Introduction
Retinoblastoma (RB), is the most common type of
primary ocular malignant tumor in infants, with a worldwide
incidence of ~1/20,000 cases (1).
Up to 1,000 new RB cases are diagnosed each year in China, which
accounts for ~20% of cases worldwide (2). Notably, pediatric patients are likely
to succumb within the 1–2 years following RB onset if no treatments
are received (3). To relieve the
symptoms of RB, various therapies have been developed, in
particular, chemotherapy followed by adjuvant therapy has been
gradually prioritized considering the severe complications caused
by other treatment strategies (4,5).
However, as drug resistance is a major reason for treatment failure
or RB recurrence, numerous studies have attempted to identify novel
biomarkers for RB (6–8).
It has been documented that the expression of
various microRNAs (miRNAs/miRs) are abnormally expressed within
neoplastic tissues and cells (9).
Among them, miR-34a was demonstrated to induce cell apoptosis,
delay cell cycle progression and encourage cell senescence, which
are considered important characteristics of tumor onset and
aggravation (10–13). Furthermore, miR-34 was demonstrated
to facilitate the onset and development of colorectal cancer
through regulating DNA methylation (14). This miRNA also blocks epidermal
growth factor receptor (EGFR) signaling by modifying
phosphoinositide 3-kinase, ultimately contributing to the
inhibition of gastric cancer progression (15). Notably, downregulated miR-34a
expression induces the multiplication of RB cells (16), though the underlying mechanisms
remain unclear.
Numerous studies have confirmed that miR-34a serves
an important role in regulating tumor cell sensitivity to
chemotherapeutic drugs, and this effect may be primarily associated
with the post-transcriptional regulation of oncogenes by miR-34a
(17–19). For instance, Vinall et al
(20) reported that miR-34a
increases the sensitivity of bladder cancer cells to cisplatin by
inhibiting the expression levels of cyclin dependent kinase 6 and
sirtuin 1. It has also suggested that miR-34a may affect the
expression of various drug-resistant proteins within tumor cells by
altering the Notch1 signaling pathway (21) Notably, the combined effect of
miR-34a and melanoma antigen-A (MAGE-A) on the chemosensitivity of
tumor cells has been confirmed (22). Furthermore, the MAGE-A family
proteins have been demonstrated to be involved in guide cell
carcinogenesis by interfering with the p53 signaling pathway and
thereby reduce the sensitivity of tumor cells to chemotherapeutic
drugs (23,24). Nevertheless, few studies have
focused on the associations among miR-34a, MAGE-A and p53
underlying the chemosensitivity of RB (1).
The present study aimed to investigate the
involvement of the miR-34a/MAGE-A/p53 signaling pathway on the
sensitivity of RB to chemotherapeutic drugs using tissue samples
and in vitro experiments, which may provide a basis for
clinically diagnosing and treating RB in the future.
Materials and methods
Collection of tissues
A total of 293 RB and adjacent tumor tissue samples
were collected from the Department of Ophthalmology, The First
Affiliated Hospital of Zhengzhou University between April 2015 and
January 2017. The mean age of patients was 23.64±17.04 months
(11–61 months), with 155 females and 138 males. Notably, only
infants <6 years old were included, and patients with family
heredity were excluded. All the tissues were fixed with 4%
paraformaldehyde at room temperature for 30 min, and were
surgically diagnosed as RB by ≥2 senior pathologists who were
blinded to the clinicopathological features of patients. The
immediate relatives of all patients with RB provided written
informed consent, and the present study was approved by the Ethics
Committee of The First Affiliated Hospital of Zhengzhou
University.
Cultivation of RB cells and cell
transfection
Human RB cell lines, HXO-Rb44, SO-Rb50, Y79 and
WERI-Rb-1, were purchased from the American Type Culture Collection
(Manassas, VA, USA). The cells were cultured with RMPI-1640
supplemented with 10% fetal calf serum (both from Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a humidified
atmosphere with 5% CO2. The culture medium was replaced
every 3–4 days, and all cell lines were maintained within 20
generations. When the confluence of RB cells reached ~50%, the
Lipofectamine™ 2000 Liposome Transfection kit (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to transfect cells. The following
oligonucleotides were used: miR-34a mimics (40 nM) forward,
5′-UGGCAGUGUCUUAGCUGGUUGU-3′ and reverse,
5′-AACCAGCUAAGACACUGCCAUU-3′; miR-34a inhibitor (40 nM) forward,
5′-ACAACCAGCUAAGACACUGCCA-3′ and reverse,
5′-UGGCAGUGUCUUAGCUGGUUGU-3′; miR-negative control (NC) (40 nM)
forward, 5′-CAGUACUUUUGUGUAGUA CAA-3′ and reverse,
5′-UUGUACUACACAAAAGUACUG-3′; pcDNA3.1-MAGE-A (30 nM); small
interfering RNA (si)-MAGE-A 3/6/12 (30 nM) forward,
5′-CUACCUGGAGUACCGGCAG-3′ and reverse,
5′-UGGCAGUGUCUUAGCUGGUUGU-3′; pcDNA3.1-p53 (30 nM) or si-p53 (30
nM) forward, 5′-GAAGAAAAUUUCCGCAAAA-3′ and reverse, 5′-CUU
UUGCGGAAAUUUUCUUC-3′ (all from Guangzhou RiboBio Co., Ltd.,
Guangzhou, China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Tissues and cells were lysed with addition of 1 ml
TRIzol solution (Invitrogen; Thermo Fisher Scientific, Inc.), and
total RNA was extracted according to the manufacturer’s protocol.
The purity and concentration of RNA were determined using
spectrophotometry. Subsequently, the extracted RNA was reverse
transcribed into cDNA, and amplified using PCR according to the
PrimeScript one-step RT-qPCR kit (Takara Bio, Inc., Otsu, Japan)
protocol. The temperature protocol for RT was as follows: 16°C for
30 min; 42°C for 30 min; and 85°C for 5 min. ABI Primer Express
software (version 2.0; Guangzhou RiboBio Co., Ltd.) was used to
design the primers (Table I). The
primers were added to the 25 µl PCR reaction system, and the
reaction conditions were as follows: Pre-denaturation at 95°C for 2
min; and 30 cycles of denaturation at 94°C for 45 sec, renaturation
at 59°C for 45 sec and extension at 72°C for 30 sec. The expression
levels of miR-34a, MAGE-A and p53 were determined using a CFX96™
thermocycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and
quantified according to the comparative Cq method
(2−∆∆Cq) (4). U6 was
designated as the internal reference for miR-34a, and β-actin was
adopted as the internal reference for MAGE-A and p53.
 | Table IPrimers for miR-34a, MAGE-A, p53, U6
and β-actin used in RT-qPCR. |
Table I
Primers for miR-34a, MAGE-A, p53, U6
and β-actin used in RT-qPCR.
| Genes | Primer
sequence |
|---|
| miR-34a | |
| Forward |
5′-CGGTATCATTTGGCAGTGTCT-3′ |
| Reverse |
5′-GTGCAGGGTCCGAGGT-3′ |
| U6 | |
| Forward |
5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse |
5′-AACGCTTCACGAATTTGCGT-3′ |
| MAGE-A | |
| Forward |
5′-ATGGAGACTCAGTTCCGAGA-3′ |
| Reverse |
5′-AAGAACTTTCATCTTGCTGG-3′ |
| p53 | |
| Forward |
5′-CTGCCCTCAACAAGATGTTTTG-3′ |
| Reverse |
5′-CTATCTGAGCAGCGCTCATGG-3′ |
| β-actin | |
| Forward |
5′-CTTAGTTGCGTTACACCCTTTCTTG-3′ |
| Reverse |
5′-CTGTCACCTTCACCGTTCCAGTTT-3′ |
Luciferase reporter gene assay
The MAGE-A fragments that contained binding sites of
miR-34a were amplified by PCR, and were then cloned into pmirGLO
double luciferase expression vector (Promega Corporation, Madison,
WI, USA) to form MAGE-A-wt. Through a similar approach, MAGE-A-mut
was constructed, except that the binding sites of miR-34a within
MAGE-A were mutated. Subsequently, RB cells that were transfected
with MAGE-A-wt, MAGE-A-mut or pRL-TK reporter gene vector were
transfected with miR-34a mimic, miR-34a inhibitor or miR-NC using
Lipofectamine 2000. The resultant firefly luciferase and Renilla
luciferase activity were measured using a Double Luciferase
Reporter assay system (Promega Corporation) following 24 h of
transfection.
Western blotting
Total protein was extracted using
immunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) and quantified using the Bradford
method. Each sample (40 µg) was separated using 8% SDS-PAGE
and then transferred onto polyvinyli-dene fluoride membranes.
Following blocking with 50 g/l skim milk powder for 1.5 h at room
temperature, bovine serum albumin was added to diluted primary
antibodies against MAGE-A (mouse anti-human; 1:500; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA; cat. no. sc-20034) and p53
(mouse anti-human; 1:500; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany; cat. no. P5813), E-cadherin, N-cadherin, vimentin, snail
(mouse anti-human; 1:1,000; Abcam, Cambridge, UK; cat. nos.
ab76055, ab98952, ab8978 and ab82846, respectively) and GAPDH
(rabbit anti-human; 1:1,000; Santa Cruz Biotechnology, Inc.; cat.
no. sc-47724). After the samples were incubated at 4°C overnight,
the membranes were washed with 1 ml/l TBS-Tween-20 three times.
Then, the rabbit anti-mouse IgG H&L (DyLight® 550)
preadsorbed secondary antibody (1:5,000; Abcam; cat. no. ab98786)
was added, and the membranes were incubated at room temperature for
2 h. Finally, GelDoc2000 Enhanced Chemiluminescence reagent
(Bio-Rad Laboratories, Inc.) was used for development in the
darkroom, and the expression levels of the aforementioned proteins
were assessed using ImageJ software (version 1.46; National
Institutes of Health, Bethesda, MD, USA) was employed to analyze
the protein bands.
Evaluation of drug susceptibility
A 100 µl cell suspension at the density of
4×103/ml was added into each well of the 96-well plates.
When cells adhered to the wall, the following drug treatments were
performed: Vincristine (0.05, 0.10, 0.50, 1.00 and 5.00
µg/ml), etoposide (0.5, 1.0, 5.0, 10.0 and 50.0
µg/ml), carboplatin (1.5, 7.5, 15.0, 75.0 and 150.0
µg/ml), and Adriamycin (1.0, 5.0, 10.0, 50.0 and 100.0
µg/ml) (all from Sigma-Aldrich; Merck KGaA). Subsequently,
the samples were agitated and cultured at 37°C for 48 h in 5%
CO2. Then, 0.5% MTT (10 µl) was added to each
well and incubated at 37°C for 4–6 h in 5% CO2.
Following the removal of the supernatant from the wells, each well
was supplemented with 100 µl acidulated isopropanol.
Following culturing at room temperature for 15–30 min, the samples
were detected based on the optical density (OD) values at 570 nm
using a Freedom Evolyzer-2200 Enzyme-Linked Immunometric meter
(Tecan Group Ltd., Männedorf, Switzerland). Finally, the inhibition
rate (IR, %) was calculated according to the formula as follows:
[1− (ODmedicine / ODcontrol)] × 100. The drug
concentration that caused death of half of the cells also known as
the half maximal inhibitory concentration (IC50), was
also calculated.
Cell apoptosis analysis
The cells were washed with 0.01 mol/l PBS, and were
centrifuged at 1,500 × g for 5 min at 4°C. The supernatant was
discarded, and 500 µl 1X binding buffer (Thermo Fisher
Scientific, Inc.) was added to adjust the concentration of cells to
1×106/ml. Subsequently, the cells in each tube were
mixed with 500 µl cell suspension, 5 µl Annexin
V-fluorescein isothiocyanate and 10 µl propidium iodide
(Invitrogen; Thermo Fisher Scientific, Inc.). Following the
incubation of the cells at room temperature for 10 min, the samples
were analyzed using a FACSCalibur flow cytometer with CellQuest
software (version 3.3; both from BD Biosciences, Franklin Lakes,
NJ, USA).
Statistical analysis
All data were analyzed using SPSS 13.0 statistical
software (SPSS, Inc., Chicago, IL, USA). The results are expressed
as the mean ± standard deviation. All the experiments were repeated
for at least three times. Differences between two groups were
analyzed using the Student’s t-test, and those of ≥3 groups were
compared using one-way analysis of variance with the
Student-Newman-Keuls post hoc test. As for the categorical data (n,
%), the χ2 test was used for analysis. Spearman’s
correlation analysis was also performed to assess the associations
among miR-34a, MAGE-A and p53 expression. The Kaplan-Meier
estimator method was applied to calculate overall survival of the
studied population, and the log-rank test was used to evaluate the
significance of differences. Cox regression model was used to
perform univariate and multivariate analyses. Furthermore, in order
to investigate the role of miR-34a and MAGE-A underlying RB
etiology, their binding sites were predicted using the Targetscan
database (http://www.targetscan.org/).
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-34a, MAGE-A and p53 expression levels
in human RB tissues and cell lines
RT-qPCR was used to detect the expression levels of
MAGE-A, p53 and miR-34a within human RB tissues and paracarcinoma
tissues. MAGE-A and p53 expression levels within RB tissues were
significantly upregulated compared with paracarcinoma tissues
(P<0.01); however, the expression of miR-34a was significantly
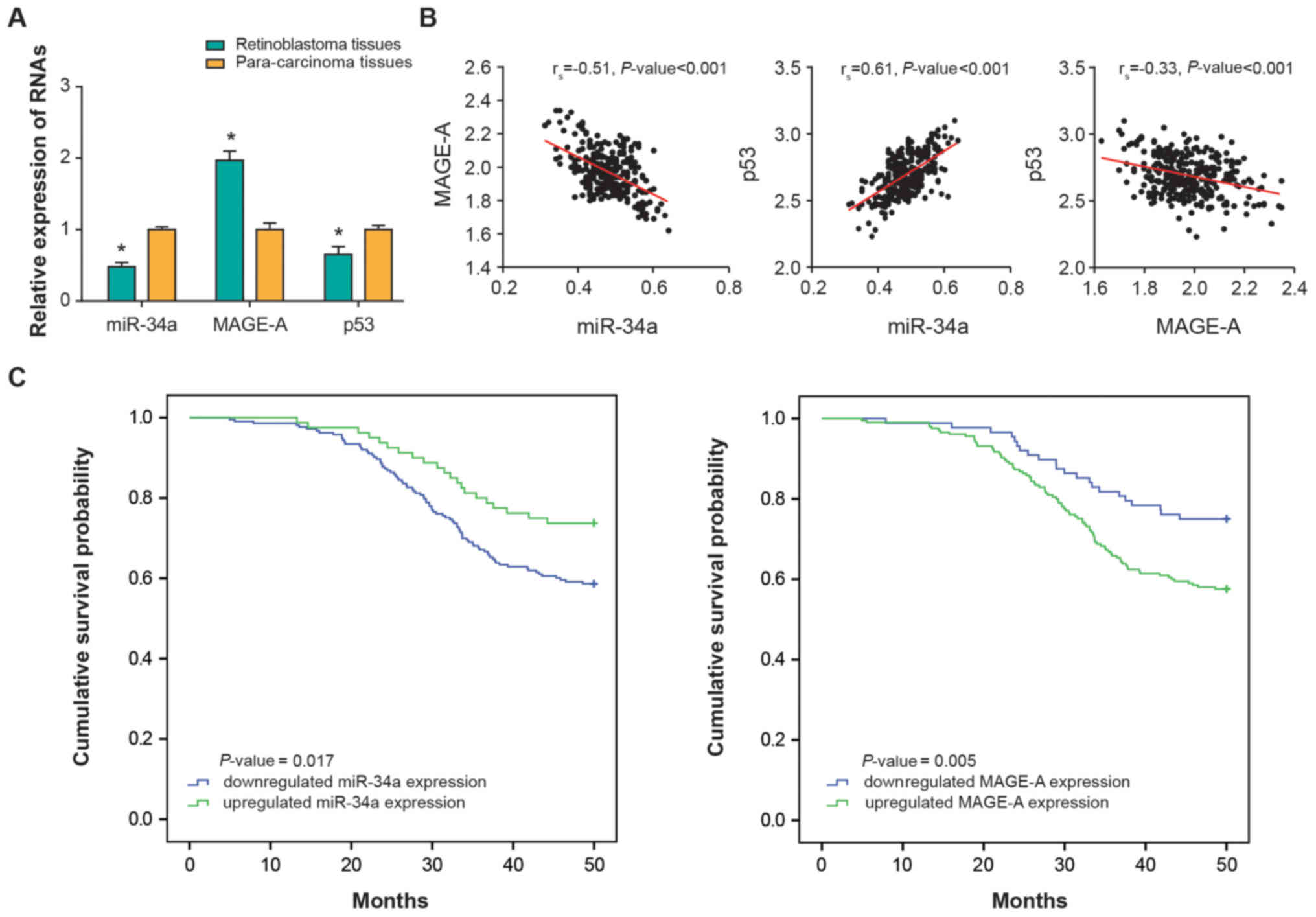
downregulated in RB tissues (P<0.01) (Fig. 1A). Further analyses demonstrated
that miR-34a expression was negatively correlated with MAGE-A
expression in the investigated RB tissues (rs −0.51;
P<0.001), and positively correlated with p53 expression
(rs, 0.61; P<0.001) (Fig. 1B). In addition, a negative
correlation was identified between MAGE-A and p53 expression levels
among the collected RB samples (rs, −0.33;
P<0.001).
The patients with RB were divided into the high
miR-34a expression (≥0.48) and low miR-34a expression groups
(<0.48) (Table II). Patients
were also divided into high and low MAGE-A expression groups using
the mean overall expression (1.97) as a threshold. It was indicated
that low miR-34a and high MAGE-A expression were significantly
associated with higher stages (D-E) of the International
Classification of Retinoblastoma (ICRB) system (5) (χ2, 4.67; P=0.031;
χ2, 58.92; P<0.001), poor differentiation
(χ2, 7.76; P=0.005; χ2, 8.87; P=0.003),
positive choroidal invasion (χ2, 6.21; P=0.013;
χ2, 4.53; P=0.033) and positive optic nerve involvement
(χ2, 7.92; P=0.005; χ2, 9.52; P=0.002), when
compared with the high miR-34a and low MAGE-A expression groups
(Table II). However, no
significant associations were identified between the two genes, and
age, sex, family history or eye effected. In addition, Kaplan-Meier
estimator analyses demonstrated that patients with RB with low
miR-or high MAGE-A expression were associated with significantly
lower overall survival rates compared with the high miR-34a or low
MAGE-A expression groups (P<0.05; Fig. 1C). Multivariate Cox regression
analysis demonstrated that low miR-34a expression [hazard ratio
(HR), 2.10; 95% confidence interval (CI), 1.10–4.02; P=0.025], high
MAGE-A expression (HR, 2.13; 95% CI, 1.04–4.17; P=0.037), stage D-E
of the ICRB system (HR, 1.89; 95% CI, 1.04–3.45; P=0.037), poor
differentiation (HR, 1.75; 95% CI, 1.02–2.94; P=0.040) and optic
nerve involvement (HR, 1.92; 95% CI, 1.11–3.23; P=0.019) were
independent prognostic factors for RB (Table III).
 | Table IIAssociation between microRNA-34a and
MAGE-A expression levels, and the clinical characteristics of
patients with RB. |
Table II
Association between microRNA-34a and
MAGE-A expression levels, and the clinical characteristics of
patients with RB.
|
Characteristics | microRNA-34a
expression
| MAGE-A expression
|
|---|
| Low | High | χ2 | P-value | Low | High | χ2 | P-value |
|---|
| Total (n=293) | 213 | 80 | | | 88 | 205 | | |
| Age, months | | | | | | | | |
| <30 | 137 | 57 | 1.25 | 0.264 | 60 | 134 | 0.22 | 0.640 |
| ≥30 | 76 | 23 | | | 28 | 71 | | |
| Sex | | | | | | | | |
| Female | 112 | 43 | 0.03 | 0.858 | 45 | 110 | 0.16 | 0.692 |
| Male | 101 | 37 | | | 43 | 95 | | |
| Family history | | | | | | | | |
| Negative | 175 | 71 | 1.88 | 0.171 | 77 | 169 | 1.17 | 0.279 |
| Positive | 38 | 9 | | | 11 | 36 | | |
| Eye affected | | | | | | | | |
| Right | 105 | 45 | 1.13 | 0.289 | 46 | 104 | 0.06 | 0.809 |
| Left | 108 | 35 | | | 42 | 101 | | |
| ICRB staging
system | | | | | | | | |
| Group A-C | 82 | 42 | 4.67 | 0.031 | 67 | 57 | 58.92 | <0.001 |
| Group D-E | 131 | 38 | | | 21 | 148 | | |
| Degree of
differentiation | | | | | | | | |
| Well and
moderately | 122 | 60 | 7.76 | 0.005 | 66 | 116 | 8.87 | 0.003 |
| Poorly | 91 | 20 | | | 22 | 89 | | |
| Choroidal
invasion | | | | | | | | |
| Negative | 101 | 51 | 6.21 | 0.013 | 54 | 98 | 4.53 | 0.033 |
| Positive | 112 | 29 | | | 34 | 107 | | |
| Scleral
invasion | | | | | | | | |
| Negative | 193 | 78 | 3.98 | 0.046 | 84 | 187 | 1.59 | 0.207 |
| Positive | 20 | 2 | | | 4 | 18 | | |
| Optic nerve
involvement | | | | | | | | |
| Negative | 86 | 47 | 7.92 | 0.005 | 52 | 81 | 9.52 | 0.002 |
| Positive | 127 | 33 | | | 36 | 124 | | |
 | Table IIIAssociation between clinical
characteristics and the overall survival of patients with RB. |
Table III
Association between clinical
characteristics and the overall survival of patients with RB.
|
Characteristics | Univariate analysis
| Multivariate
analysis
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| miRNA-34a
expression | | | | | | |
| Low vs. high | 2.57 | 1.42–4.64 | 0.002 | 2.1 | 1.10–4.02 | 0.025 |
| MAGE-A
expression | | | | | | |
| Low vs. high | 3.45 | 1.89–6.25 | <0.001 | 2.13 | 1.04–4.17 | 0.037 |
| Age, months | | | | | | |
| <30 vs.
≥30 | 1.13 | 0.68–1.87 | 0.64 | 1.33 | 0.76–2.33 | 0.320 |
| Sex | | | | | | |
| Female vs.
male | 0.96 | 0.60–1.55 | 0.873 | 1.03 | 0.61–1.75 | 0.905 |
| Family history | | | | | | |
| Positive vs.
negative | 0.76 | 0.39–1.47 | 0.414 | 0.56 | 0.27–1.19 | 0.131 |
| Eye affected | | | | | | |
| Right vs.
left | 0.85 | 0.53–1.36 | 0.498 | 0.93 | 0.55–1.58 | 0.794 |
| ICRB staging
system | | | | | | |
| Group D-E vs.
group A-C | 2.78 | 1.64–4.55 | <0.001 | 1.89 | 1.04–3.45 | 0.037 |
| Degree of
differentiation | | | | | | |
| Poorly vs.
well/moderately | 2.33 | 1.43–3.85 | 0.001 | 1.75 | 1.02–2.94 | 0.040 |
| Choroidal
invasion | | | | | | |
| Positive vs.
negative | 1.96 | 1.22–3.23 | 0.005 | 1.64 | 0.97–2.78 | 0.065 |
| Scleral
invasion | | | | | | |
| Positive vs.
negative | 1.75 | 0.75–4.17 | 0.201 | 1.19 | 0.45–3.13 | 0.730 |
| Optic nerve
involvement | | | | | | |
| Positive vs.
negative | 2.38 | 1.47–4.00 | 0.001 | 1.92 | 1.11–3.23 | 0.019 |
Sensitivities to carboplatin, etoposide,
Adriamycin and vincristine in HXO-Rb44, SO-Rb50, Y79 and WERI-RB1
cell lines
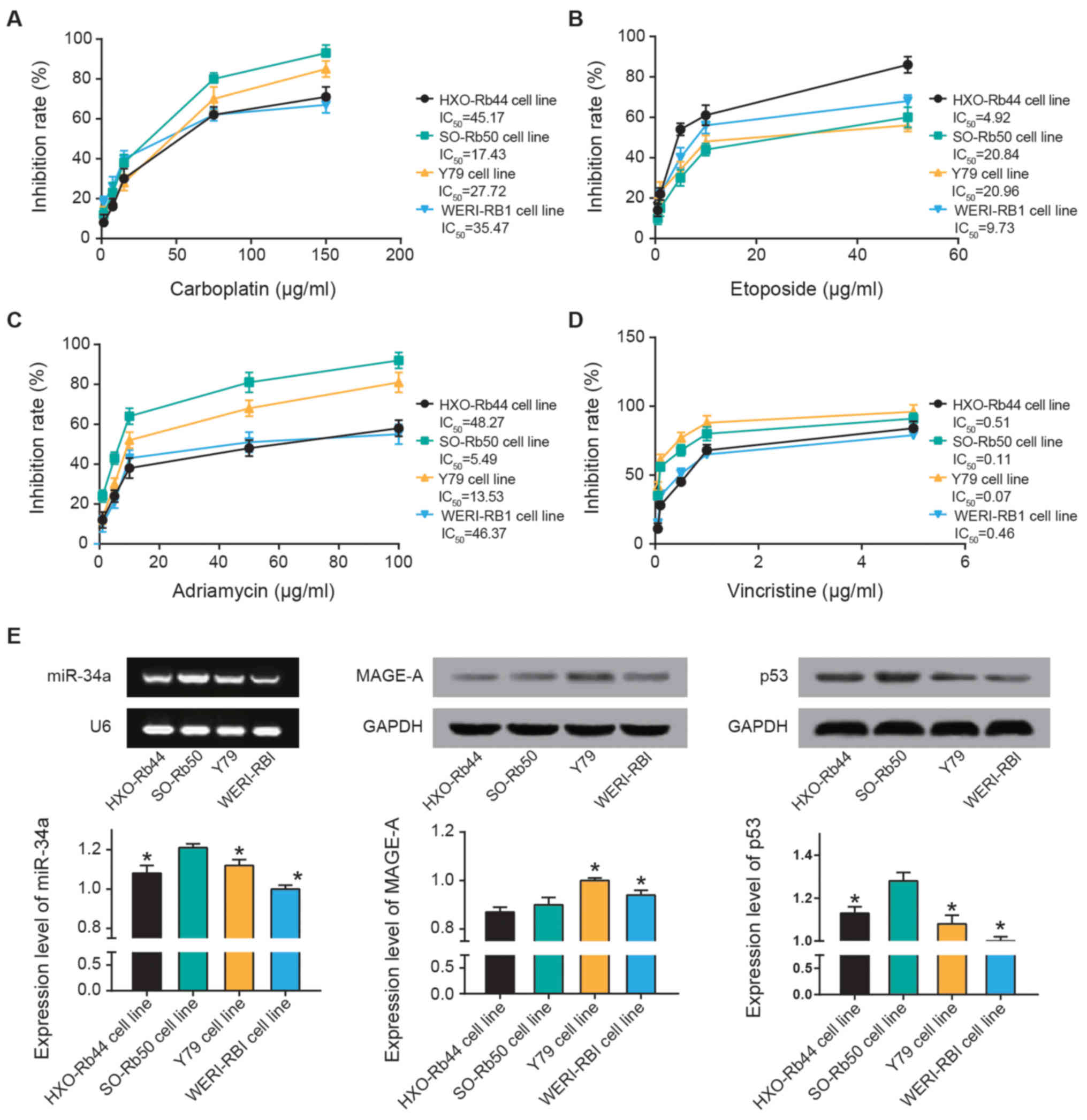
The results of the MTT assay revealed that SO-Rb50
cells exhibited a maximal sensitivity to carboplatin
(IC50, 17.43 µg/ml) and Adriamycin
(IC50, 5.49 µg/ml) (Fig. 2A-C). Furthermore, the inhibitory
response to etoposide were ranked as follows: Y79 (IC50,
20.96 µg/ml) > SO-Rb50 (IC50, 20.84
µg/ml) > WERI-RB1 (IC50, 9.73 µg/ml)
> HXO-Rb44 (IC50, 4.92 µg/ml) (Fig. 2B). Y79 (IC50, 0.07
µg/ml) and SO-Rb50 (IC50, 0.0.11 µg/ml)
exhibited higher sensitivity to vincristine compared with WERI-RBI
(IC50, 0.46 µg/ml) and HXO-Rb44 (IC50,
0.51 µg/ml) (Fig. 2D).
Furthermore, miR-34a and p53 expression levels were highest in
SO-Rb50 cells out of all four cell lines (P<0.05; Fig. 2E). MAGE-A expression in HXO-Rb50
and SO-Rb50 were the lowest among all the cell lines (P<0.05).
Thus, SO-Rb50 was selected for the following experiments.
miR-34a and MAGE-A regulate the viability
and apoptosis of RB cells
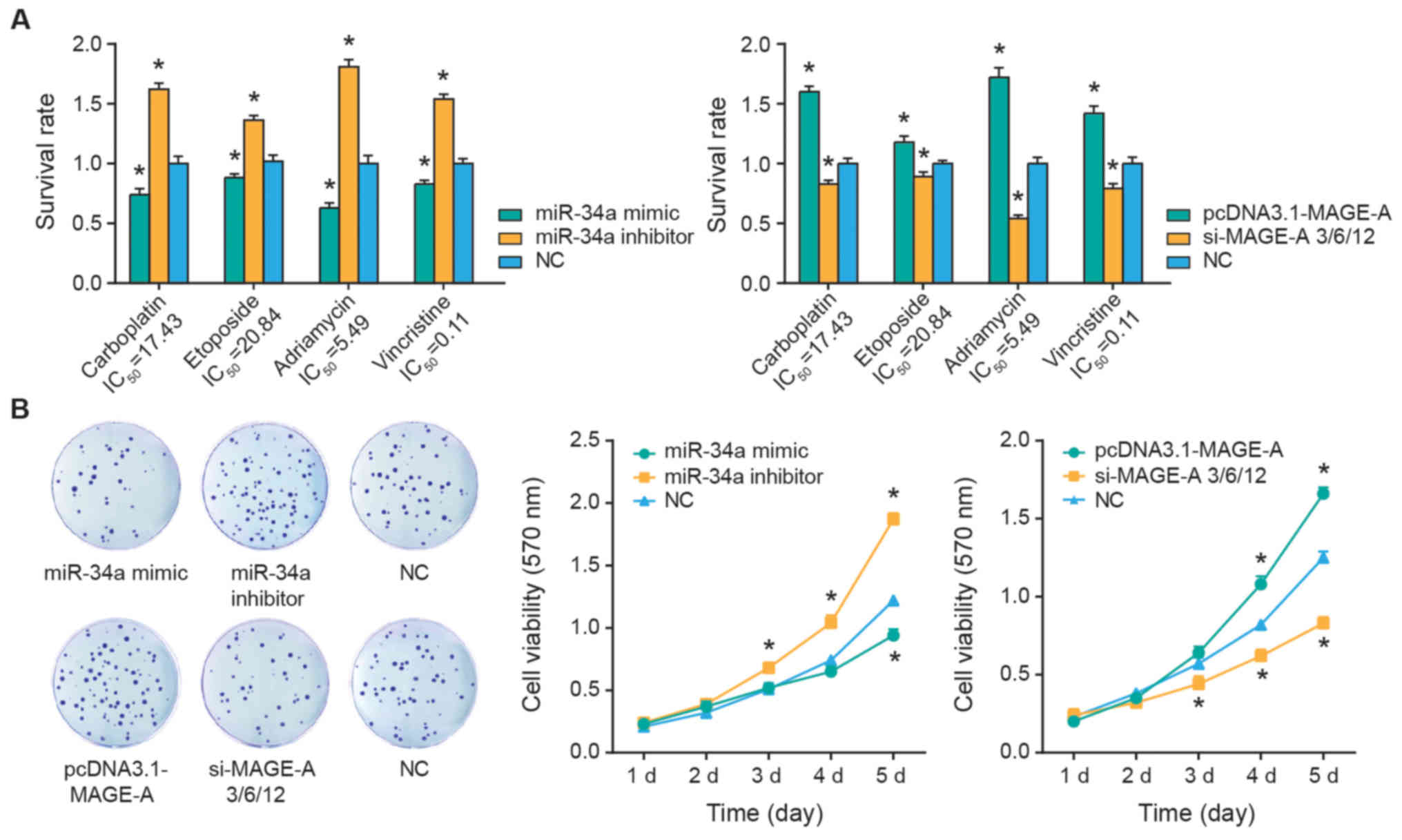
The SO-Rb50 cells were treated with 17.43
µg/ml carboplatin, 20.84 µg/ml etoposide, 5.49
µg/ml Adriamycin or 0.11 µg/ml vincristine. It was
demonstrated that the survival rates of SO-Rb50 were significantly
inhibited following miR-34a mimics transfection (P<0.05), while
silencing of miR-34a appeared to significantly increase the
survival rate of SO-Rb50 cells compared with the NC group
(P<0.05) (Fig. 3A). Contrary to
the results of miR-34a, upregulation of MAGE-A expression was
associated with significantly increased survival rates of SO-Rb50
cells in response to carboplatin, etoposide, Adriamycin and
vincristine treatment, when compared with the NC group (all
P<0.05; Fig. 3A). Concurrently,
transfection of si-MAGE-A significantly suppressed the survival
rate of SO-Rb50 cells under all four treatment conditions, when
compared with the NC group (all P<0.05). In addition, the
viability of SO-Rb50 cells was significantly attenuated in the
miR-34a mimics and si-MAGE-A groups (P<0.05), while it was
significantly improved in the miR-34a inhibitor and pcDNA3-MAGE-A
groups compared with the NC group (all P<0.05; Fig. 3B).
miR-34a and MAGE-A regulate epithelial
mesenchymal transition (EMT)-specific proteins within RB cells
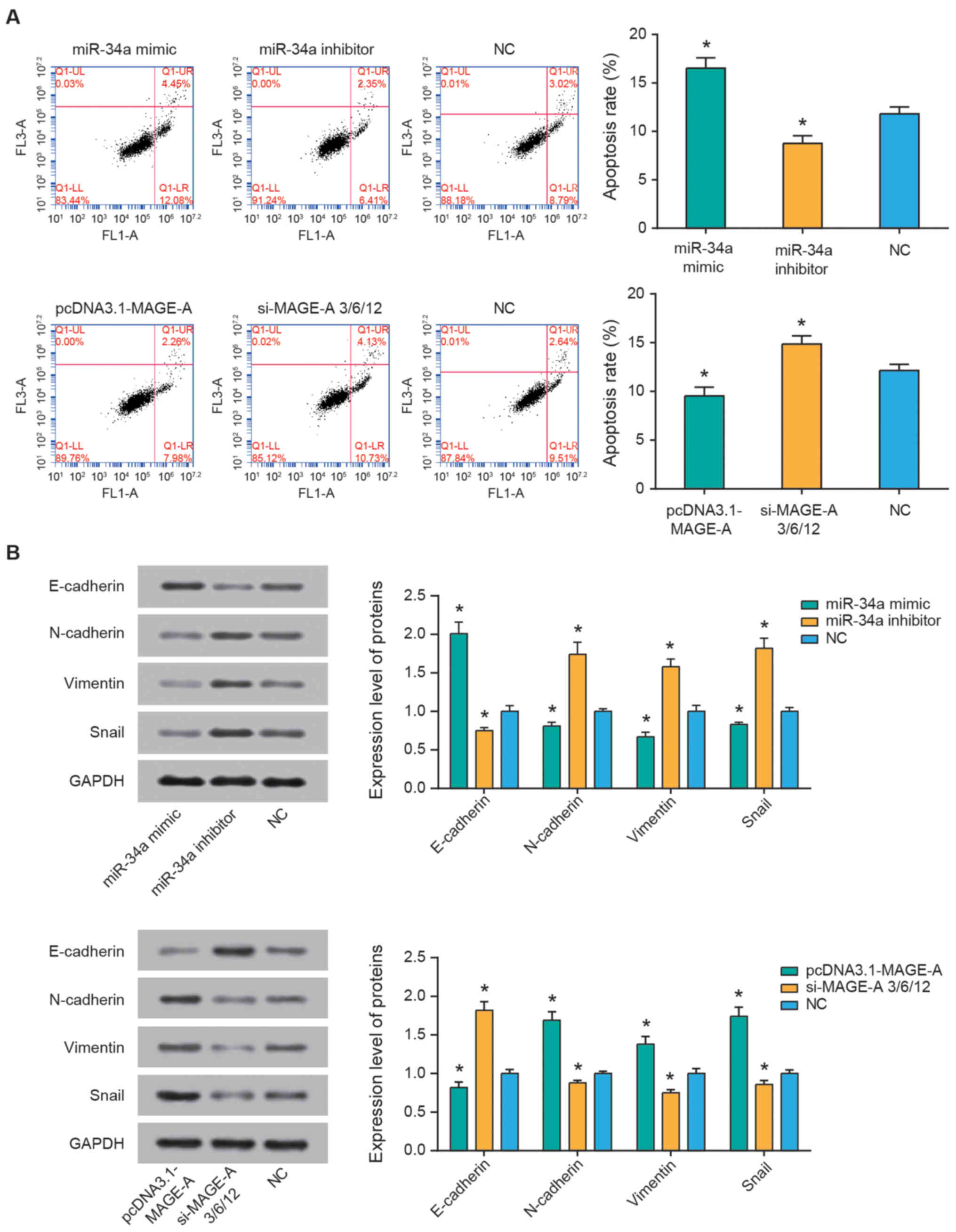
Flow cytometry was performed to assess the
percentage of apoptotic cells and explore the effect of miR-34a and
MAGE-A on the apoptotic conditions of SO-Rb50 cells. The results
revealed that miR-34a inhibitor and pcDNA3.1-MAGE-A groups
significantly reduced the apoptotic rate of SO-Rb50 cells
(P<0.05), and the apoptotic rate of miR-34a mimics and si-MAGE-A
groups was significantly higher compared with that of the NC group
(all P<0.05; Fig. 4A). In
addition, as demonstrated in Fig.
4B, miR-34a mimics transfection resulted in increased
E-cadherin expression, along with decreased N-cadherin, vimentin
and snail expression (all P<0.05), whereas miR-34a inhibitor
resulted in the opposite observations (all P<0.05). In addition,
treatment with pcDNA3.1-MAGE-A reduced E-cadherin, and increased
N-cadherin, vimentin and snail expression levels in SO-Rb50 cells
compared with the NC group (all P<0.05). Concurrently, silencing
of MAGE-A caused an increase in E-cadherin expression, and a
reduction in N-cadherin, vimentin and snail expression levels (all
P<0.05).
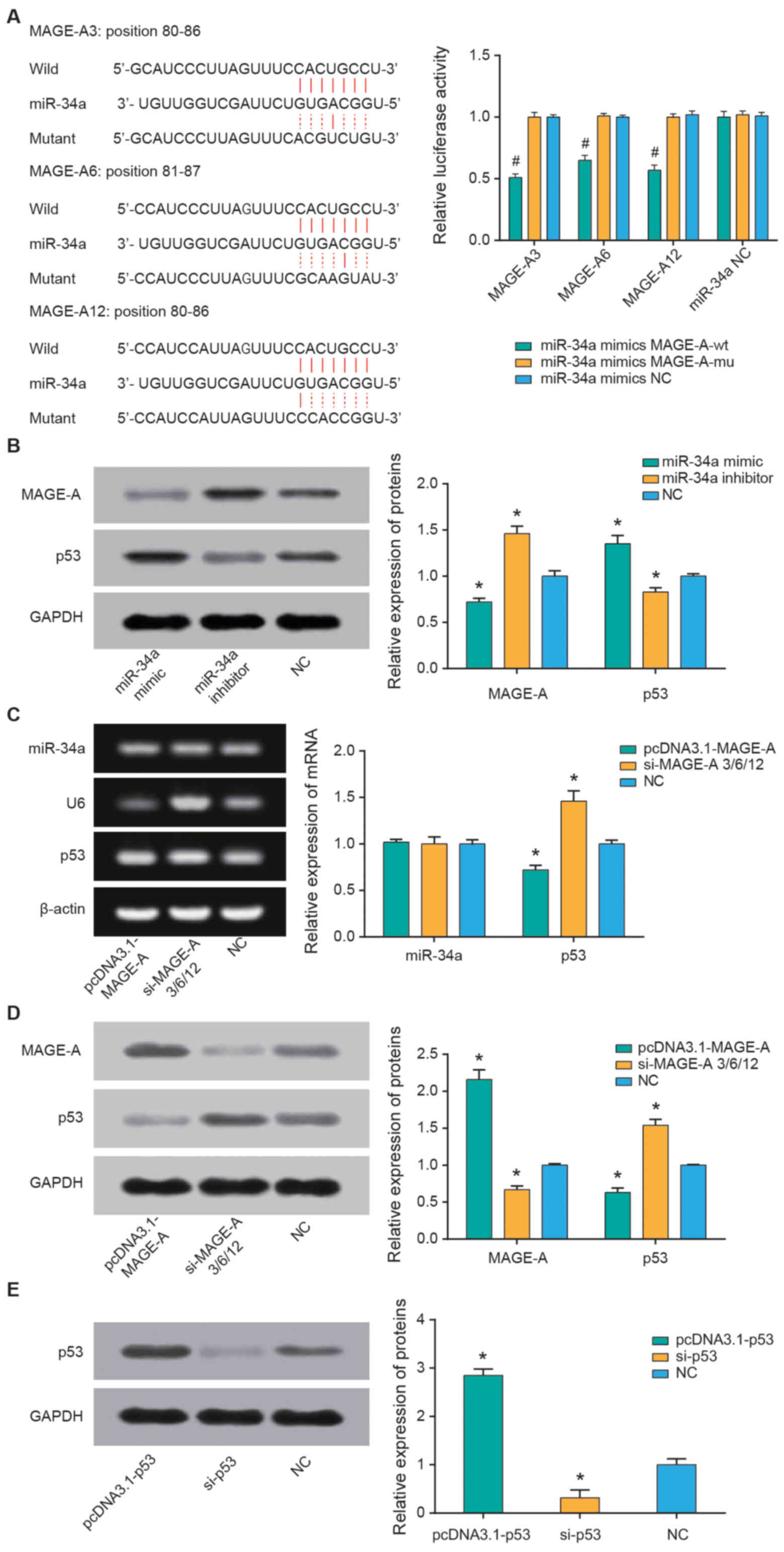
miR-34a targets MAGE-A to affect the
expression of MAGE-A and p53
Based on the TargetScan database, three members of
the MAGE-A family (MAGE-A3, MAGE-A6 and MAGE-A12) were identified
as the potential targets of miR-34a (Fig. 5A). The double luciferase reporter
gene assay was used to further explore whether miR-34a directly
binds to the 3′-untranslated region of MAGE-A. miR-34a mimics
significantly reduced the luciferase activity in the MAGE-A3,
MAGE-A6 and MAGE-A12 groups (all P<0.05), but no significant
regulatory effect was observed when MAGE-A3, MAGE-A6 and MAGE-A12
were mutated compared with the NC group (Fig. 5A). Furthermore, transfection with
miR-34a mimics significantly decreased MAGE-A and increased p53
expression levels, while miR-34a inhibitor promoted MAGE-A and
reduced p53 expression levels (all P<0.05; Fig. 5B). Additionally, overexpression of
MAGE-A significantly decreased the mRNA and protein expression of
p53 (P<0.05); however, no significant differences were observed
regarding the effect of MAGE-A on miR-34a (Fig. 5C and D).
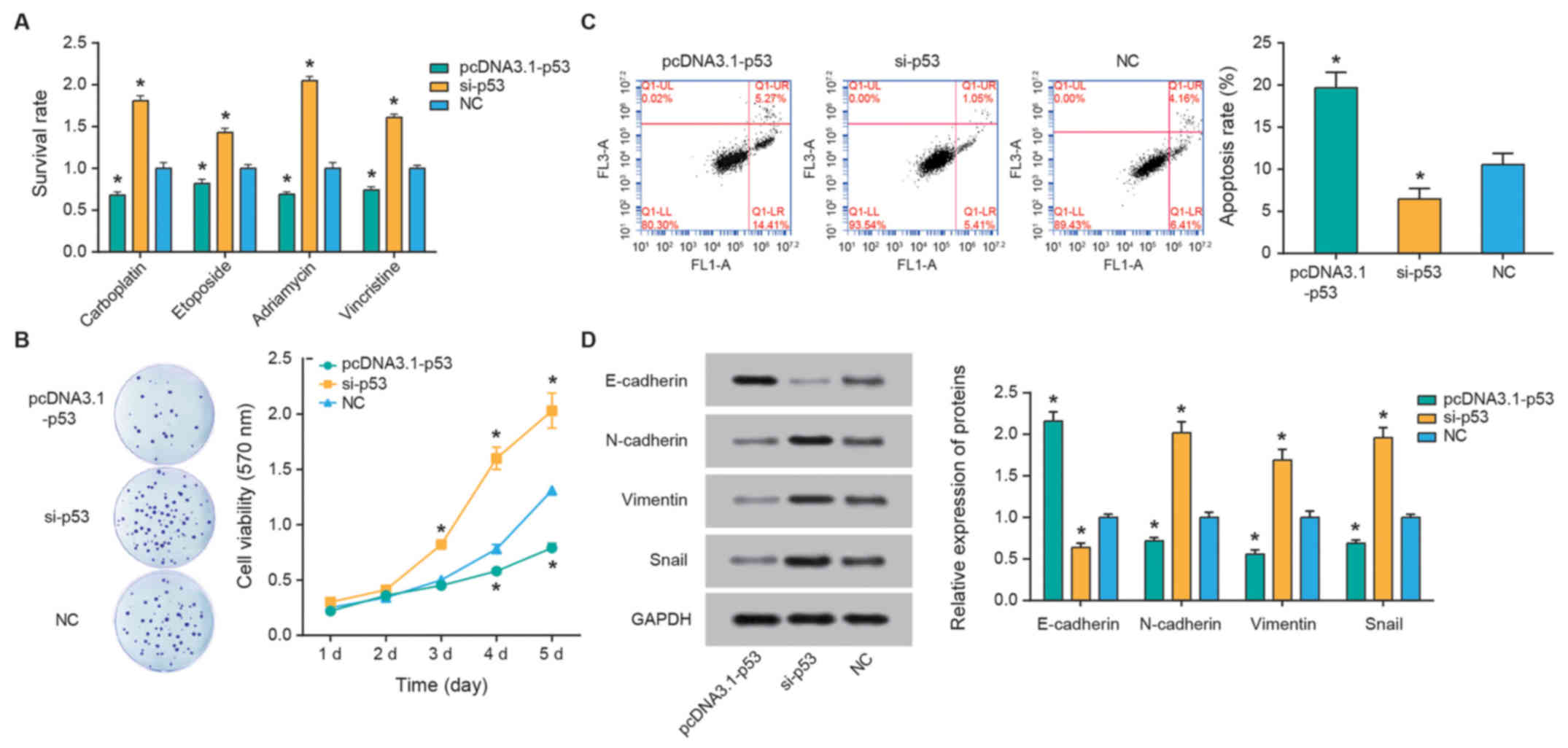
P53 alters RB cell chemosensitivity
p53 expression was significantly upregulated
following transfection with pcDNA3.1-p53 and downregulated
following si-p53 transfection compared with the NC control (both
P<0.05; Fig. 5E). Furthermore,
over-expression of p53 was associated with significantly reduced
the survival rates of SO-Rb50 cells following treatment with
carboplatin, etoposide, Adriamycin and vincristine compared with
the NC group (all P<0.05; Fig.
6A). The apoptotic rate of cells in the pcDNA3.1-p53 group was
significantly increased compared with the si-p53 group (P<0.05;
Fig. 6B), and cell viability in
the si-p53 group was increased compared with that in the
pcDNA3.1-p53 group (P<0.05; Fig.
6C). Finally, reduced p53 expression resulted in the
downregulation of E-cadherin expression, and upregulation of
N-cadherin, vimentin and snail expression levels (all P<0.05;
Fig. 6D).
Discussion
Accumulating evidence has demonstrated that aberrant
expression of miR-34a regulates the progression of various
neoplasms (25), including glioma
(26), breast cancer (27,28),
cervical cancer (29),
cholangiocarcinoma (30) and
multiple myeloma (31). The
results of the present study demonstrated that the expression level
of miR-34a was reduced in RB tissues compared with that in
paracarcinoma tissues (P<0.05). Generally, miRNAs modulate
diverse biological processes by targeting downstream mRNA (32–34).
For instance, miR-34a has been reported to regulate carcinogenesis
by targeting several mRNAs, including MAGE-A in medulloblastoma
(22), programmed cell death 1
ligand 1 or lactate dehydrogenase A in breast cancer (28), and B cell lymphoma-2 in cervical
cancer (29). The expression of
MAGE-A has been documented to be upregulated in gastric cancer
(35), epithelial ovarian cancer
(36), and lung adenocarcinoma
tissues and cells (37). In
agreement with the aforementioned studies, the results of the
RT-qPCR analysis in the present study revealed that the mRNA level
of MAGE-A was significantly upregulated in RB tissues compared with
paracarcinoma tissues (P<0.05). The clinical studies also
indicated that high MAGE-A expression and low miR-34a expression
were significantly associated with poor prognosis of patients with
RB (P<0.05). Furthermore, in order to investigate the role of
miR-34a and MAGE-A underlying RB etiology, their binding sites were
predicted using the Targetscan database, and validated their
targeted association by performing a dual luciferase reporter gene
assay. The results of the current study also suggested that MAGE-A
expression in SO-Rb50 cells was significantly reduced following
transfection with miR-34a mimics and increased following miR-34a
inhibitor transfection. Collectively, these results suggested that
the downregulation of MAGE-A by miR-34a partially mediated the
pathogenesis of RB.
Furthermore, the aforementioned carcinogenic
mechanism of miR-34a and MAGE-A in RB was hypothesized to be the
result of their contribution to enhancing cell viability and
modulating cell apoptosis (38–41).
In the present study, miR-34a mimics, miR-34a inhibitor or
pCDNA3-MAGE-A were transfected into SO-Rb50 cells, and their
effects on viability, survival rate and apoptosis of SO-Rb50 were
analyzed. The results revealed that the addition of miR-34a mimics
and silencing of MAGE-A significantly suppressed cell viability and
cell survival, and induced apoptosis. Furthermore, significantly
increased E-cadherin expression, as well as decreased N-cadherin,
vimentin and snail expression levels were observed in SO-Rb50
cells. Additionally, the transfection of miR-34a inhibitor and
pcDNA3.1-MAGE-A were able to impede the apoptosis of SO-Rb50 cells,
and encourage its viability and survival. It may be concluded that
miR-34a, through targeting MAGE-A, is involved in RB pathogenesis
by altering cell survival, cell viability, cell apoptosis and
expression levels of EMT-associated proteins.
Furthermore, miR-34a and MAGE-A were demonstrated to
regulate p53 expression, SO-Rb50 cell viability, survival and
apoptotic rates, as well as E-cadherin, N-cadherin, vimentin and
snail expression levels. p53 serves as a transcription factor that
is largely involved in cell growth, cell differentiation, cell
senescence and cell apoptosis (38,42–44).
In addition, it has been previously suggested that the inhibitory
effects of MAGE-A on p53 transactivation may lead to tumor cell
resistance to chemotherapeutic drugs (etoposide) (45). miR-34a confers chemosensitivity of
medulloblastoma cells through modulation of MAGE-A and p53
expression levels (22). Similar
to the aforementioned publications, the present study demonstrated
that miR-34a, MAGE-A and p53 were able to significantly alter the
resistance of SO-Rb50 cells to vincristine, etoposide, carboplatin
and Adriamycin. Taken together, the results suggest that miR-34a
targeted MAGE-A and thereby regulated MAGE-A to alter the
chemosensitivity of RB cells, and this phenomenon was achieved
possibly through alterations to the viability, survival, apoptosis
and EMT of RB cells.
In conclusion, miR-34a may function as a tumor
suppressor for RB by targeting MAGE-A and altering p53 expression,
indicating that the miR-34a/MAGE-A/p53 axis may serve as a
therapeutic target or diagnostic biomarker for RB. However, a
number of limitations must be addressed. Firstly, the results may
not well be generalized to other ethnicities or a larger
population, due to the limited sample size incorporated in the
present study. Secondly, only the SO-Rb50 cell line was examined,
and more cell lines should be focused on to verify the mechanisms.
Thirdly, no animal models were established, which may provide a
direct understanding regarding the influence of the
miR-34a/MAGE-A/p53 axis on RB development. Lastly, long non-coding
RNAs and circular RNAs are situated upstream of miRNAs, and may
also participate in the mechanism underlying RB carcinogenesis. As
a result, further studies are required to explore the molecular
mechanism of the miR-34a/MAGE-A/p53 axis on RB progression.
Funding
The present study was supported by Chinese National
Natural Science Foundation of Youth Fund (grant no. 81602154), the
Technology Research Projects of Henan Science and Technology
Department (grant nos. 162102410060 and 182107000054), Outstanding
Young Talent Research Fund of Zhengzhou University (grant no.
1621328002) and the Special Funding for Doctoral Team of the First
Affiliated Hospital of Zhengzhou University (grant no.
2016-BSTDJJ-11).
Availability of data and materials
All data generated or analyzed during this study are
included in this article.
Authors’ contributions
GY, YF, XL, MW, HD and QL conceived and designed the
experiments, performed the experiments. GY, YF, XL and MW analyzed
the data. HD and QL drafted the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The immediate relatives of all patients with RB
provided written informed consent, and the present study was
approved by the Ethics Committee of The First Affiliated Hospital
of Zhengzhou University.
Patient consent for publication
The immediate relatives of all patients with RB
provided written informed consent.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Aerts I, Lumbroso-Le Rouic L,
Gauthier-Villars M, Brisse H, Doz F and Desjardins L:
Retinoblastoma. Orphanet J Rare Dis. 1:312006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kivelä T: The epidemiological challenge of
the most frequent eye cancer: Retinoblastoma, an issue of birth and
death. Br J Ophthalmol. 93:1129–1131. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shields CL, Fulco EM, Arias JD, Alarcon C,
Pellegrini M, Rishi P, Kaliki S, Bianciotto CG and Shields JA:
Retinoblastoma frontiers with intravenous, intra-arterial,
periocular, and intra-vitreal chemotherapy. Eye (Lond). 27:253–264.
2013. View Article : Google Scholar
|
|
4
|
Abramson DH, Gerardi CM, Ellsworth RM,
McCormick B, Sussman D and Turner L: Radiation regression patterns
in treated retinoblastoma: 7 to 21 years later. J Pediatr
Ophthalmol Strabismus. 28:108–112. 1991.PubMed/NCBI
|
|
5
|
Fontanesi J, Pratt CB, Hustu HO, Coffey D,
Kun LE and Meyer D; Jude Children’s Research Hospital Experience
and Review of Literature: Use of irradiation for therapy of
retinoblastoma in children more than 1 year old: The St Jude
Children’s Research Hospital experience and review of literature.
Med Pediatr Oncol. 24:321–326. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shields CL, Say EA, Pointdujour-Lim R, Cao
C, Jabbour PM and Shields JA: Rescue intra-arterial chemotherapy
following retinoblastoma recurrence after initial intra-arterial
chemotherapy. J Fr Ophtalmol. 38:542–549. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ruiz del Río N, Abelairas Gómez JM, Alonso
García de la Rosa FJ, Peralta Calvo JM and de las Heras Martín A:
Genetic analysis results of patients with a retinoblastoma
refractory to systemic chemotherapy. Arch Soc Esp Oftalmol.
90:414–420. 2015.In Spanish. View Article : Google Scholar
|
|
8
|
Nalini V, Segu R, Deepa PR, Khetan V,
Vasudevan M and Krishnakumar S: Molecular insights on
post-chemotherapy retinoblastoma by microarray gene expression
analysis. Bioinform Biol Insights. 7:289–306. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Frankel LB and Lund AH: MicroRNA
regulation of autophagy. Carcinogenesis. 33:2018–2025. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tarasov V, Jung P, Verdoodt B, Lodygin D,
Epanchintsev A, Menssen A, Meister G and Hermeking H: Differential
regulation of microRNAs by p53 revealed by massively parallel
sequencing: miR-34a is a p53 target that induces apoptosis and
G1-arrest. Cell Cycle. 6:1586–1593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R,
Sun Z and Zheng X: Downregulation of CCND1 and CDK6 by miR-34a
induces cell cycle arrest. FEBS Lett. 582:1564–1568. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamakuchi M, Ferlito M and Lowenstein CJ:
miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci
USA. 105:13421–13426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma ZB, Kong XL, Cui G, Ren CC, Zhang YJ,
Fan SJ and Li YH: Expression and clinical significance of miRNA-34a
in colorectal cancer. Asian Pac J Cancer Prev. 15:9265–9270. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu G, Jiang C, Li D, Wang R and Wang W:
miRNA-34a inhibits EGFR-signaling-dependent MMP7 activation in
gastric cancer. Tumour Biol. 35:9801–9806. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dalgard CL, Gonzalez M, deNiro JE and
O’Brien JM: Differential microRNA-34a expression and tumor
suppressor function in reti-noblastoma cells. Invest Ophthalmol Vis
Sci. 50:4542–4551. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao H, Zhao H and Xiang W: Expression
level of human miR-34a correlates with glioma grade and prognosis.
J Neurooncol. 113:221–228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li L, Yuan L, Luo J, Gao J, Guo J and Xie
X: miR-34a inhibits proliferation and migration of breast cancer
through down-regulation of Bcl-2 and SIRT1. Clin Exp Med.
13:109–117. 2013. View Article : Google Scholar
|
|
19
|
Tang Y, Tang Y and Cheng YS: miR-34a
inhibits pancreatic cancer progression through Snail1-mediated
epithelial-mesenchymal transition and the Notch signaling pathway.
Sci Rep. 7:382322017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vinall RL, Ripoll AZ, Wang S, Pan CX and
deVere White RW: miR-34a chemosensitizes bladder cancer cells to
cisplatin treatment regardless of p53-Rb pathway status. Int J
Cancer. 130:2526–2538. 2012. View Article : Google Scholar
|
|
21
|
Kang L, Mao J, Tao Y, Song B, Ma W, Lu Y,
Zhao L, Li J, Yang B and Li L: MicroRNA-34a suppresses the breast
cancer stem cell-like characteristics by downregulating Notch1
pathway. Cancer Sci. 106:700–708. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Weeraratne SD, Amani V, Neiss A, Teider N,
Scott DK, Pomeroy SL and Cho YJ: miR-34a confers chemosensitivity
through modulation of MAGE-A and p53 in medulloblastoma.
Neuro-oncol. 13:165–175. 2011. View Article : Google Scholar :
|
|
23
|
Peche LY, Scolz M, Ladelfa MF, Monte M and
Schneider C: MageA2 restrains cellular senescence by targeting the
function of PMLIV/p53 axis at the PML-NBs. Cell Death Differ.
19:926–936. 2012. View Article : Google Scholar :
|
|
24
|
Nardiello T, Jungbluth AA, Mei A,
Diliberto M, Huang X, Dabrowski A, Andrade VC, Wasserstrum R, Ely
S, Niesvizky R, et al: MAGE-A inhibits apoptosis in proliferating
myeloma cells through repression of Bax and maintenance of
survivin. Clin Cancer Res. 17:4309–4319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Krauskopf J, de Kok TM, Hebels DG,
Bergdahl IA, Johansson A, Spaeth F, Kiviranta H, Rantakokko P,
Kyrtopoulos SA and Kleinjans JC: MicroRNA profile for health risk
assessment: Environmental exposure to persistent organic pollutants
strongly affects the human blood microRNA machinery. Sci Rep.
7:92622017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jin Z, Zhan T, Tao J, Xu B, Zheng H, Cheng
Y, Yan B, Wang H, Lu G, Lin Y, et al: MicroRNA-34a induces
transdifferentiation of glioma stem cells into vascular endothelial
cells by targeting Notch pathway. Biosci Biotechnol Biochem.
81:1899–1907. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Engkvist ME, Stratford EW, Lorenz S,
Meza-Zepeda LA, Myklebost O and Munthe E: Analysis of the miR-34
family functions in breast cancer reveals annotation error of
miR-34b. Sci Rep. 7:96552017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang X and Xie X, Wang H, Xiao X, Yang L,
Tian Z, Guo X, Zhang L, Tang H and Xie X: PDL1 And LDHA act as
ceRNAs in triple negative breast cancer by regulating miR-34a. J
Exp Clin Cancer Res. 36:1292017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Xie Y and Wang J: Overexpression
of microRNA-34a-5p inhibits proliferation and promotes apoptosis of
human cervical cancer cells by downregulation of Bcl-2. Oncol Res.
Aug 30–2017.Epub ahead of print. View Article : Google Scholar
|
|
30
|
Kwon H, Song K, Han C, Zhang J, Lu L, Chen
W and Wu T: Epigenetic silencing of miRNA-34a in human
cholangiocarcinoma via EZH2 and DNA methylation: Impact on
regulation of notch pathway. Am J Pathol. 187:2288–2299. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dai X, Li M and Geng F: Omega-3
polyunsaturated fatty acids eicosapentaenoic acid and
docosahexaenoic acid enhance dexamethasone sensitivity in multiple
myeloma cells by the p53/miR-34a/Bcl-2 axis. Biochemistry (Mosc).
82:826–833. 2017. View Article : Google Scholar
|
|
32
|
Ritchie W, Rasko JE and Flamant S:
MicroRNA target prediction and validation. Adv Exp Med Biol.
774:39–53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Witkos TM, Koscianska E and Krzyzosiak WJ:
Practical aspects of microRNA target prediction. Curr Mol Med.
11:93–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zheng L, Zhang Y, Liu Y, Zhou M, Lu Y,
Yuan L, Zhang C, Hong M, Wang S and Li X: miR-106b induces cell
radioresistance via the PTEN/PI3K/AKT pathways and p21 in
colorectal cancer. J Transl Med. 13:2522015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lian Y, Sang M, Gu L, Liu F, Yin D, Liu S,
Huang W, Wu Y and Shan B: MAGE-A family is involved in gastric
cancer progression and indicates poor prognosis of gastric cancer
patients. Pathol Res Pract. 213:943–948. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sang M, Wu X, Fan X, Lian Y and Sang M:
MAGE-A family serves as poor prognostic markers and potential
therapeutic targets for epithelial ovarian cancer patients: A
retrospective clinical study. Gynecol Endocrinol. 33:480–484. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sang M, Gu L, Yin D, Liu F, Lian Y, Zhang
X, Liu S, Huang W, Wu Y and Shan B: MAGE-A family expression is
correlated with poor survival of patients with lung adenocarcinoma:
A retrospective clinical study based on tissue microarray. J Clin
Pathol. 70:533–540. 2017. View Article : Google Scholar
|
|
38
|
Marcar L, Maclaine NJ, Hupp TR and Meek
DW: Mage-A cancer/testis antigens inhibit p53 function by blocking
its interaction with chromatin. Cancer Res. 70:10362–10370. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zajac P, Schultz-Thater E, Tornillo L,
Sadowski C, Trella E, Mengus C, Iezzi G and Spagnoli GC: MAGE-A
antigens and cancer immunotherapy. Front Med (Lausanne).
4:182017.
|
|
40
|
Sato F, Tsuchiya S, Meltzer SJ and Shimizu
K: MicroRNAs and epigenetics. FEBS J. 278:1598–1609. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: A review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar
|
|
42
|
Kircelli F, Akay C and Gazitt Y: Arsenic
trioxide induces p53-dependent apoptotic signals in myeloma cells
with SiRNA-silenced p53: MAP kinase pathway is preferentially
activated in cells expressing inactivated p53. Int J Oncol.
30:993–1001. 2007.PubMed/NCBI
|
|
43
|
Riley T, Sontag E, Chen P and Levine A:
Transcriptional control of human p53-regulated genes. Nat Rev Mol
Cell Biol. 9:402–412. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vousden KH and Prives C: Blinded by the
light: The growing complexity of p53. Cell. 137:413–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Monte M, Simonatto M, Peche LY, Bublik DR,
Gobessi S, Pierotti MA, Rodolfo M and Schneider C: MAGE-A tumor
antigens target p53 transactivation function through histone
deacetylase recruitment and confer resistance to chemotherapeutic
agents. Proc Natl Acad Sci USA. 103:11160–11165. 2006. View Article : Google Scholar : PubMed/NCBI
|