those targeting the necroptosis
inhibitor Casp-8 gene, i.e., miR-371-5p, miR-373 and
miR-543. Consequently, since miR-137, miR-324, miR-371-5p, miR-372,
miR-373, miR-455, miR-543, miR-595 and miR-661 did not present
regions able to bind to the 3′UTR of Casp-8 according to the
target prediction results, they were not included in our subsequent
experimental analyses. However, these miRNAs will be the subject of
our further (in-progress) studies aimed at defining other metabolic
pathways involved in necroptosis.
From the results obtained by the luciferase assay
experiments, we found that miR-371-5p, miR-373 and miR-543
suppressed the expression of Casp-8 through direct
interaction with the 3′UTR of the target (Fig. 3). Of note, emerging evidence
indicates that Casp-8 is an important negative regulator of
necroptosis (43). Treatment with
Z-VAD, a pan-caspase inhibitor, was found to promote necroptosis,
since Casp-8, when inactivated, cannot cleave the two key
inducers of necroptosis, namely RIP1 and RIP3 (43,58,59).
Moreover, the pharmacological inhibition of Casp-8 induces
the autocrine production of TNF, which triggers necroptosis
(60). The results reported herein
may shed new light into the mechanisms regulating programmed
necrosis in HCC, revealing that the three miRNAs of interest induce
necroptosis by suppressing Casp-8. Accordingly, we observed
that the pro-necroptotic effects exerted by miRNA mimics were
abolished when Casp-8 was ectopically overexpressed
(Fig. 4). Higher expression levels
of miR-371-5p, miR-373 and miR-543 were significantly associated
with lower amounts of Casp-8 mRNA in HCC tissues compared
with paired normal tissues (Figs.
1 and 2), thereby supporting
the key role of these miRNAs in controlling Casp-8
expression.
Of note, miR-373 has been previously implicated in
several types of cancers as an oncomiR (61), with its down-regulation resulting
in the growth arrest and the apoptosis of tumor cells (62). Moreover, the increased expression
levels of miR-371-5p have been associated with a shorter survival
and a poor prognosis of patients with pancreatic cancer (63). Similarly, miR-543 has been found to
be closely associated with tumor size, TNM stage and metastasis, as
well as with a poor overall and disease-free survival (64). In this context, our study may
provide new insight into the necroptotic function of this
three-miRNA signature, further providing valuable insights into its
role in HCC and its value as a potential predictor of poor
prognosis. Indeed, the present findings are in line with those of a
recent study reporting that pro-necroptotic pathways promote
inflammatory cytokine production, thereby sustaining tumor growth
(51).
Another meaningful result of this study was the
significant inverse correlation between the expression pattern of
miR-223-3p and that of the NLRP3 inflammasome found in the resected
tissue specimens from HCC patients (Fig. 5). Specifically, we demonstrated
that the downregulation of miR-223 was closely associated with
increased NLRP3 mRNA levels. This finding effectively
supports the hypothesis that miR-223 acts as a negative regulator
of NLRP3 inflammasome activity, as it was already observed in other
pathological conditions (27,29).
It is well recognized that NLRP3 is an important mediator involved
in cell necroptosis (30), and it
has been suggested that its activation may be modulated by
Casp-8 (23,26). Of note, a recent study demonstrated
that miR-223-3p prevents the ischemia/reperfusion-induced cardiac
necroptosis via targeting NLRP3 inflammasome signaling (17). Therefore, the data presented herein
may uncover a novel role for miR-223-3p and its target gene,
NLRP3, in the context of HCC, strengthening the inextricable
association between these molecular effectors involved in
programmed cell necrosis. Defining the role of miR-223-3p-mediated
NLRP3 signaling in these and other potential necroptotic
models is mandatory for the better understanding the complex
crosstalk regulating programmed cell death and for testing new
disease-specific medical treatments.
In conclusion, the results from the present study
may provide new insight into the complex miRNA-governed network of
regulated necrosis, and suggest that the newly identified signature
of necroptosis-regulating miRNAs may represent a promising
diagnostic and prognostic biomarker as well an effective
therapeutic target for HCC.
However, further studies are required to verify the
interplay between these miRNAs, their target genes and HCC
progression, and to validate our findings for applicability in the
clinical setting.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MV, FP, RMDG, MB and MA contributed to the
conception and design of the study; MB, AB and GN were involved in
the selection of the biopsy tissues. FP, RO, RA and MA acquired the
data; MV, FP and MA analyzed and interpreted the data; RMDG, MB and
GN participated in the revision of the article. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the local Institutional
Review Board (IRB) of the University Hospital of Messina, Messina,
Italy. The study protocol conforms to the ethical guidelines of the
1975 Declaration of Helsinki. Written informed consent was obtained
from all subjects at the moment of the surgical procedures. For
research purposes, no consent for retrospective de-identified data
analysis was required as deemed by the IRB.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mlynarsky L, Menachem Y and Shibolet O:
Treatment of hepatocellular carcinoma: Steps forward but still a
long way to go. World J Hepatol. 7:566–574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tang ZY: Hepatocellular carcinoma - cause,
treatment and metastasis. World J Gastroenterol. 7:445–454. 2001.
View Article : Google Scholar
|
|
4
|
Herranz H and Cohen SM: MicroRNAs and gene
regulatory networks: Managing the impact of noise in biological
systems. Genes Dev. 24:1339–1344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arner P and Kulyté A: MicroRNA regulatory
networks in human adipose tissue and obesity. Nat Rev Endocrinol.
11:276–288. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fang S, Deng Y, Gu P and Fan X: MicroRNAs
regulate bone development and regeneration. Int J Mol Sci.
16:8227–8253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kohlhapp FJ, Mitra AK, Lengyel E and Peter
ME: MicroRNAs as mediators and communicators between cancer cells
and the tumor microenvironment. Oncogene. 34:5857–5868. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Su Y, Wu H, Pavlosky A, Zou LL, Deng X,
Zhang ZX and Jevnikar AM: Regulatory non-coding RNA: New
instruments in the orchestration of cell death. Cell Death Dis.
7:e23332016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tran DDH, Kessler C, Niehus SE, Mahnkopf
M, Koch A and Tamura T: Myc target gene, long intergenic noncoding
RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and
cell survival by titrating tumor suppressor microRNAs. Oncogene.
37:75–85. 2018. View Article : Google Scholar
|
|
10
|
Allegra A, Alonci A, Campo S, Penna G,
Petrungaro A, Gerace D and Musolino C: Circulating microRNAs: New
biomarkers in diagnosis, prognosis and treatment of cancer
(Review). Int J Oncol. 41:1897–1912. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kunej T, Godnic I, Ferdin J, Horvat S,
Dovc P and Calin GA: Epigenetic regulation of microRNAs in cancer:
An integrated review of literature. Mutat Res. 717:77–84. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
13
|
Mizuguchi Y, Takizawa T, Yoshida H and
Uchida E: Dysregulated miRNA in progression of hepatocellular
carcinoma: A systematic review. Hepatol Res. 46:391–406. 2016.
View Article : Google Scholar
|
|
14
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krützfeldt J, Rajewsky N, Braich R, Rajeev
KG, Tuschl T, Manoharan M and Stoffel M: Silencing of microRNAs in
vivo with 'antagomirs'. Nature. 438:685–689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Afonso MB, Rodrigues PM, Simão AL, Gaspar
MM, Carvalho T, Borralho P, Bañales JM, Castro RE and Rodrigues
CMP: miRNA-21 ablation protects against liver injury and
necroptosis in cholestasis. Cell Death Differ. Dec 11–2017.Epub
ahead of print. PubMed/NCBI
|
|
17
|
Qin D, Wang X, Li Y, Yang L, Wang R, Peng
J, Essandoh K, Mu X, Peng T, Han Q, et al: MicroRNA-223-5p and -3p
cooperatively suppress necroptosis in ischemic/reperfused hearts. J
Biol Chem. 291:20247–20259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen D, Yu J and Zhang L: Necroptosis: An
alternative cell death program defending against cancer. Biochim
Biophys Acta. 1865:228–236. 2016.PubMed/NCBI
|
|
19
|
Wang T, Jin Y, Yang W, Zhang L, Jin X, Liu
X, He Y and Li X: Necroptosis in cancer: An angel or a demon?
Tumour Biol. 39:1010428317711539. 2017. View Article : Google Scholar
|
|
20
|
Sutterwala FS, Haasken S and Cassel SL:
Mechanism of NLRP3 inflammasome activation. Ann N Y Acad Sci.
1319:82–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Davis BK, Wen H and Ting JP: The
inflammasome NLRs in immunity, inflammation, and associated
diseases. Annu Rev Immunol. 29:707–735. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei Q, Mu K, Li T, Zhang Y, Yang Z, Jia X,
Zhao W, Huai W, Guo P and Han L: Deregulation of the NLRP3
inflammasome in hepatic parenchymal cells during liver cancer
progression. Lab Invest. 94:52–62. 2014. View Article : Google Scholar
|
|
23
|
Gurung P, Anand PK, Malireddi RK, Vande
Walle L, Van Opdenbosch N, Dillon CP, Weinlich R, Green DR,
Lamkanfi M and Kanneganti TD: FADD and caspase-8 mediate priming
and activation of the canonical and noncanonical Nlrp3
inflammasomes. J Immunol. 192:1835–1846. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Weng D, Marty-Roix R, Ganesan S, Proulx
MK, Vladimer GI, Kaiser WJ, Mocarski ES, Pouliot K, Chan FK,
Kelliher MA, et al: Caspase-8 and RIP kinases regulate
bacteria-induced innate immune responses and cell death. Proc Natl
Acad Sci USA. 111:7391–7396. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gurung P and Kanneganti TD: Novel roles
for caspase-8 in IL-1β and inflammasome regulation. Am J Pathol.
185:17–25. 2015. View Article : Google Scholar :
|
|
26
|
Kang TB, Yang SH, Toth B, Kovalenko A and
Wallach D: Caspase-8 blocks kinase RIPK3-mediated activation of the
NLRP3 inflammasome. Immunity. 38:27–40. 2013. View Article : Google Scholar
|
|
27
|
Xie XJ, Ma LG, Xi K, Fan DM, Li JG, Zhang
Q and Zhang W: Effects of microRNA-223 on morphine analgesic
tolerance by targeting NLRP3 in a rat model of neuropathic pain.
Mol Pain. 13:1744806917706582. 2017. View Article : Google Scholar
|
|
28
|
Neudecker V, Haneklaus M, Jensen O,
Khailova L, Masterson JC, Tye H, Biette K, Jedlicka P, Brodsky KS,
Gerich ME, et al: Myeloid-derived miR-223 regulates intestinal
inflammation via repression of the NLRP3 inflammasome. J Exp Med.
214:1737–1752. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang Z, Zhong L, Xian R and Yuan B:
MicroRNA-223 regulates inflammation and brain injury via feedback
to NLRP3 inflammasome after intracerebral hemorrhage. Mol Immunol.
65:267–276. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bauernfeind F, Rieger A, Schildberg FA,
Knolle PA, Schmid-Burgk JL and Hornung V: NLRP3 inflammasome
activity is negatively controlled by miR-223. J Immunol.
189:4175–4181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Haneklaus M, Gerlic M, Kurowska-Stolarska
M, Rainey AA, Pich D, McInnes IB, Hammerschmidt W, O'Neill LA and
Masters SL: Cutting edge: miR-223 and EBV miR-BART15 regulate the
NLRP3 inflammasome and IL-1β production. J Immunol. 189:3795–3799.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Venza M, Visalli M, Biondo C, Oteri R,
Agliano F, Morabito S, Teti D and Venza I: Epigenetic marks
responsible for cadmiuminduced melanoma cell overgrowth. Toxicol In
Vitro. 29:242–250. 2015. View Article : Google Scholar
|
|
34
|
Vilaysane A, Chun J, Seamone ME, Wang W,
Chin R, Hirota S, Li Y, Clark SA, Tschopp J, Trpkov K, et al: The
NLRP3 inflammasome promotes renal inflammation and contributes to
CKD. J Am Soc Nephrol. 21:1732–1744. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−∆ ∆ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
36
|
Vlachos IS, Kostoulas N, Vergoulis T,
Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD,
Prionidis K, Dalamagas T and Hatzigeorgiou AG: DIANA miRPath v.2.0:
Investigating the combinatorial effect of microRNAs in pathways.
Nucleic Acids Res. 40W:W498–W504. 2012. View Article : Google Scholar
|
|
37
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38(Suppl 2): W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Song J, Bai Z, Han W, Zhang J, Meng H, Bi
J, Ma X, Han S and Zhang Z: Identification of suitable reference
genes for qPCR analysis of serum microRNA in gastric cancer
patients. Dig Dis Sci. 57:897–904. 2012. View Article : Google Scholar
|
|
40
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M,
et al: Combinatorial microRNA target predictions. Nat Genet.
37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti
N, Castanares M and Wu M: Cleavage of RIP3 inactivates its
caspase-independent apoptosis pathway by removal of kinase domain.
Cell Signal. 19:2056–2067. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Günther C, Martini E, Wittkopf N, Amann K,
Weigmann B, Neumann H, Waldner MJ, Hedrick SM, Tenzer S, Neurath
MF, et al: Caspase-8 regulates TNF-α-induced epithelial necroptosis
and terminal ileitis. Nature. 477:335–339. 2011. View Article : Google Scholar
|
|
43
|
Oberst A, Dillon CP, Weinlich R, McCormick
LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS and Green DR:
Catalytic activity of the caspase-8-FLIP(L) complex inhibits
RIPK3-dependent necrosis. Nature. 471:363–367. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vucur M, Reisinger F, Gautheron J, Janssen
J, Roderburg C, Cardenas DV, Kreggenwinkel K, Koppe C, Hammerich L,
Hakem R, et al: RIP3 inhibits inflammatory hepatocarcinogenesis but
promotes cholestasis by controlling caspase-8- and JNK-dependent
compensatory cell proliferation. Cell Reports. 4:776–790. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang K, Liu F, Zhou LY, Ding SL, Long B,
Liu CY, Sun T, Fan YY, Sun L and Li PF: miR-874 regulates
myocardial necrosis by targeting caspase-8. Cell Death Dis.
4:e7092013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chan FK, Shisler J, Bixby JG, Felices M,
Zheng L, Appel M, Orenstein J, Moss B and Lenardo MJ: A role for
tumor necrosis factor receptor-2 and receptor-interacting protein
in programmed necrosis and antiviral responses. J Biol Chem.
278:51613–51621. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Han W, Li L, Qiu S, Lu Q, Pan Q, Gu Y, Luo
J and Hu X: Shikonin circumvents cancer drug resistance by
induction of a necroptotic death. Mol Cancer Ther. 6:1641–1649.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Newton K and Manning G: Necroptosis and
Inflammation. Annu Rev Biochem. 85:743–763. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Long JS and Ryan KM: New frontiers in
promoting tumour cell death: Targeting apoptosis, necroptosis and
autophagy. Oncogene. 31:5045–5060. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhou W and Yuan J: Necroptosis in health
and diseases. Semin Cell Dev Biol. 35:14–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu X, Zhou M, Mei L, Ruan J, Hu Q, Peng
J, Su H, Liao H, Liu S, Liu W, et al: Key roles of necroptotic
factors in promoting tumor growth. Oncotarget. 7:22219–22233.
2016.PubMed/NCBI
|
|
52
|
Seifert L, Werba G, Tiwari S, Giao Ly NN,
Alothman S, Alqunaibit D, Avanzi A, Barilla R, Daley D, Greco SH,
et al: The necrosome promotes pancreatic oncogenesis via CXCL1 and
Mincle-induced immune suppression. Nature. 532:245–249. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Strilic B, Yang L, Albarrán-Juárez J,
Wachsmuth L, Han K, Müller UC, Pasparakis M and Offermanns S:
Tumour-cell-induced endothelial cell necroptosis via death receptor
6 promotes metastasis. Nature. 536:215–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Pasparakis M and Vandenabeele P:
Necroptosis and its role in inflammation. Nature. 517:311–320.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Korniluk A, Koper O, Kemona H and
Dymicka-Piekarska V: From inflammation to cancer. Ir J Med Sci.
186:57–62. 2017. View Article : Google Scholar :
|
|
57
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget.
6:8474–8490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Geserick P, Hupe M, Moulin M, Wong WW,
Feoktistova M, Kellert B, Gollnick H, Silke J and Leverkus M:
Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting
RIP1 kinase recruitment. J Cell Biol. 187:1037–1054. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Welz PS, Wullaert A, Vlantis K, Kondylis
V, Fernández-Majada V, Ermolaeva M, Kirsch P, Sterner-Kock A, van
Loo G and Pasparakis M: FADD prevents RIP3-mediated epithelial cell
necrosis and chronic intestinal inflammation. Nature. 477:330–334.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hitomi J, Christofferson DE, Ng A, Yao J,
Degterev A, Xavier RJ and Yuan J: Identification of a molecular
signaling network that regulates a cellular necrotic cell death
pathway. Cell. 135:1311–1323. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Huang Q, Gumireddy K, Schrier M, le Sage
C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Stelzer Y, Sagi I and Benvenisty N:
Involvement of parental imprinting in the antisense regulation of
onco-miR-372-373. Nat Commun. 4:27242013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
He D, Miao H, Xu Y, Xiong L, Wang Y, Xiang
H, Zhang H and Zhang Z: MiR-371-5p facilitates pancreatic cancer
cell proliferation and decreases patient survival. PLoS One.
9:e1129302014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhai F, Cao C, Zhang L and Zhang J:
miR-543 promotes colorectal cancer proliferation and metastasis by
targeting KLF4. Oncotarget. 8:59246–59256. 2017. View Article : Google Scholar : PubMed/NCBI
|