Introduction
Multiple myeloma (MM) is a type of heterogeneous
malignancy with the biological characteristics of malignant plasma
cell (PC) proliferation in the bone marrow and complex genetic
alterations (1). Eventually, the
abnormal malignant proliferation of PCs may result in the abnormal
secretion of monoclonal immunoglobulin (M-Ig or M protein). The M
protein may cause several complications in patients with MM, such
as hyperviscosity syndrome, bone injury and renal impairment. Some
studies have indicated that the classification of M protein may be
an important indicator for the diagnosis of patients with MM.
Moreover, the precise quantification of the types and levels of M
protein is critical to monitoring the response of patients to
therapy (2,3). However, M protein classification
cannot cover all patients with MM and this disease has been found
to have a great heterogeneity at the genetic level. Therefore,
researchers have focused on the field of genetic molecular research
as regards MM in the hope of identifying novel individualized
biomarkers and treatment for this disease (4,5).
Currently, numerous researchers have demonstrate
that microRNAs (miRNAs or miRs) can play crucial roles in the
progression of a number of types of cancer, including MM (6,7).
miRNAs are a class of small non-coding RNAs with a length of
approximately 22 nucleotides. It has been extensively demonstrated
that miRNAs can play a significant role in the regulation of cell
functions by directly binding to the 3′ untranslated region (3′UTR)
of their target genes, eventually leading to the degradation of
these tumor-associated genes (8).
Certain studies have found that the dysregulated expression of
miRNAs is closely related to the genesis, progression, tumor
metastasis and drug resistance in MM (4,9,10).
Furthermore, miRNAs have been proven to participate in the
activation of several MM-associated signaling pathways, such as the
nuclear factor (NF)-κB signaling pathway, the Wnt/β-catenin
signaling pathway, the interleukin (IL)-6/signal transducer and
activator of transcription 3 (STAT3) signaling pathway and the
PI3K/Akt signaling pathway (11,12).
These results indicate that the knockdown of oncomiRNAs and the
restoration of tumor suppressor miRNAs may lead to the development
of novel treatment strategies for MM.
In recent years, an increasing number of researchers
have paid attention to the study of circulating biomarkers due to
their detectability and accessibility (13). Notably, the expression level of
circulating miRNAs can reflect the pathological state and prognosis
of patients accurately (14).
Moreover, miRNAs in human body fluids are highly stable and easy to
detect, and these advantages display their potential for use as
biomarkers for various diseases (15). Therefore, seeking effective early
biomarkers for diagnosis and targets for therapy is essential for
the diagnosis and treatment of MM.
The majority of studies on miR-30d in solid tumors
have indicated that it possesses tumor suppressor functions
(16,17). However, research on the functions
of miR-30d in MM and on its expression levels in serum is limited
(18). In the present study, we
found that the expression level of miR-30d was significantly
downregulated in the serum of patients with MM compared with the
serum of healthy donors (HDs) by reverse transcription-quantitative
polymerase chain reaction quantitative (RT-qPCR). We then examined
the association of the expression level of this miRNA with the
patient clinicopathological data, and the results revealed that the
expression level of miR-30d was significantly associated with the
concentration of hemoglobin (HGB), platelet (PLT), creatinine (Cr)
and β2-microglobulin (β2M), and bone marrow
plasma cells (BMPC) infiltration in patients with MM. Following 2
periods of treatment, the serum miR-30d expression level in
patients with primary MM was significantly increased, suggesting
that miR-30d had a great potential for use as a diagnostic
biomarker of MM. Furthermore, we examined the biological functions
of miR-30d in U266 cells by CCK-8 assay and apoptosis by flow
cytometric analysis. The results revealed that miR-30d inhibited
cell proliferation and promoted cell apoptosis in vitro.
In order to conduct a more in-depth study of the
mechanisms of action of miR-30d in MM cells, we began to search for
its target gene. By utilizing bioinformatics software, we selected
metadherin (MTDH) as a putative target of miR-30d. This
regulatory association was also confirmed in a previous study
(19). MTDH, also known as
astrocyte elevated gene-1 (AEG-1) or LYRIC, is a
novel oncogene that plays a crucial role in various human
malignancies, including prostate carcinoma (20), breast carcinoma (21), non-small cell lung cancer (22) and cervical cancer (23). Abundant functional investigations
in vitro and in vivo have indicated that MTDH is a
valuable tumor biomarker and a potential therapeutic target in
cancer. However, studies on the role of MTDH in MM are limited
(24). Firstly, in this study, we
found that MTDH promoted cell proliferation and inhibited
cell apoptosis in vitro. Furthermore, results western blot
(WB) analysis revealed that miR-30d inhibited the activation of the
downstream PI3K/Akt signaling pathway by directly binding to its
target gene, MTDH, which suggested that miR-30d plays an
important anti-carcinogenic role in the pathological process of MM
by targeting the MTDH/PI3K/Akt signaling pathway.
Materials and methods
Patient samples
In the present study, we enrolled 81 patients with
primary MM and 78 samples from HDs at the Affiliated Hospital of
Nantong University (Nantong, China) from July, 2015 to December,
2016. All patients with MM were definitively diagnosed from the
results of a bone marrow biopsy. Moreover, all patients were
divided into different stages and subtypes according to their
clinical characteristics. Serum samples of patients were collected
at the time of their first diagnosis without any treatment and
stored in RNase-free tubes at −80°C immediately until use. Normal
PCs from three HDs as controls of MM cell lines were purified from
their bone marrow specimen using CD138+ magnetic bead
separation technology (Miltenyi Biotec Corp., Gladbach, Germany) as
previously described (25). All
the protocols were approved by the Human Research Ethics Committee
of the Affiliated Hospital of Nantong University.
Detection of clinical parameters of
patients
The serum hemoglobin (HGB) concentration was
measured using an automatic blood cell analyzer Sysmex HST-N 302
(Sysmex Corp., Kobe, Japan) and determined by the colorimetry. The
serum platelet (PLT) concentration was measured using an automatic
blood cell analyzer Sysmex XE-2100 (Sysmex Corp.) and determined by
the principle of electrical impedance. The serum calcium ion
(Ca2+), albumin (ALB), creatinine (Cr), β2M
and lactate dehydrogenase (LDH) concentrations were measured using
an automatic biochemical analyzer ADVIA2400 (Siemens Corp., Berlin,
Germany) with the corresponding kit (FUJIFILM Wako Pure Chemical
Corp., Japan). The serum concentrations of light chain λ and κ were
measured using an IMMAGE specific protein analyzer (Beckman Coulter
Corp., Brea, CA, USA) with immune turbidimetry. The BMPC
infiltration percentage in the bone marrow of patients with MM was
determined by searching abnormal plasma cells under a microscope
after staining the bone marrow smears (Wright-Giemsa Stain;
Solarbio Corp., Beijing, China).
Extraction of RNA and cDNA synthesis
Serum total RNA was extracted from 400 μl
serum samples using the Ambion mirVana PARIS kit (Life
Technologies/Thermo Fisher Scientific Corp., Waltham, MA, USA)
according to the manufacturer’s instructions. Total RNA from the
cell lines was extracted using TRIzol reagent (Invitrogen/Thermo
Fisher Scientific Corp.) according to the manufacturer’s
instructions. The concentration and purity of the extracted RNA
were measured using a nanophotometer (Implen Corp., Munich,
Germany). Subsequently, 10 μg total RNA were reverse
transcribed into cDNA using the RevertAid First Strand cDNA
Synthesis kit (Thermo Fisher Scientific Corp.). The reaction
condition for the mix was at 42°C for 60 min and then at 70°C for 5
min. Reverse transcription products were stored at −20°C until
use.
Quantitative (real-time) polymerase chain
reaction (qPCR)
To examine the expression level of serum miRNAs,
qPCR was performed using the Applied Biosystems® 7500
Real-Time PCR System (Applied Biosystems/Thermo Fisher Scientific
Corp.). The reaction system included 3.0 μl cDNA (with three
technical replicates) and 10.0 μl FastStart Universal
SYBR-Green Master Mix (Roche Corp., Mannheim, Germany). The PCR
reaction condition consisted of incubation at 95°C for 10 min,
followed by denaturation at 95°C for 10 sec, annealing at 58°C for
30 sec, extension at 72°C for 30 sec, and repeating the cycle for
45 times. In serum, the expression level of miR-30d was normalized
to its internal control U6 and the relative expression level was
calculated using the 2−ΔΔCq method as previously
described (26) and using mixed
normal serum as control. In U266 cells, the expression level of
MTDH was normalized to its internal control, β-actin, and the
relative expression level was calculated using the
2−ΔΔCq method using normal PCs as a control. The
sequences of the primers used are listed as follows: miR-30d
forward, 5′-GCGTGTAAACATCCCCGAC-3′ and reverse,
5′-CAGCCACAAAAGAGCACAAT-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; MTDH forward,
5′-TAAAACAAAACTGCGGACAC-3′ and reverse, 5′-AGGGCACTGTTGTTAAACCA-3′;
and β-actin forward, 5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse,
5′-GGCACGAAGGCTCATCATT-3′.
Culture and transfection of MM cell
lines
The U266, H929, RPMI-8226 cell lines were obtained
from the Chinese Academy of Sciences Cell Bank (Shanghai, China).
The cells were cultured in Dulbecco’s modified Eagle’s medium
(DMEM) with 10% fetal bovine serum, 1% antibiotics and 1% glutamine
(both from Gibco/Thermo Fisher Scientific Corp.) in a humidified
atmosphere of 5% CO2 at 37°C. miR-30d mimics (M), its
negative control (M-), miR-30d inhibitors (I) and its negative
control (I-) were synthesized by Guangzhou RiboBio Corp.
(Guangzhou, China). MTDH-pcDNA and its negative control were
synthesized by GeneCopoeia (GeneCopoeia Corp., Guangzhou, China).
These were transfected into MM cells using Lipofectamine 3000
(Invitrogen/Thermo Fisher Scientific Corp.) according to the
manufacturer’s instructions. After 48-72 h, the transfected cells
were harvested for cell function experiments and WB analysis.
Cell proliferation assay
To detect the proliferation of MM cells, a Cell
Counting kit-8 (CCK-8; Roche Corp., Basel, Switzerland) cell
proliferation assay was performed. The MM cells were seeded at
5,000 cells per well in 96-well plates with 7 replicates.
Subsequently, 100 μl CCK-8 reagent were added to each well
and maintained in an incubator for 2 h at 37°C. OD values were
detected using a microplate reader (DeTie Corp., Nanjing, China) at
0, 24, 48 and 72 h, respectively. Finally, all data were collected
for statistical analysis.
Cell apoptosis assay
To evaluate cell apoptosis, the 7AAD-Annexin V
Apoptosis Detection kit (BD Biosciences Corp., San Jose, CA, USA)
was applied according to the manufacturer’s instructions. Briefly,
the MM cells, which were transfected and after 48 h were collected
into tubes, and then washed with cold PBS twice. Subsequently, 5
μl 7AAD and 5 μl Annexin V reagents were added to the
tubes and mixed gently, then incubated together for 15 min at 25°C
in the dark. The apoptosis of the MM cells was analyzed using a
FACSCalibur flow cytometer (BD Biosciences Corp.).
WB analysis
Total proteins were extracted from the MM cells
using RIPA buffer (high) (Solarbio Corp.) and WB analysis was
performed as previously described (27). Generally, the concentrations of
proteins in MM cells were determined using the bicinchoninic acid
(BCA) assay (Beyotime Institute of Biotechnology, Beijing, China).
A total of 20 μg protein per lane were loaded and separated
using 12% SDS-PAGE, then transferred onto nitrocellulose membranes
(Merck Millipore Corp., Darmstadt, Germany), and blocked in
PBS/Tween-20 containing 5% bovine serum albumin blocking buffer
(Solarbio Corp.) for 2 h at at 25°C. Subsequently, the membranes
with proteins were incubated overnight at 4°C with corresponding
primary antibodies. The primary antibodies used in this study and
their dilution rates are listed as follows: anti-MTDH (1:5,000;
ab124789, anti-PI3K-p85 (1:500; ab182651), anti-phospho-Akt-Ser473
(1:1,000; ab81283) (all from Abcam, Cambridge, MA, USA), anti-total
Akt (1:1,000; #9272) and anti-β-actin) (1:1,000; #4970) (all from
Cell Signaling Technology Corp., Danvers, MA, USA). The membranes
were then incubated with the secondary antibody (anti-rabbit IgG;
1:2,000; #14708; Cell Signaling Technology Corp.) for 1 h at 25°C.
Following washing 3 times with TBST buffer, the enhanced
chemiluminescent (ECL) kit (BioVision Corp., Milpitas, CA, USA) was
used to visualize the protein strips. The densitometry of protein
strips was calculated using ImageJ software version 1.6.0 and
normalized to β-actin.
Dual Luciferase reporter assay
Bioinformatics software, including TargetScan
(http://www.targetscan.org), miRanda
(http://www.microRNA.org) and PicTar (http://pictar.mdc-berlin.de/) were used to predict the
target genes of miRNAs and the results indicated that MTDH
may be the target gene of miR-30d. The wild-type (wt) MTDH
3′UTR and mutated (mut) MTDH 3′UTR were then bound to
psiCHECK-2 vectors (Promega Corp., Madison, WI, USA), respectively.
The final products were cloned and amplified into Escherichia
coli (General Microbiological Culture Collection Center,
Beijing, China). The U266 cells were then cultured in 6-well
plates, and each well was co-transfected with
wt/mut-MTDH-psiCHECK-2 and miR-30d mimics/miR-control using
Lipofectamine 3000 for 48 h. Following the lysis of the cells,
luciferase activity was measured and the fluorescence activity of
Firefly luciferase and sea kidney luciferase (as an internal
reference) was detected. The final results were calculated as the
luciferase activity/sea kidney luciferase activity.
Statistical analysis
Data were analyzed using SPSS statistical analysis
software, version 20.0 and figures were drawn using Graphpad Prism
5 software. Patient data were described by median and interquartile
as they were not in accordance with Gaussian distribution. The
Mann-Whitney U test was used to compare data differences between 2
groups. The Kruskal-Wallis H with the Tamhane’s T2 post hoc test
was used to compare differences among multiple groups. Paired data
were analyzed using the Wilcoxon paired test. The correlation
between the expression level of miR-30d with the patient
clinicopathological characteristics was examined using Spearman’s
correlation coefficient analysis. Statistically significant
differences between categorical variables were determined by the
Chi-square test. Receiver operating characteristic (ROC) curves and
area under the ROC curve (AUC) were used to assess the diagnostic
value of using miR-30d for MM. In all statistical analyses, a value
of P<0.05 was considered to indicate a statistically significant
difference.
Results
Patient characteristics
A total of 159 serum samples were used in this
study, which included 81 samples from patients with primary MM and
78 samples from HDs. The median age of the patients was 61 years of
age (range, 38-88 years), while the median age of the HDs was 59
years of age (range, 45-82 years). There were no significant
differences between the patients with MM and the HDs as regards the
composition of age and sex (P>0.05). Among these newly diagnosed
patients with MM, 38 received bortezomib-based treatment (arm A)
and 43 received thalidomide-based treatment (arm B). According to
the ISS staging system, 26 patients had stage I, 19 patients had
stage II and 36 patients had stage III of the disease. According to
the Durie-Salmon (DS) staging system, 3 patients had stage I, 39
patients had stage II and 39 patients had stage III of the disease.
According to the monoclonal component, patients were divided into
an IgG type of 28 cases, an IgA type of 17 cases, an IgM type of 1
case, a light chain type of 33 cases and a non-secretory type of 2
cases (Table I).
 | Table IBasic characteristics of the patients
with primary multiple myeloma and healthy donors. |
Table I
Basic characteristics of the patients
with primary multiple myeloma and healthy donors.
| Patients (n) | Healthy donors
(n) | P-value |
|---|
| Total | 81 | 78 | |
| Sex | | | 0.629 |
| Male | 46 | 48 | |
| Female | 35 | 30 | |
| Range of age
(years) | 38-88 | 45-82 | 0.063 |
| ISS | | | |
| Stage I | 26 | ND | ND |
| Stage II | 19 | ND | ND |
| Stage III | 36 | ND | ND |
| DS | | | |
| Stage I | 3 | ND | ND |
| Stage II | 39 | ND | ND |
| Stage III | 39 | ND | ND |
| Type of monoclonal
component | | | |
| IgG | 28 | ND | ND |
| IgA | 17 | ND | ND |
| IgM | 1 | ND | ND |
| Light chain | 33 | ND | ND |
| Non-secretory | 2 | ND | ND |
| Therapeutic
strategy | | | |
| Arm A | 38 | ND | ND |
| Arm B | 43 | ND | ND |
Expression levels and diagnostic accuracy
of serum miR-30d levels in MM
The serum expression levels of miR-30d in 81
patients with MM and 78 HDs were detected by RT-qPCR. The results
revealed that the serum expression level of miR-30d in the patients
with MM was significantly downregulated compared with that in the
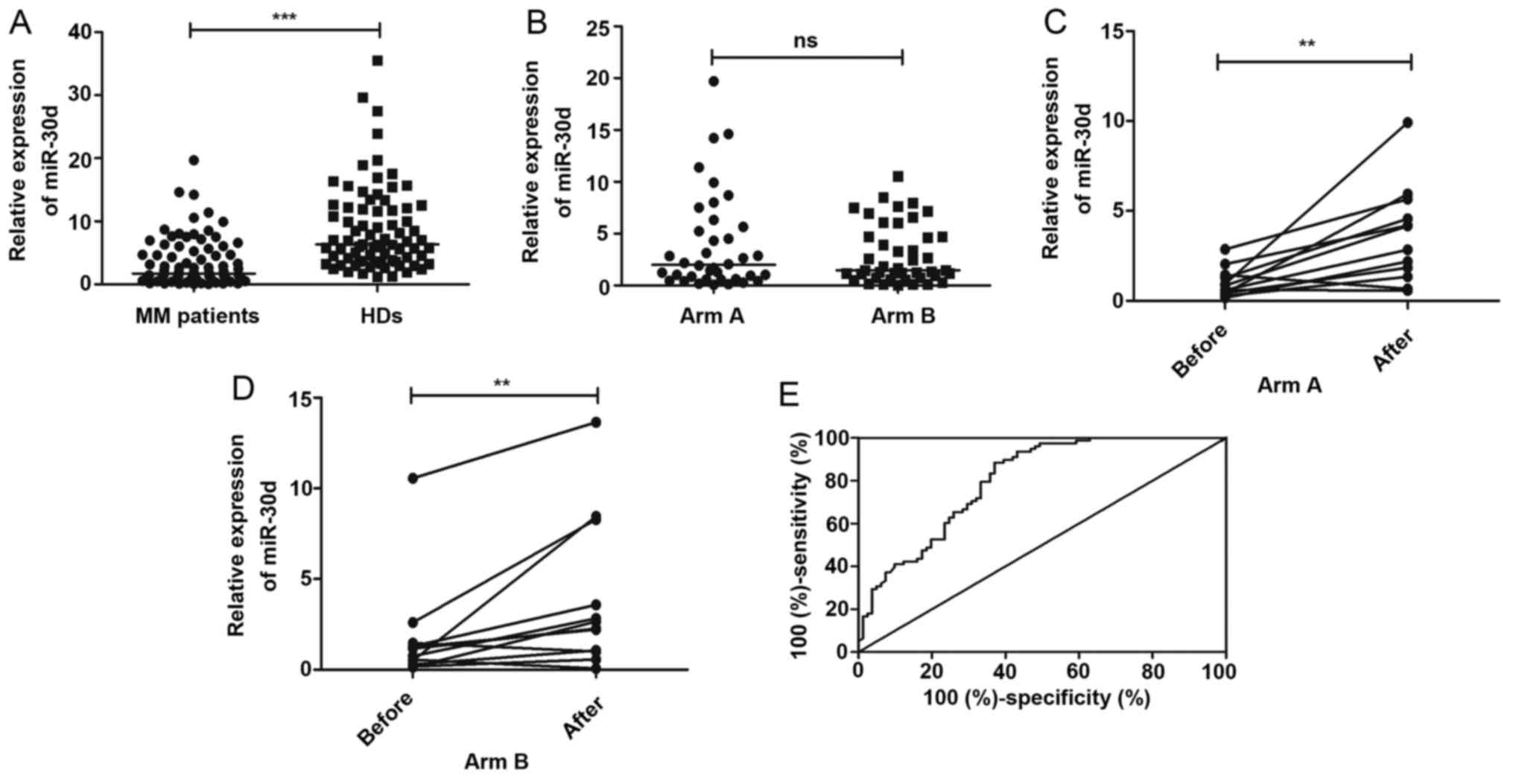
HDs (P<0.0001) (Fig. 1A). No
significant differences were observed in the serum miR-30d
expression level between the patients in arm A and arm B
(P>0.05) (Fig. 1B). Moreover,
we made a short-term (2 periods of treatment) follow-up visit to 24
patients and collected their serum samples both at the time of
diagnosis and following treatment. We concluded that the serum
expression levels of miR-30d in patients receiving both arm A and
arm B treatment were significantly improved (P=0.0360 and P=0.0304,
respectively) (Fig. 1C and D).
Subsequently, ROC curve analysis was conducted to assess the
diagnostic accuracy of miR-30d. The results revealed that the serum
level of miR-30d could differentiate patients with MM from HDs with
areas under the ROC curve (AUC) of 0.800 (95% CI, 0.733-0.868;
P<0.0001) (Fig. 1E). At the
cut-off value of 2.908 for miR-30d, the sensitivity was 88.5% and
the specificity was 63.0%.
Associations between serum levels of
miR-30d and clinical parameters of the patients with MM
We assessed the associations of the miR-30d
expression levels and the clinical parameters of patients with MM.
Firstly, we stratified the patients with MM according to the
international staging system (ISS) and Durie-Salmon (DS) staging to
investigate the association between the serum miR-30d expression
levels and the stages of MM. The results indicated that the miR-30d
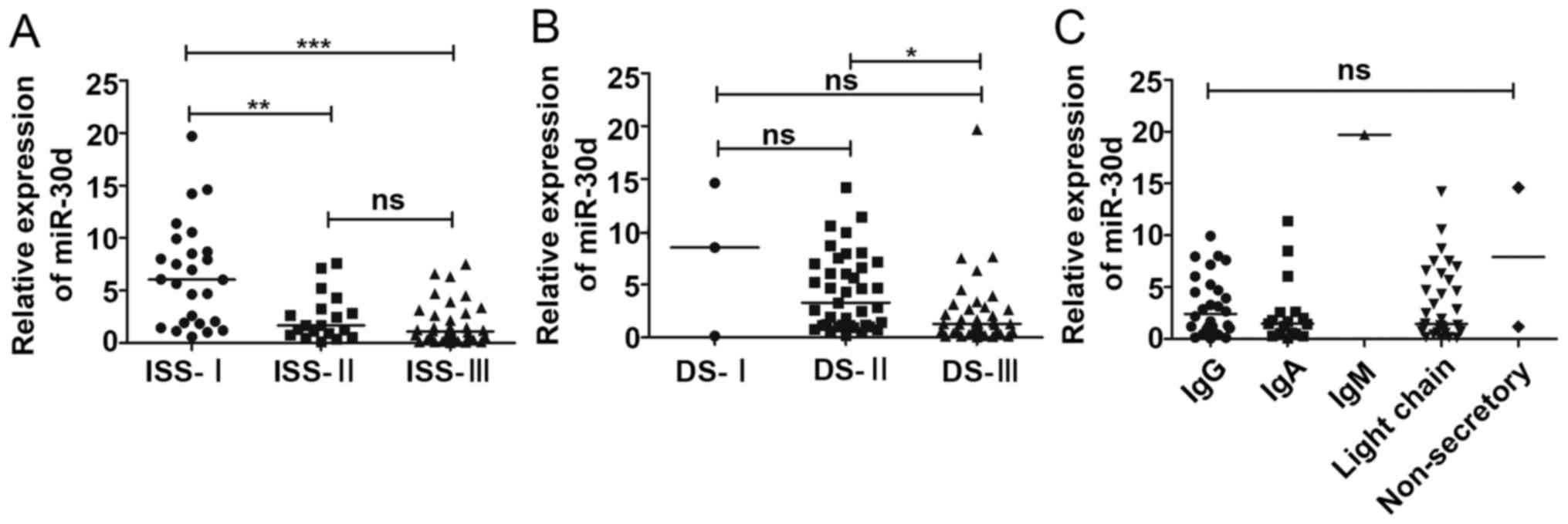
expression levels differed significantly between ISS stage I and II
(P<0.01), and stage I and III (P<0.001), while no differences
were observed between stage II and III (Fig. 2A). We also found that the miR-30d
expression levels differed significantly between DS stage II and
III (P<0.05), while miR-30d was not found to be associated with
DS stage I and II, or I and III (Fig.
2B). Moreover, we failed to associate the expression levels of
miR-30d with the types of monoclonal components of patients with MM
(P=0.364) (Fig. 2C). Furthermore,
we divided all patients into 2 groups (miR-30d low expression group
and miR-30d high expression group) according to the median of the
serum miR-30d expression levels. The results demonstrated that the
concentrations of serum HGB, PLT, Cr and β2M, and BMPC
infiltration in patients with MM differed significantly between
these 2 groups (P<0.05). Generally, the serum concentrations of
HGB and PLT in patients with MM in the miR-30d high expression
group were increased compared with those in the miR-30d low
expression group, while the serum concentrations of Cr and
β2M, and BMPC infiltration in patients with MM in the
miR-30d high expression group were decreased compared with those in
the miR-30d low expression group. However, no significant
differences were observed in the other clinical indicators,
including the concentrations of serum Ca2+, ALB, LDH,
light chain λ and light chain κ (P>0.05) (Table II).
 | Table IIComparison of clinical parameters
between the miR-30d low expression group and miR-30d high
expression group in patients with multiple myeloma. |
Table II
Comparison of clinical parameters
between the miR-30d low expression group and miR-30d high
expression group in patients with multiple myeloma.
| Parameters | Median
(interquartile range)
| P-value |
|---|
| miR-30d low
expression | miR-30d high
expression |
|---|
| HGB (g/l) | 85
(77.5-104.5) | 106 (83-130.5) |
0.0084 |
| PLT
(×109) | 127 (93-196.5) | 179
(123-237.5) |
0.0123 |
| Ca2+
(mmol/l) | 2.14
(2.02-2.235) | 2.1
(1.98-2.195) | 0.2557 |
| ALB (g/l) | 32.2
(29.1-36.2) | 35.20
(29.45-38.1) | 0.5684 |
| Cr
(μmol/l) | 76 (49-121.5) | 67 (47.5-75) |
0.0340 |
| β2M
(mg/l) | 6.4 (3-8.8) | 2.9 (1.82-4.8) |
0.0003 |
| LDH (ukat/l) | 172
(126-231.5) | 156
(126.5-209.5) | 0.3758 |
| Light chain λ
(mg/dl) | 345
(122-932.5) | 364 (192-870) | 0.9963 |
| Light chain κ
(mg/dl) | 753
(384.5-2680) | 520
(400.5-1855) | 0.5938 |
| BMPCs infiltration
(%) | 11.5
(2.875-37.5) | 2.5 (0.5-11.5) |
0.0019 |
Follow-up study of the association
between the serum levels of miR-30d and the improved clinical
parameters of the patients with MM
We collected the complete clinical indicators of 24
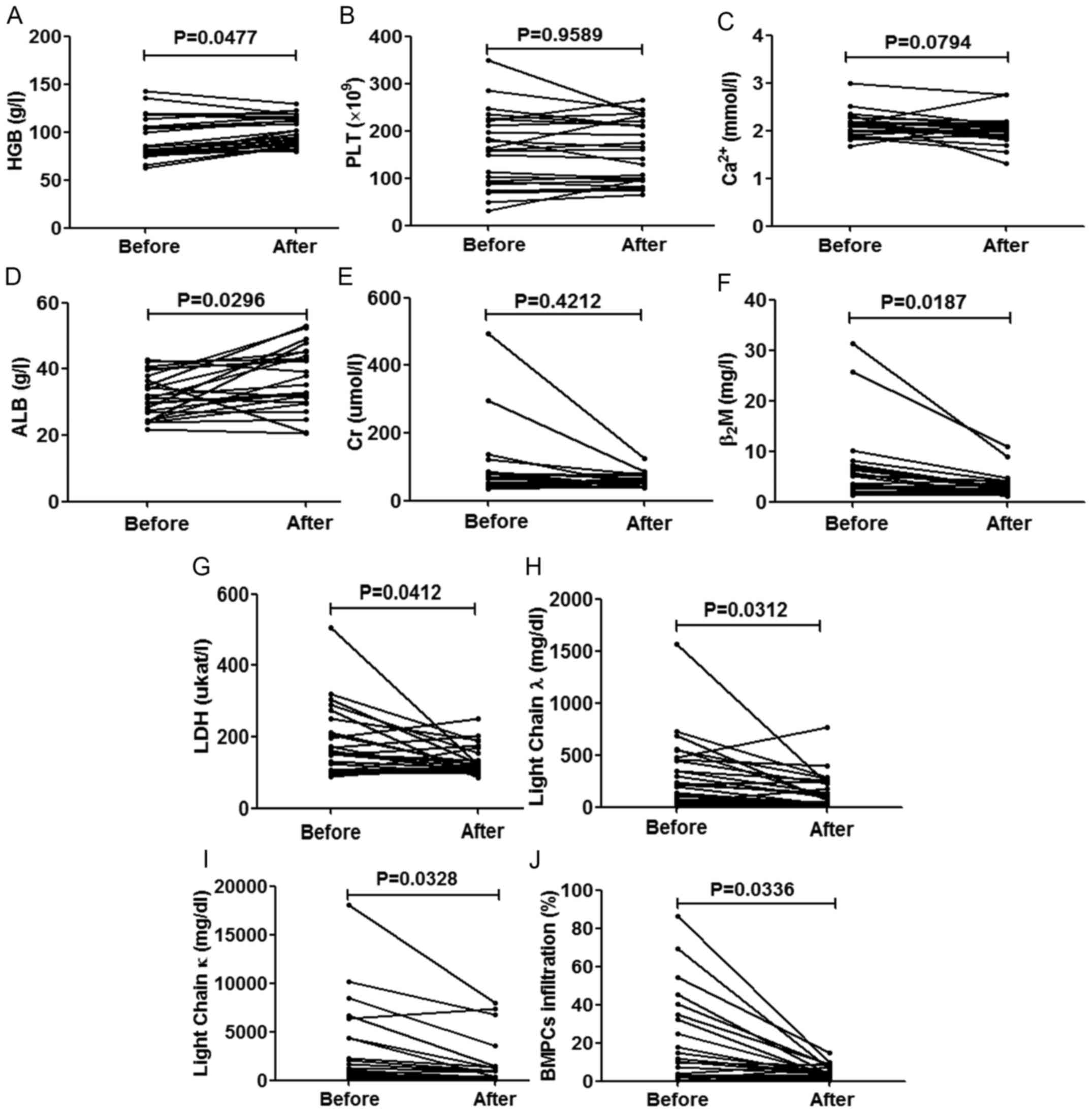
patients who were followed up. Following 2 periods of treatment,
the majority of the parameters of the patients significantly
improved, as was expected (P<0.05), apart from the expression
levels of PLT (P=0.9589), Cr (P=0.4212) (this condition may be due
to the side-effects of chemotherapy) and Ca2+ (P=0.0794)
(Fig. 3). We then compared the
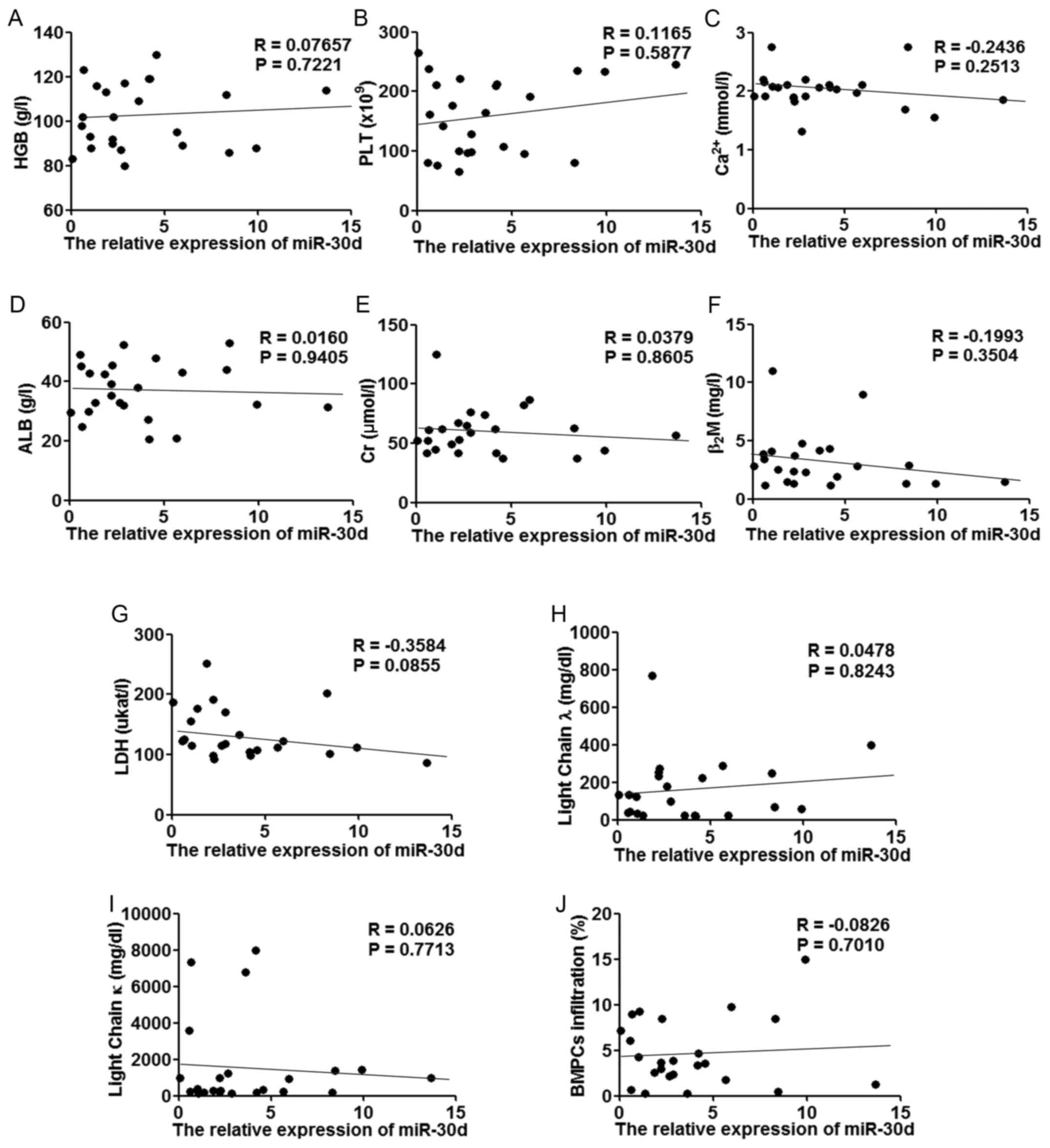
increased miR-30d levels with the improved clinical parameters;
however, we failed to find any significant correlations between
them (Fig. 4).
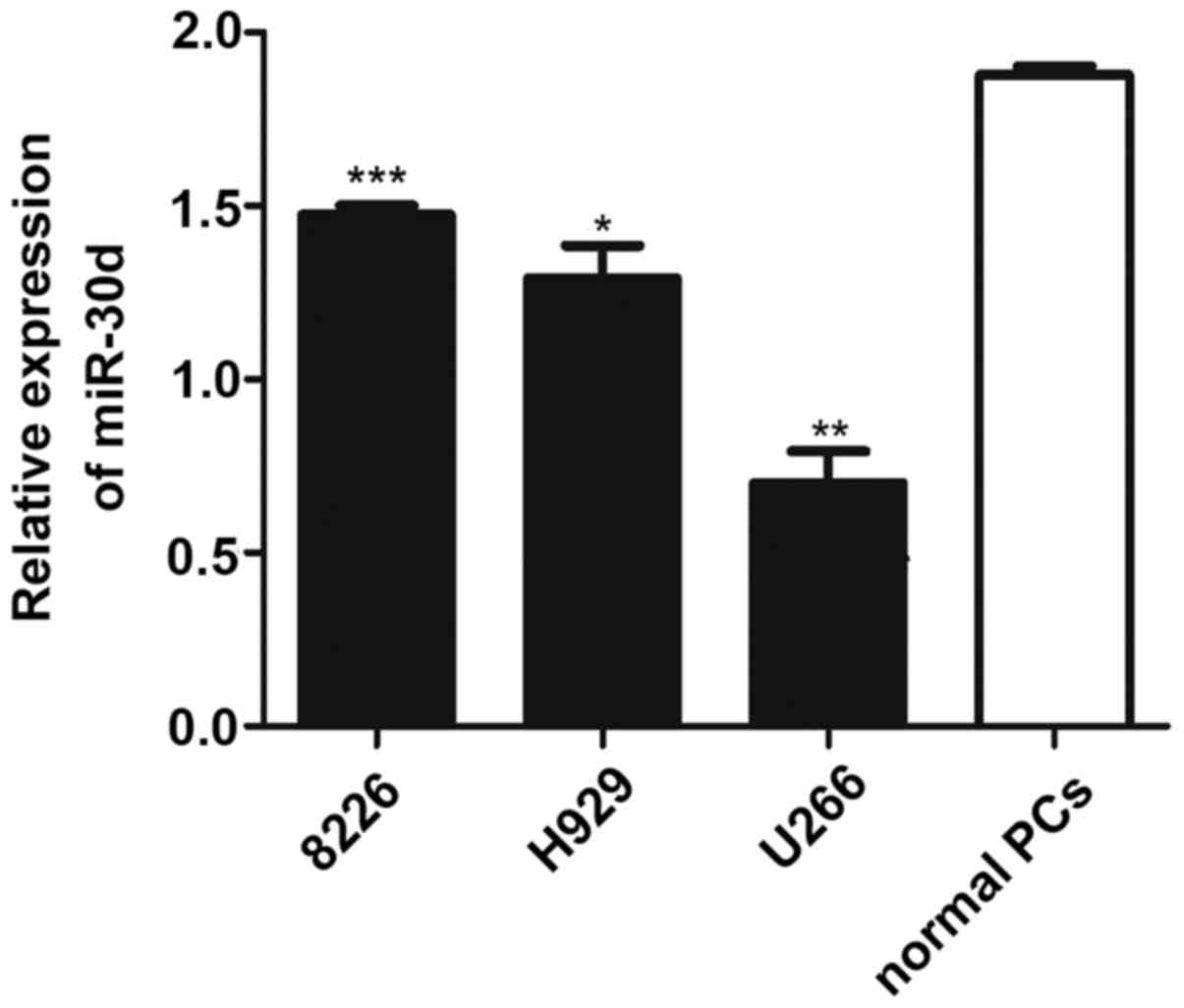
miR-30d expression is downregulated in MM
cell lines compared with normal PCs
To investigate the role of miR-30d in vitro,
we detected its expression level in MM cell lines by RT-qPCR
analysis. The results revealed that the expression level of miR-30d
was significantly decreased in MM cell lines (8226, H929 and U266
cells) compared with the normal CD138+ purified PCs
(PCs) from HDs. Moreover, the lowest miR-30d expression level was
observed in the U266 cells; thus, we conducted further cell
function experiments using this cell line (Fig. 5).
miR-30d inhibits the proliferation and
promotes the apoptosis of U266 cells
To examine the effects of miR-30d on the viability
of U266 cells, miR-30d mimics (M), mimic negative control
(M−), miR-30d inhibitor (I) and inhibitor negative
control (I−) were transfected into the U266 cells for 48
h according to the manufacturer’s instructions, respectively. The
high transfection efficiency was confirmed by RT-qPCR (Fig. 6A). The viability of the U266 cells
was then evaluated by CCK-8 assay. OD values were measured at 0,
24, 48 and 72 h, respectively. The results indicated that the cells
in the M group displayed a significantly lower cell viability rate
than those of the control group (P=0.029, 0.007, 0.003,
respectively); equally, cells in the I− group displayed
a significantly higher cell viability rate than those in the
control group (P=0.024, 0.009, 0.015, respectively). Therefore,
miR-30d significantly inhibited the proliferation of the U266 cells
(Fig. 6B and C). Moreover, at 48 h
following transfection, the apoptotic rate of these 4 groups was
detected by flow cytometry. The results revealed that the cells in
the M group exhibited a higher apoptotic rate than those in the
mimic negative control (M−) group (P=0.013); the cells
in the I group exhibited lower apoptotic rates than those in the
I− group (P=0.020). These findings confirmed that
miR-30d promoted the apoptosis of U266 cells (Fig. 6D).
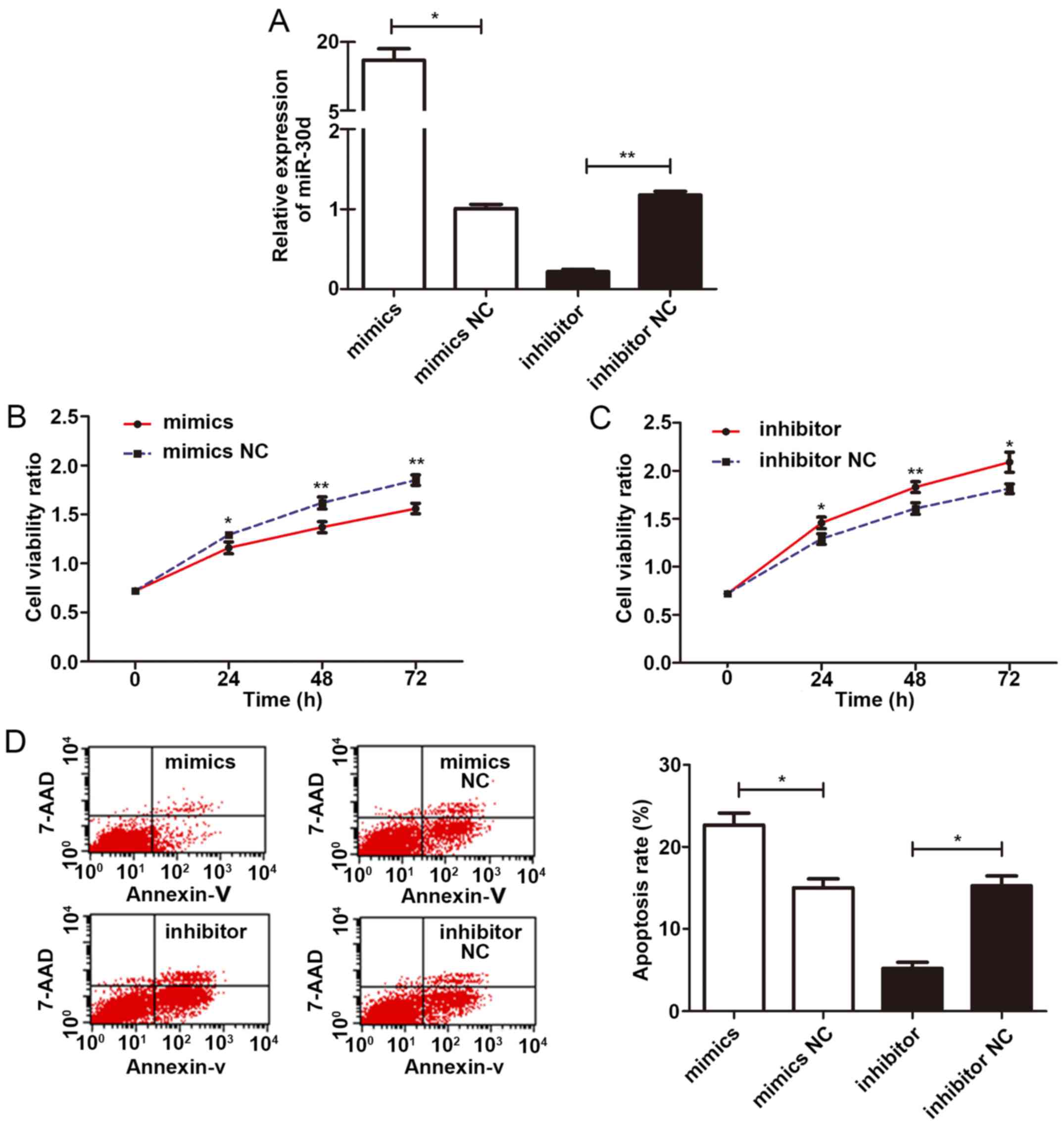 | Figure 6miR-30d inhibits the proliferation
and promotes the apoptosis of U266 cells. (A) Transfection of
miR-30d mimics significantly increased the expression level of
miR-30d, whereas miR-30d inhibitor significantly decreased the
expression level of miR-30d in U266 cells. (B) CCK-8 cell
proliferation assay demonstrated that miR-30d mimics significantly
decreased the viability rate of U266 cells compared to its negative
control at 24, 48 and 72 h (P=0.029, 0.007, 0.003, respectively).
(C) CCK-8 cell proliferation assay demonstrated that the miR-30d
inhibitor increased the viability rate of U266 cells compared to
its negative control at 24, 48 and 72 h (P=0.024, 0.009, 0.015,
respectively). (D) Flow cytometric analysis demonstrated an
increased apoptotic rate of U266 cells in the miR-30d mimics group
compared to the mimics NC group (P=0.013) and a decreased apoptotic
rate of U266 cells in the miR-30d inhibitor group compared to the
inhibitor NC group (P=0.020). All experiments were carried out in
triplicate. Statistically significant differences between groups
were determined by the Kruskal-Wallis test, followed by the
Tamhane’s T2 test as a post hoc test. *P<0.05,
**P<0.01. |
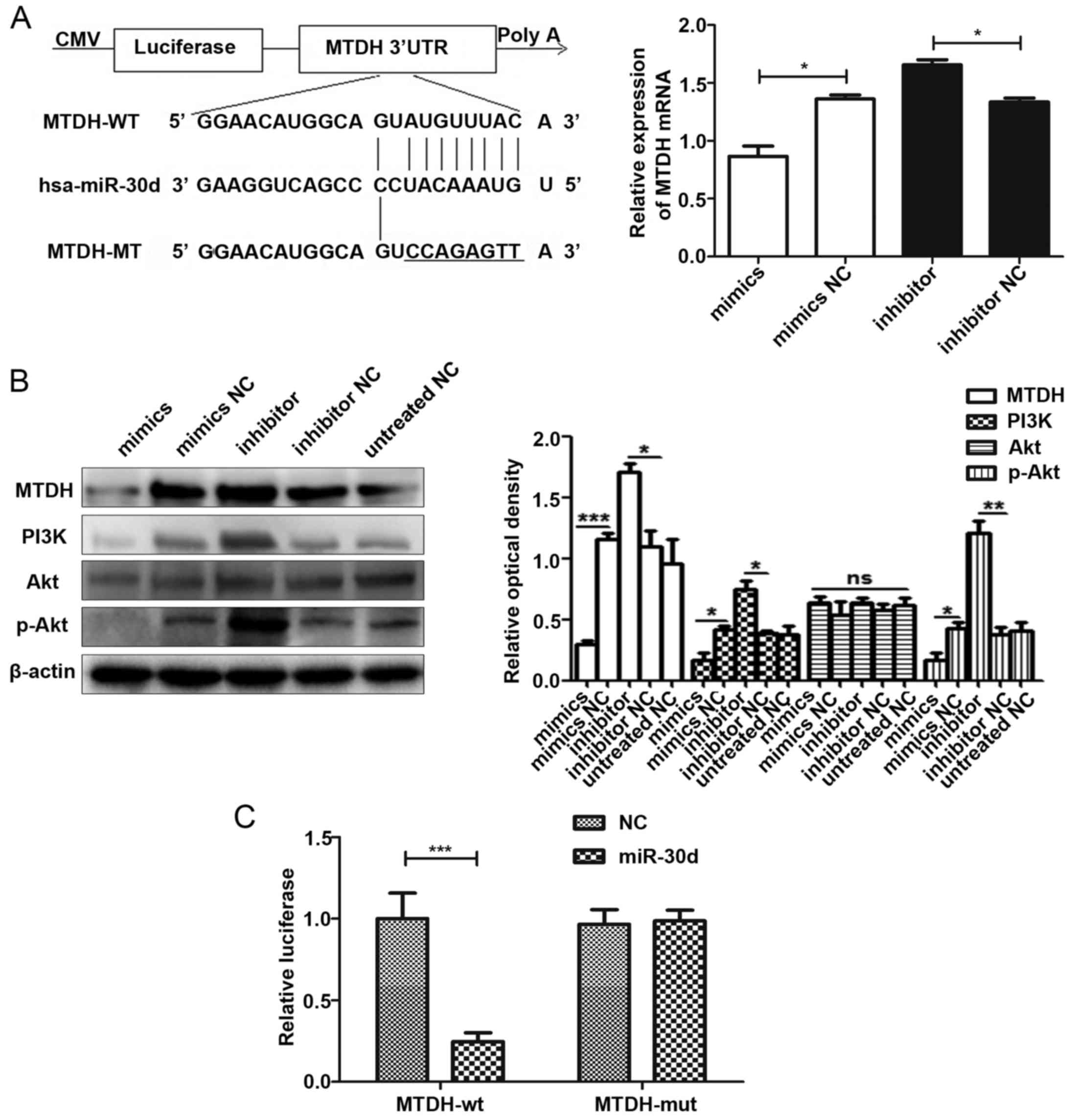
miR-30d binds to the 3′UTR of MTDH
directly and acts as an inhibitor of the PI3K/Akt signaling
pathway
Bioinformatics software was used to predict the
target genes of miR-30d and MTDH was selected as the
putative target of miR-30d. Moreover, we used RT-qPCR to
preliminarily verify the negative association between miR-30d and
the mRNA expression of MTDH (Fig. 7A). The results of WB analysis then
proved that miR-30d mimics suppressed the expression of MTDH, while
the miR-30d inhibitor promoted the expression of MTDH. At the same
time, we also detected the protein expression levels of p-Akt and
PI3K in the PI3K/Akt signaling pathway. The results indicated that
miR-30d inhibited the expression of these 2 proteins and acted as
an inhibitor of the PI3K/Akt signaling pathway (Fig. 7B). Furthermore, we conducted a
luciferase reporter assay to confirm the direct binding association
between miR-30d and MTDH. The final results indicated that
miR-30d could bind to the 3′UTR of MTDH directly (Fig. 7C).
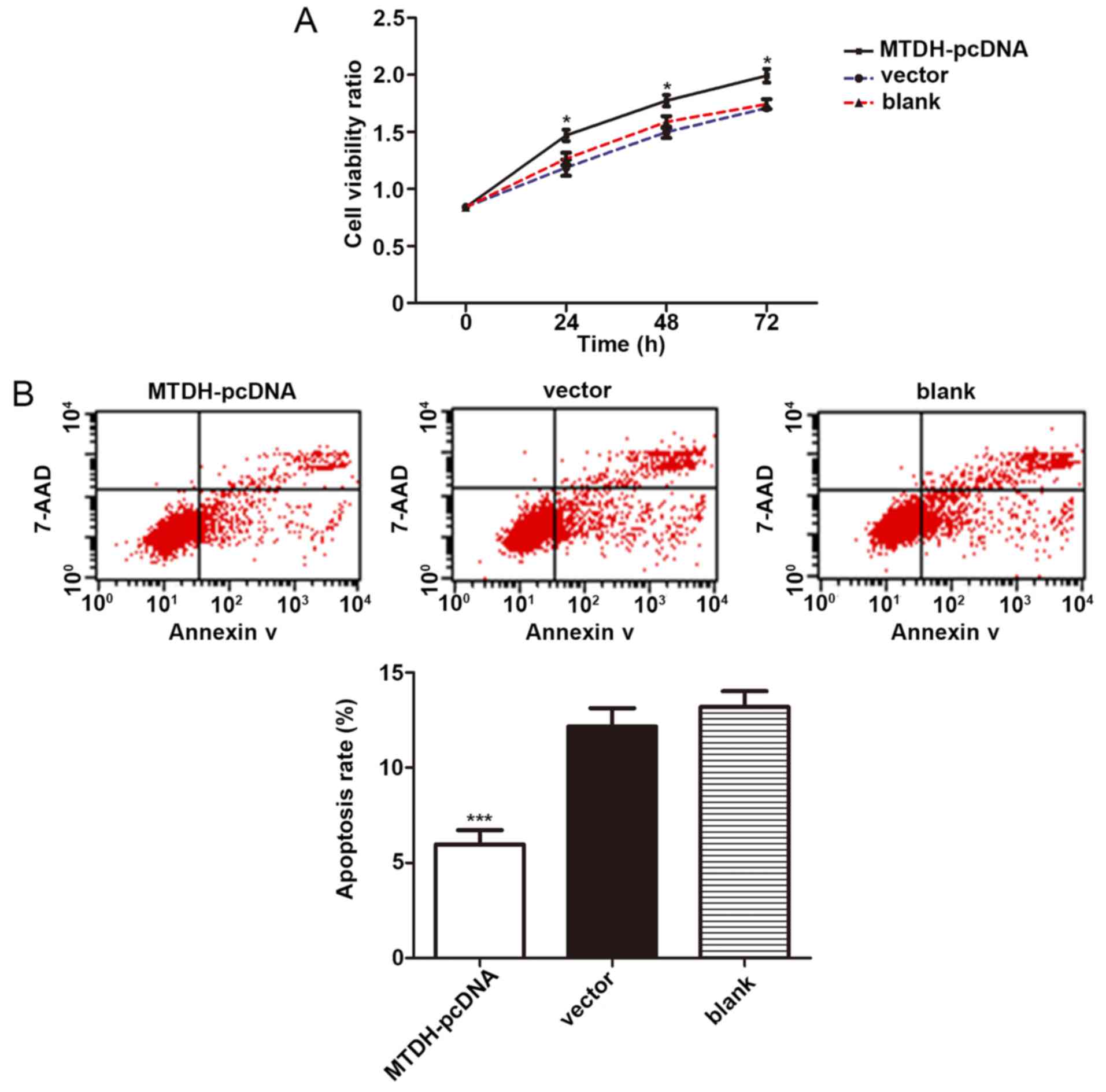
MTDH promotes the proliferation and
inhibited the apoptosis of U266 cells
To determine the effects of MTDH on the
viability of U266 cells, pcDNA of MTDH and its negative
control were transfected into the U266 cells for 48 h according to
the manufacturer’s instructions. OD values were measured by CCK-8
cell proliferation assay at 0, 24, 48 and 72 h, respectively. The
results indicated that the cells in the MTDH pcDNA group
displayed a significantly higher cell viability rate than those in
the vector control group (P=0.031, 0.017, 0.013, respectively).
Therefore, pcDNA of MTDH significantly promoted the
proliferation of U266 cells (Fig.
8A). Furthermore, at 48 h following transfection, the apoptotic
rates of the cells in these 3 groups were detected by flow
cytometry. The results revealed that the MTDH pcDNA group
had a lower apoptotic rate than the vector group (P<0.001) and
confirmed that MTDH inhibited the apoptosis of the U266
cells (Fig. 8B). These results
revealed the carcinogenic role of MTDH in U266 cells.
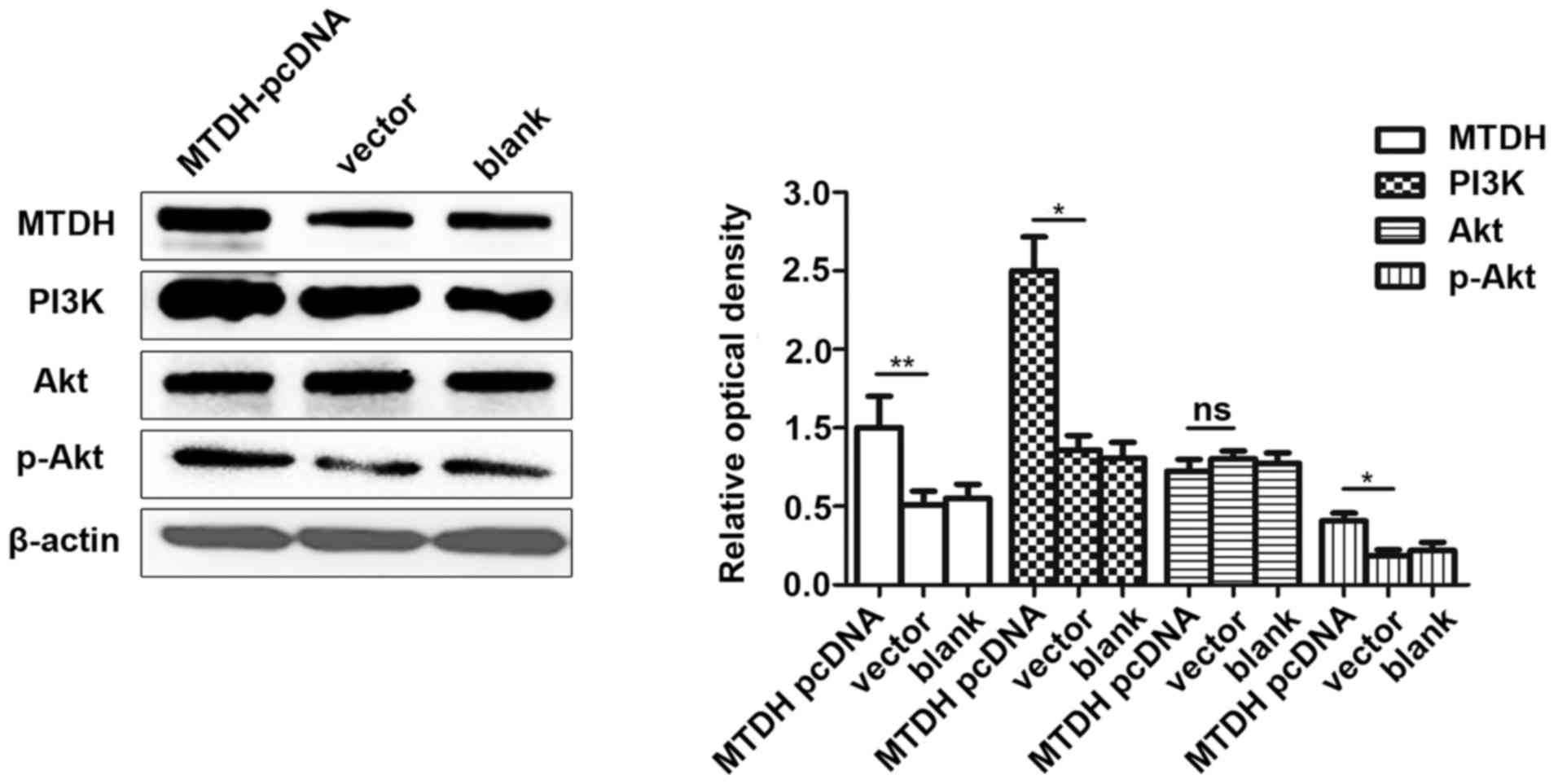
MTDH induces the activation of the
PI3K/Akt signaling pathway
To investigate the role of MTDH in the
pathogenesis of MM, MTDH-pcDNA and its negative control were
transfected into the U266 cells. The expression levels of MTDH and
the downstream PI3K/Akt signaling pathway-related proteins,
including PI3K, Akt, p-Akt were then detected by WB analysis. The
results revealed that the overexpression of MTDH induced the
expression of PI3K and p-Akt to activate the PI3K/Akt signaling
pathway (Fig. 9).
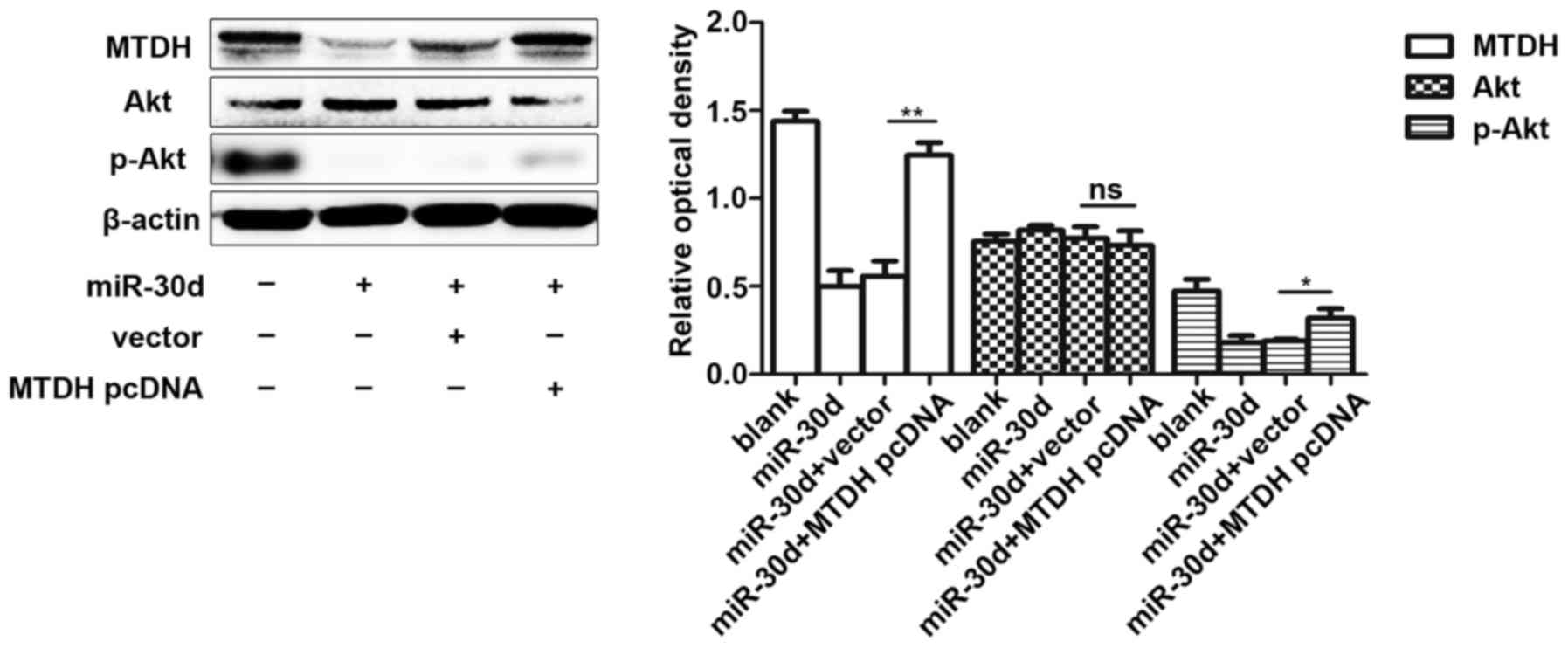
miR-30d suppresses the PI3K/Akt signaling
pathway by targeting MTDH
To investigate the mechanisms through which miR-30d
functions in MM cells by targeting MTDH, we co-transfected
miR-30d mimics and MTDH-pcDNA together into U266 cells. We
then measured the expression level of p-Akt following transfection
at 72 h, and the results demonstrated that MTDH induced the
expression level of p-Akt, while miR-30d mimics partially
alleviated this carcinogenic effect by inhibiting the expression
level of MTDH. These findings suggested that miR-30d exerts an
inhibitory effect on the PI3K/Akt signaling pathway by targeting
MTDH (Fig. 10).
Discussion
Numerous studies have indicated that genetic
abnormalities, including genomic alterations, post-transcriptional
regulation, epigenetic alterations, RNA editing and miRNA binding
site sequence variation can affect the expression levels of miRNAs
(28,29). In MM, certain pathological factors
can also lead to the dysregulation of miRNA expression. One study
using unsupervised cluster analysis revealed that CD138+
cells isolated from patients with MM and monoclonal gammopathy of
undetermined significance (MGUS) had a distinct miRNA expression
profiling compared with the normal controls (30). In general, that study suggested
that the aberrant expression level of miRNAs was associated with
the pathogenesis, diagnosis and prognosis of MM. However, few
studies to date have mentioned the specific role of miR-30d in MM
(31). Therefore, this study
focused on the exploration of the clinical value of this molecule
in MM and its in-depth mechanisms of action in MM cells.
miRNAs can mediate interactions between cells, and
exist stably in the circulating fluid. To a certain extent, some of
them can reflect the pathological condition of diseases (14). In the present study, we collected
serum of 81 newly-diagnosed patients with MM and 78 HDs, and then
detected the relative expression of miR-30d in serum by RT-qPCR. We
found that miR-30d expression was significantly downregulated in
the serum of patients with MM compared with HDs (P<0.0001).
Moreover, the diagnostic value of miR-30d in serum was assessed by
ROC curve analysis, and we found that when the cut-off value was
2.9082, sensitivity was 88.5%, specificity was 63%, and the AUC was
of 0.800. Through the followup analyses of 24 patients with primary
MM, we found that following 2 periods of treatment, the majority of
the clinical parameters of the patients had improved (P<0.05)
and the expression levels of miR-30d were increased (P<0.0001),
indicating that miR-30d can reflect the progression of MM and thus
has potential to monitor this disease. However, we failed to find
any significant correlation between miR-30d and the improved
clinical parameters of the patients with MM. Of note, it was also
found that the expression level of miR-30d in serum was
significantly associated with the percentage of BMPC infiltration,
the value of PLT, β2M, Cr, HGB and the stage of MM.
These results suggest that the miR-30d level in serum of patients
with MM possesses a certain diagnostic value in distinguishing
patients with MM from healthy controls and is mainly associated
with MM progression.
In related studies on MM, accumulating conclusions
have confirmed that circulating miRNAs are significantly associated
with the pathological processes and prognosis of diseases. miRNAs
have high clinical values in the diagnosis and prognosis of
diseases (32,33). Moreover, due to the advantages of
their stable expression in fluids, easy to collect from specimens
and high sensitivity for detection, miRNAs are expected to replace
some traditional indicators. In recent years, the rapid development
of high-throughput gene chip technology has aided researchers in
the screening of a series of differentially expressed miRNAs in
peripheral blood, and they then can select target miRNAs for
further study. Yyusnita et al (34) analyzed differential expression
profiles of miRNAs in the peripheral blood of 14 patients
newly-diagnosed with MM, 24 follow-up patients with MM and 7 HDs by
miRNA microarray technology. Ultimately, they discovered 10 miRNAs
differentially expressed both in the primary and follow-up patients
with MM compared with the HDs. In addition, they also selected 3
miRNAs only differentially expressed in patients with primary MM
and 8 miRNAs only differentially expressed in the follow-up
patients with MM. These results demonstrated that miRNAs are widely
involved in the different pathological processes of MM and may
reflect the different states of MM. Importantly, this difference
can be reflected by the expression levels of miRNAs in peripheral
blood. However, summing up some results of microarray data
(14,35), we found that profiling results were
not consistent in different laboratories. These differences may be
due to the different patient groups, methods of sample collections,
microarray platforms used and statistical methods used, etc. In
order to obtain more accurate results, it may be necessary to
standardize the whole process of the microarray analysis.
miR-30d, a member of the miR-30 family (including
miR-30a/b/c/d/e), is located on the human chromosome 8q24.22. In
recent years, a number of studies have demonstrated that this
molecule is involved in the development of many tumors (36). Xuan et al (37) found that the expression level of
miR-30d was downregulated in prostate cancer. Further studies on
cell function revealed that miR-30d inhibited the proliferation of
prostate cancer cells and this inhibitory effect may be caused by
the targeting of the Bim-1 gene. Moreover, another study on ovarian
cancer indicated that miR-30d was not only closely related to cell
proliferation, but was involved in the process of
epithelial-mesenchymal transition (EMT) in ovarian cancer cell
(17). Wu et al (19) also found that miR-30d functioned as
a tumor suppressor in renal cell carcinoma and the mechanism was
partly due to the fact that miR-30d induced apoptosis and was
regulated by the Akt/FOXO pathway in renal cell carcinoma cells. By
summarizing these discoveries, it has been identified that miR-30d
often presents a low expression level in a number of types of
cancers and its low expression probably promotes the occurrence and
development of cancers. However, studies on the role of miR-30d in
MM are limited. In this study, to clarify the role of miR-30d in
MM, we performed cell function experiments and found that miR-30d
inhibited cell proliferation and induced cell apoptosis. These
results revealed that miR-30d functions as a tumor suppressor gene
in U266 cells, which is consistent with its role in studies on
other solid tumors (19,38,39).
In order to examine the mechanisms of action of miR-30d, we
predicted the target genes of miR-30d using bioinformatics
software. We found that miR-30d had a direct binding site in the
3′UTR of MTDH. Through preliminary WB analysis experiments,
we found that the miR-30d mimics decreased the protein expression
of MTDH, while its inhibitor had the opposite effect. Subsequenlty,
luciferase reporter gene experiments were carried out to confirm
that miR-30d could bind to the 3′UTR of MTDH directly.
MTDH is a recognized oncogene in cancers and
it has been demonstratd that it plays a role in carcinogenesis
through the PI3K/Akt signaling pathway (40). Recent studies have also indicated
that some oncogenic molecules can inhibit the apoptosis of MM cells
and contribute to the progression of MM; thus, targeting these
molecules could induce the apoptosis of MM cells and may provide a
novel therapeutic strategy for MM (41,42).
After investigating the available literature, we found that
MTDH has been proven to function as an oncogene in some
solid cancers (43). However,
whether MTDH can affect the apoptosis of MM cells remains
unknown. Thus, we aim to perform further experiments in the future
to prove the association of MTDH with the apoptosis of the
U266 cell line. In this study, we found that MTDH did have a
similar mode of action in U266 cells, as the overexpression of
MTDH induce the proliferation and inhibited the apoptosis of
the U266 cell line. Subsequently, following the overexpression of
MTDH, downstream p-Akt expression was induced. In addition,
miR-30d and MTDH pcDNA were co-transfected into U266 cells,
and the results of WB analysis revealed that miR-30d attenuated the
promoting effect of MTDH on the expression of p-Akt.
Therefore, miR-30d can partially reverse the carcinogenic effects
of MTDH, suggesting that miR-30d may prove to be a novel
potential target for use in the treatment of MM.
However, our study still has some shortcomings. We
need to include a greater number of serum samples in the future to
obtain a more accurate verification. In addition, we lacked
follow-up data for the long-term survival analysis of miR-30d, and
in future studies, we aim to examine its prognostic role in MM.
Furthermore, as a serum biomarker, a single miRNA may not have
sufficient diagnostic effectiveness; the expression of a panel of
miRNAs may yield greater significance to the clinical application.
In future studies, we aim to combine miR-30d with other important
MM-associated miRNAs to evaluate the clinical diagnostic value. Our
preliminary experiments were performed only on the U266 cell line
and this may not represent the responses of all cells. Thus, in the
future, we also aim to perform the same experiments using other
cell lines to validate our findings.
In conclusion, the results of this study indicate
that the serum expression level of miR-30d is significantly
downregulated in patients with MM and it has a considerable
diagnostic value. Moreover, miR-30d exerts a significant antitumor
effect on U266 cells. The underlying mechanisms involve the binding
of this miRNA to its target gene, MTDH, and this result in
the decreased expression of this gene. The activation of
MTDH downstream the PI3K/Akt signaling pathway is then
inhibited, leading to a decrease in cell proliferation and the
induction of cell apoptosis. Our preliminary results revealed that
miR-30d has great potential for use as a novel serological marker
and therapeutic target for MM.
Funding
The present study was supported by grants from the
Jiangsu Provincial Project of Invigorating Health Through Science
and Technology (LJ201133); Jiangsu Provincial Funds for Six
Categories of Top Talents (WS-066), and the Research project of
Jiangsu provincial health and Family Planning Commission
(H201526).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
BZ and HCh wrote the initial draft, designed and
conceived the study. HCo, SJ and XW designed and conceived the
study and revised the manuscript. RJ, LS designed and performed the
experiments. XZ, YP and CJ contributed to the acquisition and
analysis of data for the work. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
All procedures performed in this study involving
human participants were in accordance with the ethical standards of
the institutional and national research committee and with the 1964
Helsinki Declaration and its later amendments or comparable ethical
standards. This study was approved by Ethics Committee of the
Hospital Affiliated to Nantong University and the committee’s
reference number was 2015-007. Informed consent was obtained from
all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Bergsagel PL, Kuehl WM, Zhan F, Sawyer J,
Barlogie B and Shaughnessy J Jr: Cyclin D dysregulation: An early
and unifying pathogenic event in multiple myeloma. Blood.
106:296–303. 2005. View Article : Google Scholar
|
|
2
|
Herrinton LJ, Demers PA, Koepsell TD,
Weiss NS, Daling JR, Taylor JW, Lyon JL, Swanson GM and Greenberg
RS: Epidemiology of the M-component immunoglobulin types of
multiple myeloma. Cancer Causes Control. 4:83–92. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lyubimova NV, Timofeev YS, Abaev VM,
Votyakova OM and Kushlinskii NE: Immunochemical Diagnosis of
Multiple Myeloma. Bull Exp Biol Med. 165:84–87. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
No authors listed. Pharmacokinetics and
pharmacodynamics of a 13-mer LNA-inhibitor-miR-221 in mice and
non-human primates. Mol Ther Nucleic Acids. 5:e3362016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo J, McKenna SL, O’Dwyer ME, Cahill MR
and O’Driscoll CM: RNA interference for multiple myeloma therapy:
Targeting signal transduction pathways. Expert Opin Ther Targets.
20:107–121. 2016. View Article : Google Scholar
|
|
6
|
Liang B, Yin JJ and Zhan XR: MiR-301a
promotes cell proliferation by directly targeting TIMP2 in multiple
myeloma. Int J Clin Exp Pathol. 8:9168–9174. 2015.PubMed/NCBI
|
|
7
|
Yang Y, Li F, Saha MN, Abdi J, Qiu L and
Chang H: miR-137 and miR-197 Induce Apoptosis and Suppress
Tumorigenicity by Targeting MCL-1 in Multiple Myeloma. Clin Cancer
Res. 21:2399–2411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calura E, Bisognin A, Manzoni M, Todoerti
K, Taiana E, Sales G, Morgan GJ, Tonon G, Amodio N, Tassone P, et
al: Disentangling the microRNA regulatory milieu in multiple
myeloma: Integrative genomics analysis outlines mixed miRNA-TF
circuits and pathway-derived networks modulated in t(4;14)
patients. Oncotarget. 7:2367–2378. 2016. View Article : Google Scholar :
|
|
9
|
Yang WC and Lin SF: Mechanisms of drug
resistance in relapse and refractory multiple myeloma. BioMed Res
Int. 2015:3414302015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Di Martino MT, Leone E, Amodio N, Foresta
U, Lionetti M, Pitari MR, Cantafio ME, Gullà A, Conforti F, Morelli
E, et al: Synthetic miR-34a mimics as a novel therapeutic agent for
multiple myeloma: In vitro and in vivo evidence. Clin Cancer Res.
18:6260–6270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Du J, Liu S, He J, Liu X, Qu Y, Yan W, Fan
J, Li R, Xi H, Fu W, et al: MicroRNA-451 regulates stemness of side
population cells via PI3K/Akt/mTOR signaling pathway in multiple
myeloma. Oncotarget. 6:14993–15007. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng P, Guo H, Li G, Han S, Luo F and Liu
Y: PSMB4 promotes multiple myeloma cell growth by activating
NF-κB-miR-21 signaling. Biochem Biophys Res Commun. 458:328–333.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grasedieck S, Sorrentino A, Langer C,
Buske C, Döhner H, Mertens D and Kuchenbauer F: Circulating
microRNAs in hematological diseases: Principles, challenges, and
perspectives. Blood. 121:4977–4984. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rocci A, Hofmeister CC, Geyer S, Stiff A,
Gambella M, Cascione L, Guan J, Benson DM, Efebera YA, Talabere T,
et al: Circulating miRNA markers show promise as new
prognosticators for multiple myeloma. Leukemia. 28:1922–1926. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li F, Xu Y, Deng S, Li Z, Zou D, Yi S, Sui
W, Hao M and Qiu L: MicroRNA-15a/16-1 cluster located at chromosome
13q14 is downregulated but displays different expression pattern
and prognostic significance in multiple myeloma. Oncotarget.
6:38270–38282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang X, Zhong X, Tanyi JL, Shen J, Xu C,
Gao P, Zheng TM, DeMichele A and Zhang L: mir-30d Regulates
multiple genes in the autophagy pathway and impairs autophagy
process in human cancer cells. Biochem Biophys Res Commun.
431:617–622. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ye Z, Zhao L, Li J, Chen W and Li X:
miR-30d blocked transforming growth factor β1-induced
epithelial-mesenchymal transition by targeting snail in ovarian
cancer cells. Int J Gynecol Cancer. 25:1574–1581. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao JJ, Lin J, Zhu D, Wang X, Brooks D,
Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S, et al: miR-30-5p
functions as a tumor suppressor and novel therapeutic tool by
targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res.
74:1801–1813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu C, Jin B, Chen L, Zhuo D, Zhang Z, Gong
K and Mao Z: MiR-30d induces apoptosis and is regulated by the
Akt/FOXO pathway in renal cell carcinoma. Cell Signal.
25:1212–1221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wan L, Hu G, Wei Y, Yuan M, Bronson RT,
Yang Q, Siddiqui J, Pienta KJ and Kang Y: Genetic ablation of
metadherin inhibits autochthonous prostate cancer progression and
metastasis. Cancer Res. 74:5336–5347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu G, Chong RA, Yang Q, Wei Y, Blanco MA,
Li F, Reiss M, Au JL, Haffty BG and Kang Y: MTDH activation by 8q22
genomic gain promotes chemoresistance and metastasis of
poor-prognosis breast cancer. Cancer Cell. 15:9–20. 2009.
View Article : Google Scholar :
|
|
22
|
Yao Y, Gu X, Liu H, Wu G, Yuan D, Yang X
and Song Y: Metadherin regulates proliferation and metastasis via
actin cytoskeletal remodelling in non-small cell lung cancer. Br J
Cancer. 111:355–364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu X, Wang D, Liu H, Feng Y, Zhu T, Zhang
L, Zhu B and Zhang Y: Knockdown of astrocyte elevated gene-1
(AEG-1) in cervical cancer cells decreases their invasiveness,
epithelial to mesenchymal transition, and chemoresistance. Cell
Cycle. 13:1702–1707. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gu C, Feng L, Peng H, Yang H, Feng Z and
Yang Y: MTDH is an oncogene in multiple myeloma, which is
suppressed by Bortezomib treatment. Oncotarget. 7:4559–4569. 2016.
View Article : Google Scholar :
|
|
25
|
Pichiorri F, Suh SS, Rocci A, De Luca L,
Taccioli C, Santhanam R, Zhou W, Benson DM Jr, Hofmainster C, Alder
H, et al: Downregulation of p53-inducible microRNAs 192, 194, and
215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma
development. Cancer Cell. 18:367–381. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Jagannathan S, Vad N, Vallabhapurapu S,
Vallabhapurapu S, Anderson KC and Driscoll JJ: MiR-29b replacement
inhibits proteasomes and disrupts aggresome+autophagosome formation
to enhance the antimyeloma benefit of bortezomib. Leukemia.
29:727–738. 2015. View Article : Google Scholar :
|
|
28
|
Misiewicz-Krzeminska I, Sarasquete ME,
Quwaider D, Krzeminski P, Ticona FV, Paíno T, Delgado M, Aires A,
Ocio EM, García-Sanz R, et al: Restoration of microRNA-214
expression reduces growth of myeloma cells through positive
regulation of P53 and inhibition of DNA replication. Haematologica.
98:640–648. 2013. View Article : Google Scholar :
|
|
29
|
Zhang Q, Wang LQ, Wong KY, Li ZY and Chim
CS: Infrequent DNA methylation of miR-9-1 and miR-9-3 in multiple
myeloma. J Clin Pathol. 68:557–561. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chi J, Ballabio E, Chen XH, Kušec R,
Taylor S, Hay D, Tramonti D, Saunders NJ, Littlewood T, Pezzella F,
et al: MicroRNA expression in multiple myeloma is associated with
genetic subtype, isotype and survival. Biol Direct. 6:232011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao JJ and Carrasco RD: Crosstalk between
microRNA30a/b/c/ d/e-5p and the canonical Wnt pathway: Implications
for multiple myeloma therapy. Cancer Res. 74:5351–5358. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kubiczkova L, Kryukov F, Slaby O,
Dementyeva E, Jarkovsky J, Nekvindova J, Radova L, Greslikova H,
Kuglik P, Vetesnikova E, et al: Circulating serum microRNAs as
novel diagnostic and prognostic biomarkers for multiple myeloma and
monoclonal gammopathy of undetermined significance. Haematologica.
99:511–518. 2014. View Article : Google Scholar :
|
|
33
|
Qu X, Zhao M, Wu S, Yu W, Xu J, Xu J, Li J
and Chen L: Circulating microRNA 483-5p as a novel biomarker for
diagnosis survival prediction in multiple myeloma. Med Oncol.
31:2192014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yyusnita N, Norsiah, Zakiah I, Chang KM,
Purushotaman VS, Zubaidah Z and Jamal R: MicroRNA (miRNA)
expression profiling of peripheral blood samples in multiple
myeloma patients using microarray. Malays J Pathol. 34:133–143.
2012.
|
|
35
|
Sevcikova S, Kubiczkova L, Sedlarikova L,
Slaby O and Hajek R: Serum miR-29a as a marker of multiple myeloma.
Leuk Lymphoma. 54:189–191. 2013. View Article : Google Scholar
|
|
36
|
Su SF, Chang YW, Andreu-Vieyra C, Fang JY,
Yang Z, Han B, Lee AS and Liang G: miR-30d, miR-181a and
miR-199a-5p cooperatively suppress the endoplasmic reticulum
chaperone and signaling regulator GRP78 in cancer. Oncogene.
32:4694–4701. 2013. View Article : Google Scholar :
|
|
37
|
Xuan H, Xue W, Pan J, Sha J, Dong B and
Huang Y: Downregulation of miR-221, -30d, and -15a contributes to
pathogenesis of prostate cancer by targeting Bmi-1. Biochemistry
(Mosc). 80:276–283. 2015. View Article : Google Scholar
|
|
38
|
Yao J, Liang L, Huang S, Ding J, Tan N,
Zhao Y, Yan M, Ge C, Zhang Z, Chen T, et al: MicroRNA-30d promotes
tumor invasion and metastasis by targeting Galphai2 in
hepatocellular carcinoma. Hepatology. 51:846–856. 2010.PubMed/NCBI
|
|
39
|
Chen D, Guo W, Qiu Z, Wang Q, Li Y, Liang
L, Liu L, Huang S, Zhao Y and He X: MicroRNA-30d-5p inhibits tumour
cell proliferation and motility by directly targeting CCNE2 in
non-small cell lung cancer. Cancer Lett. 362:208–217. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu C, Liu Y, Tan H, Li G, Su Z, Ren S, Zhu
G, Tian Y, Qiu Y and Zhang X: Metadherin regulates metastasis of
squamous cell carcinoma of the head and neck via AKT signalling
pathway-mediated epithelial-mesenchymal transition. Cancer Lett.
343:258–267. 2014. View Article : Google Scholar
|
|
41
|
Xu H, Liu C, Zhang Y, Guo X, Liu Z, Luo Z,
Chang Y, Liu S, Sun Z and Wang X: Let-7b-5p regulates proliferation
and apoptosis in multiple myeloma by targeting IGF1R. Acta Biochim
Biophys Sin (Shanghai). 46:965–972. 2014. View Article : Google Scholar
|
|
42
|
Hu Y, Lin J, Fang H, Fang J, Li C, Chen W,
Liu S, Ondrejka S, Gong Z, Reu F, et al: Targeting the
MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in
multiple myeloma. Leukemia. Mar 22–2018, Epub ahead of print.
View Article : Google Scholar
|
|
43
|
Li WF, Ou Q, Dai H and Liu CA:
Lentiviral-mediated short hairpin RNA knockdown of MTDH inhibits
cell growth and induces apoptosis by regulating the PTEN/AKT
pathway in hepatocellular carcinoma. Int J Mol Sci. 16:19419–19432.
2015. View Article : Google Scholar :
|