Introduction
Lung carcinoma has the highest cancer-associated
mortality and incident rates in the developed world (1). There were 1,800,800 new diagnosed
cases of lung cancer (12.8% in total), with 1,600,000 individuals
succumbing to mortality in 2012. The number of newly diagnosed lung
cancer increased to 14%, accounting for 1/4 cases of
cancer-associated mortality by 2016 (1). The annual increase of lung cancer
cases is almost 1,300,000, and ~80–85% of these are diagnosed as
non-small cell lung cancer (NSCLC), including lung adenocarcinoma,
squamous cell lung carcinoma and large cell lung carcinoma
(2,3). Molecular target therapy is widely
used in the treatment of lung cancer, including epidermal growth
factor receptor tyrosine kinase inhibitors and anaplastic lymphoma
kinase fusion gene inhibitors and other drugs, which can
effectively improve survival rate in specific individual patients
with lung cancer at present. However, the five-year survival rate
of NSCLC remains <20% (4). The
development of lung cancer treatment is limited due to drug
resistance and the recurrence of malignancy.
Previous studies investigating the mechanism of the
development of lung cancer have focused mainly on the Janus
kinase-signal transducer and activator of transcription, mammalian
target of rapamycin, and Wnt signaling pathways. MicroRNA (miRNA or
miR), which is ~19–25 bp in length, is a class of endogenous
non-encoding RNAs with regulatory function found in eukaryotes.
They can degrade or inhibit the expression and translation of
target genes through complementary pairing with the 3′ untranslated
region (3′UTR) of mRNAs, and are involved in the regulation of cell
proliferation, differentiation and individual development (5). Increasing evidence indicates that
miRNAs are also important in the development of NSCLC (6–8). The
serum miRNAs can be used as key biomarkers for the early diagnosis
of NSCLC (9). A numbers of reports
have shown that miRNAs are usually localized in tumor-associated
fragile sites, where they can affect the occurrence and development
of tumors by regulating the corresponding signal pathway as
oncogenes or tumor suppressor genes (10). The miRNA (miR)-200 family can be
divided into the miR-200a subfamily, including miR-200a and
miR-141, and the miR-200 subfamily, including miR200b/c and miR429
(11,12). Studies have shown that the
miR-200b/200c/429 subfamily can inhibit the migration of
hepatocellular carcinoma cells through affecting the cell
cytoskeleton and cell adhesion mediated by Rho/Rho kinase (13). Studies on miR-200 family in lung
cancer have mainly focused on their inhibitory effect. A previous
study showed that miR-200 suppressed lung adenocarcinoma cell
invasion, metastasis and lung tumorigenesis by targeting Fms
related tyrosine kinase 1 (14).
In addition, miR-200 suppressed cancer cell growth, migration and
invasion, and inhibited the tumorigenesis and metastasis of lung
cancer cells though downregulating bone morphogenetic protein
(BMP4). These findings indicated that BMP4 and miR-200s may be
suitable therapeutic targets for the treatment of lung cancer
(15). miR-200 is also important
in lung cancer resistance. The latest findings have shown that
miR-200c may be involved in regulating paclitaxel resistance
through Cathepsin L (CTSL)-mediated epithelial-mesenchymal
transition (EMT) in A549 cells, and that CTSL and micRNA-200c were
reciprocally linked in a feedback loop (16). In addition, the expression of
miR-200 was reported to be significantly lower in
nintedanib-resistant cell lines than in nintedanib-sensitive cell
lines, and the induction of miR-200 enhanced sensitivity to
nintedanib in the nintedanib-resistant A549 cells (17). However, the association between
RhoE and the miR-200 family in NSCLC remains to be elucidated.
RhoGTPase is a class of 20–30 kDa GTP binding
proteins, belonging to the main members of the Ras superfamily
(18). Several important findings
have been revealed regarding the function of the Rho family members
in tumors. RhoGTPase can induce the activation of transcription
factors, including c-Jun and nuclear factor-κB through a signal
transduction series, causing the reorganization of actin and cell
cycle progression or inhibition. Rho family GTPase 3 (RhoE) is a
member of the RhoGTPase family, belonging to the RND subfamily,
also known as RND3. Unlike other family members, it lacks GTPase
activity, so it can only be combined, but cannot be hydrolyzed to
GTP. A previous investigation of the mechanism of RhoE showed that
it can inhibit RhoA/Rho-associated protein kinase signaling through
binding to p190 GTPase-activating protein, which can reduce the
levels of RhoA-GTP (19). The
majority of studies have shown that RhoE may be a candidate tumor
suppressor gene. Previous studies, including ours, have revealed
that the tumor suppressor gene p53 can regulate the activity of
RhoE by binding to its promoter (20,21).
However, several studies have found that RhoE is relatively
overexpressed in NSCLC compared with other tumors. In addition, the
expression of RhoE may serve as an unfavorable prognostic factor in
patients with NSCLC (22,23). However, the molecular mechanism
underling this dysregulation remains to be fully elucidated.
In the present study, it was found that miRNA 200b/c
was downregulated, whereas RhoE was upregulated in NSCLC tissues.
miRNA 200b/c was found to bind to the RhoE 3′UTR and inhibit its
expression. miR-200b/c regulated the migration and invasion of
NSCLC cells through directly regulating the expression of RhoE.
Materials and methods
Cell culture
The A549 and NCI-H1299 human NSCLC cells were
obtained from the Cell Biology Institute of Chinese Academy of
Sciences (Shanghai, China). The A549 cell line was maintained in
DMEM (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.), and the NCI-H1299 cell line was maintained in RPMI-1640
medium (Invitrogen; Thermo Fisher Scientific, Inc.) with 10% FBS,
100 U/ml penicillin G sodium, and 100 µg/ml streptomycin
sulfate (Sigma; EMD Millipore, Billerica, MA, USA). The cells were
cultured in a 37°C incubator containing 5% CO2.
Plasmids and transfection
The pCDNA3.1-RhoE (pCDNA3.1 was obtained
(Invitrogen; Thermo Fisher Scientific, Inc.) and PRL-TK (Promega
Corporation, Madison, WI, USA) plasmids were stored in our
laboratory. The 3′UTR of the human RhoE gene (NM_001254738) was
amplified from human genomic DNA by polymerase chain reaction
(PCR), and cloned into the XbaI site of the pGL3-control
vector (Promega Corporation), following which its sequence was
verified and it was termed pGL3-RhoE-wt. The PCR primers used for
amplifying the RhoE 3′UTR were as follows:
5′-GCTCTAGACTCCCCTTAAATCGTG-3′ (forward) and 5′-GCTCTAGAACTGCGGA
AACATTGAC-3′ (reverse). In addition, site-directed mutagen-esis of
the miR-200b/c target site in the RhoE 3′UTR was performed using a
site-directed mutagenesis kit (Takara Bio, Inc., Otsu, Japan), with
pGL3-RhoE-wt as a template, and termed pGL3-RhoE-mut, respectively.
The pGL3-RhoE-mut primers were as follows:
5′-GAACGTACAAAATAGCTGG-3′ (forward) and 5′-CCCAAAGCATAATGCTGCAT-3′
(reverse). Small interfering (si)RNA sequences against RhoE
(Si-RhoE: 5′-AACAGATTGGAGCAGCTACdTdT-3′), control oligo, and
hsa-miR-200b/c were constructed and chemically synthesized by
Shanghai Genepharma Co., Ltd. (Shanghai, China). All plasmids and
RNA oligonucleotides were transfected into cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol.
Dual-luciferase report assay
The cells (1×105 per well) were seeded in
24-well plates 1 day prior to transfection. For the dual-luciferase
assay, the cells were transfected with 0.3 µg of
pGL3-RhoE-wt-luc or pGL3-RhoE-mut-luc plasmids, 60 nM control miRNA
or miR-200b/c, together with 0.1 µg PRL-TK in 24-well plates
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.). The cells were harvested 48 h following transfection and
lysed with lysis buffer. Firefly and Renilla were detected
by the dual-luciferase reporter assay system kit (Promega
Corporation). The lucif-erase activity was determined with the
Synergy2 instrument (BioTek Instruments, Inc., Winooski, VT, USA)
equipped with Gen5 1.04.5 software. The gene expression was
normalized by Renilla luciferase (pRL-TK) and reported as
relative luciferase activity. Three independent experiments were
performed in triplicate. The results are presented as the mean ±
standard deviation of data from three dependent experiments.
Reverse transcription (RT)-PCR and
quantitative PCR (RT-qPCR) assays
Total RNA from the cells was extracted using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. The contaminated genomic DNA was
eliminated and total RNA was reverse transcribed into cDNA with the
Prime Script RT Reagent kit with gDNA Eraser (Takara Bio, Inc.).
Subsequently, PCR was performed on a PCR system (IQ5), using
Bio-Rad iQ SYBR-Green Supermix (both from Bio-Rad Laboratories,
Inc., Hercules, CA, USA) as follows: A total of 2 µl cDNA
template was brought to a volume of 25 µl containing 12.5
µl 2X SYBR premix Ex TaqII, 1 µl of both sense and
antisense primer and 8.5 µl H2O. Amplification
was performed as the following conditions: One cycle of 95°C for 30
sec was performed to synthesize the cDNA. Forty cycles of 95°C for
5 sec, 60°C for 30 sec for PCR reaction, and a final melt curve
analysis of 65°C for 15 sec.
The primers used for specific RhoE PCR were as
follows: Forward, 5′-AAGATAGTTGTGGTGGGAGA-3′ and reverse,
5′-CATAGTAAGGAGAACCCGAA-3′. The primers used for specific GAPDH PCR
were as follows: Forward, 5′-CAAGGCCAACCGCGAGAA-3′ and reverse,
5′-CCCTCGTAGATGGGCACAGT-3′. The primers of miR-200b: Forward,
5′-TAATACTGCCTGGTAATGATGA-3′ and reverse,
5′-ATCATTACCAGGCAGTATAAAT-3′; miR-200c forward,
5′-TAATACTGCCGGGTAATGATGGA-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′ were synthesized by Genwiz,
Inc. (Suzhou, China). The relative expression levels were
determined using the 2−ΔΔCq method (24).
Cell growth assay
The A549 and NCI-H1299 cells were seeded in 96-well
plates 24 h prior to transfection. All cells were transfected
respectively with control oligos, mimics and siRhoE. Relative cell
growth was measured using the Cell Counting Kit-8 (CCK-8; Dojindo
Molecular Technologies, Inc., Kumamoto, Japan). Three independent
experiments were performed in triplicate.
Cell Transwell migration and invasion
assays
At 24 h post-transfection, the A549 cells and H1299
cells were digested by trypsin, and then collected and suspended
with serum-free culture medium. The cells (3×104) were
suspended in 0.2 ml of serum-free culture medium and plated in the
top chamber of a membrane insert (Transwell; 24-well insert; pore
size 8-µm; BD Falcon; BD Biosciences, Franklin Lakes, NJ,
USA). Ten percent FBS medium was placed in the bottom of the
24-well plate as an attractant. The cells were incubated for 48 h,
and cells that did not invade through the membrane were removed
using a cotton swab. Those cells that had invaded the membrane were
stained with crystal violet on the opposite surface of the
Transwell for the visualization (Nikon ECLIPSE 2000-U; Nikon,
Melville, NY, USA) of invasion. The assays were performed three
times.
The cell migration assay was almost the same as the
invasion assay, but used the extracellular matrix (1:4, BD
Biosciences) Matrigel-coated Transwell. The assays were repeated
three times.
Cell cycle analysis
At 48 h post-transfection, the cells were digested
by trypsin, and washed three times with ice-cold PBS, and then
suspended and fixed with 70% ice-cold ethanol overnight at 4°C. The
fixed cells were rehydrated in PBS and stained with PI/RNase at
37°C for 15 min following the manufacturer's protocol of the Cell
Cycle Detection kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing,
China). Cell cycle analyses were performing by
fluorescence-activated cell sorting analysis with a flow cytometer
(BD FACSCalibur; BD Biosciences).
Western blot analysis
The total protein samples were extracted with RIPA
buffer containing a cocktail of protease inhibitors from the
transfected cells. The protein samples were quantified by a BCA
protein assay kit (Pierce, Rockford, IL, USA). Equal amounts of
proteins (30–40 µg) were fractionated by 12% SDS-PAGE
(Bio-Rad Laboratories, Inc.), and transferred onto NC membranes
(EMD Millipore). The membranes were blocked by 5% skim milk for 1 h
and probed with antibodies against human RND3 (1:2,000, 05-723; EMD
Millipore), human N-cadherin (1:4,000, 2447-1; Epitomics),
E-cadherin (1:1,000, ab1416; Abcam), vimentin (1:4,000, 2862-1),
phosphorylated Akt (473; 1:4,000, 2118-1) (both from Epitomics),
Akt (1:1,000, ab8805), phosphorylated extracellular
signal-regulated kinase (Erk)1/2 (1:2,000, ab214362), Erk1/2
(1:2,000, ab17942) (all from Abcam) and GAPDH (1:1,000, G8795;
Sigma; AB2302; EMD Millipore). Fluorescence-conjugated anti-mouseor
rabbit IgG (P/N925-32210; P/N925-68071; 1:10,000, LI-COR
Biotechnology, Lincoln, NE, USA) were used as the secondary
antibodies, and the band intensities were quantified using the
LI-COR Odyssey infrared imaging system (LI-COR Biotechnology). The
analyses were repeated three times.
Immunohistochemistry (IHC)
Human lung adenocarcinoma tissues were obtained from
the Disease Tissue Specimen Bank of West China Hospital, Sichuan
University, from 40 cases of lung carcinoma, including normal
control and tumor tissues. All patients signed an informed consent
before tissue collection by the Disease Tissue Specimen Bank of
West China Hospital, and the study was approved by the Ethics
Committee of Sichuan University. The clinicopathological grades of
all tissues were evaluated using World Health Organization
standards. The IHC experiments were performed using the standard
avidin-biotin complex immunoperoxidase method. The slides were
incubated with anti-RhoE monoclonal antibody (1:200; Abcam;
ab79999), human N-cadherin (1:200; Epitomics; 2447-1), E-cadherin
(1:100; Abcam; ab1416), vimentin (1:200; Epitomics; 2862-1). PBS
was used as a negative control instead of RhoE antibody. The ratio
of positive cells per specimen was evaluated quantitatively and
scored using the thirteen score method: 0 for staining ≤1%, 1 for
staining of 2–25%, 2 for staining of 26–50%, 3 for staining of
51–75%, and 4 for staining >75% of the cells examined. Staining
intensity was graded as follows: 0, no signal; 1, weak; 2,
moderate; and 3, strong staining. The two scoring systems were
utilized to evaluate the levels of protein expression and a total
score of 0–12 was finally calculated and graded (20).
Bioinformatics analyses
Bioinformatic analysis was performed for searching
putative miRNAs predicted to target RhoE coding or non-coding
sequences using PicTar (https://pictar.mdc-berlin.de).
Statistical analysis
A paired-samples t-test was used to compare the
staining scores of the IHC samples. Characterizing the phenotypes
of cells were analyzed using one-way analysis of variance(ANOVA)
and Dunnett's post test. Data analysis was performed using SPSS for
Windows version 14.0 (SPSS, Inc., Chicago, IL, USA). The test level
of α was 0.05. P<0.05 was considered to indicate a statistically
significant difference.
Results
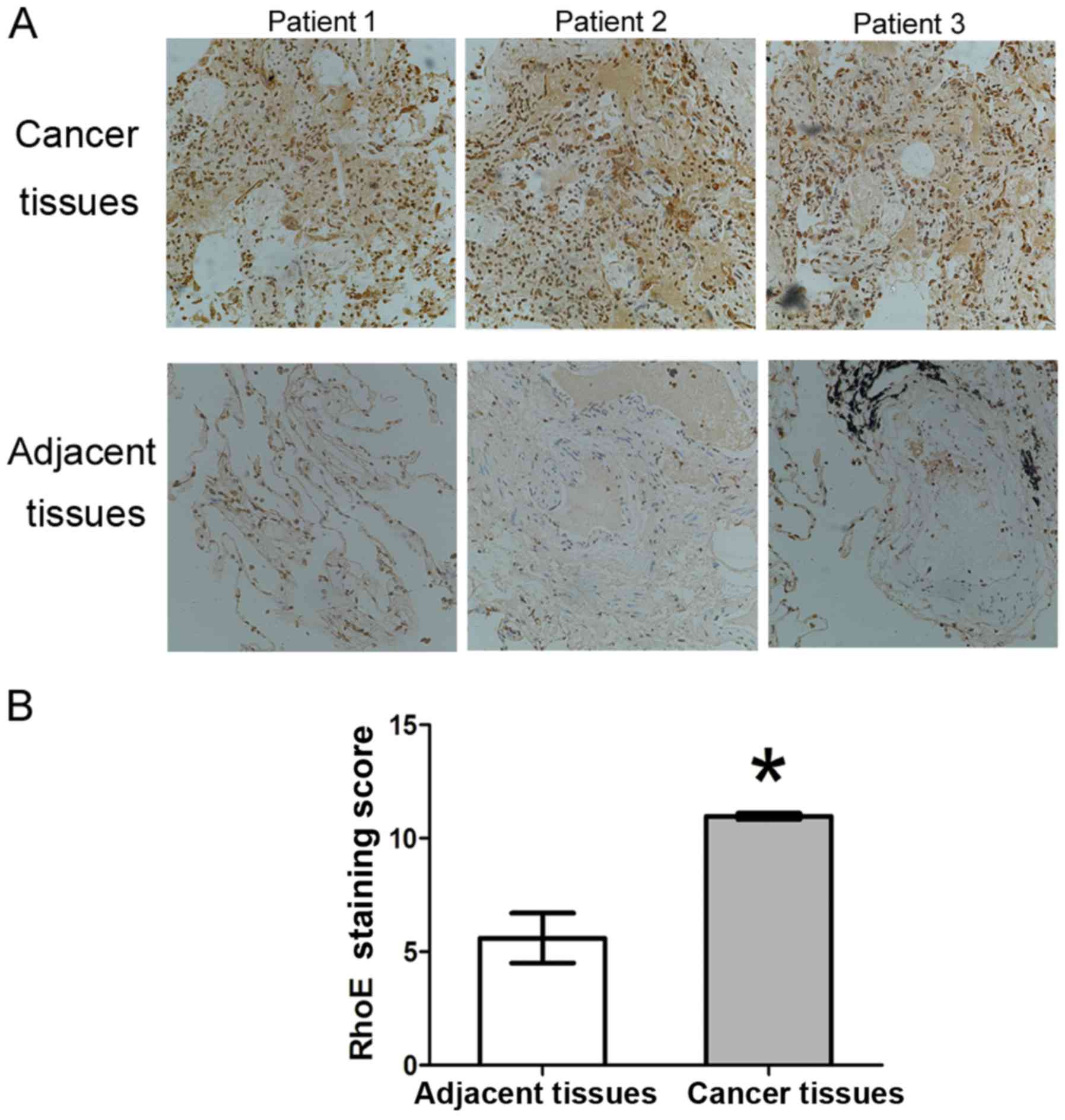
IHC analysis of the expression of RhoE in
human lung adenocarcinoma tissues
In our previous study, it was shown that the
expression of RhoE was decreased to different degrees in gastric
cancer, breast cancer and colorectal cancer tissues, and its
expression was likely regulated by epigenetic modification
(20).
In order to examine the protein expression of RhoE
in human lung adenocarcinoma tissues, 40 lung adenocarcinoma
tissues were matched with adjacent normal tissues, and the tissue
information of patients, including age, gender, tumor size,
distance-metastasis and more advanced tumor-node-metastasis stage,
are shown in Table I. IHC was used
to detect the expression of RhoE in the tissues, and the results
showed that RhoE was expressed at a higher level in tumor tissues
than that in the adjacent normal tissues (Fig. 1A). RhoE was expressed in the
tissues of all 40 cases of lung adenocarcinoma and the average
staining score was 10.91±0.18 (positive ratio of 100%), whereas the
average staining scores of the corresponding adjacent tissues was
5.86±1.27 (positive ratio of 60%). There were significant
differences in the staining score and positive rate between the
tumor tissues and adjacent tissues (P<0.005) (Fig. 1B). Therefore, RhoE was
overexpressed in the human lung adenocarcinoma tissues.
 | Table IClinicopathological features of
patients with non-small cell lung cancer. |
Table I
Clinicopathological features of
patients with non-small cell lung cancer.
| Clinical
characteristic | Total number |
|---|
| Age (years) | |
| >60 | 20 |
| ≤60 | 20 |
| Sex | |
| Male | 26 |
| Female | 14 |
| Invasion depth | |
| T1 | 10 |
| T2 | 29 |
| T3 | 1 |
| T4 | 0 |
| Lymph node
metastasis | |
| 0 | 16 |
| 1 | 7 |
| 2 | 17 |
| 3 | 0 |
| Metastasis | |
| Yes | 2 |
| No | 38 |
| TNM stage | |
| IA | 4 |
| IB | 10 |
| IIA | 7 |
| IIB | 0 |
| IIIA | 17 |
| IV | 2 |
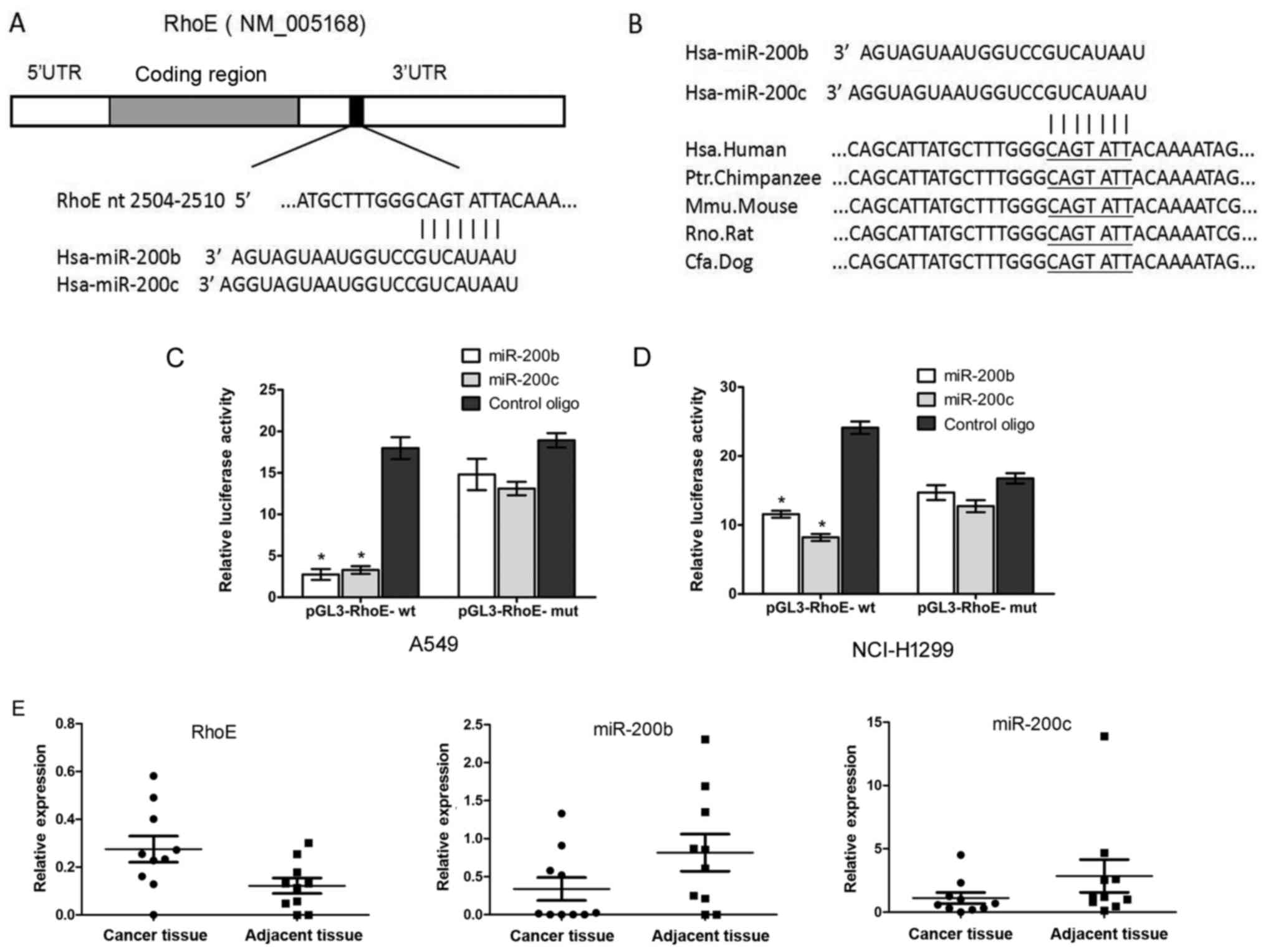
RhoE 3′UTR contains a putative target
site for miR-200b/200c, which is conserved among species and is a
direct target of miR-200b/200c
Studies have shown that miRNAs can regulate cell
apoptosis and drug sensitivity through directly targeting RhoE. To
examine and confirm the regulation of RhoE by miRNAs, the
bioinformatics tool PicTar was used to search for putative miRNAs
that are predicted to target the RhoE gene (25). Finally, hsa-miR-200b/200c was
focused on as putative miRNA binding to the 3′UTR of human RhoE,
and sequence analysis revealed that the targeting sequence for
miR-200b/200c is located at nt 2504-2510 of the RhoE 3′UTR
(Fig. 2A), and this region is
conserved across different species, as shown in Fig. 2B.
To confirm whether RhoE was a direct target of
miR-200b/200c, a dual-luciferase reporter assay was performed to
verify the sequence regions of the RhoE-3′UTR that were responsible
for the direct regulation of miR-200b/c. The 3′UTR of the human
RhoE gene was cloned into the pGL3-luciferase reporter vector
(pGL3-control) to determine whether it is a direct functional
target of miR-200b/c. This recombinant plasmid was termed
pGL3-RhoE-wt. In parallel, another luciferase reporter construct
was generated, in which the putative miR-200b/c targeting region
CAGTATT located within nt 2504–2510 was specifically mutated as
pGL3-RhoE-mut. Transient transfection with pGL3-RhoE-wt and
miR-200b/c led to a significant decrease of luciferase activity
compared with the control in A549c cells (Fig. 2C) and NCI-H1299 cells (Fig. 2D). Mutation of the miR-200b/c
putative binding site in the RhoE-3′UTR eliminated the inhibitory
effect on luciferase expression by miR-200b/c. These results
indicated that miR-200b/c directly targeted the expression of RhoE
through binding to the predicted sequence sites.
To compare the expression of miR-200b/200c and RhoE
in lung adenocarcinoma tissues and adjacent tissues, the mRNA
expression levels of miR-200b, miR-200c and RhoE in lung
adenocarcinoma tissues and paired adjacent samples were detected
using RT-qPCR analysis. The results showed that the mRNA expression
of RhoE in the tumor tissues was higher than that of the adjacent
tissues, whereas the mRNA expression levels of miR-200b and
miR-200c in the lung adenocarcinoma cancer tissues were
significantly lower than those in the adjacent tissues (Fig. 2E).
miR-200b/c can affect the expression of
RhoE in NSCLC cell lines
To examine whether miR-200b/c can affect the
expression of RhoE in NSCLC cell lines, the A549 and NCI-H1299 cell
lines, which show a relatively high expression of RhoE, were
transfected with miR-200b/c, a control oligo or Si-RhoE mixture
targeting RhoE. The protein and mRNA expression levels of RhoE were
analyzed by western blot and RT-qPCR analyses, respectively. As
shown in Fig. 3, compared with the
negative control, transient expression of miR-200b/c led to a
significant decrease of RhoE at the protein (Fig. 3A and B) and mRNA (Fig. 3C and D) levels, similar to that
caused by transfection with Si-RhoE in the two cell lines.
Furthermore, RT-qPCR analysis was performed to detect the
expression of miR-200b/c in the transfected cell lines, the results
of which showed that miR-200b/c was successfully overexpressed
(Fig. 3E and F). Based on these
data, it was concluded that RhoE was negatively regulated by
miR-200b/c, and the 3′UTR of RhoE was a functional target site for
miR-200b/c in the NSCLC cells.
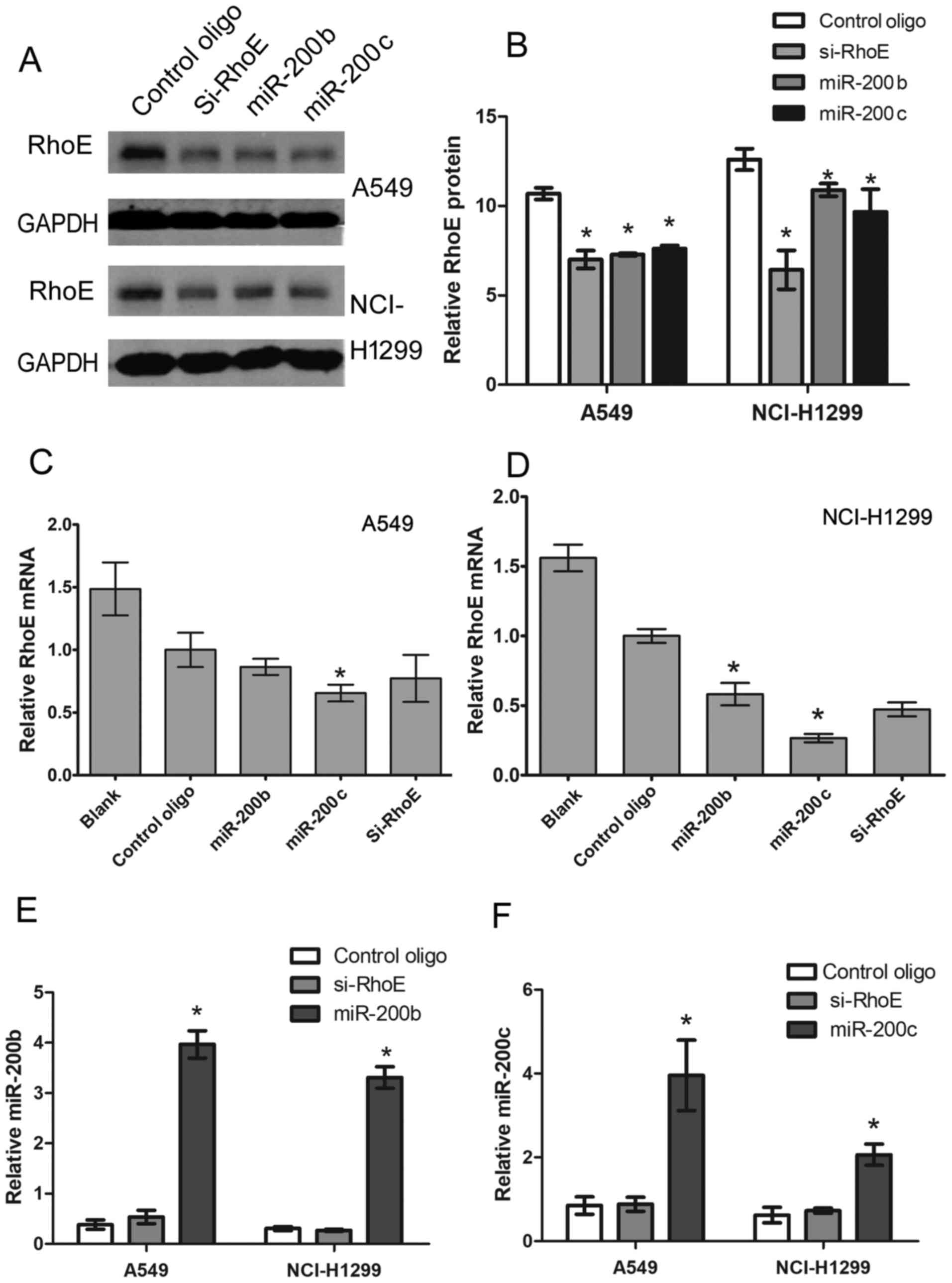 | Figure 3Protein and mRNA expression of RhoE
is regulated by miR-200b/200c. (A) Protein expression of RhoE in
A549 and NCI-H1299 cells 48 h following transfection with
miR-200b/200c, Si-RhoE or control oligo, determined with western
blot analysis. (B) Band intensities were quantified with the
Odyssey infrared imaging system. Data are presented as the mean ±
standard deviation from three independent experiments. mRNA
expression of RhoE in (C) A549 and (D) NCI-H1299 cells 48 h
following transfection with miR-200b/c, Si-RhoE or control oligo,
determined by RT-qPCR analysis. mRNA expression of RhoE was
normalized to mRNA expression of GAPDH, and data are shown as a
ratio of miR200b/200c and Si-RhoE-transfected cells to control
oligo-transfected cells using the 2−ΔΔCq method. Data
are presented as the mean ± standard deviation from three
independent experiments. *P<0.05 vs. control
oligo-transfected cells. RT-qPCR analysis of (E) miR-200b and (F)
miR-200c in A549 and NCI-H1299 cells transfected with miR-200b/c,
Si-RhoE or control oligo. Expression of miR-200b/c was normalized
to the expression of U6 using the 2−ΔΔCq method, Data
are representative of three independent experiments performed in
triplicate. RhoE, Rho family GTPase 3; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; miR,
microRNA; Si-small interfering RNA. |
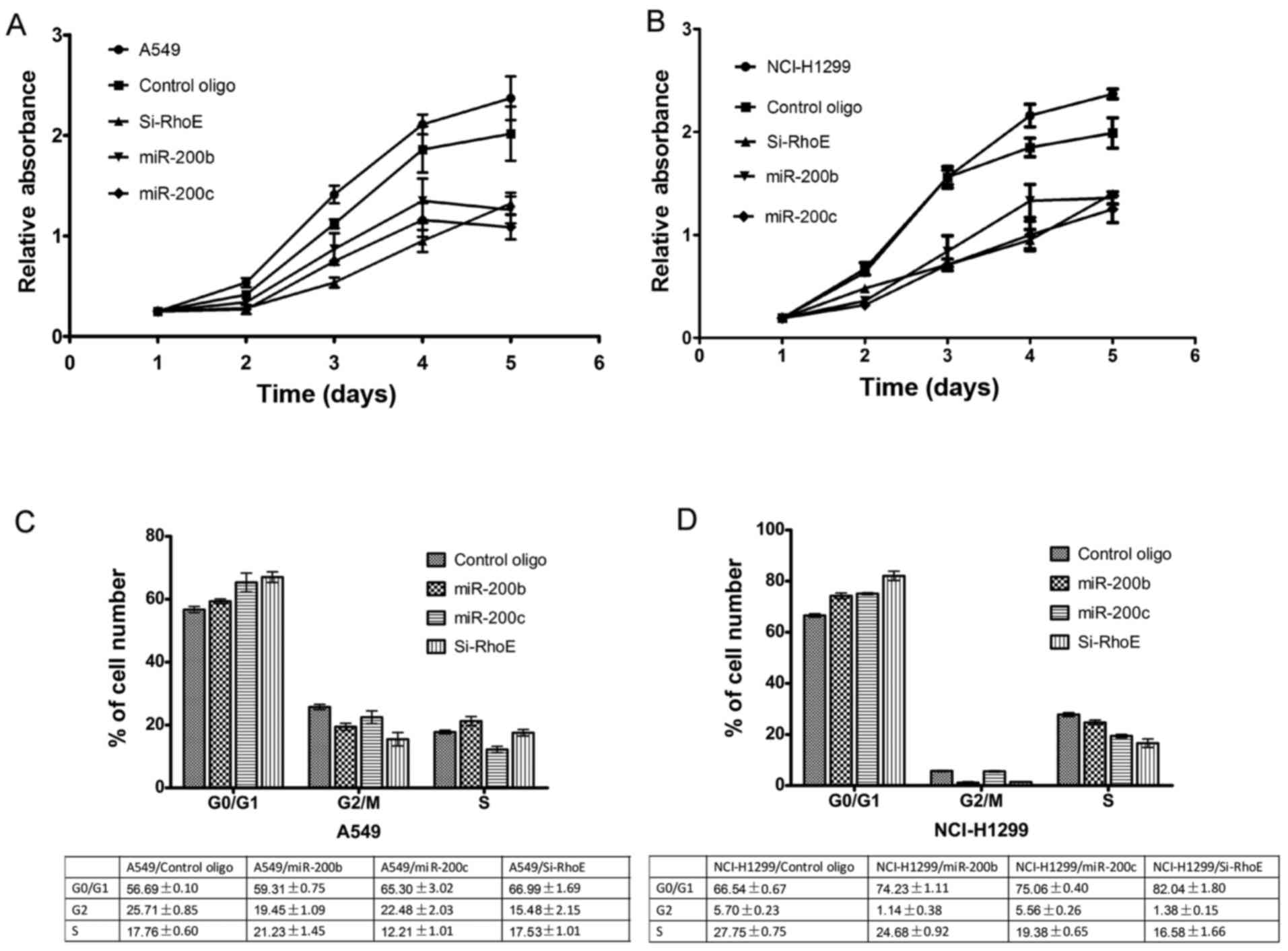
miR-200b/c inhibits proliferation,
migration, invasion and induces G0/G1 arrest in NSCLC cells
RhoE is key in the regulation of cell cycle
progression at the G1/S transition (26). The present study investigated the
effects of miR-200b/c and RhoE siRNA on cell proliferation. The
results showed that the ectopic expression of miR-200b/c markedly
inhibited the proliferation of A549 (Fig. 4A) and NCI-H1299 (Fig. 4B) cells in the CCK-8 assays. This
effect was comparable to that of RhoE siRNA. To further examine the
underlying mechanisms, the cell cycle distributions of the
miR-200b/c or RhoE siRNA transfectants were analyzed by FACS
analysis. The cell cycle progression of A549 (Fig. 4C) and NCI-H1299 (Fig. 4D) cells transfected with miR-200b/c
or the siRNA mixtures was arrested at the G1 phase, with a
concomitant decrease of cells in the S and G2/M phases, compared
with cells transfected with control oligos. These results indicated
that miR-200b/c inhibited the G1/S phase transition by
downregulating RhoE. In addition to the regulation of cell
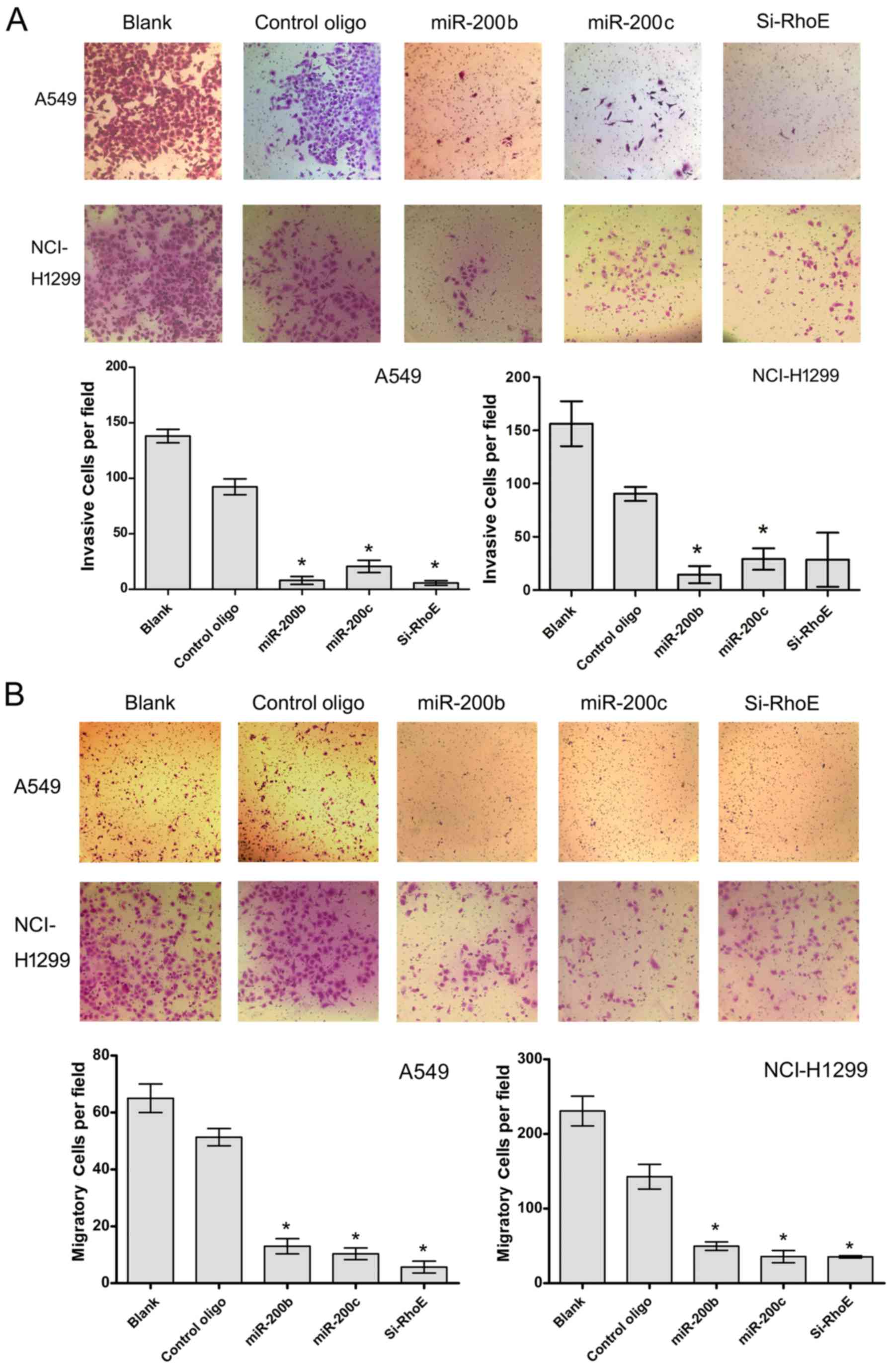
proliferation, RhoE is involved in the migration and invasion of
tumor cells, contributing to the incidence and mortality rates of
patients with cancer. In order to determine whether miR-200b/c
affects cell invasion, Matrigel invasion and migration assays were
performed. The results showed that miR-200b/c significantly
decreased the invasion and migration activity of the two cells,
similar to the effect of siRNA mixtures of transfected cells
(Fig. 5). These data suggested
that miR-200b/c negatively regulated the invasive ability of NSCLC
cells, at least in part through RhoE-mediated targeting
activity.
Expression of EMT-related transcriptors
is correlated with miR-200b/c and RhoE in NSCLC cells
To further examine the regulatory mechanism of
miR-200b/c-RhoE in NSCLC cells, the A549 and NCI-H1299 cells were
transfected with miR-200b/c, RhoE, or control oligo, and the
expression levels of AKT, ERK1/2 and EMT-related markers, including
N-cadherin, E-cadherin and vimentin, were examined by western blot
analysis. As shown in Fig. 6A–C,
the expression of E-cadherin was increased, and the expression of
N-cadherin and vimentin were decreased significantly in the cells
over-expressing miR-200b/c, and the same effect was detected in
those cells with RhoE knockdown. In addition, the expression of
EMT-related proteins N-cadherin, E-cadherin and vimentin were
examined in human lung adenocarcinoma tissues. The results showed
that the expression of E-cadherin was relatively low and that of
N-cadherin/vimentin was relatively high (Fig. 6D and E) in the cancer tissues,
compared with that in the adjacent tissues. However, no significant
change, or even the opposite expression pattern, was observed in
the expression of AKT, p-AKT, ERK1/2 or p-ERK1/2 in the miR-200b/c
group or siRhoE group compared with the controls. Together, the
above data indicated that miR-200b/c may also act through RhoE to
regulate the EMT-related markers, although the exact mechanisms
require further investigation.
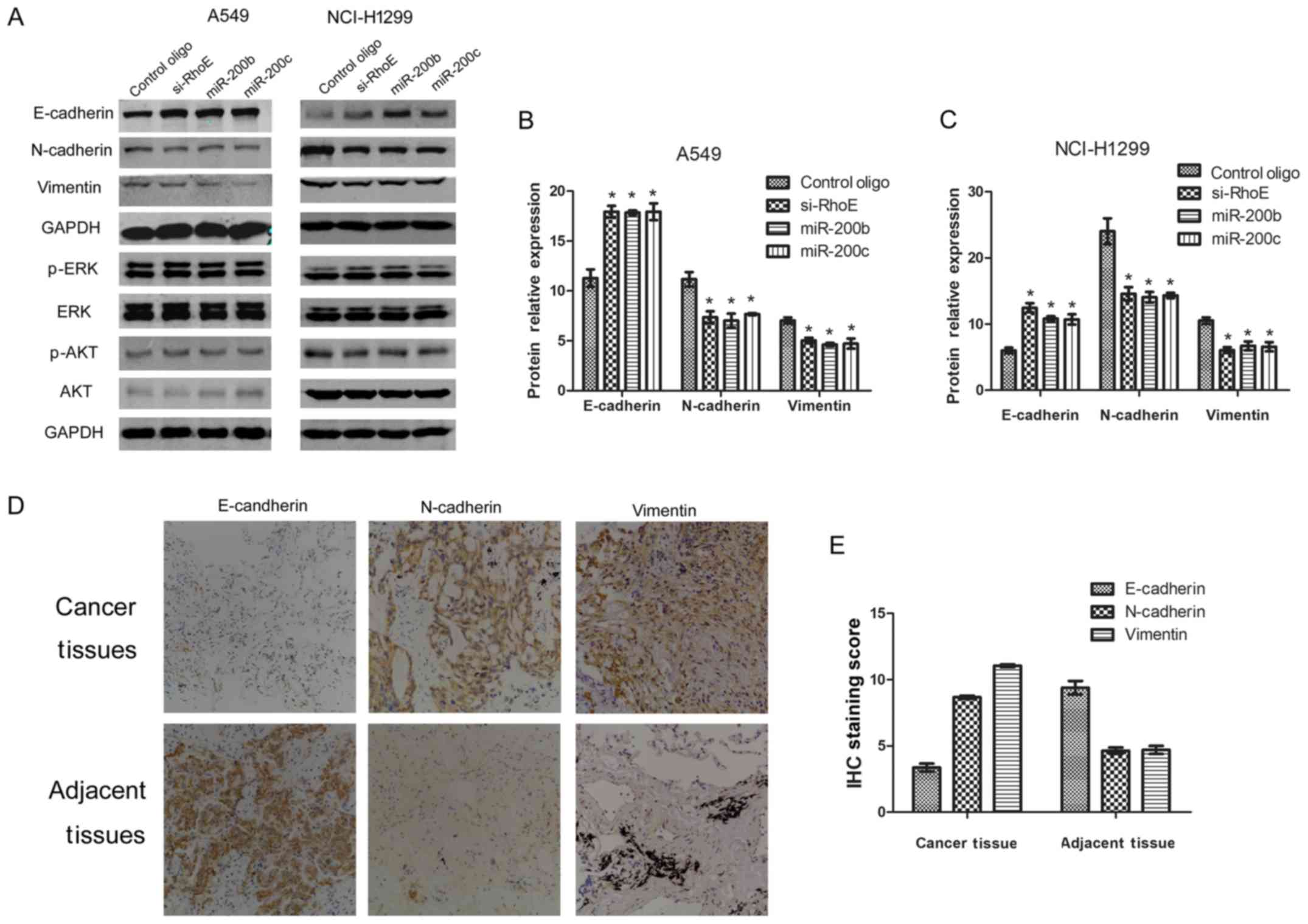 | Figure 6EMT-related protein expression. (A)
Overexpression of miR-200b/200c induces EMT signaling changes
similar to that induced by knockdown of RhoE. At 48 h
post-transfection with miR-200b/200c, Si-RhoE or control oligo in
A549 and NCI-H1299 cell line 48 h, E-cadherin, N-cadherin,
vimentin, ERK1/2, p-ERK1/2, AKT and p-AKT were examined by western
blot analysis. Band intensities in (B) A549 and (C) NCI-H1299 cells
were quantified with the Odyssey infrared imaging system. Data are
presented as the mean ± standard deviation from three independent
experiments. (D) Expression of EMT signaling pathway-related
molecules in human lung adenocarcinoma tissues (magnification, ×20;
10X objective). (E) Average staining score of EMT-related proteins
in lung adenocarcinoma tissues and adjacent tissues
(*P<0.05 vs. control). RhoE, Rho family GTPase 3;
miR, microRNA; Si-small interfering RNA; EMT,
epithelial-mesencyhmal transition; ERK, extracellular
signal-regulated kinase; p-, phosphorylated. |
Discussion
miRNAs, a group of small, non-coding RNA molecules,
act as post-transcriptional gene expression regulators in several
important physiological processes, including development, cell
differentiation and carcinogenesis. In previous years, increasing
evidence has supported the fact that miRNAs can function as
oncogenes or tumor suppressor genes in the regulation of multiple
tumorigenesis (27). The miR-200
family, reported as 'new star' miRNAs, consist of five members and
are key in a series of important physiological processes in cancer,
including the modulation of cell division and apoptosis, repression
of cancer stem cells and reversal of chemoresistance (28). miR-200s are involved in the
inhibition of EMT by targeting of transcriptional repressors of
E-cadherin and Zinc finger E-box binding homeobox 1 (ZEB1) in a
negative feedback loop (29) and
serving as a key indicator of E-cadherin-positive and
vimentin-negative cancer cell lines (12,30).
miR-200s were found to decrease the expansion of human metastatic
prostate cancer cells through inhibiting the notch signaling
pathway (31). In addition,
miR-200s can serve as a prognostic marker for patients with cancer
(32,33). It was found that the loss of
miR-200c induced significant inhibition of the invasion and
migration in NSCLC cells. In previous data, a gene set enrichment
analysis performed using the Cancer Cell Line Encyclopedia database
showed that miR-200c was suppressed, the expression level of
E-cadherin was downregulated, and the expression levels of vimentin
and ZEB1 were overexpressed among 34 NSCLC cell lines (34). It was reported that miR-200c acted
as an antitumor gene in cells with acquired EGFR-TKI resistance
(35), and that no miR-200c was
expressed in cancer stem cells (36). To date, these results suggest that
the deregulation of miR-200 is key in multiple levels as a tumor
suppressor gene. In contrast to the results of the above studies,
the results of a clinical tissue microarray showed that high
expression levels of tumor miR-200c were correlated with poor
prognosis in patients with lung cancer (32), and a high level of miR-200c has
been found in ovarian (37),
cervical (36), bile duct
(38) and nasopharyngeal (39) cancer. These results indicate that
the role of miR-200c in cancer remains controversial; in terms of
the mechanism, it may be that miR-200s has a different role in the
progression of different cancer types. However, the regulatory
mechanism of miR-200 in NSCLC remains to be fully elucidated.
In the present study, it was found that miR-200b/c
regulated the proliferation, invasion and migration in NSCLC cell
lines by targeting the Rho family protein RhoE. RhoE is a unique
and atypical member of the RND subfamily and RAS super-family,
which regulates a series of cell activities and disease progression
(40). Compared with other typical
Rho family members, RND proteins are atypical in terms of the
protein structures and functional binding, but do not hydrolyze
GTP, thus retaining constitutive activity, which is not regulated
by guanine nucleotide exchange factor (GEF) or GTP-activating
protein (GAP) (41). In addition,
the carboxy-terminal sequence of RhoE is farnesylated, whereas the
majority of Rho family proteins are modified by geranylation.
Therefore, the activity of RhoE is not regulated by GEFs, GAPs or
GDP-dissociation inhibitors, the typical regulators, but by
regulating the balance between transcription, translation and
degradation, and by post-translational modifications including
phosphorylation (42). These
results indicated that the regulation model of RhoE may be
different from other Rho family members (41).
In a previous study, RND3 was overexpressed at mRNA
and protein levels in NSCLC, and was positively correlated with the
unfavorable prognosis of patients. This suggested that the
expression of RND3 may be investigated as a marker of prognosis in
patients with NSCLC (19). In
order to translate and develop for the clinical application of
NSCLC biomarkers, selection in a large cohort of patients was
established by the long-term survival rates of patients and the
expression of several signatures, including RhoE, in the complete
resection of lung adenocarcinoma (43). The findings provided evidence for
the overexpression of RhoE, which was associated with poor
prognosis and tumor progression in patients with NSCLC as a marker
for clinical prognosis, however, this remained to be validated in a
prospective study (44). Previous
studies have shown that RhoE was downregulated in three NSCLC cell
lines, and inhibited cell proliferation through the Notch1/Notch
intracellular domain/Hes1 signaling pathway (45). However, only one cell line was
obtained from lung adenocarcinoma, which was a limitation of this
study. In addition, the mechanism of the upregulation in NSCLC
remained unclear. Therefore, evaluation of the impact of the
dysregulation of RhoE on NSCLC requires further investigation.
In the present study, RhoE was overexpressed in
NSCLC, which was different from other types of tumor. It was found
that human RhoE harbored two putative sites recognized by miR-200b
and miR-200c, which suggested that miR-200b/c may be directly
regulated by RhoE at the post-transcription level. A
dual-luciferase-based reporter assay confirmed the binding sites of
miR-200b/c on the 3′UTR of RhoE, and miR-200b/c inhibited the
protein and mRNA expression levels of RhoE. miR-200b/c
significantly inhibited proliferation and invasion activities and
induced G0/G1 arrest in the A549 and NCI-H1299 NSCLC cells.
Previous studies have shown that miR-200b/c is involved in the
regulation of EMT, which can be used in tumor diagnosis and as a
treatment target, however, the specific regulatory mechanism
remained to be elucidated. In the present study, the suppression of
RhoE and overexpression of miR-200b/c had a similar effect on the
EMT signaling pathway, indicating that miR-200b/c regulated EMT and
that this regulation may be regulated by RhoE.
Acknowledgments
The authors would like to thank the technician Cui
Yiyuan (Neuromolecular Research Laboratory of West China Hospital)
and the technician Zhang Yi (Core Facility of West China Hospital)
for their assistance with experiments and equipment in the
pathological and RT-qPCR analyses.
Funding
The study was supported by the National Natural
Science Foundation of China (grant no. 81702403); the National Key
R&D Program of China (grant no. 2016YFC1303200/2016 YFC1303203)
and the National Natural Science Foundation (grant no.
81621003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QT and HX designed the experiments. QT performed the
majority of the experiments and analyzed the data. QT and HX wrote
the manuscript. ML carried out the construction of plasmids and
performed Transwell assay. LC performed the real-time PCR assay.
ML, LC and HX helped to revise the manuscript. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
All patients signed an informed consent when
collecting the pathological tissue by Disease Tissue Specimen Bank
of West China Hospital, Sichuan University and the study was
approved by the Ethics Committee of Sichuan University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moss EG: MicroRNAs: Hidden in the genome.
Curr Biol. 12:R138–R140. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ceppi P, Mudduluru G, Kumarswamy R, Rapa
I, Scagliotti GV, Papotti M and Allgayer H: Loss of miR-200c
expression induces an aggressive, invasive, and chemoresistant
phenotype in non-small cell lung cancer. Mol Cancer Res.
8:1207–1216. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F,
Bao G, Kong H, Ge C, Zhang F, et al: miRNA-200c inhibits invasion
and metastasis of human non-small cell lung cancer by directly
targeting ubiquitin specific peptidase 25. Mol Cancer. 13:1662014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi L, Zhang S, Wu H, Zhang L, Dai X, Hu
J, Xue J, Liu T, Liang Y and Wu G: MiR-200c increases the
radiosensitivity of non-small-cell lung cancer cell line A549 by
targeting VEGF-VEGFR2 pathway. PLoS One. 8:e783442013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang C, Ding M, Xia M, Chen S, Van Le A,
Soto-Gil R, Shen Y, Wang N, Wang J, Gu W, et al: A five-miRNA panel
identified from a multicentric case-control study serves as a novel
diagnostic tool for ethnically diverse non-small-cell lung cancer
patients. EBioMedicine. 2:1377–1385. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lujambio A, Calin GA, Villanueva A, Ropero
S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso
MS, Faller WJ, et al: A microRNA DNA methylation signature for
human cancer metastasis. Proc Natl Acad Sci USA. 105:13556–13561.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wong CM, Wei L, Au SL, Fan DN, Zhou Y,
Tsang FH, Law CT, Lee JM, He X, Shi J, et al: MiR-200b/200c/429
subfamily negatively regulates Rho/ROCK signaling pathway to
suppress hepatocellular carcinoma metastasis. Oncotarget.
6:13658–13670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Roybal JD, Zang Y, Ahn YH, Yang Y, Gibbons
DL, Baird BN, Alvarez C, Thilaganathan N, Liu DD, Saintigny P, et
al: miR-200 Inhibits lung adenocarcinoma cell invasion and
metastasis by targeting Flt1/VEGFR1. Mol Cancer Res. 9:25–35. 2011.
View Article : Google Scholar :
|
|
15
|
Kim JS, Kurie JM and Ahn YH: BMP4
depletion by miR-200 inhibits tumorigenesis and metastasis of lung
adenocarcinoma cells. Mol Cancer. 14:1732015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao YF, Han ML, Xiong YJ, Wang L, Fei Y,
Shen X, Zhu Y and Liang ZQ: A micRNA-200c/cathepsin L feedback loop
determines paclitaxel resistance in human lung cancer A549 cells in
vitro through regulating epithelial-mesenchymal transition. Acta
Pharmacol Sin. 39:1034–1047. 2018. View Article : Google Scholar
|
|
17
|
Nishijima N, Seike M, Soeno C, Chiba M,
Miyanaga A, Noro R, Sugano T, Matsumoto M, Kubota K and Gemma A:
miR-200/ZEB axis regulates sensitivity to nintedanib in non-small
cell lung cancer cells. Int J Oncol. 48:937–944. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu M, Bi F, Zhou X and Zheng Y: Rho
GTPase regulation by miRNAs and covalent modifications. Trends Cell
Biol. 22:365–373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wennerberg K, Forget MA, Ellerbroek SM,
Arthur WT, Burridge K, Settleman J, Der CJ and Hansen SH: Rnd
proteins function as RhoA antagonists by activating p190 RhoGAP.
Curr Biol. 13:1106–1115. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ongusaha PP, Kim HG, Boswell SA, Ridley
AJ, Der CJ, Dotto GP, Kim YB, Aaronson SA and Lee SW: RhoE is a
pro-survival p53 target gene that inhibits ROCK I-mediated
apoptosis in response to genotoxic stress. Curr Biol. 16:2466–2472.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu Y, Zhou J, Xia H, Chen X, Qiu M, Huang
J, Liu S, Tang Q, Lang N, Liu Z, et al: The Rho GTPase RhoE is a
p53-regulated candidate tumor suppressor in cancer cells. Int J
Oncol. 44:896–904. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang C, Zhou F, Li N, Shi S, Feng X, Chen
Z, Hang J, Qiu B, Li B, Chang S, et al: Overexpression of RhoE has
a prognostic value in non-small cell lung cancer. Ann Surg Oncol.
14:2628–2635. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cuiyan Z, Jie H, Fang Z, Kezhi Z, Junting
W, Susheng S, Xiaoli F, Ning L, Xinhua M, Zhaoli C, et al:
Overexpression of RhoE in non-small cell lung cancer (NSCLC) is
associated with smoking and correlates with DNA copy number
changes. Cancer Biol Ther. 6:335–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
25
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M,
et al: Combinatorial microRNA target predictions. Nat Genet.
37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bektic J, Pfeil K, Berger AP, Ramoner R,
Pelzer A, Schäfer G, Kofler K, Bartsch G and Klocker H: Small
G-protein RhoE is underexpressed in prostate cancer and induces
cell cycle arrest and apoptosis. Prostate. 64:332–340. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu M, Lang N, Qiu M, Xu F, Li Q, Tang Q,
Chen J, Chen X, Zhang S, Liu Z, et al: miR-137 targets Cdc42
expression, induces cell cycle G1 arrest and inhibits invasion in
colorectal cancer cells. Int J Cancer. 128:1269–1279. 2011.
View Article : Google Scholar
|
|
28
|
Feng X, Wang Z, Fillmore R and Xi Y:
MiR-200, a new star miRNA in human cancer. Cancer Lett.
344:166–173. 2014. View Article : Google Scholar :
|
|
29
|
Brabletz S and Brabletz T: The ZEB/miR-200
feedback loop - a motor of cellular plasticity in development and
cancer? EMBO Rep. 11:670–677. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vallejo DM, Caparros E and Dominguez M:
Targeting Notch signalling by the conserved miR-8/200 microRNA
family in development and cancer cells. EMBO J. 30:756–769. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ,
Wang YK, Zeng F, Zhou JH and Zhang YK: High expression of serum
miR-21 and tumor miR-200c associated with poor prognosis in
patients with lung cancer. Med Oncol. 29:618–626. 2012. View Article : Google Scholar
|
|
33
|
Yu J, Ohuchida K, Mizumoto K, Sato N,
Kayashima T, Fujita H, Nakata K and Tanaka M: MicroRNA,
hsa-miR-200c, is an independent prognostic factor in pancreatic
cancer and its upregulation inhibits pancreatic cancer invasion but
increases cell proliferation. Mol Cancer. 9:1692010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sato H, Shien K, Tomida S, Okayasu K,
Suzawa K, Hashida S, Torigoe H, Watanabe M, Yamamoto H, Soh J, et
al: Targeting the miR-200c/LIN28B axis in acquired EGFR-TKI
resistance non-small cell lung cancer cells harboring EMT features.
Sci Rep. 7:408472017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shimono Y, Zabala M, Cho RW, Lobo N,
Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ,
Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG, et al: Altered MicroRNA
expression in cervical carcinomas. Clin Cancer Res. 14:2535–2542.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Meng F, Henson R, Lang M, Wehbe H,
Maheshwari S, Mendell JT, Jiang J, Schmittgen TD and Patel T:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang L, Deng T, Li X, Liu H, Zhou H, Ma
J, Wu M, Zhou M, Shen S, Li X, et al: microRNA-141 is involved in a
nasopha-ryngeal carcinoma-related genes network. Carcinogenesis.
31:559–566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nobes CD, Lauritzen I, Mattei MG, Paris S,
Hall A and Chardin P: A new member of the Rho family, Rnd1,
promotes disassembly of actin filament structures and loss of cell
adhesion. J Cell Biol. 141:187–197. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J
and Settleman J: Identification of a novel human Rho protein with
unusual properties: GTPase deficiency and in vivo farnesylation.
Mol Cell Biol. 16:2689–2699. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Riou P, Villalonga P and Ridley AJ: Rnd
proteins: Multifunctional regulators of the cytoskeleton and cell
cycle progression. BioEssays. 32:986–992. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Raz DJ, Ray MR, Kim JY, He B, Taron M,
Skrzypski M, Segal M, Gandara DR, Rosell R and Jablons DM: A
multigene assay is prognostic of survival in patients with
early-stage lung adenocarcinoma. Clin Cancer Res. 14:5565–5570.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Paysan L, Piquet L, Saltel F and Moreau V:
Rnd3 in cancer: A Review of the evidence for tumor promoter or
suppressor. Mol Cancer Res. 14:1033–1044. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang Y, Hu C, Yang H, Cao L, Li Y, Deng P
and Huang L: Rnd3 regulates lung cancer cell proliferation through
notch signaling. PLoS One. 9:e1118972014. View Article : Google Scholar : PubMed/NCBI
|