Introduction
Chemotherapy is usually given systemically to
patients with cancer following surgical intervention or
radiotherapy (1). However, certain
patients are unresponsive to chemotherapy whereby the cancer cells
become resistance to chemotherapeutic drugs, a phenomenon known as
multidrug resistance (MDR). MDR is resistance to multiple
structurally and mechanistically unassociated classes of anticancer
drugs by the cells (1), and has
become a major impediment to cancer treatment due to cancer relapse
which eventually leads to mortality.
Numerous reports have suggested a plausible
association between MDR and Y-box binding protein-1 (YB-1), an
evolutionary conserved DNA or RNA binding protein. YB-1 was
initially discovered as a transcription factor that binds to the
Y-box sequence (inverted CCAAT), present in the promoter region of
the major histocompatibility complex class (MHC) II (2). It has pleotropic functions and is
implicated in fundamental processes, including transcriptional
regulation, DNA repair, mRNA splicing, translation and stability
(3). The presence of nuclear YB-1
has been demonstrated as associated with enhanced transcription of
the multidrug resistance 1 (MDR1) gene, eventually leading
to the upregulation of P-glycoprotein, which is indicative of
chemoresistance, cancer progression and poor prognosis (4). YB-1 was also suggested to be a
promising prognostic biomarker for patients with breast cancer
(5). A recent report has further
suggested the involvement of YB-1 in cisplatin resistance in
gastric cancer (6).
Janus kinase (JAK)/signal transducer and activator
of transcription (STAT) signaling is an important pathway involved
in fundamental cellular processes, including tumourigenesis, immune
responses and sex determination (7-9). In
particular, aberrant expression of STAT3 is associated with cancer
progression through the facilitation of cell proliferation,
metastasis and chemoresistance (10). Dysregulated STAT3 signaling has
been implicated in gastric carcinogenesis (11,12).
Immunohistochemical staining of STAT3 in gastric adenocarcinoma
tissues demonstrated that STAT3 serves as a predictor of poor
prognosis and is associated with lymph node metastasis (13,14).
Disruption of STAT3 leads to a decrease in gastric cancer cell
survival through a reduction in the expression of the
anti-apoptotic molecule, survivin (15). Furthermore, inhibition of STAT3 has
been reported to increase apoptosis in gastric cancer cells treated
with cisplatin (16). Notably, the
expression levels of STAT3 downstream targets, including B cell
lymphoma-2 (Bcl-2) and c-Myc, were reported to be down-regulated,
while Bax and p53 levels were increased, elevating the sensitivity
of gastric cancer cells to apoptosis (16).
As YB-1 and JAK/STAT signaling are known to enhance
chemoresistance in gastric cancer, the present study aimed to
determine whether they interact with each other to promote
chemoresistance. The results of the current study suggested the
involvement of YB-1 and STAT3 in the chemoresistance of gastric
cancer NUGC3 cells, whereby the expression of multi-drug
resistance-associated proteins, ABBC3 and ABCC2, were
transcriptionally regulated by YB-1 and STAT3, respectively.
Notably, the inhibition of YB-1 and STAT3 activities were
demonstrated to exhibit a synergistic effect on suppressing
chemoresistance and cell invasion, suggesting that the combination
of YB-1 antagonists with small molecule inhibitors of STAT3 may
serve as an effective therapy for chemoresistance in gastric
cancer.
Methods and materials
Cell culture
The poorly differentiated gastric adenocarcinoma
cell line NUGC3 (American Type Culture Collection, Manassas, VA,
USA) was routinely cultured in RPMI-1640 supplemented with 10%
fetal bovine serum (FBS) (both form HyClone; GE Healthcare Life
Sciences, Logan, UT, USA). When the cells were 80-90% confluent,
subculturing of the cells were performed.
Short interfering RNA (siRNA/si)
transfection
NUGC3 cells were plated in 6-well plates at a
density of 2×105 cells/well. YBX1, ABCC2
and ABBC3 ON-TARGETplus SMARTpool siRNAs, as well as
non-targeting pool siRNA were obtained from GE Healthcare
Dharmacon, Inc. (Lafayette, CO, USA), while siRNA for STAT3
was obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
Table I list the sequences of
siRNAs used in the present study. NUGC3 cells were then transfected
with 20 nM of siNegative, siYB-1, siSTAT3, siABCC2 or siABCC3 using
DharmaFECT1 Transfection reagent (GE Healthcare Dharmacon, Inc.).
Following an overnight incubation with siRNAs, culture medium was
replaced prior to further incubation. The transfected cells were
then collected at 48 h post transfection for RNA isolation and 72 h
post transfection for protein isolation. For double knockdown of
STAT3 and YB-1, each siRNA was used at a final
concentration of 20 nM.
 | Table IList of siRNA sequences used. |
Table I
List of siRNA sequences used.
| siRNA | Target sequence
(5′-3′) |
|---|
| NT siRNA | |
| D-001810-10 |
UGGUUUACAUGUCGACUAA |
| D-001810-11 |
UGGUUUACAUGUUGUGUGA |
| D-001810-12 |
UGGUUUACAUGUUUUCUGA |
| D-001810-13 |
UGGUUUACAUGUUUUCCUA |
| YB-1
siRNA | |
| J-010213-06 |
CUGAGUAAAUGCCGGCUUA |
| J-010213-07 |
CGACGCAGACGCCCAGAAA |
| J-010213-08 |
GUAAGGAACGGAUAUGGUU |
| J-010213-09 |
GCGGAGGCAGCAAAUGUUA |
| ABCC2
siRNA | |
| J-004225-05 |
GGAUGAAUCUCGACCCUUU |
| J-004225-06 |
GUAUCAGGUUUGCCAGUUA |
| J-004225-07 |
GGUCAAGUGUUCUACAGAU |
| J-004225-08 |
GAACCUGACUGUCUUCUUU |
| ABCC3
siRNA | |
| J-007312-05 |
GGACAAAGGAGUAGUAGCU |
| J-007312-06 |
GCACACCGGCUUAACACUA |
| J-007312-07 |
CCACCCUGCUGAUACAGUA |
| J-007312-08 |
ACAAUGAUCCAGUCACCUA |
RT-qPCR
For RT-qPCR, 48 h following siRNA transfection,
total RNA was extracted using the RNeasy Mini kit (Qiagen GmbH,
Hilden, Germany). A total of 1 µg of total RNA was then used
to synthesize first strand cDNA with the SuperScript III
First-Strand Synthesis system (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) as described previously
(17). The HT7900 Fast Real-Time
PCR system was used to examine alterations in gene expression using
Fast SYBR-Green Master mix (both from Applied Biosystems; Thermo
Fisher Scientific, Inc.). For the housekeeping gene, GAPDH was used
for normalization. The thermocycling conditions used were as
follows: initial activation at 95°C for 40 sec; and then 40 cycles
consisting of melting at 95°C for 1 sec and annealing/extension at
60°C for 20 sec. The 2−∆∆Cq method (18) was utilized to express the changes
in gene expression and each sample was run in triplicate. Table II contains the list of primers
used.
 | Table IIList of primers used for RT-qPCR. |
Table II
List of primers used for RT-qPCR.
| Gene | Forward primer
sequence (5′→3′) | Reverse primer
sequence (5′→3′) |
|---|
| YB-1 |
AAGTGATGGAGGGTGCTGAC |
TTCTTCATTGCCGTCCTCTC |
| STAT3 |
ACCAGCAGTATAGCCGCTTC |
GCCACAATCCGGGCAATCT |
| GAPDH |
GAAGGTGAAGGTCGGAGTCAACG |
TGCCATGGGTGGAATCATATTGG |
| ABCC1 |
ATGCTCACTTTCTGGCTGGTA |
AGCGATCTGAGAAACAGGACA |
| ABCC2 |
TATCCCACGAAGTGACAGAGG |
ATGGTCGTCTGAATGAGGTTG |
| ABCC3 |
GGACTTCCAGTGCTCAGAGG |
TGGATGAGGTTGTCAGTCTCC |
| ABCC4 |
TGTTCTTCTGGTGGCTCAATC |
AGAACCCTTGCAACTCCTCTC |
| ABCC5 |
CATCCGGACTACTTCCAAACA |
CAGAGACCACACGTCTTCCAT |
| ABCG2 |
AACTTCTGCCCAGGACTCAAT |
CAGGTAGGCAATTGTGAGGAA |
Western blot analysis
Total protein extraction and protein measurement
were performed as previously described (17). Radioimmunoprecipitation lysis
buffer (Pierce; Thermo Fisher Scientific, Inc.) was used to lyse
the cells. Following Bradford protein assay quantification of the
isolated proteins, 30 µg of proteins were separated using a
8 or 10% SDS-PAGE gel, then transferred onto a polyvinyl difluoride
membrane with a semi-dry system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Next, membranes were incubated with 5% bovine
serum albumin (BSA) (Santa Cruz Biotechnology, Inc.), at room
temperature for 1 h, followed by incubation at 4°C overnight with
the following primary antibodies: rabbit poly-clonal anti-YB-1
[1:1,000; RIKEN, Wako-shi, Japan; (17,19),
the antibody was obtained from Dr Ken Matsumoto, who is stationed
at RIKEN, Wakoshi.]; rabbit polyclonal anti-STAT3 (1:2,000; cat.
no. 12640); rabbit polyclonal anti-phospho-STAT3 (Ser727) (1:1,000;
cat. no. 9134); rabbit polyclonal anti-phospho-STAT3 (Tyr705)
(1:1,000; cat. no. 9145) (all from Cell Signaling Technology, Inc.,
Danvers, MA, USA); and mouse monoclonal anti-β-actin (1:6,000)
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany; cat. no. A2228).
Subsequent incubation of the membranes was performed with
horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000;
Sigma-Aldrich, Merck KgaA; cat. no. A0545) or goat anti-mouse IgG
(1:6,000; Sigma-Aldrich, Merck KgaA; cat. no. A9044) secondary
antibodies at room temperature for 1 h. Proteins were identified
with SuperSignal West Pico Chemiluminescent ECL substrate (Pierce;
Thermo Fisher Scientific, Inc.) and quantified by GS-800 optical
densitometry with Quantity-One Image Analysis software version 4.62
(both from Bio-Rad Laboratories, Inc.).
Cell migration and invasion assays
Migration assays were performed with 6.5-mm
polycarbonate membrane Transwell inserts (Corning Incorporated,
Corning, NY, USA), while BD BioCoat™ Matrigel™ Invasion chambers
(BD Biosciences, Franklin Lakes, NJ, USA) were used for the
invasion assays. A total of 8×104 NUGC3 cells were
seeded in 200 µl of RPM1-1640 serum-free media in the upper
part of the Transwell migration or invasion inserts, 48 h post
siRNA transfection. Subsequently, 600 µl of RPMI-1640 with
20% FBS was placed in the lower chamber of each well, which acted
as a chemoattractant for the cells. The cells were then allowed to
migrate or invade for 20 or 48 h, respectively, prior to fixation
with absolute methanol for 3 min and stained with 0.5% (w/v)
crystal violet in 20% methanol for 30 min at room temperature.
Inserts were then washed with distilled water. Next, the upper
membrane of the inserts were wiped with wet cotton swabs to remove
cells that did not migrate or invade. To visualize migrated or
invaded NUGC3 cells, images of five different fields were captured
with a Nikon SMZ 1500 stereo microscope (Nikon Corporation, Tokyo,
Japan) at ×100 magnification and stained cells were counted. The
assay was performed in triplicates.
Determination of inhibitory concentration
(IC50) of chemotherapy drugs
NUGC3 cells were grown at 4×104
cells/well in a 24-well plate, and transfected with siNegative or
siYB-1 after 24 h post seeding using the aforementioned protocol.
Then, following transfection for 48 h, treatment with varying
concentrations of doxorubicin hydrochloride (0, 0.3125, 0.625,
1.25, 2.5 and 5 µM) or epirubicin hydrochloride (0, 0.3125,
0.625, 1.25, 2.5, 10 and 20 µM) (both from Sigma-Aldrich;
Merck KGaA) was performed. The corresponding percentages of cell
viability at different concentrations of drugs were determined by
MTS assay, after 24 h incubation. Subsequently, 600 µl of
MTS mixture (diluted 1:5 in RPMI-1640 with 10% FBS; Promega
Corporation, Madison, WI, USA) were added to each of the wells. A
negative control set of blank wells containing only MTS solution
with no cells was also prepared. Incubation of cells was performed
in the dark at 37°C for 4 h. CellTiter 96® AQueous One
Solution Cell Proliferation assay reagent (Promega Corporation) was
added to cells to measure cell viability. A wavelength of 490 nm
was used for measuring of the absorbance with an ELISA plate
reader. The IC50 value of each drug was then determined
by plotting the survival rate after 24 h of drug treatment against
varying drug concentrations.
Transmission electron microscopy
(TEM)
The effect of doxorubicin treatment in NUGC3 cells
was evaluated using TEM. NUGC3 cells were treated with
IC50 of doxorubicin for 48 h. Cells in the supernatant
were obtained by centrifugation at 1,000 × g for 5 min at room
temperature and adherent cells were detached and then collected by
centrifuging at 300 × g for 5 min at room temperature. Then, 2.5%
glutaraldehyde was used to fix the cell pellets for 1 h at room
temperature, prior to washing with 1X PBS thrice and post-fixation
with 1% osmium tetroxide (Agar Scientific Ltd., Stansted, UK) for 1
h at room temperature. Following dehydration with increasing
concentrations of ethanol, the pellet was embedded in araldite. The
fixation in araldite was performed at 40°C for 45 min, then 50°C
for 30 min and finally 55°C or 30 min. The samples were sectioned
and mounted on a copper grid coated with formvar. Sections of the
samples were stained with uranyl acetate for 10 min and lead
citrate for 8 min, both at room temperature and finally viewed
under a Philips CM120 BioTwin TEM (FEI; Thermo Fisher Scientific,
Inc.) at ×2,800 or ×4,400 magnifications.
Immunofluorescence staining
NUGC3 cells were grown on a Lab-Tek 4-Chambered
coverglass (Nalge Nunc International; Thermo Fisher Scientific,
Inc.) until 60-70% confluency was achieved. For cell fixation,
cells were incubated with 4% para-formaldehyde for 20 min at room
temperature. Subsequently, 0.05% Tween-20/PBS was used for washing
twice, followed by permeabilisation with 0.2% Triton-X/PBS for 5
min and blocking with 1% (w/v) BSA at room temperature for 30 min.
The YB-1 primary antibody (1:250) (17) was diluted in 1% BSA (w/v) 1X PBS
and incubated at 4°C overnight. Washing was performed thrice with
0.05% Tween-20/PBS. Next, cells were incubated with goat
anti-rabbit IgG Alexa Fluor 594 secondary antibody (1:400; cat. no.
A11012; Thermo Fisher Scientific, Inc.) at room temperature in the
dark for 1 h. The cover glass was removed following washing thrice
again with 0.05% Tween-20/PBS and mounted on a coverslip with
VECTASHIELD fluorescent mounting medium containing DAPI (Vector
Laboratories., Burlingame, CA, USA), which counterstains the
nucleus. Subsequently, an Olympus Fluoview FV1000 Laser Scanning
confocal microscope (Olympus Corporation, Tokyo, Japan) was used to
visualize the stained cells at ×400 or ×600 magnifications.
For siYB-1, siSTAT3 or siNegative transfected-NUGC3
cells, immunofluorescence staining was performed on the cover slips
with seeded cells at 48 h post transfection. The primary antibodies
used were anti-phospho-STAT3 (Tyr705) (1:50), anti-total STAT3
(1:1,000) (both from Cell Signaling Technology, Inc.) and anti-YB-1
(1:250) (17), followed by the use
of goat anti-rabbit IgG Alexa Fluor 594 (1:400) (cat. no. A11012)
or goat anti-rabbit IgG Alexa Fluor 488 (1:400) (both from Thermo
Fisher Scientific, Inc.; cat. no. A11008) secondary antibodies. The
procedures were the same as aforementioned with the exception that
the cells were fixed with 100% methanol for 20 min for
immunofluorescence staining of phospho-STAT3 (Tyr705) and total
STAT3 at room temperature.
Cotreatment of the JAK inhibitor AG490
and doxorubicin hydrochloride, following YB-1 knockdown in NUGC3
cells
Cell were transfected as aforementioned for 48 h and
0 or 50 µM AG490 was used for the 48 h treatment, prior to
exposure to medium with or without IC50 of doxorubicin
hydrochloride for 24 h. Subsequently, an MTS assay was performed to
determine cell viability.
Doxorubicin hydrochloride treatment in
siABCC2 and siABCC3 NUGC3 cells
NUGC3 cells were grown in a 24-well plate at a
density of 4×104 cells/well, followed by treatment with
siABCC2, siABBC3 or siNegative, to a final concentration of 20 nM,
with the aforementioned transfection procedures. At 48 h post
transfection, treatment with doxorubicin hydrochloride for 24 h was
performed, then cell viability was evaluated using an MTS
assay.
Statistical analysis
The GraphPad Prism 5.0 (GraphPad Software, Inc., La
Jolla, CA, USA) statistical package was used. For the means between
two groups, a two tailed Student’s t-test was used; whereas for
comparison between ≥3 groups, one-way analysis of variance was used
followed by Tukey’s post-hoc test. Data are presented as mean ±
standard error of the mean. P<0.05 was considered to indicate a
statistically significant difference. All experiments were repeated
in triplicates.
Results
YB-1 protein translocalizes to the
nucleus upon chemotherapy treatment in NUGC3 cells
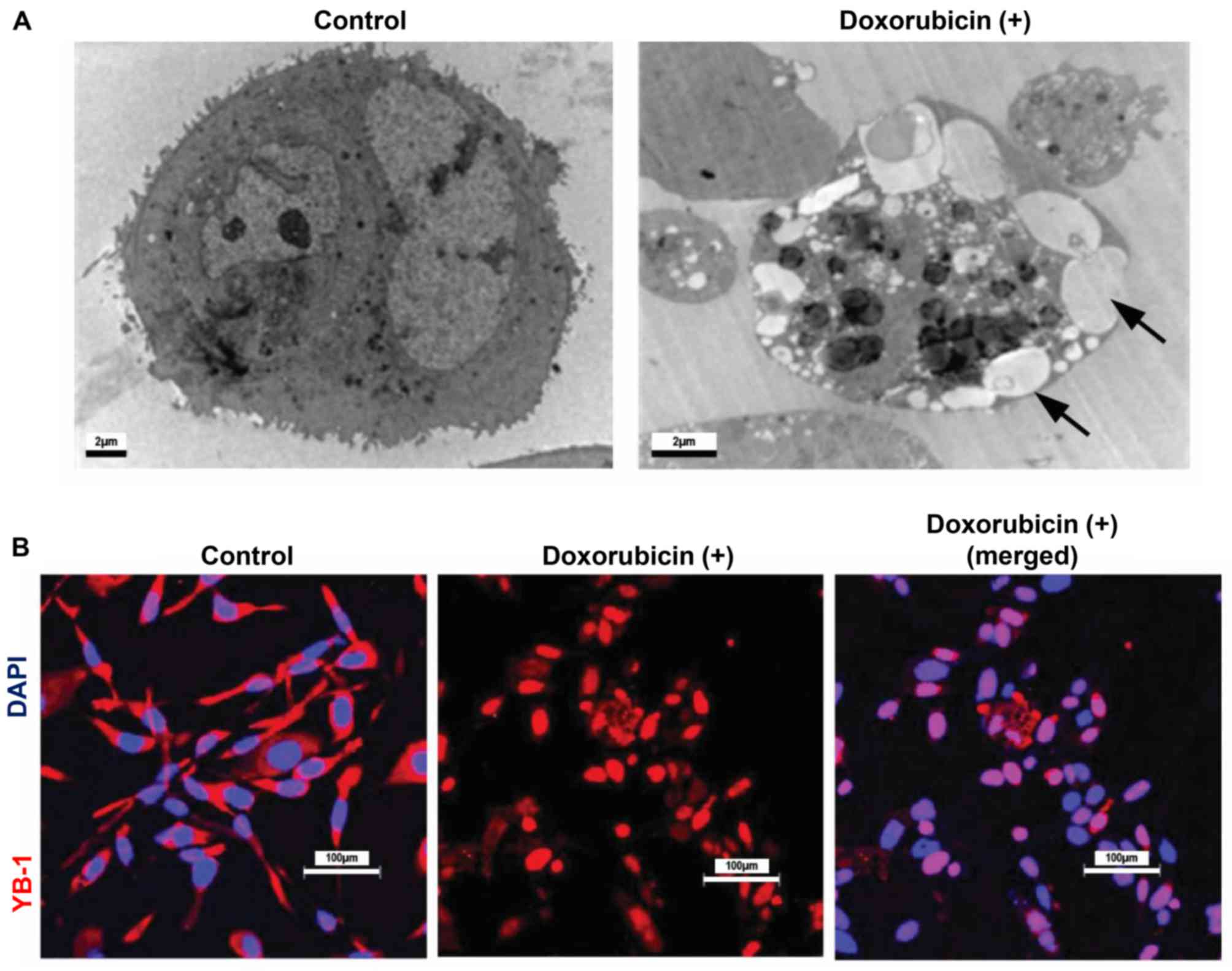
Several reports have suggested that chemotherapeutic
drugs kill cancer cells susceptible to the treatment through
promoting apoptosis, and have implicated nuclear YB-1 in
chemoresistance (19-21). Hence, whether the treatment of
NUGC3 cells with chemotherapy drugs, including doxorubicin
hydrochloride were able to induce cell death was examined. NUGC3
cells treated with doxorubicin hydrochloride at a concentration of
4.65 µM (IC50) (data not shown) for 48 h
exhibited morphological changes, including cytoplasmic blebbing
(Fig. 1A; black arrows),
indicative of apoptosis. Subsequently, whether nuclear
translocation of YB-1 increased upon doxorubicin hydrochloride
treatment was investigated. Notably, a marked increase in the
levels of nuclear YB-1 was detected upon doxorubicin hydrochloride
compared with the control, suggesting the potential role of YB-1 in
chemoresistance (Fig. 1B).
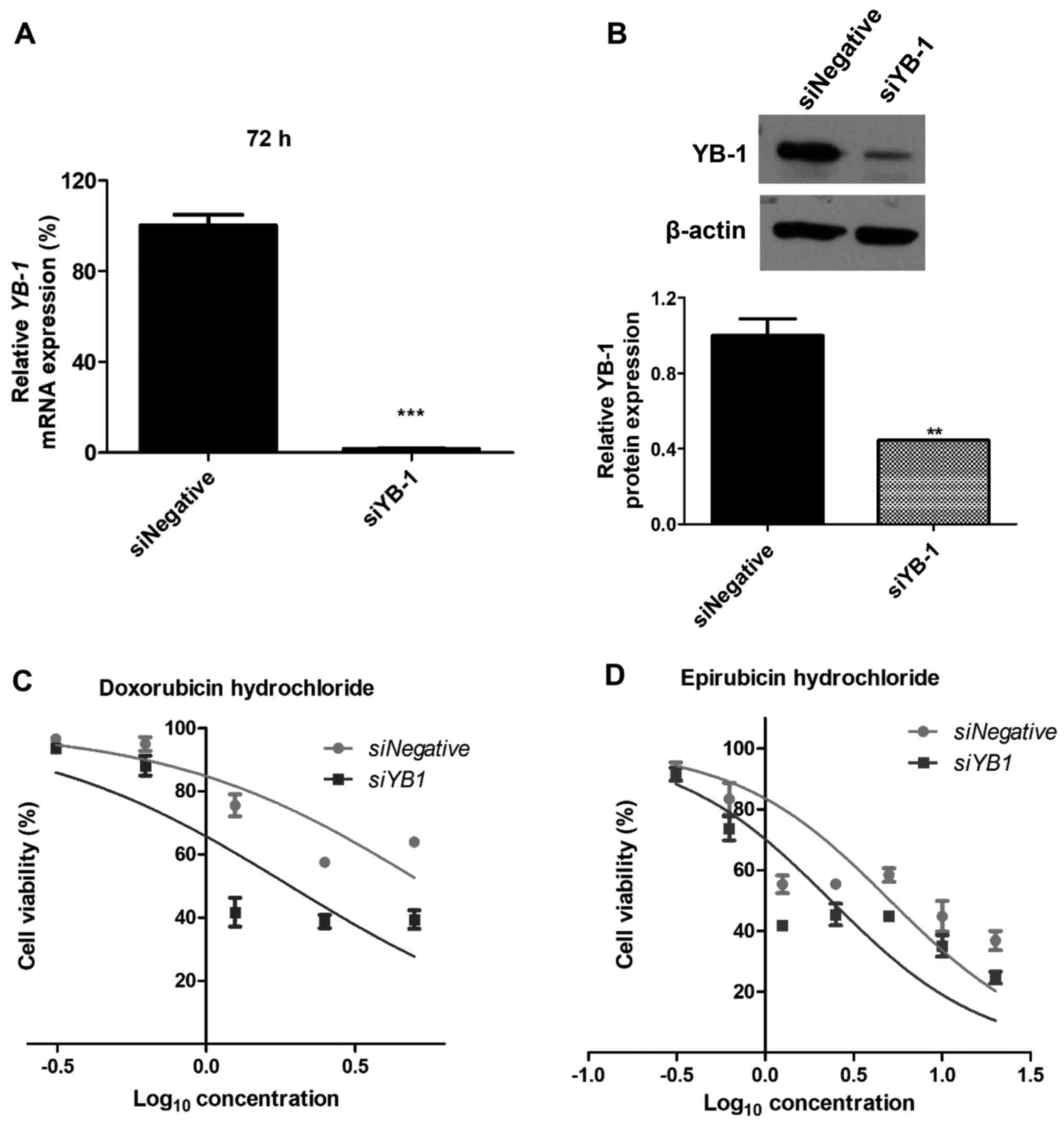
Knockdown of YB-1 decreases
chemoresistance of NUGC3 cells
To investigate whether YB-1 functions in
chemoresistance of gastric cancer NUGC3 cells, the inhibitory
effects of YB-1 on cell viability, following exposure to
chemotherapy drugs were evaluated. The siRNA-mediated silencing
efficiency of YB-1 at the mRNA and protein levels was ~99 and 56%,
respectively (Fig. 2A and B).
Notably, inhibition of YB-1 induced a decrease in chemoresistance
of the cells to doxorubicin hydrochloride and epirubicin
hydrochloride, two of the commonly used chemotherapy drugs for
cancer treatment. While the IC50 of doxorubicin
hydrochloride in NUGC3 cells transfected with siYB-1 was 2.8
µM, the IC50 of doxorubicin hydrochloride in
control cells was 7.2 µM (Fig.
2C). Similarly, the IC50 of epirubicin hydrochloride
in cells transfected with siYB-1 was decreased compared with
control cells (2.1 vs. 6.3 µM); however, this was not
statistically significant (Fig.
2D).
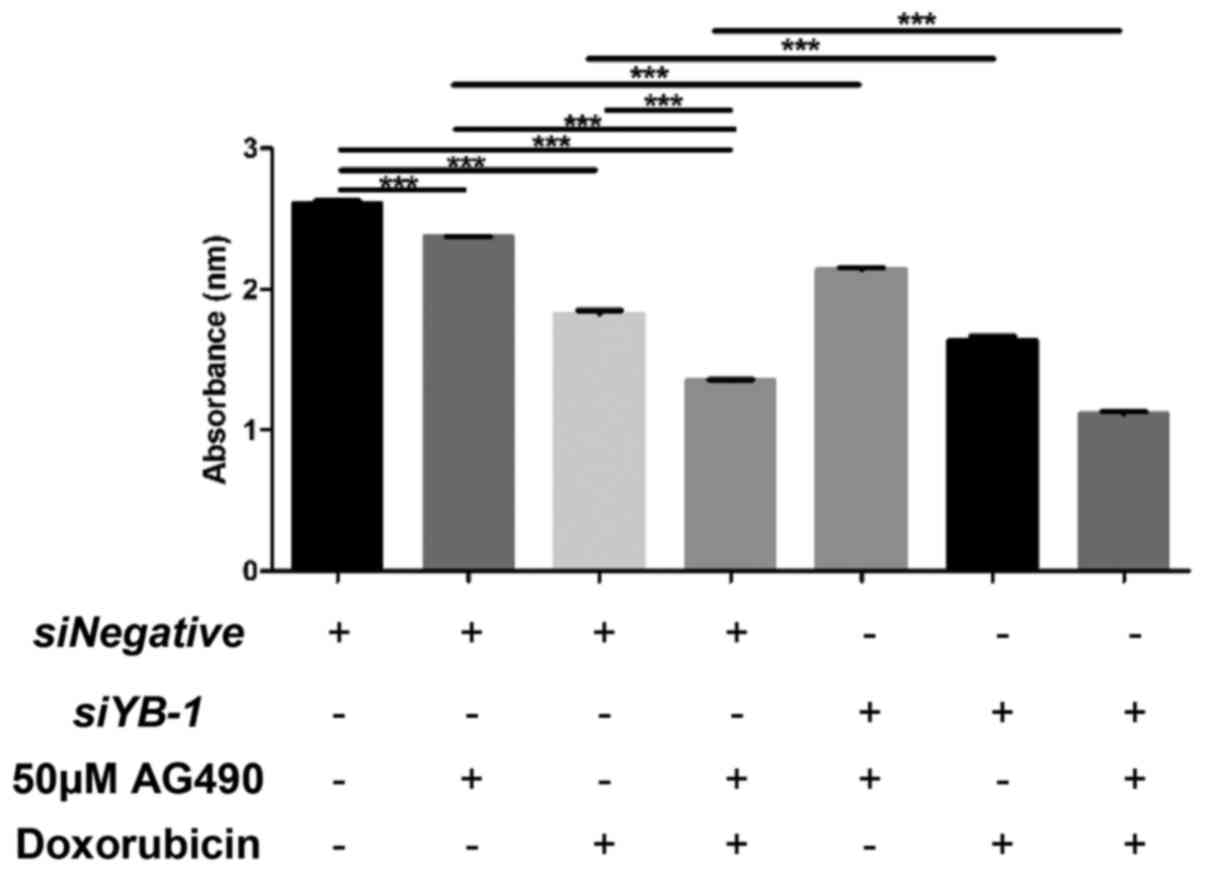
Synergistic inhibitory effects of YB-1
and JAK/STAT signaling on chemoresistance
The JAK/STAT signaling pathway has also been
implicated in the chemoresistance of cancer cells (10,22,23).
For example, STAT3-dependent myeloid cell leukemia-1 (Mcl-1)
expression was reported to affect chemo-sensitivity in melanoma,
whereby Mcl-1 confers survival advantage against B-Raf
proto-oncogene serine/threonine kinase inhibitors (24). Therefore, the present study
examined the potential interaction between JAK/STAT signaling and
YB-1 in the chemoresistance of NUGC3 cells. The viability of
siNegative-transfected control cells co-treated with the JAK2
inhibitor AG490 and doxorubicin hydrochloride compared with that of
the control cells treated with AG490 or doxorubicin hydrochloride
alone was examined. As shown in Fig.
3, AG490 or doxorubicin hydrochloride treatment alone reduced
the viability of cells compared with the untreated control group.
Notably, co-treatment of AG490 and doxorubicin hydrochloride
further enhanced the decreased viability induced by AG490 or
doxorubicin hydrochloride. The effects of co-treatment with AG490
and doxorubicin hydrochloride on cell viability were examined in
YB-1 knockdown cells. Cell death in YB-1 knockdown
cells treated with AG490, doxorubicin hydrochloride, or AG490 and
doxorubicin hydrochloride, was further enhanced compared with that
in siNegative-transfected control cells treated with AG490,
doxorubicin hydrochloride, or AG490 and doxorubicin hydrochloride,
respectively. These suggest that JAK/STAT signaling and YB-1
function synergistically in promoting chemoresistance of gastric
cancer cells to doxorubicin hydrochloride.
YB-1 and STAT3 regulate the expression of
different ABC transporters
To obtain insight into the possible interaction
between YB-1 and JAK/STAT signaling in chemoresistance, the effect
of YB-1 knockdown on STAT3 expression and vice versa was
evaluated. However, no significant changes were observed in total
STAT3 and phosphorylated STAT3 (Tyr705 and Ser727) levels upon
YB-1 knockdown in NUGC3 cells (Fig. 4A, upper panel). Furthermore,
immunofluorescence staining of siYB-1-transfected NUGC3 cells
revealed no significant changes in the subcellular localization of
phosphorylated STAT3 (Tyr705) and total STAT3 in the nucleus
(Fig. 4A, lower panel). Similarly,
STAT3 knockdown did not significantly affect YB-1 expression
(Fig. 4B, upper panel) or the
subcellular localization of YB-1 in the nucleus (Fig. 4B, lower panel).
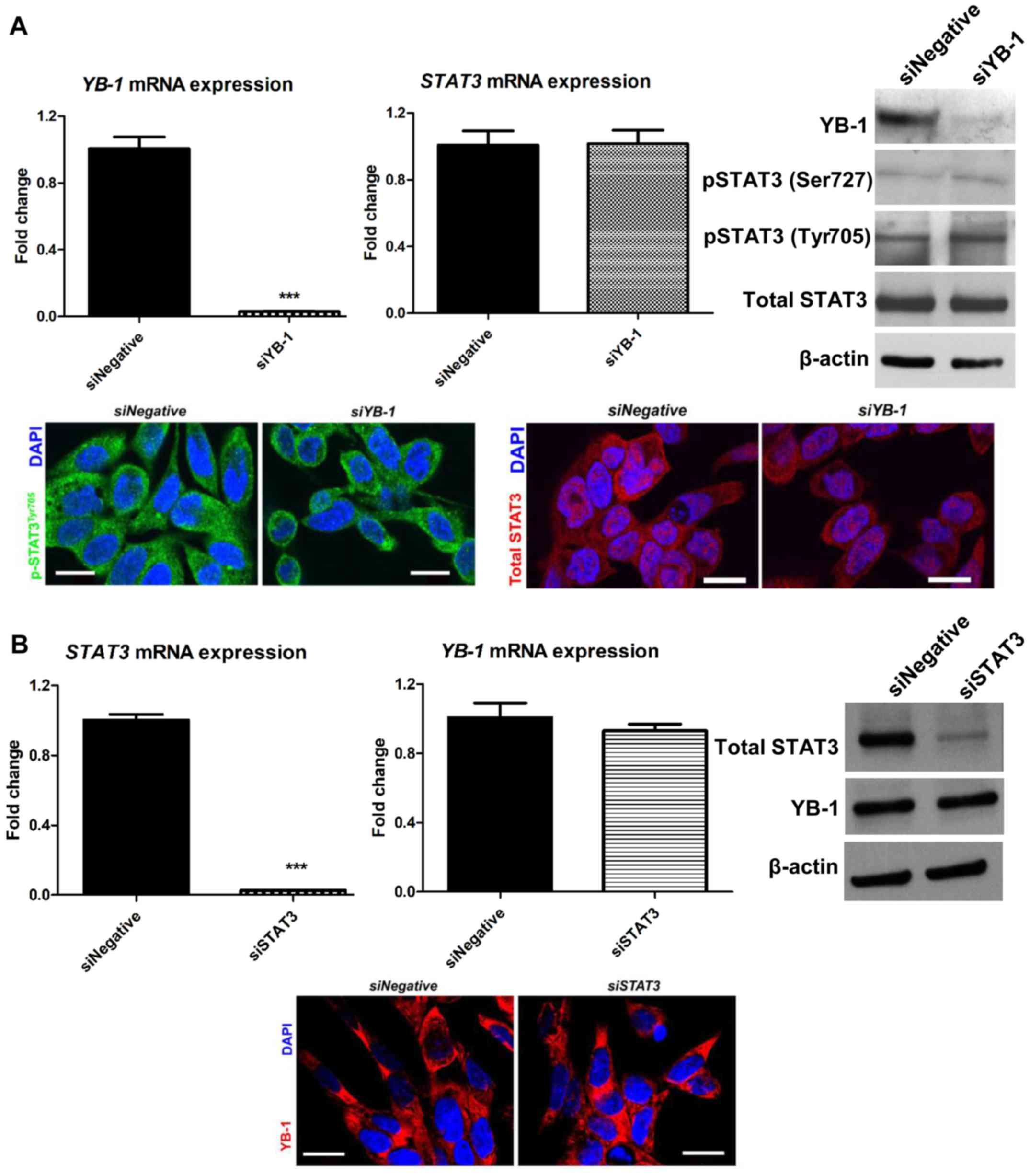 | Figure 4Expression of YB-1 and STAT3 is not
mutually regulated. (A) YB-1 and STAT3 mRNA levels
upon YB-1 knockdown (upper left and middle panels). No
significant changes in STAT3 mRNA were detected when YB-1
was inhibited in NUGC3 cells. YB-1 knockdown also failed to
affect total and phosphorylated STAT3 levels (upper right panel).
Similarly, immunofluorescence staining of phospho-STAT3 (Tyr705)
and total STAT3 demonstrated no changes in their subcellular
localization upon YB-1 knockdown (lower panel). (B)
STAT3 knockdown resulted in a significant decrease in
STAT3 mRNA levels, but not YB-1 mRNA levels (upper
left and middle panels). Consistently, STAT3 inhibition did not
affect YB-1 expression (upper right panel). Immunofluorescence
staining revealed no changes in the subcellular localization of
YB-1 upon STAT3 knockdown (lower panel). Data are presented
as the mean ± standard error of the mean from three independent
experiments, ***P<0.001. Scale bar, 20 µm.
YB-1, Y-box binding protein-1; si, small interfering RNA; JAK,
Janus kinase; STAT, signal transducer and activator of
transcription; p, phosphorylated. |
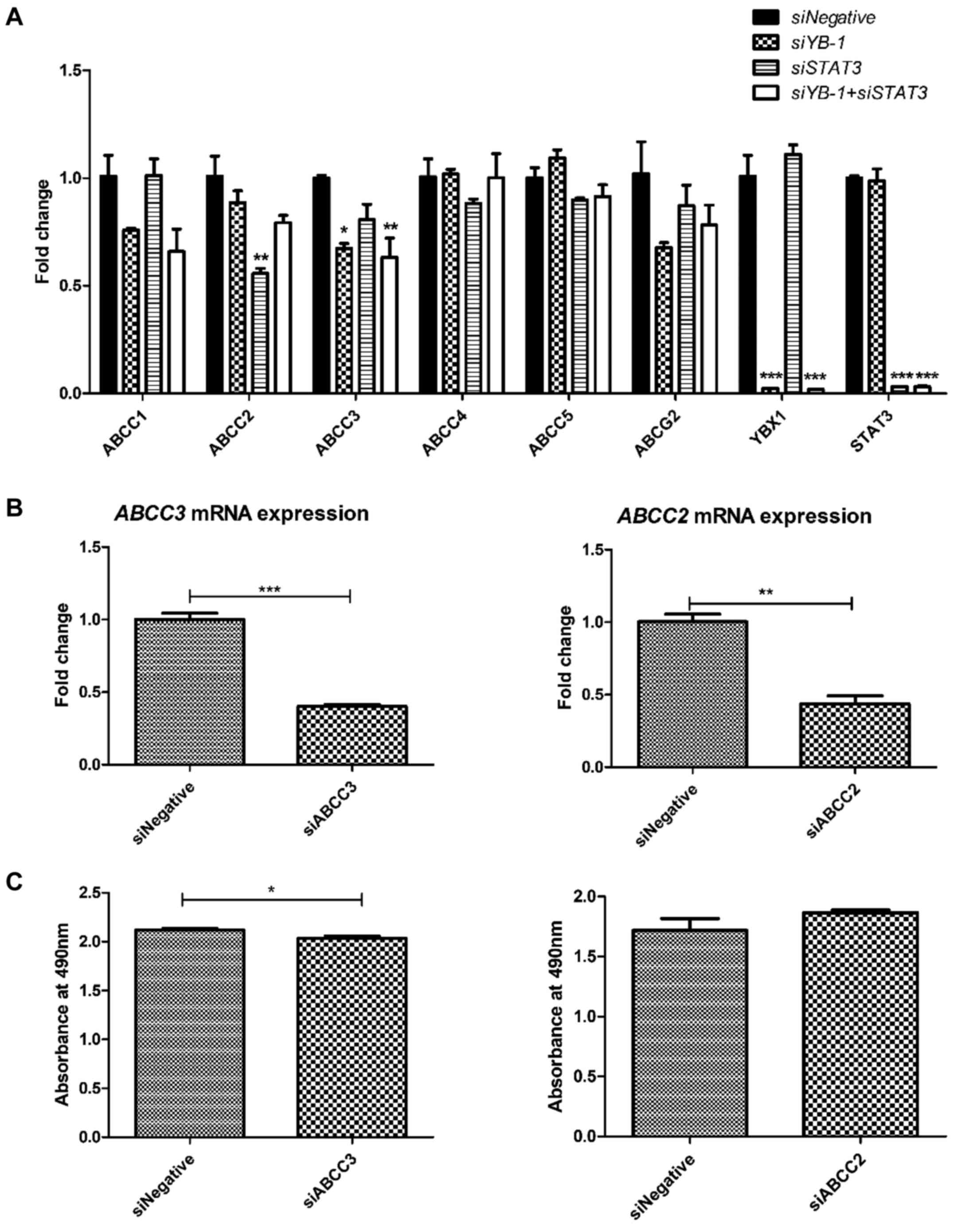
As ABC transporters are implicated in multidrug
resistance in various cancer types (25-28),
the present study aimed to evaluate the regulatory effect of YB-1
and STAT3 on ABC transporters, as the two proteins act as
transcriptional factors. Upon YB-1 knockdown in NUGC3 cells,
a signifi-cant reduction in the ABBC3 transporter gene was
observed out of the six different ABC transporters (ABCC1,
ABCC2, ABCC3, ABCC4, ABCC5 and
ABCG2) evaluated (Fig. 5A).
However, STAT3 knockdown only led to a significant decrease
in ABCC2 expression (Fig.
5A). These results suggest that YB-1 and STAT3 act
independently in chemoresistance by regulating the expression of
different ABC transporters, which serve essential roles in
multidrug resistance in various cancer types. The possible
synergistic effects of YB-1 and STAT3 on ABC transporter expression
were also examined. However, no significant synergistic effects
were observed, confirming that YB-1 and STAT3 functions
independently in regulating the expression of ABC transporters.
The role of ABCC2 and ABCC3 on the viability of
NUGC3 cells treated with doxorubicin hydrochloride was then
examined. The knockdown efficiencies of siABCC3 and siABCC2 were
59.9% (Fig. 5B, left panel) and
56.5% (Fig. 5B, right panel),
respectively. ABCC3 knockdown in NUGC3 cells led to
decreased cell viability upon doxorubicin hydrochloride treatment,
as compared with the negative control (Fig. 5C, left panel). However, no
significant reduction in the viability of ABCC2 knockdown
cells was observed upon treatment with doxorubicin hydrochloride
(Fig. 5C, right panel).
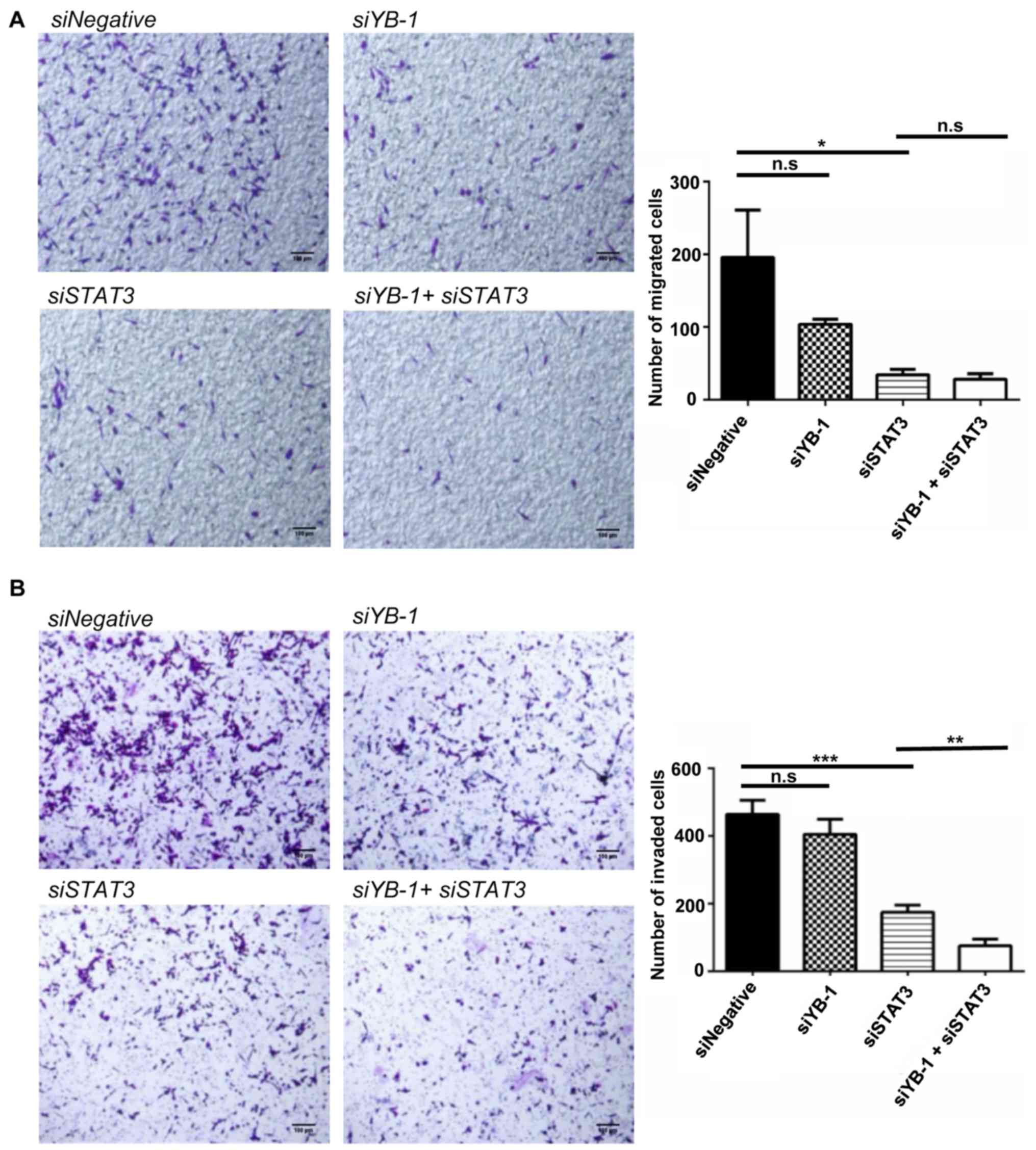
YB-1 and STAT3 exhibit synergistic
effects on facilitating cell invasion in NUGC3 cells
As previously mentioned, YB-1 and STAT3 exhibited a
synergistic effect in mediating chemoresistance of NUGC3 cells
(Fig. 3). Hence, the effects of
their combination on cell migration and invasion were investigated.
A double knockdown of YB-1 and STAT3 was performed in
NUGC3 cells. STAT3 knockdown alone significantly decreased
cell migration as compared with the control. YB-1 knockdown
alone also led to a decline in cell migration, albeit statistically
insignificant. The double knockdown of YB-1 and STAT3
failed to further enhance the decrease caused by STAT3
knockdown alone, suggesting that there was no synergistic effect on
cell migration (Fig. 6A). In
contrast, the double knockdown of YB-1 and STAT3 in
NUGC3 cells further inhibited cell invasion caused by YB-1
or STAT3 knockdown alone (Fig.
6B), suggesting the synergistic role of YB-1 and STAT3 in cell
invasion.
Discussion
YB-1 is a pleotropic protein which is upregulated in
numerous human malignancies, including gastric cancer (29), nasopharyrngeal carcinoma (19) and breast cancer (17). YB-1 has been reported to serve a
vital role in MDR in patients with cancer (4,30).
Under normal conditions, YB-1 is predominantly expressed in the
cytoplasm (31), but translocates
to the nucleus upon exposure to environmental stress, including UV
light, drug treatment and hyperthemia (4,31).
Furthermore, the YB-1 nuclear localization is associated with
MDR1 gene expression, whose product mediates chemoresistance
in various cancer types, including breast, synovial sarcoma and
osteosarcoma cancer (3). In line
with the potential role of YB-1 in chemo-resistance in various
cancer types, including nasopharyngeal cancer (19), breast cancer (21), melanoma (32) and ovarian cancer (33), the present study demonstrated the
cytoplasmic to nuclear translocation of YB-1 in the NUGC3 gastric
cancer cell line upon doxorubicin hydrochloride treatment. In
support of this, YB-1 reduction led to a significant
decrease in the chemoresistance of NUGC3 cells to doxorubicin
hydrochloride and epirubicin hydrochloride.
Notably, constitutively-active JAK/STAT signaling
has also been demonstrated to be associated with gastric cancer
development and progression (34).
In particular, it has been reported that STAT3 is phosphorylated by
JAK, which subsequently translocates to the nucleus, and acts as a
transcription factor to modulate the expression of a wide variety
of genes that participate in proliferation, migration as well as
invasion in gastric cancer cells (35,36).
STAT3 also promotes anti-apoptotic molecule expression, including
Bcl-2 and Mcl-1, thereby conferring pro-survival signals in gastric
cancer cells (16). Furthermore,
it serves as a useful biomarker in gastric cancer prognosis
(37), with a high expression of
STAT3 identified in gastric cancer (15).
YB-1 and JAK/STAT signaling have been implicated in
chemoresistance (7,20). Hence, a possible synergistic effect
of YB-1 and JAK/STAT signaling on chemoresistance was examined
using siYB-1 and the JAK2 inhibitor AG490 in gastric cancer NUGC3
cells. Notably, an enhanced reduction in the viability of NUGC3
cells co-treated with siYB-1 and AG490 in the presence of
doxorubicin hydrochloride was observed, as compared with the
viability in cells treated with siYB-1 or AG490 alone upon
doxorubicin hydrochloride treatment. Thus, these results suggest
that YB-1 and JAK/STAT signaling are involved in chemoresistance,
and may act synergistically to promote chemoresistance of gastric
cancer. To obtain more insight into the mechanism underlying their
synergistic effects on chemoresistance, the inhibitory effect of
YB-1 on STAT3 levels was ascertained. However, no alterations in
total STAT3 and phosphorylated STAT3 levels upon YB-1
knockdown were detected. Similarly, STAT3 knockdown did not
lead to any striking changes in YB-1 expression. YB-1 inhibition
was shown to decrease phosphorylated STAT3 (Ser727) levels, but not
phosphorylated STAT3 (Tyr705) or total STAT3 levels in breast
cancer cells (38). This may
suggest the cell type-dependent effect of YB-1 on STAT3 activity.
Nonetheless, gene expression analysis of MDR-associated genes and
ABC transporters upon siYB-1 or siSTAT3 revealed differential gene
regulations, whereby YB-1 regulates ABCC3 expression while
STAT3 regulates ABCC2 expression. In addition, no
synergistic effects were observed for YB-1 and STAT3 on the ABC
transporters, further suggesting that YB-1 and STAT3 regulate the
expression of ABC transporters independently.
Regarding the role of ABCC2 and ABCC3
in chemo-resistance, siRNA mediated knockdown of the two ABC
transporters was then performed. Cell viability was reduced to an
extent in doxorubicin hydrochloride treated siABCC3 NUGC3 cells,
but not in siABCC2 cells. The possible explanations for these
observations may be i) the low knockdown efficiency of ABCC2; or
ii) ABCC2 is not the sole target of STAT3 in chemoresistance. The
ABCC2 siRNAs used in the present experiment consisted of four
individual siRNAs that were pooled together, which would minimise
the off-target effects of transfection. However, the knockdown
efficiency was relatively low and therefore no significant
difference in cell viability was observed when compared with the
control cells. Alternatively, STAT3 may regulate chemoresistance
via other targets, besides ABCC2. For example, STAT3 is also
involved in the regulation of anti-apoptotic Bcl-2 and Mcl-1
proteins, which may confer chemoresistance in gastric cancer
(16). Future experiments should
investigate the role of ABCC2 and ABCC3 in mediating
chemoresistance by determining whether ABCC2 and ABCC3
overexpression reverse the effects of YB-1 and STAT3 knockdown on
NUGC3 cells. Furthermore, determining how YB-1 and STAT3 regulate
the expression of ABCC3 and ABCC2, respectively, may be of
importance. Using the more advanced clustered regularly
inter-spaced short palindromic repeat/CRISPR-associated protein 9
system to knockout ABCC2 or ABCC3 gene expression may be performed
to determine the effects on chemoresistance, as the knockdown
efficiencies using siABCC2 and siABCC3 are relatively low.
The results demonstrated that YB-1 and STAT3 do not
mutually regulate the expression of one another, but the two
proteins act synergistically in the chemoresistance of gastric
cancer cells, suggesting that they may also act together in other
biological processes. A previous study reported that STAT3
expression is associated with spread to the lymph nodes in gastric
cancer tissues, thus enhancing gastric cancer progression (39). Consistently, the knockdown of
STAT3 led to a significant reduction in cell migratory and
invasive properties. However, YB-1 has been implicated in the
migratory property of NUGC3 cells, but not cell invasion and
epithelial-to-mesenchymal transition, complex processes essential
to the initial metastatic cascade (40). No significant inhibitory effects of
YB-1 were detected with regards to the migratory and invasive
properties of NUGC3 cells. Notably, STAT3 knockdown-mediated
decrease in cell invasion was further enhanced upon YB-1
knockdown. It was reported that YB-1 prevents apoptosis in
Her2-overexpressing breast cancer cells in vitro and
promotes tumour growth in mice via the mammalian target of
rapamycin/STAT3 signaling pathway (41), indicating a possible cross-talk
between YB-1 and STAT3 signaling underlying the pathogenesis of
breast cancer. Future experiments to determine the effects of YB-1
and STAT3 downregulation on gastric cancer in vivo may aid
in determining the associated signaling pathways of these two
molecules.
The present results indicate that YB-1 and STAT3 act
synergistically in chemoresistance and cell invasion in NUGC3
gastric cancer cells, thus providing the potential for the
development of novel therapeutic interventions targeting these two
molecules in gastric cancer.
Funding
The present study was supported by the Ministry of
Education grants (grant no. MOE2013-T2-1-129 to BHB) and (grant no.
MOE T1-2016 Sep-13 to QH). JPL was a recipient of the Ong Hin Tiang
Scholarship in Cancer Research.
Availability of data and materials
All data generated or analysed during the present
study are included in this published article.
Authors’ contributions
PJC and TTG designed and performed the experiments.
JPL wrote the manuscript and performed the experiments. PK
performed the experiments. QH, BHB and GHB conceived the experiment
designs and revised the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: Role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Didier DK, Schiffenbauer J, Woulfe SL,
Zacheis M and Schwartz BD: Characterization of the cDNA encoding a
protein binding to the major histocompatibility complex class II Y
box. Proc Natl Acad Sci USA. 85:7322–7326. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eliseeva IA, Kim ER, Guryanov SG,
Ovchinnikov LP and Lyabin DN: Y-box-binding protein 1 (YB-1) and
its functions. Biochemistry (Mosc). 76:1402–1433. 2011. View Article : Google Scholar
|
|
4
|
Bargou RC, Jürchott K, Wagener C, Bergmann
S, Metzner S, Bommert K, Mapara MY, Winzer KJ, Dietel M, Dörken B,
et al: Nuclear localization and increased levels of transcription
factor YB-1 in primary human breast cancers are associated with
intrinsic MDR1 gene expression. Nat Med. 3:447–450. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Janz M, Harbeck N, Dettmar P, Berger U,
Schmidt A, Jürchott K, Schmitt M and Royer HD: Y-box factor YB-1
predicts drug resistance and patient outcome in breast cancer
independent of clinically relevant tumor biologic factors HER2, uPA
and PAI-1. Int J Cancer. 97:278–282. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mo D, Fang H, Niu K, Liu J, Wu M, Li S,
Zhu T, Aleskandarany MA, Arora A, Lobo DN, et al: Human helicase
RECQL4 drives cisplatin resistance in gastric cancer by activating
an AKT-YB1-MDR1 signaling pathway. Cancer Res. 76:3057–3066. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khanna P, Chua PJ, Bay BH and Baeg GH: The
JAK/STAT signaling cascade in gastric carcinoma (Review). Int J
Oncol. 47:1617–1626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kiu H and Nicholson SE: Biology and
significance of the JAK/STAT signalling pathways. Growth Factors.
30:88–106. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Darnell JE Jr, Kerr IM and Stark GR:
Jak-STAT pathways and transcriptional activation in response to
IFNs and other extracellular signaling proteins. Science.
264:1415–1421. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Catlett-Falcone R, Dalton WS and Jove R:
STAT proteins as novel targets for cancer therapy. Signal
transducer an activator of transcription. Curr Opin Oncol.
11:490–496. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Z, Si X, Xu A, Meng X, Gao S, Qi Y,
Zhu L, Li T, Li W and Dong L: Activation of STAT3 in human gastric
cancer cells via interleukin (IL)-6-type cytokine signaling
correlates with clinical implications. PLoS One. 8:e757882013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giraud AS, Menheniott TR and Judd LM:
Targeting STAT3 in gastric cancer. Expert Opin Ther Targets.
16:889–901. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim DY, Cha ST, Ahn DH, Kang HY, Kwon CI,
Ko KH, Hwang SG, Park PW, Rim KS and Hong SP: STAT3 expression in
gastric cancer indicates a poor prognosis. J Gastroenterol Hepatol.
24:646–651. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Deng JY, Sun D, Liu XY, Pan Y and Liang H:
STAT-3 correlates with lymph node metastasis and cell survival in
gastric cancer. World J Gastroenterol. 16:5380–5387. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanda N, Seno H, Konda Y, Marusawa H,
Kanai M, Nakajima T, Kawashima T, Nanakin A, Sawabu T, Uenoyama Y,
et al: STAT3 is constitutively activated and supports cell survival
in association with survivin expression in gastric cancer cells.
Oncogene. 23:4921–4929. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang S, Chen M, Shen Y, Shen W, Guo H,
Gao Q and Zou X: Inhibition of activated Stat3 reverses drug
resistance to chemotherapeutic agents in gastric cancer cells.
Cancer Lett. 315:198–205. 2012. View Article : Google Scholar
|
|
17
|
Lim JP, Shyamasundar S, Gunaratne J,
Scully OJ, Matsumoto K and Bay BH: YBX1 gene silencing inhibits
migratory and invasive potential via CORO1C in breast cancer in
vitro. BMC Cancer. 17:2012017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Tay WL, Yip GW, Tan PH, Matsumoto K, Yeo
R, Ng TP, Kumar SD, Tsujimoto M and Bay BH: Y-Box-binding protein-1
is a promising predictive marker of radioresistance and
chemoradioresistance in nasopharyngeal cancer. Mod Pathol.
22:282–290. 2009. View Article : Google Scholar
|
|
20
|
Kosnopfel C, Sinnberg T and Schittek B:
Y-box binding protein 1 - a prognostic marker and target in tumour
therapy. Eur J Cell Biol. 93:61–70. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang J, Tan PH, Li KB, Matsumoto K,
Tsujimoto M and Bay BH: Y-box binding protein, YB-1, as a marker of
tumor aggressiveness and response to adjuvant chemotherapy in
breast cancer. Int J Oncol. 26:607–613. 2005.PubMed/NCBI
|
|
22
|
Stephanou A, Brar BK, Knight RA and
Latchman DS: Opposing actions of STAT-1 and STAT-3 on the Bcl-2 and
Bcl-x promoters. Cell Death Differ. 7:329–330. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nascimento AS, Peres LL, Fari AVS, Milani
R, Silva RA, da Costa Fernandes CJ, Peppelenbosch MP,
Ferreira-Halder CV and Zambuzzi WF: Phosphoproteome profiling
reveals critical role of JAK-STAT signaling in maintaining
chemoresistance in breast cancer. Oncotarget. 8:114756–114768.
2017. View Article : Google Scholar
|
|
24
|
Becker TM, Boyd SC, Mijatov B,
Gowrishankar K, Snoyman S, Pupo GM, Scolyer RA, Mann GJ, Kefford
RF, Zhang XD, et al: Mutant B-RAF-Mcl-1 survival signaling depends
on the STAT3 transcription factor. Oncogene. 33:1158–1166. 2014.
View Article : Google Scholar
|
|
25
|
Saneja A, Khare V, Alam N, Dubey RD and
Gupta PN: Advances in P-glycoprotein-based approaches for
delivering anticancer drugs: Pharmacokinetic perspective and
clinical relevance. Expert Opin Drug Deliv. 11:121–138. 2014.
View Article : Google Scholar
|
|
26
|
Noguchi K, Katayama K, Mitsuhashi J and
Sugimoto Y: Functions of the breast cancer resistance protein
(BCRP/ABCG2) in chemotherapy. Adv Drug Deliv Rev. 61:26–33. 2009.
View Article : Google Scholar
|
|
27
|
Zhang W, Meng Y, Liu N, Wen X-F and Yang
T: Insights into chemo-resistance of prostate cancer. Int J Biol
Sci. 11:1160–1170. 2015. View Article : Google Scholar :
|
|
28
|
Zhang G, Wang Z, Qian F, Zhao C and Sun C:
Silencing of the ABCC4 gene by RNA interference reverses multidrug
resistance in human gastric cancer. Oncol Rep. 33:1147–1154. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo T, Yu Y, Yip GW, Baeg GH, Thike AA,
Lim TK, Tan PH, Matsumoto K and Bay BH: Y-box binding protein 1 is
correlated with lymph node metastasis in intestinal-type gastric
cancer. Histopathology. 66:491–499. 2015. View Article : Google Scholar
|
|
30
|
Oda Y, Sakamoto A, Shinohara N, Ohga T,
Uchiumi T, Kohno K, Tsuneyoshi M, Kuwano M and Iwamoto Y: Nuclear
expression of YB-1 protein correlates with P-glycoprotein
expression in human osteosarcoma. Clin Cancer Res. 4:2273–2277.
1998.PubMed/NCBI
|
|
31
|
Koike K, Uchiumi T, Ohga T, Toh S, Wada M,
Kohno K and Kuwano M: Nuclear translocation of the Y-box binding
protein by ultraviolet irradiation. FEBS Lett. 417:390–394. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schittek B, Psenner K, Sauer B, Meier F,
Iftner T and Garbe C: The increased expression of Y box-binding
protein 1 in melanoma stimulates proliferation and tumor invasion,
antagonizes apoptosis and enhances chemoresistance. Int J Cancer.
120:2110–2118. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yahata H, Kobayashi H, Kamura T, Amada S,
Hirakawa T, Kohno K, Kuwano M and Nakano H: Increased nuclear
localization of transcription factor YB-1 in acquired
cisplatin-resistant ovarian cancer. J Cancer Res Clin Oncol.
128:621–626. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jackson CB and Giraud AS: STAT3 as a
prognostic marker in human gastric cancer. J Gastroenterol Hepatol.
24:505–507. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signalling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Joo MK, Park JJ and Chun HJ: Recent
updates of precision therapy for gastric cancer: Towards optimal
tailored management. World J Gastroenterol. 22:4638–4650. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gong W, Wang L, Yao JC, Ajani JA, Wei D,
Aldape KD, Xie K, Sawaya R and Huang S: Expression of activated
signal transducer and activator of transcription 3 predicts
expression of vascular endothelial growth factor in and angiogenic
phenotype of human gastric cancer. Clin Cancer Res. 11:1386–1393.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fujii T, Seki N, Namoto-Matsubayashi R,
Takahashi H, Inoue Y, Toh U, Kage M and Shirouzu K: YB-1 prevents
apoptosis via the mTOR/STAT3 pathway in HER-2-overexpressing breast
cancer cells. Future Oncol. 5:153–156. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Deng J, Liang H, Zhang R, Sun D, Pan Y,
Liu Y, Zhang L and Hao X: STAT3 is associated with lymph node
metastasis in gastric cancer. Tumour Biol. 34:2791–2800. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo TT, Yu YN, Yip GW, Matsumoto K and Bay
BH: Silencing the YB-1 gene inhibits cell migration in gastric
cancer in vitro. Anat Rec (Hoboken). 296:891–898. 2013. View Article : Google Scholar
|
|
41
|
Lee C, Dhillon J, Wang MY, Gao Y, Hu K,
Park E, Astanehe A, Hung MC, Eirew P, Eaves CJ, et al: Targeting
YB-1 in HER-2 overexpressing breast cancer cells induces apoptosis
via the mTOR/STAT3 pathway and suppresses tumor growth in mice.
Cancer Res. 68:8661–8666. 2008. View Article : Google Scholar : PubMed/NCBI
|