Introduction
Breast cancer has become a leading cause of
cancer-associated mortality in the world, and the most common
cancer among women (1). The
majority of these mortalities are caused by distant metastasis and
resistance to the currently available therapeutics (2). An estimated 15-20% of patients with
breast cancer exhibit overexpression of human epidermal growth
factor receptor 2 (HER2), leading to a poorer prognosis and
survival (3,4). At present, therapy with anti-HER2
mono-antibodies, including trastuzumab, is applied to treat
patients with HER2-positive breast cancer (5,6).
Trastuzumab is designed to target HER2 and silence its function,
and is primarily used for early stage or metastatic gastric and
breast cancer with positive HER2 mutations. Trastuzumab may be
effective for initial treatment, although resistance increases
substantially following a period of exposure. In addition, there is
a clear need for useful therapeutic biomarkers that may be used for
predicting chemoresponses to treatment with trastuzumab (7). Therefore, there is an urgent
necessity to reveal the mechanism of trastuzumab resistance and
identify useful molecular markers and therapeutic targets for
patients with breast cancer.
With the advanced development of whole genome and
transcriptome sequencing technologies and the ENCODE project
(8), researchers have drawn the
conclusion that the majority of genomic DNA is represented in
processed transcripts that may not be translated into functional
proteins, namely non-coding RNAs (ncRNAs) (9). Long ncRNAs (lncRNAs) are an important
group of ncRNAs that contain >200 nucleotides (10). During recent years, numerous
reports have demonstrated that lncRNAs may serve as critical
biological regulators in the functions of cellular and molecular
signaling pathways; for example, they may exert their important
functional roles at the post-transcriptional level by sponging
microRNAs (11), and mediate
transcriptional regulation via chromatin modification (12,13).
Exosomes have attracted considerable attention in
the field of biomarker discovery. The release of exosomes into the
extracellular space affords an opportunity to exchange cellular
contents, membranes, proteins and gene fragments (14). Exosomes are membrane-derived
vesicles and have a size range of 20-200 nm when released into
bodily fluids, including blood, urine and malignant ascites. These
vesicles contain DNA, protein fragments, and coding or ncRNAs
secreted by their parental cell cytoplasm, and may be absorbed into
recipient cells (15). A recent
study indicated that the exosomes from chemosensitive/resistant
cells were able to markedly influence the chemoresponse of
recipient cells through the transfer of specific genes, including
lncRNAs (16). However, this
conclusion requires further validation. Most importantly, exosomes
contain genes and proteins, reflecting the features of cancer
cells, which may facilitate the development of highly sensitive
diagnostic strategies for monitoring the therapeutic response
conditions of cancer in a rapid and non-invasive manner (17).
The present study determined the roles of
exosome-transmitted lncRNAs on trastuzumab resistance, and further
investigated the therapeutic options for patients with
trastuzumab-resistant breast cancer. The present verified the
involvement of the lncRNA-small nucleolar RNA host gene 14 (SNHG14)
in the mediation of trastuzumab responses via tumor cell
extracellular exosomes. The results indicated that exosomal SNHG14
may be a novel biomarker for breast cancer treatment and
monitoring.
Materials and methods
Clinical samples
In total, 72 serum samples were collected from
patients with advanced ER2+ breast cancer [male/female
ratio, 0/72; age range, 35-68 years (median age, 55 years)] who
received treatment with trastuzumab at Hainan General Hospital
(Haikou, China) between January 2013 and June 2017. Samples of 5 ml
venous blood from each participant were collected by venipuncture
prior to starting chemotherapy. Serum was segregated via
centrifugation at 1,600 x g for 10 min at room temperature within 2
h following collection, followed by a second centrifugation at
12,000 x g for 10 min at 4°C to remove the residual cells debris.
Each serum supernatant was transferred into RNase-free tubes and
stored at −80°C until use. Tumor response was confirmed through
computed tomography and evaluated according to the Response
Evaluation Criteria In Solid Tumors (RECIST; version 1.1) (18) as complete response (CR), partial
response (PR), stable disease (SD) and progressive disease (PD).
All the patients were pathologically confirmed and the clinical
tissue samples were collected prior to the commencement of
chemotherapy at Hainan General Hospital. Patients with other types
of cancer, breast benign disease and autoimmune diseases were
excluded. Written informed consent was obtained from each
participant prior to blood collection. The study protocol was
approved by the Clinical Research Ethics Committee of Hainan
General Hospital.
Cell culture
The human breast cancer cell lines SKBR-3 and BT474,
which harbor HER2 activating mutations, were purchased from the
Chinese Type Culture Collection, Chinese Academy of Sciences
(Shanghai, China). The two cell lines were cultured in RPMI-1640
medium (BioWhittaker; Lonza Group, Ltd., Basel, Switzerland)
supplemented with 10 mM HEPES, 1 mM L-glutamine, 100 U/ml
penicillin/streptomycin (BioWhittaker; Lonza Group, Ltd.) and
heat-inactivated 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), at 37°C in a humidified
incubator with 5% CO2. Trastuzumab (Herceptin) was
obtained from Roche Diagnostics (Basel, Switzerland) and dissolved
in sterile water. Trastuzumab-resistant SKBR-3/Tr and BT474/Tr
cells were obtained via continuous culture with 5 mg/ml trastuzumab
for 6 months, as previously reported (19,20),
and were cultured in RPMI-1640 medium with 250 μg/ml
trastuzumab.
Cell invasion assay
Cell invasion ability was tested using a Matrigel
Transwell assay. A total of 100 μl Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA) was first added onto the bottom of the
Transwell chamber (24-well insert; 8-mm pore size; Corning
Incorporated, Corning, NY, USA), and 1×105 breast cancer
cells in reduced serum medium (5% serum; Opti-MEM; Gibco, Thermo
Fisher Scientific, Inc.) were placed on the coated membrane in the
chamber. Cells that migrated through the permeable membrane were
fixed in methanol for 20 min at room temperature, stained with
crystal violet for 5 min at room temperature, and counted under a
microscope at ×20 magnification (DMI4000B; Leica Microsystems GmbH,
Wetzlar, Germany) in random fields in each well.
Exosome isolation and labelling
Exosomes were extracted from breast cancer cell
culture medium or serum samples using an ExoQuick precipitation kit
(System Biosciences, LLC, Palo Alto, CA, USA), according to the
manufacturer's protocol. The culture medium or serum was thawed on
ice and centrifuged at 3,000 x g for 15 min (4°C) to remove cells
and cell debris. Subsequently, 250 μl supernatant was mixed
with 63 μl ExoQuick precipitation kit and incubated at 4°C
for 30 min following a brief up-and-down mix, followed by
centrifugation at 1,500 × g for 30 min (4°C). The supernatant was
removed via careful aspiration, followed by a further 5 min of
centrifugation at 1,500 × g (4°C) to remove the residual liquid.
The exosome-containing pellet was subsequently resuspended in 250
μl PBS. The final pellets, containing exosomes, were
collected for characterization and RNA isolation. Purified exosomes
were labeled with PKH26 Red Fluorescent Cell Linker kit for General
Cell Membrane Labeling (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), according to the manufacturer's protocol.
RNA extraction
Extraction of RNA from exosomes was performed using
the commercial miRNeasy Serum/Plasma kit (Qiagen, Inc., Valencia,
CA, USA), and RNA extraction from the cell fraction was performed
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. All RNA elution steps
were performed at 12,000 × g for 15 sec (4°C), and the RNA was
finally eluted in 15 μl RNase-free ultra-pure water.
Transmission electron microscopy
The exosome pellets were resuspended in 50 μl
PBS and a drop of the suspension was placed on a sheet of Parafilm.
A carbon-coated copper grid was floated on the drop for 5 min at
room temperature. The grid was removed, and the excess liquid was
drained by touching the grid edge against a piece of clean filter
paper. The grid was subsequently placed onto a drop of 2%
phosphotungstic acid at pH 7.0 for ~5 sec, and the excess liquid
was drained off. The grid was allowed to dry for 5 min and examined
using a JEM-1200 EX microscope (JEOL, Ltd., Tokyo, Japan) with a
voltage of 80 keV and a resolution of 0.2 nm.
Nanoparticle tracking analysis (NTA)
A total of ~0.3 ml supernatant was loaded into the
sample chamber of an LM10 Nanosight unit (Nanosight, Ltd.,
Salisbury, UK) and three videos of either 30 or 60 sec were
recorded of each sample. Data analysis was performed using NTA 2.1
software (Nanosight, Ltd.). In NTA, the paths of unlabeled
particles (i.e. microvesicles) acting as point scatterers,
undergoing Brownian motion in a 0.25 ml chamber through which a
635-nm laser beam is passed, is determined from a video recording,
with the mean squared displacement determined for each possible
particle. The diffusion coefficient and sphere-equivalent
hydrodynamic radius are subsequently determined using the
Stokes-Einstein equation, and results are displayed as a particle
size distribution (21).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was reverse transcribed using SuperScript
III® (Invitrogen; Thermo Fisher Scientific, Inc.) and
amplified by RT-qPCR based on the TaqMan method using a TaqMan
Human RNA Assay kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) on a Bio-Rad CFX96 Sequence Detection System (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The gene expression levels
were normalized to GAPDH expression (22). RT-qPCR results were analyzed and
expressed relative to quantification cycle values (23), and subsequently converted to fold
changes. All the primers were synthesized by Guangzhou RiboBio Co.,
Ltd. (Guangzhou, China), and their sequences were as follows:
lncRNA-SNHG14 forward, 5′-GGGTGTTTACGTAGACCAGAACC-3′ and reverse,
5′-CTTCCAAAAGCCTTCTGCCTTAG-3′; and GAPDH forward,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
The thermocycling conditions were 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min.
RNA oligoribonucleotides and cell
transfection
The small interfering (si)RNA against lncRNA-SNHG14
was synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China).
The apoptosis regulator Bcl-2 (Bcl-2) inhibitor venetoclax was
purchased from Roche Diagnostics. Cells were plated at
5×104 cells/well in 24-well plates ~24 h prior to
transfection or treatment. When the cells had reached 30-50%
confluence, the cells were treated with venetoclax at 0.5
μM. Transfection was performed using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific), following the manufacturer's protocol. Transfection
efficiency was evaluated in every experiment by RT-qPCR 24 h
subsequently to ensure that the cells were successfully
transfected. Functional experiments were performed 48 h
post-transfection. The final concentration of the RNA
oligoribonucleotides was 100 nM. The sequences of the siRNAs were
as follows (5′→3′): si-SNHG14 #1 sense, GCAAAUGAAAGCUACCAAU and
antisense, AUUGGUAGCU UUCAUUUGC; si-SNHG14 #2 sense,
GCACAAUAUCUUUGAACUA and antisense, UAGUUCAAAGAUAUUGUGC; and
si-SNHG14 #3 sense, CUAGAAUCCUAAAGGCAAA and antisense,
UUUGCCUUUAGGAUUCUAG. The si-Negative Control_05815 (cat. no.
siN05815122147) was obtained from Guangzhou RiboBio Co., Ltd.
Expression profile analysis of
lncRNAs
RNA expression profiling was performed using an
Agilent human lncRNA microarray V.2.0 platform (GPL18109; Agilent
Technologies, Inc., Santa Clara, CA, USA). Quantile normalization
and subsequent data processing were performed using Agilent Gene
Spring Software 11.5 (Agilent Technologies, Inc.). Heat maps
representing differentially regulated genes were generated using
Cluster 3.0 software (developed by Professor Michiel de Hoon,
Center for Computational Biology and Bioinformatics, Columbia
University, New York, NY, USA). Following the establishment of a
cDNA library by extracting the total RNA from exosomes,
hybridization and washing the samples, four breast cancer cell
types were analyzed. Exogenous RNAs developed by External RNA
Controls Consortium (24) were
used as controls. The exosomal lncRNA microarray process was
performed by KangChen BioTech Co., Ltd. (Shanghai, China).
Fluorescence in situ hybridization
analysis (FISH)
Nuclear and cytosolic fraction separation was
performed using a PARIS kit (Life Technologies; Thermo Fisher
Scientific, Inc.), and RNA FISH probes were designed and
synthesized by Guangzhou RiboBio Co., Ltd., according to the
manufacturer's protocol. Cells were fixed in 4% formaldehyde for 15
min at room temperature and subsequently washed with PBS. The fixed
cells were treated with pepsin and dehydrated in ethanol. The
air-dried cells were incubated further with 40 nM FISH probe in
hybridization buffer. Following hybridization, the slide was
washed, dehydrated and mounted with Prolong Gold Antifade Reagent
with DAPI (Life Technologies; Thermo Fisher Scientific, Inc.) for
detection. The slides were visualized for immunofluorescence with a
×40 fluorescence microscope (DMI4000B; Leica Microsystems
GmbH).
Terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labelling
(TUNEL) assay
TUNEL staining was performed to evaluate cellular
apoptosis. Cells were treated with extracted exosomes or combined
with 0.5 μM venetoclax for 24 h and fixed using 4%
formaldehyde at room temperature for 15 min. Cells were fixed and
stained with a TUNEL kit, according to the manufacturer's protocol
(Vazyme, Piscataway, NJ, USA; TUNEL Bright-Red Apoptosis Detection
kit; cat. no. A113). TUNEL-positive cells were counted under a ×20
fluorescence microscopy (DMI4000B; Leica Microsystems GmbH).
Signal transduction reporter array
Cignal Finder Transduction Reporter Array (Qiagen,
Inc.) was used to simultaneously investigate alterations in the
activities of 50 canonical signaling pathways in response to
treatment with exosomal lncRNA-SNHG14. Cells were treated with
SNHG14-overexpressing exosomes for 24 h and were subsequently
transfected with a mixture of a transcription factor-responsive
firefly luciferase reporter and a constitutively expressing
Renilla construct using Attractene Transfection Reagent
(Qiagen, Inc.). A total of 24 h subsequently, the relative activity
of each pathway was determined by firefly luciferase/Renilla
and normalized to untreated controls. Experiments were performed in
triplicate.
Western blotting
Cell lysates were prepared with
radioimmunoprecipitation assay buffer containing protease
inhibitors (Sigma-Aldrich; Merck KGaA). Protein concentrations were
measured using a Bicinchoninic Acid Protein Assay kit, according to
the manufacturer's protocol (Beyotime Institute of Biotechnology,
Haimen, China). Equal amounts of protein (25 μg) were
separated by 10% SDS-PAGE and transferred onto polyvinylidene
fluoride membranes (EMD Millipore, Billerica, MA, USA). The
membrane was blocked with 5% (5 g/100 ml) non-fat dry milk in TBS
with Tween (TBS-T) for 2 h at room temperature. The membranes were
incubated overnight at 4°C with a 1:1,000 solution of primary
rabbit antibodies: Anti-E-cadherin (Abcam, Cambridge, UK; cat. no.
ab15148), anti-β-catenin (Abcam; cat. no. ab16051), anti-vimentin
(Abcam; cat. no. ab92547), anti-cluster of differentiation (CD)63
(Abcam; cat. no. ab134045), anti-CD81 (Abcam; cat. no. ab109201),
anti-cleaved poly(ADP-ribose) polymerase (PARP; cat. no. 5625; Cell
Signaling Technology, Inc., Danvers, MA, USA), anti-cleaved
caspase-3 (cat. no. 9664; Cell Signaling Technology, Inc.),
anti-apoptosis regulator Bcl-2 (Bcl-2; cat. no. 4223, Cell
Signaling Technology, Inc.), anti-apoptosis regulator BAX (Bax;
cat. no. 5023; Cell Signaling Technology, Inc.), and anti-β-actin
(Abcam; cat. no. ab8227). The horseradish peroxidase-conjugated
anti-rabbit antibody (cat. no. 7074; 1:5,000; Cell Signaling
Technology, Inc.) was used as secondary antibody for immunostaining
for 1 h at room temperature. Densitometry was performed using
ImageJ version 1.51r (25).
Statistical analysis
The Kolmogorov-Smirnov test was used to determine
the normality of the distribution of the data in each group. Count
data are described as a frequency and were examined using Fisher's
exact test. The Mann-Whitney U test or Kruskal-Wallis test (post
hoc Mann-Whitney U test with Bonferroni's correction) was used for
evaluating the differences among clinical cohort groups or cell
groups. Receiver operating characteristic (ROC) curves and the area
under the curve (AUC) were established to discriminate between
patients with breast cancer responding to trastuzumab and those not
responding by using MedCalc version 11.4.2.0 software (MedCalc
Software bvba, Ostend, Belgium). All statistical analyses were
performed with SPSS 17.0 software (SPSS Inc., Chicago, IL, USA).
Data are presented as the median ± inter-quartile range. P<0.05
was considered to indicate a statistically significant
difference.
Results
Characterization of trastuzumab-resistant
breast cancer cell lines
To investigate the underlying regulatory mechanism
of trastuzumab resistance, two trastuzumab-resistant sublines
derived from the HER2+ parental cell lines SKBR-3
(SKBR-3/Pr) and BT474 (BT474/Pr) were established (SKBR-3/Tr and
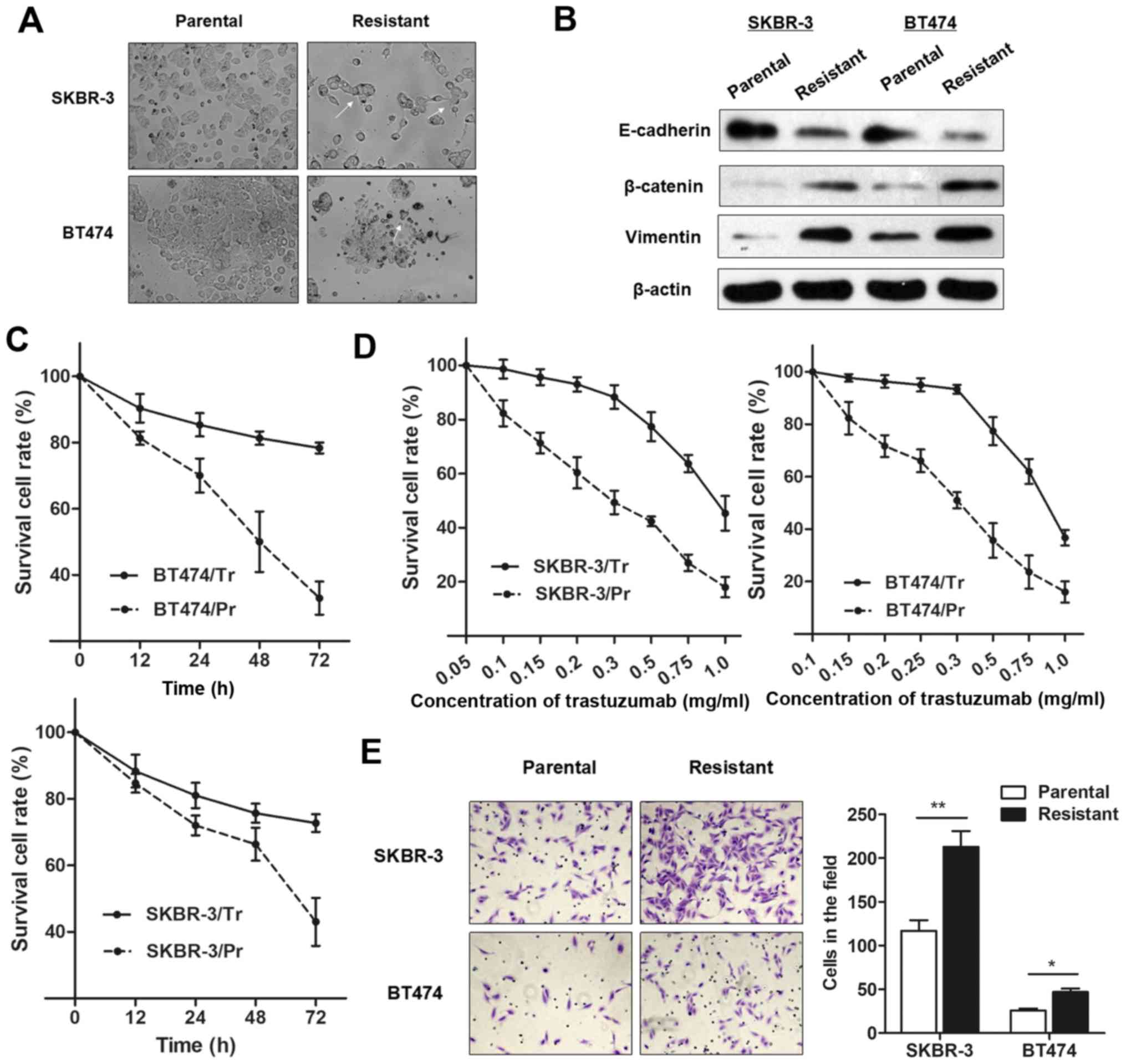
BT474/Tr, respectively), as described above. As presented in
Fig. 1A, the trastuzumab-resistant
cells exhibited specific morphological alterations consistent with
EMT, including the loss of cell polarity causing a spindle-cell
morphology, increased intercellular separation signifying the loss
of intercellular adhesion, and increased formation of pseudopodia.
Western blot analysis demonstrated an increase in EMT-relevant
protein expression in trastuzumab-resistant cells compared with
parental cells (Fig. 1B).
Furthermore, SKBR-3/Tr and BT474/Tr cells exhibited elevated cell
viability, in contrast to the parental cells, when incubated with
culture medium containing 0.25 mg/ml trastuzumab for 48 h (Fig. 1C). However, the
concentration-effect curve indicated that the half-maximal
inhibitory concentration (IC50) of trastuzumab (48 h)
for SKBR-3/Tr cells was 0.83 mg/ml, while the IC50 of
trastuzumab in SKBR-3/Pr was 0.29 mg/ml, meaning that the SKBR-3/Tr
had 2.86 times the trastuzumab resistance of SKBR-3 parental cells.
Similarly, the BT474/Tr cell line had 3.29 times the trastuzumab
resistance of BT474 parental cells (IC50 values were
0.79 and 0.24 mg/ml, respectively; Fig. 1D). More importantly, a
significantly increased number of chemoresistant cells were
observed to migrate through the collagen membrane compared with
parental cells (Fig. 1E),
indicating an increased cell migratory ability.
Characterization of exosomes secreted
from trastuzumab-resistant and -sensitive cells
Exosomes may be actively secreted from a variety of
cell types, including cancer cells. To determine whether exosomes
may be secreted from breast cancer cells, and whether the secreted
exosomes are able to regulate trastuzumab resistance, SKBR-3/Tr and
SKBR-3/Pr parental cells were incubated in exosome-free medium
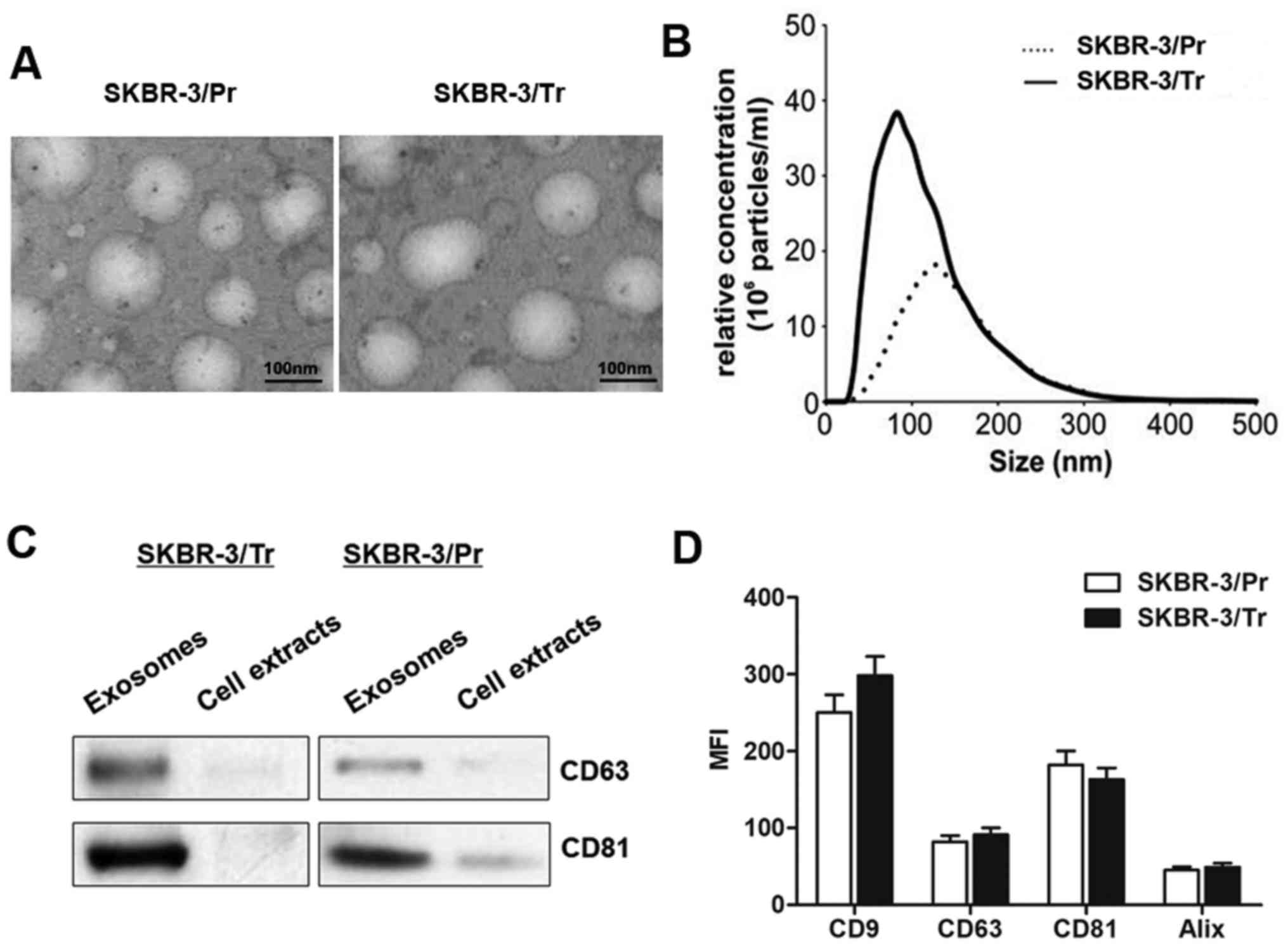
containing exosome-free FBS. As presented in Fig. 2A, transmission electron microscopy
revealed similar morphological characteristics between the
SKBR-3/Tr and SKBR-3/Pr exosomes, with a homogeneous structure. In
addition, their size was within the characteristic diameter range
of 40-120 nm, with a median diameter of ~100 nm. NTA of the
isolated exosomes revealed that the SKBR-3/Tr cells released 2.73
times more exosomes compared with SKBR-3/Pr cells (Fig. 2B). The presence of exosome protein
markers, including CD63 and CD81, was further confirmed by western
blot analysis. The results demonstrated specific bands in isolated
exosomal fraction, and not in the whole cell lysate (Fig. 2C). Further characterization of the
exosomes by flow cytometry for these exosomal markers did not
reveal any significant differences between the SKBR-3/Tr and
SKBR-3/Pr cell-derived exosomes (Fig.
2D), though increased expression of CD9 was noted in SKBR-3/Tr
cells, in contrast to SKBR-3/Pr cells, without statistical
significance. Taken together, the present data confirmed the
release of exosomes from SKBR-3/Tr and SKBR-3/Pr cells, and no
protein markers were identified to differentiate between the two
sublines.
Microarray profiling for exosomal lncRNAs
associated with trastuzumab resistance
Having verified the release of exosomes from
trastuzumab-resistant cells, the present study sought to define the
specific exosomal lncRNA(s) that may regulate trastuzumab
resistance. The trastuzumab resistance-associated exosomal lncRNAs
released in the conditioned media in these two chemoresistant cell
lines, the and coupled parental cell lines, were characterized by
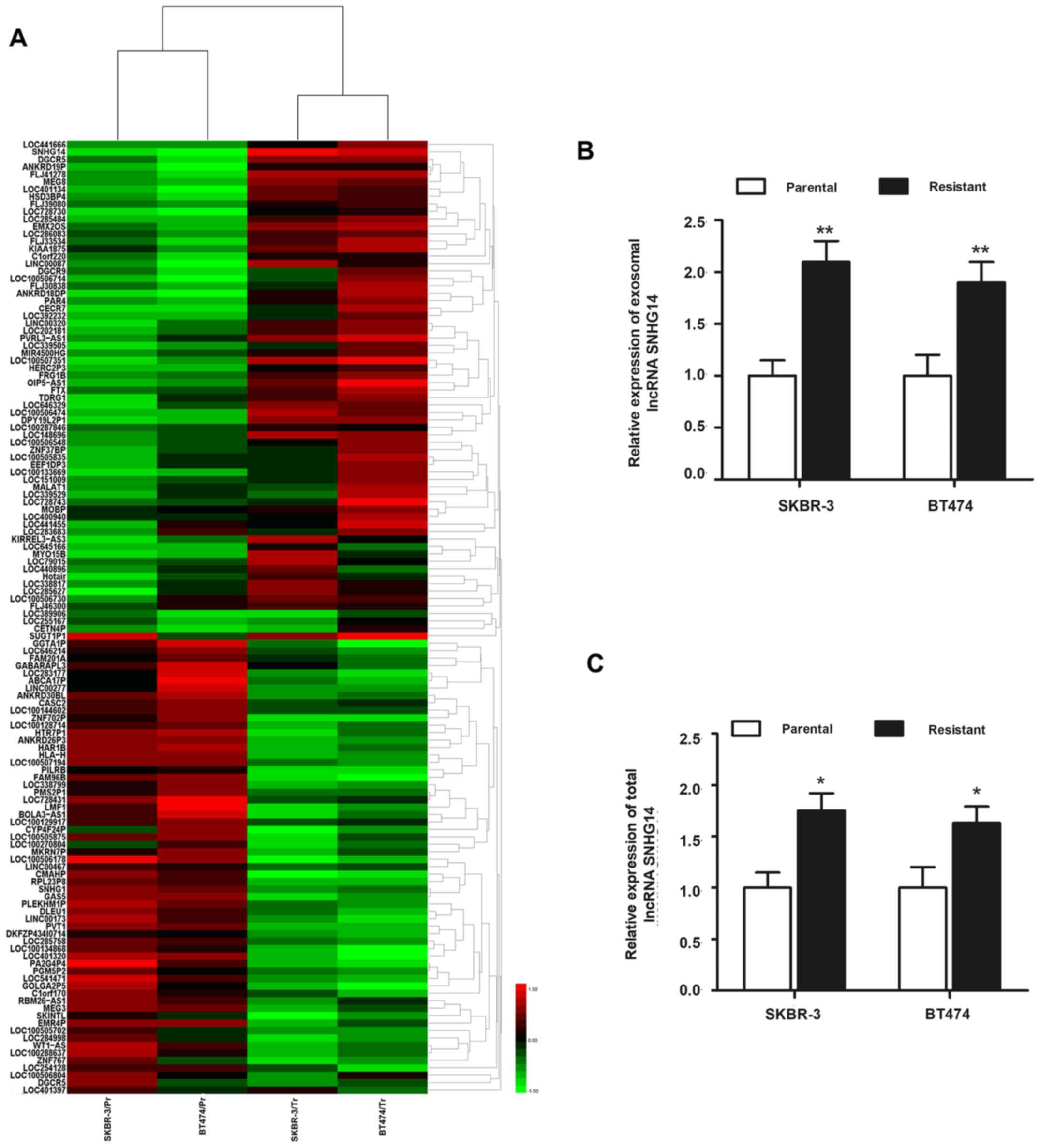
microarray analysis. A total of 126 exosomal lncRNAs were
identified to be dysregulated (>2-fold) in resistant cell lines
compared with the respective parental cells (Fig. 3A). Among those, exosomal
lncRNA-SNHG14 was markedly and most highly expressed in SKBR-3/Tr
and BT474/Tr cells, while its expression was significantly lower in
SKBR-3/Pr and BT474/Pr cells, suggesting that SNHG14 may be
essential for the formation of trastuzumab resistance. An RT-qPCR
experiment was subsequently performed to validate the upregulation
of lncRNA-SNHG14. As presented in Fig.
3B, markedly elevated expression of exosomal SNHG14 was
verified in the two trastuzumab-resistant cell lines compared with
the respective parental cells. The top five exosomal lncRNAs whose
expression was higher in the two trastuzumab-resistant cell lines
compared with the parental cells are listed in Tables I and II. In addition, the present study also
detected the intracellular (total) expression of lncRNA-SNHG14 in
the two cell lines, and a similar expression pattern was observed
in the intracellular fraction (Fig.
3C). Since lncRNA-SNHG14 was dysregulated at the intracellular
and extracellular (exosome) level in trastuzumab-resistant cells
compared with parental cells, lncRNA-SNHG14 was the focus of the
following experiments.
 | Table ITop 5 exosomal lncRNAs in SKBR-3/Tr
cells compared with SKBR-3/Pr cells. |
Table I
Top 5 exosomal lncRNAs in SKBR-3/Tr
cells compared with SKBR-3/Pr cells.
| Sampleno. | lncRNA | Fold-change | Standard
deviation | Location |
|---|
| 1 | CECR7 | 11.36 | 1.08 | 22q11.1 |
| 2 | SNHG14 | 10.18 | 2.34 | 15q11.2 |
| 3 | ANKRD18DP | 8.34 | 0.75 | 3q29 |
| 4 | LOC100507351 | 6.16 | 3.29 | 17q25.3 |
| 5 | LOC728743 | 4.94 | 1.23 | 7q36.1 |
 | Table IITop 5 exosomal lncRNAs in BT474/Tr
cells as compared to BT474/Pr cells. |
Table II
Top 5 exosomal lncRNAs in BT474/Tr
cells as compared to BT474/Pr cells.
| Sample no. | lncRNA | Fold-change | Standard
deviation | Location |
|---|
| 1 | SNHG14 | 13.65 | 1.85 | 15q11.2 |
| 2 | KIRREL3-AS3 | 9.37 | 1.23 | 11q24.2 |
| 3 | LOC100507351 | 7.41 | 0.96 | 17q25.3 |
| 4 | LINC00087 | 5.81 | 1.17 | Xq26.3 |
| 5 | LOC285627 | 4.49 | 0.86 | 5q33.3 |
Exosomal lncRNA-SNHG14 is required for
trastuzumab resistance in breast cancer cells
With the verification of the aberrant expression of
exosomal lncRNA-SNHG14 in chemoresistant cell lines, the present
study sought to determine whether lncRNA-SNHG14 is essential for
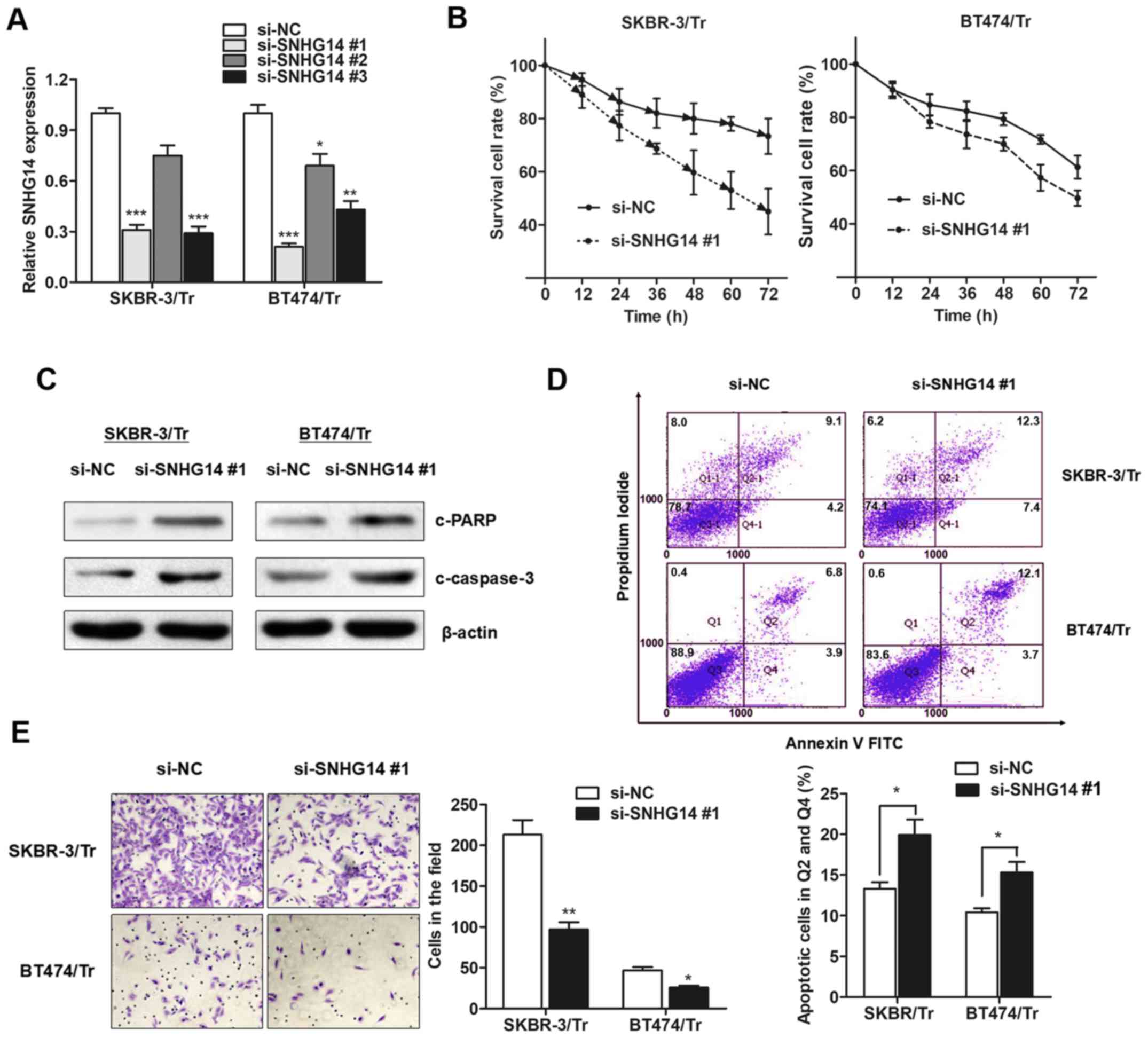
trastuzumab resistance. A total of three siRNAs against
lncRNA-SNHG14 were generated, and it was observed that
lncRNA-SNHG14 expression was primarily decreased in exosomes
derived from SKBR-3/Tr and BT474/Tr cells incubated with si-SNHG14
#1 (Fig. 4A), which was used for
the following experiments. Compared with the response of the
control group, silencing lncRNA-SNHG14 resensitized breast cancer
cells to treatment with trastuzumab (Fig. 4B). Furthermore, increased cleavage
of PARP and caspase-3 was observed in SNHG14-knockdown resistant
breast cancer cells following treatment with trastuzumab (Fig. 4C). Consistently, flow cytometry
demonstrated that trastuzumab exposure resulted in an increased
proportion of apoptotic cells among SNHG14-knockdown cells
(Fig. 4D), indicating that SNHG14
promotes trastuzumab resistance by suppressing cellular apoptosis.
In addition, knockdown of SNHG14 partially abrogated the obtained
invasive ability of the two cell lines (Fig. 4E). Collectively, the results of the
present study demonstrated that lncRNA-SNHG14 may be essential for
trastuzumab resistance in breast cancer.
Exosome-mediated transfer of
lncRNA-SNHG14 may result in trastuzumab chemoresistance
To examine whether lncRNA-SNHG14 regulates
trastuzumab resistance through the delivery of exosomes, it was
demonstrated the SNHG14-containing exosomes may be taken up by
recipient cells via a two-pronged approach. Firstly, the present
study examined whether secreted exosomes may be taken up by
recipient cells by labeling isolated exosomes with PKH26 dye from
SKBR-3/Tr cells. The labeled exosomes were subsequently added and
incubated with SKBR-3/Pr and BT474/Pr cells. As presented in
Fig. 5A, the majority of the
recipient cells exhibited a red signal under the confocal
microscope. Secondly, the present study examined whether these
exosomes were able to deliver lncRNA-SNHG14 to recipient cells,
similar to the intercellular transfer of other non-coding RNAs, as
previously reported (26,27). As expected, RT-qPCR analysis
demonstrated an increase in lncRNA-SNHG14 in the two recipient cell
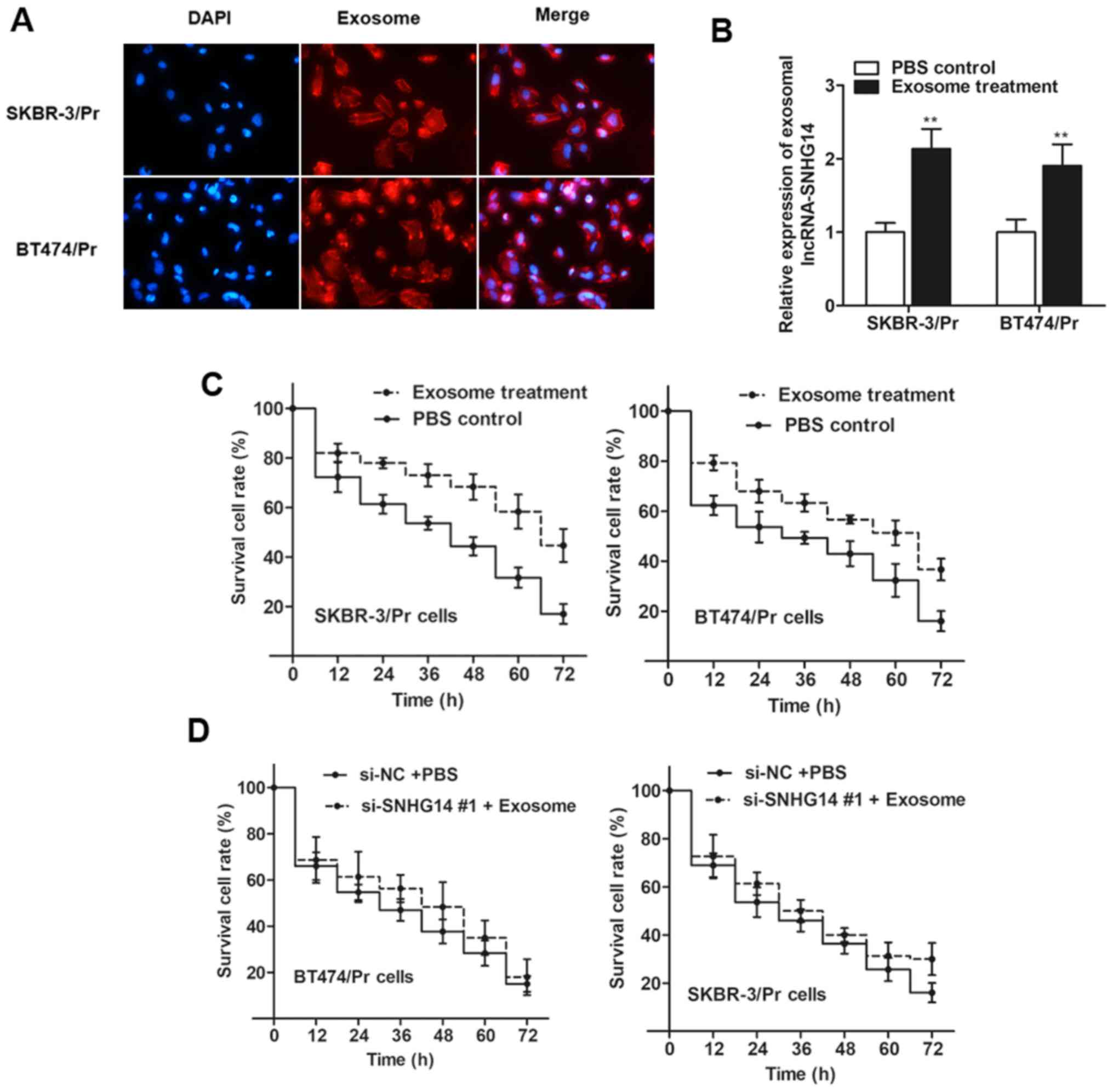
lines incubated with exosomes from SKBR-3/Tr cells (Fig. 5B). Thus, it was verified that
lncRNA-SNHG14-containing exosomes may be taken up by recipient
cells.
Subsequently, the present study sought to determine
whether SKBR-3/Pr and BT474/Pr cells with an elevated exosomal
SNHG14 level displayed increased resistance to trastuzumab compared
with the response of control cells. As expected, the two recipient
cell lines exhibited increased cell viability following treatment
with exosomes compared with control cells (Fig. 5C). However, cell viability
exhibited little difference when recipient cells were transfected
with si-SNHG14 #1 prior to exosomal treatment (Fig. 5D), suggesting that it was the
exosomal SNHG14 that induced trastuzumab resistance.
Exosomal lncRNA-SNHG14 promotes
trastuzumab resistance by activating Bcl-2/apoptosis regulator BAX
(Bax) signaling
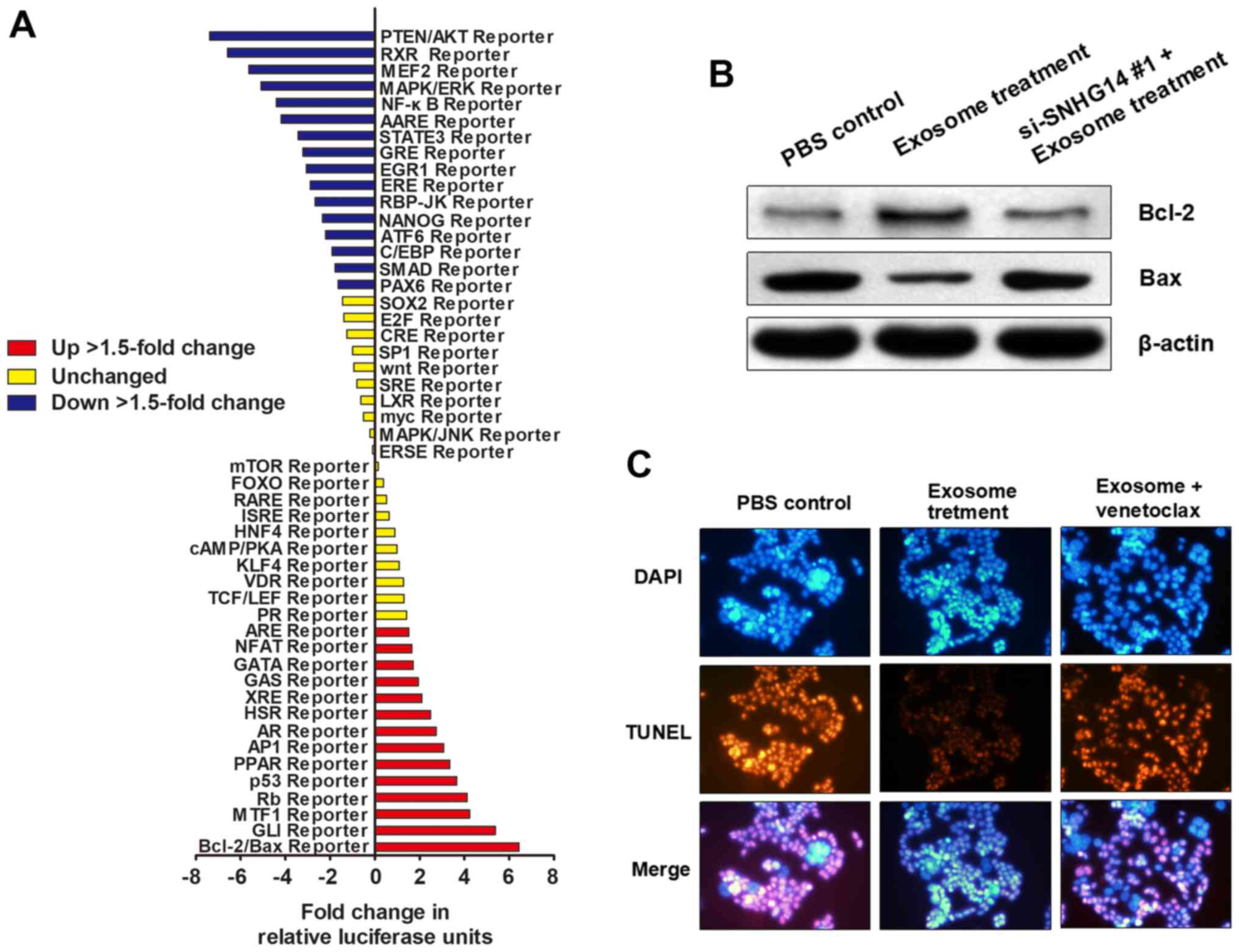
To determine how exosomal lncRNA-SNHG14 contributes
to trastuzumab resistance, a Signal Transduction Reporter Array was
used to simultaneously investigate the activity alterations of 50
canonical signaling pathways in SKBR-3/Pr upon treatment with
exosomes derived from SKBR-3/Tr cells. Notably, Bcl-2/Bax signaling
was identified to be one of the most significantly activated
pathways following treatment with exosomes containing lncRNA-SNHG14
(Fig. 6A), suggesting that
exosomal lncRNA-SNHG14 may promote trastuzumab resistance by
regulating Bcl-2/Bax pathway-mediated cellular apoptosis. Western
blotting demonstrated that exosomal treatment promoted Bcl-2
expression and inhibited Bax expression in SKBR-3/Pr; however,
transfection with si-SNHG14 #1 prior to exosomal treatment markedly
reversed the effect of exosomal treatment, indicating that this
apoptosis-inhibiting effect was induced by exosomal SNHG14
(Fig. 6B). Furthermore, TUNEL
assay showed that exosome treatment inhibited cell apoptosis
levels, but this effect was abrogated by subsequent treatment of
Bcl-2 inhibitor venetoclax (Fig.
6C). Therefore, exosomal lncRNA-SNHG14 may induce trastuzumab
resistance through inhibiting apoptotic proteins and cell apoptosis
via Bcl-2/Bax pathway, however, this needs a further investigation
to reveal specific regulating mode.
Serum exosomal SNHG14 levels are
upregulated in trastuzumab-resistant patients
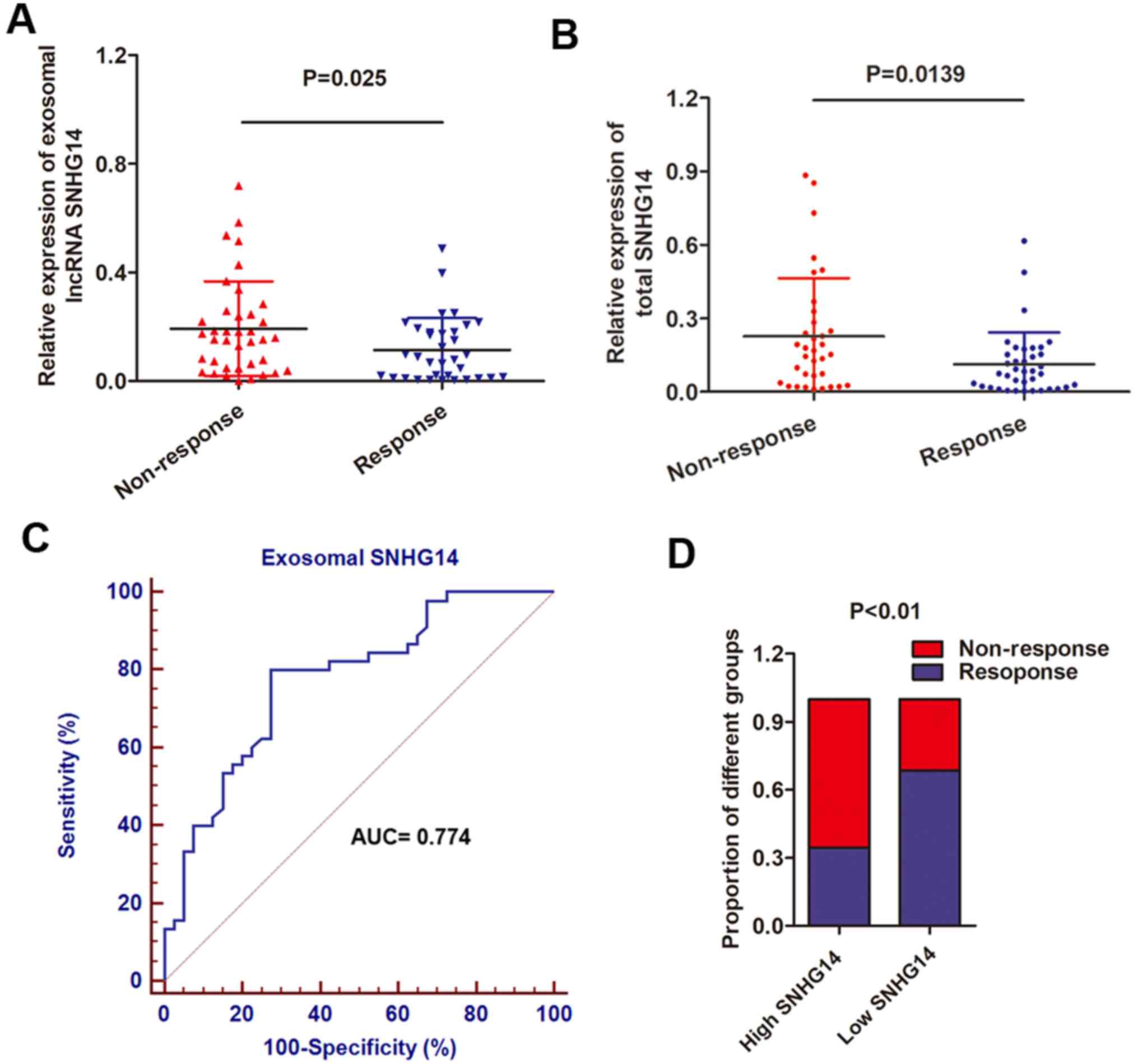
To determine the serum exosomal lncRNA-SNHG14
expression, exosomes were obtained from 72 serum samples from
patients with advanced HER2+ breast cancer who received
single-agent treatment with trastuzumab. Patients were divided into
responding (CR + PR; 38 patients) and non-responding (SD + PD; 34
patients) groups, according to RECIST (version 1.1) (18). It was observed that the serum
exosomal SNHG14 expression level was increased in patients who did
not respond to treatment with trastuzumab compared with those who
experienced a response to trastuzumab (Fig. 7A). In addition, total SNHG14 was
also upregulated in serum samples from non-responding patients when
compared with responding patients (Fig. 7B), further confirming the
upregulation of exosomal SNHG14. To further examine the potential
of circulating exosomal lncRNAs as predictors for breast cancer,
the present study evaluated the association between exosomal SNHG14
levels and clinical characteristics. As presented in Table III, increased expression of
exosomal SNHG14 was markedly associated with distant metastasis,
lymph node metastasis and cardiac toxicity (P<0.01 for all). In
addition, the ROC curve was used to evaluate the diagnostic value
of exosomal lncRNA-SNHG14 in the serum. The ROC analysis
demonstrated an AUC of 0.774, with a diagnostic sensitivity and
specificity reaching 80.0% and 72.5% (95% confidence interval,
0.670-0.857) with the established cut-offs (3.09), respectively
(Fig. 7C). Under this
stratification criteria (3.09), the proportion of patients not
responding to chemotherapy was significantly increased in the high
exosomal SNHG14-expressing group compared with the low group
(Fig. 7D). These results indicated
that exosomal lncRNA-SNHG14 in serum may serve as a potential
diagnostic biomarker for breast cancer.
 | Table IIIClinical characteristics of 72 human
epidermal growth factor receptor-positive patients and the
expression of serum exosomal SNHG14. |
Table III
Clinical characteristics of 72 human
epidermal growth factor receptor-positive patients and the
expression of serum exosomal SNHG14.
| Factors | No. of cases | Serum exosomal
lncRNA SNHG14 (2−∆∆Cq; median ± interquartile
range) | P-value |
|---|
| Age, years | | | 0.887 |
| <50 | 43 | 4.32
(1.24–7.55) | |
| ≥50 | 29 | 4.57
(1.45–7.50) | |
| Tumor size | | | 0.026 |
| <6 cm | 31 | 3.75
(1.24–6.66) | |
| ≥6 cm | 41 | 4.64
(1.66–8.17) | |
| Weight loss | | | 0.154 |
| <3 kg | 42 | 3.98
(2.07–6.66) | |
| ≥3 kg | 30 | 4.15
(1.69–8.65) | |
| Histological
grade | | | 0.037 |
| Well
differentiated | 16 | 3.77
(2.12–6.36) | |
| Moderately
differentiated | 38 | 4.06
(2.07–7.45) | |
| Poorly
differentiated | 18 | 4.37
(2.78–8.65) | |
| Lymph node
metastasis | | | 0.002 |
| No | 16 | 3.36
(0.29–7.38) | |
| Yes | 56 | 4.37
(2.07–8.65) | |
| Distant
metastasis | | | 0.000 |
| No | 40 | 3.77
(1.55–7.46) | |
| Yes | 32 | 4.88
(1.69–8.65) | |
| Cardiac
toxicity | | | 0.006 |
| Yes | 25 | 4.82
(1.08–7.94) | |
| No | 47 | 3.95
(1.37–6.78) | |
In conclusion, lncRNA-SNHG14 promotes trastuzumab
resistance by targeting Bcl-2/Bax signaling, inducing the
suppression of apoptotic proteins expression and inhibition of cell
apoptosis. Moreover, lncRNA-SNHG14 can be packaged into exosomes
and secreted from trastuzumab-resistant breast cancer cells,
transferring resistance to recipient-sensitive cells (Fig. 8).
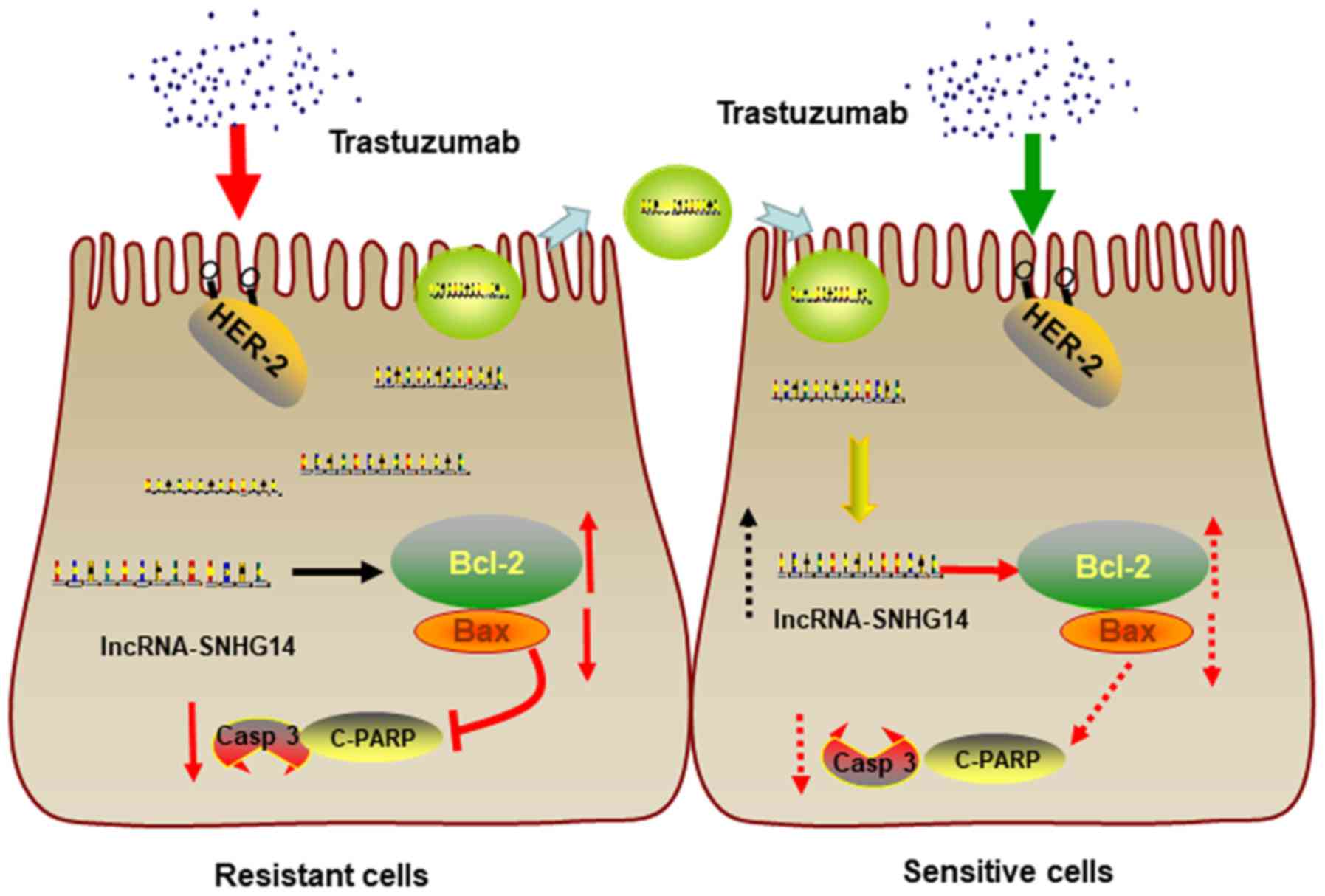 | Figure 8Schematic diagram of exosomal
lncRNA-SNHG14 in breast cancer trastuzumab resistance. In
trastuzumab-resistant breast cancer cells, lncRNA-SNHG14 promotes
trastuzumab resistance by targeting Bcl-2/Bax signaling, inducing
the suppression of apoptotic proteins expression and inhibition of
cell apoptosis. Moreover, lncRNA-SNHG14 can be packaged into
exosomes and secreted from trastuzumab-resistant breast cancer
cells, transferring resistance to recipient-sensitive cells.
SNGH14, small nucleolar RNA host gene 14; lncRNA, long non-coding
RNA; Bcl-2, apoptosis regulator Bcl-2; Bax, apoptosis regulator
BAX; HER-2, human epidermal growth factor receptor 2; Casp 3,
caspase-3; c-PARP, cleaved poly(ADP-ribose) polymerase. |
Discussion
Extensive efforts in the past have contributed to
the understanding of molecular and cellular mechanisms of action of
chemoresistance, one of the principal causes of the failure of
treatment for advanced cancer types. However, little progress has
been made in recent years (28).
Thus, novel molecular signatures appear to hold promise for tumor
characterization and may be used as potential prognostic markers
and treatment targets. To identify potential molecular therapeutic
markers for treatment with trastuzumab, trastuzumab-resistant cell
lines were established, and the functional association between
trastuzumab resistance and specific exosomal lncRNAs was further
investigated. The present data demonstrated that exosomal
lncRNA-SNHG14 was upregulated in trastuzumab-resistant cells.
Treatment with exosomes extracted from trastuzumab-resistant cells
promoted trastuzumab resistance in parental cells.
It is known that patients with breast cancer who
overexpress HER2 are associated with a poor prognosis (29). HER2-targeted therapy for patients
with HER2-positive breast cancer has markedly improved survival
rates and reduced mortality in recent years (30). HER2 gene amplification was first
associated with worse clinical outcomes in the late 1980s by Pegram
et al (31); it was advised
that HER2 overexpression may be an effective biomarker for early
diagnostic monitoring and clinical prognosis. Additional studies in
the USA further revealed that residents in Asian-Pacific regions
had high occurrence rates of diagnosis with HER2-positive status,
with a poorer prognosis in comparison with other regions (32-34).
Trastuzumab has proven efficacy as first-line treatment of advanced
HER2+ breast cancer. However, the initial benefit does
not last long prior to the occurrence of conversion, resulting in
acquired resistance (35).
Therefore, breakthroughs are required in the search for effective
therapeutic targets to overcome acquired trastuzumab resistance,
particularly for patients with HER2+ sites.
Exosomes are nano-sized vesicles secreted upon the
fusion of vesicular-like entities with plasma membranes in a large
number of cell types (36).
Emerging evidence has revealed the unique properties of exosomes,
including their ability to embed specific microRNAs, circular RNAs
or lncRNAs, their stability and easy detection in the circulatory
system (37). Exosomes have been
identified to be a means of information exchange between different
types of cells, through the transfer of constituents, including
lncRNAs (38), which are known to
function as important activators or inhibitors to regulate gene
expression, and to be involved in a variety of biological processes
(39).
In recent years, it has been accepted that lncRNAs
may be protected by exosomes from degradation in the circulation
and may be useful for monitoring cancer in the early stages
(40,41). Therefore, microarray analysis was
performed to identify the dysregulated exosomal lncRNAs in
trastuzumab-resistant cells compared with parental cells. By using
a two-steps approach, exosomal lncRNA-SNHG14 was identified to have
potential involvement in trastuzumab resistance. lncRNA-SNHG14,
alternatively termed UBE3A-ATS, is located on chromosome 15q11.2.
SNHG14 may overlap with the entire UBE3A gene and promoter, thus
inhibiting the expression of UBE3A and causing neurogenetic
disorders, including Angelman syndrome (42). In cancer research, Liu et al
(43) reported that lncRNA-SNHG14
may act as a competing endogenous RNA to promote the migration and
invasion of clear cell renal cell carcinoma by regulating neural
Wiskott-Aldrich syndrome protein. For the other seven preliminarily
identified lncRNAs, their roles are largely unknown in cancer
progression. In one case, Zhang et al (44) demonstrated that cat eye syndrome
chromosome region, candidate 7 was significantly associated with
overall survival in hepatocellular carcinoma; another study
reported by Yue et al (45)
suggested that LOC285627 was highly expressed in ankylosing
spondylitis and Vogt-Koyanagi-Harada disease.
To determine whether the ectopic expression of
exosomal lncRNA-SNHG14 mediates trastuzumab resistance, gain and
loss-function experiments were performed in the present study. As
expected, it was observed that knockdown of lncRNA-SNHG14 in
trastuzumab-resistant cells promoted cellular apoptosis, while
treatment with exosomes extracted from the culture medium of
resistant cells markedly reduced trastuzumab-induced cell death.
Furthermore, it was demonstrated that exosomal lncRNA-SNHG14
induced trastuzumab resistance by targeting the Bcl-2/Bax apoptosis
signaling pathway. These results suggested that exosomal
lncRNA-SNHG14 may promote trastuzumab resistance in breast cancer
cells primarily by regulating apoptosis-associated proteins. Based
on the functional observations, the exosomal SNHG14 level in
clinical serum samples was subsequently determined. A number of
attempts have been made to use lncRNAs in serum or plasma as useful
predictors in breast cancer (46,47).
However, these potential tumor biomarkers are frequently in
relatively low abundance and degradation occurs easily. lncRNAs are
enriched and more stable in the circulatory exosome system and are
protected from RNase degradation. The identification of exosomal
lncRNAs in bodily fluids suggested their predictive application in
clinical diagnosis or prognosis for different types of cancer
(48). Emerging evidence has
uncovered the unique properties of exosomes, including their
ability to embed specific lncRNAs, their stability and their easy
detection in the circulatory system (49,50).
As expected, the present data clearly demonstrated that the
exosomal lncRNA-SNHG14 level was upregulated in
trastuzumab-resistant patients, and was associated with the
trastuzumab response. Notably, exosomal SNHG14 was correlated with
cardiac toxicity. Cardiac toxicity is an important side effect of
treatment with trastuzumab; the upregulation of SNHG14 in patients
with resistance to trastuzumab accompanied by cardiac toxicity
suggested that SNHG14 may be involved in trastuzumab resistance and
cardiac toxicity, and furthermore, that cardiac toxicity may
contribute to trastuzumab resistance. However, this requires
further investigation.
There are a number of limitations to the present
study. First, the functional roles of the seven preliminarily
identified lncRNAs by microarray screening were not deeply
investigated. In the future, the present study may be extended to
identify their functions during cancer progression and
chemo-resistance. Second, a deeper understanding of the regulatory
roles of exosomal SNHG14 in Bcl-2/Bax pathway and trastuzumab
resistance is required. Trastuzumab resistance has been widely
studied and various pathways have been indicated to be involved
(51); however, whether these
pathways are associated with each other and co-regulated remains
unknown. Third, a trastuzumab-resistant xenograft model in nude
mice was not produced to validate the data obtained from the in
vitro studies. This is in development at present and the
present results may be tested in a trastuzumab-resistant model in
the future, increasing the validity of these data.
In conclusion, the present findings suggested that
the exosome-mediated transfer of lncRNA-SNHG14 induces trastuzumab
resistance in breast cancer cells, and exosomal lncRNA-SNHG14 in
human serum may be considered to be a potential diagnostic
biomarker for breast cancer, enhancing the clinical benefits of
trastuzumab therapy.
Acknowledgments
Not applicable.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vu T and Claret FX: Trastuzumab: Updated
mechanisms of action and resistance in breast cancer. Front Oncol.
2:622012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Robidoux A, Tang G, Rastogi P, Geyer CE
Jr, Azar CA, Atkins JN, Fehrenbacher L, Bear HD, Baez-Diaz L,
Sarwar S, et al: Lapatinib as a component of neoadjuvant therapy
for HER2-positive operable breast cancer (NSABP protocol B-41): An
open-label, randomised phase 3 trial. Lancet Oncol. 14:1183–1192.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Narayan M, Wilken JA, Harris LN, Baron AT,
Kimbler KD and Maihle NJ: Trastuzumab-induced HER reprogramming in
'resistant' breast carcinoma cells. Cancer Res. 69:2191–2194. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wolff AC, Hammond ME, Schwartz JN, Hagerty
KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer
A, et al American Society of Clinical Oncology/College of American
Pathologists: American Society of Clinical Oncology/College of
American Pathologists guideline recommendations for human epidermal
growth factor receptor 2 testing in breast cancer. Arch Pathol Lab
Med. 131:18–43. 2007.PubMed/NCBI
|
|
7
|
Donepudi MS, Kondapalli K, Amos SJ and
Venkanteshan P: Breast cancer statistics and markers. J Cancer Res
Ther. 10:506–511. 2014.PubMed/NCBI
|
|
8
|
ENCODE Project Consortium: An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang S, Qin C, Cao G, Xin W, Feng C and
Zhang W: Systematic analysis of long non-coding RNAs in the
senescence-accelerated mouse prone 8 brain using RNA sequencing.
Mol Ther Nucleic Acids. 5:e3432016. View Article : Google Scholar
|
|
11
|
Brockdorff N: Non-coding RNA and Polycomb
recruitment. RNA. 19:429–442. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li P, Zhang X, Wang H, Wang L, Liu T, Du
L, Yang Y and Wang C: MALAT1 is associated with poor response to
oxaliplatin-based chemotherapy in colorectal cancer patients and
promotes chemo-resistance through EZH2. Mol Cancer Ther.
16:739–751. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee TH, D'Asti E, Magnus N, Al-Nedawi K,
Meehan B and Rak J: Microvesicles as mediators of intercellular
communication in cancer - the emerging science of cellular
'debris'. Semin Immunopathol. 33:455–467. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Théry C, Ostrowski M and Segura E:
Membrane vesicles as conveyors of immune responses. Nat Rev
Immunol. 9:581–593. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pefanis E, Wang J, Rothschild G, Lim J,
Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, et al:
RNA exosome-regulated long non-coding RNA transcription controls
super-enhancer activity. Cell. 161:774–789. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kourembanas S: Exosomes: Vehicles of
intercellular signaling, biomarkers, and vectors of cell therapy.
Annu Rev Physiol. 77:13–27. 2015. View Article : Google Scholar
|
|
18
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar
|
|
19
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi SJ, Wang LJ, Yu B, Li YH, Jin Y and
Bai XZ: LncRNA-ATB promotes trastuzumab resistance and
invasion-metastasis cascade in breast cancer. Oncotarget.
6:11652–11663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Soo CY, Song Y, Zheng Y, Campbell EC,
Riches AC, Gunn-Moore F and Powis SJ: Nanoparticle tracking
analysis monitors microvesicle and exosome secretion from immune
cells. Immunology. 136:192–197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu SY, Huang X and Cheong KL: Recent
advances in marine algae polysaccharides: Isolation, structure, and
activities. Mar Drugs. 15:152017. View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Baker SC, Bauer SR, Beyer RP, Brenton JD,
Bromley B, Burrill J, Causton H, Conley MP, Elespuru R, Fero M, et
al: The External RNA Controls Consortium: A progress report. Nat
Methods. 2:731–734. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schindelin J, Rueden CT, Hiner MC and
Eliceiri KW: The ImageJ ecosystem: An open platform for biomedical
image analysis. Mol Reprod Dev. 82:518–529. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kogure T, Lin WL, Yan IK, Braconi C and
Patel T: Intercellular nanovesicle-mediated microRNA transfer: A
mechanism of environmental modulation of hepatocellular cancer cell
growth. Hepatology. 54:1237–1248. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xin H, Li Y, Buller B, Katakowski M, Zhang
Y, Wang X, Shang X, Zhang ZG and Chopp M: Exosome-mediated transfer
of miR-133b from multipotent mesenchymal stromal cells to neural
cells contributes to neurite outgrowth. Stem Cells. 30:1556–1564.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Z, Wang N, Li W, Liu P, Chen Q, Situ
H, Zhong S, Guo L, Lin Y, Shen J, et al: Caveolin-1 mediates
chemoresistance in breast cancer stem cells via β-catenin/ABCG2
signaling pathway. Carcinogenesis. 35:2346–2356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu D and Hung MC: Overexpression of ErbB2
in cancer and ErbB2-targeting strategies. Oncogene. 19:6115–6121.
2000. View Article : Google Scholar
|
|
30
|
Berry DA, Cronin KA, Plevritis SK, Fryback
DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD and
Feuer EJ; Cancer Intervention and Surveillance Modeling Network
(CISNET) Collaborators: Effect of screening and adjuvant therapy on
mortality from breast cancer. N Engl J Med. 353:1784–1792. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pegram MD, Finn RS, Arzoo K, Beryt M,
Pietras RJ and Slamon DJ: The effect of HER-2/neu overexpression on
chemotherapeutic drug sensitivity in human breast and ovarian
cancer cells. Oncogene. 15:537–547. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Parise C and Caggiano V: Disparities in
the risk of the ER/PR/HER2 breast cancer subtypes among Asian
Americans in California. Cancer Epidemiol. 38:556–562. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Telli ML, Chang ET, Kurian AW, Keegan TH,
McClure LA, Lichtensztajn D, Ford JM and Gomez SL: Asian ethnicity
and breast cancer subtypes: A study from the California Cancer
Registry. Breast Cancer Res Treat. 127:471–478. 2011. View Article : Google Scholar
|
|
34
|
Kurian AW, Fish K, Shema SJ and Clarke CA:
Lifetime risks of specific breast cancer subtypes among women in
four racial/ethnic groups. Breast Cancer Res. 12:R992010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Denzer K, Kleijmeer MJ, Heijnen HF,
Stoorvogel W and Geuze HJ: Exosome: From internal vesicle of the
multivesicular body to intercellular signaling device. J Cell Sci.
113:3365–3374. 2000.PubMed/NCBI
|
|
37
|
van den Boorn JG, Dassler J, Coch C,
Schlee M and Hartmann G: Exosomes as nucleic acid nanocarriers. Adv
Drug Deliv Rev. 65:331–335. 2013. View Article : Google Scholar
|
|
38
|
Kucharzewska P and Belting M: Emerging
roles of extracellular vesicles in the adaptive response of tumour
cells to microenvironmental stress. J Extracell Vesicles. 2:22013.
View Article : Google Scholar
|
|
39
|
Mercer TR and Mattick JS: Structure and
function of long non-coding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang H, Fu H, Xu W and Zhang X: Exosomal
non-coding RNAs: A promising cancer biomarker. Clin Chem Lab Med.
54:1871–1879. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Q, Shao Y, Zhang X, Zheng T, Miao M,
Qin L, Wang B, Ye G, Xiao B and Guo J: Plasma long non-coding RNA
protected by exosomes as a potential stable biomarker for gastric
cancer. Tumour Biol. 36:2007–2012. 2015. View Article : Google Scholar
|
|
42
|
Sadikovic B, Fernandes P, Zhang VW, Ward
PA, Miloslavskaya I, Rhead W, Rosenbaum R, Gin R, Roa B and Fang P:
Mutation Update for UBE3A variants in Angelman syndrome. Hum Mutat.
35:1407–1417. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu G, Ye Z, Zhao X and Ji Z: SP1-induced
up-regulation of lncRNA SNHG14 as a ceRNA promotes migration and
invasion of clear cell renal cell carcinoma by regulating N-WASP.
Am J Cancer Res. 7:2515–2525. 2017.
|
|
44
|
Zhang J, Fan D, Jian Z, Chen GG and Lai
PB: Cancer specific long non-coding RNAs show differential
expression patterns and competing endogenous RNA potential in
hepatocellular carcinoma. PLoS One. 10:e01410422015. View Article : Google Scholar
|
|
45
|
Yue Y, Zhang J, Yang L, Liu S, Qi J, Cao
Q, Zhou C, Wang Y, Kijlstra A, Yang P, et al: Association of long
non-coding RNAs polymorphisms with ankylosing spondylitis,
Vogt-Koyanagi-Harada disease, and Behcet's disease. Invest
Ophthalmol Vis Sci. 59:1158–1166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tan Q, Yu Y, Li N, Jing W, Zhou H, Qiu S,
Liang C, Yu M and Tu J: Identification of long non-coding RNA 00312
and 00673 in human NSCLC tissues. Mol Med Rep. 16:4721–4729. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tan Q, Zuo J, Qiu S, Yu Y, Zhou H, Li N,
Wang H, Liang C, Yu M and Tu J: Identification of circulating long
non-coding RNA GAS5 as a potential biomarker for non-small cell
lung cancer diagnosisnon-small cell lung cancer, long non-coding
RNA, plasma, GAS5, biomarker. Int J Oncol. 50:1729–1738. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kim KM, Abdelmohsen K, Mustapic M,
Kapogiannis D and Gorospe M: RNA in extracellular vesicles. Wiley
Interdiscip Rev RNA. 8:82017. View Article : Google Scholar
|
|
49
|
Boukouris S and Mathivanan S: Exosomes in
bodily fluids are a highly stable resource of disease biomarkers.
Proteomics Clin Appl. 9:358–367. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Keller S, Ridinger J, Rupp AK, Janssen JW
and Altevogt P: Body fluid derived exosomes as a novel template for
clinical diagnostics. J Transl Med. 9:862011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Barok M, Joensuu H and Isola J:
Trastuzumab emtansine: Mechanisms of action and drug resistance.
Breast Cancer Res. 16:2092014. View Article : Google Scholar : PubMed/NCBI
|