Introduction
Chronic lymphocytic leukemia (CLL) is the most
frequent and wide-spread type of leukemia affecting adults
(1). Each year, 15,000 new
patients are diagnosed with CLL, which is approximately 1.1% of the
cancer-related incidence, and approximately 5,000 patients succumb
to the disease, which is approximately 0.8% of cancer-associated
mortality in the United States (1). CLL has a higher probability of
occurring in the elderly. The majority of patients with CLL are 65
years old or older, and the incidence and mortality of CLL in China
has gradually increased in recent years. This could be attributed
to the huge population base and an aging society (2). As a type of slow-growing B-cell
lymphoma, CLL usually begins with the aggregation of large numbers
of malignant and mature B-lymphocytes in the peripheral blood, bone
marrow and secondary lymphoid tissues. This subsequently leads to a
disorder in or to the collapse of hematopoietic function (1). However, the diagnosis of CLL always
occurs in the early-intermediate stages of the disease and 90% of
the patients are asymptomatic (3,4).
Moreover, there is no clinical evidence that early therapy would
improve the survival rate of patients with CLL. Actually, many
patients with CLL exhibit indolent symptoms and have a good life
expectancy, whereas minorities face the rapid development of
disease and a poor prognosis (3).
Currently, the preferred treatment for CLL is chemoimmunotherapy;
however, the pharmaceutical effects do not achieve the desired
efficiency, which may be blocked by the still unclear pathogenesis
of CLL (5). Therefore, it is of
utmost importance to intensively explore the molecular mechanisms
responsible for CLL initiation and development and to identify
novel targets for the precise diagnosis and treatment of CLL.
Small ubiquitin related modifier (SUMO)-specific
protease 2 (SENP2) is a member of the SUMO-specific protease (SENP)
family that is essential to the processes of SUMOylation and
de-SUMOylaton (6,7). Among the three subfamilies of SENPs,
SENP2 can be classified as a member of the first subfamily, which
is characterized by broad substrate specificity (8). SENP2 is localized in the nuclear pore
through binding with the Nup153 nucleoporin and a nuclear
envelope-related protease. It has been demonstrated that SENP2
plays a critical role in embryonic erythropoiesis when
overexpressed, which is similar to SENP1 (9). Previously, some researchers reported
that SENP2can promote the development of embryonic erythropoiesis
by regulating the p53-Mdm2 signaling pathway in the placenta
(10,11). However, it has also been reported
that SENP2 plays a potential role in the tumorigenesis of various
types of cancer, such as hepatocellular carcinoma, breast cancer
and bladder cancer (12-15). For example, the study by Shen et
al reported that the overexpression of SENP2 in hepatocellular
carcinoma cells inhibited cell proliferation through the regulation
of β-catenin stability, while the opposite effect was observed by
the silencing of SENP2 (14).
Moreover, the study by Tan et al also illustrated the
downregulation of SENP2 in bladder cancer tissues and the
inhibition of the migratory and invasive ability of bladder cancer
cells by the overexpression of SENP2 through the blocking if the
activation of matrix metalloproteinase (MMP)13 in vitro
(13). The study by Nait Achour
et al verified that SENP2 suppressed the proliferation of
estrogen-dependent or-independent MCF7 breast cancer cells by
preventing the interaction between the SENP2 and ERα proteins
(12). However, whether SENP2 is
involved in the development and occurrence of CLL has not been
extensively explored and warrants further investigation.
The Notch signaling pathway plays important roles in
the proliferation, differentiation, apoptosis, and other
physiological activities of normal cells and has been identified as
an evolutionarily conserved signaling pathway (16). However, the abnormal activation of
the Notch signaling pathway in CLL has also been reported by a
number of studies and the overexpression and mutation of some Notch
molecules has been reported to be associated with drug resistance,
a poor prognosis, and other issues in CLL (17-23).
Nwabo Kamdje et al and Rosati et al found that some
Notch receptors such as Notchl and Notch2, and ligands such as
Jaggedl and Jagged2 have a high expression in patients with CLL and
in primary CLL cells (17,18). In addition, the activation of the
Notch signaling pathway is associated with the nuclear factor
(NF)-κB signaling pathway and NF-κB can upregulate the expression
of Jagged1, which interacts with Notch to continually activate the
Notch signaling pathway in CLL cells (24,25).
Notably, Sun et al identified Wnt/β-catenin signaling as the
signaling pathway downstream of Notch and the mechanism of the
promoting effect of hepatocarcinogenesis by Notch1 (26). Jiang et al also reported
that SENP2 inhibited the growth of hepatocellular carcinoma cells
by the modulation of β-catenin stability through WW
domain-containing oxidoreductase (WWOX), a novel inhibitor of the
Wnt/β-catenin pathway (15).
Therefore, we inferred that SENP2 may also inhibit the occurrence
and development of CLL via the regulation of β-catenin to affect
the Notch signaling pathway.
In this study, we first detected the protein and
mRNA expression levels of SENP2 in patients with CLL. We then
established CLL cells in which SENP2 was overexpressed or silenced
to determine their invasive and chemotactic ability, their
sensitivity to cytarabine and dexamethasone, the cell apoptotic
state, the expression level of β-catenin, the activation state of
the Notch and NF-κB signaling pathways, and other processes. This
study aimed to clearly determine whether SENP2 functions as a tumor
suppressor in CLL through the modulation of the Notch and NF-κB
signaling pathways.
Materials and methods
Samples, cells, antibodies and
reagents
Peripheral blood from 43 patients with CLL (26/43
before treatment and 17/43 post-treatment; 15 female and 28 male
patients; age range, 47-80 years) and 21 healthy volunteers (8
female and 13 male healthy volunteers; age range, 50-74 years) was
collected (January, 2016 to July, 2017) at the Fujian Medical
University Union Hospital (Fuzhou, China). Sex, age, stage and the
expression level of serum lactate dehydrogenase (LDH),
β2-microglobulin (β2-MG), and cell genetics of the CLL patients
were also collected from the Fujian Medical University Union
Hospital (Fuzhou, China). The study was approved by the Ethics
Committee of the Fujian Medical University Union Hospital and
written consent was obtained from all participants in this
study.
The human CLL cell line, MEC2, was purchased from
the American Type Culture Collection (ATCC, Manassas, VA, USA). All
cells were maintained in RPMI-1640 medium supplemented with 10%
fetal bovine serum (FBS) (both from Gibco/Thermo Fisher Scientific,
Waltham, MA, USA), at 37°C in an atmosphere containing 5%
CO2 in 10 cm2 plates at
0.3-0.5×106 cells/ml. The primary antibodies used for
western blot analysis and flow cytometry were purchased from
various companies. The anti-Bcl-2 (1:1,000; cat. no. 2870),
anti-Bax (1:1,000; cat. no. 5023), anti-cleaved caspase-9 (1:1,000;
cat. no. 7237), anti-caspase-9 (1:1,000; cat. no. 9508),
anti-cleaved caspase-3 (1:1,000; cat. no. 9664), anti-caspase-3
(1:1,000; cat. no. 9665), anti-Cyld (1:1,000; cat. no. 12797),
anti-c-Myc (1:1,000; cat. no. 5605), anti-Notch1 (1:3,000; cat. no.
3268), anti-β-catenin (1:1,000; cat. no. 8480), anti-IKKβ (1:1,000;
cat. no. 8943), anti-IκBα (1:1,000; cat. no. 4812), anti-p65
(1:1,000; cat. no. 8242), anti-p50 (1:1,000; cat. no. 12540) and
anti-p53 (1:1,000; cat. no. 2524) antibodies were from Cell
Signaling Technology (Danvers, MA, USA). The anti-IKKα (1:1,000;
cat. no. ab32041) and anti-GAPDH (1:1,000; cat. no. ab37168)
antibodies were from Abcam (Cambridge, UK). The anti-SENP2
(1:5,000; cat. no. sc-46638) antibody was obtained from Santa Cruz
Biotechnology (Santa Cruz, CA, USA). The Cell Counting kit-8 was
from Dojindo Laboratories (Kumamoto, Japan). Cytarabine and
dexamethasone were purchased from Selleck Chemicals (Houston, TX,
USA).
Establishment of CLL cells in which SENP2
was overexpressed or silenced
The primer for SENP2 was designed using Primer 5.0
software and provided by Sangon Biotech (Shanghai) Co., Ltd.
(Shanghai, China). The cDNA sequence of SENP2 was amplified from
pGEM-SENP2 and constructed into the vector pBABE-hygro to generate
pBABE-SENP2 by BioSCIRes Biotech Co., Ltd. (Shanghai, China). The
vector pBABE-SENP2 was used to prepare retroviral preparations for
co-transfection of 293T cells (ATCC) with liposomes. Subsequently,
shRNA sequences targeting SENP2 were designed by RNAi designer, and
synthesized by Sangon Biotech (Shanghai) Co., Ltd. The shRNA
sequences were inserted into the PLVX vector to generate
PLVX-shRNA-SENP2 BioSCIRes Biotech Co., Ltd.). The mixture of
PLVX-shRNA-SENP2, psPAX2 and pMDG2 was transfected into 293T cells
using Lipofectamine 2000 reagent (Invitrogen/Thermo Fisher
Scientific) to generate lentivirus. Eventually, the MEC2 cells, at
a density of 8×104 cells/ml, were infected with the
recombinant of lentivirus plus 8 µg/ml polybrene (Sigma, St.
Louis, MO, USA).
Western blot analysis
The CLL cells in which SENP2 was overexpressed or
silenced were seeded in 6-cm dishes. After reaching 70% confluence,
cytarabine (0.128 µg/ml) or dexamethasone (630 µg/ml)
or DMSO were added and the cells were further incubated for 24 h in
a humidified atmosphere at 37°C with 5% CO2.
Subsequently, all CLL cells were collected by centrifugation (1,000
rpm) at 4°C for 10 min and kept in RIPA lysis buffer (Beyotime,
China) for 30 min at 4°C for cell lysis, then collected by
centrifugation (12,000 rpm) at 4°C for 15 min. The supernatant was
collected and the acquired total protein was measured using a BCA
kit (Beyotime, China). Next, 20 µg protein was separated by
10-12% SDS-PAGE and then transferred onto PVDF Transfer Membranes
(Millipore, Billerica, MA, USA). Following the blocking of these
membranes by 5% non-fat milk in TBST (0.1% Tris-buffered saline
with 1 ml/l Tween-20) for 1 h at room temperature, they were
incubated with the corresponding primary antibodies at 4°C
overnight. After washing with TBST 3 times, the membranes were
further incubated with HRP-conjugated secondary antibodies
(anti-goat, 1:2000, cat. no. A0181; anti-rabbit, 1:2000, cat. no.
A0208; anti-mouse, 1:2000, cat. no. A0216) (all from Beyotime,
China) for 1 h at room temperature. Finally, the bands were
displayed by a SuperSignal West Pico Chemiluminescent Substrate
(Thermo Fisher Scientific, Massachusetts, MA, USA) according to the
protocol provided by the manufacturer. Western blot quantification
was carried out using ImageJ software. Signals were adjusted for
background and normalized to the signal for GAPDH.
Reverse transcription-quantitative PCR
(RT-qPCR)
In brief, we first designed the primers for SENP2
and GAPDH as shown in Table I;
GAPDH was used as the reference gene. Second, peripheral blood
lymphocytes (PBLCs) of patients with CLL were obtained using the
lymphocyte isolation kit (cat. no. LTS1077; Tian Jin Hao Yang
Biological Manufacture Co., Ltd., Tianjin, China). Total RNA was
then extracted from the samples obtained from the PBLCs of patients
with CLL according to the one-step TRIzol extraction. Third, the
measurement of total RNA quality was conducted on a Nano-100 Micro
Spectrophotometer (Hangzhou Allsheng Instruments Co., Ltd.,
Hangzhou, China) and the cDNA of SENP2 and GAPDH were then reverse
transcribed through a general PCR step according to the PrimeScript
RT reagent kit (Takara Biomedical Technology Co., Ltd., Beijing,
China). Finally, the levels of SENP2 and GAPDH cDNA were determined
by qPCR according to the instructions provided with the SYBR Premix
Ex Taq kit (Takara Biomedical Technology Co., Ltd, Beijing, China)
to calculate the relative mRNA expression level of SENP2. The
relative expression level of SENP2 was determined according to the
2−ΔΔCq method, as previously described (27).
 | Table IThe primer sequences of the related
genes. |
Table I
The primer sequences of the related
genes.
| Gene | | Primer sequence
(5'-3') |
|---|
| SENP2 | Forward |
CATTGGAGCCTGGTGGTGAT |
| SENP2 | Reverse |
TGTTGAGGAATCTCGTGTGGTT |
| GAPDH | Forward |
CGGATTTGGTCGTATTGGG |
| GAPDH | Reverse |
GATTTTGGAGGGATCTCGC |
Transwell assay
The Transwell system (24-wells, 8 µm pore
size with polycarbonate membrane; Corning Costar, Lowell, MA, USA)
was used for the cell invasion assay. Briefly, 60 µl of
Matrigel were diluted by RPMI-1640 at a ratio of 1:7 then added
into the upper wells of pre-cooled Transwell plates, and the plates
were then incubated at 37°C for 4 h. Subsequently, the CLL cells in
which SENP2 was overexpressed or silenced were harvested and
suspended in RPMI-1640 medium without FBS, and then
1×106 cells were added per well to the upper wells.
Following incubation in a humidified atmosphere at 37°C with 5%
CO2 for 24 h, the number of cells that crossed the
Matrigel and moved into the lower wells were counted using a
hemocytometer.
CCK-8 assay
The CLL cells in which SENP2 was overexpressed or
silenced were seeded in 96-well plates at a density of 2×105
cells/ml and incubated with at 37°C for 48 h. These cells were then
incubated with concentration gradients of cytarabine or
dexamethasone or DMSO from 0 to 10 mg/ml and for each
concentration, there were 5 parallel control wells. The cells were
then further incubated for 24, 48 or 72 h before the CCK-8 reagents
(10 µl) were added. All plates were shaken for a further 15
min on a shaker and the absorbance was measured at 450 nm using a
Varioskan Flash multimode reader (Thermo Fisher Scientific).
Chemotaxis chamber assay
The chemotactic response of the CLL cells in which
SENP2 was overexpressed or silenced was detected in96-well
chemotaxis chambers with polycarbonate filters (5 µm pore
size, 3.2 mm diameter size, 30 µl well size) (Chemo TX
Disposable Chemotaxis System, NeuroProbe, Gaithersburg, MD, USA).
In brief, the CLL cells in which SENP2 was overexpressed or
silenced were cultured under standard conditions and then suspended
in RPMI-1640. The suspended CLL cells (2×105 cells/ml)
were added to the upper chamber. The dilution of CXCL12 (200 ng/ml)
were added to the lower chamber and the cells were incubated for 8
h in a humidified atmosphere at 37°C with 5% CO2.
Subsequently, the CLL cells that did not migrate were removed from
the membrane surface in the upper chamber using a cotton swab, and
the CLL cells that had migrated to the lower surface of the
membrane were fixed with 4% paraformaldehyde (PFA) for 10 min and
then labeled with DAPI (Sigma) to mark the cell nuclei. The number
of migrating cells in 5 random fields was counted at ×50
magnification by using a double-blinded approach, and images were
captured at ×200 magnification with a fluorescent microscope (LEICA
DM 4000B; Leica, Wetzlar, Germany).
Flow cytometric analysis
The CLL cells in which SENP2 was overexpressed or
silenced were seeded into 6 cm dishes (1×105 cells/ml).
Cytarabine (0.128 µg/ml) or dexamethasone (630 µg/ml)
were used to treat the CLL cells that were further cultured for 24,
48 or 72 h. Subsequently, an Annexin V-FITC Apoptosis Detection kit
(BD Biosciences, San Jose, CA, USA) was used for apoptosis
detection. In brief, the cells were harvested and suspended in
binding buffer (100 µl), achieving a final concentration of
1×106/ml. Annexin V-FITC (5 µl) and propidium
iodide (PI, 20 µg/ml, 5 µl) were added and the cells
were further cultured for 15 min at room temperature in the dark.
The cells were analyzed by flow cytometry (FacsCalibur flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA) following the
addition of binding buffer (400 µl). The obtained data were
processed using FlowJo 7.6 software.
Statistical analysis
In this study, all data are expressed as the means ±
SD (n≥3) without special instructions. The Kruskal-Wallis test was
used for multiple group comparisons; the Mann-Whitney test was used
for group comparisons with the P-values and significance threshold
corrected by the Bonferroni correction. A paired-samples t-test or
the Wilcoxon signed rank test were used for paired-samples
analysis. Statistical analysis was carried out using SPSS15.0
software. Statistical differences with a value of P<0.05 were
considered significant.
Results
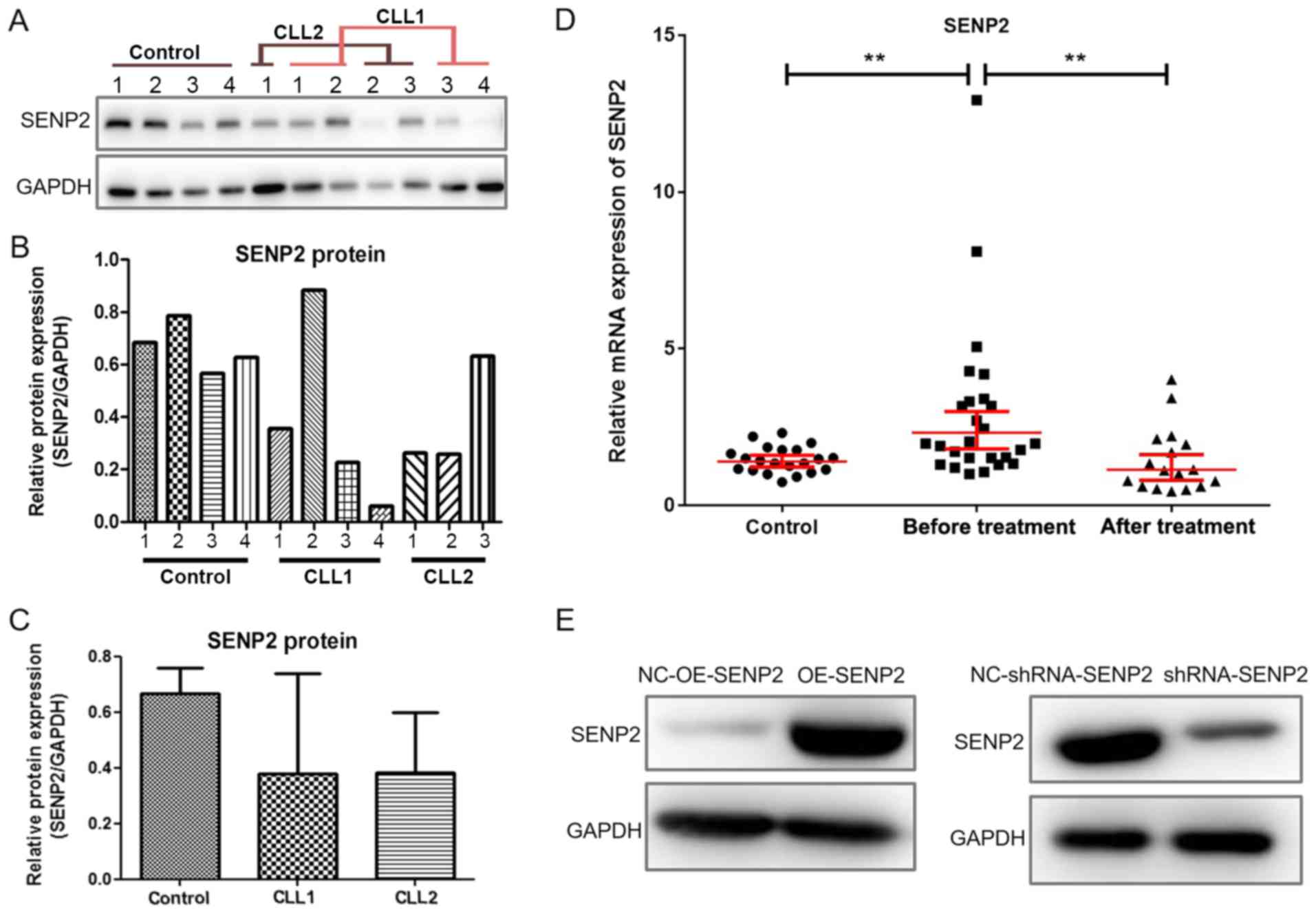
SENP2 is downregulated in the peripheral
blood of patients with CLL
Peripheral blood was collected from 43 patients with
CLL (26/43 before treatment and 17/43 post-treatment) and 21
healthy volunteers, and western blot analysis was used to detect
the protein levels of SENP2 in PBLCs. The results of some randomly
selected samples are shown in Fig.
1A-C. It was found that the protein level of SENP2 was
generally lower in the peripheral blood of patients with CLL when
compared with that of the healthy volunteers.
We further utilized RT-qPCR to detect the mRNA
levels of SENP2 in peripheral blood mononuclear cells of 43
patients with CLL(26/43 before treatment and 17/43post R-FC
chemotherapy treatment) and 21 healthy volunteers. As shown in
Fig. 1D, patients with CLL without
treatment exhibited higher mRNA levels of SENP2 than the healthy
volunteers (control) group and the post-treatment group (P=0.0034).
The association between the mRNA expression of SENP2 and the
clinicopathological characteristics including sex, age, stage, LDH
expression and cell genetics of the 26/43 patients with CLL before
treatment was also evaluated. As shown in Table II, the mRNA level of SENP2 was
associated with the expression of LDH (P<0.05); however, no
significant association was observed with the other
clinicopathological characteristics examined.
 | Table IIAssociation between the mRNA
expression of SENP2 and the clinical or biological characteristics
of the 26 patients CLL (untreated). |
Table II
Association between the mRNA
expression of SENP2 and the clinical or biological characteristics
of the 26 patients CLL (untreated).
| Characteristic | Median of
2−ΔΔcq (range) | P-value |
|---|
| Sex | | |
| Male | 1.90
(1.07-12.94) | 0.535 |
| Female | 2.46
(1.01-5.06) | |
| Age (years) | | |
| ≥60 | 2.21
(1.07-12.94) | 0.607 |
| <60 | 1.93
(1.01-8.10) | |
| Binet staging | | |
| A and B | 1.84
(1.07-8.10) | 0.382 |
| C | 2.81
(1.01-12.94) | |
| LDH expression | | |
| > 250U/l | 3.32
(1.97-8.10) | 0.025 |
| ≤ 250U/l | 1.74
(1.01-12.94) | |
| β2-MG | | |
| >3 mg/l | 1.97
(1.21-8.10) | 0.462 |
| ≤3 mg/l | 1.65
(1.01-3.40) | |
| Cell genetics | | |
| With a poor
prognosis index | 2.46
(1.52-4.19) | 0.322 |
| Without a poor
prognosis index | 1.77
(1.01-5.06) | |
Establishment of CLL cells in which SENP2
was overexpressed or silenced
To further explore the role of SENP2 in CLL cells
in vitro, cell models in which SENP2 was overexpressed or
silenced were established based on the human CLL cell line, MEC2.
The overexpression and silencing of SENP2 were verified by western
blot analysis and the results are shown in Fig. 1E. The results demonstrated that the
expression of SENP2 was upregulated and downregulated in the cells
in which SENP2 was overexpressed or silenced by shRNA,
respectively.
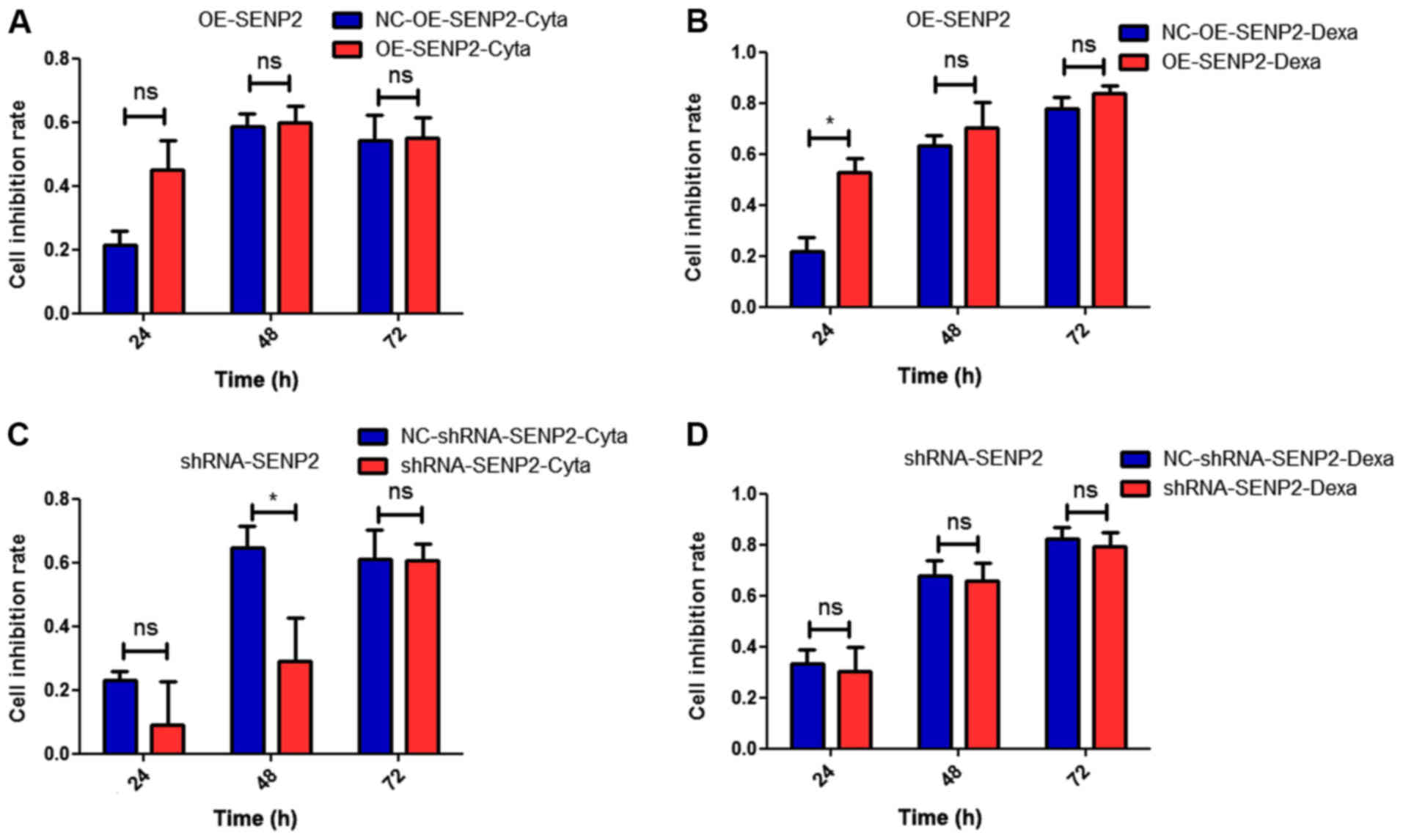
Overexpression of SENP2 has the tendency to enhance
the sensitivity of CLL cells to cytarabine and dexamethasone at an
early stage. Cytarabine and dexamethasone are anticancer drugs
commonly used in the treatment of leukemia. Thus, in this study, we
examined the sensitivity of CLL cells, in which SENP2 was
overexpressed or silenced, to chemotherapy, as this is also an
important factor for determining the role of SENP2 in cancer.
ACCK-8 assay was conducted to detect the response of the CLL cells,
in which SENP2 was overexpressed or silenced, to cytarabine or
dexamethasone. As shown in Fig. 2,
with the overexpression of SENP2, we observed a trend towards an
increased the sensitivity of CLL cells to cytarabine and
dexamethasone compared with the controls. The silencing of SENP2
was associated with a tendency towards increased drug resistance.
However, with the prolongation of the treatment time with
cytarabine and dexamethasone, the difference in the proliferation
of the CLL cells in which SENP2 was overexpressed or silenced and
the control group became inconspicuous. Collectively, it was
hypothesized that SENP2 may enhance the sensitivity of CLL cells to
chemotherapy at an early stage.
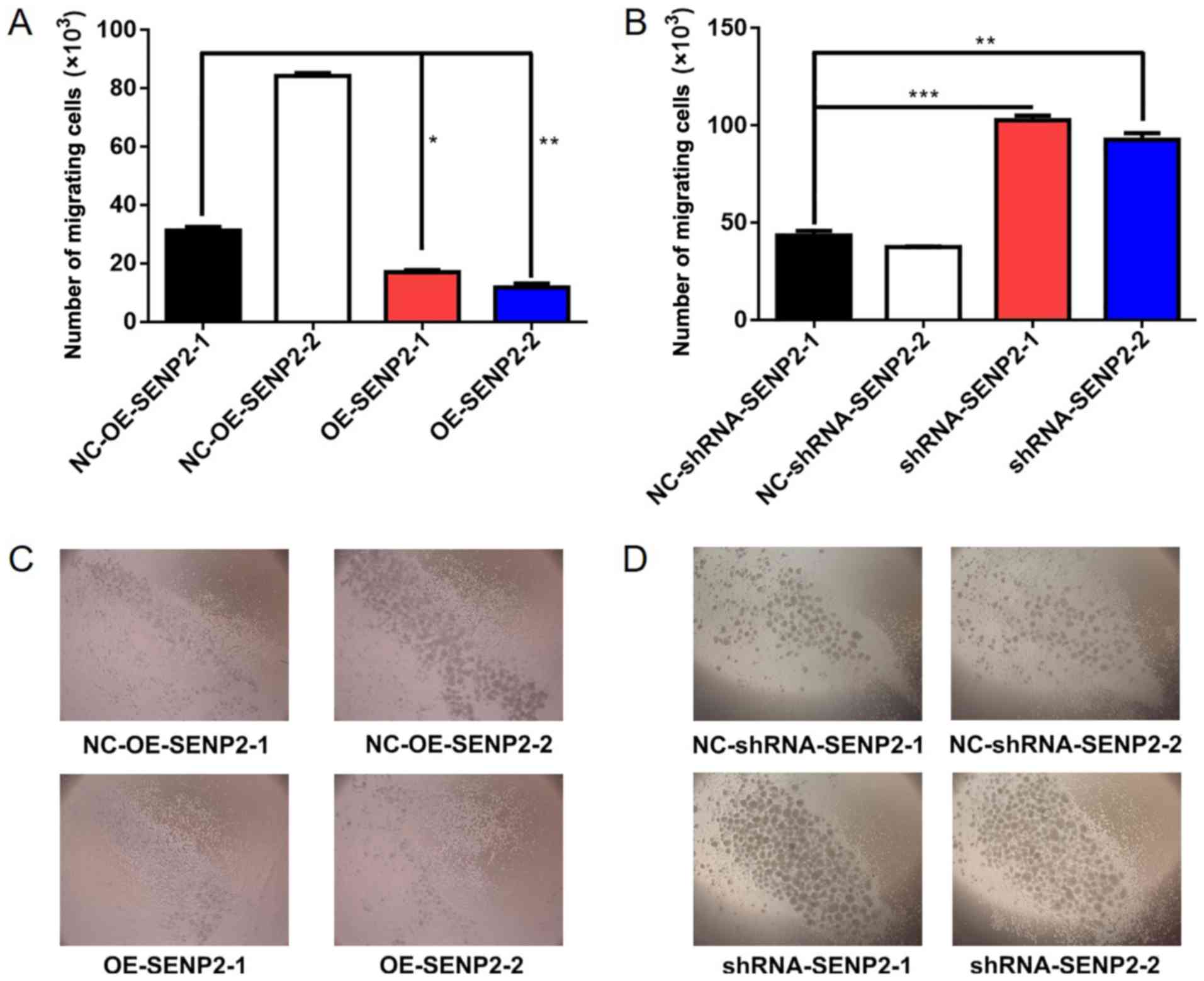
Overexpression of SENP2 decreases the
invasive and chemotactic ability of CLL cells
Metastasis and invasion are crucial factors for
grading malignancy in cancers; thus, we considered it essential to
examine the effect of SENP2 on the invasive ability of CLL cells.
Transwell assays were performed to examine the effect of SENP2
overexpression or silencing on the invasive ability of CLL cells
and the results demonstrated that SENP2 overexpression resulted in
fewer cells migrating from the upper chamber to the lower chamber,
while the silencing of SENP2 in CLL cells had the opposite effect
(P<0.01 and P<0.001, Fig.
3).
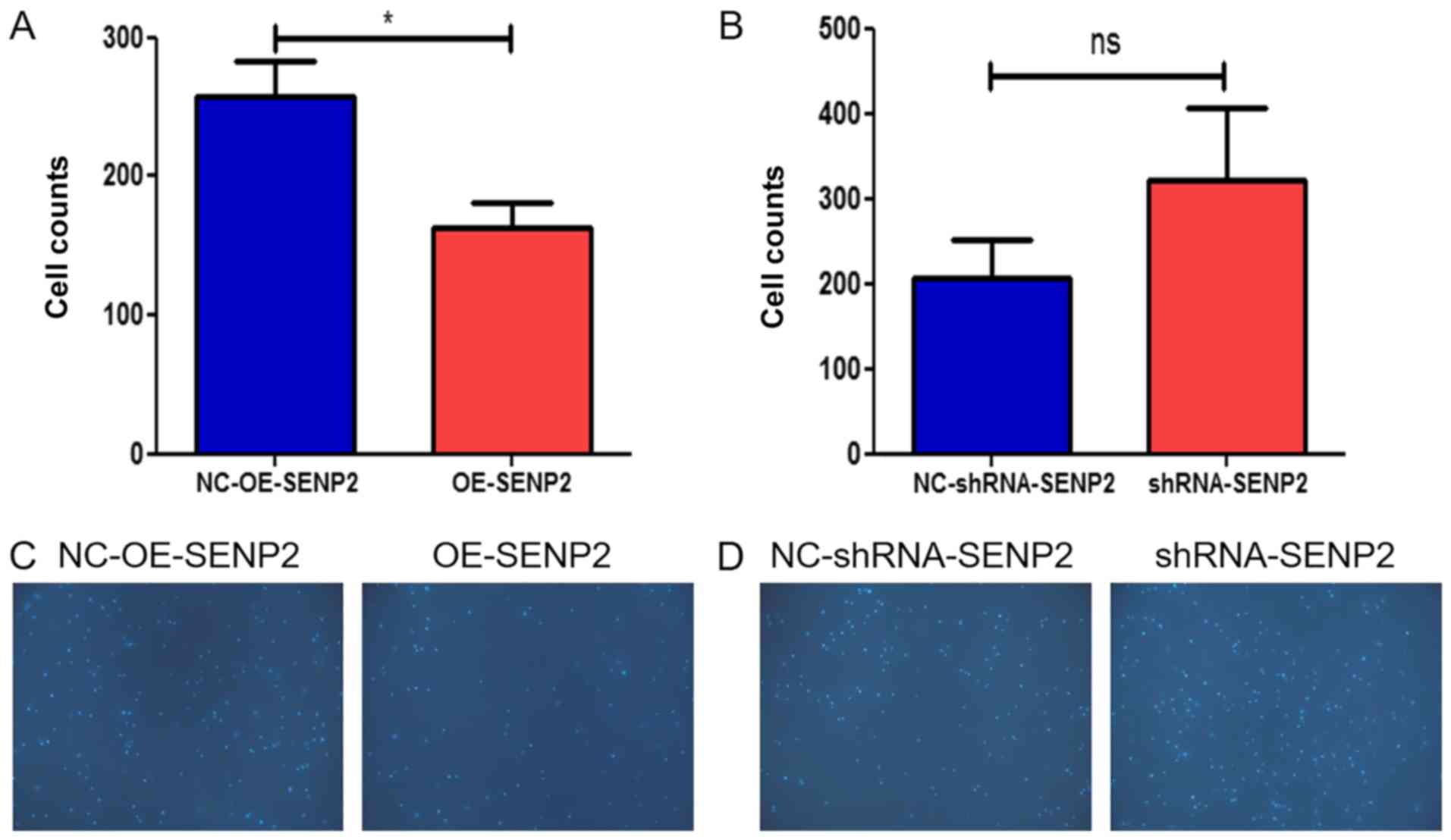
In addition, the chemotactic response of leukocytes
may reflect the prognosis for CLL patients; thus, we also examined
the effect of SENP2 on the response of CLL cells to CXCL12 in a
chemotaxis chamber. The data from the chemotaxis chamber assay
revealed that the overexpression of SENP2 in the CLL cells
inhibited the migration of the MEC2 cells into the lower chamber
(Fig. 4). When SENP2 was silenced,
the CLL cells exhibited an increased chemotactic response. The
increased expression of SENP2 decreased both the migration and
chemotactic ability of the CLL cells.
Overexpression of SENP2 promotes the
apoptosis of CLL cells
To investigate whether SENP2 promotes the apoptosis
of CLL cells, western blot analysis and flow cytometry were used to
detect the apoptosis rate of the CLL cells in which SENP2 was
overexpressed following treatment with cytarabine or dexamethasone.
The results of flow cytometry are shown in Fig. 5. It was found that following
treatment with dexamethasone, the SENP2-overexpressing MEC2 cells
exhibited more late-stage apoptosis than the control MEC2 cells.
These results prove that SENP2 not only promotes the apoptosis of
CLL cells, but also the ability of SENP2 to enhance the sensitivity
of the cells to drug treatment with dexamethasone. When the
SENP2-overexpressing MEC2 cells were treated with dexamethasone for
24, 48 or 72 h, the cell apoptosis increased in a time-dependent
manner, and the cells also exhibited a time-dependent sensitivity
to dexamethasone (Fig. 5).
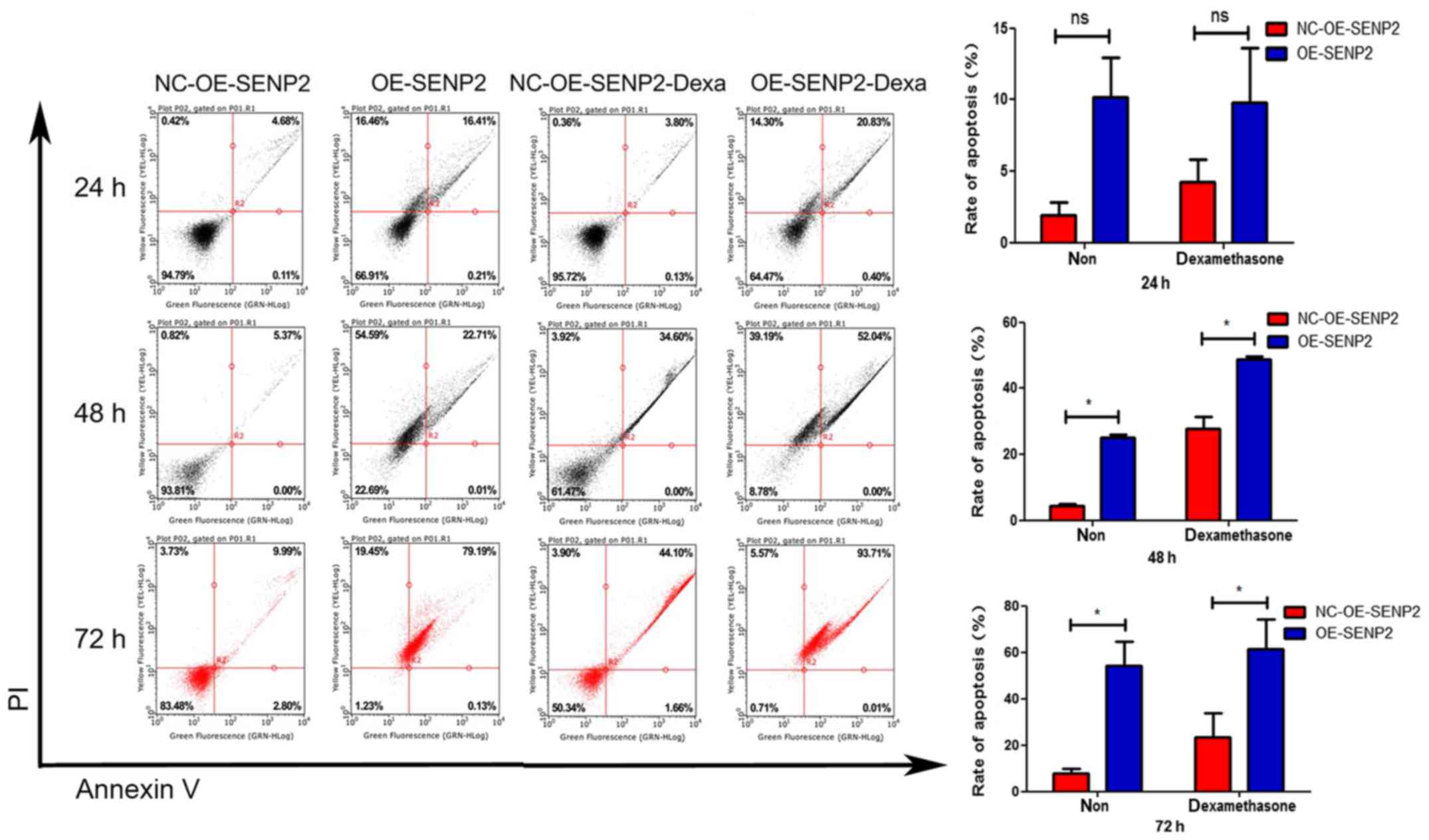 | Figure 5Overexpression of SENP2 promotes the
apoptosis of CLL cells. The CLL cells in which SENP2 was
overexpressed or silenced were seeded into 6 cm dishes. Upon
reaching 60% confluence, dexamethasone or DMSO were used to treat
the CLL cells the cells were then further cultured for 24, 48 or 72
h. Subsequently, an Annexin V-FITC Apoptosis Detection kit was used
to examine cell apoptosis. Finally, the cells were analyzed by flow
cytometry following the addition of binding buffer (400 µl).
The statistical method used was a paired samples t-test.
*P<0.05; ns, no statistical significance. CLL,
chronic lymphocytic leukemia; NC-OE-SENP2, MEC2 cells transfected
with null (control) overexpression vector; OE-SENP2, MEC2 cells
transfected with SENP2 overexpression vector; NC-shRNA-SENP2, MEC2
cells transfected with null control shRNA; shRNA-SENP2, MEC2 cells
transfected with shRNA against SENP2. Dexa, dexamethasone. |
In addition, SENP2 overexpression and treatment with
dexamethasone or cytarabine appeared to increase the expression of
the pro-apoptotic proteins, p53 compared with NC-OE-SENP2 but
without statistical significance. Furthermore, SENP2 overexpression
and treatment with dexamethasone or cytarabine did not
significantly affect the expression of other pro-apoptotic
proteins, such as cleaved caspase-3, cleaved caspase-9 and Bax, and
on the expression of the anti-apoptotic protein, Bcl-2, compared
with NC-OE-SENP2 (Figs. 6 and
7). The above-mentioned results
indicate that SENP2 can increase the apoptosis of CLL cells in
response to chemotherapeutics; however, the mechanisms involved
warrant further investigation.
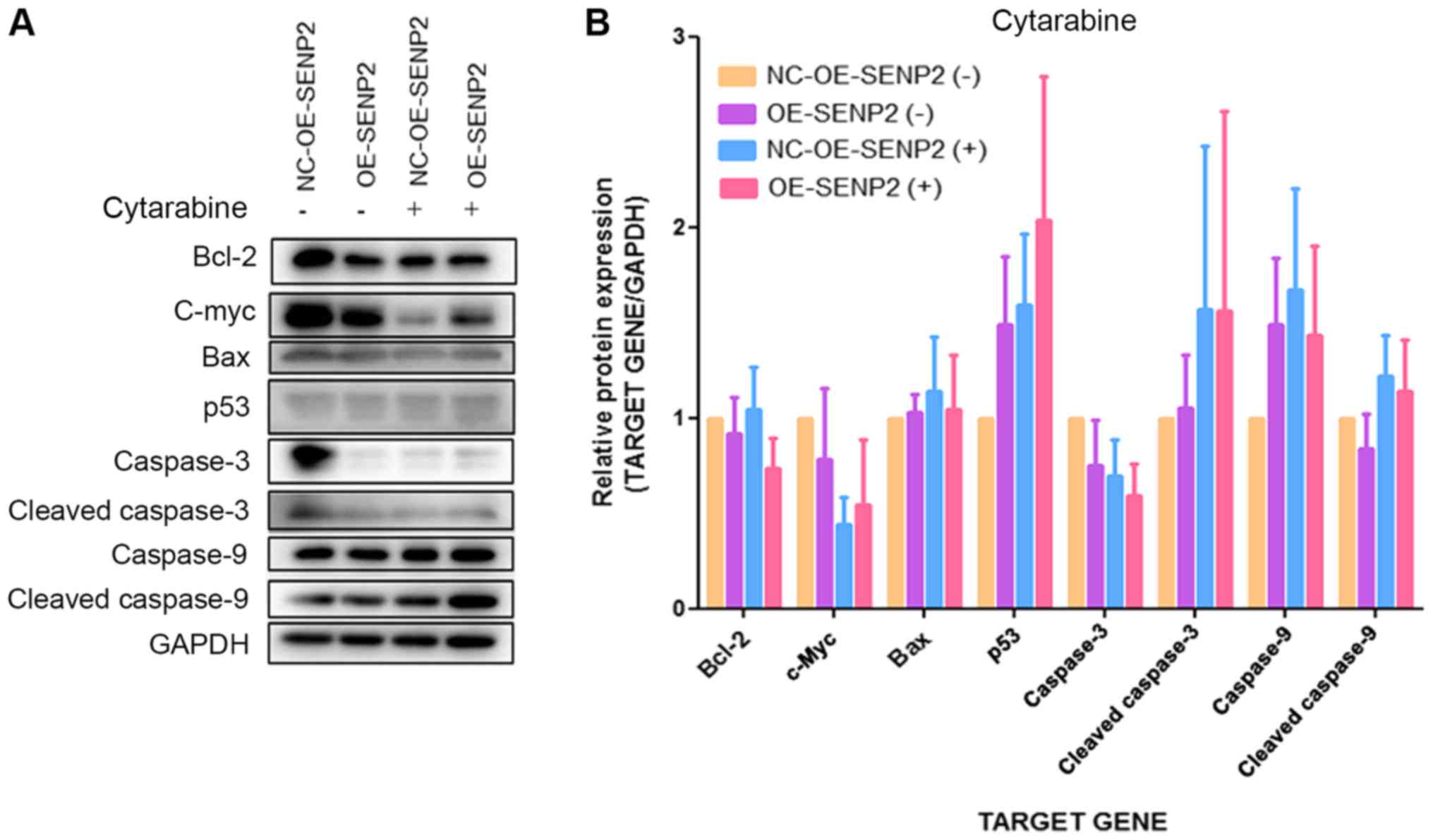 | Figure 6Effect of SENP2 overexpression on
apoptosis-related proteins in CCL cells treated with or without
cytarabine. (A and B) MEC2 cells in whichSENP2 was overexpressed
and the control cells were cultured in 6 cm dishes and then treated
with cytarabine for 24 h after reaching 70% confluence. Western
blot analysis was then performed to inspect the expression levels
of cleaved caspase-9, caspase-9, cleaved caspase-3, caspase-3,
c-Myc, p53, Bax, and Bcl-2 in these cells. GAPDH was used to
confirm equal amount of proteins loaded in each lane. CLL, chronic
lymphocytic leukemia; NC-OE-SENP2, MEC2 cells transfected with null
(control) overexpression vector; OE-SENP2, MEC2 cells transfected
with SENP2 overexpression vector; NC-shRNA-SENP2, MEC2 cells
transfected with null control shRNA; shRNA-SENP2, MEC2 cells
transfected with shRNA against SENP2; +, cytarabine treatment, -,
no cytarabine treatment. |
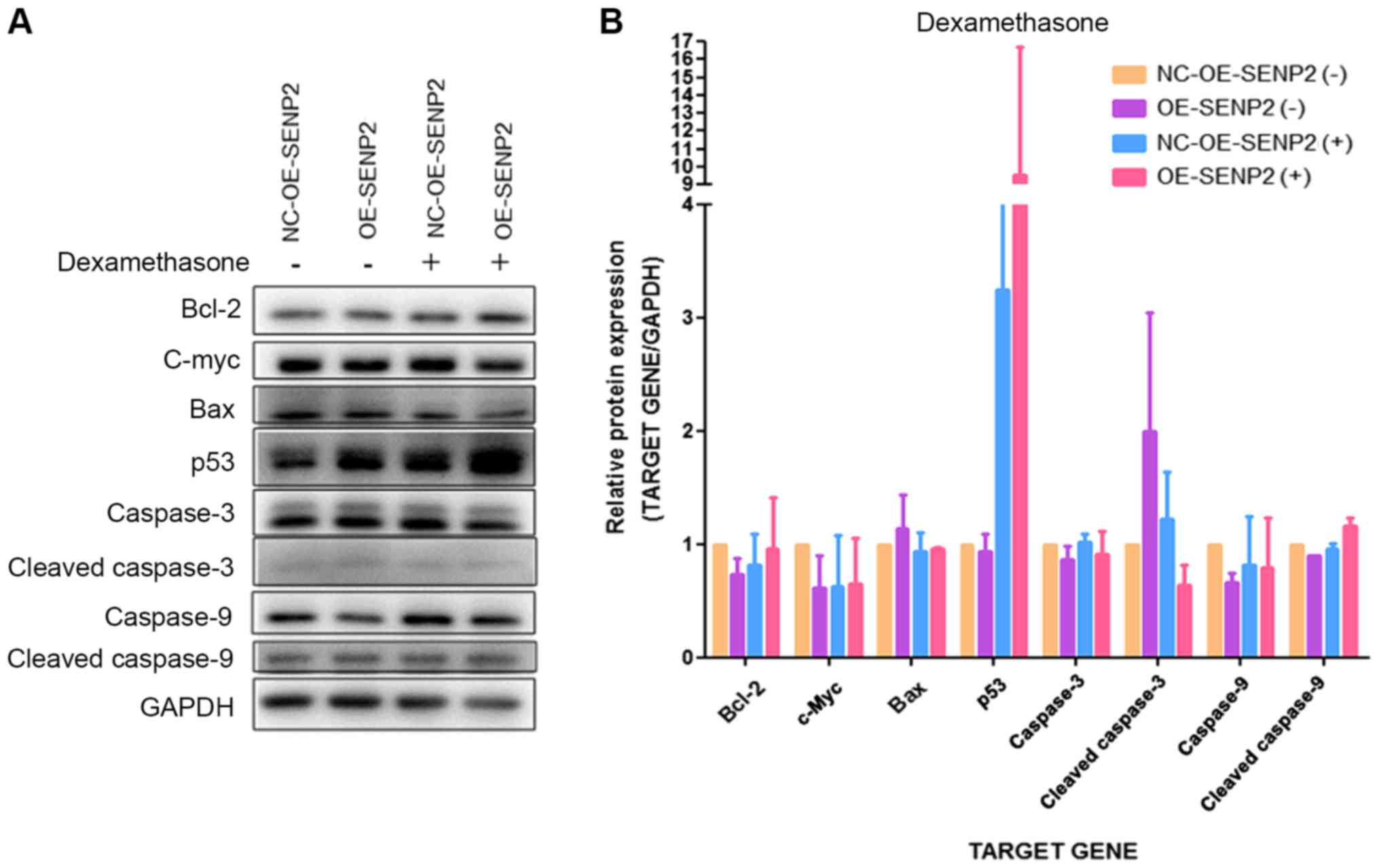 | Figure 7Effect of SENP2 overexpression on
apoptosis-related proteins in CLL cells with or without
dexamethasone treatment. (A and B) MEC2 cells in which SENP2 was
overexpressed and the control cells were cultured in 6 cm dishes
and then treated with dexamethasone for 24 h after reaching 70%
confluence. Western blot analysis was then performed to inspect the
expression levels of cleaved caspase-9, caspase-9, cleaved
caspase-3, caspase-3, c-Myc, p53, Bax and Bcl-2 in these cells.
GAPDH was used to confirm equal amount of proteins loaded in each
lane. CLL, chronic lymphocytic leukemia; NC-OE-SENP2, MEC2 cells
transfected with null (control) overexpression vector; OE-SENP2,
MEC2 cells transfected with SENP2 overexpression vector;
NC-shRNA-SENP2, MEC2 cells transfected with null control shRNA;
shRNA-SENP2, MEC2 cells transfected with shRNA against SENP2; +,
dexamethasone treatment, −, no dexamethasone treatment. |
SENP2 suppresses the Notch and NF-κB
signaling pathways
A number of studies have reported that the Notch
signaling pathway is aberrantly activated in CLL cells and the
activation of the Notch and NF-κB signaling pathway can increase
the survival of CLL cells (17,18,20,22,24,25).
Therefore, in order to further illustrate the molecular mechanisms
of action of SENP2 in CLL cells, the expression of the Notch and
NF-κB signaling pathways in CLL cells in which SENP2 was
overexpressed or silenced was evaluated. As shown in Fig. 8A, the overexpression of SENP2
downregulated the expression of Cyld, IKKβ, IκBα, IKKα, p65 and
p50, all of which are proteins in the NF-κB signaling pathway. The
overexpression of SENP2 also downregulated the expression of Notch1
in the Notch signaling pathway. It has also been reported that
SENP2 can regulate the stability of β-catenin (14). In this study, the MEC2 cells in
which SENP2 was overexpressed also exhibited a downregulation in
the expression of β-catenin (Fig.
8A). However, the silencing of SENP2 upregulated both the Notch
signaling pathway and the NF-κB signaling pathway (Fig. 8B). Based on these results, we can
infer that SENP2 acts as a tumor suppressor in CLL cells through
the regulation of the expression of β-catenin to suppress the Notch
and NF-κB signaling pathways.
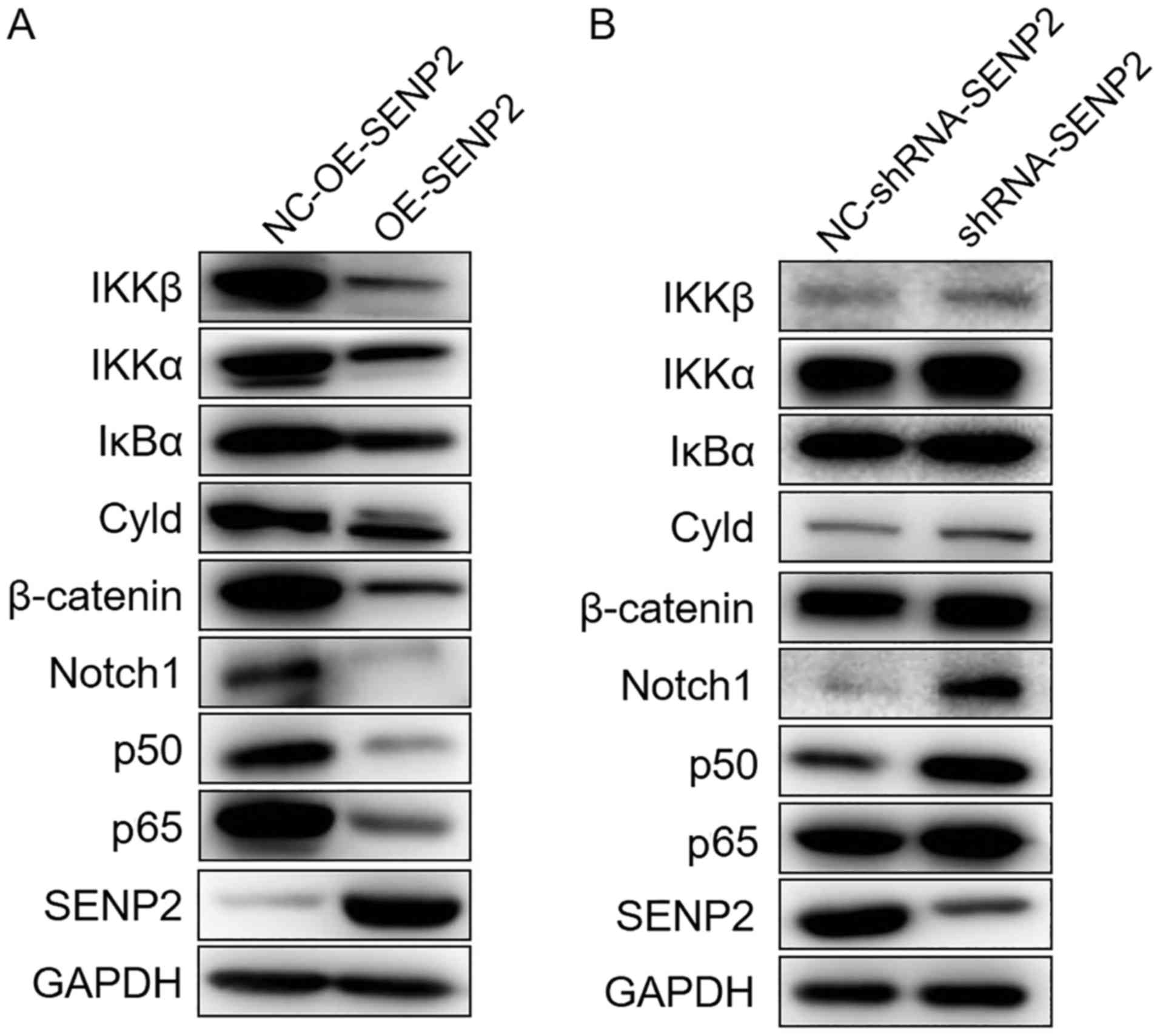 | Figure 8SENP2 suppresses the Notch and NF-κB
signaling pathways. (A) SENP2-overexpressingMEC2 cells and the
control cells were cultured in 6 cm dishes and they were harvested
after reaching 80-90% confluence, and these cells were then examind
by western blot analysis to detect the expression levels of IKKβ,
IKKα, IκBα, Cyld, C-myc, β-catenin, Notch1, p50, p65 and SENP2. (B)
MEC2 cells in which SENP2 was silenced and the control cells were
treated as described in (A). GAPDH was used to confirm equal amount
of proteins loaded in each lane. NC-OE-SENP2, MEC2 cells
transfected with null (control) overexpression vector; OE-SENP2,
MEC2 cells transfected with SENP2 overexpression vector;
NC-shRNA-SENP2, MEC2 cells transfected with null control shRNA;
shRNA-SENP2, MEC2 cells transfected with shRNA against SENP2. |
Discussion
Recently, SENP2 was identified as a tumor suppressor
in hepatocellular carcinoma, bladder cancer and breast cancer. It
has been shown that SENP2 can inhibit hepatocellular carcinoma cell
growth by regulating the stability of WWOX that is essential for
β-catenin stability (14,15). In addition, the invasion and
migration of bladder cancer cells has also been shown to be
suppressed by SENP2 through the blocking of MMP13 expression
(13). In 2014, Nait Achour et
al reported that SENP2 inhibited the proliferation of breast
cancer cells via the suppression of the transcription of estrogen
receptor α (ERα) signaling (12).
Therefore, we wished to determine whether the anti-tumor effect of
SENP2 can be applied to other malignant tumors and whether SENP2
may be a potential target for cancer treatment if the underlying
mechanisms are determined. In this study, we selected a CLL model,
which is the main subject of our research group, as the study
object to explore the potential anti-tumor effects of SENP2 on
CLL.
To date, at least to the best of our knowledge,
there are no reports of the role of SENP2 in CLL. Therefore, we
wished to determine whether the SENP2 is associated with the
occurrence and development of CLL in situ. The peripheral
blood of 43 patients with CLL and 21 healthy volunteers was
collected to analyze the protein expression level of SENP2, and the
results revealed that SENP2 was generally downregulated in the
peripheral blood of patients with CLL with or without treatment
compared with the healthy volunteers, although there was no
statistically significant difference. As the sample size used for
western blot analysis was limited, it would affect the results of
statistical analysis which are prone to a type II error, leading to
false-negative results. However, among the 4 patients before
treatment and 3 patients after treatment who were randomly
selected, 3 and 2 patients exhibited a significantly lower SENP2
expression compared to the control group in our western blot
analysis. Thus, we explored the role of SENP2 in MEC2 cells using
different experiments, although no statistically significant
difference was observed between the healthy controls and patients
with CLL. The differential mRNA expression levels of SENP2 in the
patients with CLL and healthy volunteers revealed that the mRNA
expression level of SENP2 in the patients with CLL was higher than
that in both the healthy volunteers and post-treatment patients,
indicating the differential expression of SENP2 in CLL cells which
may be associated with chemotherapy. Notably, the results of
western blot analysis and RT-qPCR appear to be contradictory, which
may be attributed to the inconsistency between the transcriptional
level and the protein expression level caused by the complex
processes from mRNA translation to the formation of functional
proteins.
It is well known that a poor prognosis of patients
with CLL is associated with a greater age and later disease stage
(1). Moreover, LDH, β2-MG and
cytogenetic abnormalities are also commonly used factors for
evaluating the prognosis of patients with CLL (28-31).
Therefore, in this study, the association between the mRNA
expression level of SENP2 and the clinicopathological
characteristics of patients with CLL before treatment were analyzed
for the first time, at least to the best of our knowledge. Among
the variety of clinicopathological characteristics of CLL, the
expression of LDH was demonstrated to be associated with the mRNA
expression level of SENP2, indicating the possible involvement of
SENP2 in the clinical course of disease. Considering that the
number of samples in this study is small and the collected samples
are mostly from inpatients and that we did not have early CLL
specimens, the association between SENP2 and the prognostic factors
of CLL warrants further investigation.
Since the aforementioned results provide the
preliminary proofs of the potential involvement of SENP2 in CLL,
whether SENP2 can affect the development of CLL needs to be
determined. For this purpose, in this study, CLL cell models in
which SENP2 was overexpressed or silenced were established to
determine the effect of the expression level of SENP2 on the
sensitivity of the cells to chemotherapy, and on cell viability,
cell invasion and the apoptotic state following treatment with
cytarabine and dexamethasone. Furthermore, the association between
the expression of SENP2 and the activation state of the Notch and
NF-κB signaling pathways was also investigated to explore the
underlying mechanisms. The successful construction of CLL cell
lines in which SENP2 was overexpressed or silenced was verified by
western blot analysis. Subsequently, the trend of chemosensitivity
shown in Fig. 2 is consistent with
our hypothesis that SENP2 enhances the sensitivity of MEC2 cells to
cytarabine and dexamethasone, although without statistical
significance. Thus, we may need to further improve our experimental
methods. At the same time, we noted that when the cells were
treated with cytarabine or dexamethasone for 24 h, obvious
differences between the groups were observed, but at 48 and 72 h,
these differences were not obvious. It can thus be hypothesized
that the overexpression of SENP2 has the tendency to enhance the
sensitivity of CLL cells to cytarabine and dexamethasone at an
early stage. However, as the time of drug action increases, tumor
cells can antagonize this effect through other mechanisms. The
mechanism of tumor drug resistance has been an important issue for
researchers, which will also be the focus of our future research.
Moreover, the results of Transwell and chemotaxis chamber assay in
this study demonstrated that the overexpression of SENP2 decreased
cell invasion and the cell chemotactic response, whereas the
silencing of SENP2 led to the opposite results. Cell invasion
reflects the invasive potential of cancer cells. These results also
proved that SENP2 may exert anti-tumor effects on CLL cells by
disrupting their chemotactic ability. In addition, the
overexpression of SENP2 combined with cytarabine or dexamethasone
treatment synergistically decreased the viability and the promoted
apoptosis of CLL cells. In conclusion, these results completely
verified that SENP2 exerts an anti-tumor effect on CLL by
suppressing cell invasion and chemotactic ability, and inducing
cell apoptosis and enhancing the sensitivity of CLL cells to
chemotherapy.
Although we had decided that the anti-tumor effect
of SENP2 could be applied to CLL from various routes and that the
Notch signaling pathway may be involved, the underlying molecular
mechanisms needed to be more fully examined. In our previous study,
we reported that the Notch signaling pathway regulated the NF-κB
signaling pathway in CLL cells, by disrupting the expression of
RelA and RelB, key members in the NF-κB family (24). Therefore, we inferred that the
NF-κB signaling pathway may also be involved in the regulation of
SENP2 in CLL cells. In this study, western blot analysis was
performed to analyze both the NF-κB and Notch signaling pathway in
the CLL cells in which SENP2 was overexpressed or silenced. The
final results revealed that the overexpression of SENP2
simultaneously suppressed the NF-κB and Notch signaling pathways,
which may be a result of the reversible de-SUMOylation of proteins
by SENP2. Notably, the silencing of SENP2 upregulated the Notch
signaling pathway and NF-κB signaling pathways. The study by Lee
et al (32) suggested that
the SUMO modification of the key molecule NEMO of the NF-κB
signaling pathway is the key step for the activation of the NF-κB
signaling pathway following DNA damage. Therefore, in CLL, SENP2
may inhibit the activity of the NF-κB signaling pathway by NEMO
SUMO modification, while the activity of the NF-κB signaling
pathway reduces the expression of IκB. Based on these results, we
summarized that the Notch signaling pathway and NF-κB signaling
pathway may be regulated by SENP2. To explain this phenomenon, we
inferred that there must be some other regulatory mechanisms
between the SENP2 and the NF-κB signaling pathway which need to be
investigated in future studies.
Collectively, this study first explored the
expression of SENP2 in primary CLL cells and the association
between SENP2 and CLL-related prognostic factors. We further
examined the effect of SENP2 on CLL cell functions and the effect
of SENP2 on key signaling pathways in CLL, including the Notch,
Wnt/β-catenin and NF-κB pathways. However, the number of CLL cases
collected in this study was relatively small, and thus some of the
results may not be convincing. In addition, we did not further
explore the mechanisms of action of SENP2 and its association with
SUMOylation and de-SUMOylation. Therefore, in future studies, we
aim to expand the sample size to further clarify the association
between SENP2 and CLL prognosis, as well as with chemotherapy, as
well as to illustrate the functional location of SENP2, and to
explore the mechanisms of SENP2 as regards the regulation of the
Notch, Wnt/β-catenin, and NF-κB signaling pathways and its
association with SUMOylation and de-SUMOylation.
In conclusion, the results of this study clearly
illustrate that SENP2 expression is generally downregulated in
patients with CLL and may inhibit cell invasion and the cell
chemotactic ability, and promote cell chemosensitivity and cell
apoptosis in CLL by suppressing the NF-κB and Notch signaling
pathways. This study may provide a theoretical basis for the
development of novel therapeutic strategies for CLL treatment.
Funding
This study was supported by the Natural Science
Foundation of Fujian Province of China (No. 2016J01458), Joint
Funds for the Innovation of Science and Technology of Fujian
Province of China (No. 2017Y9005), the Construction Project of
Fujian Medical Center of Hematology (Min201704), and the National
and Fujian Provincial Key Clinical Specialty Discipline
Construction Program, P.R. China.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. The datasets used and/or
analyzed during the current study are available from the
corresponding author on reasonable request.
Authors' contributions
XLC, SFW and ZSX conceived and designed and
supervised the study; XLC, SFW, XTL, HXL and TTW performed the
experiments; SQW, ZJQ and RZ analyzed the data; XLC, SFW, RZ and
ZSX wrote the manuscript. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
The use of human samples in this study was approved
by the Ethics Committee of the Fujian Medical University Union
Hospital and written consent was obtained from all participants in
this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Nabhan C and Rosen ST: Chronic lymphocytic
leukemia: A clinical review. JAMA. 312:2265–2276. 2014. View Article : Google Scholar
|
|
2
|
Barrientos JC: Management of chronic
lymphocytic leukemia in the elderly. Cancer Contr. 22(Suppl):
S17–S23. 2015. View Article : Google Scholar
|
|
3
|
Stilgenbauer S, Furman RR and Zent CS:
Management of chronic lymphocytic leukemia. Am Soc Clin Oncol Educ
Book. 35:164–175. 2015. View Article : Google Scholar
|
|
4
|
Xu Z, Zhang J, Wu S, Zheng Z, Chen Z and
Zhan R: Younger patients with chronic lymphocytic leukemia benefit
from rituximab treatment: A single center study in China. Oncol
Lett. 5:1266–1272. 2013. View Article : Google Scholar
|
|
5
|
Jamroziak K, Puła B and Walewski J:
Current treatment of chronic lymphocytic leukemia. Curr Treat
Options Oncol. 18:52017. View Article : Google Scholar
|
|
6
|
Chow KH, Elgort S, Dasso M, Powers MA and
Ullman KS: The SUMO proteases SENP1 and SENP2 play a critical role
in nucleoporin homeostasis and nuclear pore complex function. Mol
Biol Cell. 25:160–168. 2014. View Article : Google Scholar
|
|
7
|
Goeres J, Chan PK, Mukhopadhyay D, Zhang
H, Raught B and Matunis MJ: The SUMO-specific isopeptidase SENP2
associates dynamically with nuclear pore complexes through
interactions with karyopherins and the Nup107-160 nucleoporin
subcomplex. Mol Biol Cell. 22:4868–4882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim JH and Baek SH: Emerging roles of
desumoylating enzymes. Biochim Biophys Acta. 1792:155–162. 2009.
View Article : Google Scholar
|
|
9
|
Kang X, Qi Y, Zuo Y, Wang Q, Zou Y,
Schwartz RJ, Cheng J and Yeh ET: SUMO-specific protease 2 is
essential for suppression of polycomb group protein-mediated gene
silencing during embryonic development. Mol Cell. 38:191–201. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang M, Chiu SY and Hsu W: SUMO-specific
protease 2 in Mdm2-mediated regulation of p53. Cell Death Differ.
18:1005–1015. 2011. View Article : Google Scholar
|
|
11
|
Chiu SY, Asai N, Costantini F and Hsu W:
SUMO-specific protease 2 is essential for modulating p53-Mdm2 in
development of trophoblast stem cell niches and lineages. PLoS
Biol. 6:e3102008. View Article : Google Scholar
|
|
12
|
Nait Achour T, Sentis S, Teyssier C,
Philippat A, Lucas A, Corbo L, Cavaillès V and Jalaguier S:
Transcriptional repression of estrogen receptor α signaling by
SENP2 in breast cancer cells. Mol Endocrinol. 28:183–196. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tan M, Gong H, Wang J, Tao L, Xu D, Bao E,
Liu Z and Qiu J: SENP2 regulates MMP13 expression in a bladder
cancer cell line through SUMOylation of TBL1/TBLR1. Sci Rep.
5:139962015. View Article : Google Scholar
|
|
14
|
Shen HJ, Zhu HY, Yang C and Ji F: SENP2
regulates hepatocellular carcinoma cell growth by modulating the
stability of β-catenin. Asian Pac J Cancer Prev. 13:3583–3587.
2012. View Article : Google Scholar
|
|
15
|
Jiang QF, Tian YW, Shen Q, Xue HZ and Li
K: SENP2 regulated the stability of β-catenin through WWOX in
hepatocellular carcinoma cell. Tumour Biol. 35:9677–9682. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar
|
|
17
|
Nwabo Kamdje AH, Bassi G, Pacelli L,
Malpeli G, Amati E, Nichele I, Pizzolo G and Krampera M: Role of
stromal cell-mediated Notch signaling in CLL resistance to
chemotherapy. Blood Cancer J. 2:e732012. View Article : Google Scholar
|
|
18
|
Rosati E, Sabatini R, Rampino G, Tabilio
A, Di Ianni M, Fettucciari K, Bartoli A, Coaccioli S, Screpanti I
and Marconi P: Constitutively activated Notch signaling is involved
in survival and apoptosis resistance of B-CLL cells. Blood.
113:856–865. 2009. View Article : Google Scholar
|
|
19
|
Balatti V, Lerner S, Rizzotto L, Rassenti
LZ, Bottoni A, Palamarchuk A, Cascione L, Alder H, Keating MJ,
Kipps TJ, et al: Trisomy 12 CLLs progress through NOTCH1 mutations.
Leukemia. 27:740–743. 2013. View Article : Google Scholar
|
|
20
|
Rossi D, Rasi S, Fabbri G, Spina V,
Fangazio M, Forconi F, Marasca R, Laurenti L, Bruscaggin A, Cerri
M, et al: Mutations of NOTCH1 are an independent predictor of
survival in chronic lymphocytic leukemia. Blood. 119:521–529. 2012.
View Article : Google Scholar
|
|
21
|
Herishanu Y, Pérez-Galán P, Liu D,
Biancotto A, Pittaluga S, Vire B, Gibellini F, Njuguna N, Lee E,
Stennett L, et al: The lymph node microenvironment promotes B-cell
receptor signaling, NF-kappaB activation, and tumor proliferation
in chronic lymphocytic leukemia. Blood. 117:563–574. 2011.
View Article : Google Scholar
|
|
22
|
Fabbri G, Rasi S, Rossi D, Trifonov V,
Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S, et al:
Analysis of the chronic lymphocytic leukemia coding genome: Role of
NOTCH1 mutational activation. J Exp Med. 208:1389–1401. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Puente XS, Pinyol M, Quesada V, Conde L,
Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz
M, et al: Whole-genome sequencing identifies recurrent mutations in
chronic lymphocytic leukaemia. Nature. 475:101–105. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu ZS, Zhang JS, Zhang JY, Wu SQ, Xiong
DL, Chen HJ, Chen ZZ and Zhan R: Constitutive activation of NF-κB
signaling by NOTCH1 mutations in chronic lymphocytic leukemia.
Oncol Rep. 33:1609–1614. 2015. View Article : Google Scholar
|
|
25
|
Baldoni S, Sportoletti P, Del Papa B,
Aureli P, Dorillo E, Rosati E, Ciurnelli R, Marconi P, Falzetti F
and Di Ianni M: NOTCH and NF-κB interplay in chronic lymphocytic
leukemia is independent of genetic lesion. Int J Hematol.
98:153–157. 2013. View Article : Google Scholar
|
|
26
|
Sun Q, Wang R, Luo J, Wang P, Xiong S, Liu
M and Cheng B: Notch1 promotes hepatitis B virus X protein-induced
hepatocarcinogenesis via Wnt/β-catenin pathway. Int J Oncol.
45:1638–1648. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
28
|
Parikh SA and Shanafelt TD: Prognostic
factors and risk stratification in chronic lymphocytic leukemia.
Semin Oncol. 43:233–240. 2016. View Article : Google Scholar
|
|
29
|
Farah R, Al Danaf J, Braiteh N, Costa JM,
Farhat H, Mariani G and Giansily-Blaizot M: Life-threatening
bleeding in factor VII deficiency: The role of prenatal diagnosis
and primary prophylaxis. Br J Haematol. 168:452–455. 2015.
View Article : Google Scholar
|
|
30
|
Zenz T, Mertens D, Küppers R, Döhner H and
Stilgenbauer S: From pathogenesis to treatment of chronic
lymphocytic leukaemia. Nat Rev Cancer. 10:37–50. 2010. View Article : Google Scholar
|
|
31
|
Seiffert M, Dietrich S, Jethwa A, Glimm H,
Lichter P and Zenz T: Exploiting biological diversity and genomic
aberrations in chronic lymphocytic leukemia. Leuk Lymphoma.
53:1023–1031. 2012. View Article : Google Scholar
|
|
32
|
Lee MH, Mabb AM, Gill GB, Yeh ET and
Miyamoto S: NF-κB induction of the SUMO protease SENP2: A negative
feedback loop to attenuate cell survival response to genotoxic
stress. Mol Cell. 43:180–191. 2011. View Article : Google Scholar
|