Introduction
The principal issue in the therapy of glioma is the
recurrence, even following surgical debulking combined with radio-
or chemotherapy, of newly formed solid masses in other distant
areas of the brain. These recurrences are due to glioma cells that
detach from the original tumor mass and disseminate in the brain
parenchyma to establish new tumoral niches. The relatively poor
advancement in glioma therapy in the last decades may be partly due
to the lack of suitable in vitro and in vivo glioma
models. The most reliable in vitro glioma model is currently
provided by glioma stem cells (GSC), a subpopulation of glioma
cells that, when propagated in culture in the absence of serum as
neurospheres or adherent cells plated over laminin, retain their
original genotype and phenotype (1). GSC are characterized by
tumorigenicity, chemoresistance, radioresistance and infiltrative
ability, making them crucial targets for therapeutic
strategies.
Since glioma recurrence primarily involves
mechanisms associated with cell detachment and attachment (2), extracellular matrix (ECM)-binding
proteins expressed on cell membranes, such as integrins, have been
intensively exploited as potential targets to counteract glioma
malignancy.
Certain RGD-binding integrins, particularly αvβ3,
αvβ5 and α5β1, are overexpressed in glioma cells compared with
normal brain tissues. They have also been identified to be
expressed by non-tumoral cell types present in the tumoral niche,
such as proliferating vascular endothelial cells and pericytes
(2). In in vitro and in
vivo animal models, the prototype RGD-integrin antagonist
cilengitide and other structurally related small-molecule integrin
antagonists (SMIA) were identified to modulate migration and
apoptotic processes in glioma cell lines (3-5).
However, the promising results obtained using cilengitide were not
confirmed in clinical trials, prompting efforts to synthesize SMIA
with different chemical structures and pharmacological
properties.
These compounds have primarily been tested on glioma
and other cancer cell lines grown in the presence of serum, and,
although the function of integrins and periostin in gliomas has
been well-characterized (6), the
expression pattern of RGD-binding integrins and their function in
regulating glioma cell infiltration in the GSC model remain
unclear.
In an attempt to rectify this deficit in our
knowledge, the functional effects elicited by SMIA-integrin binding
in modulating the ECM-integrin interaction and the subsequent
effects elicited by 1a-RGD on integrin-dependent signal
transduction pathways in three human GSC lines grown in serum-free
medium and plated on laminin coated dishes were investigated.
The results of the present study indicated that
1a-RGD decreases cell migration and induces cell detachment and
caspase-dependent anoikis in detached GSC, thus highlighting the
important potential of SMIA to decrease the malignant dissemination
of GSC.
Materials and methods
Synthesis of 1a-RGD
1a-RGD was synthesized as described previously
(7), by means of a solution-phase
method that exploited the benzyloxycarbonyl protection strategy.
The binding of 1a-RGD to integrin receptors was determined using
in vitro binding assays, and its half-maximal inhibitory
concentration (IC50) values for αvβ3 and αvβ5 were
identified to be 6.4 and 7.7 nM, respectively (7). The integrin antagonist 1a-RGD was
solubilized in PBS at a concentration of 2 mM, and for the
treatments it was diluted in the GSC growth medium to a final
concentration of 25 µM.
Cell culture
Samples designated GSC3, GSC4 and GSC7 were isolated
from tumor tissue in the Neurosurgery Department at the Institute
for Research, Hospitalization and Care-University Hospital
(IRCCS-AOU) San Martino-Cancer Research Institute (IST) (Genoa,
Italy) following informed consent, according to European Union
legislation on informed consent and The Declaration of Helsinki,
from the patients and Institutional Ethical Committee (IRCCS San
Martino-IST) approval. The donor patients were undergoing brain
surgery for the first time. Patients had never received previous
radio- or chemotherapy and their tumors were classified by the
pathologists as glioblastoma (GB) grade IV (World Health
Organization classification). Clinicopathological characteristics
are presented in Table I.
 | Table IClinicopathological characteristics
of patients and GSC tumorigenic potential in mice. |
Table I
Clinicopathological characteristics
of patients and GSC tumorigenic potential in mice.
| GSC line | Sex | Age, years | WHO grade | Type | Subtype | OS, months | Mouse survival,
days |
|---|
| 3 | Male | 48 | IV | Primary | Neural | 14.4 | 120 |
| 4 | Male | 78 | IV | Secondary | Classical | 13.5 | 80 |
| 7 | Male | 71 | IV | Primary | ND | 3.6 | 75 |
Primary cell cultures were established as described
previously (8). Briefly, tumor
samples were collected immediately following surgery, washed and
mechanically dissociated. Primary cultures were grown in Dulbecco’s
modified Eagle’s medium (DMEM)/Ham’s F12 and neurobasal medium
(1:1) (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
with added GlutaMAX™ (Gibco; Thermo Fisher Scientific, Inc.), 100
µg/ml penicillin/streptomycin, 1% B-27 and N2 supplements
(Gibco; Thermo Fisher Scientific, Inc.), 10 ng/ml basic fibroblast
growth factor (bFGF) and 20 ng/ ml epidermal growth factor (EGF)
(PeproTech, Inc., Rocky Hill, NJ, USA). For routine culture, cells
were plated at 20,000 cells/cm2 on laminin-coated
vessels [10 µg/ml laminin-1 (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) in PBS for 6-12 h], passaged at 70% confluence
using Accutase dissociation reagent (Sigma; Merck KGaA) and split
at a 1:3 ratio. The cells were used in experiments up to but not
beyond the fifth passage in vitro. The GSC lines used were
characterized at the outset for their tumorigenic properties by
orthotopically xenografting 10,000 cells into the striatum of
6-8-week-old female non-obese diabetic/severe combined
immunodeficient mice (average weight, 30 g each). For each
glioblastoma cell line, 4 mice were used, under standard housing
conditions (27°C, 50% humidity,12-h light/12-h dark cycle), with
free access to food and water (Table
I).
All experiments involving animals were performed at
IRCCS-AOU San Martino-IST in compliance with the guidelines
approved by the Italian Ministry of Health and the Committee for
Animal Well-Being in Cancer Research.
Normal human astrocytes (NHA) were purchased from
Thermo Fisher Scientific, Inc. and grown in Astrocyte Medium
(Gibco; Thermo Fisher Scientific, Inc.) in the presence of 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.),
according to the manufacturer’s protocol. NHA were used for
experiments not beyond the fifth in vitro passage and, 24 h
before the experiments, the medium was replaced with the same
medium used for GSC cultures to standardize experimental
conditions.
Phenotypic characterization of GSC and
their modification following in vitro differentiation: GSC
differentiation in vitro
GSC cultures were seeded on Matrigel-coated glass
coverslips and maintained for 2 weeks in DMEM/Ham’s F12
supplemented with 2 mM L-glutamine, 50 IU/ml penicillin/50
µg/ml streptomycin and 10% FBS.
Phenotypic characterization of GSC and
their modification following in vitro differentiation:
Immunocytochemistry
GSC and their differentiated counterparts plated
onto laminin-coated glass coverslips were fixed with 4%
paraformaldehyde, permeabilized with PBS/0.1% Triton X-100, and
stained with the following primary antibodies: Mouse monoclonal
anti-nestin (1:1,000; cat. no. MAB1259; Novus Biologicals, Ltd.,
Cambridge, UK), rabbit anti-microtubule-associated protein 2 (MAP2;
1:1,000; cat. no. PA5-17646; Chemicon International; Thermo Fisher
Scientific, Inc.), rabbit anti-glial fibrillary acidic protein
(GFAP; 1:10,000; cat. no. Z0334; Dako; Agilent Technologies, Inc.,
Santa Clara, CA, USA) and rabbit anti-sex-determining region Y box
2 (SOX2; 1:500; cat. no. AB5603; EMD Millipore, Billerica, MA,
USA). Immunocomplexes were detected with secondary fluorescent
antibodies DyLight 488-conjugated goat anti-mouse immunoglobulin G
(IgG) (cat. no. 111-545-003) and DyLight 594-conjugated goat
anti-rabbit IgG (cat. no. 111-585-003) (Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA). Cells were counterstained
with Hoechst 33342 dye (Sigma-Aldrich; Merck KGaA) to identify all
nuclei. Images were captured by automated Zeiss AxioImager M2
equipped with an Axiocam MRM (Zeiss GmbH, Jena, Germany). Results
are presented as the percentage of stained cells from randomly
selected fields.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
For mRNA expression analysis, RNA was extracted from
GSC and NHA using QIAzol (Qiagen, Inc., Valencia, CA, USA),
followed by digestion with DNase I (cat. no. D4263; Sigma-Aldrich;
Merck KGaA) digestion step. RNA quality was assessed by determining
the A260/A280 ratio and the
concentration was estimated at 260 nm. The primers were designed
using Primer3 Input software (version 0.4.0; National Center for
Biotechnology Information, Bethesda, MD, USA) and the specificity
of each primer was checked by BLAST analysis (www.ncbi.nlm.nih.gov/tools/primer-blast). Primers used
for integrin subunits and for the housekeeping gene ribosomal
protein L6 (RPL6) in RT-qPCR have been reported previously
(3) and are presented in Table II; primers used to amplify
stemness-related mRNAs (9) are
presented in Table III. PCR
experiments were performed using the QuantiTect SYBR Green kit
(Qiagen, Inc.) containing 10 µl 2X master mix, 1 µl
each of 10 µM (0.4 µM) forward and reverse primer, 1
µl template cDNA and 7 µl nuclease-free water to a
total volume of 20 µl, according to the manufacturer’s
protocol. Each assay was run with no template control. Cycling
conditions were 95°C for 15 min; then 40 cycles of 95°C for 15 sec,
60°C for 30 sec and 72°C for 30 sec. At the end of the PCR, a
melting curve analysis was performed to check for the presence of
primer-dimers. Experiments were performed on three different cell
preparations and each run was analyzed in duplicate.
 | Table IIPrimers used to amplify RGD-binding
integrin mRNA in reverse transcription-quantitative polymerase
chain reaction experiments. |
Table II
Primers used to amplify RGD-binding
integrin mRNA in reverse transcription-quantitative polymerase
chain reaction experiments.
| Gene | Accession no. | Primer sequence
(5′-3′) |
|---|
| αv | NM_002210 | F:
actggcttaagagagggctgtg |
| R:
tgccttacaaaaatcgctga |
| β3 | NM_000212 | R:
tcctcaggaaaggtccaatg |
| R:
tcctcaggaaaggtccaatg |
| β5 | NM_002213 | F:
agcctatctccacgcacact |
| R:
cctcggagaaggaaacatca |
| α5 | NM_002205 | F:
cctgctgtccaccatgtcta |
| R:
ttaatggggtgattggtggt |
| β1 | NM_133376 | F:
tccaatggcttaatttgtgg |
| R:
cgttgctggcttcacaagta |
| RPL6 | NM_001024662.1 | F:
agattacggagcagcagcgcaagattg |
| R:
gcaaacacagatcgcaggtagccc |
 | Table IIIPrimers used to amplify
stemness-associated mRNAs in reverse transcription-quantitative
polymerase chain reaction assays. |
Table III
Primers used to amplify
stemness-associated mRNAs in reverse transcription-quantitative
polymerase chain reaction assays.
| Gene | Accession no. | Primer sequence
(5′-3′) |
|---|
| CD133 | NM_006017.2 | F:
ccaccgctctagatactgctg |
| R:
cctatgccaaaccaaaacaaa |
| OCT4 | NM_002701.4 | F:
ggtccgagtgtggttctgtaa |
| R:
atagcctggggtaccaaaatg |
| MUSASHI | NM_002442.3 | F:
actgaagtttcccaccaggat |
| R:
actgttcatgaaggtccaacg |
| NANOG | NM_024865.2 | F:
cagtctggacactggctgaa |
| R:
ctcgctgattaggctccaac |
| BMI1 | NM_005180.6 | F:
ggaaagcaggcaagacttttt |
| R:
caaactatggcccaatgctta |
| NESTIN | NM_006617.1 | F:
gggacaagagaacctggaaac |
| R:
ggttcacttccacagactcca |
| SOX2 | NM_003106 | F:
ggacttctttttgggggacta |
| R:
gcaaacttcctgcaaagctc |
| EZH2 | NM_152998.1 | F:
tgccattgctaggttaattgg |
| R:
acaaccggtgtttcctcttct |
Data are expressed as the fold increase in each gene
in GSC compared with NHA, using the 2−ΔΔCq method
(10) following normalization to
the RPL6 housekeeping gene.
Fluorescence-activated cell sorting
(FACS) analysis
The expression of αvβ3, αvβ5 and α5β1 on cell
membranes was determined using FACS analysis with antibodies
directed against the integrin receptors. Briefly, following gentle
detachment of the cells using a 1 mM EDTA/PBS solution to preserve
the integrity of membrane proteins, cells were pelleted at 800 × g
for 10 min and resuspended in 1 ml PBS to obtain a suspension of
20,000 cells/ml. The cell suspension was then incubated with the
following antibodies (1 µg/ml): Mouse monoclonal
anti-integrin αvβ3 antibody (cat. no. MAB1976; Merck KGaA), Alexa
Fluor 633-conjugated goat anti-mouse (cat. no. A21050; Thermo
Fisher Scientific, Inc.), fluorescein isothiocyanate
(FITC)-conjugated mouse monoclonal anti-integrin αvβ5 antibody
(cat. no. MAB1961F; Merck KGaA) and FITC-conjugated mouse
monoclonal anti-integrin α5 antibody (cat. no. CBL497F; Merck
KGaA).
To determine apoptosis, an Annexin V-binding assay
was used (Annexin V-FITC Apoptosis Detection kit; eBio-science;
Thermo Fisher Scientific, Inc.), according to the manufacturer’s
protocol. Following treatments, spontaneously detached cells were
recovered and stored separately, whereas attached cells were
detached using 1 mM EDTA/PBS solution as aforementioned. The
suspensions of attached and detached cells were centrifuged at 800
× g for 10 min, and the pellets were suspended in 1 ml Annexin V
buffer, according to the manufacturer’s protocol. FITC-conjugated
Annexin V was added and cells were incubated for 15 min at room
temperature. Following the addition of propidium iodide (PI),
samples were acquired using a FACSVantage SE instrument (BD
Biosciences, Franklin Lakes, NJ, USA) and analyzed using CellQuest
software (version 5.1; BD Biosciences). At least 10,000 events per
sample were recorded. Each experiment was performed three
times.
Cell viability assays
Cells were plated in a 96-well plate (10,000
cells/100 µl per well) and treated with DMEM/Ham’s F12 and
neurobasal medium (1:1) containing 25 µM 1a-RGD for 8, 24
and 48 h. Following treatment, 20 µl MTS reagent (CellTiter
96 AQueous One Solution Cell Proliferation assay; Promega
Corporation, Madison, WI, USA) was added to each well. After 3 h of
incubation at 37°C, the absorbance was determined in a multi-well
plate reader at 450 nm. For each experimental point, eight wells
were used and each independent experiment was performed three
times.
The cell viability results were normalized to
time-point-matched controls; 1a-RGD stock solution (200 mM in PBS)
was diluted in the growth medium and added to the wells. In control
wells, only the growth medium was added.
Cell migration assays
GSC (20,000 cells/well) were plated in culture
medium lacking growth factors on a MaxGel (Sigma-Aldrich; Merck
KGaA)-coated Transwell (Costar; Corning Incorporated, Corning, NY,
USA). As chemoattractants, 500 µl EGF (20 ng/ml) and bFGF
(10 ng/ ml)-containing medium were placed under the Transwell
membranes. The migration assay was performed for 8 h in the
presence or absence of 25 µM 1a-RGD. Following removal of
the MaxGel, the cells present on the lower side of the membrane
were stained with DAPI (Sigma-Aldrich; Merck KGaA) and enumerated
using a fluorescence microscope. For each membrane, 10 fields were
observed. Each experiment was performed at least three times.
Western blot analysis
Cells grown in 60 mm diameter dishes were treated
for the indicated times with 25 µM 1a-RGD. The cells were
then rinsed twice in ice-cold PBS, and 200 ml cell lysis buffer [50
mM Tris/HCl pH 7.4, 1% (v/v) NP40, 0.25% (w/v) sodium deoxycholate,
1 mM phenylmethylsulfonyl fluoride, 1 mM
Na3VO4, 1 mM EDTA, 30 mM sodium
pyrophosphate, 1 mM NaF, 1 mg/ml leupeptin, 1 mg/ml pepstatin A, 1
mg/ml aprotinin and 1 mg/ml microcystin] was added to the dishes.
Following scraping, the cells were sonicated for 10 min and
centrifuged at 12,000 × g for 5 min at 4°C. The amount of proteins
in the supernatant was then determined using a Bicinchoninic Acid
Protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). For
western blot analysis, 30 µg proteins were separated by
SDS-PAGE (10% gel) at 150 V for 2 h and blotted onto 0.22 mm
nitrocellulose membranes at 50 mA for 16 h. The membranes were
first blocked for 2 h in Tris-buffered saline containing Tween-20
(TBST; 10 mM Tris/HCl, 150 mM NaCl and 0.1% Tween-20) containing 5%
non-fat dry milk powder (TBSTM) and then incubated with the
appropriate antibody [anti-phospho-FAK rabbit polyclonal antibody
(cat. no. 3283); anti-phospho-Akt rabbit polyclonal antibody (cat.
no. 9271); Cell Signaling Technology, Inc., Danvers, MA, USA]
diluted 1:1,000 in TBSTM at 4°C for 16 h with gentle agitation. The
membranes were rinsed three times in TBST and then incubated at
21°C for 2 h with a horseradish peroxidase-conjugated goat
anti-rabbit IgG secondary antibody (cat. no. 12-348; Upstate
Biotechnology, Inc., Lake Placid, NY, USA) diluted 1:10,000 in
TBSTM. The membranes were rinsed three times in TBST and the
luminescence signal was captured using an ImageQuant LAS4000, GE
Healthcare (Chicago, IL, USA). Each experiment was performed at
least twice.
ELISA nucleosome assay
For the relative quantification of cell death, a
colorimetric ELISA sandwich immunoassay was used to detect
nucleosomes (Cell Death Detection ELISA; Roche Diagnostics GmbH,
Mannheim, Germany). Cells were plated in a 12-well plate and
treated with cell culture medium containing 25 µM 1a-RGD for
48 h, in the presence and absence of the caspase inhibitor
carbobenzoxy-Val-Ala-Asp-fluoromethylketone (zVAD-fmk; 1
µM). Following incubation, cell lysates were prepared using
the lysis buffer supplied in the kit and the assay was performed
according to the manufacturer’s protocol. Finally, the samples were
measured in a multi-well reader at 405 nm. Eight wells were used
for each experiment and each experiment was performed three
times.
Caspase activity assay
Cells were plated on clear-bottomed 96-well black
plates (Costar; Corning Incorporated) at a density of 5,000
cells/well. Caspase-3/7 and -9 activities were determined following
the addition of growth medium containing 25 µM 1a-RGD for 48
h, in the presence and absence of the caspase inhibitor zVAD-fmk (1
µM), using Caspase Glo 3/7 and Caspase Glo 9 kits (Promega
Corporation). Each experiment was performed in triplicate.
Statistical analysis
Results are expressed as the mean ± standard
deviation and analyzed using Student’s t-test when comparing two
groups or ANOVA and a Tukey’s post hoc test for comparing more than
two groups. Statistical analyses were performed using GraphPad
Prism software (version 5.0; GraphPad Software, Inc., La Jolla, CA,
USA). P<0.05 was considered to indicate a significant
difference.
Results
Characterization of GSC stemness
To characterize and confirm the stemness status of
the GSC obtained from specimens from patients with GB, a series of
experiments was performed. First, the determination of the stemness
markers nestin, MAP2, GFAP and SOX2, in undifferentiated and
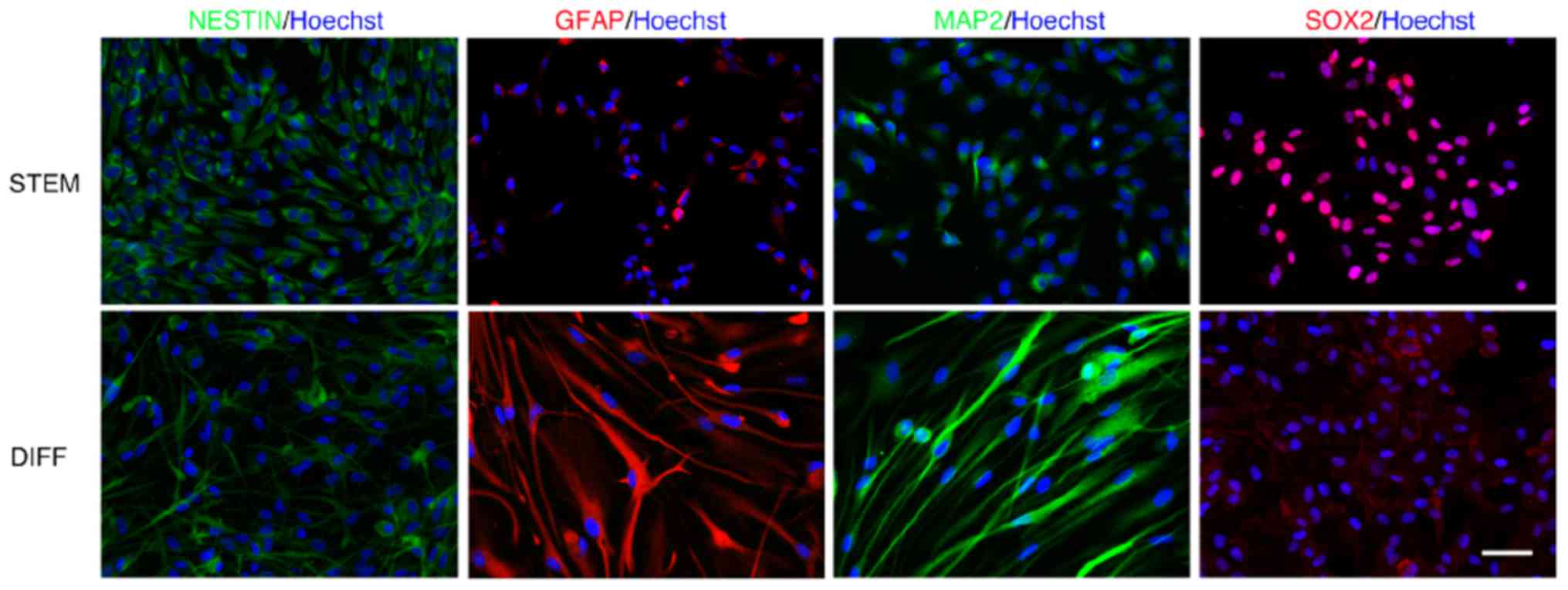
differentiated GSC was performed by immunostaining (Fig. 1).
When GSC3, normally grown in a serum-free medium,
were differentiated using 10% FBS, a statistically significant
increase (P<0.001) was observed in MAP2- and GFAP-positive cells
in comparison with undifferentiated GSC. Conversely,
undifferentiated GSC were positive for nestin and SOX2 staining
(Table IV). It cannot be excluded
that the adhesion conditions on the microscopy glasses may serve a
function in inducing slight morphological alterations. However,
staining of the markers was consistent and reproducible under all
conditions. The immunostaining was also performed in GSC4 and GSC7
lines with similar results (data not shown).
 | Table IVCells positive for nestin, MAP2, GFAP
and SOX2. |
Table IV
Cells positive for nestin, MAP2, GFAP
and SOX2.
| GSC line | Nestin | MAP2 | GFAP | SOX2 |
|---|
| 3 | Stem | 96 (±2) | 18 (±3) | 25 (±5) | 96 (±5) |
| Differentiated | 68 (±4) | 80 (±4) | 89 (±8) | 44 (±6) |
| 4 | Stem | 55 (±5) | 5 (±2) | 18 (±5) | 70 (±5) |
| Differentiated | 28 (±6) | 32 (±8) | 34 (±8) | 39 (±8) |
| 7 | Stem | 95 (±3) | 44 (±5) | 19 (±2) | 88 (±6) |
| Differentiated | 75 (±5) | 28 (±5) | 74 (±13) | 39 (±4) |
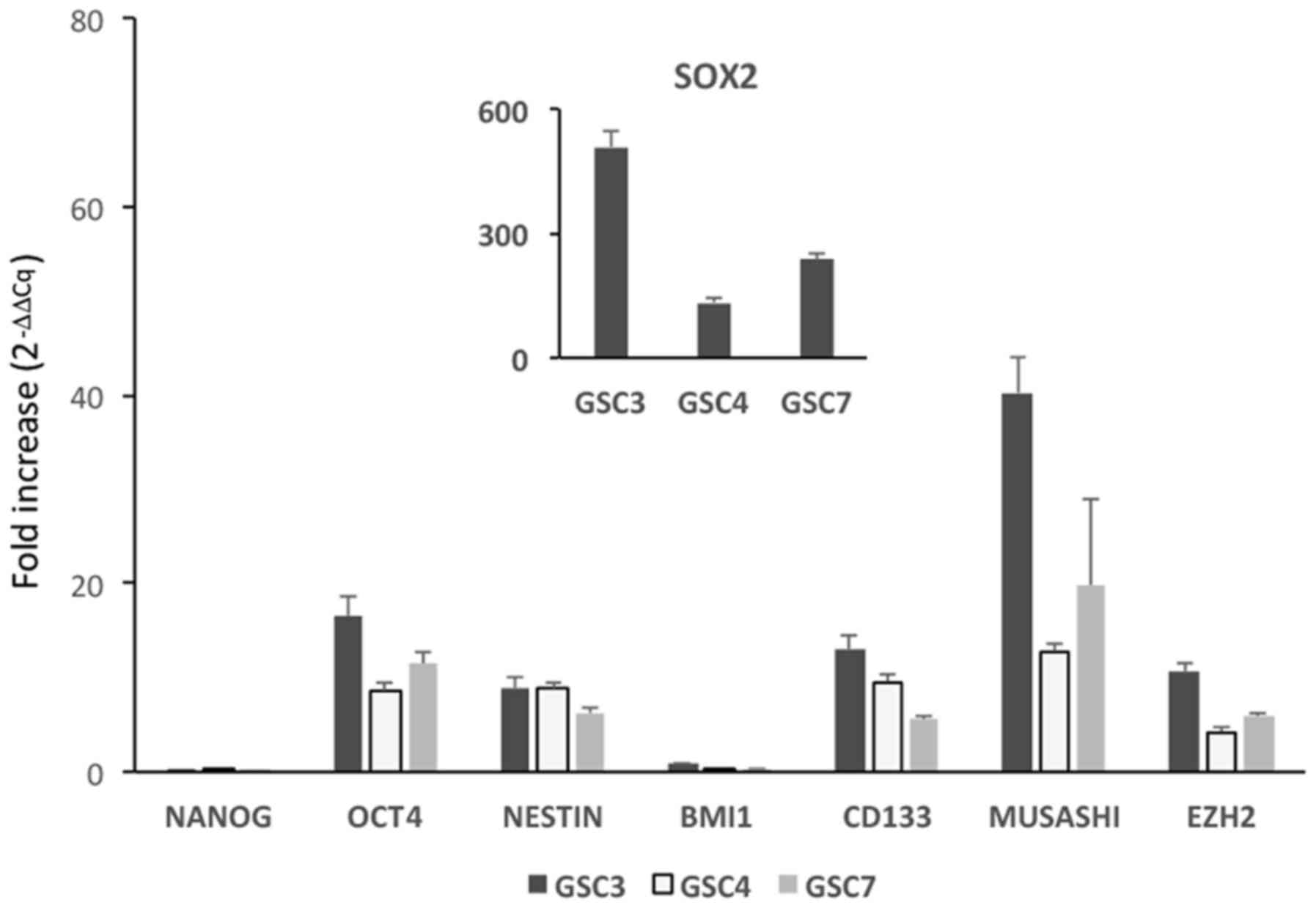
Stemness of GSC confirmed by RT-qPCR
The stemness status of the GSC was confirmed further
by the transcriptomic analysis of six stemness markers (9,11) in
addition to SOX2 and nestin, in undifferentiated GSC and NHA, used
in all experiments as non-tumor reference cells. Except for BMI1
and Nanog, whose expression was similar in the two cell
populations, all other stemness marker transcripts exhibited
significantly increased expression in GSC compared with in NHA,
thus demonstrating that, at least in under the culture conditions
used, GSC retained their original stemness status (Fig. 2). In addition, when the analysis
was repeated in undifferentiated GSC and in differentiated GSC
grown with 10% FBS, the expression of all the transcripts in the
latter was significantly lower (data not shown), thus confirming
the immunocytochemistry results.
GSC express αv, β3, β5 and α5 integrin
subunits
Since previous studies have indicated that some
RGD-binding integrins are overexpressed in GB (12), the αvβ3, αvβ5 and α5β1 integrin
expression pattern in GSC were investigated by determining the mRNA
amounts of αv, β3, β5, α5 and β1 subunits. The RT-qPCR experiments
clearly identified that αv, α5 and β5 subunits are overexpressed in
all three cell lines examined, compared with the expression profile
for NHA (Fig. 3A), with the fold
increase ranging between 5 and 500, whereas the expression of β1
and β3 was less pronounced compared with their relative expression
in NHA.
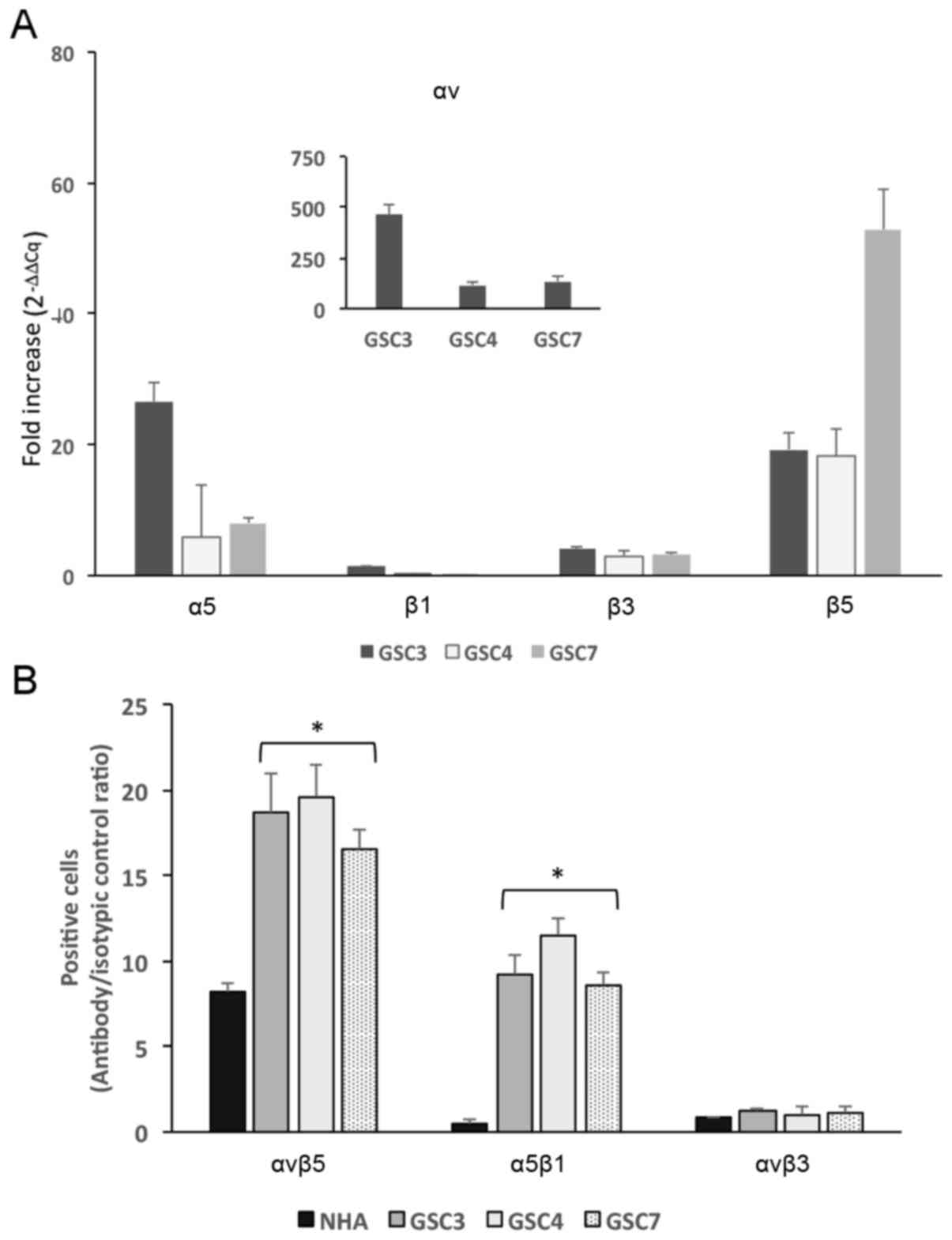 | Figure 3Integrin expression in GSC3, GSC4 and
GSC7 lines. (A) mRNA extracted from GSC3, GSC4, GSC7 and NHA was
amplified using the reverse transcription-quantitative polymerase
chain reaction. Results are expressed as the fold increase
(2−ΔΔCq) determined using RPL6 as a housekeeping gene.
The mean RPL6 Cq values were 20.44 for NHA, 16.22 for GSC3, 16.374
for GSC4 and 16.389 for GSC7. (B) Flow cytometric analysis was
performed on GSC3, GSC4, GSC7 and NHA to assess integrin receptor
expression on the cell surface. The cell suspensions were incubated
with antibodies recognizing the integrin receptors and >10,000
cells for each experimental point were assessed. Results are
expressed as antibody signal/isotypic control ratio.
*P<0.05 vs. NHA. GSC, glioma cancer stem cell; NHA,
normal human astrocyte; RPL6, ribosomal protein L6. |
To further confirm the integrin receptor expression
on GSC membranes, FACS experiments were performed using conjugated
fluorescent antibodies recognizing αvβ3, αvβ5 and α5β1 integrins.
The expression results, reported as specific signal/ isotypic
control ratio, are presented in Fig.
3B. The surface expression of αvβ5 and α5β1 integrins was more
abundant in GSC compared with in NHA, whereas for αvβ3, no
significant difference was identified between GSC and NHA.
These results are in good agreement with the mRNA
results from the RT-qPCR assay, and the poor expression of β3
subunit may account for the different integrin expression on the
cell surface.
1a-RGD induces cell death in GSC
Since certain discrepancies concerning the effect of
RGD antagonists on cancer cells were identified previously
(2), the effect of 1a-RGD
treatment on GSC viability was determined. In the present study,
1a-RGD was used at a concentration of 25 µM on the basis of
its IC50 (10.2±0.8 µM), deduced from
concentration-response curves performed in GB cell lines (3).
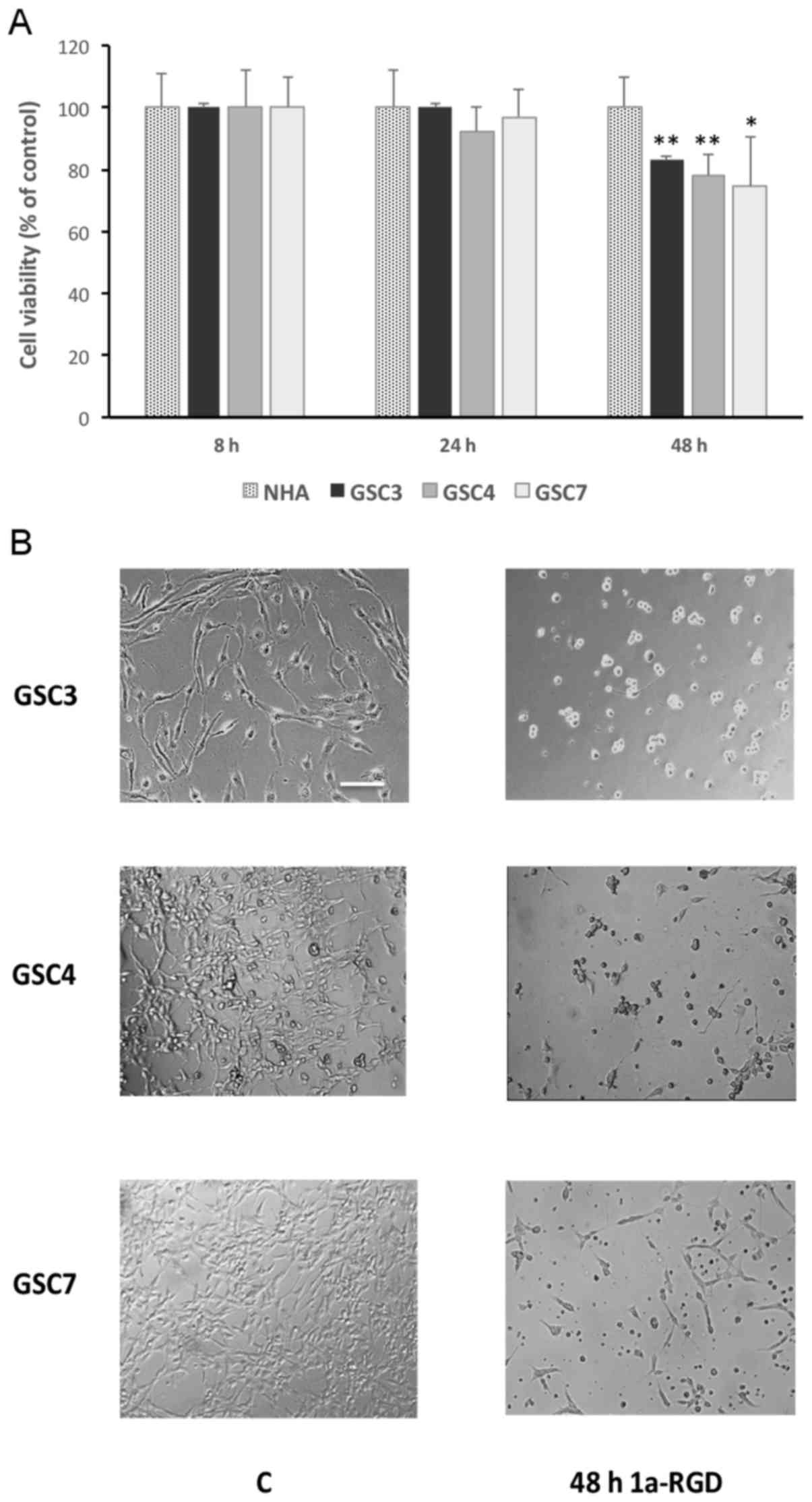
After 48 h of treatment with 1a-RGD, a significant
decrease was observed in the viability of GSC that was not
replicated in the non-tumorigenic NHA control cells (Fig. 4A). More specifically, following
treatment, only GSC were observed to have detached from the plastic
dishes. They also exhibited altered morphology, assuming the round
shape typical of cells undergoing cell death (Fig. 4B); in contrast, neither cell
detachment nor a change in morphology was observed in
1a-RGD-treated NHA (data not shown). These results clearly indicate
that 1a-RGD was responsible for the loss of GSC viability, in a
process possibly mediated by integrin inhibition.
To verify whether 1a-RGD was responsible for the
loss of GSC viability, the extent and type of cell death was
investigated separately in adherent and detached cells from the
same wells following 1a-RGD treatment (25 µM for 48 h) using
an Annexin V/PI FACS assay (Fig. 5A
and B). The floating cells and the still adherent cells were
collected from each well. Following enumeration, the two fractions
were subjected to FACS analysis. Although the three GSC lines
exhibited differential sensitivity to the induction of cell death
elicited by 1a-RGD, the viability in adherent cells was not
significantly different from that observed in untreated cells
(controls). In contrast, in detached cells, a marked increase in
the number of apoptotic cells was observed, indicating that 1a-RGD
causes detachment-induced anoikis in GSC.
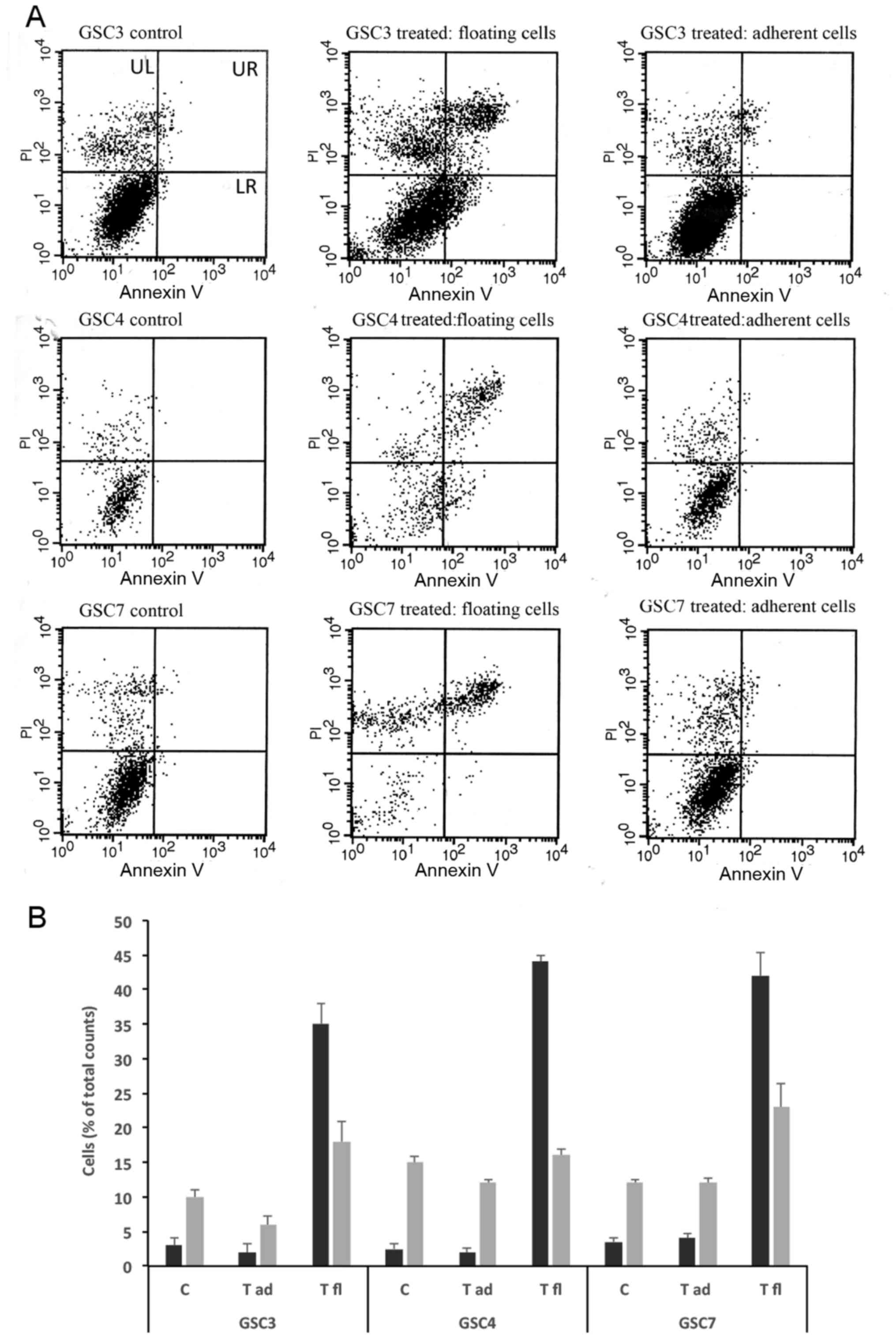 | Figure 51a-RGD treatment induces
detachment-dependent anoikis. (A) Following treatment with 25
µM 1a-RGD for 48 h, detached and adherent GSC lines from the
same wells were separately analyzed by fluorescence-activated cell
sorting to assess cell death. (B) Results from flow cytometric
analysis. Apoptotic cells were present in the floating fraction,
whereas no difference was observed between controls and still
adherent treated cells. GSC, glioma cancer stem cell; PI, propidium
iodide; UL, upper left; UR, upper right; LL, lower left; LR, lower
right; C, controls; T, treated; fl, floating; ad, adherent. |
1a-RGD induces anoikis in GSC via a
caspase-dependent mechanism
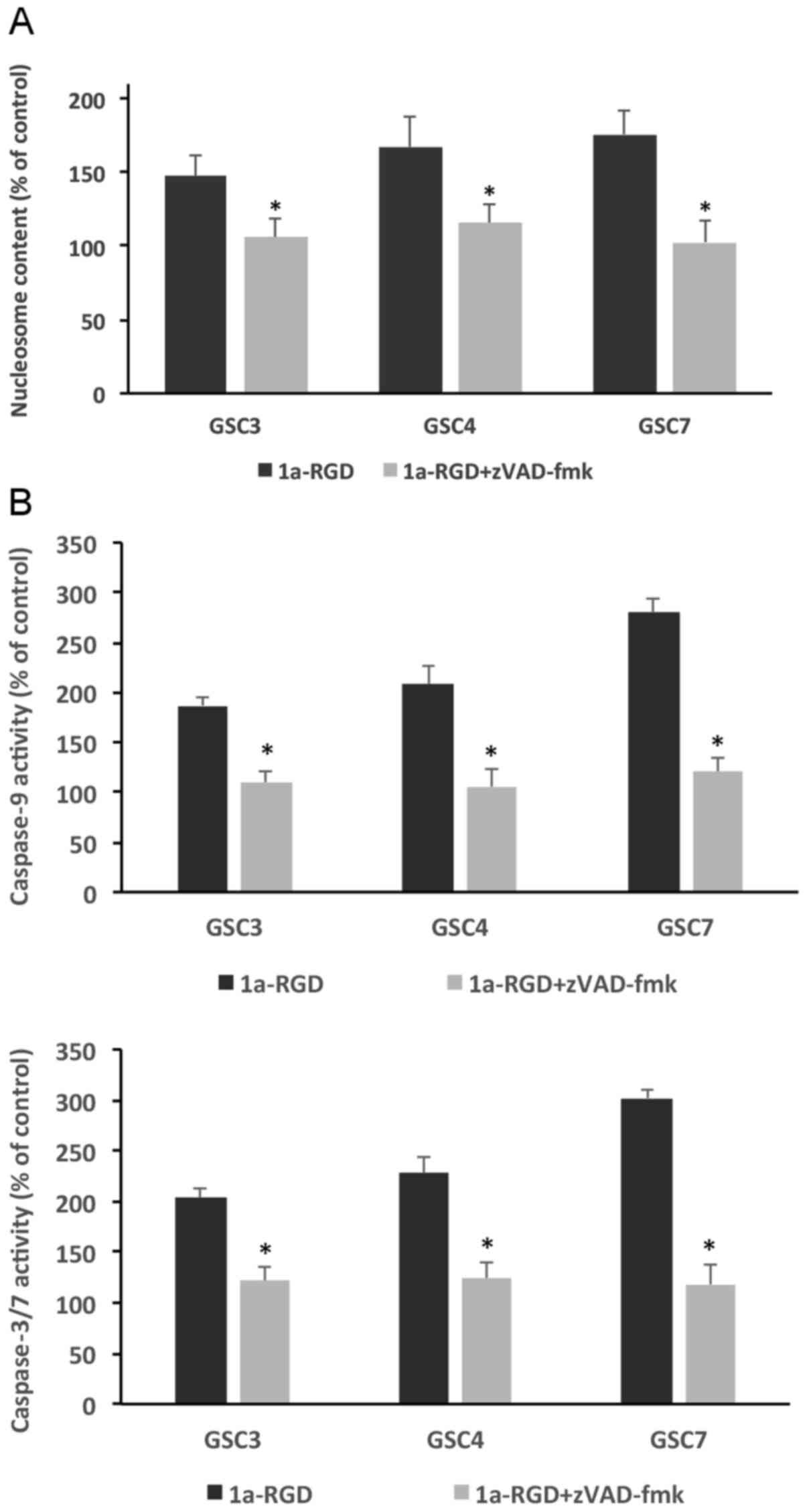
Under the same experimental conditions, anoikis
onset was determined using two different assays: A nucleosome ELISA
assay and a caspase-3/7 activity assay. Treating the cells for 48 h
with 25 µM 1a-RGD induced a significant increase in
nucleosome content compared with control samples in all three GSC
examined; notably, this increase was partially rescued by
incubating GSC with 1 µM zVAD-fmk, thus suggesting that
caspase-3/7 mediate, at least in part, this anoikis-associated
process (Fig. 6A).
This supposed function of caspase-3/7 in sustaining
anoikis was further confirmed using a fluorimetric caspase activity
assay performed under the same experimental conditions. Similar to
that observed using the nucleosome assay, the increase in
caspase-3/7 activity observed in 1a-RGD-treated GSC was
significantly blunted by the caspase inhibitor zVAD-fmk (Fig. 6B), thus confirming that the
observed anoikis was indeed dependent on caspase-3/7
activation.
1a-RGD-induced anoikis is not sustained
by autophagy
Since integrin inhibition has been identified to
induce atypical anoikis in glioma cells (13), the possibility that anoikis
elicited by integrin inhibition via 1a-RGD may involve processes
associated with autophagy in GSC was investigated.
GSC were exposed to 25 µM 1a-RGD for 48 h,
and cell lysates were subjected to electrophoresis and western
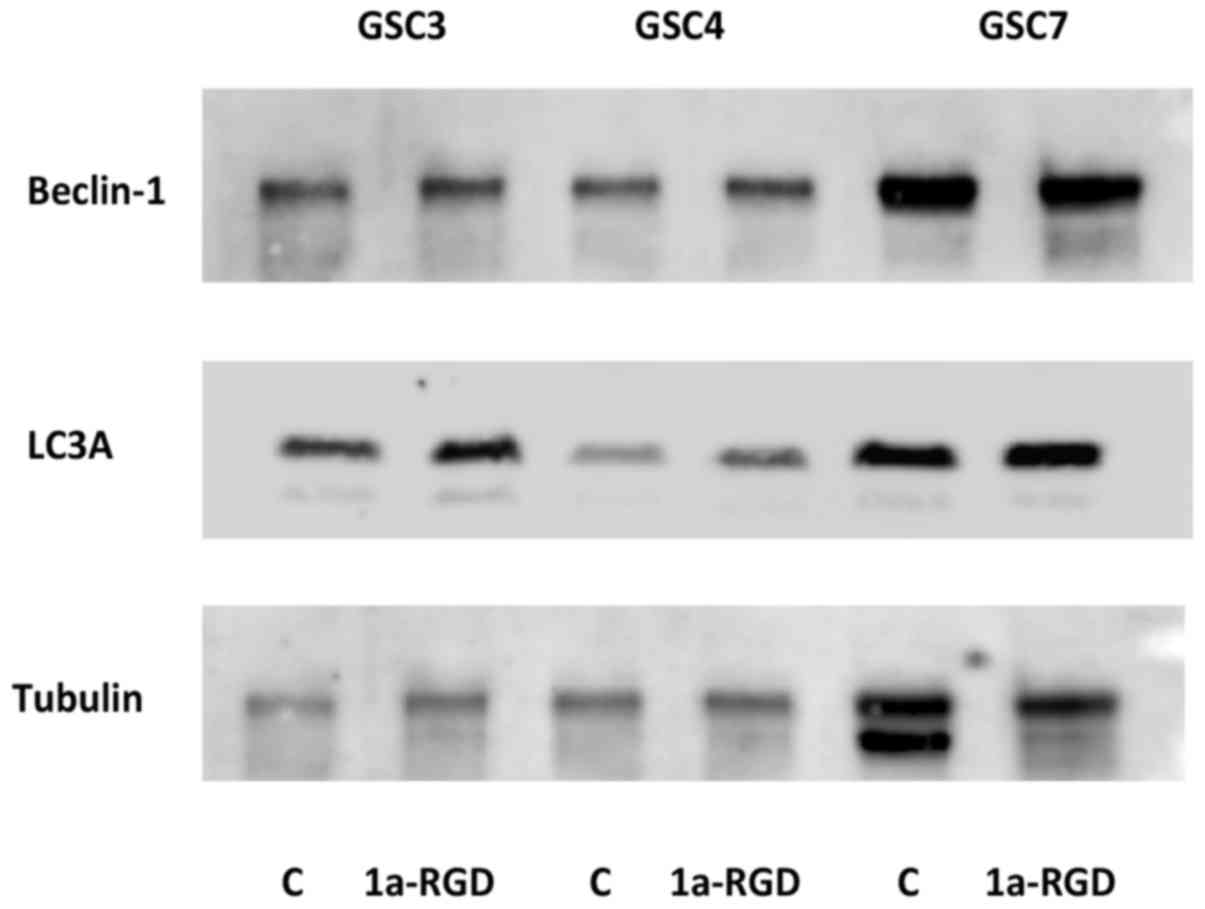
blotting (Fig. 7). There were no
changes in total levels of light chain 3 (LC3)-II and beclin-1, two
proteins associated with the induction of autophagy (14), suggesting that, in the present
study, cell death was due to caspase-dependent anoikis, with no
apparent contribution from autophagy-associated processes.
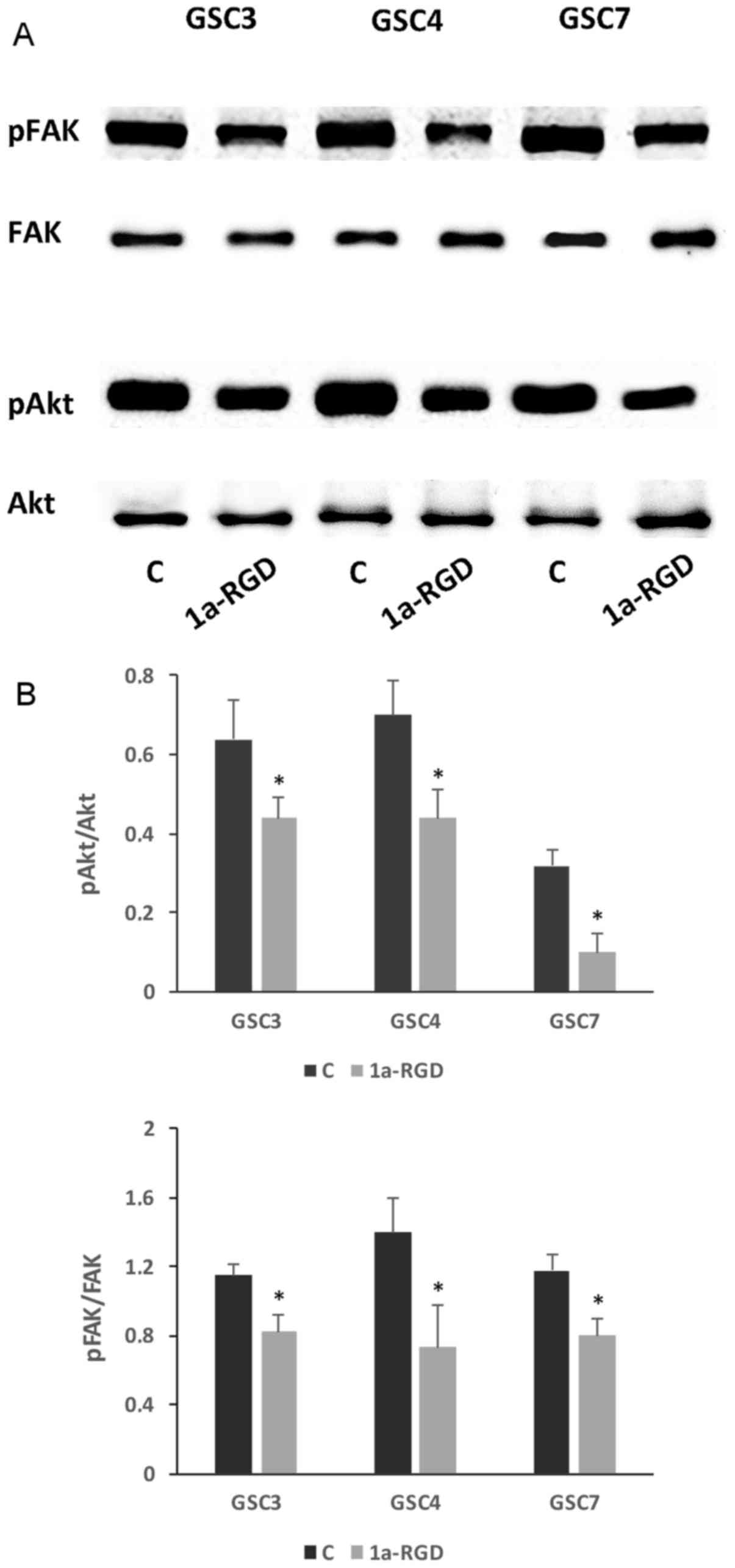
1a-RGD inhibits focal adhesion kinase
(FAK) and protein kinase B (Akt) phosphorylation
The interaction with integrins of several endogenous
ECM proteins leads to the activation of a variety of downstream
protein kinases, such as FAK, extracellular-signal-regulated kinase
(ERK) and Akt (2,3). To investigate whether 1a-RGD
interferes with the activation of these signaling pathways, western
blot analysis was used to determine the effect of 25 µM
1a-RGD treatment on the phosphorylation state of these downstream
kinases. It was identified that 1a-RGD treatment for 48 h
significantly decreases FAK and Akt phosphorylation (Fig. 8A and B), whereas ERK
phosphorylation was not affected (data not shown); a shorter
treatment (8 h) of GSC with 1a-RGD did not affect the
phosphorylation state of the two kinases (data not shown).
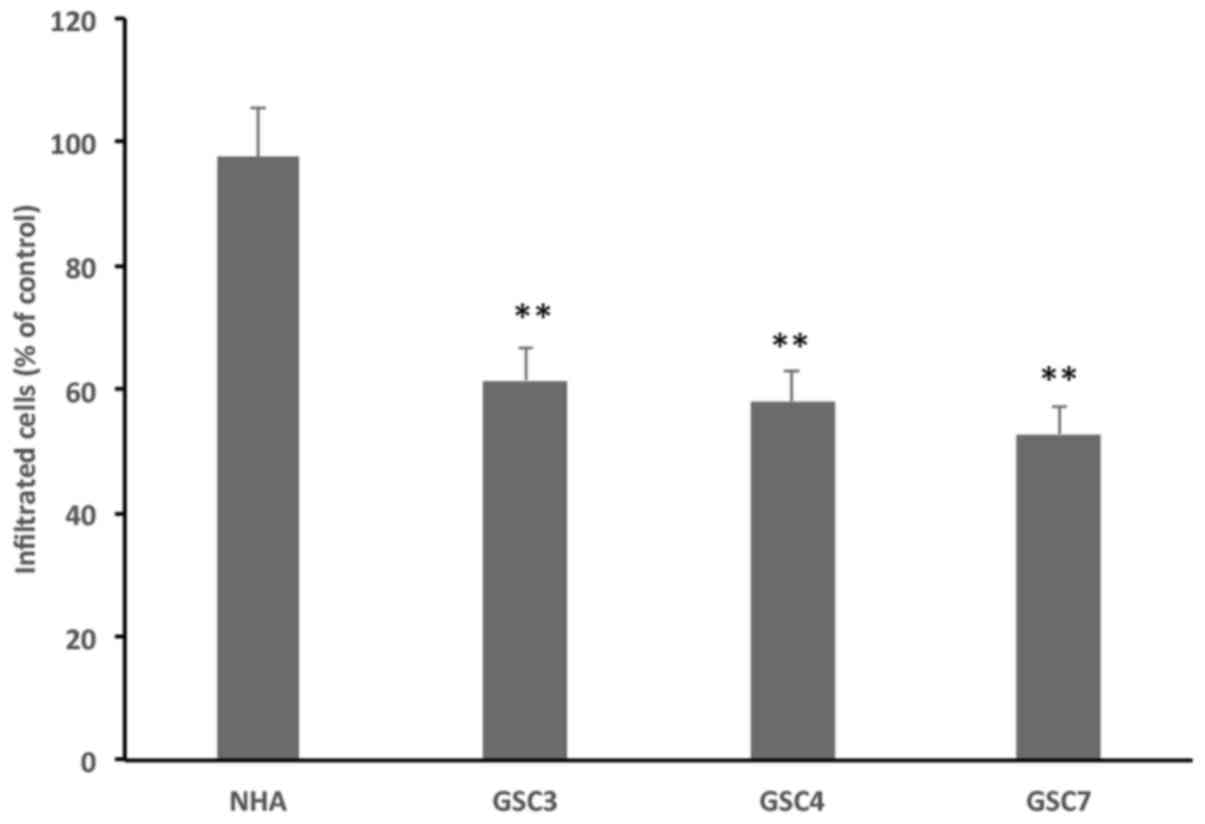
1a-RGD decreases GSC migration
The features of GSC most implicated in their
malignancy are their ability to disseminate, along with their
potential invasiveness into the surrounding parenchyma. Since, in
some cellular models, the integrin-dependent inhibition of
downstream FAK and Akt pathways is functionally associated with a
decrease in cell motility, it was investigated whether 1a-RGD
affects GSC migration in the experimental model.
A Transwell chamber assay was performed using EGF
and bFGF as chemoattractant. Treatment with 25 µM 1a-RGD for
8 h resulted in significantly decreased numbers of migratory cells
in all three GSC tested, compared with controls. Notably, this
inhibitory effect was not observed in NHA (Fig. 9), possibly due to the lower
integrin expression in these non-tumoral cells, as
aforementioned.
Discussion
Integrins are an appealing potential target in
cancer biology because they mediate crucial features of tumor
malignancy, i.e. detachment from the original tumoral mass,
metastatic dissemination and the invasion of distant sites, which
culminate in the formation of a new tumoral niche (15,16).
Disappointing results from early clinical studies, employing the
canonical prototype integrin-binding molecule cilengitide have
stimulated attempts to synthesize and exploit the pharmacological
properties of other RGD-containing molecules with different
chemical structures (2). This
approach appears to be particularly suitable for GB because tumor
cells themselves and those belonging to the tumor niche overexpress
RGD-binding integrins. In addition, the strategy of targeting
integrins may impair the dissemination of glioma cancer stem cells,
responsible for the fatal relapses observed following surgery in
patients with glioma (17). Also,
in a recent study, αvβ3 integrin has been identified as an
effective target for cilengitide in a subset of GBM (18).
Studies performed using in vitro models of
mouse and human glioma cell lines have identified that several
RGD-containing integrin antagonists affect cell viability,
apoptosis and sensitize glioma cells to alkylating drugs (3,19).
However, in these studies, glioma cell lines were grown and
maintained in the presence of serum, a condition that may alter the
original phenotype of the tumor tissue. To partially overcome this
drawback in the present study, the stemness properties were
initially characterized, together with the expression of
RGD-binding integrin subunits, of GSC grown under adherent
conditions and in the absence of serum. Following validation of the
model, the effects of an RGD integrin antagonist, 1a-RGD, on cell
viability, cell migration and cell death mechanisms in GSC were
investigated.
In agreement with a previous study (20), the results of the present study
identified that GSC grown on laminin-coated plastic, in the
presence of the growth factors EGF and bFGF and neurobasal medium,
retain their stemness features in vitro and give rise to
brain tumors when implanted in a rodent model. Quantitative
analysis of stemness markers by RT-qPCR clearly demonstrated that
the transcript levels of these markers were more abundant than in
differentiated NHA, confirming that stemness of the brain tumor
cell population was retained under the experimental conditions
used.
The use of suitable and reliable control cells when
comparing the expression of specific biomarkers in GSC is
currently, for several theoretical and technical reasons, a
much-debated and controversial issue that has been poorly addressed
in the literature.
Primary cultures of astrocytes represent a valuable
tool to study their function in health and disease (21). Human astrocytes grown and
propagated in vitro may be obtained primarily from three
sources: i) Primary fetal astrocytes sometimes considered the
current ‘gold standard’ (22); ii)
human induced pluripotent stem cell-derived astrocytes (23); and iii) acutely purified astrocytes
from fetal and adult human brain obtained using an
immunopanning-based technique using an antibody targeting hepatic
and glial cell adhesion molecule (or glial cell adhesion molecule),
a surface protein expressed by human astrocytes, to generate highly
purified (>95%) cultures of primary human astrocytes (24) maintained under serum-free
conditions. However, currently, a detailed comparison among human
in vitro astrocytes obtained from different sources remains
lacking and it is therefore impossible to determine which model is
preferable. Furthermore, all cell culture systems have intrinsic
problems, since no culture system can fully replicate the in
vivo conditions. It is remarkable that Zhang et al
(24) identified that human
astrocytes exist in two distinct developmental stages: A fetal
highly proliferative astrocyte precursor cell and a mature
post-mitotic astrocyte. The advantage of using fetal astrocytes is
associated with the fact that they can be purchased commercially,
and can be frozen, stored and defrosted at a later time for
subsequent experiments. However, on the basis of these premises, we
are well aware of the limitations of using fetal astrocytes (NHA)
as normal control in studies with GSC, but we also consider that
they currently represent the most accessible source of human
astrocytes. Therefore, future studies aimed at comparing the
features of human astrocytes propagated in vitro and
obtained from different sources are mandatory to assess which
cellular model should be adopted as a proper control in studies
involving GSC.
Another notable result obtained in the present study
concerns the expression of integrins in GSC: The majority of data
available in the literature concerning the expression of
RGD-binding integrins were obtained using glioma cell lines grown
in the presence of serum, a condition that may not reflect the
genotype of cell populations present in the original brain tumor
(25). In agreement with previous
studies describing the overexpression of RGD-binding integrins in
glioma cells and tumor specimens (12), the results of the present study
indicated that GSC overexpressed the αv, β5 and α5 integrin subunit
transcripts compared with NHA, whereas no difference was identified
in the level of β3 transcript. These results were confirmed by FACS
analysis of integrin expression on GSC membranes, suggesting that
the low β3 expression may be the limiting step for αvβ3 receptor
expression.
Previous studies identified that cilengitide induced
detachment-mediated anoikis in pediatric glioma and neuroblastoma
cell lines (26), inhibited
proliferation and promoted apoptosis via downregulation of FAK-,
Src- and Akt-dependent pathways in glioma cells (27), and in glioma cancer cells triggered
antiapoptotic autophagy mechanisms that sustained
detachment-dependent atypical anoikis (13). However, the function of autophagy
in regulating cell death in glioma cells is controversial and
appears to be strictly associated with the type of cell examined
and to the culture conditions, because it has been identified that,
in glioma cell lines, autophagy was enhanced and supported cell
death, acting as a pro-apoptotic element (28).
The results of the presents study clearly indicate
that 1a-RGD induces detachment-mediated anoikis in GSC and that the
contribution, if any, of other cell-death-associated mechanisms,
such as necrosis and autophagy, appears to be marginal and
limited.
Three separate lines of evidence from the present
study support this conclusion. First, the cells that remained
attached to the wells after 48 h of treatment with 1a-RGD were
viable, whereas most of the detached cells had undergone cell
death. Secondly, analysis of the detached cells by FACS/Annexin V
staining exhibited no increase in the necrotic index in treated
cells compared with untreated cells. Finally, the levels of two
autophagy markers, LC3 and Beclin-1 (14,28),
were not modified in detached cells following 1a-RGD treatment.
It was also identified that caspase activity,
together with nucleosome formation, was markedly decreased when GSC
were simultaneously treated with the pan-caspase inhibitor
zVAD-fmk, clearly indicating that the type of anoikis observed in
GSC required caspase-3/7 and caspase-9 activation. This result is
in contrast with those of Silginer et al (13), which described atypical
caspase-independent anoikis induced by cilengitide in human and
mouse glioma cancer cells (13).
Several experimental factors, such as different integrin subtype
expression patterns and the pharmacological profile of the integrin
antagonists used, may account for the differences in the reported
mechanisms leading to cell death. The results of the present study
indicate that autophagy is not involved in 1a-RGD-induced anoikis;
nevertheless, no firm conclusions can be drawn, and the function of
autophagy in cell death processes and GB dissemination clearly
requires further investigation.
Integrin-dependent regulation of cellular effects
such as survival, growth, migration and resistance to anoikis are
mediated by FAK activation by two distinct, but not mutually
exclusive, mechanisms: Activation of phosphoinositide 3-kinase
(PI3K)/Akt- and ERK-dependent signaling pathways and regulation of
the crosstalk between integrins and growth factor receptor
signaling (29).
The results of western blot experiments, obtained
using anti-phospho-specific antibodies, indicate that the FAK- and
Akt-dependent pathways are likely to serve a significant function
in 1a-RGD-dependent anoikis induction and inhibition of cell
migration. The experiments of the present study were performed in
the presence of 25 µM 1a-RGD for 24 h: In other experiments,
it has not been possible to detect significant p (phospho-)Akt and
pFAK signal decreases in GSC following shorter treatment times (for
example, 8-h treatment; data not shown). We hypothesize that a
contact time <24 h is insufficient to reveal a change in the
phosphorylation status of these proteins; indeed, FAK and Akt are
the convergence point for a number of distinct signaling pathways
and therefore a slight decrease in their phosphorylation state may
be difficult to detect, being masked by concomitant stimulation
from other receptors, such as growth factor receptors.
Similar mechanisms have been described for other
cell types: In fibroblasts, the activation of β1 integrin triggers
a viability signal, mediated by the activation of a FAK/PI3K/
Akt-dependent signaling pathway, that protects cells from apoptosis
(30), and in human intestinal
epithelial cells, the suppression of anoikis requires a selective
repertoire of integrins to activate the FAK/Src-dependent pathway
(31). The relevance of FAK
inhibition as a key event in anoikis resistance has recently been
highlighted in an elegant study (32) in which it was revealed that
internalized integrins trigger the assembly of endosomal signaling
complexes which, in turn, recruit FAK and induce anoikis
resistance, thereby promoting cancer cell survival and metastatic
growth. Further studies on GSC, aimed at dissecting the specific
mechanism by which integrin antagonists counteract anoikis
resistance, are therefore required to understand the potential
function of SMIA in limiting GSC dissemination.
The inhibition of GSC migration is an intriguing
result, particularly because targeting the ability of GSC to
infiltrate distant brain areas, via brain parenchyma or brain
vessels (2), is possibly the
primary issue in limiting glioma malignancy. This issue has been
investigated previously (33)
Although we did not investigate in detail the molecular mechanisms
involved in the observed inhibition of cell migration induced by
1a-RGD, a previous study identified that an RGD-integrin antagonist
and a blocking antibody directed against αβ subunits decreased the
pro-migratory effect elicited by transforming growth factor β
(TGF-β) in human glioma cell lines (34). This mechanism is possibly the one
that occurred in our experimental model: In other experiments of
the present study not described, GSC were identified to express
abundant levels of TGF-β and TGF-β-receptor mRNAs, suggesting the
existence of an autocrine TGF-β-dependent mechanism mediating
several important cellular effects.
In this scenario, we hypothesize that, in our model,
1a-RGD may therefore decrease cell migration by inhibiting the
release of free active TGF-β from its ternary complex, as has been
identified to occur in other cellular systems (35).
In conclusion, the results of the present study
indicate that GSC grown under adherent conditions are a suitable
model for investigating the interactions between integrins and SMIA
as well as the molecular mechanisms that underlie the functional
consequences elicited by this interaction. In addition, the results
of the present study identified that the integrin antagonist 1a-RGD
induces detachment-mediated caspase-dependent anoikis and markedly
inhibits the migration of GSC, supporting the possibility, to be
investigated in future studies in in vivo and in
vitro models, that SMIA may be a useful tool for promoting
anoikis in GSC, and thus for limiting the intracranial
dissemination of glioma cells.
Funding
The present study was supported by the Italian
Ministry for University and Research by Project of Relevant
National Interest 2015 (grant no. 20157WW5EH_006).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
MP designed the research project and performed part
of the experiments; MCG performed part of the experiments and
contributed to the experimental design; AD isolated GSC from glioma
specimen and characterized the cells performing tumorigenicity and
immunocytochemistry experiments; EC performed flow cytometry
experiments; MS and LC synthesized 1a-RGD; SS co-ordinated the
research activity, contributed to the experimental design and was
involved in drafting the manuscript and revising it critically.
Ethics approval and consent to
participate
Tumor tissues were obtained from the Neurosurgery
Department at the Institute for Research, Hospitalization and
Care-University Hospital (IRCCS) San Martino-Cancer Research
Institute (IST) (Genoa, Italy) following informed consent,
according to European Union legislation on informed consent and The
Declaration of Helsinki, from the patients and Institutional
Ethical Committee (IRCCS San Martino-IST) approval. All experiments
involving animals were performed at IRCCS-AOU San Martino-IST in
compliance with the guidelines approved by the Italian Ministry of
Health and the Committee for Animal Well-Being in Cancer
Research.
Patient consent for publication
Not applicable.
Competing interests
The authors have no conflict of interests to
declare.
Acknowledgments
Not applicable.
References
|
1
|
Pollard SM, Yoshikawa K, Clarke ID, Danovi
D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, et
al: Glioma stem cell lines expanded in adherent culture have
tumor-specific phenotypes and are suitable for chemical and genetic
screens. Cell Stem Cell. 4:568–580. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paolillo M, Serra M and Schinelli S:
Integrins in glioblastoma: Still an attractive target? Pharmacol
Res. 113A:55–61. 2016. View Article : Google Scholar
|
|
3
|
Russo MA, Paolillo M, Sanchez-Hernandez Y,
Curti D, Ciusani E, Serra M, Colombo L and Schinelli S: A
small-molecule RGD-integrin antagonist inhibits cell adhesion, cell
migration and induces anoikis in glioblastoma cells. Int J Oncol.
42:83–92. 2013. View Article : Google Scholar :
|
|
4
|
Panzeri S, Zanella S, Arosio D, Vahdati L,
Dal Corso A, Pignataro L, Paolillo M, Schinelli S, Belvisi L,
Gennari C, et al: Cyclic isoDGR and RGD peptidomimetics containing
bifunctional diketopiperazine scaffolds are integrin antagonists.
Chemistry. 21:6265–6271. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng NC, van Zandwijk N and Reid G:
Cilengitide inhibits attachment and invasion of malignant pleural
mesothelioma cells through antagonism of integrins αvβ3 and αvβ5.
PLoS One. 9:e903742014. View Article : Google Scholar
|
|
6
|
Mikheev AM, Mikheeva SA, Trister AD,
Tokita MJ, Emerson SN, Parada CA, Born DE, Carnemolla B, Frankel S,
Kim DH, et al: Periostin is a novel therapeutic target that
predicts and regulates glioma malignancy. Neuro-oncol. 17:372–382.
2015. View Article : Google Scholar :
|
|
7
|
Arosio D, Belvisi L, Colombo L, Colombo M,
Invernizzi D, Manzoni L, Potenza D, Serra M, Castorina M, Pisano C,
et al: A potent integrin antagonist from a small library of cyclic
RGD pentapeptide mimics including benzyl-substituted
azabicycloalkane amino acids. ChemMedChem. 3:1589–1603. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Carra E, Barbieri F, Marubbi D, Pattarozzi
A, Favoni RE, Florio T and Daga A: Sorafenib selectively depletes
human glioblastoma tumor-initiating cells from primary cultures.
Cell Cycle. 12:491–500. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee KH, Ahn EJ, Oh SJ, Kim O, Joo YE, Bae
JA, Yoon S, Ryu HH, Jung S, Kim KK, et al: KITENIN promotes glioma
invasiveness and progression, associated with the induction of EMT
and stemness markers. Oncotarget. 6:3240–3253. 2015.PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Δ Δ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
11
|
Hadjimichael C, Chanoumidou K,
Papadopoulou N, Arampatzi P, Papamatheakis J and Kretsovali A:
Common stemness regulators of embryonic and cancer stem cells.
World J Stem Cells. 7:1150–1184. 2015.PubMed/NCBI
|
|
12
|
Desgrosellier JS and Cheresh DA: Integrins
in cancer: Biological implications and therapeutic opportunities.
Nat Rev Cancer. 10:9–22. 2010. View
Article : Google Scholar
|
|
13
|
Silginer M, Weller M, Ziegler U and Roth
P: Integrin inhibition promotes atypical anoikis in glioma cells.
Cell Death Dis. 5:e10122014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mizushima N and Yoshimori T: How to
interpret LC3 immunob-lotting. Autophagy. 3:542–545. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Seguin L, Desgrosellier JS, Weis SM and
Cheresh DA: Integrins and cancer: Regulators of cancer stemness,
metastasis, and drug resistance. Trends Cell Biol. 25:234–240.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Buchheit CL, Weigel KJ and Schafer ZT:
Cancer cell survival during detachment from the ECM: Multiple
barriers to tumour progression. Nat Rev Cancer. 14:632–641. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Beier D, Schulz JB and Beier CP:
Chemoresistance of glioblastoma cancer stem cells--much more
complex than expected. Mol Cancer. 10:1282011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cosset É, Ilmjärv S, Dutoit V, Elliott K,
von Schalscha T, Camargo MF, Reiss A, Moroishi T, Seguin L, Gomez
G, et al: Glut3 addiction is a druggable vulnerability for a
molecularly defined subpopulation of glioblastoma. Cancer Cell.
32:856–868.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Christmann M, Diesler K, Majhen D,
Steigerwald C, Berte N, Freund H, Stojanović N, Kaina B, Osmak M,
Ambriović-Ristov A, et al: Integrin αVβ3 silencing sensitizes
malignant glioma cells to temozolomide by suppression of homologous
recombination repair. Oncotarget. 8:27754–27771. 2017. View Article : Google Scholar
|
|
20
|
Rahman M, Reyner K, Deleyrolle L, Millette
S, Azari H, Day BW, Stringer BW, Boyd AW, Johns TG, Blot V, et al:
Neurosphere and adherent culture conditions are equivalent for
malignant glioma stem cell lines. Anat Cell Biol. 48:25–35. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lange SC, Bak LK, Waagepetersen HS,
Schousboe A and Norenberg MD: Primary cultures of astrocytes: Their
value in understanding astrocytes in health and disease. Neurochem
Res. 37:2569–2588. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Malik N, Wang X, Shah S, Efthymiou AG, Yan
B, Heman-Ackah S, Zhan M and Rao M: Comparison of the gene
expression profiles of human fetal cortical astrocytes with
pluripotent stem cell derived neural stem cells identifies human
astrocyte markers and signaling pathways and transcription factors
active in human astrocytes. PLoS One. 9:e961392014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lundin A, Delsing L, Clausen M, Ricchiuto
P, Sanchez J, Sabirsh A, Ding M, Synnergren J, Zetterberg H, Brolén
G, et al: Human iPS-derived astroglia from a stable neural
precursor state show improved functionality compared with
conventional astrocytic models. Stem Cell Reports. 10:1030–1045.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Sloan SA, Clarke LE, Caneda C,
Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G,
et al: Purification and characterization of progenitor and mature
human astrocytes reveals transcriptional and functional differences
with mouse. Neuron. 89:37–53. 2016. View Article : Google Scholar :
|
|
25
|
Lee J, Kotliarova S, Kotliarov Y, Li A, Su
Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al:
Tumor stem cells derived from glioblastomas cultured in bFGF and
EGF more closely mirror the phenotype and genotype of primary
tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Leblond P, Dewitte A, Le Tinier F,
Bal-Mahieu C, Baroncini M, Sarrazin T, Lartigau E, Lansiaux A and
Meignan S: Cilengitide targets pediatric glioma and neuroblastoma
cells through cell detachment and anoikis induction. Anticancer
Drugs. 24:818–825. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Oliveira-Ferrer L, Hauschild J, Fiedler W,
Bokemeyer C, Nippgen J, Celik I and Schuch G: Cilengitide induces
cellular detachment and apoptosis in endothelial and glioma cells
mediated by inhibition of FAK/src/AKT pathway. J Exp Clin Cancer
Res. 27:862008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Antonioli M, Di Rienzo M, Piacentini M and
Fimia GM: Emerging mechanisms in initiating and terminating
autophagy. Trends Biochem Sci. 42:28–41. 2017. View Article : Google Scholar
|
|
29
|
Bianconi D, Unseld M and Prager GW:
Integrins in the spotlight of cancer. Int J Mol Sci. 17:2037–2064.
2016. View Article : Google Scholar
|
|
30
|
Xia H, Nho RS, Kahm J, Kleidon J and Henke
CA: Focal adhesion kinase is upstream of phosphatidylinositol
3-kinase/ Akt in regulating fibroblast survival in response to
contraction of type I collagen matrices via a beta 1 integrin
viability signaling pathway. J Biol Chem. 279:33024–33034. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Beauséjour M, Thibodeau S, Demers MJ,
Bouchard V, Gauthier R, Beaulieu JF and Vachon PH: Suppression of
anoikis in human intestinal epithelial cells: Differentiation
state-selective roles of α2β1, α3β1, α5β1, and α6β4 integrins. BMC
Cell Biol. 14:532013. View Article : Google Scholar
|
|
32
|
Alanko J, Mai A, Jacquemet G, Schauer K,
Kaukonen R, Saari M, Goud B and Ivaska J: Integrin endosomal
signalling suppresses anoikis. Nat Cell Biol. 17:1412–1421. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Niola F, Zhao X, Singh D, Sullivan R,
Castano A, Verrico A, Zoppoli P, Friedmann-Morvinski D, Sulman E,
Barrett L, et al: Mesenchymal high-grade glioma is maintained by
the ID-RAP1 axis. J Clin Invest. 123:405–417. 2013. View Article : Google Scholar :
|
|
34
|
Platten M, Wick W, Wild-Bode C, Aulwurm S,
Dichgans J and Weller M: Transforming growth factors beta(1)
(TGF-beta(1)) and TGF-beta(2) promote glioma cell migration via
Up-regulation of alpha(V)beta(3) integrin expression. Biochem
Biophys Res Commun. 268:607–611. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Parvani JG, Galliher-Beckley AJ, Schiemann
BJ and Schiemann WP: Targeted inactivation of β1 integrin induces
β3 integrin switching, which drives breast cancer metastasis by
TGF-β. Mol Biol Cell. 24:3449–3459. 2013. View Article : Google Scholar : PubMed/NCBI
|