Introduction
Neuroblastoma is the most common type of
extracranial solid tumor in children. It arises from cells of
sympathoadrenal lineage from the neural crest, which migrate away
from their place of origin during embryogenesis. Primary tumors
typically occur in the adrenal glands, chest, head, neck and pelvis
(1). The clinical presentation of
neuroblastoma varies from spontaneous regression to rapid
progression despite multimodal therapies. This depends on the age
of the patient at diagnosis, as well as genetic prognostic factors,
with the most important being N-Myc (MYCN) gene
amplification, which occurs in 25% of tumors (2). Annually, >600 novel cases of
neuroblastoma are recognized in the USA, with ~50% of the patients
being diagnosed at the advanced stages of the disease (1). The outcome of therapy of high-risk
neuroblastoma cases remains poor, with the long-term survival rate
being <50% (3).
Recently, assessments of serum microRNAs (miRNAs)
have developed into promising non-invasive diagnostic tools for
patients, including those with neuroblastoma (4). miRNAs are small non-coding RNA
molecules that function as negative regulators of gene expression
at the post-transcriptional level. The expression of >60% of
protein-coding genes is considered to be regulated by miRNAs. One
transcript can be recognized by numerous miRNAs, whereas one miRNA
is able to regulate a number of mRNAs. Therefore, the expression of
miRNAs must be tightly regulated to ensure normal growth and
development of an organism (5).
Dysregulation of miRNA expression contributes to tumor progression
in numerous types of cancer, including neuroblastoma (5). Certain miRNA molecules, such as
miRNA-34a and miRNA-17-92, have been described as potent tumor
suppressors and oncogenes in neuroblastoma, respectively (6). This is due to their involvement in
the regulation of the tumor phenotype via control of cellular
processes, including proliferation, differentiation and apoptosis
(6). Although the function of
several individual miRNA molecules has been elucidated in
neuroblastoma (5), the functions
of numerous miRNAs require experimental delineation.
miRNA-3613-3p is a novel miRNA molecule, which
attracted interest in our previous study during screening for
differentially expressed miRNAs in BE(2)-C neuroblastoma cells with
monocyte chemoattractant protein-induced protein 1 (MCPIP1)
overexpression (7). It was
identified that the overexpression of MCPIP1 causes significant
downregulation of miRNA-3613-3p expression in BE(2)-C cells as well
as the upregulation of several putative target genes of
miRNA-3613-3p (7). To date, the
expression of miRNA-3613-3p has been demonstrated by several other
groups (8–13). However, the effect of miRNA-3613-3p
over-expression on cell viability has only been studied in one type
of cell. Zhang et al (14)
identified that miRNA-3613-3p led to a significant downregulation
of cell proliferation via negative regulation of genes encoding
cyclin-dependent kinase 1, NUF2 NDC80 kinetochore complex
component, baculoviral inhibitor of apoptosis protein
repeat-containing 5, ZW10-interacting kinetochore protein and SPC24
NDC80 kinetochore complex component in human HepG2 hepatoblastoma
cells. Furthermore, miRNA-3613-3p was identified among five miRNA
molecules as a central regulatory factor in RNA
interference-dependent control of p53 expression in the
aforementioned cell line (15).
Additionally, the expression of miRNA-3613-3p was low in the
hepatocytes and serum of patients with acute viral hepatitis caused
by hepatitis B virus and was associated with possible
downregulation of signal transducer and activator of transcription
3 expression due to specific binding with miRNA-3613-3p (13). Another premise of involvement of
miRNA-3616-3p in cancer is its potential biomarker value in
adenocarcinoma (11) and the
enrichment of the complementary strand of the mature miRNA-3613-3p
in exosomes derived from a colon cancer cell line (9). Deregulated expression of
miRNA-3613-3p has also been identified in several other
pathological states (8,10,12).
Nevertheless, the involvement of miRNA-3613-3p in the pathogenesis
of neuroblastoma remains to be elucidated.
On the basis of our previous study (7), it was decided to investigate the
potential functions of miRNA-3613-3p in neuroblastoma. First, the
differential expression of miRNA-3613-3p was assessed in a range of
human neuroblastoma cell lines. Furthermore, the seven putative
target genes of miRNA-3613-3p were investigated using extensive
bioinformatics analysis. In order to verify the interaction between
miRNA-3613-3p and its predicted target mRNAs, three steps were
performed. The expression of the seven proposed target genes was
investigated in BE(2)-C human neuroblastoma cells transfected with
miRNA-3613-3p mimic using the reverse transcription-quantitative
polymerase chain reaction (RT-qPCR). The expression of the selected
putative target genes was analyzed at the protein level using
western blotting. The predicted binding sites in the apoptotic
protease-activating factor 1 (APAF1) 3′ untranslated region
(3′UTR) were verified using a luciferase reporter assay.
Additionally, the effect of enforced miRNA-3613-3p expression on
BE(2)-C human neuroblastoma cell viability. was assessed.
Furthermore, the expression and activation levels of several
proteins involved in apoptosis in BE(2)-C cells overexpressing
miRNA-3613-3p were investigated.
Materials and methods
Cell culture
Seven human neuroblastoma cell lines, BE(2)-C
[American Type Culture Collection (ATCC), Manassas, VA, USA], Kelly
[Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH
(DSMZ), Leibniz, Germany], IMR-32 (ATCC), SK-N-SH (ATCC), CHP-134
[European Collection of Authenticated Cell Cultures (ECACC), Porton
Down, UK], LAN-1 (DSMZ) and LAN-5 (DSMZ), and a prostate cancer
cell line PC3 (ECACC) were cultured at 37°C in an incubator with 5%
CO2. Cells were routinely screened for Mycoplasma
contamination and all tests were negative according to a MycoAlert™
Mycoplasma Detection kit (Lonza Group, Ltd., Basel,
Switzerland). LAN-1 cells were cultured in a 1:1 mixture of Eagle's
minimal essential medium and Ham's F12 medium (both from
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). BE(2)-C cells were cultured in a 1:1 mixture of
Eagle's minimal essential medium and Ham's F12 medium supplemented
with non-essential amino acids (NEAA), 1 mM sodium pyruvate and 10%
fetal bovine serum. Kelly and CHP-134 cells were cultured in
RPMI-1640 (Sigma-Aldrich; Merck KGaA) with 10% fetal bovine serum.
IMR-32 and SK-N-SH cells were cultured in Eagle's minimal essential
medium supplemented with 10% fetal bovine serum, 1% NEAA solution
and 1 mM sodium pyruvate. LAN-5 cells were cultured in RPMI-1640
supplemented with 20% fetal bovine serum. The PC3 cell line was
cultured in a 1:1 mixture of Ham's F12 medium and Dulbecco's
modified Eagle's medium (Sigma-Aldrich; Merck KGaA) with 10% fetal
bovine serum. All neuroblastoma cell culture media were
additionally supplemented with 50 µg/ml gentamicin
(Sigma-Aldrich; Merck KGaA), whereas the PC3 cell culture was
supplemented with 40 µg/ml gentamicin.
Generation of the genetic construct
A fragment of APAF1 (NCBI accession no.
NM_013229.2) 3′UTR containing putative binding sites for
miRNA-3613-3p was amplified from BE(2)-C cells by PCR using primers
with recognition sites for XhoI and SalI restriction
enzymes added at the 5′ end (forward,
5′-ACCTGCTCGAGAAATTGGTATTTTAATACTG-3′ and reverse,
5′-AGTAAGTCGACAGCAAGACTCTGTCTC CAA-3′). For the amplification of
the 3′UTR fragment DyNAzyme II DNA Polymerase (Thermo Fisher
Scientific, Inc.) was used following the manufacturer's protocol.
The thermocycling conditions were: Pre-incubation at 94°C for 3
min; 35 cycles of denaturation at 94°C for 30 sec, annealing at
55°C for 30 sec and elongation at 72°C for 30 sec; final elongation
at 72°C for 10 min. Next, the products were separated by agarose
gel electrophoresis using a 1% gel, visualized using SimplySafe™
(Eurx, Gdańsk, Poland) and isolated using the Gel-Out kit (A&A
Biotechnology, Gdynia, Poland) according to the manufacturer's
protocol. Isolated products and pmiRGlo plasmid vector (E1330;
Promega Corporation, Madison, WI, USA) were digested using
XhoI and SalI restriction enzymes. Following
digestion, the pmiRGlo plasmid was additionally dephosphorylated
using calf intestinal alkaline phosphatase. Next, DNA was purified
using a QIAquick Nucleotide Removal kit (Qiagen GmbH, Hilden,
Germany). The APAF1 3′UTR fragment was incorporated into the
pmiRGlo plasmid vector using T4 DNA ligase. The genetic construct
obtained was verified using DNA sequencing (Genomed, Warszawa,
Poland).
Cell transfection with miRNA-3613-3p
mimic
miRNA-3613-3p mimic (cat. no. 4464066) and scrambled
oligonucleotide (cat. no. 4464058), lacking complementarity to
sequences in the human transcriptome, as a negative control were
purchased from Ambion; Thermo Fisher Scientific, Inc. For enforced
overexpression of miRNA-3613-3p, BE(2)-C cells were seeded at a
density of 3×104 cells/cm2 in complete medium
as aforementioned. After 24 h of culture, the medium was changed.
miRNA mimic or negative control and Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.) were diluted in Opti-MEM™ (Gibco;
Thermo Fisher Scientific, Inc.), and added to the cell culture at
final concentrations of 30, 40 and 50 nM. Transfected cells were
passaged after 24 h. For prolonged overexpression of miRNA-3613-3p,
miRNA-3613-3p mimic or negative control and Lipofectamine 2000
diluted in Opti-MEM were added to the transfected cells culture
after 48 h. Overexpression of miRNA-3613-3p was verified using
RT-qPCR.
LAN-1 cell transfection with
miRNA-3613-3p inhibitors
miRNA-3613-3p inhibitor (cat. no. 4464084) and
corresponding negative control (cat. no. 4464076) were purchased
from Ambion; Thermo Fisher Scientific, Inc. For the silencing of
miRNA-3613-3p, LAN-1 neuroblastoma cells were seeded at a density
of 2.6×104 cells/cm2 in complete medium as
aforementioned. After 24 h of culture, the medium was changed. The
miRNA-3613-3p inhibitor, negative control and Lipofectamine 2000
transfection agent were diluted in Opti-MEM. The inhibitor and
negative control were added to the cell culture at a final
concentration of 30 nM. Cells were collected 48 h after
transfection.
Kelly cell transfection with genetic
constructs containing an expression cassette for wild-type or
mutated MCPIP1 protein
Plasmid vectors used to obtain enforced expression
of MCPIP1 protein were as described previously (16). The transfection procedure for Kelly
cells was as described in our previous study (17). Overexpression of wild-type or
mutated MCPIP1 protein was verified by western blotting.
RNA isolation
RNA was extracted from cells using
TRI-REAGENT® (Lab Empire, Rzeszów, Poland) according to
the manufacturer's protocol. The concentration and purity of the
isolated RNA were determined using a NanoDrop ND-1000
spectrophotometer (Thermo Fisher Scientific, Inc.). The integrity
of the RNA was verified by agarose gel electrophoresis using a 1%
gel.
mRNA RT-qPCR
For each sample, 500 ng of RNA was
reverse-transcribed using Moloney murine leukemia virus reverse
transcriptase (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol, using a GenAmp thermocycler (PerkinElmer,
Inc., Waltham, MA, USA). qPCR was performed using an Eco
thermocycler (Illumina, San Diego, CA, USA) with KAPA
SYBR® FAST qPCR Master mix (Kapa Biosystems, Inc.,
Wilmington, MA, USA). The thermocycling conditions were:
Pre-incubation at 95°C for 10 min; 40 cycles of denaturation at
95°C for 15 sec, and annealing and elongation at 60°C for 30 sec; a
melting cycle at 95°C for 15 sec, 55°C for 15 sec and 95°C for 15
sec. The cDNA used for qPCR was diluted 50-fold, and the reference
gene 40S ribosomal protein S13 (RPS13) was used. The primers
for DICER, APAF1, DNA fragmentation factor subunit β
(DFFB), neurofibromin 1 (NF1), retinoic acid-related
orphan receptor α (RORA), kinesin family member 3A
(KIF3A) and RPS13 used in these experiments were
described previously (7). The
sequences of von Hippel-Lindau protein (VHL) primers were as
follows: Forward, 5′-TCCACAGCTACCGAGGTCA-3′ and reverse,
5′-GGCAAAAATAGGCTGTCCGT-3′. All experiments were performed in
triplicate at least three times. For quantification of the relative
mRNA level, the ΔΔCq method was used (18).
miRNA RT-qPCR
RT of miRNAs was performed using miRCURY LNA™
Universal RT microRNA PCR (Exiqon; Qiagen GmbH). For each reaction,
100 ng total RNA was used. miRNA RT-qPCR experiments were performed
using miRCURY LNA microRNA PCR and ExiLENT SYBR Green master mix
(Exiqon; Qiagen GmbH), according to the manufacturer's protocol.
For the PCR, the cDNA was diluted 80-fold and U6 small nuclear RNA
(snRNA) was used as the reference. Primers for miRNA-3613-3p (cat.
no. 2100646) and the reference U6 snRNA (cat. no. 203907) were
provided by Exiqon; Qiagen GmbH. All RT-qPCR experiments were
performed in triplicate at least three times, and the ΔΔCq method
(18) was used to quantify the
relative level of miRNA-3613-3p.
Protein extraction and western blot
analysis
Protein extraction was performed using TRI-REAGENT,
according to the manufacturer's protocol. Protein concentration was
assessed using the bicinchoninic acid method (19). Protein extracts were resolved using
SDS-PAGE (8, 12 or 15% gel, depending on the protein weight).
Subsequently, the western blotting procedure was performed as
described previously (20). The
following primary antibodies purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA) were used: Anti-DICER rabbit
monoclonal antibody (mAb) (cat. no. 5362, 1:1,000); anti-APAF1
rabbit mAb (cat. no. 8723, 1:1,000); anti-caspase-9 mouse mAb (cat.
no. 9508, 1:1,000); anti-cleaved caspase-9 rabbit mAb (cat. no.
7237, 1:10,00); anti-caspase-7 rabbit mAb (cat. no. 12827,
1:1,000); anti-cleaved caspase-7 rabbit mAb (cat. no. 8438,
1:1,000); anti-poly(ADP-ribose) polymerase (PARP) rabbit mAb (cat.
no. 9542, 1:1,000); anti-caspase-3 rabbit mAb (cat. no. 9665,
1:1,000); anti-cleaved caspase-3 rabbit mAb (cat. no. 9664,
1:1,000); and anti-α-tubulin rabbit mAb (cat. no. 2144, 1:1,000).
Anti-rabbit IgG goat mAb (cat. no. 7074, 1:2,000; Cell Signaling
Technology, Inc.) and anti-mouse IgG rabbit polyclonal antibody
(cat. no. A-9044, 1:40,000; Sigma-Aldrich; Merck KGaA) conjugated
to horseradish peroxidase were used as the secondary antibodies.
The relative protein levels were determined using densitometric
scanning of immunoreactive bands using Quantity One software
(version 4.6.9; Bio-Rad Laboratories, Inc., Hercules, CA, USA) and
normalized to α-tubulin levels.
Cell viability assay
BE(2)-C cells were cultured in 96-well plates and
transfected with miRNA-3613-3p mimic or negative control. After 48
h, the transfection with miRNA-3613-3p mimic or negative control
was repeated as aforementioned. Cellular ATP levels were assessed
using an ATPlite Luminescence ATP Detection assay system
(PerkinElmer, Inc.), according to the manufacturer's protocol.
Luminescence signals were determined using an Infinite M200
luminescence reader (Tecan Group, Ltd., Mannedorf, Switzerland).
The experiment was performed three times.
Luciferase reporter assay
BE(2)-C cells were seeded in the 96-well plates and
cultured in complete medium for 24 h. Next, the cells were
co-transfected with 40 nM miRNA-3613-3p mimic or negative control,
and 300 ng pmiRGlo plasmid vector with the cloned fragment of APAF1
3′UTR or empty pmiRGlo plasmid vector. Luciferase activity was
assessed using the Dual-Luciferase Reporter assay system (Promega
Corporation), according to the manufacturer's protocol 48 h after
co-transfection. Determination of luminescence signals from firefly
and Renilla luciferases was performed using an Infinite M200
luminescence reader. The experiment was performed three times.
Bioinformatics analysis
Visualization of the precursor form of the
miRNA-3613-3p was generated using the Mfold algorithm (unafold.rna.albany.edu/?q=mfold/download-mfold).
Additionally, miRNA-3613-3p was subjected to analysis using the
miRPath version 3.0 web server algorithm (21). Biological processes potentially
regulated by the miRNA were examined using microT-CDS (22). For the Kyoto Encyclopedia or Genes
and Genomes (www.genome.jp/kegg) pathway enrichment analysis,
P<0.05 was considered to indicate a statistically significant
difference and the threshold for microT-CDS was 0.8. Furthermore,
the predicted target genes of miRNA-3613-3p were determined using
the miRWalk2.0 database (23), and
the enriched functional patterns from the PANTHER (www.pantherdb.org/about.jsp) and Gene Ontology
algorithms (geneontology.org) were analyzed.
Furthermore, the visualization of miRNA-mRNA duplexes and
calculation of free energies of each binding was performed using
the RNAhybrid tool (24). In
addition, the difference in the free energy of each base-pairing
interaction between miRNA and mRNA was determined using the PITA
algorithm (25). In order to
determine the cross-species conservation of putative binding sites
of miRNA-3613-3p, the TargetScan algorithm was used (26).
Statistical analysis
Data are presented as the mean ± standard error of
the mean. All experiments were performed at least three times.
Experiments were performed on Kelly cells transfected with an empty
vector, wild-type or the mutated ribonuclease domain-deprived
MCPIP1, BE(2)-C cells transfected with negative control or
miRNA-3613-3p mimic and LAN-1 cells transfected with negative
control or miRNA-3613-3p inhibitor. Pairwise t-tests were performed
to compare the differences between the means of the NC vs. 3613-3p.
For comparison of more than two means, one-way analysis of variance
with Tukey's post hoc test implemented in Statistica (version 13;
StatSoft, Inc., Tulsa, OK, USA) was performed. P<0.05 was
considered to indicate a statistically significant difference.
Results
miRNA-3613-3p is differentially expressed
in a range of human neuroblastoma cell lines
The expression levels of miRNA-3613-3p were
determined in seven human neuroblas-toma cell lines and a prostate
cancer cell line PC3, which was used as a positive control, as the
expression of miRNA-3613-3p in the PC3 cell line has been
identified previously (27–29).
In total, six of the neuroblastoma cell lines analyzed were
char-acterized by MYCN amplification [Kelly, CHP-134,
IMR-32, LAN-1, LAN-5 and BE(2)-C], whereas SK-N-SH exhibited only
two copies of the oncogene. The highest endogenous expression of
miRNA-3613-3p was observed in LAN-1 cells, whereby the mean level
of miRNA-3613-3p was increased ~6-fold compared with that in PC3
prostate cancer cells (Fig. 1A).
In Kelly, IMR-32 and LAN-5 cell lines, the mean expression level of
miRNA-3613-3p was only slightly higher (between 1.5- and 2-fold
increased) compared with that in PC3 cells (Fig. 1A). In three human neuroblastoma
cell lines, SK-N-SH, CHP-134 and BE(2)-C cells, the expression
levels of miRNA-3613-3p were decreased compared with that in PC3
cells. The lowest mean expression of miRNA-3613-3p was observed in
BE(2)-C cells, with a 60% decrease compared with PC3 cells.
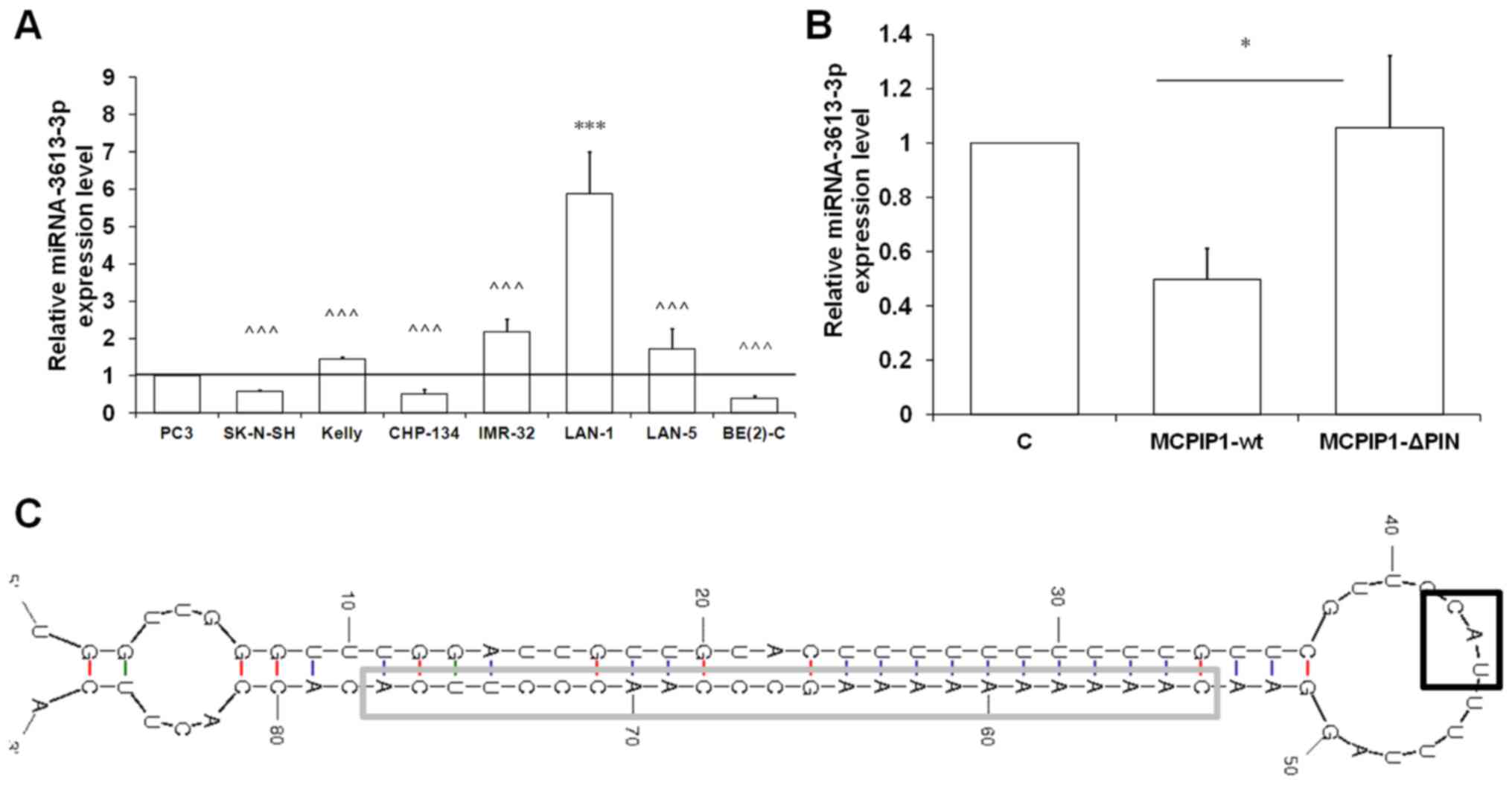 | Figure 1miRNA-3613-3p is differentially
expressed in a range of human neuroblastoma cell lines. (A)
miRNA-3613-3p expression levels in seven human neuroblastoma cell
lines and PC3 prostate cancer cell line determined using RT-qPCR
and U6 snRNA as the reference gene. (B) miRNA-3613-3p expression
level in Kelly human neuroblastoma cells with enforced expression
of the wild-type (MCPIP1-wt) or mutated (MCPIP1-ΔPIN) form of
MCPIP1 protein assessed by RT-qPCR. U6 snRNA was used as the
reference gene. Results are presented as the mean ± standard error
of the mean. To compare more than two means, analysis of variance
was performed [(A) F(1,7)=15.0046, P=0.000006; (B) F(1,2)=7.7228, P=0.0291]. (C)
Pre-miRNA-3613-3p secondary structure. The CAU sequence is shown
in-frame. *P<0.05 and ***P<0.001 vs.
control; ^^^P<0.001 vs. LAN-1. miRNA, microRNA;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; snRNA, small nuclear RNA; MCPIP1, monocyte
chemoattractant protein-induced protein 1; C, control; MCPIP1-wt,
wild-type MCPIP1; MCPIP1-ΔPIN, mutated ribonuclease domain-deprived
MCPIP1. |
In our previous study, it was identified that
enforced expression of the ribonuclease MCPIP1 caused significant
downregulation of miRNA-3616-3p in human neuroblastoma BE(2)-C
cells (7). To investigate this
observation further, the expression of miRNA-3613-3p was determined
in another neuroblastoma cell line with high MYCN
amplification, Kelly, with enforced MCPIP1 expression. The mean
levels of miRNA-3613-3p were significantly decreased in cells with
enforced MCPIP1 expression (50% decreased), compared with the
control cells (Fig. 1B). However,
in Kelly cells over-expressing the mutated form of MCPIP1 protein,
deprived of ribonucleolytic activity, no significant differences
were observed in miRNA-3613-3p expression (Fig. 1B). These results indicated that the
precursor form of miRNA-3613-3p may be cleaved directly by the
MCPIP1 protein. Visualization of the precursor form of
miRNA-3613-3p uncovered a characteristic
pyrimidine-purine-pyrimidine loop sequence (CAU) (Fig. 1C) that has been identified in all
MCPIP1 target genes (30).
miRNA-3613-3p potentially regulates
pathways involved in neuroblastoma pathogenesis
As the differential gene expression across the cell
lines suggests a potential involvement of miRNA-3613-3p in
neuroblastoma pathogenesis, miRNA-3613-3p was investigated by
bioinformatics analysis using the DIANA TOOLS-miRPath algorithm
(21) to determine its possible
function in this process. KEGG pathway analysis revealed 44
pathways that may be regulated by miRNA-3613-3p (all P<0.05).
Following a thorough review of the literature, 16 biological
processes involved in growth, metastasis or chemoresistance of
neuroblastoma associated with miRNA-3613-3p were identified
(Fig. 2A). The most important and
statistically significant signaling pathways were as follows: Hippo
signaling pathway, ubiquitin-mediated proteolysis, Wnt signaling
pathway, forkhead box O signaling pathway and transforming growth
factor β (TGFβ) signaling pathway (Fig. 2A). Furthermore, miRNA-3613-3p was
predicted to be involved in various cellular processes including
proteoglycan turnover in cancer, tumorigenesis-associated
transcriptional dysregulation and regulation of cellular signaling
in cancer (Fig. 2A).
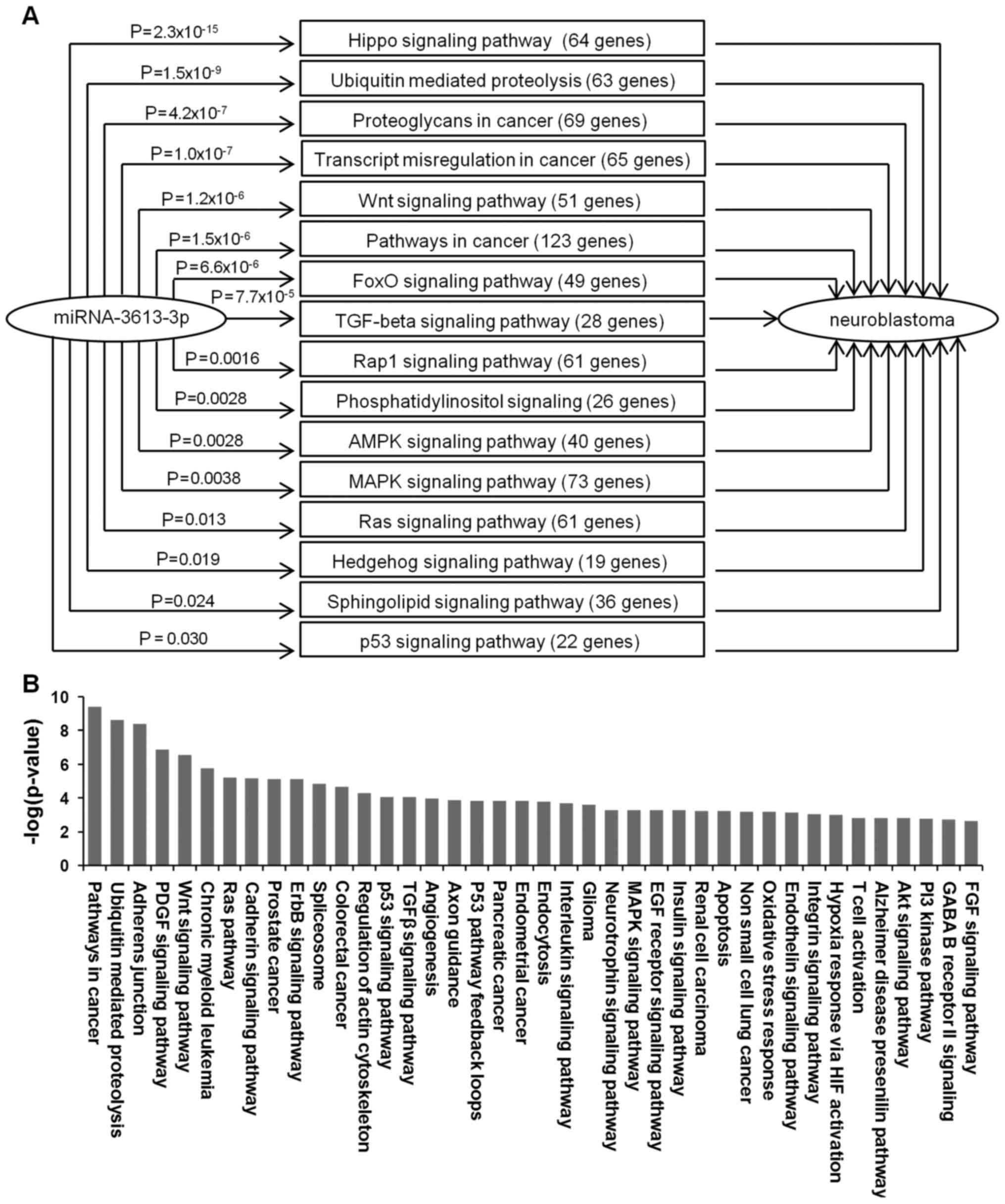 | Figure 2Bioinformatic analysis of the
potential involvement of miRNA-3613-3p in neuroblastoma
pathogenesis. (A) Potential Kyoto Encyclopedia of Genes and Genomes
miRNA-3613-3p target pathways identified through literature
research with involvement in neuroblastoma growth, metastasis or
chemoresistance. (B) Bioinformatic functional analysis of the
processes regulated by predicted miRNA-3613-3p target genes
generated using Gene Ontology and the PANTHER algorithm. miRNA,
microRNA; FoxO, forkhead box O; TGF, transforming growth factor;
AMPK, AMP-activated protein kinase; MAPK, mitogen-activated protein
kinase; FGF, fibroblast growth factor; GABA, γ-aminobutyric acid;
PI3 kinase, phosphoinositide 3-kinase; Akt, protein kinase B; EGF,
epidermal growth factor; PDGF, platelet-derived growth factor. |
Additionally, the predicted targets of miRNA-3613-3p
identified on the miRWalk2.0 database were investigated using
bioinformatics functional analysis using Gene Ontology and PANTHER
algorithms. The examination showed a substantial overlap with the
aforementioned KEGG pathway analysis. According to the Gene
Ontology algorithm, the majority of the putative miRNA-3613-3p
target genes regulate tumorigenesis; however, they may also be
involved in apoptosis and p53 pathway regulation (Fig. 2B). Bioinformatics functional
analysis using PANTHER algorithm revealed a potential involvement
of miRNA-3613-3p in the regulation of several signaling pathways
important in neuroblastoma pathogenesis, such as the Wnt, TGFβ or
protein kinase B (Akt) signaling pathways (Fig. 2B).
Bioinformatics analysis of predicted
miRNA-3613-3p target genes
The identification of mRNAs that interact with a
miRNA of interest usually starts by searching multiple databases
for predicted miRNA target genes. The algorithms developed for
prediction of miRNA-mRNA interactions take into account the miRNA
complementarity to the 3′UTR of the transcript. However, certain
algorithms additionally use conservation comparison, such as
TargetScan or DIANA-microT, whereas others consider binding site
accessibility (e.g. PITA and rna22) (31). In our previous study (7), eight putative target genes of
miRNA-3613-3p were proposed. In the present study, miRNA-3613-3p
was resubjected to thorough bioinformatics analysis using different
algorithms and seven of the originally proposed predicted target
genes were selected by identifying those that were present in all
databases with high probability values, and are involved in
neuroblastoma pathogenesis. The genes selected were as follows:
APAF1, DFFB, DICER, NF1, RORA,
KIF3A and VHL. Additionally, visualization of
miRNA-mRNA duplexes and analysis of the free energy of their
binding were performed. Putative binding sites with the free energy
of the binding <−20 kcal/mol (1 kcal=4.184 kJ) were identified
for all seven predicted target genes (Fig. 3A and B) (7). Furthermore, the free energies
calculated for estimated binding between DFFB or NF1
transcripts and miRNA-3613-3p were <−25 kcal/mol (7). The visualization of complementarity
between miRNA-3613-3p and the predicted binding sites at the target
mRNA 3′UTRs allowed the differentiation of well-defined seed
regions at the 5′ end of miRNA-3613-3p for all the putative target
genes with the exception of DICER. However, the fitting of
the DICER transcript and miRNA-3613-3p contained a distinct
3′-supplementary site (Fig.
3A).
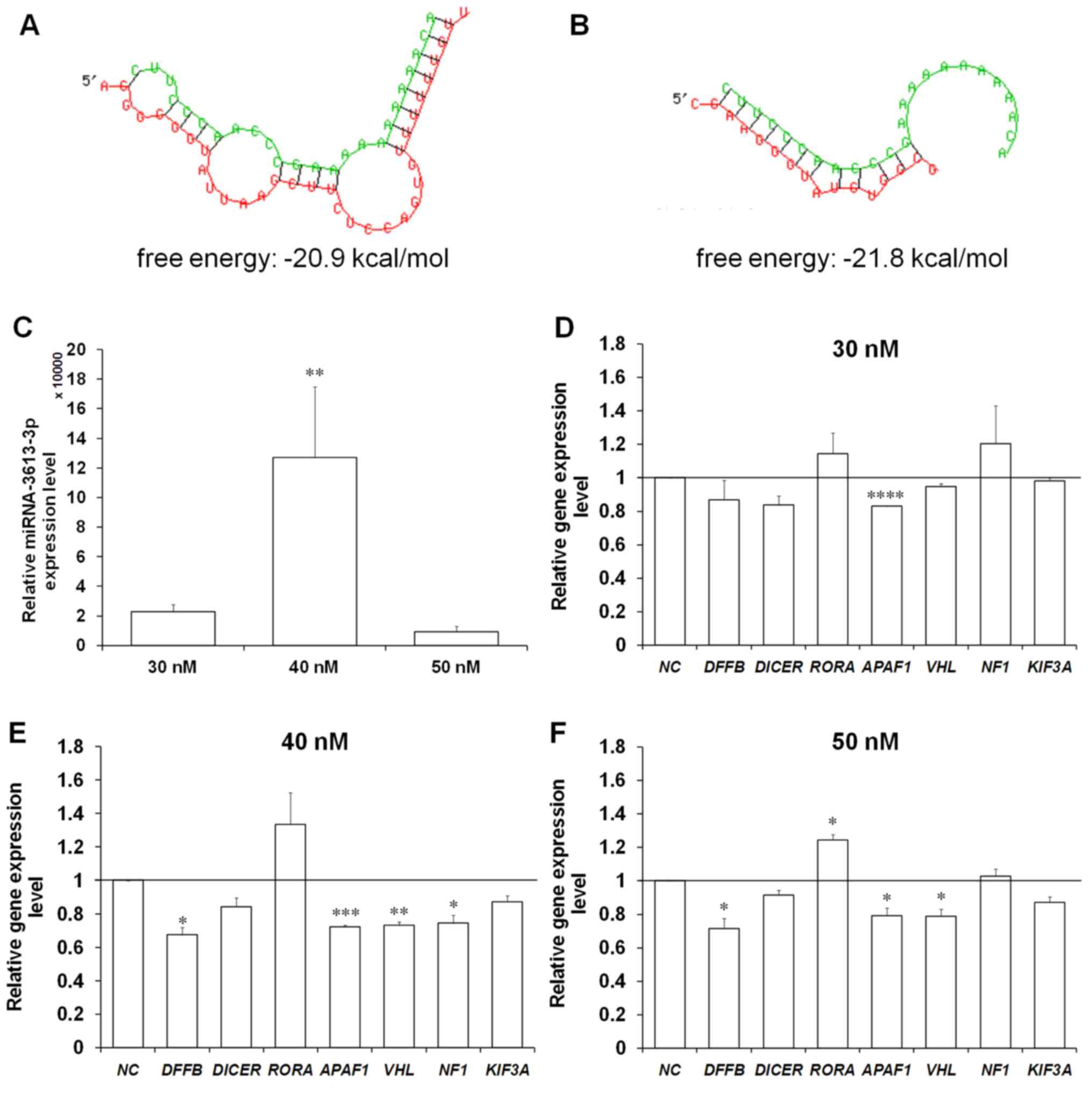 | Figure 3Enforced expression of miRNA-3613-3p
causes a significant decrease in the expression of certain of its
predicted target genes in the BE(2)-C human neuroblastoma cell
line. The visualization of complementarity regions of miRNA-3613-3p
and (A) DICER or (B) APAF1 mRNA, and calculation of
free energy of the binding. The miRNA sequence is indicated in
green and the target mRNA sequence is indicated in red. (C)
Expression levels of miRNA-3613-3p in the transfected cells
measured using RT-qPCR with U6 small nuclear RNA used as the
reference. Expression levels of seven predicted miRNA-3613-3p
target genes assessed using RT-qPCR in BE(2)-C cells transfected
with miRNA-3613-3p mimic at (D) 30, (E) 40 and (F) 50 nM. 40S
ribosomal protein S13 was used as the reference gene. Results are
presented as the mean ± standard error of the mean. To compare more
than two means, analysis of variance was performed [(A) F(1,3)=9.36334, P= 0.01111].
*P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 vs. NC.
miRNA, microRNA; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; NC, negative control (scrambled
oligonucleotide); DFFB, DNA fragmentation factor subunit β;
RORA, retinoic acid-related orphan receptor α; APAF1,
apoptotic protease-activating factor 1; VHL, von
Hippel-Lindau protein; NF1, neurofibromin 1; KIF3A,
kinesin family member 3A. |
Another important factor of miRNA-mediated
regulation of gene expression is binding site accessibility, which
depends on the secondary structure of a target RNA (31). The difference in the free energy of
the base-pairing interactions between miRNA-3613-3p and the mRNAs,
and within the mRNA sequence itself, were calculated and are
presented as the PITA score (Table
I). The general consensus is that predicted targets with PITA
scores <−10 kcal/mol are functional
(genie.weiz-mann.ac.il/pubs/mir07/mir07_notes.html). Such
differences in free energy of base pairing interactions between
miRNAs and mRNAs, and within the target sequence were identified
for DFFB, NF1 and RORA (Table I). Although the effect of
miRNA-mediated gene silencing depends on the miRNA concentration,
even sites with PITA scores >−10 kcal/mol may be functional in
cells with high expression of a certain miRNA (29). Therefore, it was decided not to
exclude any of the predicted target genes from further experimental
verification on the basis of the PITA score.
 | Table IBioinformatics analysis of putative
binding sites of target mRNAs. |
Table I
Bioinformatics analysis of putative
binding sites of target mRNAs.
| Gene | No. of sites | PITA score |
|---|
| APAF1 | 55 | −7.4 |
| DFFB | 53 | −11.53 |
| DICER | 151 | −7.78 |
| NF1 | 247 | −14.9 |
| RORA | 225 | −11.88 |
| KIF3A | 127 | −9.37 |
| VHL | 56 | −9.16 |
Enforced expression of miRNA-3613-3p
causes a significant decrease in the expression of several
predicted target genes
Human BE(2)-C neuroblastoma cells were transfected
with miRNA-3613-3p mimic to increase the level of miRNA-3613-3p.
This cell line was selected for transfection with miRNA mimic as it
expressed the lowest endogenous level of miRNA-3613-3p (Fig. 1A). As miRNA-mediated gene silencing
depends on the concentration of a certain miRNA in the cell
(32), three different
concentrations of the miRNA mimic were used for transfection (30,
40 and 50 nM). The mean level of enforced expression in BE(2)-C
cells transfected with the 40 nM concentration of the mimic was
increased ~6-fold compared with that in the cells transfected with
miRNA mimic at the 30 nM concentration (Fig. 3C). Of note, the mean expression
level of miRNA-3613-3p in the cells transfected with the highest
concentration of the mimic (50 nM) was lower compared with cells
transfected with the two other miRNA-3613-3p mimic concentrations
(30 and 40 nM) (Fig. 3C). This
phenomenon may have been caused by the toxic effect of high
concentrations of mimic in the cells. Therefore, the results
obtained for the cells transfected with 50 nM mimic should be
interpreted with caution.
The first approach to verify the aforementioned
predicted target genes of miRNA-3613-3p was to assess their
expression levels in BE(2)-C cells transfected with miRNA-3613-3p
mimic. In cells transfected with the lowest concentration of mimic,
a decrease in mRNA levels to 80% was observed only for APAF1
compared with the control cells transfected with a scrambled
oligonucleotide (Fig. 3D). The
mean levels of expression of other putative target genes
(DICER, DFFB, VHL, KIF3A, RORA
and NF1) remained unchanged compared with the control cells
(Fig. 3D). BE(2)-C cells
transfected with 40 nM miRNA mimic were characterized by a
significant downregulation in 4/7 predicted target genes, namely
DFFB, APAF1, VHL and NF1 to between 65
and 75% of control levels (Fig.
3E). As the mean overexpression of the miRNA in cells
transfected with 40 nM mimic was increased 6-fold compared with
cells transfected with the 30 nM group (Fig. 3C), it was hypothesized that an
increased level of enforced miRNA-3613-3p expression leads to more
potent silencing of the putative target genes. Although the level
of miRNA-3613-3p in the cells transfected with 50 nM miRNA mimic
was decreased compared with the cells transfected with 30 nM miRNA
mimic (Fig. 3C), a significant
decrease in the mean expression levels of DFFB, APAF1
and VHL, to 80% of the control, was observed (Fig. 3F).
To examine further the possible regulatory
mechanisms by miRNA-3613-3p on the predicted target genes, the
relative protein levels of two of the aforementioned seven putative
targets were determined. APAF1 was selected as its gene
expression was significantly decreased in the cells transfected
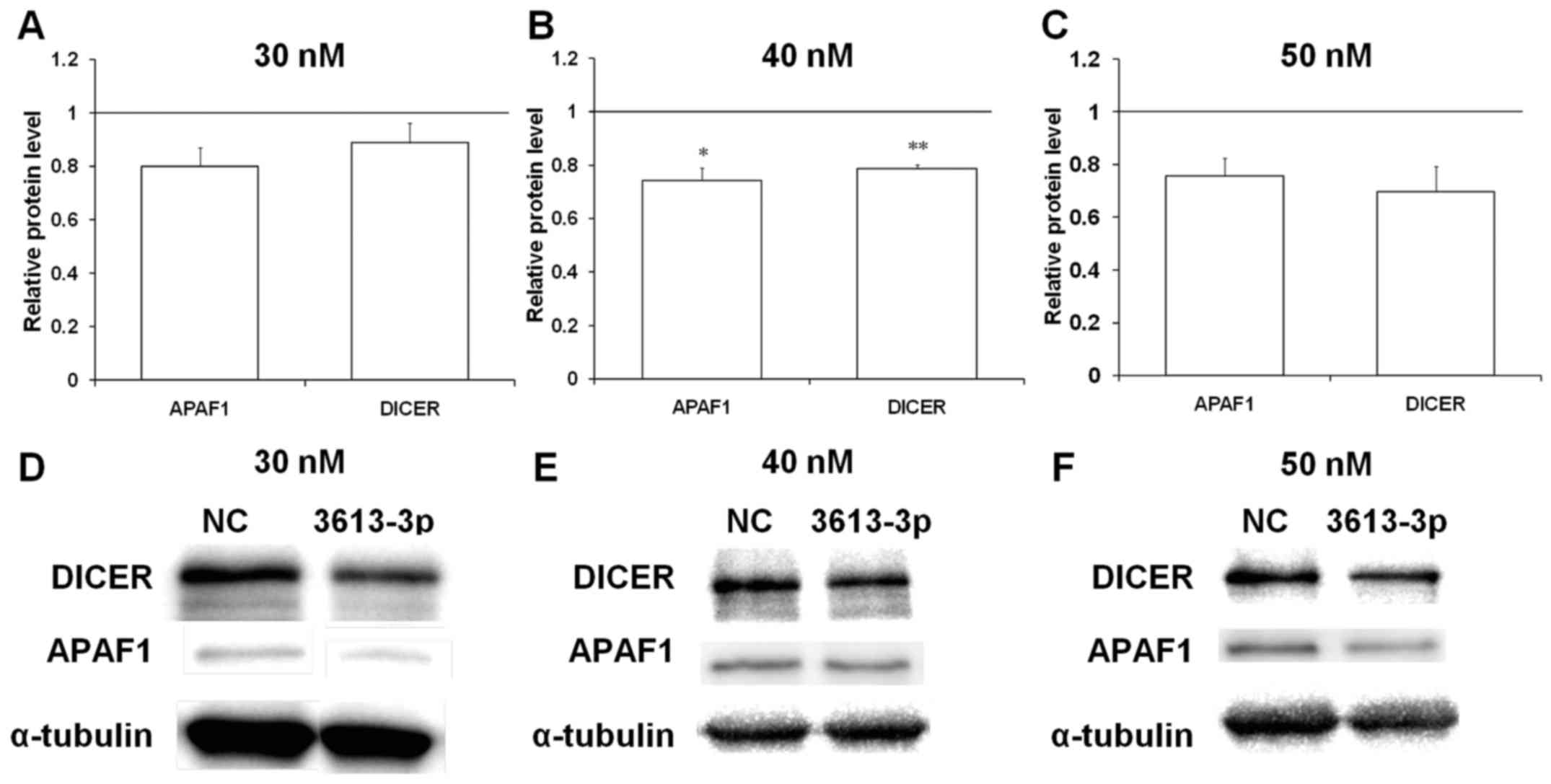
with miRNA mimic at all concentrations (Fig. 3D–F). Furthermore, the relative
protein levels of DICER in the transfected cells were
determined, as it has a crucial function in miRNA biogenesis
(33). Examination of the possible
binding between miRNA-3613-3p and APAF1 or DICER
mRNAs uncovered putative binding sites with low free energies in
the base pairing of the two transcripts (Fig. 3A and B). No significant alterations
were identified in the protein levels of APAF1 and DICER in cells
transfected with the mimic at 30 nM compared with in the control
cells (Fig. 4A). Western blot
analysis of APAF1 levels in the cells transfected with 40 nM
miRNA-3613-3p mimic revealed that its signal was downregulated to
between 70 and 80% of that of the control (Fig. 4B). Similarly, DICER protein
expression was downregulated only in cells transfected with 40 nM
mimic (Fig. 4B). These results
confirmed that a higher concentration of miRNA-3613-3p in BE(2)-C
neuroblastoma cells provided more potent silencing of the predicted
target genes at the protein level.
On the basis of the aforementioned results,
APAF1 was selected as the most likely target gene of
miRNA-3613-3p and subjected to further verification. The expression
of this gene was decreased upon enforced expression of
miRNA-3613-3p in all tested conditions at the mRNA and protein
levels (Figs. 3D–F and 4).
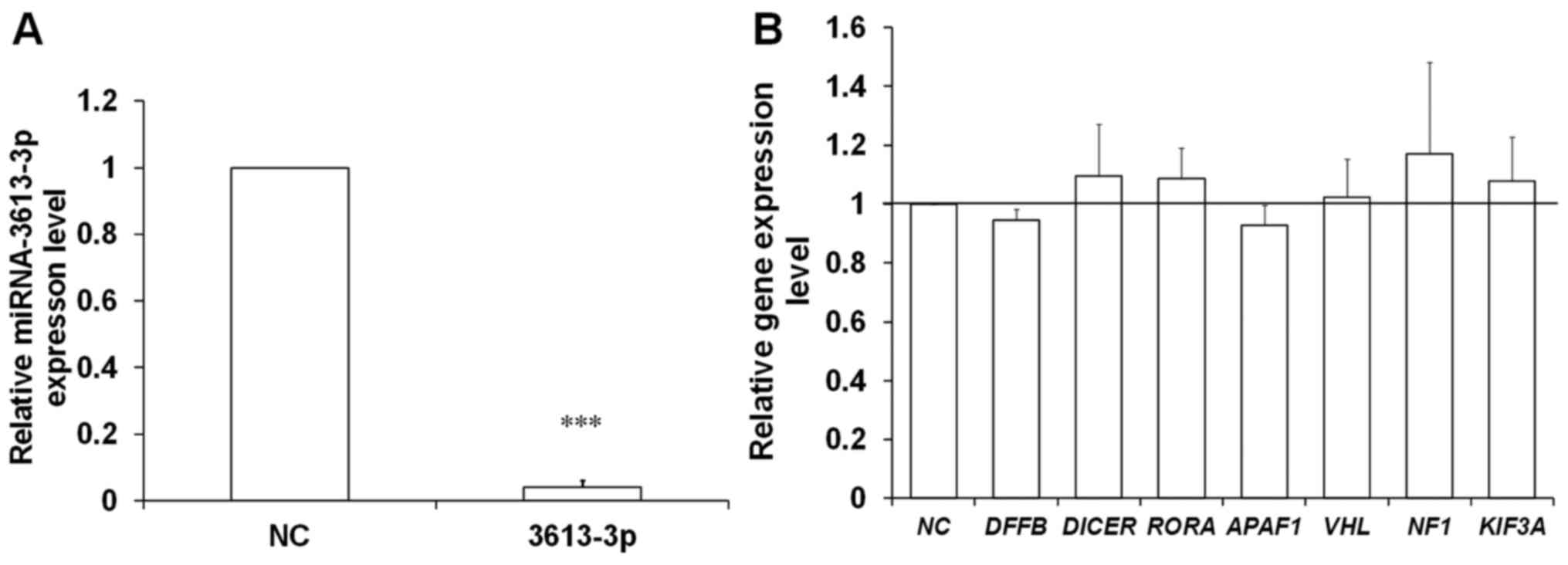
Downregulation of miRNA-3613-3p in LAN-1
neuroblastoma cells does not affect the expression of putative
target genes
Additionally, in order to obtain the downregulation
of the miRNA investigated, LAN-1 neuroblastoma cells were
transfected with miRNA-3613-3p inhibitor. This cell line was
selected for the transfection with the inhibitors owing to its
having the highest endogenous miRNA-3613-3p expression (Fig. 1A). The level of silencing achieved
of the miRNA investigated was significantly potent, at ~7% of the
control (Fig. 5A).
To examine further the effect of the altered
miRNA-3613-3p levels on the putative target gene expression, the
mRNA levels of all seven predicted target transcripts were assessed
in LAN-1 cells transfected with the inhibitors. As expected, the
expression of DFFB, APAF1, VHL and NF1
genes was not decreased in this model. In addition, an increase in
the expression of the putative miRNA-3613-3p target genes was not
observed (Fig. 5B). However, the
endogenous level of miRNA-3613-3p in LAN-1 neuroblastoma cells was
four orders of magnitude lower compared with in BE(2)-C cells
transfected with the mimic. Furthermore, the decrease in the
predicted target genes in the BE(2)-C cells upon upregulation of
the miRNA investigated was ~30% compared with the control (Fig. 3D–F). Therefore, the lack of
enhanced putative miRNA-3613-3p target gene expression in LAN-1
cells trans-fected with the inhibitors was not unforeseen.
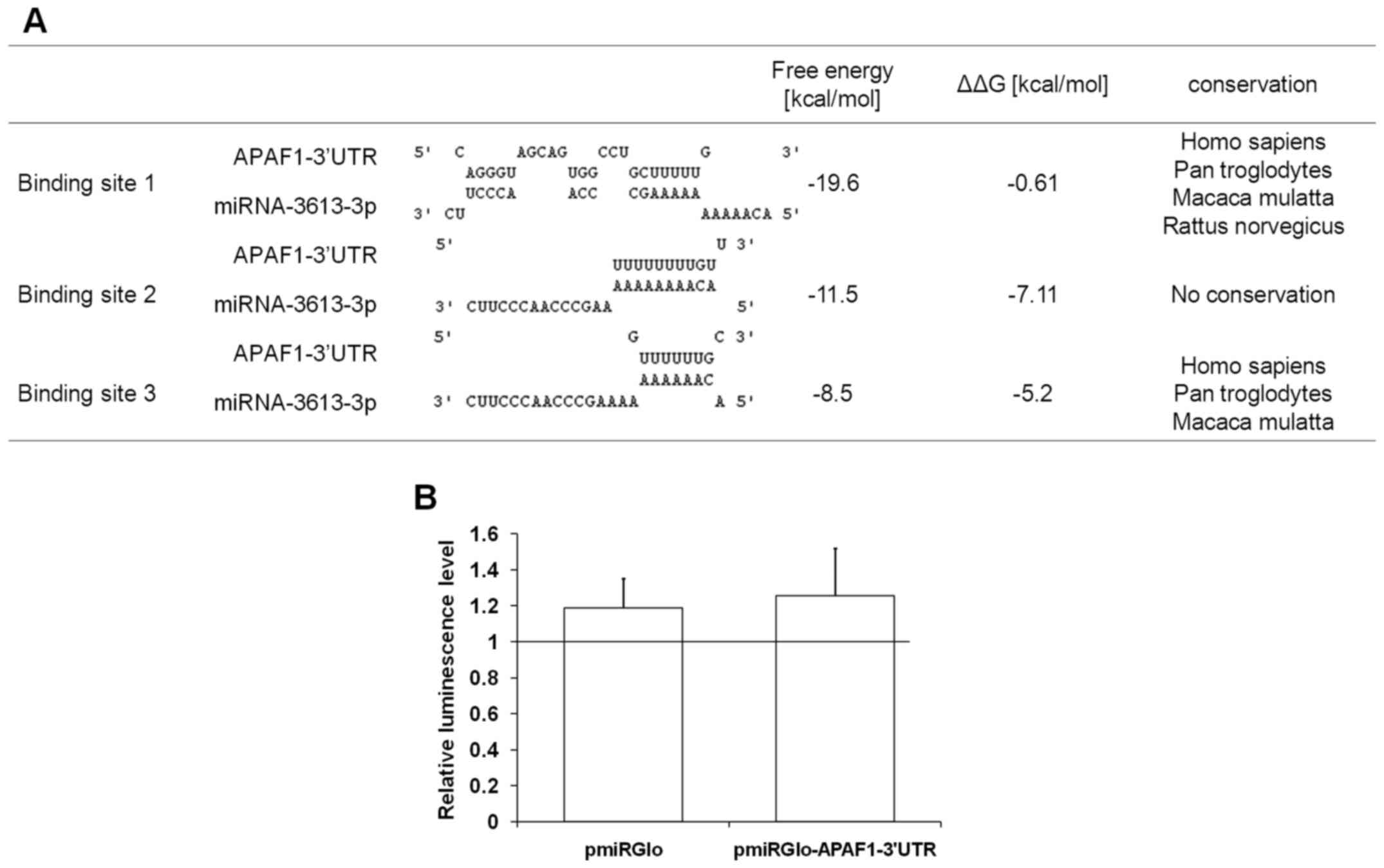
Putative binding sites for miRNA-3613-3p
predicted in the APAF1 sequence are not functional
Three putative binding sites for miRNA-3613-3p were
predicted in the APAF1 3′UTR (Fig. 6A). The free energies of the binding
between miRNA-3613-3p and APAF1 mRNA varied between −8.5 and
−20 kcal/mol (Fig. 6A).
Additionally, the differences between the free energies of the
base-pairing interactions between miRNA-3613-3p and APAF1
mRNA, and within the mRNA, were calculated using the PITA
algorithm. The analysis revealed lower free energies of the
miRNA-mRNA binding compared with base-pairing interactions within
the mRNA sequence, although for binding site 1, the difference was
minimal (Fig. 6A). Taking into
account the fact that cross-species conservation is usually
observed for functional miRNA-binding sites (31), the conservation of the three
predicted binding sites was investigated. Bioinformatics analysis
revealed that predicted binding sites 1 and 3, but not 2, are
conserved across species (Fig.
6A).
For experimental verification of the interaction
between the putative binding sites in the APAF1 3′UTR
sequence and miRNA-3613-3p, luciferase reporter assays were
performed. It was identified that miRNA-3613-3p did not affect the
luciferase activity in cells transfected with the pmiRGlo vector
containing the predicted binding sites of miRNA-3613-3p cloned from
the APAF1 3′UTR (Fig. 6B).
This result provides evidence for the lack of specific binding of
miRNA-3613-3p to the predicted binding sites in the APAF1
3′UTR. Therefore, the observed downregulation of APAF1
expression following miRNA-3613-3p overexpression (Figs. 3D–F and 4) may not have been caused by the direct
interactions between the APAF1 and miRNA-3613-3p
sequences.
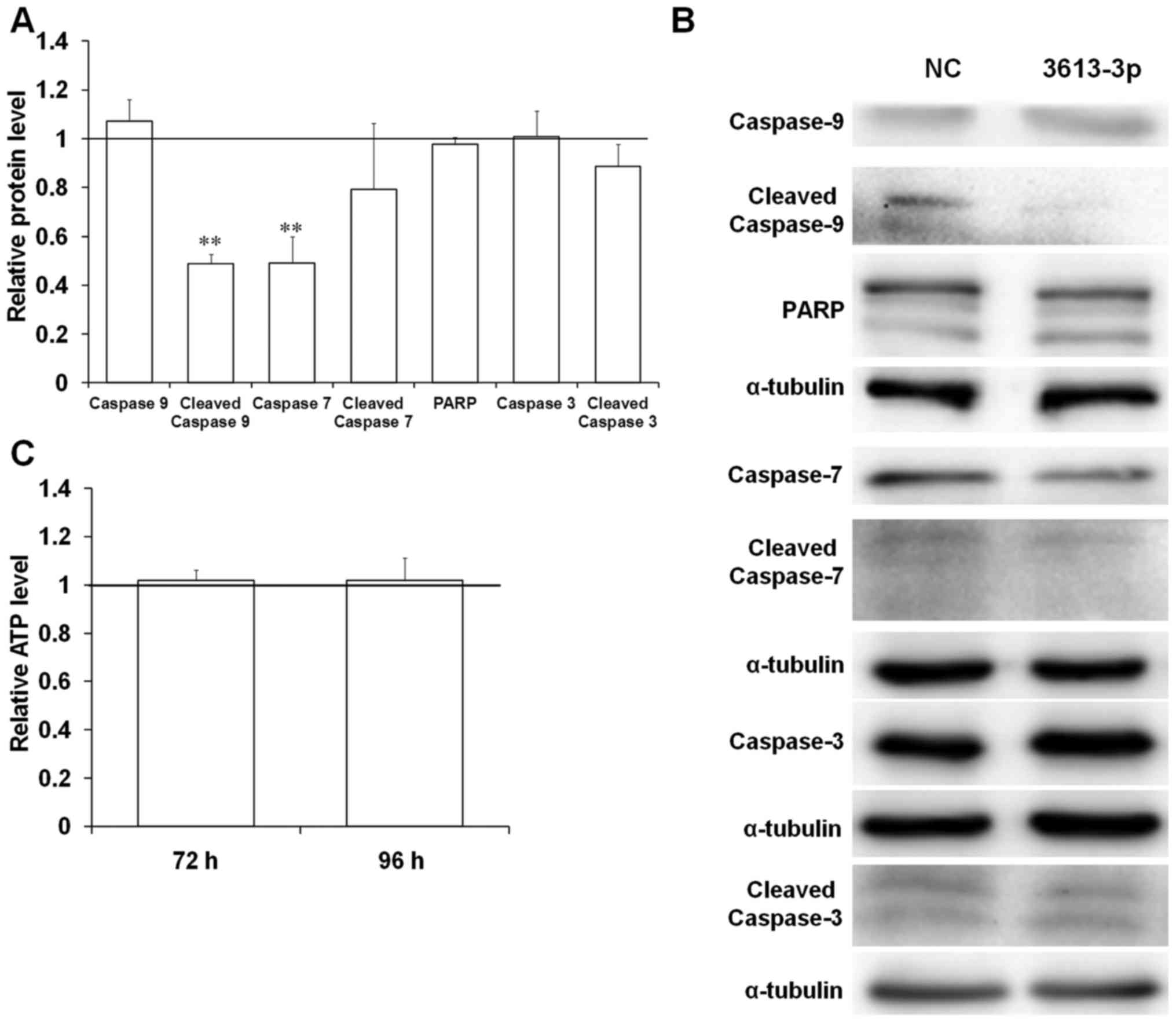
Enforced expression of miRNA-3613-3p
affects the levels and activation of selected proteins involved in
apoptosis in the BE(2)-C human neuroblastoma cell line
As the APAF1 protein serves a crucial function in
the triggering of caspase cascades via the intracellular apoptotic
pathway (34), the levels and
activation of the important proteins involved in this process was
investigated in cells with miRNA-3613-3p overexpression. The
western blot analysis results revealed that the protein expression
of PARP, caspase-3, cleaved caspase-3 and caspase-9 remained
unchanged in cells transfected with miRNA mimic (Fig. 7A and B). However, the relative
protein levels of caspase-7 significantly decreased to 50% of the
control group upon miRNA-3613-3p overexpression (Fig. 7A and B). APAF1 protein facilitates
proteolytic cleavage and activation of caspase-9 following the
release of cytochrome c from mitochondria (34). As the cells transfected with
miRNA-3613-3p mimic were characterized by a decrease in
APAF1 expression levels, it was expected that the activation
of the caspase-9 protein would be affected. A significant
downregulation in cleaved caspase-9 expression in the cells with
miRNA-3613-3p overexpression to 50% of the control was observed
(Fig. 7A and B). Furthermore, the
level of inactive caspase-9 remained unchanged in cells with
miRNA-3613-3p overexpression (Fig. 7A
and B). Thus, miRNA-3613-3p may have disrupted caspase-9
activation via APAF1 downregulation.
Apoptosis activation in cells serves a crucial
function in the regulation of cellular growth and survival.
Measurement of cellular ATP levels is a sensitive and precise
method for the assessment of cell viability (35). Therefore, the ATP content in the
cells transfected with miRNA-3613-3p mimic was determined to
investigate whether miRNA-3613-3p overexpression affects the
viability of BE(2)-C cells. The transfection of 40 nM miRNA-3613-3p
mimic did not significantly change the ATP level in cells after 72
and 96 h (Fig. 7C). This result
indicates that miRNA-3613-3p overexpression does not affect the
viability and growth of BE(2)-C cells.
Discussion
Patients with low- and intermediate-risk
neuroblastoma may be successfully treated with surgical resection
and neo-adjuvant chemotherapy. However, the treatment of high-risk
neuroblastoma cases requires aggressive multimodal therapies,
including chemotherapy, abscission of the tumor, radiotherapy,
13-cis-retinoic acid-induced cellular differentiation and
immunotherapy with anti-GD2 ganglioside antibodies (1). Despite the application of the
aforementioned treatment strategies, the event-free survival rate
of patients with high-risk neuroblastoma remains 50% (3). Thus, there is a requirement for the
development of novel therapeutic strategies.
miRNA molecules are considered a potential source of
novel clinical tools in neuroblastoma (36,37).
Differential miRNA expression in distinct subtypes of neuroblastoma
as well as the development of miRNA microarrays has enabled the use
of miRNA expression patterns as specific biomarkers for disease
risk stratification (36).
Furthermore, miRNA molecules are present in bodily fluids,
including plasma, serum and saliva. Assessment of circulating miRNA
expression patterns revealed significant differences between
healthy individuals and patients with neuroblastoma (37). The lack of invasiveness of the mode
of collection of circulating miRNA samples makes them a likely
novel class of precise biomarkers (37). In addition, a number of individual
miRNAs have been identified to function as potent oncogenes or
tumor suppressors, which provides opportunities for the development
of miRNA-based therapies (5). One
of the strategies is to restore the normal levels of
tumor-suppressive miRNA using miRNA mimics, chemically modified
oligonucleotides that have the sequence of the naturally occurring
miRNA molecule (37). Targeted
delivery of miRNA-34a mimic to neuroblastoma cells was demonstrated
to inhibit tumor growth in vivo (38). Another potential strategy for
miRNA-based therapies is silencing of specific oncogenic miRNA via
antagomirs, chemically modified oligonucleotides that have
complementary sequences to an endogenous miRNA (39). Injection of miRNA-17-5p antagomirs
leads to complete neuroblastoma tumor regression in vivo in
30% of cases (40). With the
increasing possibility of the clinical application of miRNAs in
neuroblastoma treatment, it is important to understand the
involvement of miRNA-dependent gene regulation networks in disease
pathogenesis.
In our previous study, it was demonstrated that
MCPIP1 ribonuclease overexpression significantly inhibits the
growth and proliferation rate of BE(2)-C human neuroblastoma cells
(41). Application of miRNA
microarrays allowed for the delineation of miRNA expression
patterns in BE(2)-C cells following MCPIP1 overexpression. Of note,
the most signifi-cantly downregulated miRNA upon MCPIP1
overexpression was a novel miRNA, miRNA-3613-3p (7), which, to the best of our knowledge,
had not been previously investigated in neuroblastoma. Deregulation
of miRNA-3613-3p expression in Kelly and BE(2)-C cells with MCPIP1
overexpression, characterized by inhibition of pro-proliferative
pathways, suggests its involvement in the regulation of
neuroblastoma cell biology. Additionally, it was identified that
the overexpression of MCPIP1 protein causes the downregulation of
the most potent oncogene in neuroblastoma, MYCN, in two
highly tumorigenic cell lines, BE(2)-C and Kelly, as well as
inhibition of the Akt/mammalian target of rapamycin signaling
pathway (17). Of note, the
PANTHER algorithm revealed the potential involvement of
miRNA-3613-3p in the regulation of the Akt signaling pathway.
Further evidence of the involvement of miRNA-3613-3p in
neuroblastoma cell biology is the differential expression of
miRNA-3613-3p demonstrated in our study in a variety of human
neuroblastoma cell lines. Of note, the cell lines characterized by
increased miRNA-3613-3p expression (LAN-1, LAN-5, IMR-32 and
Kelly), compared with the PC3 cell line, are N-type neuroblastoma
cells (42). They adhere poorly to
the cell culture plate and exhibit a tendency to aggregate.
Furthermore, the cell lines with the lowest endogenous expression
of miRNA-3613-3p [BE(2)-C and CHP-134] are I-type neuroblastoma
cells, identified by marked adhesion to the cell culture plate and
extensive migration (43,44). Thus, there may be an association
between the expression level of miRNA-3613-3p and neuroblastoma
cell phenotype. Comparison of the expression of the miRNA
investigated in neuroblastoma cell lines and the normal cells of
the same origin would be worthwhile and may shed more light on the
possible oncogenic function of miRNA-3613-3p in the pathogenesis of
this type of cancer. Neuroblastoma is an embryonal tumor that
arises from the sympathoadrenal cells in the neural crest of the
embryo. Owing to technical difficulties and ethical considerations,
human embryonic neural crest cells are not available for research
purposes. Deriving neural crest cells from human induced
pluripotent stem cells appears to be a promising alternative and is
currently being developed by several research groups (45).
An extensive search of the databases and analysis
using various bioinformatics software allowed us to select seven
putative target genes of miRNA-3613-3p. Of note, all the predicted
target genes that were downregulated in cells with miRNA-3613-3p
overexpression (APAF1, DFFB, DICER, VHL
and NF1) exhibit a tumor-suppressor potential. APAF1
encodes a protein crucial for the activation of the caspase cascade
in the programmed cell death intracellular pathway (34). DFFB, another potential
miRNA-3613-3p target gene, encodes a nuclease involved in DNA
fragmentation during apoptosis (46). As one of the hallmarks of cancer is
resistance to programmed cell death, pro-apoptotic proteins exhibit
tumor-suppressive potential in numerous types of cancer (47). Additionally, APAF1 was identified
to be a suppressor of another type of tumor of neuroectodermal
origin: Melanoma (48).
Furthermore, DICER, a putative target gene of miRNA-3613-3p,
encodes a key ribonuclease involved in miRNA biogenesis, which
supports the finding of global miRNA downregulation in the
pathogenesis of neuroblastoma (5).
Furthermore, a low expression of this gene was identified to be a
prognostic factor for stage 4 neuroblastoma (33). VHL serves a function in
neuronal cell differentiation (49). In addition, the expression level of
this gene could serve as a biomarker in neuroblastoma, and its
downregulation points to a high-risk subtype of the disease
(50). NF1 encodes a
negative regulator of the mitogen-activated protein kinase
signaling pathway and is an important prognostic factor of retinoic
acid therapy outcome (51).
Additionally, BE(2)-C cells with MCPIP1 protein overexpression and
miRNA-3613-3p downregulation are characterized by significant
increases in APAF1 and DFFB levels at the
transcriptional and translational levels (7). These results support the data from
the present study that demonstrated the downregulation of the
expression of the aforementioned genes in BE(2)-C cells transfected
with miRNA-3613-3p mimic, and may indicate an oncogenic function of
miRNA-3613-3p. However, owing to high heterogeneity of
neuroblastoma cell lines, it may be difficult to draw general
conclusions concerning the function of the gene in the pathogenesis
of this type of cancer. Silencing of the miRNA investigated in
another neuroblastoma cell line, LAN-1, did not lead to an increase
in expression of any of the seven predicted target genes. This may
suggest that the action of miRNA-3613-3p depends strongly on the
cellular context.
Of the seven putative target genes of miRNA-3613-3p,
the downregulation of APAF1 in cells with miRNA-3613-3p
overexpression was the most significant compared with control
cells. Despite the presence of the three putative binding sites for
miRNA-3613-3p at the 3′UTR of APAF1 mRNA, the reporter gene
assay did not signal an interaction between the predicted sequences
in the transcript and miRNA-3613-3p. Nevertheless, the APAF1
gene may be regulated by miRNA-3613-3p through binding sites at the
5′UTR or coding sequence in the transcript. Alternatively, the
downregulation of the gene in cells with miRNA-3613-3p
overexpression may be the result of miRNA-3613-3p targeting an
activator of APAF1, subsequently downregulating APAF1
expression indirectly.
The protein product of the APAF1 gene serves
a key function in the activation of the caspase cascade in the
intracellular apoptotic pathway. Following release from the
mitochondria, cytochrome c forms a complex with the APAF1
oligomer. This interaction allows caspase-9 to be cleaved and
activated, which consequently leads to the proteolytic activation
of executive caspases, such as caspases-7 and -3, resulting in
apoptosis (34). Analysis of the
expression and activation of several proteins involved in
programmed cell death in cells transfected with the miRNA-3613-3p
mimic produced notable results. A significant decrease was observed
in the level of activated cleaved caspase-9 in human BE(2)-C
neuroblastoma cells. This is in accordance with the aforementioned
decrease in the expression of the activator of this caspase, APAF1
protein, in cells transfected with the miRNA-3613-3p mimic.
Inhibition of caspase-9 proteolysis in cells with ectopic
miRNA-3613-3p expression may limit the possibility of activating
the apop-tosis process. Furthermore, in cells with miRNA-3613-3p
overexpression, no alterations in the levels and activation of the
executive caspase-3 and PARP protein, responsible for DNA
fragmentation during the last phase of the apoptosis process
(52), were identified. This
confirms further a lack of activation of the programmed cell death
process following miRNA-3613-3p overexpression in human BE(2)-C
neuroblastoma cells.
In conclusion, the results of the present study
identified that miRNA-3613-3p may directly or indirectly regulate
the expression of several genes with tumor suppressor potential
(APAF1, DFFB, NF1, VHL and
DICER) in human neuroblastoma cells. The most likely target
gene of miRNA-3613-3p appears to be APAF1; however, this
interaction might not be direct. Transfection with miRNA mimic did
not result in the activation of BE(2)-C cell apoptosis after 96 h.
However, it may inhibit this process by lowering caspase-9
proteolysis via downregulation of APAF1 protein. Additionally, it
was identified that enforced expression of miRNA-3613-3p does not
affect the viability of BE(2)-C cells. The results obtained
indicate a possible tumor-promoting function of miRNA-3613-3p in
BE(2)-C neuroblastoma cells.
Acknowledgments
Not applicable.
Funding
The present study was supported by a grant from the
Research Project Competition for Young Researchers and PhD Students
of the Faculty of Biochemistry, Biophysics and Biotechnology
Jagiellonian University (grant no. 12/2016) and a grant from the
Polish National Science (grant no. 2011/03/B/NZ1/00024). The
Faculty of Biochemistry, Biophysics and Biotechnology of
Jagiellonian University is a partner of the Leading National
Research Center (KNOW) supported by the Ministry of Science and
Higher Education.
Availability of data and materials
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
Authors' contributions
IN, EB and MD performed experiments and acquired
data; IN interpreted the data; IN designed experiments and drafted
the manuscript; HR and IH edited the manuscript. All authors
approved the final content for journal submission and
publication.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Louis CU and Shohet JM: Neuroblastoma:
Molecular pathogenesis and therapy. Annu Rev Med. 66:49–63. 2015.
View Article : Google Scholar :
|
|
2
|
Maris JM, Hogarty MD, Bagatell R and Cohn
SL: Neuroblastoma. Lancet. 369:2106–2120. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pinto NR, Applebaum MA, Volchenboum SL,
Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F,
Schleiermacher G, Park JR, et al: Advances in risk classification
and treatment strategies for neuroblastoma. J Clin Oncol.
33:3008–3017. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
de Carvalho IN, de Freitas RM and Vargas
FR: Translating microRNAs into biomarkers: What is new for
pediatric cancer? Med Oncol. 33:492016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhi F, Wang R, Wang Q, Xue L, Deng D, Wang
S and Yang Y: MicroRNAs in neuroblastoma: Small-sized players with
a large impact. Neurochem Res. 39:613–623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mei H, Lin ZY and Tong QS: The roles of
microRNAs in neuroblastoma. World J Pediatr. 10:10–16. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boratyn E, Nowak I, Horwacik I, Durbas M,
Mistarz A, Kukla M, Kaczówka P, Łastowska M, Jura J and Rokita H:
Monocyte chemoattractant protein-induced protein 1 overexpression
modulates transcriptome, including microRNA, in human neuroblastoma
cells. J Cell Biochem. 117:694–707. 2016. View Article : Google Scholar
|
|
8
|
Liu H, Chen G, Liang M, Qin H, Rong J, Yao
J and Wu Z: Atrial fibrillation alters the microRNA expression
profiles of the left atria of patients with mitral stenosis. BMC
Cardiovasc Disord. 14:102014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ji H, Chen M, Greening DW, He W, Rai A,
Zhang W and Simpson RJ: Deep sequencing of RNA from three different
extracellular vesicle (EV) subtypes released from the human LIM1863
colon cancer cell line uncovers distinct miRNA-enrichment
signatures. PLoS One. 9:e1103142014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang N, Bu R, Duan Z, Zhang X, Chen P, Li
Z, Wu J, Cai G and Chen X: Profiling and initial validation of
urinary microRNAs as biomarkers in IgA nephropathy. PeerJ.
3:e9902015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pu Q, Huang Y, Lu Y, Peng Y, Zhang J, Feng
G, Wang C, Liu L and Dai Y: Tissue-specific and plasma microRNA
profiles could be promising biomarkers of histological
classification and TNM stage in non-small cell lung cancer. Thorac
Cancer. 7:348–354. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kumar S, Vijayan M and Reddy PH:
MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer's
disease. Hum Mol Genet. 26:3808–3822. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh A, Rooge S, Varshney A, Vasudevan M,
Bhardwaj A, Venugopal S, Trehanpati N, Kumar M, Geffers R, Kumar V,
et al: Global micro RNA expression profiling in the liver biopsies
of Hepatitis B virus infected patients suggests specific miRNA
signatures for viral persistence and hepatocellular injury.
Hepatology. 67:1695–1709. 2017. View Article : Google Scholar
|
|
14
|
Zhang D, Liu E, Kang J, Yang X and Liu H:
MiR-3613-3p affects cell proliferation and cell cycle in
hepatocellular carcinoma. Oncotarget. 8:93014–93028.
2017.PubMed/NCBI
|
|
15
|
Zhang Y, Kang R, Liu W, Yang Y, Ding R,
Huang Q, Meng J, Xiong L and Guo Z: Identification and analysis of
p53-mediated competing endogenous RNA network in human
hepatocellular carcinoma. Int J Biol Sci. 13:1213–1221. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mizgalska D, Wegrzyn P, Murzyn K, Kasza A,
Koj A and Jura J, Jarzab B and Jura J: Interleukin-1-inducible
MCPIP protein has structural and functional properties of RNase and
participates in degradation of IL-1beta mRNA. FEBS J.
276:7386–7399. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Boratyn E, Nowak I, Durbas M, Horwacik I,
Sawicka A and Rokita H: MCPIP1 exogenous overexpression inhibits
pathways regulating MYCN oncoprotein stability in neuroblastoma. J
Cell Biochem. 118:1741–1755. 2017. View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
19
|
Smith PK, Krohn RI, Hermanson GT, Mallia
AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ and
Klenk DC: Measurement of protein using bicinchoninic acid. Anal
Biochem. 150:76–85. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Horwacik I, Durbas M, Boratyn E, Węgrzyn P
and Rokita H: Targeting GD2 ganglioside and aurora A kinase as a
dual strategy leading to cell death in cultures of human
neuroblastoma cells. Cancer Lett. 341:248–264. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vlachos IS, Zagganas K, Paraskevopoulou
MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T and
Hatzigeorgiou AG: DIANA-miRPath v3.0: Deciphering microRNA function
with experimental support. Nucleic Acids Res. 43:W460–6. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paraskevopoulou M, Georgakilas G,
Kostoulas N, Vlachos I, Vergoulis T, Reczko M, Filippidis C,
Dalamagas T and Hatzigeorgiou A: DIANA-microT web server v5.0:
service integration into miRNA functional analysis workflows.
Nucleic Acids Res. 41:W169–W173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:e050052015. View Article : Google Scholar :
|
|
27
|
Hessvik NP, Phuyal S, Brech A, Sandvig K
and Llorente A: Profiling of microRNAs in exosomes released from
PC-3 prostate cancer cells. Biochim Biophys Acta. 1819:1154–1163.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chiyomaru T, Yamamura S, Fukuhara S,
Hidaka H, Majid S, Saini S, Arora S, Deng G, Shahryari V, Chang I,
et al: Genistein up-regulates tumor suppressor microRNA-574–3p in
prostate cancer. PLoS One. 8:e589292013. View Article : Google Scholar
|
|
29
|
Sohn EJ, Won G, Lee J, Lee S and Kim SH:
Upregulation of miRNA3195 and miRNA374b mediates the
anti-angiogenic properties of melatonin in hypoxic PC-3 prostate
cancer cells. J Cancer. 6:19–28. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mino T, Murakawa Y, Fukao A, Vandenbon A,
Wessels HH, Ori D, Uehata T, Tartey S, Akira S, Suzuki Y, et al:
Regnase-1 and Roquin regulate a common element in inflammatory
mRNAs by spatiotemporally distinct mechanisms. Cell. 161:1058–1073.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Witkos TM, Koscianska E and Krzyzosiak WJ:
Practical aspects of microRNA target prediction. Curr Mol Med.
11:93–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shu J, Xia Z, Li L, Liang ET, Slipek N,
Shen D, Foo J, Subramanian S and Steer CJ: Dose-dependent
differential mRNA target selection and regulation by let-7a-7f and
miR-17-92 cluster microRNAs. RNA Biol. 9:1275–1287. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY,
Diccianni MB, London WB, Chang CH and Yu AL: microRNA signature and
expression of Dicer and Drosha can predict prognosis and delineate
risk groups in neuroblastoma. Cancer Res. 70:7841–7850. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cecconi F: Apaf1 and the apoptotic
machinery. Cell Death Differ. 6:1087–1098. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Riss T, Moravec R, Niles A, Duellman S,
Benink H, Worzella T and Minor L: Cell Viability Assays in Assay
Guidance Manual. Sittampalam G, et al: Eli Lilly & Company and
the National Center for Advancing Translational Sciences Bethesta,
MD: 2013
|
|
36
|
Stallings RL: MicroRNA involvement in the
pathogenesis of neuroblastoma: Potential for microRNA mediated
therapeutics. Curr Pharm Des. 15:456–462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shalaby T, Fiaschetti G, Baumgartner M and
Grotzer MA: Significance and therapeutic value of miRNAs in
embryonal neural tumors. Molecules. 19:5821–5862. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tivnan A, Orr WS, Gubala V, Nooney R,
Williams DE, McDonagh C, Prenter S, Harvey H, Domingo-Fernández R,
Bray IM, et al: Inhibition of neuroblastoma tumor growth by
targeted delivery of microRNA-34a using anti-disialoganglioside GD2
coated nanoparticles. PLoS One. 7:e381292012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Verissimo CS, Molenaar JJ, Fitzsimons CP
and Vreugdenhil E: Neuroblastoma therapy: What is in the pipeline?
Endocr Relat Cancer. 18:R213–R231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fontana L, Fiori ME, Albini S, Cifaldi L,
Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V,
Giacomini P, et al: Antagomir-17-5p abolishes the growth of
therapy-resistant neuroblastoma through p21 and BIM. PLoS One.
3:e22362008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Skalniak A, Boratyn E, Tyrkalska SD,
Horwacik I, Durbas M, Lastowska M, Jura J and Rokita H: Expression
of the monocyte chemotactic protein-1-induced protein 1 decreases
human neuro-blastoma cell survival. Oncol Rep. 31:2385–2392. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Baumann Kubetzko FB, Di Paolo C, Maag C,
Meier R, Schäfer BW, Betts DR, Stahel RA and Himmelmann A: The PAX5
oncogene is expressed in N-type neuroblastoma cells and increases
tumorigenicity of a S-type cell line. Carcinogenesis. 25:1839–1846.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Voigt A, Hartmann P and Zintl F:
Differentiation, proliferation and adhesion of human neuroblastoma
cells after treatment with retinoic acid. Cell Adhes Commun.
7:423–440. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ross R: Cellular heterogeneity.
Neuroblastoma. Cheung N and Cohn S: Springer; Berlin: pp. 55–60.
2005, View Article : Google Scholar
|
|
45
|
Liu JA and Cheung M: Neural crest stem
cells and their potential therapeutic applications. Dev Biol.
419:199–216. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Judson H, van Roy N, Strain L,
Vandesompele J, Van Gele M, Speleman F and Bonthron DT: Structure
and mutation analysis of the gene encoding DNA fragmentation factor
40 (caspase-activated nuclease), a candidate neuroblastoma tumour
suppressor gene. Hum Genet. 106:406–413. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Campioni M, Santini D, Tonini G, Murace R,
Dragonetti E, Spugnini EP and Baldi A: Role of Apaf-1, a key
regulator of apoptosis, in melanoma progression and
chemoresistance. Exp Dermatol. 14:811–818. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Murata H, Tajima N, Nagashima Y, Yao M,
Baba M, Goto M, Kawamoto S, Yamamoto I, Okuda K and Kanno H: Von
Hippel-Lindau tumor suppressor protein transforms human
neuroblastoma cells into functional neuron-like cells. Cancer Res.
62:7004–7011. 2002.PubMed/NCBI
|
|
50
|
Hoebeeck J, Vandesompele J, Nilsson H, De
Preter K, Van Roy N, De Smet E, Yigit N, De Paepe A, Laureys G,
Påhlman S, et al: The von Hippel-Lindau tumor suppressor gene
expression level has prognostic value in neuroblastoma. Int J
Cancer. 119:624–629. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hölzel M, Huang S, Koster J, Ora I,
Lakeman A, Caron H, Nijkamp W, Xie J, Callens T, Asgharzadeh S, et
al: NF1 is a tumor suppressor in neuroblastoma that determines
retinoic acid response and disease outcome. Cell. 142:218–229.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Savitskaya MA and Onishchenko GE:
Mechanisms of apoptosis. biochemistry (Mosc). 80:1393–1405. 2015.
View Article : Google Scholar
|