Introduction
Renal cell carcinoma (RCC) is a type of cancer found
in the lining of the kidney tubules, and it is among the 10 most
frequently occurring human cancers (1-3). RCC
is the second leading cause of mortality associated with urological
malignant neoplasms (1-3). The prognosis of patients with RCC
remains poor, with the 5-year survival rate remaining between 5 and
12% (1,4). RCC results in a number of symptoms,
including weight loss, fever, hypertension, hypercalcemia, night
sweats and malaise (5,6). The most common histological subtype
is clear cell RCC, accounting for approximately 80-90% of all RCC
cases (3). Approximately 30% of
patients with RCC have metastatic lesions (7). Smoking tobacco, hypertension and
obesity are considered as risk factors for RCC (8).
Advances in the treatment of RCC have been derived
from agents approved by the Food and Drug Administration (8). These agents target several pathways,
including mammalian target of rapamycin (mTOR), multiple
pro-angiogenic growth factors such as vascular endothelial growth
factor (VEGF) and platelet-derived growth factor (PDGF), and their
receptors, VEGFR and PDGFR (8,9).
Despite the development of therapeutic regimens (8), the prognosis of patients with RCC
remains poor, mainly due to delayed diagnosis and a relatively high
incidence of metastasis (10,11).
Although the vast majority of patients exhibit a marked clinical
response, the therapeutic effects of these inhibitors are limited
due to the development of drug-resistant phenotypes (10,11).
Therefore, more potent and specific therapeutic strategies are
urgently required in order to identify novel diagnostic and
therapeutic targets for RCC (8,12-14).
However, the molecular mechanisms underlying RCC tumorigenesis
remain elusive (12-14).
The gene for regucalcin, which was discovered as a
calcium-binding protein (15,16),
is localized on the X chromosome (17-19).
Regucalcin is expressed in various types of cells and tissues
(20,21), and has been demonstrated to play
multifunctional roles in the regulation of manifold cells (21-24).
Regucalcin has been shown to maintain calcium homeostasis, inhibit
various signaling pathways involving various protein kinases and
protein phosphatases, suppress cytosolic protein synthesis and
nuclear DNA and RNA synthesis, and regulate nuclear gene expression
in cells (22-25). Moreover, regucalcin has been shown
to suppress proliferation (26)
and apoptosis (27) mediated
through various signaling factors in various types of cells. Thus,
regucalcin has been shown to play a pivotal role in maintaining
cell homeostasis (22,23,28).
Importantly, the gene expression and protein levels
of regucalcin have been shown to be downregulated in various tumor
tissues of mammalian and human subjects (29,30).
We previously demonstrated that regucalcin gene expression was
decreased in the tumor tissues of human cancer patients, including
those with pancreatic cancer (31), breast cancer (32), liver cancer (33), lung cancer (34) and colorectal cancer (35). The prolonged survival of these
cancer patients was shown to be associated with a higher regucalcin
expression as compared with a lower regucalcin expression in their
tumor tissues (31-35). In addition, the overexpression of
regucalcin was shown to exert suppressive effects on the
proliferation of human pancreatic cancer MIA PaCa-2 cells (31), MDA-MB-231 breast cancer cells
(32), liver cancer HepG2 cells
(33), lung adenocarcinoma A549
cells (34) and colorectal cancer
RKO cells (35) in vitro.
These findings support the view that regucalcin plays a crucial
role as a suppressor in human cancer cells, and that its
downregulated gene expression leads to the development of
carcinogenesis in various tissues of human subjects. Regucalcin may
therefore be a novel target molecule in the diagnosis and therapy
of human cancer.
In the present study, furthermore, we investigated
whether regucalcin plays a role as a suppressor in human RCC.
Regucalcin is expressed in rat kidney proximal tubular epithelial
cells (20,21) and plays a physiological and
pathophysiological role in cell regulation and metabolic disorder
in the kidney (24). Of note, the
gene expression and protein levels of regucalcin are downregulated
in the kidney tumor tissues of human subjects (29,30).
Regucalcin may thus be a novel target molecule in the diagnosis and
therapy of RCC. The involvement of regucalcin in human RCC has not
yet been investigated, to the best of our knowledge. Therefore, the
involvement of regucalcin in human RCC was investigated in the
current study. Of note, it was demonstrated that the survival of
patients with clear cell RCC with a higher regucalcin gene
expression in their tumor tissues was prolonged, as evaluated by
the analysis of gene expression using the Gene Expression Omnibus
(GEO) database (GSE36895). Moreover, the over-expression of
regucalcin was found to suppress the growth of clear cell human RCC
A498 cells in vitro. The current findings furthermore
support the view that the downregulation of regucalcin gene
expression predisposes patients to various types of cancer.
Targeting regucalcin may thus prove to be of clinical significance
in the suppression of cancer development. A delivery system with
the regucalcin gene may provide a novel therapeutic strategy for
human renal cancer.
Materials and methods
Materials and reagents
Dulbecco’s modified Eagle’s medium (DMEM) with 4.5
g/l glucose, L-glutamine and sodium pyruvate and antibiotics [100
µg/ml penicillin and 100 µg/ml streptomycin (P/S)]
were purchased from Corning (Mediatech, Inc. Manassas, VA, USA).
Fetal bovine serum (FBS) was from HyClone (Logan, UT, USA).
Lipofectamine reagent was obtained from Promega (Madison, WI, USA).
Tumor necrosis factor-α (TNF-α) was from R&D Systems
(Minneapolis, MN, USA). Sodium butyrate, roscovitine, sulforaphane,
wortmannin, PD98059, staurosporine, 5, 6-dichloro-1-β-D-ri
bofuranosylbenzimidazole (DRB), lipopolysacchaide (LPS), caspase-3
inhibitor and all other reagents were purchased from Sigma-Aldrich
(St. Louis, MO, USA) unless otherwise specified. Gemcytabine was
obtained from Hospira, Inc. (Lake Forest, IL, USA). Gemcitabine and
caspase-3 inhibitor were diluted in phosphate-buffered saline (PBS)
and the other reagents were dissolved in 100% ethanol prior to
use.
Patient datasets
A curated gene expression dataset comprising 23
normal and 29 tumor samples of the kidney cortex tissues of
patients with clear cell RCC were obtained through the GEO database
(GSE36895) for the analysis of regucalcin expression (36). These datasets contained gene
expression data derived from the Affymetrix U133_plus2 platform.
For microarray analysis, the expression and raw expression data
(CEL files) were summarized and normalized using the Robust
Multi-array Average algorithm and the Bioconductor package affy
(http://www.bioconductor.org/packages/2.0/bioc/html/affy.html).
The Spotfire Decision Site for Functional Genomics software package
(TIBCO Software, Palo Alto, CA, USA) was used for visualization of
the microarray data. For protein expression analysis, a dataset of
immunohistochemistry was obtained from the Human Protein Atlas
(HPA; www.proteinatlas.org), which is a
database of proteins in human normal tissues and cancers (37,38).
These were non-normally distributed data. We also evaluated
regucalcin expression in 3 normal tissues and 12 renal
adenocarcinoma tissues from the kidneys of patients by using the
dataset of 2 antibodies (HPA029102 and HPA029103) for regucalcin.
Moreover, we used the TCGA dataset of 468 patients with clear cell
RCC (39) for outcome analysis.
The data for regucalcin expression and clinical annotation were
obtained by SurvExpress (40).
Human renal cell carcinoma cells
We used A498 cells obtained from the American Type
Culture Collection (ATCC; Rockville, MD, USA). A498 cells are a
renal proximal tubular epithelial cell line originating from a male
adult patient with clear cell renal cell carcinoma (RCC), and are
non-metastatic cells (41). The
A498 cells are suitable as a transfection host. The cells were
cultured in a DMEM containing 10% FBS and 1% P/S.
Transfection of regucalcin cDNA
The A498 cells were transfected with the empty pCXN2
vector or pCXN2 vector expressing cDNA encoding human full length
(900 bp) regucalcin (regucalcin cDNA/pCXN2). These vectors were
prepared as used in our previous study (42). For transient transfection assays,
the A498 cells were grown on 24-well plates to approximately 70–80%
confluence. The regucalcin cDNA/pCXN2 or empty pCXN2 vector were
transfected into A498 cells using the synthetic cationic lipid,
Lipofectamine reagent, according to the manufacture’s instructions
(Promega, Madison, WI, USA) (42).
Following overnight incubation, Geneticin (600 µg/ml G418;
Sigma-Aldrich) was added to the wells for selection, and the cells
were cultured for 3 weeks. The surviving cells were plated at
limiting dilution to isolate transfectants. Multiple surviving
clones were isolated, transferred to 35-mm dishes, and grown in
medium without geneticin. We obtained transfectant clones 1 and 2
exhibiting stable expression of regucalcin. The regucalcin levels
in these clones were markedly increased as compared with those in
the wild-type cells as shown in Fig.
2A and B. The regucalcin levels in clone 1 was higher than that
of clone 2. Clone 1 was used in the following experiments.
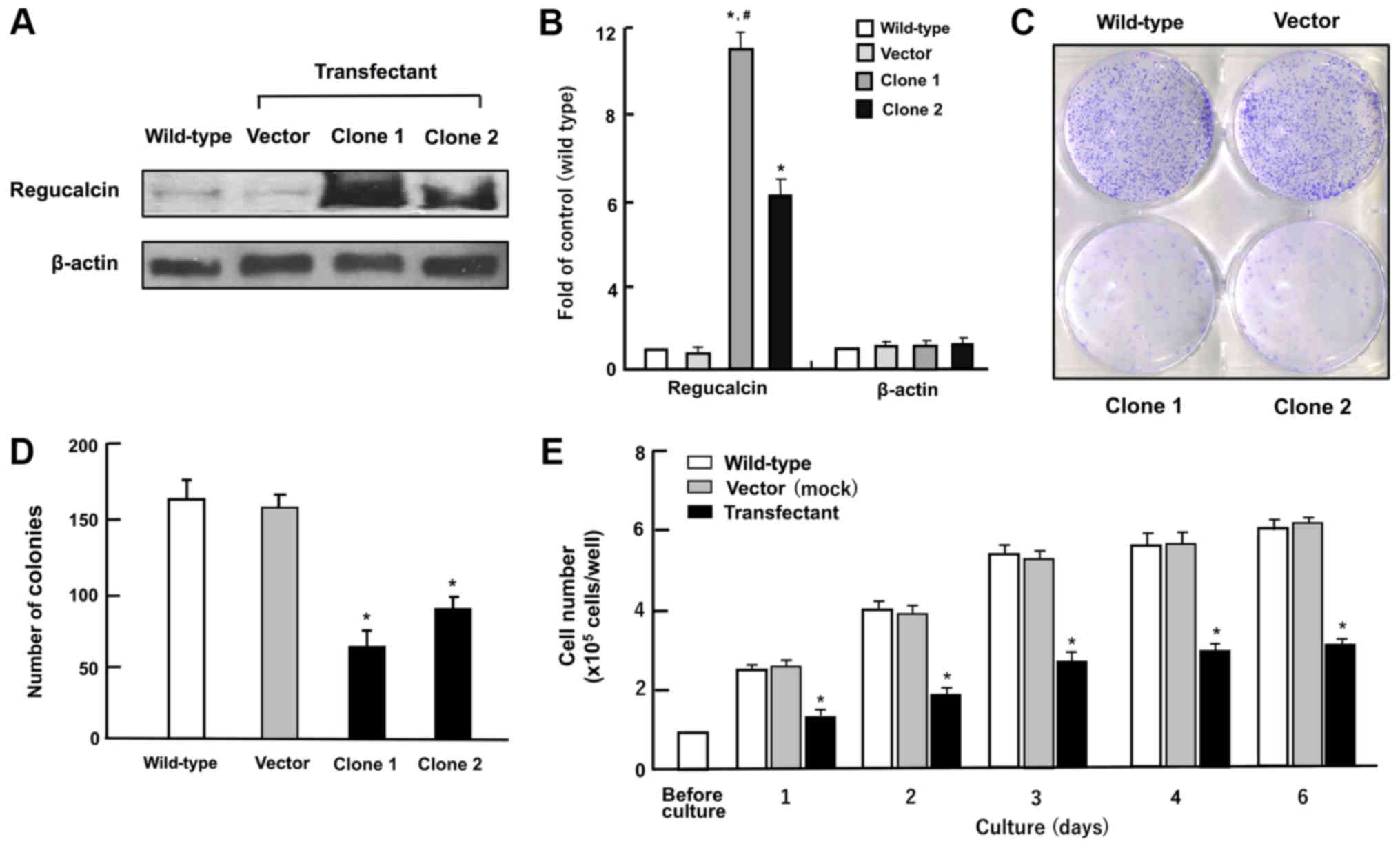 | Figure 2Overexpression of regucalcin
suppresses colony formation and proliferation in human clear cell
RCC A498 cells in vitro. (A and B) Regucalcin content in the
cells cultured in DMEM containing 10% FBS and 1% P/S for 3 days as
analyzed by western blot analysis with an anti-regucalcin antibody.
Lane 1, wild-type cells; lane 2, cells transfected with empty
vector/pCXN2 (designated as vector); lanes 3 or 4, cells (clone 1
or 2) transfected with the human regucalcin cDNA /pCXN2. A
representative of 5 films is shown. (A) Representative image. (B)
Fold of control; *P<0.001 versus wild-type cells, and
#P<0.001 versus clone 2, determined by the Student’s
t-test. (C and D) Colony formation. The A498 wild-type cells and
transfectants (vector, clone 1 or 2) were cultured for 8 days, and
the colonies then stained with 0.5% crystal violet and counted. (C)
Representative image. (D) Colonies containing >50 cells were
counted under a microscope. (E) In the cell proliferation assay,
A498 wild-type cells or clone 1 were cultured in DMEM for 1, 2, 3,
4 or 6 days, and the numbers of attached cells were counted. Data
are presented as the means ± SD obtained from 8 wells of 2
replicate plates per data set using different dishes and cell
preparations. *P<0.001 versus wild-type cells (white
bar) or control vector (grey bar), determined by one-way ANOVA with
the Tukey-Kramer post hoc test. |
Colony formation assay
The A498 wild-type cells or transfectants were
seeded into 6-well dishes at a density of 1×103/well and
cultured in medium containing 10% FBS and 1% P/S under conditions
of 5% CO2 and 37°C for 8 days, when visible clones were
formed on the plates (43). The
colonies were washed with PBS and fixed with methanol (0.5 ml per
well) for 20 min at room temperature, and then washed 3 times with
PBS. The colonies were then stained with 0.5% crystal violet for 30
min at room temperature. Stained cells were washed 4 times with
PBS. The plates were air-dried for 2 h at room temperature. The
colonies containing >50 cells were counted under a microscope
(Olympus MTV-3; Olympus Corporation, Tokyo, Japan).
Cell proliferation and growth assays
The A498 wild-type cells (1×105/ml per
well) and A498 cells transfected with the regucalcin cDNA
(1×105/ml per well) were cultured using a 24-well plate
in DMEM containing 10% FBS and 1% P/S for 1, 2, 3, 4 or 6 days in a
water-saturated atmosphere containing 5% CO2 and 95% air
at 37°C (42,44). In separate experiments, the A498
wild-type cells or transfectants were cultured in DMEM containing
10% FBS and 1% P/S in the presence of either sodium butyrate (10
and 100 µM), roscovitine (10 and 100 nM), sulphoraphan (1
and 10 nM), wortmannin (0.1 or 1 µM), PD98059 (1 or 10
µM), staurosporine (10 or 100 nM), TNF-α (0.1 or 1 ng/ml),
DRB (0.1 or 1 µM), or gemcitabine (1 or 10 nM) for 3 days.
Following culture, the cells were detached from each culture dishes
by adding a sterile solution (0.1 ml per well) of 0.05% trypsin
plus EDTA in Ca2+/Mg –free PBS (Thermo Fisher
Scientific, Waltham, MA, USA) with incubation for 2 min at 37°C.
Each well was then supplemented with 0.9 ml of DMEM containing 10%
FBS and 1% P/S. The cell number in the cell suspension was counted
as described below in the section ‘Cell counting’.
Cell death assay
The A498 wild-type cells (1×105/ml per
well) cells and A498 cells transfected with the regucalcin cDNA
(1×105/ml per well) were cultured using a 24-well plate
in DMEM containing 10% FBS and 1% P/S for 3 days. Upon reaching
subconfluency, they were cultured for an additional 24 h in the
presence or absence of either TNF-α (0.1 or 1 ng/ml) or LPS (0.1 or
1 µg/ml) (45). In separate
experiments, the A498 wild-type cells (1×105/ml per
well) or transfectants were cultured for 3 days, and upon reaching
subconfluency, then cultured for an additional 24 h in the presence
or absence of either TNF-α (1 ng/ml) or LPS (1 µg/ml) with
or without caspase-3 inhibitor (10 µM) for 24 h (45). Following culture, the cells were
detached by the addition of a sterile solution (0.1 ml per well) of
0.05% trypsin plus EDTA in Ca2+/Mg2+-free PBS
per well as described above in the section of ‘Cell proliferation
assay’, and the cell number was counted as described below in the
section ‘Cell counting’.
Cell counting
To detach cells on each well, the culture dishes
were incubated for 2 min at 37°C following the addition of a
solution (0.1 ml per well) of 0.05% trypsin plus EDTA in
Ca2+/Mg2+-free PBS, and the cells were
detached through pipetting after the addition of DMEM (0.9 ml)
containing 10% FBS and 1% P/S (31,42,44,45).
Medium containing the suspended cells (0.1 ml) was mixed by the
addition of 0.1 ml of 0.5% trypan blue staining solution. The
number of viable cells was counted under a microscope (Olympus
MTV-3; Olympus Corporation) with a hemocytomete (Sigma-Aldrich)
using a cell counter (Line Seiki H-102P, Tokyo, Japan). For each
dish, we took the average of 2 counts. Cell numbers are shown as
number per well.
Western blot analysis
The A498 wild-type cells, control vector
cDNA-transfected cells, or regucalcin cDNA-transfected cells were
plated in 100 mm dishes at a density of 1×106 cells/well
in 10 ml of DMEM containing 10% FBS and 1% P/S. Following culture
for 3 days, the cells were washed 3 times with cold PBS and removed
from the dish by scraping using cell lysis buffer (Cell Signaling
Technology, Inc., Danvers, MA, USA) with the addition of protease
and protein phosphatase inhibitors (Roche Diagnostics,
Indianapolis, IN, USA). The lysates were then centrifuged at 17,000
× g, at 4°C for 10 min. The protein concentration of the
supernatant was determined for western blotting using the Bio-Rad
Protein Assay Dye (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
with bovine serum albumin as a standard. The supernatant was stored
at −80°C until used. Samples of 40 µg of supernatant protein
per lane were separated by SDS polyacrylamide gel electrophoresis
(12%, SDS-PAGE) and transferred onto nylon membranes for
immunoblotting using specific antibodies against various proteins
obtained from Cell Signaling Technology, Inc. including Ras (#14429
rabbit), PI3 p1100 α (#4255, rabbit), Akt (#9272, rabbit),
phosphor-Akt (#9271, rabbit), mitogen-activated protein kinase
(MAPK; #4695, rabbit), phosphor-MAPK (#4370, rabbit), Rb (#9309,
mouse), p21 (#2947, rabbit), c-jun (#9165, rabbit), signal
transducer and activator of transcription 3 (Stat3; #12640,
rabbit), phospho-Stat3 (#9131, rabbit) and β-actin (#3700, mouse)
and Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA) including
p53 (sc-126, mouse), c-fos (sc-52, rabbit), nuclear factor (NF)-κB
p65 (sc-109, rabbit) and β-catenin (sc-39350, mouse). Rabbit
anti-regucalcin antibody was obtained from Abcam (Cambridge, MA,
USA; ab213459, rabbit), as has been used previously (22,28,32).
Target proteins were incubated with one of the primary antibodies
(1:1,000) overnight at 4℃, followed by horseradish
peroxidase-conjugated secondary antibodies (Santa Cruz
Biotechnology, Inc., mouse sc-2005 or rabbit sc-2305; diluted
1:2,000). The immunoreactive blots were visualized with a
SuperSignal West Pico Chemiluminescent Substrate detection system
(Thermo Scientific, Rockford, IL, USA) according to the
manufacturer’s instructions. β-actin (diluted 1:2,000; Cell
Signaling Biotechnology, Inc.; #3700, mouse) was used as a loading
control. Three blots from independent experiments were scanned on
an Epson Perfection 1660 Photo scanner, and bands quantified using
Image J software.
Statistical analysis
Statistical significance was determined using
GraphPad InStat version 3 for Windows XP (GraphPad Software Inc.,
La Jolla, CA, USA). Multiple comparisons were performed by one-way
analysis of variance (ANOVA) with the Tukey-Kramer multiple
comparisons post test for parametric data as indicated. Survival
curves were constructed by Kaplan-Meier analysis and were compared
with the log-rank test as performed with IBM SPSS. The non-normally
distributed data in Fig. 1B were
analyzed by the Mann-Whitney test and the rest of the data were
analyzed with the Student’s t-test to compare the means as
performed with IBM SPSS Statistics 18 software (IBM, Chicago, IL,
USA; http://www.ibm.com). P<0.05 was considered to
indicate a statistically significant difference.
Results
Survival of patients with renal cancer with a
higher regucalcin gene expression is prolonged
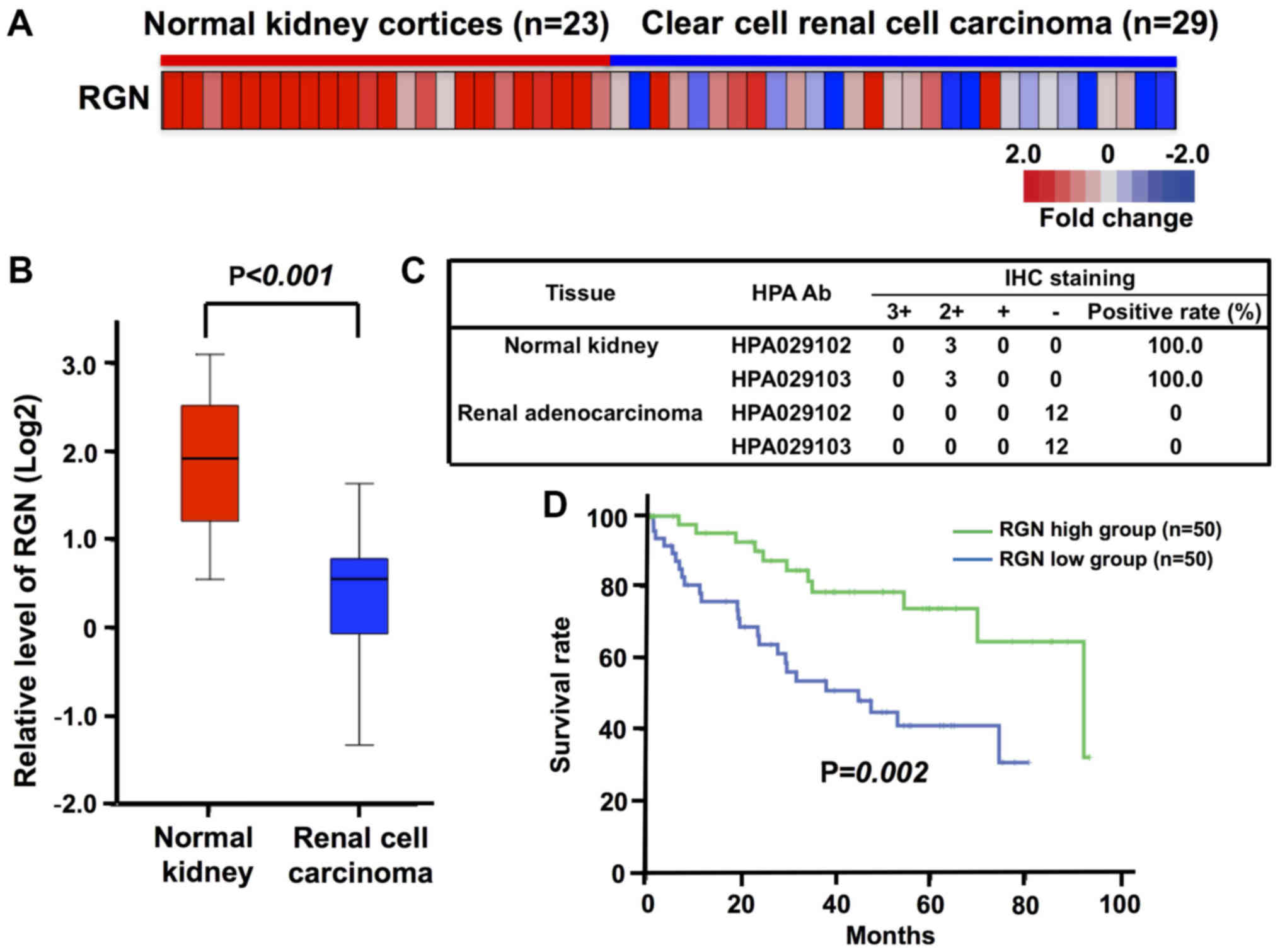
To evaluate the potential involvement of regucalcin
in human clear cell RCC, a curated gene expression dataset
comprising 23 normal and 29 tumor samples in the kidney cortex
tissues of patients with clear cell RCC was obtained through the
GEO database (GSE36895) for the analysis of regucalcin expression
(36). We compared regucalcin gene
expression in the tumor tissues of patients with clear cell RCC
using microarray data from the GEO database. Overall, regucalcin
expression was found to be visually decreased in the tumor tissues
as compared with that in the normal tissues derived from the kidney
cortex of patients with clear cell RCC (Fig. 1A). Quantitative analysis confirmed
that the expression of regucalcin in the tumor tissues of the
kidney cortex of patients with clear cell RCC was markedly
decreased as compared with that in the normal tissue of the patient
kidneys (Fig. 1B). To confirm the
reduction in regucalcin levels, we analyzed the expression of
regucalcin in 3 tissues of normal kidneys and 12 tissues of renal
adenocarcinoma in the immunohistochemistry database (The Human
Protein Atlas). The results from 2 independent regucalcin
antibodies revealed that the expression of regucalcin in renal
cancer patients was clearly suppressed as compared with that in the
normal kidneys (Fig. 1C).
Moreover, to determine whether the reduced regucalcin expression is
associated with prognosis, we compared the outcome of patients with
clear cell RCC with a high or low level of regucalcin mRNA
expression in the tumor tissues using Kaplan-Meier curve analysis.
To this end, we analyzed the outcome of 468 patients with clear
cell RCC using the TCGA dataset, and used data with higher (50
patients)/lower (50 patients) groups defined as top/bottom 10%,
respectively (Fig. 1D). A reduced
regucalcin expression was found to be associated with a poor
prognosis of patients with clear cell RCC (Fig. 1D). These results support the view
that the reduced regucalcin gene expression significantly
contributes to the development of carcinogenesis in human clear
cell RCC cells, leading to a worse clinical outcome.
Overexpression of regucalcin suppresses
the growth of A498 cells
To generate regucalcin-overexpressing cells, human
clear cell RCC A498 cells were transiently transfected with the
empty pCXN2 vector or the vector containing full length (33 kDa
protein) human regucalcin using lipofection. We obtained
transfectant clone 1 or 2 with stable expression of regucalcin. The
regucalcin levels in these clones were increased 11.2- or 6.1-fold
as compared with wild-type cells, respectively (Fig. 2A and B). To determine the effects
of the overexpression of regucalcin on the growth of A498 cells
in vitro, clone 1 was used in the following experiments.
First, to determine the effects of the overexpression of regucalcin
on colony formation, the A498 wild-type cells and transfectants
were cultured for 8 days when colony formation clearly appeared
(Fig. 2C). The number of colonies
was found to be decreased in the regucalcin-overexpressing
trans-fectants (vector, clone 1 or 2) as compared with that of the
wild-type cells (Fig. 2D). The
mass growth of the wild-type A498 cells was enhanced with the
increasing days of culture periods (Fig. 2E). This enhancement was clearly
suppressed in the transfectants (Fig.
2E). Thus, the overexpression of regucalcin suppressed the
colony formation and proliferation of human renal cancer A498
cells.
Suppressive effects of the overexpression
of regucalcin on cell growth are independent of cell death
The effect of the overexpression of regucalcin on
the death of A498 cells was then investigated. The A498 wild-type
cells and transfectants were cultured for 3 days to reach
subconfluency. They were then cultured for a further 24 h following
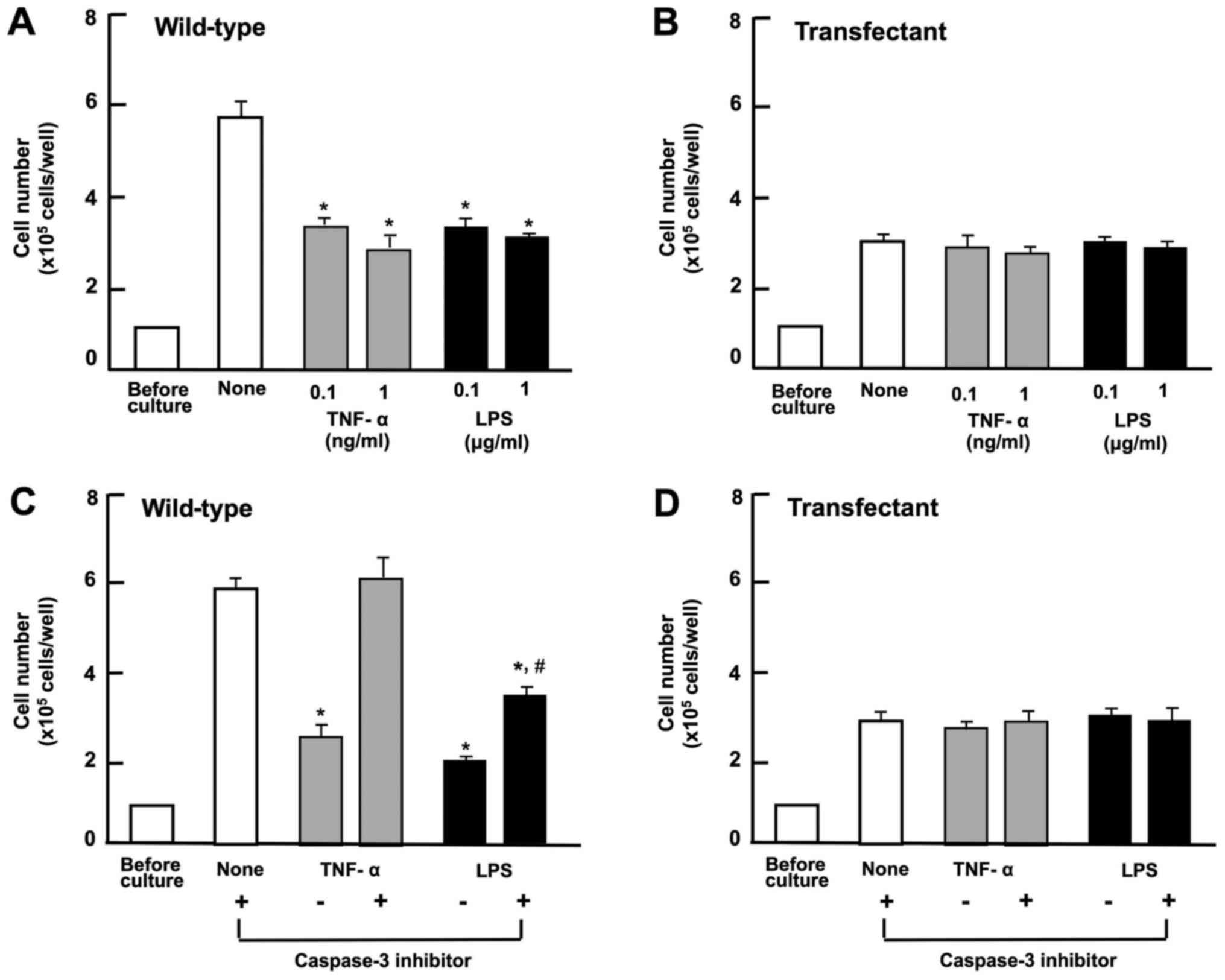
the addition of various factor known to induce apoptotic cell death
(27,45). The number of wild-type cells was
decreased by culture with TNF-α (0.1 or 1 ng/ml) or LPS (0.1 or 1
µg/ml) (Fig. 3A). The
overexpression of regucalcin did not lead to the death of the
wild-type cells, and the apoptotic cell death-inducing factors did
not cause the cell death of the transfectants (Fig. 3B). This suggests that the
suppressive effects of regucalcin over-expression on cell growth do
not result from the death of A498 cells.
We then investigated whether the effects of
overexpressed regucalcin on cell death are mediated via caspase-3,
which activates nuclear DNA fragmentation, inducing apoptotic cell
death (16). Regucalcin has been
demonstrated to suppress nuclear DNA fragmentation due to the
inhibition of caspase-3 activity in isolated rat liver nuclei
(16) and to induce apoptotic cell
death in cloned rat hepatoma H4-II-E cells induced by TNF-α or
thapsigargin (45). The A498
wild-type cells and transfectants, upon reaching subconfluency were
cultured in the presence of TNF-α (1 ng/ml) or LPS (1 µg/ml)
with or without caspase-3 inhibitor (10 µM) for 24 h
(Fig. 3C and D). The effects of
LPS or TNF-α on cell death were either not observed or diminished
in the presence of caspase-3 inhibitor (Fig. 3C). The stimulatory effects of TNF-α
or LPS on cell death were either not observed or diminished in
transfectants cultured with or without caspase-3 inhibitor
(Fig. 3D). These findings suggest
that the suppressive effects of regucalcin overexpression on cell
growth are likely due, at in least part, to the inhibition of
caspase-3 activity in A498 cells. Thus, the suppressive effects of
regucalcin overexpression on the proliferation of A498 cells are
not mediated by cell death.
Suppressive effects of regucalcin overexpression
on the proliferation of A498 cells are mediated through various
signaling pathways.
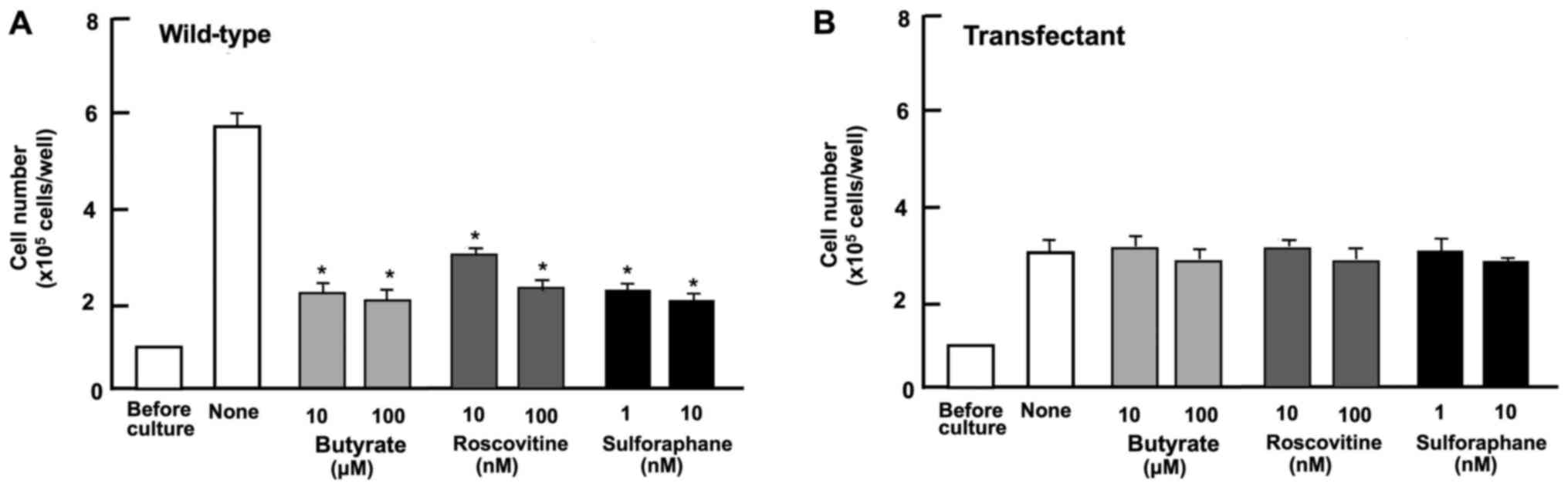
To determine the mechanisms through which regucalcin
overexpression suppresses the proliferation of A498 cells, we
investigated whether the suppressive effects of regucalcin
overexpression are attenuated in the presence of various inhibitors
that induce cell cycle arrest in vitro (Fig. 4). Wild-type cells were cultured for
3 days in the presence of butyrate (10 and 100 µM) (46), roscovitine (10 and 100 nM)
(47) or sulforaphane (1 and 10
nM) (48). The proliferation of
the wild-type cells was suppressed in the presence of these
inhibitors (Fig. 4A). The effects
of these inhibitors were not potentiated in the transfectants
(Fig. 4B). These results suggest
that the overexpression of regucalcin induces G1 and G2/M phase
cell cycle arrest in the A498 cells.
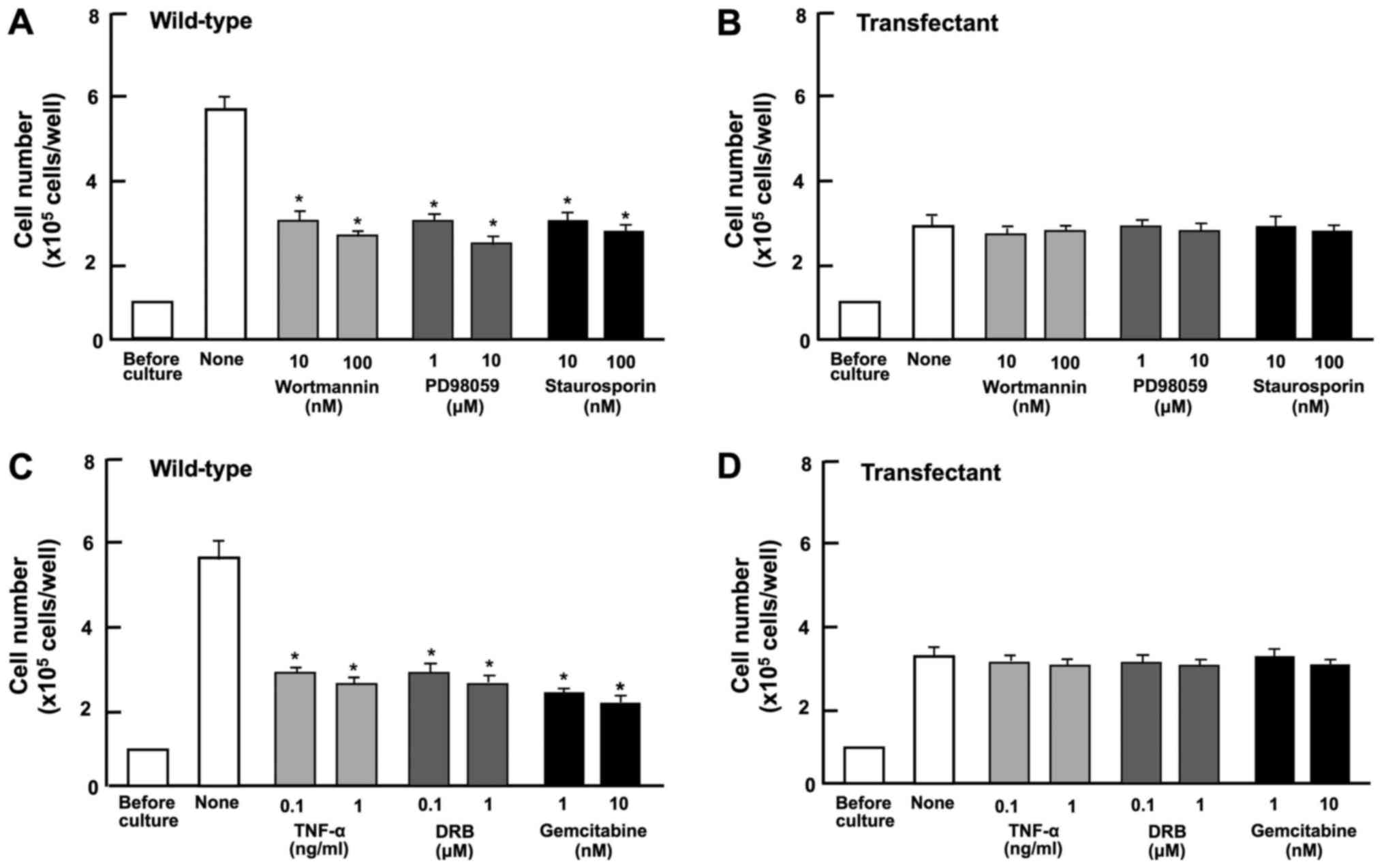
Subsequently, we determined the involvement of
signaling factors in the suppressive effect on cell proliferation
induced by the overexpression of regucalcin. The proliferation of
the A498 wild-type cells was suppressed in the presence of
wortmannin (0.1 or 1 µM), an inhibitor of PI3 kinase
(50), PD98059 (1 or 10
µM), an inhibitor of extracellular signal-regulated kinase
(ERK) and MAP kinase (51), and
staurosporine (10 or 100 nM), a calcium signaling-related inhibitor
(52) (Fig. 5A). The blocking of these pathways
did not potentiate the suppressive effects of regucalcin
overexpression of cell proliferation (Fig. 5B).
DRB is an inhibitor of RNA polymerase II-dependent
transcriptional activity (53).
Gemcitabine is a potent antitumor agent that induces nuclear DNA
damage (54). In the current
study, these inhibitors inhibited the proliferation of wild-type
cells (Fig. 5C). However, these
effects did not occur in the trans-fectants (Fig. 5D). These results suggest that the
overexpression of regucalcin suppresses various signaling processes
linked to cell proliferation, and that the
regucalcin-overexpressing cells exhibit a lack of responses to the
above-mentioned inhibitors of these pathways.
Overexpression of regucalcin regulates the
expression of various proteins linked to cell signaling and
transcriptional activity. We then investigated whether the
overexpression of regucalcin affects the expression of key protein
involved in signaling pathways and transcriptional activity. The
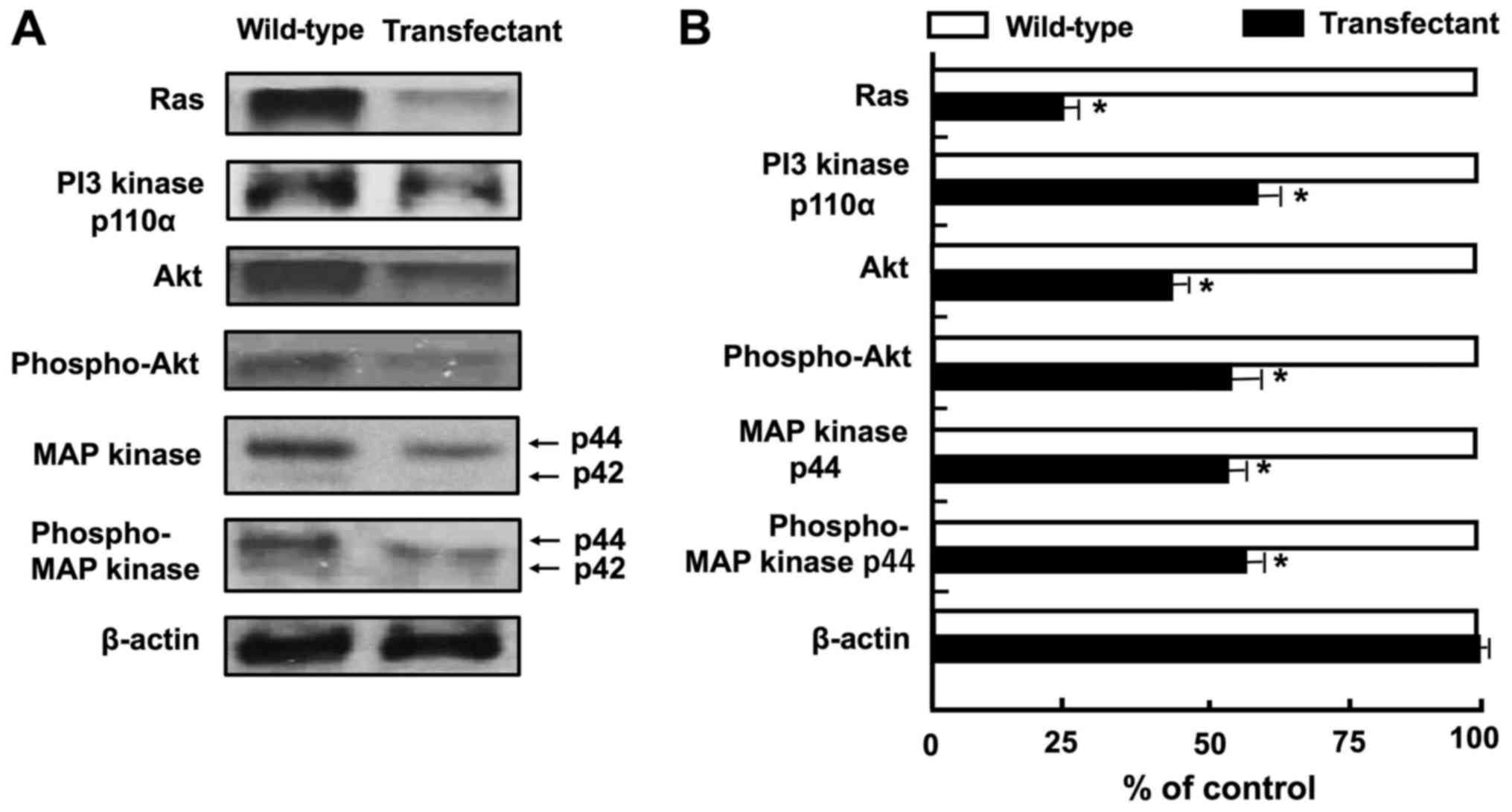
results of western blot analysis revealed that the levels of Ras,
PI3 kinase, Akt, phospho-Akt, MAP kinase and phospho-MAP kinase
were diminished by the overexpression of regucalcin (Fig. 6A and B). These results suggest that
the overexpression of regucalcin suppresses the activation of
Ras-linked signaling pathways in A498 cells. By contrast, the
overexpression of regucalcin elevated the protein levels of tumor
suppressors, p53 and Rb, and p21, an inhibitor of the cell cycle
(Fig. 6C and D). In addition, the
overexpression of regucalcin diminished the levels of c-fos, c-jun,
Stat3, β-catenin and NF-κB p65, which are transcription factors
linked to the proliferation of A498 cells (23,25,55)
(Fig. 6C and D). We determined the
changes in the levels of 14 proteins, which may be major signaling
proteins related to the proliferation of cancer cells, although
various other proteins are also implicated in the proliferation of
cancer cells.
Discussion
In this study, we performed the profiling of gene
expression and survival analysis of 52 patients with clear cell RCC
using the GEO database (GSE36895) for outcome analysis. The data
obtained demonstrated that the prolonged survival of patients with
RCC was associated with a higher regucalcin gene expression, and
that the diminished regucalcin gene expression was accompanied by
the poor prognosis of patients with RCC. This suggests that the
diminished regucalcin gene expression may partly contribute to the
development or aggressiveness of carcinogenesis in human RCC, and
may lead to a worse clinical outcome for patients with RCC.
Moreover, to determine a mechanism for this clinical finding, we
investigated whether regucalcin overexpression suppresses the
proliferation of the RCC A498 human clear cell line in
vitro. The overexpression of regucalcin was shown to suppress
colony formation and the proliferation of A498 cells without
inducing necrosis or apoptotic cell death in vitro. Thus,
this study demonstrated a crucial role of regucalcin in suppressing
the growth of human clear cell RCC cells. Endogenous regucalcin may
therefore play a suppressive role in the development of human RCC.
However, further studies using multi-datasets are warranted to
corroborate the results of this study.
The mechanistic characterization of the suppressive
effects of regucalcin overexpression on the proliferation of A498
cells was investigated using various inhibitors that regulate cell
signaling pathways. This suppressive effect was not potentiated by
butyrate, roscovitine or sulphoraphan, which induce cell cycle
arrest. Butyrate induces the inhibition of G1 progression (46). Roscovitine is a potent and
selective inhibitor of the cyclin-dependent kinase cdc2, cdk2m and
cdk5 (47). Sulforaphane induces
G2/M phase cell cycle arrest (48). Our data suggest that regucalcin
overexpression causes G1 and G2/M phase cell cycle arrest in A498
cells. Further experiments are required to confirm this finding
using other analysis of cell cycle. Similar effects of regucalcin
have been shown in various other types of cells, including normal
rat kidney proximal tubular epithelial NRK52E cells (49), cloned rat hepatoma H4-II-E cells
(44), human pancreatic cancer MIA
PaCa-2 cells (31), MDA-MB-231
breast cancer cells (32), liver
cancer HepG2 cells (33), lung
adenocarcinoma A549 cells (34)
and colorectal cancer RKO cells (35) in vitro. Notably, the
overexpression of regucalcin has been shown to increase the
expression of p21, a cell cycle inhibitor, supporting the view that
regucalcin plays a role in cell cycle arrest (22-26),
and we demonstrate herein that thus is also the case in A498
cells.
It was then investigated whether regucalcin
regulates cell signaling pathways using various inhibitors. The
suppressive effects of regucalcin overexpression on the growth of
A498 cells were not potentiated by staurosporine, an inhibitor of
protein kinase C (52),
wortmannin, an inhibitor of the PI3 kinase (PI3K)/Akt signaling
pathway (50), or PD98059, an
inhibitor of extracellular signal-regulated kinase (ERK)/MAP kinase
(also termed MAPK) (51). The
overexpression of regucalcin and its suppressive effects on cell
proliferation were associated with the inhibition of various
signaling pathways, namely Ca2+-dependent kinases,
PI3K/Akt and ERK/MAPK, in A498 cells. Thus, it is suggested that
regucalcin is a suppressor of diverse signaling pathways in human
RCC cells. Furthermore, the results of western blot analysis
revealed that regucalcin overexpression induced a decrease in the
levels of various proteins that are involved in signaling pathways
linked to Ras, PI3K, Akt and MAPK, in A498 cells (51). The suppressive effects of
regucalcin, and its regulation of various signaling pathways, have
also been observed in various other types of human cancer cells
in vitro (31-35).
The suppressive effects of regucalcin overexpression
on the proliferation of A498 cells were not altered by culture with
DRB, an inhibitor of RNA polymerase II-dependent transcriptional
activity (53), and were not
potentiated by culture with gemcitabine, which is used in the
therapy of human cancer as an antitumor agent that induces nuclear
DNA damage (54). This drug
inhibits the proliferation and stimulates apoptotic cell death in
various types of cancer cells (54). The results of this study suggest
that regucalcin partly regulates pathways implicated in the mode of
action of DRB and gemcitabine. Regucalcin has been demonstrated to
directly suppress DNA and RNA synthesis using isolated rat liver
nuclei (25).
Regucalcin has been shown to play a role in the
regulation of cell nuclear function (25). Importantly, the overexpression of
regucalcin has been demonstrated to enhance the gene expression
levels of the tumor suppressor p53 and Rb, and that of p21, an
inhibitor of the cell cycle, and to suppress the gene expression of
ras, c-fos and c-myc, oncogenes, due to binding to nuclear DNA in
cloned rat hepatoma H4-II-E cells in vitro (55). Similarly, in this study, regucalcin
overexpression was found to elevate the protein levels of the tumor
suppressors p53, Rb and p21 (44,56),
and diminish those of ras, c-fos, c-jun, Stat3, β-catenin and NF-κB
p65, which are transcription factors linked to cancer cell
proliferation, in A498 cells. These findings suggest that
endogenous regucalcin plays a pivotal role in suppressing the
growth of cancer cells due to regulation of the expression of
various proteins linked to transcription factors, tumor suppressors
and oncogenes involved in tumor development. Regucalcin binds to
DNA (55) and regulates the gene
expressions of various proteins in the nucleus of normal and cancer
cells (25).
In conclusion, the current study demonstrates that
the prolonged survival of patients with clear cell RCC is
associated with a higher regucalcin gene expression in the tumor
tissues, and that the overexpression of regucalcin suppresses
colony formation and proliferation in human clear cell RCC A498
cells in vitro. Endogenous regucalcin may play a potential
role as a suppressor in the development of human renal cancer. Our
previous studies have demonstrated that survival is prolonged in
patients with pancreatic cancer (31), breast cancer (32), liver cancer (33), lung adenocarcinoma (34), and colorectal cancer (35) exhibiting higher regucalcin gene
expression in their tumor tissues. Thus, endogenous regucalcin may
play a pivotal role as a suppressor of carcinogenesis in human
cancer of various types. The downregulation of the regucalcin gene
expression may lead to the development of carcinogenesis in human
subjects. Targeting regucalcin may be clinically significant in the
diagnosis and potential therapy for human cancer of various types.
The delivery of the regucalcin gene, which is overexpressed in
tumor tissues, may constitute a novel therapeutic approach to
treating human cancer.
Acknowledgments
Not applicable.
Funding
The present study was supported in part by the
Foundation for Biomedical Research on Regucalcin, Japan.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors’ contributions
MY conceived and designed the study. MY, SO, OH and
TM performed the experiments and discussed the findings. MY wrote
the manuscript, and SO, OH and TM reviewed and edited the
manuscript. All authors have read and approved the manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols used databases or cell
culture in vitro.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shroff EH, Eberlin LS, Dang VM, Gouw AM,
Gabay M, Adam SJ, Bellovin DI, Tran PT, Philbrick WM, Garcia-Ocana
A, et al: MYC oncogene overexpression drives renal cell carcinoma
in a mouse model through glutamine metabolism. Proc Natl Acad Sci
USA. 112:6539–6544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Juengel E, Afschar M, Makarević J, Rutz J,
Tsaur I, Mani J, Nelson K, Haferkamp A and Blaheta RA: Amygdalin
blocks the in vitro adhesion and invasion of renal cell carcinoma
cells by an integrin-dependent mechanism. Int J Mol Med.
37:843–850. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He YH, Chen C and Shi Z: The biological
roles and clinical implications of microRNAs in clear cell renal
cell carcinoma. J Cell Physiol. 233:4458–4465. 2018. View Article : Google Scholar
|
|
6
|
Sullivan S: Paraneoplastic cough and renal
cell carcinoma. Can Respir J. 2016:59385362016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Flanigan RC, Campbell SC, Clark JI and
Picken MM: Metastatic renal cell carcinoma. Curr Treat Options
Oncol. 4:385–390. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Capitanio U and Montorsi F: Renal cancer.
Lancet. 387:894–906. 2016. View Article : Google Scholar
|
|
9
|
Thakur A and Jain SK: Kidney cancer:
Current progress in treatment. World J Oncol. 2:158–165.
2011.PubMed/NCBI
|
|
10
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Yuan Y, Liang P, Guo X, Ying Y,
Shu X-S, Gao M Jr and Cheng Y: OSR1 is a novel epigenetic silenced
tumor suppressor regulating invasion and proliferation in renal
cell carcinoma. Oncotarget. 8:30008–30018. 2017.PubMed/NCBI
|
|
12
|
Rini BI and Atkins MB: Resistance to
targeted therapy in renal-cell carcinoma. Lancet Oncol.
10:992–1000. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rini BI: New strategies in kidney cancer:
Therapeutic advances through understanding the molecular basis of
response and resistance. Clin Cancer Res. 16:1348–1354. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Z, Zhu R and Zheng J, Chen C, Huang
C, Ma J, Xu C, Zhai W and Zheng J: Cryptotanshinone inhibits
proliferation yet induces apoptosis by suppressing STAT3 signals in
renal cell carcinoma. Oncotarget. 8:50023–50033. 2017.PubMed/NCBI
|
|
15
|
Yamaguchi M and Yamamoto T: Purification
of calcium binding substance from soluble fraction of normal rat
liver. Chem Pharm Bull (Tokyo). 26:1915–1918. 1978. View Article : Google Scholar
|
|
16
|
Yamaguchi M and Sakurai T: Inhibitory
effect of calcium-binding protein regucalcin on
Ca2+-activated DNA fragmentation in rat liver nuclei.
FEBS Lett. 279:281–284. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shimokawa N and Yamaguchi M: Molecular
cloning and sequencing of the cDNA coding for a calcium-binding
protein regucalcin from rat liver. FEBS Lett. 327:251–255. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shimokawa N, Matsuda Y and Yamaguchi M:
Genomic cloning and chromosomal assignment of rat regucalcin gene.
Mol Cell Biochem. 151:157–163. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thiselton DL, McDowall J, Brandau O,
Ramser J, d’Esposito F, Bhattacharya SS, Ross MT, Hardcastle AJ and
Meindl A: An integrated, functionally annotated gene map of the
DXS8026-ELK1 interval on human Xp11.3-Xp11.23: Potential hotspot
for neurogenetic disorders. Genomics. 79:560–572. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamaguchi M and Isogai M: Tissue
concentration of calcium-binding protein regucalcin in rats by
enzyme-linked immunoadsorbent assay. Mol Cell Biochem. 122:65–68.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakagawa T and Yamaguchi M: Hormonal
regulation on regucalcin mRNA expression in cloned normal rat
kidney proximal tubular epithelial NRK52E cells. J Cell Biochem.
95:589–597. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamaguchi M: Role of regucalcin in
maintaining cell homeostasis and function (Review). Int J Mol Med.
15:371–389. 2005.PubMed/NCBI
|
|
23
|
Yamaguchi M: Regucalcin and cell
regulation: role as a suppressor in cell signaling. Mol Cell
Biochem. 353:101–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamaguchi M: The potential role of
regucalcin in kidney cell regulation: Involvement in renal failure
(Review). Int J Mol Med. 36:1191–1199. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamaguchi M: Role of regucalcin in cell
nuclear regulation: Involvement as a transcription factor. Cell
Tissue Res. 354:331–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamaguchi M: Suppressive role of
regucalcin in liver cell proliferation: Involvement in
carcinogenesis. Cell Prolif. 46:243–253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yamaguchi M: The anti-apoptotic effect of
regucalcin is mediated through multisignaling pathways. Apoptosis.
18:1145–1153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yamaguchi M: The Role of Regucalcin in
Cell Homeostasis and Disorder. Nova Science Publishers Inc.; New
York, NY: pp. 1–288. 2017
|
|
29
|
Yamaguchi M: Involvement of regucalcin as
a suppressor protein in human carcinogenesis: Insight into the gene
therapy. J Cancer Res Clin Oncol. 141:1333–1341. 2015. View Article : Google Scholar
|
|
30
|
Murata T and Yamaguchi M: Alternatively
spliced variants of the regucalcin gene in various human normal and
tumor tissues. Int J Mol Med. 34:1141–1146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yamaguchi M, Osuka S, Weitzmann MN,
El-Rayes BF, Shoji M and Murata T: Prolonged survival in pancreatic
cancer patients with increased regucalcin gene expression:
Overexpression of regucalcin suppresses the proliferation in human
pancreatic cancer MIA PaCa-2 cells in vitro. Int J Oncol.
48:1955–1964. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yamaguchi M, Osuka S, Weitzmann MN, Shoji
M and Murata T: Increased regucalcin gene expression extends
survival in breast cancer patients: Overexpression of regucalcin
suppresses the proliferation and metastatic bone activity in
MDA-MB-231 human breast cancer cells in vitro. Int J Oncol.
49:812–822. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yamaguchi M, Osuka S, Weitzmann MN,
El-Rayes BF, Shoji M and Murata T: Prolonged survival in
hepatocarcinoma patients with increased regucalcin gene expression:
HepG2 cell proliferation is suppressed by overexpression of
regucalcin in vitro. Int J Oncol. 49:1686–1694. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamaguchi M, Osuka S, Shoji M, Weitzmann
MN and Murata T: Survival of lung cancer patients is prolonged with
higher regucalcin gene expression: Suppressed proliferation of lung
adenocarcinoma A549 cells in vitro. Mol Cell Biochem. 430:37–46.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamaguchi M, Osuka S and Murata T:
Prolonged survival of colorectal cancer patients is associated with
higher regucalcin gene expression: Overexpressed regucalcin
suppresses growth of human colorectal carcinoma cells in vitro. Int
J Oncol. 53:1313–1322. 2018.PubMed/NCBI
|
|
36
|
Peña-Llopis S, Vega-Rubín-de-Celis S, Liao
A, Leng N, Pavía-Jiménez A, Wang S, Yamasaki T, Zhrebker L,
Sivanand S, Spence P, et al: BAP1 loss defines a new class of renal
cell carcinoma. Nat Genet. 44:751–759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Uhlen M, Oksvold P, Fagerberg L, Lundberg
E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S,
et al: Towards a knowledge-based Human Protein Atlas. Nat
Biotechnol. 28:1248–1250. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar
|
|
39
|
Cancer Genome Atlas Research Network:
Comprehensive molecular characterization of clear cell renal cell
carcinoma. Nature. 499:43–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Aguirre-Gamboa R, Gomez-Rueda H,
Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R,
Rodriguez-Barrientos A, Tamez-Peña JG and Treviño V: SurvExpress:
An online biomarker validation tool and database for cancer gene
expression data using survival analysis. PLoS One. 8:e742502013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma X, Gu L, Li H, Gao Y, Li X, Shen D,
Gong H, Li S, Niu S, Zhang Y, et al: Hypoxia-induced overexpression
of stanniocalcin-1 is associated with the metastasis of early stage
clear cell renal cell carcinoma. J Transl Med. 13:562015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Misawa H, Inagaki S and Yamaguchi M:
Suppression of cell proliferation and deoxyribonucleic acid
synthesis in the cloned rat hepatoma H4-II-E cells overexpressing
regucalcin. J Cell Biochem. 84:143–149. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fang Z, Tang Y, Fang J, Zhou Z, Xing Z,
Guo Z, Guo X, Wang W, Jiao W, Xu Z and Liu Z: Simvastatin inhibits
renal cancer cell growth and metastasis via AKT/mTOR, ERK and
JAK2/STAT3 pathway. PLoS One. 8:e628232013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yamaguchi M and Daimon Y: Overexpression
of regucalcin suppresses cell proliferation in cloned rat hepatoma
H4-II-E cells: Involvement of intracellular signaling factors and
cell cycle-related genes. J Cell Biochem. 95:1169–1177. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Izumi T and Yamaguchi M: Overexpression of
regucalcin suppresses cell death in cloned rat hepatoma H4-II-E
cells induced by tumor necrosis factor-alpha or thapsigargin. J
Cell Biochem. 92:296–306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Charollais RH, Buquet C and Mester J:
Butyrate blocks the accumulation of CDC2 mRNA in late G1 phase but
inhibits both the early and late G1 progression in chemically
transformed mouse fibroblasts BP–A31. J Cell Physiol. 145:46–52.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Meijer L, Borgne A, Mulner O, Chong JP,
Blow JJ, Inagaki N, Inagaki M, Delcros JG and Moulinoux JP:
Biochemical and cellular effects of roscovitine, a potent and
selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and
cdk5. Eur J Biochem. 243:527–536. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Singh SV, Herman-Antosiewicz A, Singh AV,
Lew KL, Srivastava SK, Kamath R, Brown KD, Zhang L and Baskaran R:
Sulforaphane-induced G2/M phase cell cycle arrest involves
checkpoint kinase 2-mediated phosphorylation of cell division cycle
25C. J Biol Chem. 279:25813–25822. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nakagawa T, Sawada N and Yamaguchi M:
Overexpression of regucalcin suppresses cell proliferation of
cloned normal rat kidney proximal tubular epithelial NRK52E cells.
Int J Mol Med. 16:637–643. 2005.PubMed/NCBI
|
|
50
|
Serrano-Nascimento C, da Silva Teixeira S,
Nicola JP, Nachbar RT, Masini-Repiso AM and Nunes MT: The acute
inhibitory effect of iodide excess on sodium/iodide symporter
expression and activity involves the PI3K/Akt signaling pathway.
Endocrinology. 155:1145–1156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pelech SL, Charest DL, Mordret GP, Siow
YL, Palaty C, Campbell D, Charlton L, Samiei M and Sanghera JS:
Networking with mitogen-activated protein kinases. Mol Cell
Biochem. 127–128. 157–169. 1993.
|
|
52
|
Tamaoki T, Nomoto H, Takahashi I, Kato Y,
Morimoto M and Tomita F: Staurosporine, a potent inhibitor of
phospholipid/Ca++ dependent protein kinase. Biochem
Biophys Res Commun. 135:397–402. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Palangat M, Grass JA, Langelier MF,
Coulombe B and Landick R: The RPB2 flap loop of human RNA
polymerase II is dispensable for transcription initiation and
elongation. Mol Cell Biol. 31:3312–3325. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tang SC and Chen YC: Novel therapeutic
targets for pancreatic cancer. World J Gastroenterol.
20:10825–10844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tsurusaki Y and Yamaguchi M: Role of
regucalcin in liver nuclear function: Binding of regucalcin to
nuclear protein or DNA and modulation of tumor-related gene
expression. Int J Mol Med. 14:277–281. 2004.PubMed/NCBI
|
|
56
|
Tsurusaki Y and Yamaguchi M:
Overexpression of regucalcin modulates tumor-related gene
expression in cloned rat hepatoma H4-II-E cells. J Cell Biochem.
90:619–626. 2003. View Article : Google Scholar : PubMed/NCBI
|