Introduction
Cervical cancer is the fourth most common malignancy
in women worldwide, caused by high-risk human papilloma virus (HPV)
strains, mainly HPV16 and HPV18 (1–4), and
if not treated promptly, could eventually lead to the development
of malignancy. Cervical cancer incidence and mortality are
declining in developing countries, mainly due to the introduction
of effective screening tests (5,6). The
treatment of cervical cancer relies on surgical interventions
combined with radiotherapy methods that may cause fertility
impairment (7,8). Despite significant efforts in the
last 20 years, novel therapeutic approaches have not been
introduced in clinical practice (9,10).
The reason for this delay is the paucity of comprehensive studies
on the actual molecular determinants of cervical malignancy that
could open the way for the identification of novel therapeutic
targets. Such an approach can actually be effective, as is
illustrated by the recent paradigm of bladder cancer. The
characterization of the molecular landscape of bladder cancer led
to the introduction of the first targeted therapy for the disease,
namely employing antibodies that inactivate programmed death
receptor 1/programmed death receptor ligand 1 (11). This paradigm indicates that
membrane proteins are excellent therapeutic targets, and that an
efficient approach to characterize them in depth is actually
offered by proteomic methods (12,13).
Membrane proteins are involved in important processes, including
signal transduction, cell communication and molecule transport.
Additionally, their systematic study could eventually lead to the
identification of biomarkers for prognostic or diagnostic
purposes.
The systematic analysis of the proteomic studies of
cervical cancer cell lines by our group, involving their secretome
and the total cell extract (14–16),
offered several exciting insights on the biological processes
relevant to cervical cancer pathology, including cytoskeletal
remodeling and redox signaling. However, the membrane proteome has
not been investigated thus far due to its particular biochemical
properties, such as hydrophobicity (12). Thus, the aim of the present study
was to develop an efficient and reproducible membrane protein
enrichment protocol, and to the best our knowledge, to compare for
the first time the expression of membrane proteins of four
informative cervical cell lines, one derived from normal cervical
keratinocytes (HCK1T) and three cancerous types, C33A
(HPV−), SiHa (HPV16+) and HeLa
(HPV18+).
Materials and methods
Cell culture
The cancer cell lines SiHa (HPV16+), HeLa
(HPV18+) and C33A (HPV−), commercially
available from the American Type Culture Collection (Manassas, VA,
USA), were cultured as previously described (17). HCK1T cells were kindly provided by
Dr Tohru Kiyono (Division of Carcinogenesis and Cancer Prevention,
National Cancer Center Research Institute, Tokyo, Japan) and were
cultured as previously described (18). A cell pellet of ~107
cells/75−cm2 flask was stored at −20°C until use. Four
biological replicates were used for each cell line.
Total protein isolation
The collection of the total cell extract was
performed as previously described (14).
Membrane protein isolation
Initially, the cell pellet was resolubilized in 1 ml
of solution 1 [25 mM Tris-HCl (pH 7.5) (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA), 45% sucrose (Fluka, Munich, Germany), 1 mM EDTA
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 150 mM
NaCl (AppliChem GmbH, Darmstadt, Germany) and 10 mM
MgCl2 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany),
with protease inhibitors (Roche Diagnostics, Basel, Switzerland) at
3.6% v/v final concentration], incubated for 20 min on ice, and
homogenized with Dounce homogenizer 10–20 times. Sonication was
used to achieve cell lysis, followed by centrifugation for 10 min
at 2,000 × g at 4°C, with aspiration of the supernatant and removal
of the pellet. Solution 2 [25 mM Tris-HCl (pH 7.5), 150 mM NaCl and
10 mM MgCl2] was added to the supernatant to a final
volume of 2 ml. Furthermore, ultra-centrifugation at 100,000 × g
was performed for 1 h at 4°C to collect the pellet containing the
membranes. The pellet was solubilized in 2 ml of solution 2.
Ultra-centrifugation was performed at 100,000 × g for 1 h at 4°C.
The resulting pellet was solubilized in 432 µl of solution 3
[25 mM Tris-HCl (pH 7.5), 150 mM NaCl and 3.6% v/v protease
inhibitors]. This step was followed by the addition of SDS (Bio-Rad
Laboratories, Inc.) to a final concentration of 2% w/v, and the
solution was incubated for 30 min at room temperature.
Ultracentrifugation was performed at 100,000 × g for 1 h at room
temperature. The supernatant was transferred to centrifugal filter
units with a 30-kDa cutoff (Merck KGaA), which contained 3.5 ml of
solution 3, followed by further centrifugation at 2,500 × g at
12°C, until a final volume of 100 µl was obtained. Total
protein quantitation was performed using the Pierce BCA protein
assay (Thermo Fisher Scientific, Inc.).
Sample preparation for liquid
chromatography coupled with tandem mass spectrometry
(LC-MS/MS)
From each cell membrane protein solution, 10
µg was analyzed by 12% SDS-PAGE, and stained with Coomassie
Colloidal Blue (Fluka) overnight. Bands were excised from the gels
and cut in small pieces (1–2 mm). Gel pieces were destained in 40%
acetonitrile (Fisher Chemical, Loughbrough, UK) and 50 mM
NH4HCO3, reduced in 10 mM dithioerythritol
and 100 mM NH4HCO3 (all from Sigma-Aldrich;
Merck KGaA), and alkylated in 50 mM iodoacetamide (Applichem GmbH)
and 100 mM NH4HCO3 (Sigma-Aldrich; Merck
KGaA). Samples were dried using the Savant Speedvac™ concentrator
(Thermo Fisher Scientific, Inc.) and trypsinized overnight with 600
ng trypsin (Roche Diagnostics) in 10 mM
NH4HCO3. Peptide extraction was performed by
two washes of the trypsinized gel pieces with 50 mM
NH4HCO3, followed by two washes with 50%
acetonitrile and 5% formic acid (Sigma-Aldrich; Merck KGaA) for 15
min at room temperature, with agitation. Extracted peptides were
dried using the Savant Speedvac concentrator and analyzed by
LC-MS/MS. The same protocol was also followed for the total cell
extract.
LC-MS/MS
Dried peptides were solubilized in 100 µl
ultra-pure water and separated on an UltiMate 3000 Nano HPLC Dionex
Ultimate® 3000 RSLS system (Dionex™, Camberly, UK).
Initially, 5 µl from each sample was loaded on a Dionex
0.1×20 mm, 5-µm C18 nanotrap column, at a flow rate of 5
µl/min in 98% mobile phase A (0.1% formic acid) and 2%
mobile phase B (100% acetonitrile and 0.1% formic acid). In the
next step, the sample was eluted into an Acclaim PepMap C18
nanocolumn, 75 µm × 50 cm (Dionex), at a flow rate of 0.3
µl/min. The trap and the nanoflow column were maintained at
35°C. The samples were eluted with a gradient of solvent B,
starting at 1% B for 5 min, and rising to 5% B at 10 min, 25% B at
180 min and 65% B at 240 min. The column was then washed and
re-equilibrated prior to injection of the next sample. The eluant
was ionized using a Proxeon Nano Spray Electron Spray Ionization
source (nebulizer pressure, 100 psi), operating in positive ion
mode into an Orbitrap Elite FTMS (ThermoFinnigan MAT GmbH, Bremen,
Germany). Ionization voltage was at 2.6 kV and the capillary
temperature was at 200°C. The mass spectrometer was operated in
MS/MS mode scanning from 380 to 2,000 atomic mass units. The
resolution of ions in MS1 was 60,000, and 7,500 for higher-energy
C-trap dissociation (HCD) in MS2. The top 20 multiply charged ions
were selected from each scan for MS/MS analysis using HCD at 35%
collision energy. Data analysis was performed with Proteome
Discoverer 1.4 software package (Thermo Fisher Scientific, Inc.),
using the SEQUEST search engine (19) and the UniProt Universal Protein
Knowledgebase (http://www.uniprot. org/). The search
was performed using carbamidomethylation of cysteine as static
modifications and oxidation of methionine as dynamic modifications.
Two missed cleavage sites, a precursor mass tolerance of 10 ppm and
a fragment mass tolerance of 0.05 Da were allowed. SEQUEST results
were filtered for false-positive identifications. The procedure of
LC-MS/MS was followed for the analysis of the membrane fraction and
the total cell extract.
Classification of membrane proteins and
validation of the enrichment membrane protocol
Subsequent to selecting the reliable protein
identifications of membrane fractions, which were present in 75% of
cancer biological replicates, they were categorized based on their
subcellular location. This was achieved by employing the UniProt
database (http://www.uniprot.org/). To confirm the
successful enrichment of the isolation protocol, the number and the
net percentages of membrane and transmembrane proteins were defined
and compared with the membrane fraction and the total cell
extract.
Quantification and statistical
analysis
Quantification analysis was performed at the peptide
level. The intensity for each protein in each biological replicate
was normalized, i.e., quotient of intensity for the particular
protein to the sum of all intensities of all proteins of the
specific biological sample, multiplied by 106. The mean
normalized intensity for each protein was then determined for all
biological replicates. The aforementioned procedure was repeated
for all cell lines.
Only proteins present in 75% of the samples in at
least one group were further processed for quantification and
statistical analysis (Mann-Whitney). Differentially expressed
proteins selected for further analysis were considered as those
with a fold-change value of either of >2 (upregulated in a
cancer cell line compared with the normal HCK1T cell line) or
<0.5 (downregulated in a cancer cell line compared with the
normal HCK1T cell line), and with a P-value of <0.05. This
comparison resulted in three lists of differentially expressed
proteins (C33A vs. HCK1T, HeLa vs. HCK1T and SiHa vs. HCK1T).
Ingenuity® Pathway Analysis
(IPA)
The identified differentially expressed proteins
were subjected to IPA® analysis (Qiagen, Inc., Valencia,
CA, USA; www.qiagen.com/ingenuity). Entry names of
differentially expressed proteins were converted to gene names
following their entry in the Retrieve/ID mapping of the UniProt
database (http://www.uniprot.org/). The processed
gene names were imported into IPA to map the canonical pathways and
generate biological networks. Data were submitted as fold-change
values, i.e., ratios, calculated against the control group (HCK1T).
Hypothetical networks were built among the experimental proteins
and the IPA database proteins. Only statistically significant
(P≤0.05, Fisher’s Exact Test performed by the Ingenuity®
Pathway Analysis platform) canonical pathways were selected.
Canonical pathways were classified according to the P-value and the
expression ratio in the cervical cancer cell line compared with
that in the HCK1T cell line. The ratio of the canonical pathways
was calculated based on the number of molecules from the input
dataset divided by the total number of molecules in the pathway
that was predicted by IPA. Molecules participating in the important
canonical pathways according to IPA analysis were listed by their
gene names.
Protein-protein interaction (PPI)
network
A PPI network of the 174 common differentially
expressed proteins among the three cancer cell lines and the normal
HCK1T cell line was constructed using the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) v10.5 database
(http://string-db.org/). The confidence score was
set to >0.4.
Results
Overview
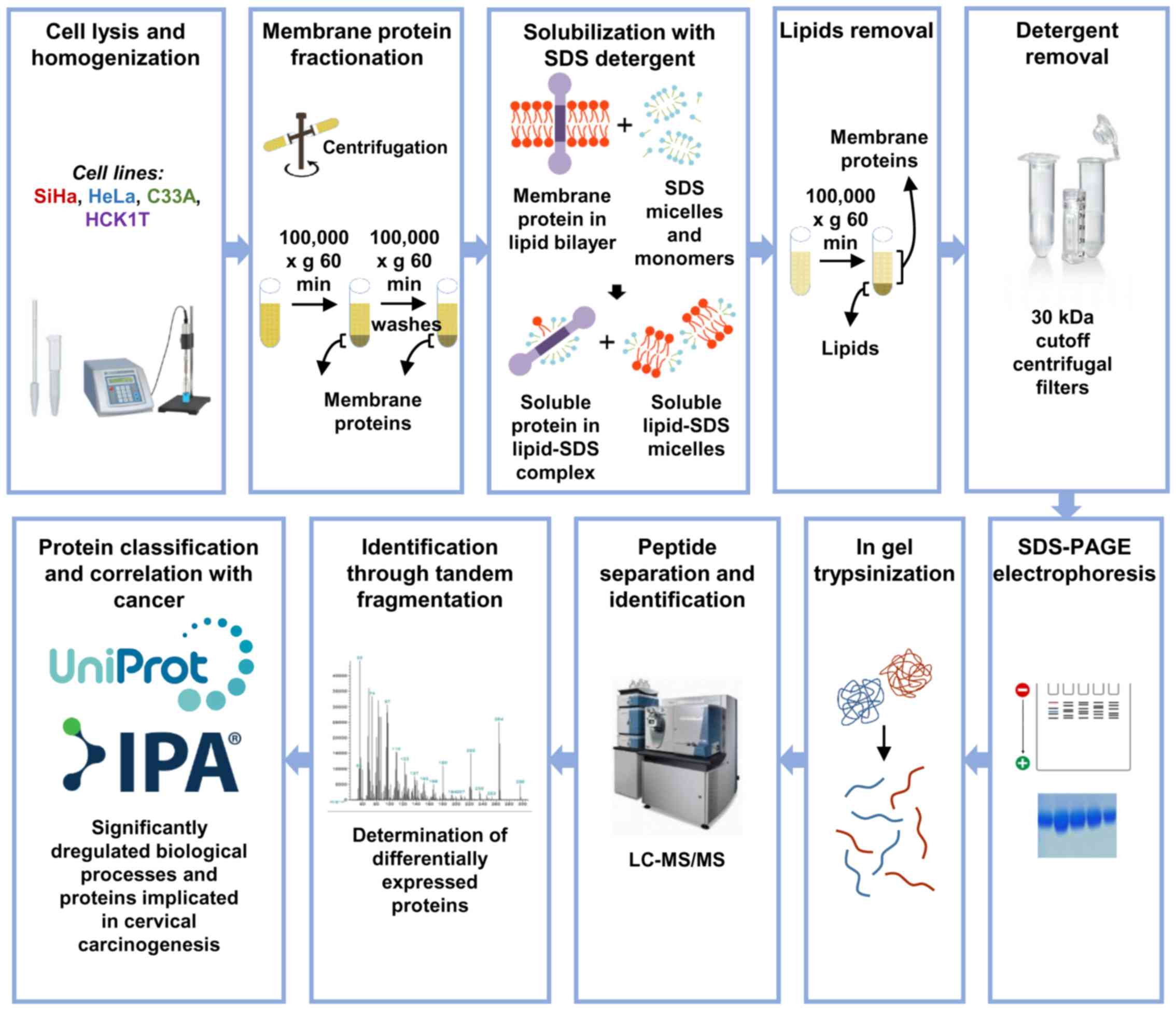
The overview of the present study approach is
presented in Fig. 1. The aim was
to develop a novel reproducible protocol for the isolation and
enrichment of membrane proteins, and their subsequent
characterization by mass spectrometry and bioinformatics
analysis.
Reproducibility
The membrane protein enrichment protocol exhibited
satisfactory reproducibility, since 40–62% of the total
identifications were present in at least 75% of the biological
replicates of the four cell lines studied. The numbers of reliable
protein identifications are shown in Table I.
 | Table IProtein identifications in four
biological replicates and identifications present in 75% of
biological replicates. |
Table I
Protein identifications in four
biological replicates and identifications present in 75% of
biological replicates.
| Cell line | Total
identifications obtained from four biological replicates | Identifications
present in 75% of biological replicates, n (%) |
|---|
| C33A | 3,213 | 1,961 (61) |
| HeLa | 2,975 | 1,827 (61) |
| SiHa | 2,477 | 1,522 (62) |
| HCK1T | 2,132 | 854 (40) |
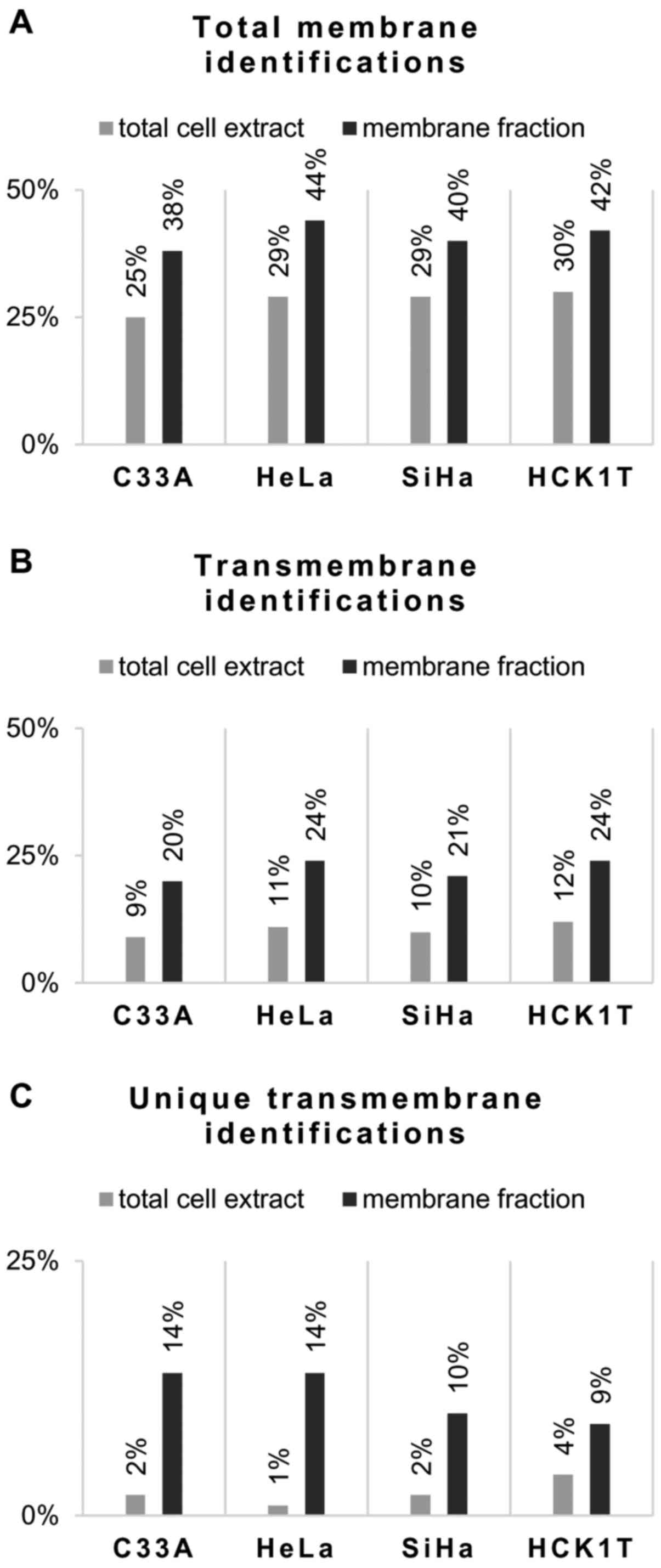
Comparison of membrane fraction vs
total cell extract. The percentage of membrane
proteins identified in the four cell lines ranged from 38 to 44%
and the percentage of transmembrane proteins from 20 to 24%
(Table II). These percentages
were higher compared with the corresponding data obtained from
total cell extract analysis following the same workflow (Fig. 2; and Pappa et al,
unpublished data). The mean enrichment ratio of membrane proteins
identified by this approach compared with that of the total cell
extracts was 1.5, and 2.1 for the transmembrane proteins. In
addition, the number of unique transmembrane proteins identified by
the present protocol was higher compared with the transmembrane
protein identifications obtained from the total cell extract, as
shown in Table II.
 | Table IIReliable protein identifications
present in 75% of biological replicates of the four cell lines. |
Table II
Reliable protein identifications
present in 75% of biological replicates of the four cell lines.
| Cellular
fraction | All protein
identifications, n | Unique
identifications, n | Membrane protein
identifications, n (%) | Unique membrane
protein identifications, n | Transmembrane
protein identifications, n (%) | Unique
transmembrane protein identifications, n |
|---|
| C33A total cell
extract | 2183 | 891 | 544 (25) | 163 | 186 (9) | 52 |
| C33A membrane
fraction | 1961 | 669 | 746 (38) | 367 | 398 (20) | 263 |
| HeLa total cell
extract | 1912 | 664 | 552 (29) | 95 | 202 (11) | 20 |
| HeLa membrane
fraction | 1827 | 579 | 794 (44) | 337 | 444 (24) | 262 |
| SiHa total cell
extract | 1961 | 826 | 563 (29) | 170 | 199 (10) | 32 |
| SiHa membrane
fraction | 1522 | 388 | 607 (40) | 214 | 319 (21) | 152 |
| HCK1T total cell
extract | 1712 | 1005 | 514 (30) | 248 | 200 (12) | 73 |
| HCK1T membrane
fraction | 854 | 147 | 361 (42) | 95 | 200 (23) | 73 |
Analysis of protein differential
expression in the SiHa, HeLa, C33A and HCK1T membrane
fractions
Comparison of the expression levels of the proteins
in the membrane fraction of the four cell lines was conducted using
a total of four samples per category, corresponding to different
biological replicates. Each cancer cell line was compared with the
normal HCK1T cell line. Differentially expressed and statistically
significant proteins (fold-change >2 or <0.5; P<0.05,
Mann-Whitney) were identified by this analysis. Upregulated
proteins in cancer were considered as those proteins either with a
fold-change value >2 or being unique in a cancer cell line.
Downregulated proteins in cancer were considered those with a
fold-change value <0.5 or being unique in the normal HCK1T cell
line. The following results were obtained from the comparisons of
the HCK1T control cell line with the three cervical cancer cell
lines. A total of 102 differentially expressed proteins (54
downregulated and 48 upregulated in SiHa) were detected from the
SiHa vs. HCK1T comparison, whereas 264 were identified only in SiHa
(upregulated in SiHa) and 61 only in HCK1T (downregulated in SiHa).
Furthermore, 162 differentially expressed proteins (59
downregulated and 103 upregulated in HeLa) were detected from the
HeLa vs. HCK1T comparison, whereas 430 were identified only in HeLa
(upregulated in HeLa) and 43 only in HCK1T (downregulated in HeLa).
Finally, 169 differentially expressed proteins (90 upregulated and
79 downregulated in C33A) were detected from the C33A vs. HCK1T
comparison, whereas 600 were identified only in C33A (upregulated
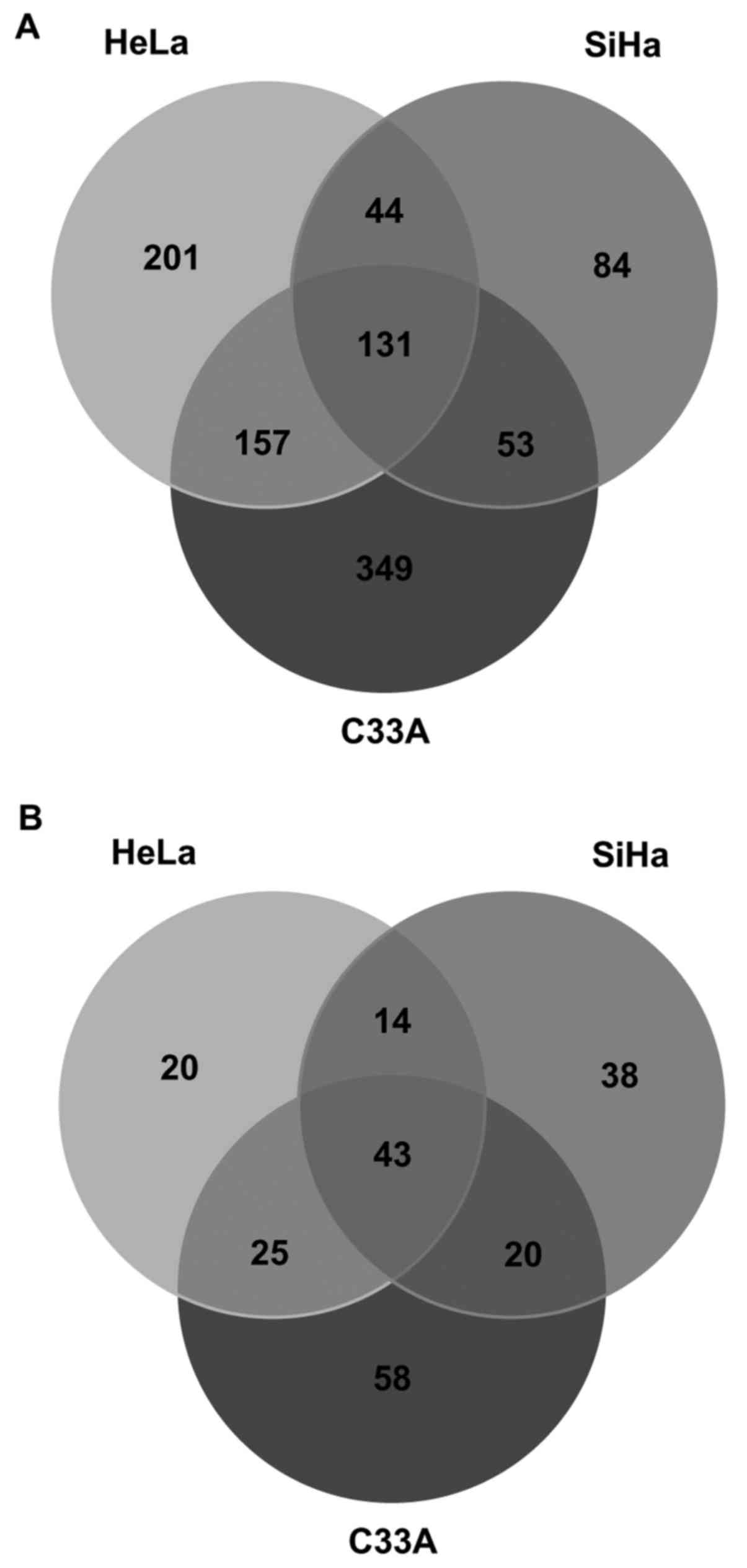
in C33A) and 67 only in HCK1T (downregulated in C33A). The overlap
between the upregulated proteins among the three cancer cell lines
compared with the normal HCK1T cell line is presented by a Venn
diagram in Fig. 3A. There were 131
common upregulated identifications that correspond to 42% of
proteins in the SiHa vs. HCK1T comparison, 25% of proteins in the
HeLa vs. HCK1T comparison and 19% of proteins in the C33A vs. HCK1T
comparison (Fig. 3A).
Additionally, there were unique differentially expressed proteins
identified in all comparisons (84 in SiHa vs. HCK1T, 201 in HeLa
vs. HCK1T and 349 in C33A vs. HCK1T) corresponding to 27% in SiHa
vs. HCK1T, 38% in HeLa vs. HCK1T and 51% in C33A vs. HCK1T.
Furthermore, the overlap between the downregulated proteins among
the three cancer cell lines compared with the normal HCK1T cell
line is presented by a Venn diagram in Fig. 3B. There were 43 common
identifications that correspond to 37% of proteins in the SiHa vs.
HCK1T comparison, 42% of proteins in the HeLa vs. HCK1T comparison
and 30% of proteins in the C33A vs. HCK1T comparison. Also, there
were unique differentially expressed proteins identified in all
comparisons (38 in SiHa vs. HCK1T, 20 in HeLa vs. HCK1T and 58 in
C33A vs. HCK1T) corresponding to 33% in SiHa vs. HCK1T, 20% in HeLa
vs. HCK1T and 40% in C33A vs. HCK1T.
The most prominent differentially expressed proteins
(TMX2, FAM120A, CLPTM1, CKAP5 and NCSTN) that followed the same
pattern of expression in all cancer cell lines are presented in
Table III, along with
information on their molecular function. Moreover, for these
specific proteins, a literature search was performed on previous
relevant studies describing their role in cancer biology (20–26),
which was incorporated in Table
III.
 | Table IIIProminent membrane proteins involved
in cancer pathology. |
Table III
Prominent membrane proteins involved
in cancer pathology.
| Gene name | Protein name | Regulation in
cancer (C33A, cell lines HeLa and SiHa) vs. HCK1T | Molecular
function | Studies performed
in other types of cancer (Ref.) |
|---|
| TMX2 | Thioredoxin-related
transmembrane protein 2 | Upregulated | Cell redox
homeostasis | Upregulated in
breast cancer (20) |
| FAM120A | Constitutive
coactivator of PPAR-γ-like protein 1 | Upregulated | Oxidative
stress-induced survival signaling | Upregulated in
gastric cancer (21,22) |
| CLPTM1 | Cleft lip and
palate transmembrane protein 1 | Upregulated | May play a role in
T-cell development | Upregulated in
colon cancer (23) |
| CKAP5 |
Cytoskeleton-associated protein 5 | Upregulated | Cadherin binding,
microtubule plus-end binding | Upregulated mRNA
levels in cervical cancer (24) |
| NCSTN | Nicastrin | Upregulated | Membrane protease
cleaving Notch receptors | Upregulated in
non-small cell lung cancer (25,26) |
IPA and PPI network
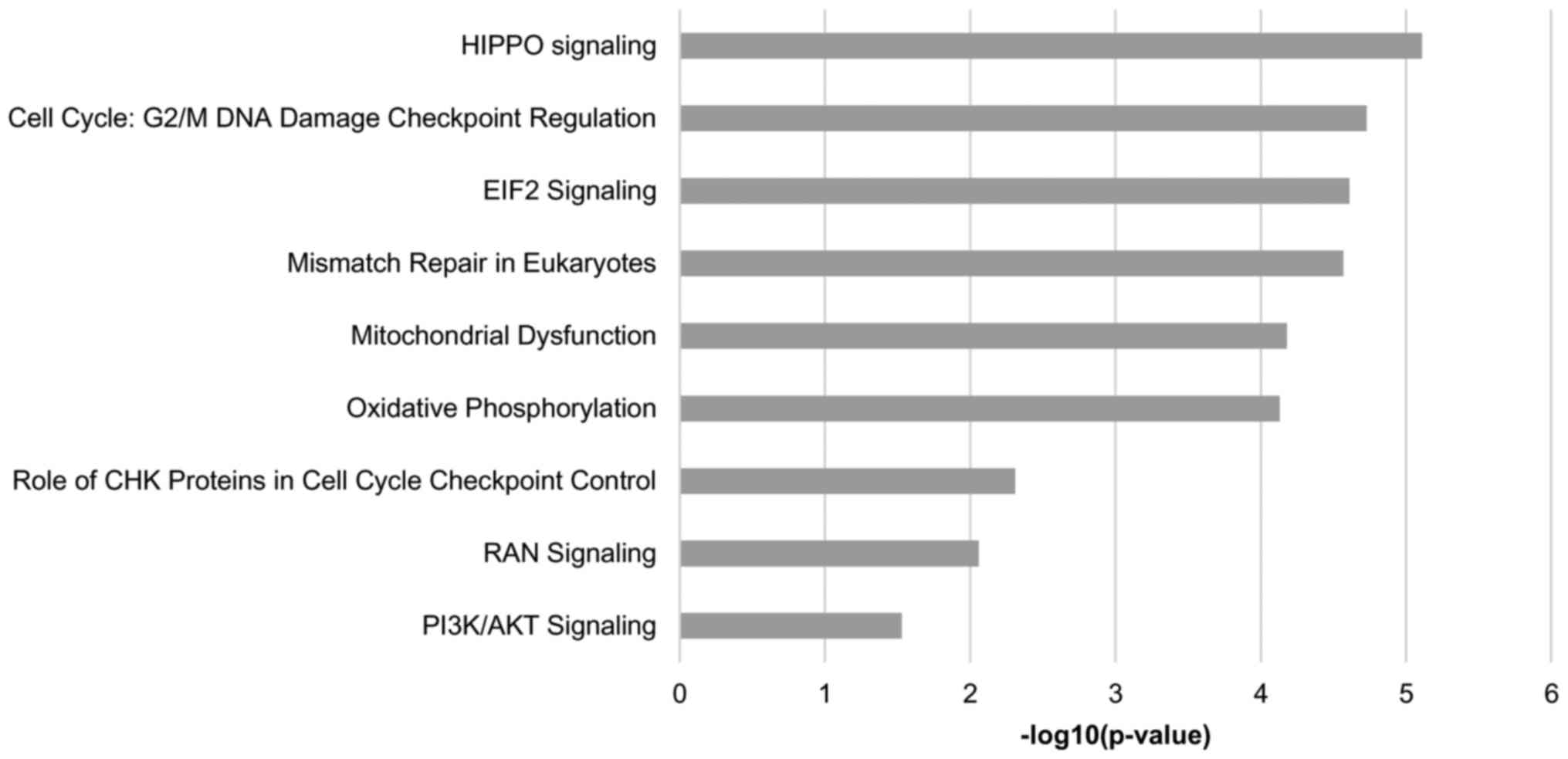
To further characterize the biological functions and
pathways relevant to the differentially expressed proteins, IPA
software was employed. A total of nine pathways were found to be
common among the three cancer cell lines compared with those in
HCK1T cells, and are presented in Fig.
4. Among the common predicted pathways are ‘HIPPO signaling’,
‘RAN signaling’, ‘PI3K/AKT signaling’, ‘cell cycle: G2/M DNA damage
checkpoint regulation’ and ‘EIF2 signaling’. Furthermore, there
were unique predicted pathways in all comparisons. Specifically, 19
pathways were predicted in SiHa vs. HCK1T (Table IV), 4 pathways in HeLa vs. HCK1T
(Table V) and 33 pathways in C33A
vs. HCK1T (Table VI).
Transmembrane proteins are indicated in the tables.
 | Table IVCanonical pathways and involved
molecules in the SiHa cell line, as predicted by IPAa. |
Table IV
Canonical pathways and involved
molecules in the SiHa cell line, as predicted by IPAa.
| Ingenuity canonical
pathways | P-value | Ratio | Molecules |
|---|
| Common predicted
pathways in SiHa, HeLa and C33A | | | |
| HIPPO
signaling |
2.69×10−5 |
1.30×10−1 | PPP1CC, YWHAQ,
YWHAG, PPP1CA, SKP1, PPP2R1A, YWHAZ, YWHAH, YWHAB |
| Cell cycle: G2/M
DNA damage checkpoint regulation |
4.27×10−6 |
1.90×10−1 | TOP2A, YWHAQ,
YWHAG, SKP1, YWHAZ, YWHAH, YWHAB, TOP2B |
| Mismatch repair in
eukaryotes |
2.69×10−5 |
4.55×10−1 | PCNA, RFC2,
RFC1b, RPA1, RFC5 |
| Mitochondrial
dysfunction |
6.61×10−5 |
1.19×10−1 | FIS1b, NDUFA5, NDUFB9, SDHB, COX6B1, ATP5H,
PRDX5, ATP5A1, NDUFB6b,
NCSTNb, COX7A2L, CYB5Ab, ATP5F1, NDUFB3b, MAOAb |
| Oxidative
phosphorylation |
7.41×10−5 |
1.51×10−1 | NDUFA5, COX6B1,
NDUFB9, SDHB, ATP5H, ATP5A1, NDUFB6b, COX7A2L, CYB5Ab, ATP5F1, NDUFB3b |
| PI3K/AKT
signaling |
4.17×10−4 |
9.18×10−2 | YWHAQ, YWHAG,
CTNNB1, HSP90B1, PPP2R1A, YWHAZ, YWHAH, YWHAB, GYS2 |
| Role of CHK
proteins in cell cycle checkpoint control |
4.90×10−3 |
1.40×10−1 | PCNA, PPP2R1A,
RFC2, RFC1b, RPA1, RFC5 |
| EIF2 signaling |
1.82×10−2 |
5.56×10−2 | RPS27L, PPP1CC,
EIF3G, RPLP2, PPP1CA, RPS7, RPS8, RPS14 |
| RAN signaling |
3.00×10−2 |
1.67×10−1 | RANGAP1, KPNA2 |
| Unique predicted
pathways in SiHa | | | |
| Dopamine receptor
signaling |
1.15×10−2 |
8.47×10−2 | PPP1CC, PPP1CA,
PPP2R1A, SPR, MAOAb |
| Granzyme B
signaling |
9.12×10−4 |
3.33×10−1 | BID, NUMA1 |
| Cysteine
biosynthesis III mammalia |
1.45×10−3 |
1.11×10−1 | AHCYL1, AHCYL2,
AHCY |
| ERK5 signaling |
1.74×10−3 |
2.73×10−1 | YWHAQ, EGFRb, YWHAG, YWHAZ, YWHAH, YWHAB |
| Methionine
degradation I to homocysteine |
2.29×10−3 |
1.02×10−1 | AHCYL1, AHCYL2,
AHCY |
| L-cysteine
degradation III |
2.63×10−3 |
1.19×10−1 | GOT1 |
| Interferon
signaling |
2.82×10−3 |
6.54×10−2 | IFITM2b, ISG15, IFITM3b |
| Aspartate
biosynthesis |
4.79×10−3 |
7.02×10−2 | GOT1 |
| Glutamate
degradation II |
5.01×10−3 |
7.69×10−2 | GOT1 |
| Unfolded protein
response |
5.37×10−3 |
1.88×10−1 | HSPA9, DNAJC3,
HSP90B1, HSPA5, ERO1B |
| ERK/MAPK
signaling |
2.09×10−2 |
2.01×10−1 | HSPB1, PPP1CC,
YWHAQ, STAT3, YWHAG, PPP1CA, PPP2R1A, YWHAZ, YWHAH, YWHAB |
| Myc-mediated
apoptosis signaling |
2.29×10−2 |
1.10×10−1 | YWHAQ, YWHAG,
YWHAZ, BID, YWHAH, YWHAB |
| Aldosterone
signaling in epithelial cells |
2.29×10−2 |
5.03×10−2 | HSPB1, HSPA9,
DNAJC3, DNAJC9, HSP90B1, HSPA5, ICMTb, AHCY |
| Protein
ubiquitination pathway |
2.34×10−2 |
1.11×10−1 | HSPB1, PSMD4,
HSPA9, DNAJC3, DNAJC9, HSP90B1, HSPA5, SKP1, B2M |
| Phenylalanine
degradation IV mammalian, via side chain |
2.51×10−2 |
1.82×10−1 | GOT1, MAOAb |
| Methylglyoxal
degradation III |
2.69×10−2 |
6.06×10−2 | AKR1C3, AKR1B1 |
| Endoplasmic
reticulum stress pathway |
3.47×10−2 |
1.54×10−1 | DNAJC3, HSP90B1,
HSPA5 |
| IGF-1
signaling |
4.57×10−2 |
5.04×10−1 | YWHAQ, STAT3,
YWHAG, CSNK2A2, YWHAZ, YWHAH, YWHAB |
| 14-3-3-mediated
signaling |
4.57×10−2 |
5.10×10−1 | VIM, YWHAQ, YWHAG,
YWHAZ, YWHAH, YWHAB |
 | Table VCanonical pathways and involved
molecules in the HeLa cell line, as predicted by IPAa. |
Table V
Canonical pathways and involved
molecules in the HeLa cell line, as predicted by IPAa.
| Ingenuity canonical
pathways | P-value | Ratio | Molecules |
|---|
| Common predicted
pathways in SiHa, HeLa, C33A | | | |
| Mismatch repair in
eukaryotes |
2.69×10−5 |
4.55×10−1 | PCNA, RFC2, RFC1,
RPA1, RFC5 |
| HIPPO
signaling |
7.76×10−6 |
1.74×10−1 | YWHAQ, SMAD2,
PPP2R1A, PPP1CC, YWHAG, YWHAH, YWHAB, SMAD3, YWHAZ, SCRIB, SKP1,
PPP1CA |
| Cell cycle: G2/M
DNA damage checkpoint regulation |
1.86×10−5 |
2.14×10−1 | YWHAQ, TOP2B,
YWHAG, YWHAH, YWHAB, PTPMT1, YWHAZ, TOP2A, SKP1 |
| EIF2 signaling |
2.45×10−5 |
1.18×10−1 | PABPC1, RPL4, RPS8,
RPLP2, RPL30, RPL39, EIF2A, EIF3G, PPP1CC, RPS27L, EIF2B5, EIF3A,
RPL6, PPP1CA, RPS5, RPS3, RPS14 |
| Mitochondrial
dysfunction |
6.61×10−5 |
1.19×10−1 | FIS1b, NDUFA5b, NDUFB9b, SDHBb, COX6B1b, ATP5Hb, PRDX5, ATP5A1, NDUFB6, NCSTN,
COX7A2L, CYB5Ab, ATP5F1b, NDUFB3b, MAOAb |
| Oxidative
phosphorylation |
7.41×10−5 |
1.51×10−1 | NDUFA5, COX6B1,
NDUFB9, SDHB, ATP5H, ATP5A1, NDUFB6, COX7A2L, CYB5A, ATP5F1,
NDUFB3 |
| Role of CHK
proteins in cell cycle checkpoint control |
4.90×10−3 |
1.40×10−1 | PCNA, PPP2R1A,
RFC2, RFC1, RPA1, RFC5 |
| RAN signaling |
8.71×10−3 |
2.50×10−1 | KPNB1, RANGAP1,
KPNA2 |
| PI3K/AKT
signaling |
2.95×10−2 |
8.16×10−2 | YWHAQ, MTOR,
PPP2R1A, YWHAG, YWHAH, YWHAB, YWHAZ, PTGS2 |
| Unique predicted
pathways in HeLa | | | |
| Leukotriene
biosynthesis |
3.31×10−2 |
2.50×10−1 | MGST2b, GGT1b |
| nNOS signaling in
skeletal muscle cells |
6.76×10−3 |
2.73×10−1 | SNTB2, DMD,
DAG1 |
| Glutaryl-CoA
degradation |
3.31×10−2 |
2.50×10−1 | HADHB, HADH |
| Telomere extension
by telomerase |
4.17×10−2 |
2.22×10−1 | XRCC6, XRCC5 |
 | Table VICanonical pathways and involved
molecules in the C33A cell line, as predicted by IPAa. |
Table VI
Canonical pathways and involved
molecules in the C33A cell line, as predicted by IPAa.
| Ingenuity canonical
pathways | P-value | Ratio | Molecules |
|---|
| Common predicted
pathways in SiHa, HeLa, C33A | | | |
| HIPPO
signaling |
2.29×10−4 |
1.59×10−1 | YWHAQ, SMAD2,
PPP1CC, YWHAE, YWHAB, SMAD3, CD44, YWHAZ, SCRIB, PPP2R5E,
PPP1CA |
| EIF2 signaling |
6.31×10−12 |
2.01×10−1 | RPS18, RPLP2,
RPL39, EIF2A, RPS11, RPS7, RPL27A, PPP1CC, EIF3B, EIF3A, GSK3B,
RPS2, RPS5, RPS3, PPP1CA, RPL18, PABPC1, RPS19, RPL4, RPL3, RPS8,
RPL30, RPS10, RPL12, EIF3G, RPL6, RPSA, RPS14, RPLP0 |
| Cell cycle: G2/M
DNA damage checkpoint regulation |
1.20×10−5 |
2.38×10−1 | YWHAQ, CDKN2A,
TOP2B, YWHAE, YWHAB, YWHAZ, TRIP12, TOP2A, PLK1, CCNB1 |
| Mismatch repair in
eukaryotes |
2.69×10−5 |
4.55×10−1 | PCNA, RFC2, RFC1,
RPA1, RFC5 |
| Mitochondrial
dysfunction |
6.61×10−5 |
1.19×10−1 | FIS1b, NDUFA5b, NDUFB9b, SDHBb, COX6B1b, ATP5Hb, PRDX5b, ATP5A1b, NDUFB6b, NCSTNb, COX7A2Lb, CYB5Ab, ATP5F1b, NDUFB3b, MAOAb |
| Oxidative
phosphorylation |
7.41×10−5 |
1.51×10−1 | NDUFA5, COX6B1,
NDUFB9, SDHB, ATP5H, ATP5A1, NDUFB6, COX7A2L, CYB5A, ATP5F1,
NDUFB3 |
| Role of CHK
proteins in cell cycle checkpoint control |
4.90×10−3 |
1.40×10−1 | PCNA, PPP2R1A,
RFC2, RFC1, RPA1, RFC5 |
| PI3K/AKT
signaling |
1.23×10−2 |
1.02×10−1 | YWHAQ, ITGB1, MTOR,
ITGA3, YWHAE, YWHAB, YWHAZ, GSK3B, PPP2R5E, GYS2 |
| RAN signaling |
1.45×10−2 |
2.50×10−1 | KPNB1, RANGAP1,
KPNA2 |
| Unique predicted
pathways in C33A | | | |
| Sertoli
cell-Sertoli cell junction signaling |
7.94×10−3 |
9.56×10−2 | ITGB1b, ACTBb, YBX3b, F11Rb, ITGA3b, ACTA2b, ACTN4b, ACTG2b, GSK3Bb, ACTC1, ACTG1, ACTN1, ACTA1 |
| Caveolar-mediated
endocytosis signaling |
1.35×10−5 |
1.97×10−1 | ITGB1b, ITGA3b, ACTA2b, ACTBb, CAV1b, ITGB4b, ACTG2b, PTRFb, ACTC1b, ACTG1, ACTA1, EGFR |
| Remodeling of
epithelial adherens junctions |
1.82×10−5 |
2.08×10−1 | ARPC1B, ACTA2,
ACTB, ACTG2, ACTN4, IQGAP1, ACTC1, ACTG1, ACTA1, ACTN1, ARPC4 |
| Agrin interactions
at neuromuscular junction |
7.24×10−5 |
1.96×10−1 | ITGB1b, ITGA3b, PKLRb, ACTA2b, ACTBb, ACTG2b, ACTC1b, ACTG1b, ACTA1b, EGFRb |
| RhoGDI
signaling |
7.94×10−5 |
1.34×10−1 | ITGB1b, ARPC1Bb, ACTBb, GNA11b, RACK1b, ITGA3b, ACTA2b, EZRb, CD44b, ARHGEF2, ACTG2, ACTC1, ACTG1, ARPC4,
ACTA1, MSN |
| NRF2-mediated
oxidative stress response |
2.40×10−4 |
1.18×10−1 | DNAJB12b, PPIBb, ACTBb, DNAJC3b, CUL3b, ACTA2b, SCARB1b, MGST2b, CCT7, GSK3B, ACTG2, DNAJB6, ACTC1,
HACD3, ACTG1, ACTA1, CBR1 |
| Regulation of
actin-based motility by Rho |
4.47×10−4 |
1.59×10−1 | ITGB1b, ITGA3b, ARPC1Bb, ACTA2b, ACTBb, ACTG2b, GSNb, ACTC1b, ACTA1b, ARPC4b |
| ILK signaling |
8.32×10−4 |
1.10×10−1 | ITGB1b, ACTBb, FERMT2b, VIMb, MTORb, ACTA2b, KRT18b, ACTN4b, PPP2R5E, ITGB4, ACTG2, GSK3B, ACTC1,
ACTG1, ACTN1, ACTA1 |
| Mechanisms of viral
exit from host cells |
8.51×10−4 |
2.31×10−1 | ACTA2, ACTB, ACTG2,
ACTC1, ACTG1, ACTA1 |
| Glycolysis I |
1.23×10−3 |
2.63×10−1 | PKLR, ENO3, ENO2,
GAPDH, PFKP |
| Integrin
signaling |
1.51×10−3 |
1.01×10−1 | ITGB1b, ARPC1Bb, ACTBb, GSNb, ITGA3b, ACTA2b, CAV1b, CAPN2b, ACTN4b, GSK3Bb, ITGB4b, ACTG2b, ACTC1b, ACTG1b, ARPC4b, ACTN1, ACTA1b |
| Paxillin
signaling |
2.04×10−3 |
1.24×10−1 | ITGB1b, ITGA3b, ACTA2b, ACTBb, ITGB4b, ACTG2b, ACTN4b, ACTC1b, ACTG1b, ACTA1b, ACTN1b |
| FAK signaling |
3.24×10−3 |
1.23×10−1 | ITGB1b, ITGA3b, ACTA2b, ACTBb, CAPN2b, ACTG2b, ACTC1b, ACTG1b, ACTA1, EGFR |
| Actin cytoskeleton
signaling |
3.39×10−3 |
9.58×10−2 | ITGB1b, ARPC1Bb, ACTBb, IQGAP1b, GSNb, ITGA3b, ACTA2b, EZRb, ACTN4b, ACTG2, ACTG1, ACTC1, ARPC4, ACTN1,
ACTA1, MSN |
| Death receptor
signaling |
4.27×10−3 |
1.27×10−1 | ACIN1, ACTA2, ACTB,
LMNA, ACTG2, ACTC1, ACTG1, ACTA1, HSPB1 |
| Regulation of
cellular mechanics by Calpain protease |
4.68×10−3 |
1.49×10−1 | ITGB1b, ITGA3b, EZRb, CAPN2b, ACTN4b, ACTN1b, EGFRb |
| Leukocyte
extravasation signaling |
5.13×10−3 |
9.43×10−2 | ITGB1b, MMP14b, ACTBb, F11Rb, ITGA3b, ACTA2, EZRb, CD44b, ACTN4b, ACTG2, ACTG1, ACTC1, ACTN1, ACTA1,
MSN |
| 2-Ketoglutarate
dehydrogenase complex |
5.89×10−3 |
6.67×10−1 | DLST, DLD |
| Fcγ
receptor-mediated phagocytosis in macrophages and monocytes |
6.17×10−3 |
1.20×10−1 | ARPC1B, ACTA2, EZR,
ACTB, ACTG2, ACTC1, ACTG1, ACTA1, ARPC4 |
| Epithelial adherens
junction signaling |
7.08×10−3 |
1.01×10−1 | ARPC1B, ACTA2,
ACTB, ACTG2, ACTN4, IQGAP1, ACTC1, ACTG1, ACTA1, ACTN1, ARPC4,
EGFR |
| TCA cycle II
eukaryotic |
7.41×10−3 |
2.22×10−1 | CS, DLST, DLD,
FH |
| VEGF signaling |
1.26×10−2 |
1.07×10−1 | YWHAE, ACTA2, ACTB,
ACTG2, ACTN4, ACTC1, ACTG1, ACTA1, ACTN1 |
| Tec kinase
signaling |
1.38×10−2 |
9.23×10−2 | ITGB1b, ITGA3b, ACTA2b, GTF2Ib, ACTBb, RACK1b, GNA11b, STAT3b, ACTG2b, ACTC1b, ACTG1b, ACTA1b |
| γ-Linolenate
biosynthesis II animals |
1.45×10−2 |
2.50×10−1 | CYB5Ab, ACSL1b, FADS1b |
| MSP-RON signaling
pathway |
1.62×10−2 |
1.30×10−1 | ACTA2, ACTB, ACTG2,
ACTC1, ACTG1, ACTA1 |
| Germ cell-Sertoli
cell junction signaling |
1.82×10−2 |
8.89×10−2 | ITGB1b, ITGA3b, ACTA2b, ACTBb, ACTG2b, ACTN4b, GSNb, IQGAP1b, ACTC1b, ACTG1b, ACTA1b, ACTNb |
| Clathrin-mediated
endocytosis signaling |
3.55×10−2 |
8.05×10−2 | ITGB1b, ARPC1Bb, ACTA2b, ACTBb, USP9Xb, PPP3CCb, ITGB4b, ACTG2b, ACTC1, ACTG1, ACTA1, ARPC4 |
| Agranulocyte
adhesion and diapedesis |
3.63×10−2 |
8.27×10−2 | ITGB1b, ITGA3b, ACTA2b, EZRb, MMP14b, ACTBb, ACTG2b, ACTC1b, ACTG1b, ACTA1b, MSNb |
| RAR activation |
3.72×10−2 |
8.00×10−2 | NR2F1, SMAD2,
SMAD9, SMAD3, ACTB, SMARCB1, NR2F2, SMARCD2, ARID2, SNW1, SMARCC1,
PRMT1 |
|
Glycoaminoglycan-protein linkage region
biosynthesis |
4.47×10−2 |
1.18×10−3 | GXYLT1b |
|
Phosphatidylethanolamine biosynthesis
III |
4.47×10−2 |
1.02×10−2 | PTDSS2b |
| Branched-chain
α-keto acid dehydrogenase complex |
4.47×10−2 |
1.07×10−1 | DLD |
| Assembly of RNA
polymerase III complex |
4.68×10−2 |
2.50×10−1 | GTF3C1, SF3A1 |
Moreover, using the STRING database, a PPI network
of the 174 common differentially expressed proteins among the three
cancer cell lines and the normal HCK1T cell line was created. The
network is presented in Fig. 5 and
illustrates that 142 out of the 174 proteins have at least one
functional connection.
Discussion
In the present study, an efficient and reproducible
enrichment protocol for membrane proteins was developed, resulting
in the identification of a significant number of unique membrane
proteins relevant to cervical cancer. Moreover, the number of
unique transmembrane proteins identified in the membrane fractions
was higher compared with the total cell extracts. These unique
transmembrane protein identifications offer novel insights on a
previously inaccessible region of the cell proteome. The findings
of this study can be further validated through functional analysis,
utilizing techniques such as in vitro gene editing employing
the CRISPR/Cas9 system [as reviewed in (27,28)].
Few studies exist on the development of a membrane
protein enrichment protocol compatible with proteomics platforms.
Previous membrane proteomic studies reported a lower number of
membrane protein identifications in comparison with the present
study (29–31). Besides the total cell extract and
membrane cell extract protocols in the present study, we have also
developed in recent studies an enrichment protocol for secreted
proteome (15,16). Therefore, the three protocols for
total cell extract, membrane cell extract and secretome extract
reveal unique identifications and thus offer complementary views of
the cellular compartments of the cervical cell lines proteome
(14–16).
The current study also contributes to the
identification of molecular pathways deregulated in cervical
cancer. The most interesting of these pathways are ‘HIPPO
signaling’, ‘RAN signaling’, ‘PI3K/AKT signaling’, ‘cell cycle:
G2/M DNA damage checkpoint regulation’ and ‘EIF2 signaling’, which
are common in all cancer cell lines. Specifically, the predicted
involvement of the HIPPO signaling pathway in cervical cancer is
supported by a recent report on the central role of the HIPPO
regulator yes-associated protein 1 in HPV-positive oropharyngeal
malignancies (32). The common 174
deregulated proteins in cervical cancer cell lines identified by
the present approach exhibit informative associations, as
illustrated by the PPI network results (Fig. 5). Among these 174 proteins, 142
(82%) are functionally associated. The overwhelming majority of the
interactions in the network (>90%) consist of experimentally
validated findings.
The comparison of cancer cell lines (C33A, HeLa and
SiHa) with the normal HCK1T cell line provided a significant number
of differentially expressed proteins. Among these, the most notable
membrane proteins that were upregulated in all three cancer cell
lines, regardless of the presence of HPV, include
thioredoxin-related transmembrane protein 2, constitutive
coactivator of PPAR-γ-like protein 1, cleft lip and palate
transmembrane protein 1, nicastrin and cytoskeleton-associated
protein 5. These membrane proteins have not been reported in
previous cervical cancer studies; however, published reports
support their involvement in other cancer types, as shown in
Table III. Nicastrin in
particular is considered as a putative therapeutic target in breast
cancer (25,26), and further investigation of its
potential for cervical cancer therapy is warranted. Thus, the
identified membrane proteins reveal novel perspectives for the
molecular characterization of cervical cancer. Overall, the
successful fractionation of membrane proteins provides a
significant pool of potential cervical cancer therapeutic targets
for further functional validation and characterization, and
eventually for their therapeutic exploitation.
The present study achieved successful isolation of
membrane proteins by employing a novel protocol involving
differential ultracentrifugation and detergent-based
solubilization, generating a significant pool of promising drug
targets.
Funding
This study was funded by the European Union’s
European Social Fund and Greek National Funds through the Program
THALIS, under the Operational Program Education and Lifelong
Learning of the National Strategic Reference Framework (grant no.
70-3-11830), and by the Oncology Program of the Central Council of
Health of the Ministry of Health (grant no. 70-3-9209).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
KIP, JZ and NPA participated in the acquisition of
funding, and study design and supervision. KIP interpreted the
results and drafted the manuscript. PC, AX and GM performed the
experiments and statistical analysis, and contributed to the
writing of the manuscript. GK performed the cell culture and
Ingenuity® Pathway Analysis. VL created the protein-protein
interactions network. MM performed the mass spectrometry analysis.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Dr Tohru Kiyono
(National Cancer Centre Research Institute, Tokyo, Japan) for the
generous gift of the normal cervical HCK1T cell line.
References
|
1
|
Feichtinger M and Rodriguez-Wallberg KA:
Fertility preservation in women with cervical, endometrial or
ovarian cancers. Gynecol Oncol Res Pract. 3:82016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li Y and Xu C: Human
papillomavirus-related cancers. Adv Exp Med Biol. 1018:23–34. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Serrano B, Brotons M, Bosch FX and Bruni
L: Epidemiology and burden of HPV-related disease. Best Pract Res
Clin Obstet Gynaecol. 47:14–26. 2018. View Article : Google Scholar
|
|
4
|
Pappa KI, Kontostathi G, Lygirou V,
Zoidakis J and Anagnou NP: Novel structural approaches concerning
HPV proteins: Insight into targeted therapies for cervical cancer
(Review). Oncol Rep. 39:1547–1554. 2018.PubMed/NCBI
|
|
5
|
Willows K, Lennox G and Covens A:
Fertility-sparing management in cervical cancer: Balancing
oncologic outcomes with reproductive success. Gynecol Oncol Res
Pract. 3:92016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parkin DM: Cancer in developing countries.
Cancer Surv. 19-20:519–561. 1994.PubMed/NCBI
|
|
7
|
Bentivegna E, Maulard A, Pautier P,
Chargari C, Gouy S and Morice P: Fertility results and pregnancy
outcomes after conservative treatment of cervical cancer: A
systematic review of the literature. Fertil Steril. 106:1195–1211
e1195. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lawrenz B, Mahajan N and Fatemi HM: The
effects of cancer therapy on women’s fertility: What do we know
now? . Future Oncol. 12:1721–1729. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barra F, Lorusso D, Leone Roberti,
Maggiore U, Ditto A, Bogani G, Raspagliesi F and Ferrero S:
Investigational drugs for the treatment of cervical cancer. Expert
Opin Investig Drugs. 26:389–402. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hampson L, Martin-Hirsch P and Hampson IN:
An overview of early investigational drugs for the treatment of
human papilloma virus infection and associated dysplasia. Expert
Opin Investig Drugs. 24:1529–1537. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Campbell MT, Siefker-Radtke AO and Gao J:
The state of immune checkpoint inhibition in urothelial carcinoma:
Current evidence and future areas of exploration. Cancer J.
22:96–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mermelekas G and Zoidakis J: Mass
spectrometry-based membrane proteomics in cancer biomarker
discovery. Expert Rev Mol Diagn. 14:549–563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tomonaga T and Kume H: Biomarker discovery
of colorectal cancer using membrane proteins extracted from cancer
tissues. Rinsho Byori. 63:322–327. 2015.Japanese. PubMed/NCBI
|
|
14
|
Pappa KI, Lygirou V, Kontostathi G,
Zoidakis J, Makridakis M, Vougas K, Daskalakis G, Polyzos A and
Anagnou NP: Proteomic analysis of normal and cancer cervical cell
lines reveals deregulation of cytoskeleton-associated proteins.
Cancer Genomics Proteomics. 14:253–266. 2017. View Article : Google Scholar :
|
|
15
|
Kontostathi G, Zoidakis J, Makridakis M,
Lygirou V, Mermelekas G, Papadopoulos T, Vougas K, Vlamis-Gardikas
A, Drakakis P, Loutradis D, et al: Cervical cancer cell line
secretome highlights the roles of transforming growth
factor-beta-induced protein ig-h3, peroxiredoxin-2, and NRF2 on
cervical carcinogenesis. Biomed Res Int. 2017:41807032017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pappa KI, Kontostathi G, Makridakis M,
Lygirou V, Zoidakis J, Daskalakis G and Anagnou NP: High resolution
proteomic analysis of the cervical cancer cell lines secretome
documents deregulation of multiple proteases. Cancer Genomics
Proteomics. 14:507–521. 2017.PubMed/NCBI
|
|
17
|
Makridakis M, Gagos S, Petrolekas A,
Roubelakis MG, Bitsika V, Stravodimos K, Pavlakis K, Anagnou NP,
Coleman J and Vlahou A: Chromosomal and proteome analysis of a new
T24-based cell line model for aggressive bladder cancer.
Proteomics. 9:287–298. 2009. View Article : Google Scholar
|
|
18
|
Narisawa-Saito M, Handa K, Yugawa T, Ohno
S, Fujita M and Kiyono T: HPV16 E6-mediated stabilization of ErbB2
in neoplastic transformation of human cervical keratinocytes.
Oncogene. 26:2988–2996. 2007. View Article : Google Scholar
|
|
19
|
Eng JK, Fischer B, Grossmann J and Maccoss
MJ: A fast SEQUEST cross correlation algorithm. J Proteome Res.
7:4598–4602. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Apostolou P, Toloudi M and Papasotiriou I:
Identification of genes involved in breast cancer and breast cancer
stem cells. Breast Cancer (Dove Med Press). 7:183–191. 2015.
|
|
21
|
Holden S and Raymond FL: The human gene
CXorf17 encodes a member of a novel family of putative
transmembrane proteins: cDNA cloning and characterization of
CXorf17 and its mouse ortholog orf34. Gene. 318:149–161. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bartolomé RA, García-Palmero I, Torres S,
López-Lucendo M, Balyasnikova IV and Casal JI: IL13 Receptor α2
signaling requires a scaffold protein, FAM120A, to activate the FAK
and PI3K pathways in colon cancer metastasis. Cancer Res.
75:2434–2444. 2015. View Article : Google Scholar
|
|
23
|
Zang Y, Nie W, Fang Z and Li B: Cleft lip
and palate transmembrane protein 1 rs31489 polymorphism is
associated with lung cancer risk: A meta-analysis. Tumour Biol.
35:5583–5588. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schneider MA, Christopoulos P, Muley T,
Warth A, Klingmueller U, Thomas M, Herth FJ, Dienemann H, Mueller
NS, Theis F, et al: AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five
specific mitosis-associated genes correlate with poor prognosis for
non-small cell lung cancer patients. Int J Oncol. 50:365–372. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sarajlić A, Filipović A, Janjić V, Coombes
RC and Pržulj N: The role of genes co-amplified with nicastrin in
breast invasive carcinoma. Breast Cancer Res Treat. 143:393–401.
2014. View Article : Google Scholar
|
|
26
|
Lombardo Y, Filipović A, Molyneux G,
Periyasamy M, Giamas G, Hu Y, Trivedi PS, Wang J, Yagüe E, Michel
L, et al: Nicastrin regulates breast cancer stem cell properties
and tumor growth in vitro and in vivo. Proc Natl Acad Sci USA.
109:16558–16563. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sander JD and Joung JK: CRISPR-Cas systems
for editing, regulating and targeting genomes. Nat Biotechnol.
32:347–355. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Doudna JA and Charpentier E: Genome
editing. The new frontier of genome engineering with CRISPR-Cas9 .
Science. 346:12580962014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hua Y, Jia X, Sun M, Zheng L, Yin L, Zhang
L and Cai Z: Plasma membrane proteomic analysis of human
osteosarcoma and osteoblastic cells: Revealing NDRG1 as a marker
for osteosarcoma. Tumour Biol. 32:1013–1021. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qu Z, Gao F, Li L, Zhang Y, Jiang Y, Yu L,
Zhou Y, Zheng H, Tong W, Li G, et al: Label-free quantitative
proteomic analysis of differentially expressed membrane proteins of
pulmonary alveolar macrophages infected with highly pathogenic
porcine reproductive and respiratory syndrome virus and its
attenuated strain. Proteomics. 17:17001012017. View Article : Google Scholar
|
|
31
|
Zhang Z, Zhang L, Hua Y, Jia X, Li J, Hu
S, Peng X, Yang P, Sun M, Ma F, et al: Comparative proteomic
analysis of plasma membrane proteins between human osteosarcoma and
normal osteoblastic cell lines. BMC Cancer. 10:2062010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Alzahrani F, Clattenburg L, Muruganandan
S, Bullock M, MacIsaac K, Wigerius M, Williams BA, Graham ME, Rigby
MH, Trites JR, et al: The hippo component YAP localizes in the
nucleus of human papilloma virus positive oropharyngeal squamous
cell carcinoma. J Otolaryngol Head Neck Surg. 46:152017. View Article : Google Scholar : PubMed/NCBI
|