Introduction
Lung cancer remains the leading cause of cancer
mortality and has become an increasingly serious public health
burden globally (1). Lung cancer
is categorized into small cell lung cancer (SCLC) and non-SCLC
(NSCLC) following pathological pattern. NSCLC accounts for ~80% of
all lung cancer cases, including squamous cell carcinoma,
adenocarcinoma, large cell carcinoma and adenosquamous cell
carcinoma (2). NSCLC is often
diagnosed at an advanced stage and the 5-year survival rate remains
~15% (3). Therefore, there is an
urgent requirement to identify and screen for tumor-associated
genes, further investigate the molecular mechanisms of NSCLC
tumorigenesis and development, and identify novel biomarkers for
early diagnosis and therapeutic targets of NSCLC. Tumor-associated
genes rely on diverse signaling pathways to influence the
proliferation, migration and invasion of tumor cells, and
subsequently disseminate from primary tumors to metastasize through
lymphatic and blood vessels. Understanding the molecular mechanisms
underlying tumor metastasis has a critical significance in
formulating treatment and early diagnosis. Receptor tyrosine
kinases (RTKs) serve a vital function in regulating cellular
process, including cell migration, proliferation, differentiation,
survival and cell cycle regulation (4,5).
Deregulation of RTKs includes chromosomal translocations, autocrine
activation, overexpression of RTKs or gain-of-function mutations
which occurs in cancer (6). The
downstream of tyrosine kinase (DOK) protein family consists of
seven members, mediating intracellular signaling transduction
downstream of RTKs (7-9). DOK proteins share a topology domain
characterized by a central phosphotyrosine-binding (PTB) domain, an
N-terminal pleckstrin homology (PH) domain and Src homology 2
target motifs in the C-terminus (7,10).
On the basis of their location, DOK proteins are divided into three
subgroups: DOKs 1-3 are mainly expressed in hematopoietic tissues
(11), DOKs 4-6 are primarily
present within the nervous system (12,13),
and DOK7 is predominantly in skeletal muscle and the heart, which
exhibits a distinct function compared with other members (14). Aberrant expression of DOK proteins
has been identified in multiple malignancies. DOKs 1-3
co-operatively suppress aggressive histiocytic sarcoma (11). DOK2 was regarded as a biomarker of
patients with gastric cancer with poor prognosis (15,16).
In a mouse model, deletion of DOKs 1-3 resulted in abnormal
proliferation of bronchoalveolar stem cells and alveolar type II
cells, which subsequently progressed to lung cancer (7). In addition, a decreased expression
level of DOK2 was observed in human lung cancer tissues, and forced
DOK2 overexpression repressed proliferation of lung cancer cells
(7).
DOK7, as a non-catalytic adaptor protein, is
essential for the activation of the tyrosine kinase muscle-specific
kinase (MuSK) and acetylcholine receptor clustering, which are
indispensable for neuromuscular junctions (17,18).
The function of DOK7 in cancer was revealed gradually.
Demethylation of DOK7 decreased the proliferation and invasion of
esophageal squamous cell carcinoma cells (19). The PTB domain is the region through
which DOK7 interacts with the juxtamembrane region of MuSK,
although the PTB and PH domains are required for the activation of
MuSK (18,20,21).
The PH domain consists of ~120 amino acids, mediating intracellular
and extracellular signaling, also acting as important constituents
of the cytoskeleton (22-24). PTB was also able to contact the
cell membrane, and regulate intracellular and extracellular
signaling. For example, PH and PTB domains of DOK4 are required for
DOK4 localization at the membrane (10). The DOK1 PH domain appears to be
required for tyrosine phosphorylation of the protein and its normal
localization to a subcellular membrane component (10).
In our previous study, we identified that DOK7
transcripts were decreased in lung cancer, and the lower DOK7 level
was associated with poorer overall survival and progression-free
survival (25). Overexpression of
the DOK 7 transcript variant 1 (DOK7V1) limited the proliferation
and migration, but enhanced the adhesion to the extracellular
matrix, of lung cancer cells (25). However, the underlying molecular
mechanism of DOK7V1 and the functions of its domains in lung cancer
cells remain unknown. Therefore, the aim of the present study was
to investigate the functions of its domains and the molecular
mechanism underlying its involvement in lung cancer cells.
Materials and methods
Cell culture
Lung cancer A549, SKMES-1 and H3122 cell lines were
purchased from the American Type Culture Collection (Manassas, VA,
USA). A549 and SKMES-1 cells were routinely cultured in Dulbecco’s
modified Eagle’s medium (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) and H3122 cells were cultured in RPMI-1640 medium (Thermo
Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum
(Shanghai ExCellBio, Inc., Shanghai, China), in an incubator at
37°C with a 95% humidified atmosphere containing 5%
CO2.
Construction of DOK7V1 plasmid and
transfection
DOK7V1-Flag and pEnter (Vector) plasmids were
purchased from Vigene Biosciences, Inc. (Rockville, MD, USA).
DOK7V1Δ-PH and DOK7V1Δ-PTB were constructed using a Q5®
Site-Directed Mutagenesis kit (Without Competent Cells) (New
England BioLabs, Inc., Ipswich, MA, USA). Purified DOK7V1 and
control plasmids were transfected into A549 and SKMES-1 cells using
an Easyject Plus electroporator (EquiBio Ltd., Altrincham, UK).
H3122 cells were transfected with DOK7V1, DOK7V1Δ-PH and
DOK7V1Δ-PTB, respectively, using Neofect™ DNA transfection reagent
(Neofectbiotech Co., Ltd., Beijing, China).
RNA isolation and reverse
transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from A549, SKMES-1 and H3122
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). cDNA was generated from each 1 µg RNA
sample using a QuantiTect Reverse Transcription kit (Qiagen GmbH,
Hilden, Germany), according to the manufacturer’s protocol. PCR was
performed as described previously (26) with the following primer sequences:
DOK7, 5′-ACTGGGCTGGCGTCTTCTTCC-3′ (forward) and
5′-TCGGACGATGCAGTCGAACAG-3′ (reverse); and GAPDH,
5′-GGCTGCTTTTAACTCTGGTA-3′ (forward) and 5′-GACTGTGGTCATGAGTCCTT-3′
(reverse).
Western blot analysis
Cells were collected and lysed using
Radioimmunoprecipitation Assay Lysis and Extraction Buffer (Thermo
Fisher Scientific, Inc.), followed by centrifugation at 13,000 × g
for 15 min at 4°C. Total protein concentrations in the supernatant
were determined using a DC Protein Assay kit (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) and an ELx800 spectrophotometer (Bio-Tek
Instruments, Inc., Winooski, VT, USA). Equal amounts (30 µg)
of protein samples were separated by SDS-PAGE (10% gel) and blotted
onto a nitrocellulose membrane. The membrane was blocked using 5%
skimmed milk in Tris-buffered saline for 1 h at room temperature.
Proteins were then probed using anti-human FLAG (1:1,000; cat. no.
F3040; Sigma; Merck KGaA, Darmstadt, Germany), anti-DOK7 antibody
(1:1,000; cat. no. ab75049; Abcam, Cambridge, MA, USA),
anti-protein kinase B (AKT) antibody (1:1,000; cat. no. 2920; Cell
Signaling Technology, Inc., Danvers, MA, USA), anti-phospho (p)-AKT
(1:1,000; cat. no. 4051; Cell Signaling Technology, Inc.),
anti-phosphoinositide 3-kinase (PI3K) antibody (1:500; cat. no.
sc-365290; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
anti-p-PI3K antibody (1:500; cat. no. sc-374534; Santa Cruz
Biotechnology, Inc.), anti-mammalian target of rapamycin (mTOR)
antibody (1:1,000; cat. no. ab2732; Abcam), anti-p-mTOR antibody
(1:1,000; cat. no. 2971; Cell Signaling Technology, Inc.),
anti-Rho-associated protein kinase (Rock) antibody (1:500; cat. no.
sc-17794; Santa Cruz Biotechnology, Inc.), anti-p-Rock antibody
(1:1,000; cat. no. ab2732; Abcam), anti-focal adhesion kinase (FAK)
antibody (1:500; cat. no. sc-1688; Santa Cruz Biotechnology, Inc.),
anti-p-FAK antibody (1:500; cat. no. sc-11766; Santa Cruz
Biotechnology, Inc.), anti-paxillin antibody (1:500; cat. no.
sc-365174; Santa Cruz Biotechnology, Inc.), anti-p-paxillin
antibody (1:500; cat. no. sc-365020; Santa Cruz Biotechnology,
Inc.), anti-human GAPDH antibody (1:500; cat. no. sc-47724; Santa
Cruz Biotechnology, Inc.) and corresponding horseradish peroxidase
(HRP)-conjugated goat anti-rabbit and goat anti-mouse
immunoglobulin G secondary antibodies (1:3,000; cat. nos. ZB-2301M
and ZB-2305; OriGene Technologies, Inc., Beijing, China),
respectively. Protein bands were visualized and analyzed using
Luminata Forte western blot HRP substrate (Merck KGaA) and a UV
imager (Uvitec, Inc., Cambridge, UK).
Immunofluorescence
Cells on glass coverslips were rinsed with PBS three
times, and then fixed with 4% paraformaldehyde for 20 min at room
temperature. Anti-human FLAG was diluted 1:1,000 in blocking buffer
(1% bovine serum albumin in PBS), prior to adding to the coverslips
and incubating for 1 h at room temperature. Following three washes
with PBS, the coverslips were incubated using Alexa
Fluor® 488-conjugated secondary antibodies (1:100; cat.
no. A-21205; Thermo Fisher Scientific, Inc.) for 45 min at room
temperature. Following three further washes with PBS, nuclei were
stained with DAPI (1:500,000; cat. no. D-1306; Thermo Fisher
Scientific, Inc.) for 2 min at room temperature, followed by three
further washes with PBS. The coverslips were mounted on glass
slides and incubated at room temperature overnight.
Immunofluorescence images were visualized using a confocal
microscopy system (Leica TCS SP8; Leica Microsystems GmbH, Wetzlar,
Germany) with a 63X oil-immersion objective.
In vitro cell proliferation assays
A total of 3,000 cells in 200 µl culture
medium was added to each well of a 96-well plate. A total of five
plates were used to obtain cell density readings following
incubation at 37°C for up to 4 days. Following incubation, the
medium was removed and 100 µl 10% Cell Counting Kit-8
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added
for 1 h at 37°C. Subsequently, the absorbance was determined at a
wavelength of 450 nm using a spectrophotometer (BioTek Instruments,
Inc.).
In vitro tumor cell migration assay
Cells (1×106 cells/well) were seeded in a
6-well plate and cultured in an incubator at 37°C overnight. The
cell monolayer was scratched with a 200-µl pipette tip to
create an artificial wound prior to washing twice with PBS to
remove floating cells. The migration of cells was monitored and
recorded every 6 h using an inverted light microscope at ×10 and
×20 magnification for 24 h. The wound distance was determined and
analyzed using ImageJ software (version 1.62; National Institutes
of Health, Bethesda, MD, USA).
In vitro tumor cell Matrigel adhesion
assay
A 96-well plate was coated with 5 µg
Matrigel/100 µl/well and air-dried overnight at room
temperature. Following rehydration, 20,000 cells/200 µl/well
were seeded into each well for 40 min. Following incubation, medium
was removed and the plate was washed three times with PBS to remove
the non-adherent cells. Adherent cells were fixed with 4%
formaldehyde for 30 min prior to being stained with 0.5% crystal
violet solution for 10 min at room temperature. Subsequently,
crystal violet was dissolved in 10% acetic acid and the absorbance
at 570 nm was determined using a spectrophotometer (BioTek
Instruments, Inc.).
Gene set enrichment analysis (GSEA)
The association between biological
processes/pathway, phenotypes and DOK7 expression level was
analyzed using the GSEA program (version 2.2; www.broad.mit.edu/gsea). Samples from The Cancer
Genome Atlas (TCGA) datasets were separated into high or low DOK7
expression groups using the median as the cut-off. GSEA was used to
calculate a pathway Enrichment Score that assessed whether genes
from a predefined gene set of PI3K/AKT/mTOR and FAK/paxillin
signaling pathways were enriched among the high- (or low-) ranked
genes or distributed randomly. Default settings were utilized when
using software applications. Significance was determined by
permutation analysis (1,000 permutations) and calculation of the
false discovery rate (FDR). A gene set was considered to be
significantly enriched when the FDR score was <0.05.
Statistical analysis
Data are presented as the mean ± standard deviation.
Experimental procedures were repeated independently at least three
times. Statistical analysis was performed using a two-sided
Student’s t-test for two-group comparisons and by one-way ANOVA,
followed by a Bonferroni post hoc test, for multiple group
comparisons. All statistical analyses were performed using Prism
(version 5; GraphPad Software, Inc., La Jolla, CA, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
Overexpression of DOK7V1 has an
inhibitory effect on proliferation and migration, but a positive
effect on adhesion in lung cancer cells
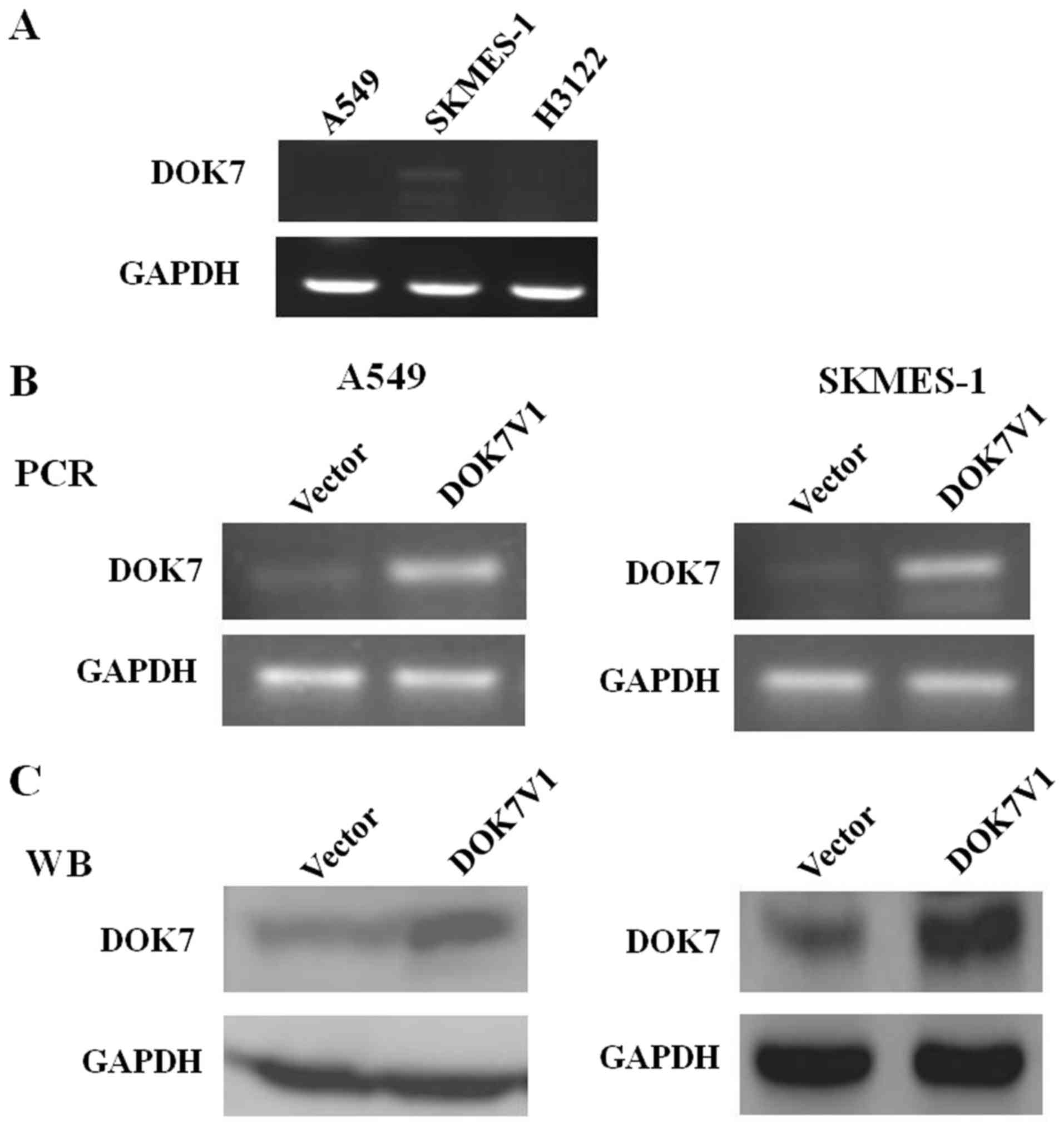
To select the optimal cell lines for further
investigation, the expression of DOK7V1 in A549, SKMES-1 and H3122
cells was determined using RT-PCR analysis. The stable transfection
of DOK7V1 in the lung cancer cell lines was verified by RT-PCR and
western blot analysis (Fig. 1). To
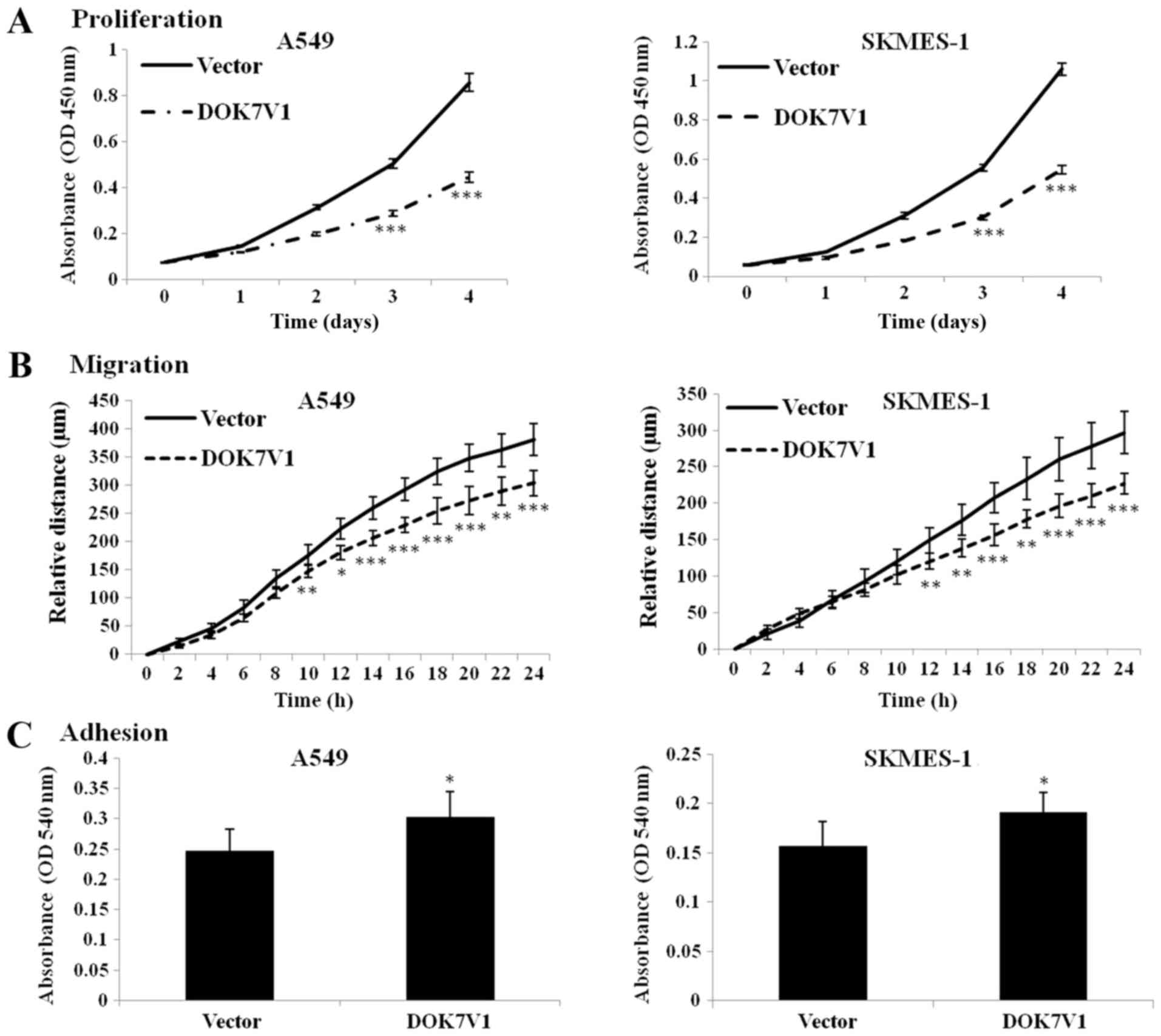
further confirm the effect of DOK7V1 on malignant phenotypes of
lung cancer cells, the experiments were performed in A549 and
SKMES-1 cells. The results indicated that overexpression of DOK7V1
significantly decreased the proliferation and migration, but
significantly increased the adhesion, of A549 and SKMES-1 cells
(Fig. 2).
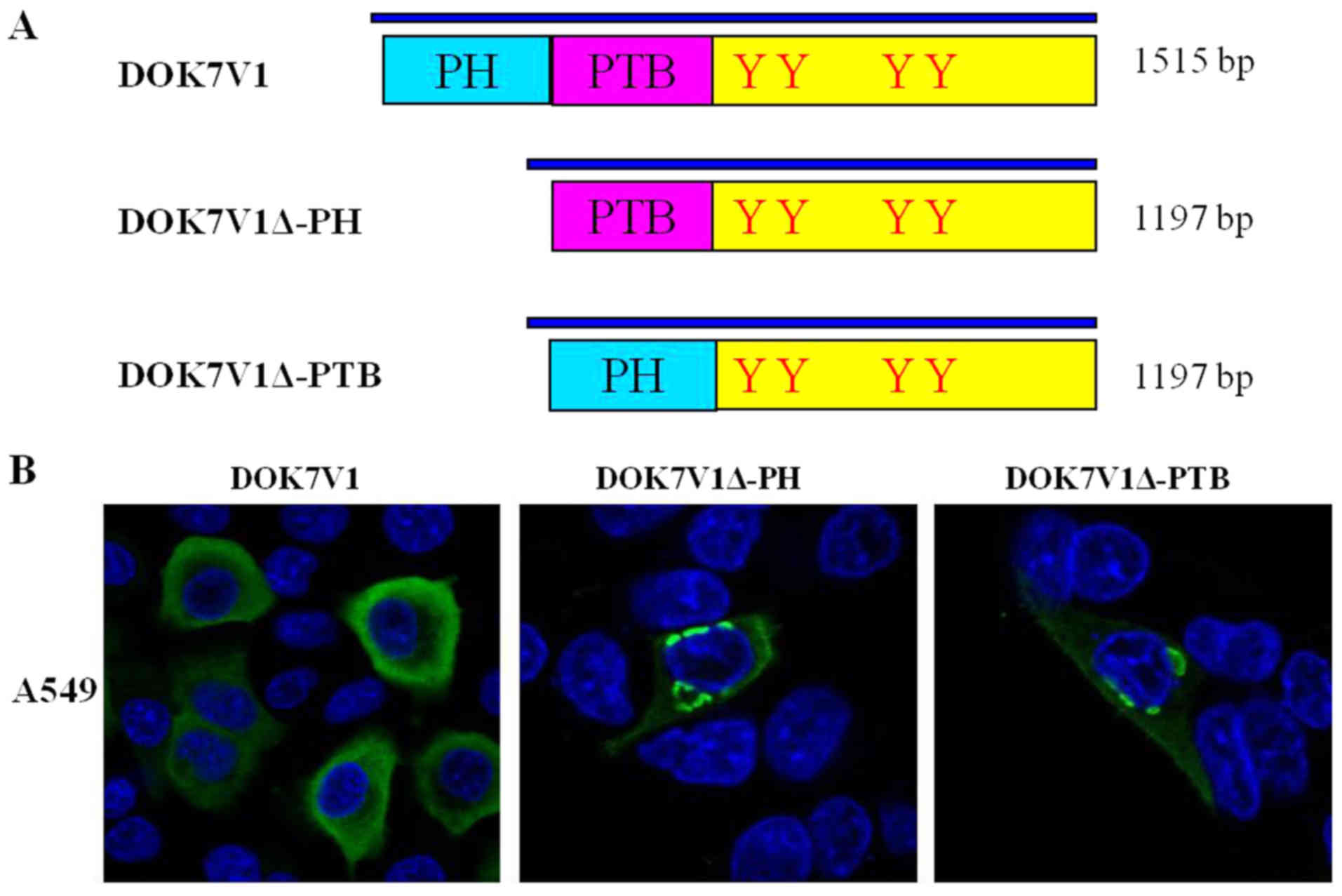
PH domain and PTB domain determined the
distribution of DOK7V1 in membrane and cytoplasm
DOK7V1 contains two functional domains: A PH domain
and a PTB domain. To investigate the structural determinant
regulating the intracellular distribution and expression of DOK7V1,
as well as the biological function, two DOK7V1 truncated fragments
were constructed based on the wild-type protein, DOK7V1Δ-PH and
DOK7V1Δ-PTB (Fig. 3A). The
immunofluorescence imaging results indicated that wild-type DOK7V1
was located and expressed in the membrane and cytoplasm. The
truncated versions of DOK7V1 without the two functional domains led
to a shift in the localization of DOK7V1 protein from the cytoplasm
to be perinuclear (Fig. 3B). The
results indicated that these two functional domains determined the
distribution of DOK7V1 in the membrane and cytoplasm, which may
influence the function of DOK7V1 in lung cancer cells.
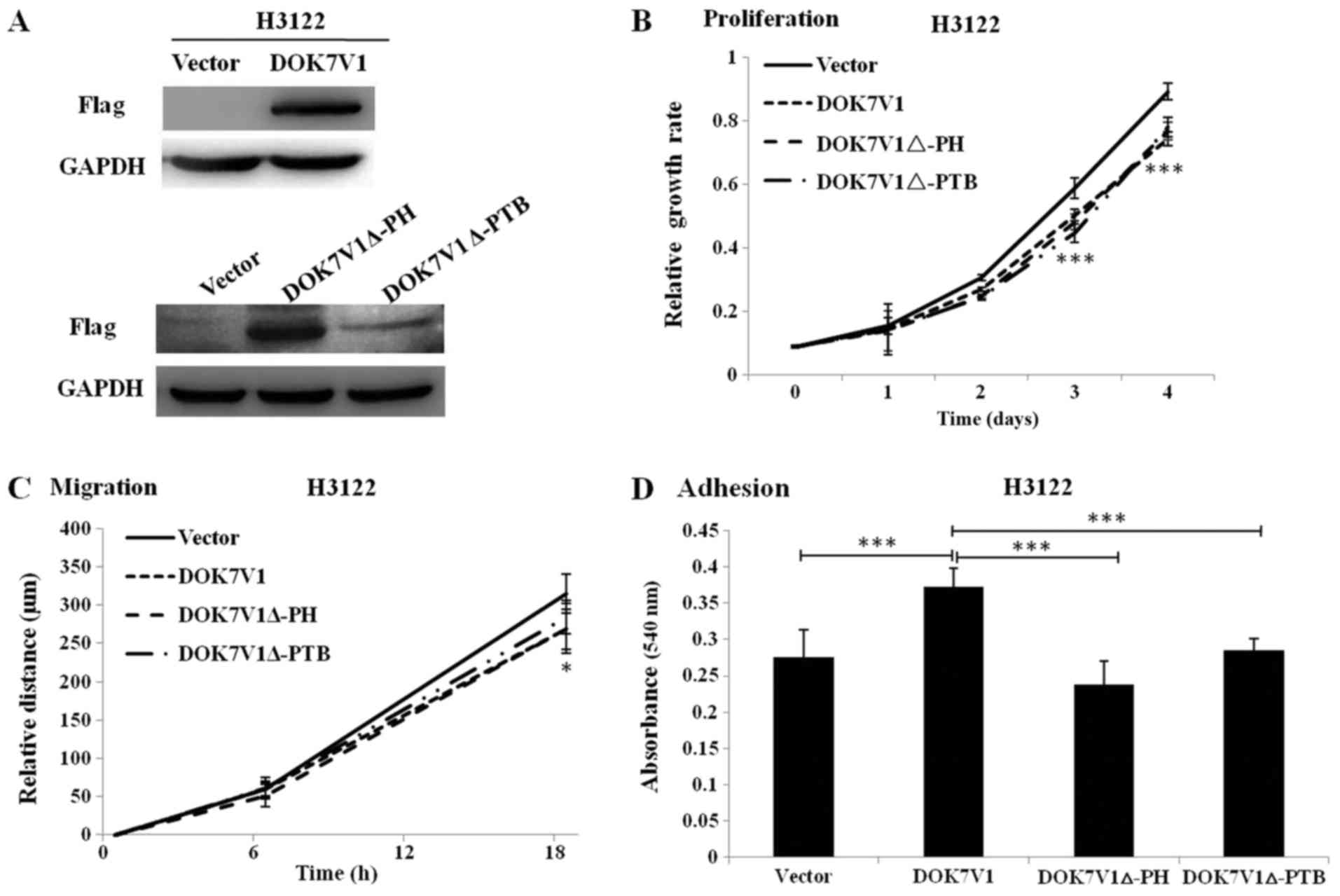
Effect of the PH domain and PTB domain of
DOK7V1 on the proliferation, migration and adhesion of lung cancer
cells
To further assess the biological function of
DOK7V1Δ-PH and DOK7V1Δ-PTB, these truncated versions were stably
transfected into H3122 cells and their expression was determined by
western blotting. DOK7V1, DOK7V1Δ-PH and DOK7V1Δ-PTB were
identified to exhibit increased expression in stably transfected
cells compared with in the corresponding control cells (Fig. 4A). It was identified that cell
proliferation was significantly decreased from the third day, and
migration was significantly decreased at 18 h, in cells singly
overexpressing DOK7V1Δ-PH and DOK7V1Δ-PTB compared with vector
control cells, which exhibited the same trend as DOK7V1 (Fig. 4B and 4C). The adhesion assay revealed an
increased rate in DOK7V1-overexpressing cells compared with the
control group; however, the DOK7V1Δ-PH and DOK7V1Δ-PTB truncated
versions lost the tumor-promoting effects identified in DOK7V1
(Fig. 4D).
No differences exist between the two
truncated versions and DOK7V1 in inhibiting PI3K/AKT/mTOR signal
pathway phosphorylation
To further study the signal pathways in which DOK7V1
is involved, the protein levels and phosphorylation levels of AKT,
PI3K, mTOR and Rock in A549 cells were determined. Decreased levels
of AKT, PI3K and Rock phosphorylation were observed in A549 cells
with DOK7V1 overexpression, whereas the total AKT, PI3K and Rock
protein levels were similar in comparison with their corresponding
controls (Fig. 5A). mTOR
phosphorylation and the total mTOR protein levels were decreased in
A549 cell lines with DOK7V1 overexpression (Fig. 5A). The AKT phosphorylation status
was investigated in H3122 cells transfected with various DOK7V1
constructs. Wild-type DOK7V1 overexpression significantly arrested
AKT activation, and this inhibition was similar in cells expressing
the DOK7V1Δ-PH and DOK7V1Δ-PTB truncated versions (Fig. 5B). Taken together, these results
indicated that the two truncated version of DOK7V1 did not
eliminate the inhibitory function of DOK7V1 in activating AKT.
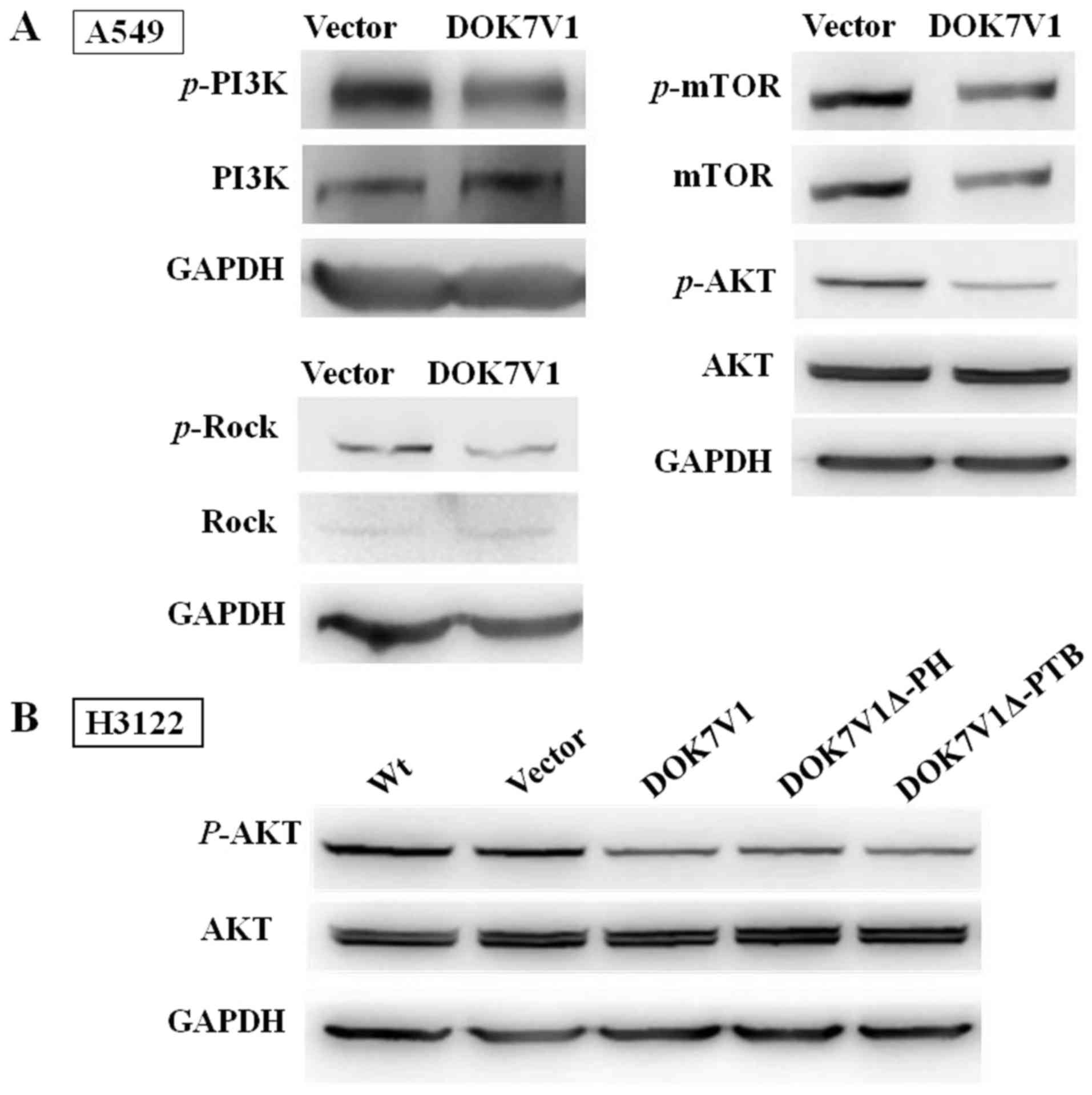 | Figure 5No differences exist between the two
truncated versions and DOK7V1 in inhibiting PI3K/AKT/mTOR signal
pathway phosphorylation. (A) Immunoblotting was performed using
anti-p-AKT and anti-AKT, p-mTOR and mTOR, p-PI3K and PI3K, Rock and
p-Rock. GAPDH, AKT and mTOR were separated in the same lane of the
gel. (B) DOK7V1Δ-PH and DOK7V1Δ-PTB did not result in the
loss-of-function of the DOK7V1-mediated inhibition of AKT
activation. DOK7V1, downstream of tyrosine kinase 7 transcript
variant 1; PI3K, phosphoinositide 3-kinase; AKT, protein kinase B;
mTOR, mammalian target of rapamycin; p-, phospho-; Rock,
Rho-associated protein kinase; PH, pleckstrin homology; PTB,
phosphotyrosine-binding; Wt, wild-type (cells without
treatment). |
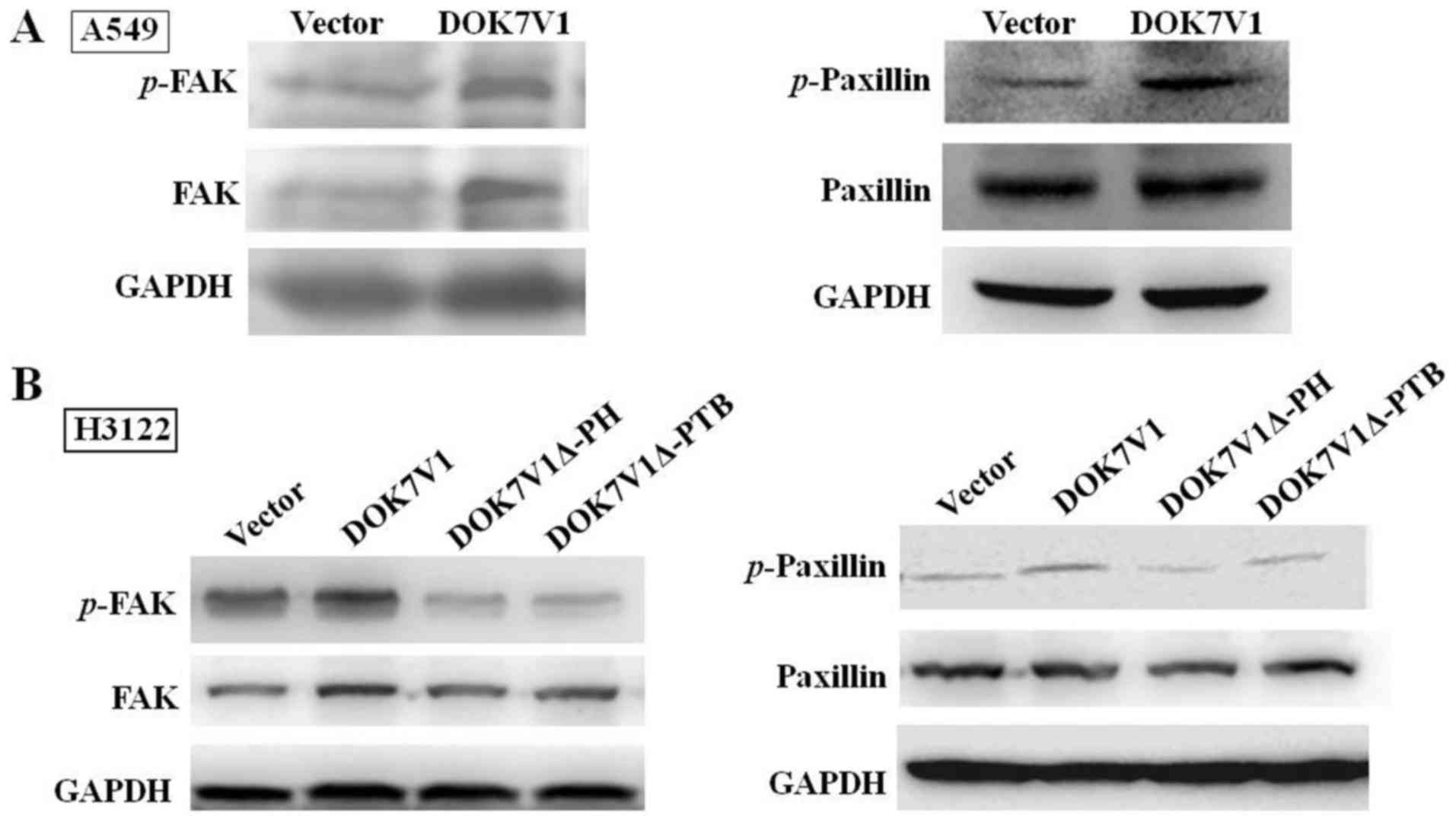
DOK7V1Δ-PH and DOK7V1Δ-PTB eliminate the
activating effect of DOK7V1 on FAK and paxillin
FAK and paxillin are important proteins in focal
adhesion, by connecting the cytoskeleton to the extracellular
matrix. In view of DOK7V1 increasing the adhesion of lung cancer
cells, the changes in associated factors in the FAK/paxillin
signaling pathway were investigated. The overexpression of DOK7V1
significantly increased FAK and paxillin phosphorylation (Fig. 6A) in A549 cells; however, this
influence was partially eliminated in H3122 cells expressing
DOK7V1Δ-PH and DOK7V1Δ-PTB in cells (Fig. 6B). These results indicated that the
PH domain and PTB domain serve a key function in regulating cell
adhesion.
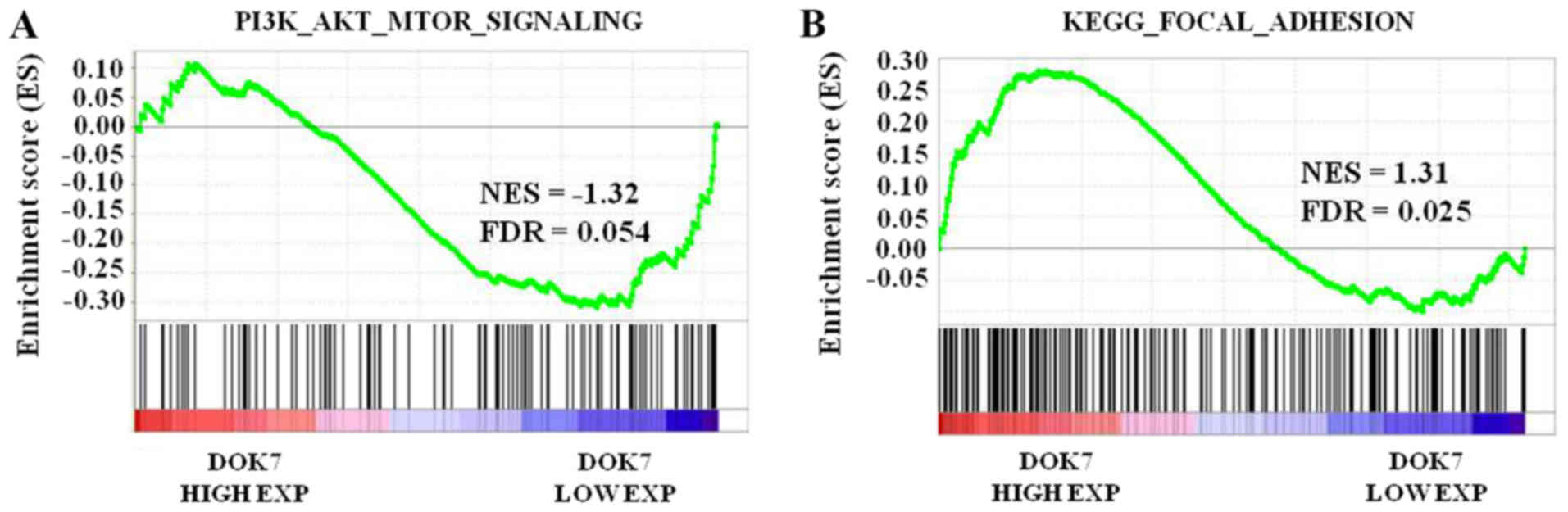
DOK7 overexpression is negatively
associated with activation of the PI3K/AKT/mTOR signaling pathway
and is positively associated with adherent ability
To further analyze the function of DOK7 expression
in PI3K/AKT/mTOR signaling in lung cancer, GSEA was performed using
a lung cancer dataset from TCGA. The lung cancer samples were
separated according to the median DOK7 level in the specimens into
high or low expression groups. Enrichment plots of GSEA
demonstrated that the gene set of PI3K/AKT/mTOR signaling was
primarily enriched in the low DOK7 expression group (Fig. 7A). The association between DOK7
expression and activation of FAK/paxillin signaling in lung cancer
was identified using GSEA. As presented in Fig. 7B, the gene signatures of adhesion,
which emphasized the importance of FAK in its description, were
enriched in patients with high DOK7 expression. These results
further suggested that, in lung cancer samples, DOK7 expression was
negatively associated with PI3K/AKT/mTOR signaling activation and
is positively associated with activation of the FAK/paxillin
signaling pathway.
Discussion
In our previous study, we preliminarily studied the
function of DOK7V1 in lung cancer cells and the association with
the prognosis of patients with lung cancer (25). In the present study, the focus was
on the mechanisms and functions of DOK7V1 and its domains in the
lung cancer cells. The results of the present study suggested a
hitherto unknown regulatory mechanism of DOK7V1 in lung cancer
cells, which influences cell adhesion, but not the proliferation
and migration by its primary domain.
The functions of the DOK protein family in lung
cancer have gradually been identified. DOKs 1-3 have been
identified to be absent from or decreased in lung cancer (23). In mouse models, a lack of DOKs 1-3
promoted the proliferation of bronchoalveolar stem cells and
alveolar type II cells, which resulted in the development of lung
cancer, particularly in triple knockout mouse models (11). Decreased DOK2 levels were
identified in primary lung adenocarcinomas and lymph node
metastases, whereas no difference in expression of DOK1 and DOK3
was identified in lung cancer compared with normal tissues,
although a decreased DOK3 level was observed in lymph node
metastasis tissues when compared with its expression in primary
tumor (7). DOK2 was identified as
a tumor suppressor gene of lung cancer, since DOK2 was associated
with epidermal growth factor receptor mutations to promote lung
cancer (27). DOK7 was the primary
focus in the present study. In breast cancer, DOK7 is downregulated
owing to the hypermethylation of its CpG islands (22). The results of this study indicated
that DOK proteins may serve an inhibitory function in cancer
development, and gene deletion or hypermethylation of DOK proteins
may occur in cancer cells. In addition, our previous study
identified a lower expression level of DOK7 in lung cancer and
DOK7V1 overexpression, resulting in a decrease in proliferation and
migration, and an increase in adhesion in lung cancer cells
(25). In the present study, the
experiments in which DOK7V1 inhibited proliferation and migration,
but promoted adhesion in other lung cancer cell lines were repeated
to confirm the effect of DOK7V1 on the malignant phenotype.
Furthermore, the change of invasion following transfection of lung
cancer cells with DOK7V1 was investigated, but no difference
between the test group and control group was identified.
DOK7V1 is a 1,515-residue protein, containing a PH
domain, a PTB domain and C-terminal sites of tyrosine
phosphorylation. DOK7 is a substrate of MuSK and also an activator
of its kinase activity, and is therefore an important protein for
forming the vertebrate neuromuscular junction, and may facilitate
trans-autophosphorylation of the kinase activation loop via a
dimeric arrangement of its PH and PTB domains (28). A truncation of the domains may lead
to changes in protein localization. A recent study revealed that
DOK7 bound to membranes containing phosphatidylinositol phosphates
(PIPs) through PH domain binding, facilitating local clustering of
PIP molecules in the bilayer (29). In the present study, the PH and PTB
domains were identified to determine the localization of DOK7V1 in
the membrane and cytoplasm, as revealed by the nuclear membrane
distribution of PH- and PTB-truncated DOK7V1 proteins (DOK7V1Δ-PH
and DOK7V1Δ-PTB). Does this change in location have an effect on
the function of the protein? In the present study, in vitro
functional experiments indicated that DOK7V1 over-expression
inhibited H3122 cell proliferation, motility and adhesion, and
DOK7V1Δ-PH and DOK7V1Δ-PTB have the same effect on proliferation
and motility as DOK7V1, but with the enhancement of adhesion
eliminated.
Dysregulation of the PI3K/AKT/mTOR signaling pathway
has been implicated in the cancerous migration, proliferation and
poor prognosis of various types of cancer, including lung cancer
(30-32). Tumor suppressor genes decreased the
proliferation and migration of NSCLC by inhibiting the
PI3K/AKT/mTOR signaling pathway (33). The in vitro results of the present
study indicated that DOK7V1 inhibited the phosphorylation of AKT,
PI3K, mTOR and ROCK in A549 cells. The two truncated versions of
DOK7V1 also could not active AKT in H3122 cells. PI3K/AKT signaling
appears to rely on DOK7 in the cytoplasm or associated with the
RTK. The weakened PI3K/AKT signaling may be caused by the retention
of truncated DOK7V1 at the perinuclear area. Further analysis
indicated that an increased level of DOK7V1 was negatively
associated with the activation of PI3K/AKT/mTOR signaling.
According to these results, we hypothesize that DOK7V1
overexpression may inhibit the proliferation and migration by
suppressing the activation of PI3K/AKT/mTOR signaling pathway.
However, a clear conclusion cannot be made. Investigation of
whether DOK7V1 decreases the proliferation and migration of lung
cancer cells following inhibition of the PI3K/AKT/mTOR signaling
pathway is required in future studies.
Hyperactivation of the FAK/paxillin signaling
pathway was identified to be significantly associated with cell
adhesion (34-36). FAK and paxillin dynamics serve an
essential function in regulating the adhesion of various cells
(37). In the present study, an
increased adhesion rate of cells overexpressing DOK7V1 compared
with that of the control group was revealed; however, the
DOK7V1Δ-PH and DOK7V1Δ-PTB truncated versions eliminated the
tumor-promoting effects observed in DOK7V1. Conversely, FAK and
paxillin were also weakened, which provides further evidence of the
association between its location and function. Furthermore, it was
confirmed that the gene signatures associated with FAK/paxillin
signaling activation were enriched in patients with high DOK7
expression. These results further suggested that in lung cancer
specimens, DOK7 expression was positively associated with
activation of the FAK/paxillin signaling pathway. Certainly,
further research is required to reach a clear conclusion.
In summary, it was identified that DOK7V1
downregulation is associated with poor prognosis of patients with
lung cancer. The truncations of the DOK7V1 domains appeared to have
effect on the localization of the protein. DOK7V1 overexpression
reversed the malignant phenotypes of H3122 cells, including
proliferation, migration and adhesion. DOK7V1Δ-PH and DOK7V1Δ-PTB
were able to eliminate the function of DOK7 in adhesion, but not in
proliferation and migration. The two truncated version retained the
inhibitory effect of DOK7V1 on AKT activation, but inactivated the
enhanced effect of DOK7V1 on FAK and paxillin. We hypothesize that
DOK7V1 may inhibit proliferation and migration via negatively
regulating the PI3K/AKT/mTOR signaling pathway and increasing
adhesion by upregulating the FAK/paxillin signaling pathway in lung
cancer cells. The results of the present study provide a novel
basis to improve our understanding of the pathogenesis of lung
cancer.
Funding
The present study was supported by Shandong
Provincial Natural Science Foundation, China (grant no.
ZR2017PH047) and the Research Foundation of Yantai Yuhuangding
Hospital (grant no. 201604).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors’ contributions
WGJ, GC, HZ and LY designed the study. GC, HY, LY
and SL performed the experiments. HZ and GC analyzed the data, and
prepared and revised the paper. All authors had final approval of
the submitted and published versions of the paper.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldstraw P, Ball D, Jett JR, Le Chevalier
T, Lim E, Nicholson AG and Shepherd FA: Non-small-cell lung cancer.
Lancet. 378:1727–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang M, Lewinska M, Fan X, Zhu J and Yuan
ZM: PRR14 is a novel activator of the PI3K pathway promoting lung
carcinogenesis. Oncogene. 35:5527–5538. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Blume-Jensen P and Hunter T: Oncogenic
kinase signalling. Nature. 411:355–365. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ullrich A and Schlessinger J: Signal
transduction by receptors with tyrosine kinase activity. Cell.
61:203–212. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lemmon MA and Schlessinger J: Cell
signaling by receptor tyrosine kinases. Cell. 141:1117–1134. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Berger AH, Niki M, Morotti A, Taylor BS,
Socci ND, Viale A, Brennan C, Szoke J, Motoi N, Rothman PB, et al:
Identification of DOK genes as lung tumor suppressors. Nat Genet.
42:216–223. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Simister PC and Feller SM: Order and
disorder in large multi-site docking proteins of the Gab
family–implications for signalling complex formation and inhibitor
design strategies. Mol Biosyst. 8:33–46. 2012. View Article : Google Scholar
|
|
9
|
Jones N and Dumont DJ: Recruitment of
Dok-R to the EGF receptor through its PTB domain is required for
attenuation of Erk MAP kinase activation. Curr Biol. 9:1057–1060.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bedirian A, Baldwin C, Abe J, Takano T and
Lemay S: Pleckstrin homology and phosphotyrosine-binding
domain-dependent membrane association and tyrosine phosphorylation
of Dok-4, an inhibitory adapter molecule expressed in epithelial
cells. J Biol Chem. 279:19335–19349. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mashima R, Honda K, Yang Y, Morita Y,
Inoue A, Arimura S, Nishina H, Ema H, Nakauchi H, Seed B, et al:
Mice lacking Dok-1, Dok-2, and Dok-3 succumb to aggressive
histiocytic sarcoma. Lab Invest. 90:1357–1364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Crowder RJ, Enomoto H, Yang M, Johnson EM
Jr and Milbrandt J: Dok-6, a Novel p62 Dok family member, promotes
Ret-mediated neurite outgrowth. J Biol Chem. 279:42072–42081. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grimm J, Sachs M, Britsch S, Di Cesare S,
Schwarz-Romond T, Alitalo K and Birchmeier W: Novel p62dok family
members, dok-4 and dok-5, are substrates of the c-Ret receptor
tyrosine kinase and mediate neuronal differentiation. J Cell Biol.
154:345–354. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cossins J, Liu WW, Belaya K, Maxwell S,
Oldridge M, Lester T, Robb S and Beeson D: The spectrum of
mutations that underlie the neuromuscular junction synaptopathy in
DOK7 congenital myasthenic syndrome. Hum Mol Genet. 21:3765–3775.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
An CH, Kim MS, Yoo NJ and Lee SH:
Mutational and expressional analysis of a haploinsufficient tumor
suppressor gene DOK2 in gastric and colorectal cancers. APMIS.
119:562–564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miyagaki H, Yamasaki M, Takahashi T,
Kurokawa Y, Miyata H, Nakajima K, Takiguchi S, Fujiwara Y, Mori M
and Doki Y: DOK2 as a marker of poor prognosis of patients with
gastric adenocarcinoma after curative resection. Ann Surg Oncol.
19:1560–1567. 2012. View Article : Google Scholar
|
|
17
|
Yamanashi Y, Higuch O and Beeson D:
Dok-7/MuSK signaling and a congenital myasthenic syndrome. Acta
Myol. 27:25–29. 2008.PubMed/NCBI
|
|
18
|
Okada K, Inoue A, Okada M, Murata Y,
Kakuta S, Jigami T, Kubo S, Shiraishi H, Eguchi K, Motomura M, et
al: The muscle protein Dok-7 is essential for neuromuscular
synaptogenesis. Science. 312:1802–1805. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang SM, Li SY, Yu HB, Li JR and Sun LL:
Repression of DOK7 mediated by DNMT3A promotes the proliferation
and invasion of KYSE410 and TE-12 ESCC cells. Biomed Pharmacother.
90:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hamuro J, Higuchi O, Okada K, Ueno M,
Iemura S, Natsume T, Spearman H, Beeson D and Yamanashi Y:
Mutations causing DOK7 congenital myasthenia ablate functional
motifs in Dok-7. J Biol Chem. 283:5518–5524. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hallock PT, Xu CF, Park TJ, Neubert TA,
Curran T and Burden SJ: Dok-7 regulates neuromuscular synapse
formation by recruiting Crk and Crk-L. Genes Dev. 24:2451–2461.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mayer BJ, Ren R, Clark KL and Baltimore D:
A putative modular domain present in diverse signaling proteins.
Cell. 73:629–630. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pawson T: Protein modules and signalling
networks. Nature. 373:573–580. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saraste M and Hyvönen M: Pleckstrin
homology domains: A fact file. Curr Opin Struct Biol. 5:403–408.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen G, Yu H, Satherley L, Zabkiewicz C,
Resaul J, Zhao H, Mu H, Zhi X, He J, Ye L, et al: The downstream of
tyrosine kinase 7 is reduced in lung cancer and is associated with
poor survival of patients with lung cancer. Oncol Rep.
37:2695–2701. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao H, Yu H, Martin TA, Zhang Y, Chen G
and Jiang WG: Effect of junctional adhesion molecule-2 expression
on cell growth, invasion and migration in human colorectal cancer.
Int J Oncol. 48:929–936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Berger AH, Chen M, Morotti A, Janas JA,
Niki M, Bronson RT, Taylor BS, Ladanyi M, Van Aelst L, Politi K, et
al: DOK2 inhibits EGFR-mutated lung adenocarcinoma. PLoS One.
8:e795262013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bergamin E, Hallock PT, Burden SJ and
Hubbard SR: The cytoplasmic adaptor protein Dok7 activates the
receptor tyrosine kinase MuSK via dimerization. Mol Cell.
39:100–109. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Buyan A, Kalli AC and Sansom MS:
Multiscale Simulations Suggest a Mechanism for the Association of
the Dok 7 PH Domain with PIP-Containing Membranes. PLOS Comput
Biol. 12:e10050282016. View Article : Google Scholar
|
|
30
|
Chen QY and Costa M: PI3K/Akt/mTOR
Signaling Pathway and the Biphasic Effect of Arsenic in
Carcinogenesis. Mol Pharmacol. 94:784–792. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi H, Pu J, Zhou XL, Ning YY and Bai C:
Silencing long non-coding RNA ROR improves sensitivity of
non-small-cell lung cancer to cisplatin resistance by inhibiting
PI3K/Akt/mTOR signaling pathway. Tumour Biol.
39:10104283176975682017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu J, Yao J, Huang R, Wang Y, Jia M and
Huang Y: Ghrelin promotes human non-small cell lung cancer A549
cell proliferation through PI3K/Akt/mTOR/P70S6K and ERK signaling
pathways. Biochem Biophys Res Commun. 498:616–620. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lv X, Li CY, Han P and Xu XY:
MicroRNA-520a-3p inhibits cell growth and metastasis of non-small
cell lung cancer through PI3K/AKT/mTOR signaling pathway. Eur Rev
Med Pharmacol Sci. 22:2321–2327. 2018.PubMed/NCBI
|
|
34
|
Cui S, Wang J, Wu Q, Qian J, Yang C and Bo
P: Genistein inhibits the growth and regulates the migration and
invasion abilities of melanoma cells via the FAK/paxillin and MAPK
pathways. Oncotarget. 8:21674–21691. 2017.PubMed/NCBI
|
|
35
|
Du T, Qu Y, Li J, Li H, Su L, Zhou Q, Yan
M, Li C, Zhu Z and Liu B: Maternal embryonic leucine zipper kinase
enhances gastric cancer progression via the FAK/Paxillin pathway.
Mol Cancer. 13:1002014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu XS, Sun W, Ge CY, Zhang WZ and Fan YZ:
Contribution of the PI3K/MMPs/Ln-5γ2 and EphA2/FAK/Paxillin
signaling pathways to tumor growth and vasculogenic mimicry of
gallbladder carcinomas. Int J Oncol. 42:2103–2115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu YL, Lu S, Szeto KW, Sun J, Wang Y,
Lasheras JC and Chien S: FAK and paxillin dynamics at focal
adhesions in the protrusions of migrating cells. Sci Rep.
4:60242014. View Article : Google Scholar : PubMed/NCBI
|