Introduction
Lung cancer leads to high levels of cancer morbidity
and cancer-associated mortality worldwide (1). Lung cancers are clinically classified
as non-small-cell lung cancer (NSCLC; accounts for 80-85% of cases)
and small cell lung cancer (SCLC) (2). Surgical resection is the most
potentially curative therapeutic modality for this disease.
Cis-diammine-dichloro-platinum II-based adjuvant chemotherapy
significantly improves the prognosis of patients with advanced
NSCLC (3), particularly in those
with Stage II-IIIA (4,5). However, innate non-sensitiveness to
or acquired resistance to cisplatin is a major challenge in the
management of patients with lung cancer (6,7).
Therefore, the identification of mechanisms underlining cisplatin
chemoresistance in NSCLC is urgently required.
Advances in technology, including DNA sequencing,
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and microarray methods, enable the discovery of
predictive markers and the identification of significant expression
at the transcriptional level of chemoresistance-associated genes
(6,7). In particular, the profiling by
microarray screening is highly effective in predicting
chemotherapeutic sensitivity, with thousands of genes being
simultaneously evaluated (8-10).
However, the relatively low sensitivity and poor lower thresholds
of microarray detection reduce its accuracy; thus, follow-up
quantitative methods are required to confirm the results.
Stable isotope labeling by amino acids in cell
culture (SILAC) is effective in distinguishing the protein
profiling from one group to the other (11-13).
Five passages would transform ~97% of 12C-labeled amino
acids in A549 cells into 13C-labeled amino acids
[1-(1/2)5 = 97%] and thus, cells only contain 'heavy'
proteins (11,13). The incorporation of stable isotopes
facilitates the quantitative recognition of the differences in
expression profiles by tandem mass spectrometry between the
12C- and 13C-labeled A549 cells. SILAC has
also been useful in the identification of cancer biomarkers, and
chemoresistance-associated biomarkers in hepatocellular carcinoma
(14), breast cancer (15) and lung cancer (16,17).
In the present study, a cisplatin-resistant A549
cell clone (A549R) was isolated from A549 cells post-serial
passages under cisplatin pressure. The differences in
proliferation, apoptosis and autophagy were investigated between
A549R and A549 cells under cisplatin treatment. Then the SILAC
method was utilized to profile A549R specific proteomics under
cisplatin treatment. The results implied that autophagy may be an
important mechanism underlining the cisplatin resistance of NSCLC
A549 cells.
Materials and methods
Reagents, cell culture,
cisplatin-resistant clone selection and treatment
Human NSCLC A549 cells were purchased from American
Type Culture Collection (Manassas, VA, USA) and were cultured in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C, under 5% CO2. For the selection of
cisplatin-resistant clone, A549 (A549R) cells were seeded in
12-well plates (Corning Incorporated, Corning, NY, USA), with
<200 cells per well, and were then incubated at 37°C for 3-5
days with 1 µM cisplatin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). The larger cell colonies were picked and
propagated with DMEM + 10% FBS. Another 9 passages of selection
were performed via colony forming assays with 1 µM cisplatin
treatment, which were followed by a further 10 passages of
selection with 2 µM cisplatin treatment. For A549R
selection, A549 cells were cultured with 1 µM cisplatin for
5 passages (without selection/purification of larger colonies), and
then larger colonies were isolated after each passage for a further
5 passages with 1 µM cisplatin. A similar selection process
was performed for the isolation of colonies following treatment
with 2 µM cisplatin. For the stability examinations, A549R
cells were cultured for an additional 20 passages in DMEM without
cisplatin, then the colony forming and growth assays were
performed; A549R cells prior to serial passaging were used as the
control cells.
For heavy (H)- or light (L)-Lysine labeling
experiments, A549 or A549R cells were cultured serially for 5
passages in SILAC™ DMEM (Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS without Lysine, which was then
respectively supplemented with
13C6H14N2O2-Lysine-HCL
(H-labeled) or
12C6H14N2O2-Lysine-HCL
(L-labeled). A total of 10 µM cisplatin was added to each
group of cells, which were incubated for 24 h at 37°C with 5%
CO2 in a T75 cell flask. For SILAC proteomics analysis,
~1×107 H- or L-labeled A549R/A549 cells with 80-90%
confluence were collected for further analysis.
L- or H-labeled A549/A549R cells were
collected and washed four times with 10 ml ice-cold
phosphate-buffered saline (PBS) and counted
A total of 1×107 L- or H-labeled cells
were lysed with 0.5% 4-Nonylphenol Ethoxylate (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) containing 1.1 µM
pepstatin A (Sigma-Aldrich; Merck KGaA) on ice for 30 min. Nuclei
and other organelles were removed following centrifugation at 5,000
× g for 10 min at 4°C. The supernatant protein samples were
transferred to fresh tubes and then the protein concentration was
quantified with a Bicinchoninic Acid protein assay (Thermo Fisher
Scientific, Inc.). For SILAC analysis, the H- and L-labeled protein
samples were mixed in a ratio of 1:1; the remaining samples were
stored at −80°C prior to subsequent use.
Colony forming, cell proliferation and
migration assays
For the colony forming assay, ~200 A549 or A549R
cells were seeded in 12-well plates and incubated at 37°C with DMEM
containing 0 or 10 µM cisplatin for 3-5 days. The cell
colonies were stained at room temperature with 0.005% crystal
violet for 10 min and observed with a UVP imaging system (UVP; LLC,
Phoenix, AZ, USA). The colony number and size were quantified,
respectively. To generate the growth curves of A549 or A549R cells,
103 cells were incubated with DMEM containing 0 or 10
µM cisplatin for 0, 24, 48 or 72 h at 37°C, under 5%
CO2. Then the cell number in each group was counted with
the Olympus BX60 light microscope (Olympus Corporation, Tokyo,
Japan). For the cell migration assay, A549 or A549R cells were
cultured in 25 cm cell dishes with DMEM + 10% FBS to ~85%
confluence, and were then scratched with a cell scratcher (Costar;
Corning Incorporated). Cells were cultured for a further 24 h at
37°C with DMEM + 10% FBS, containing 2 µM cisplatin. The
number of cells that crossed the baseline was then counted as the
number of migrating cells using the Olympus BX60 light microscope
(Olympus Corporation).
Fluorescence-activated cell sorting
(FACS) can flow analysis of apoptotic cells
A total of 1×106 A549 or A549R cells were
treated at 37°C with or without 10 µM cisplatin for 24 h;
then cells in each group were collected for flow cytometry analysis
with a Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide
(PI) Apoptosis Detection kit (Abcam, Cambridge, UK). A549 or A549R
cells were trypsinized with 0.125% trypsin and then suspended in 1
ml binding buffer, to which 10 µl Annexin V-FITC and 10
µl PI were added successively for incubation at room
temperature in the dark for 15 min. The number of apoptotic cells
was then determined using a FACScan flow cytometer (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and analyzed using FlowJo
version 10 (FlowJo LLC, Ashland, OR, USA).
Imaging of autophagic puncta with green
fluorescence protein (GFP)-light chain (LC)-3 reporter
For the imaging of autophagic vesicles (puncta),
A549 or A549R cells were transfected with a GFP-LC3 reporter
plasmid (1 µg per well of a 12-well plate; Biovector Science
Laboratory, Beijing, China) for 6 h using Lipofectamine 3000™
(Invitrogen; Thermo Fisher Scientific, Inc.). Fresh DMEM containing
2% FBS was added to the cells, which were then treated with or
without 10 µM cisplatin at 37°C for 24 h. Treatment with 3
µM Rapamycin (Sigma-Aldrich; Merck KGaA) was taken as the
positive autophagy induction control, and blank A549 or A549R cells
(cells transfected with the GFP-LC3 reporter plasmid only) with
fresh DMEM containing 2% FBS was used as the blank control. A total
of 5 nM 3-methyladenine (3MA; an autophagy inhibitor;
Sigma-Aldrich; Merck KGaA) was utilized to inhibit
cisplatin-induced autophagy in A549 or A549R cells via treatment
for 24 h at 37°C. Autophagic puncta were imaged and counted by
confocal laser microscopy, and analyzed using FluoView software
version 5.0 (both from Olympus Corporation).
Protein digestion, identification and
quantification
The mixed H-/L-labeled protein sample was added into
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) loading buffer and incubated in pre-boiled water for 3
min. Proteins were then separated by electrophoresis with 12%
SDS-PAGE (as described below) and stained with Coomassie Brilliant
Blue at 26°C for 3 h. The whole gel lane was sliced into 40 pieces
according to Sun et al (18). The excised sections were
homogenized and de-stained twice with a 1:1 ratio of 50 mM Tris
acetonitrile and 50 mM ammonium bicarbonate solution (both from
Sigma-Aldrich; Merck KGaA). The extraction of tryptic peptides from
the gel was sequentially performed with 5% Trifluoroacetate
(Beijing Chemical Co., Ltd., Beijing, China) in the microwave oven
at 750 W for 8 min, and with 2.5% Trifluoroacetate and with 50%
Tris acetonitrile in the microwave oven at 750 W for 8 min. The
extracts were pooled and dried completely by centrifugal
lyophilization.
A mobile phase of 90 min at a flow rate of 300
nl/min was performed to separate each peptide mixture sample from
the sliced gel, which were then subjected analysis with a Linear
Trap Quadruple-Fourier Transform (LTQ-FT) mass spectrometer (Thermo
Fisher Scientific, Inc.), which was equipped with a nanospray
source and Agilent 1100 high-performance liquid chromatography
system (Agilent Technologies, Inc., Santa Clara, CA, USA). The
peptide eluent was introduced directly to an LTQ-FT mass
spectrometer via electrospray ionization. Positively identified
proteins were considered when at least two reliable peptides were
matched and a protein score >64 was observed. The false positive
rate of identified peptides was calculated as the ratio of total
peptide hits in the reverse database to the number of peptide hits
in the forward database above the same threshold. Identified
proteins were quantified by SILAC-specific software (MSQuant 1.4.1;
msquant.sourceforge.net) and inspected
manually. Peptide abundances were calculated as ratios of the areas
of the mono-isotopic peaks of the H-labeled versus the L-labeled
peptides, and the protein ratios were calculated from the average
of all quantified peptides of it.
Western blotting
Nuclear and cytosol fractions of the protein samples
were isolated from A549 or A549R cells using a Nuclear/Cytosol
Fractionation kit (BioVision, Inc., Milpitas, CA, USA) and then a
protease inhibitor (Sigma-Aldrich; Merck KGaA) was added. The
concentration of each protein sample was determined using a BCA
Protein Assay Reagent kit (Pierce; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. Proteins (8
µg/lane) were separated by 12% SDS-PAGE and transferred to a
nitrocellulose membrane (EMD Millipore, Billerica, MA, USA) in
order to separate the proteins in each sample by molecular weight.
Then the membrane was blocked with 2% bovine serum albumin
(Sigma-Aldrich; Merck KGaA) at 4°C overnight, and then incubated
with the rabbit or mouse anti-human LC3 (cat. no. sc-28266; 1:500),
autophagy-related protein (Atg) 7 (cat. no. sc-517310; 1:500) or
β-actin primary antibodies (cat. no. sc-517582; 1:1,000; all from
Santa Cruz Biotechnology, Inc.) for 2 h at room temperature (26°C).
Membranes were then incubated with horseradish peroxidase
(HRP)-conjugated anti-rabbit secondary antibodies [bovine
anti-rabbit immunoglobulin G (IgG)-HRP: cat. no. sc-2379, 1:1,000;
or bovine anti-mouse IgG-HRP: cat. no. sc-2380, 1:1,000; Santa Cruz
Biotechnology, Inc.] for 1 h at room temperature (26°C). Membranes
were washed 4 times with 1X PBS-Tween-20 (0.1% final concentration)
prior to each incubation. The antigen-antibody binding was
visualized with Enhanced chemiluminescence (Thermo Fisher
Scientific, Inc.) using the UVP BioSpectrum 500 imaging system
(UVP, LLC, Phoenix, AZ, USA) and ImageJ version 1.43b (National
Institutes of Health, Bethesda, MD, USA).
Gene ontology (GO) analysis
GO analyses were performed using DAVID 6.7
(david.ncifcrf.gov/). Apoptosis- and
autophagy-associated genes were selected for analysis when the
P-value of the correlation was <0.05.
Statistical analysis
SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA)
was utilized for statistical analysis. Quantitative results were
presented as the mean ± standard error of 3 or 4 repeated
experiments. Statistical differences were analyzed with Student's
t-test or one-way analysis of variance with Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Acquisition of cisplatin resistance in
human lung cancer A549 cells following serial passages with
cisplatin treatment
Cisplatin-resistant human lung cancer A549 cells
(A549R cells) were obtained following serial passages under 1
µM cisplatin (5 blind passages, then purification for
another 5 passages) and then 2 µM cisplatin (5 blind
passages, then purification for another 5 passages) treatment via
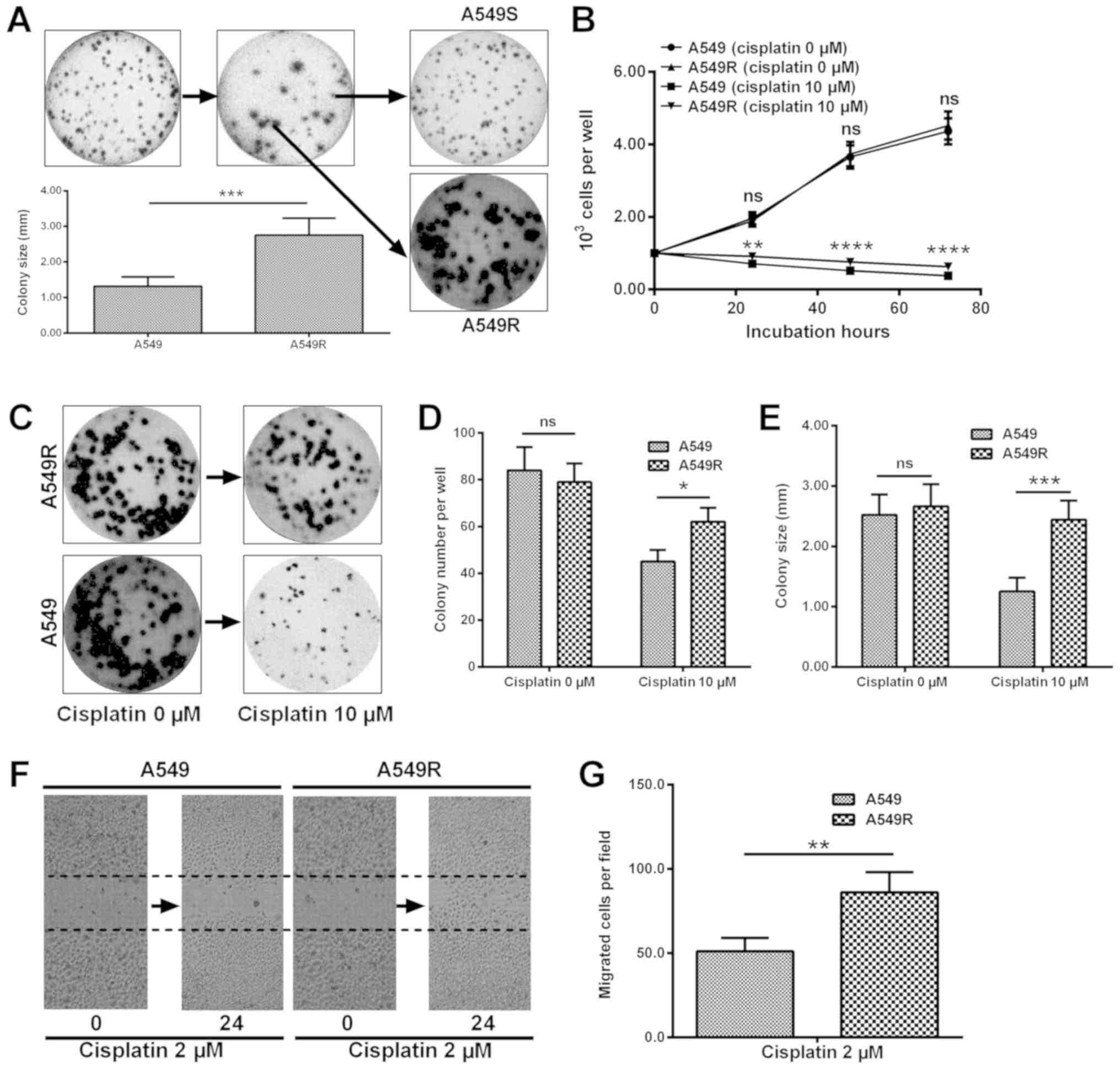
colony forming assays. As indicated in Fig. 1A, a phenotype with a larger colony
size of A549 cells was obtained (2.75±0.48 vs. 1.32±0.26;
P<0.001). The level of proliferation in A549R cells was
significantly higher than that of A549 cells, under treatment with
10 µM cisplatin for 24, 48 or 72 h (P<0.05, P<0.01 or
P<0.001; Fig. 1B). Colony
formation results also confirmed the difference in the level of
proliferation between A549R and A549 cells (Fig. 1C). The colony number (Fig. 1D) and colony size (Fig. 1E) were greater in A549R cells
post-treatment with 10 µM cisplatin (P<0.05 or
P<0.001). In addition, a migration assay was performed for A549
and A549R cells in the presence of 2 µM cisplatin. It was
demonstrated in Fig. 1F and G that
more cells crossed the baseline in the A549R cell group
(P<0.01). Taken together, the results indicate that cisplatin
resistance was acquired in A549 cells post 10 passages under
cisplatin treatment.
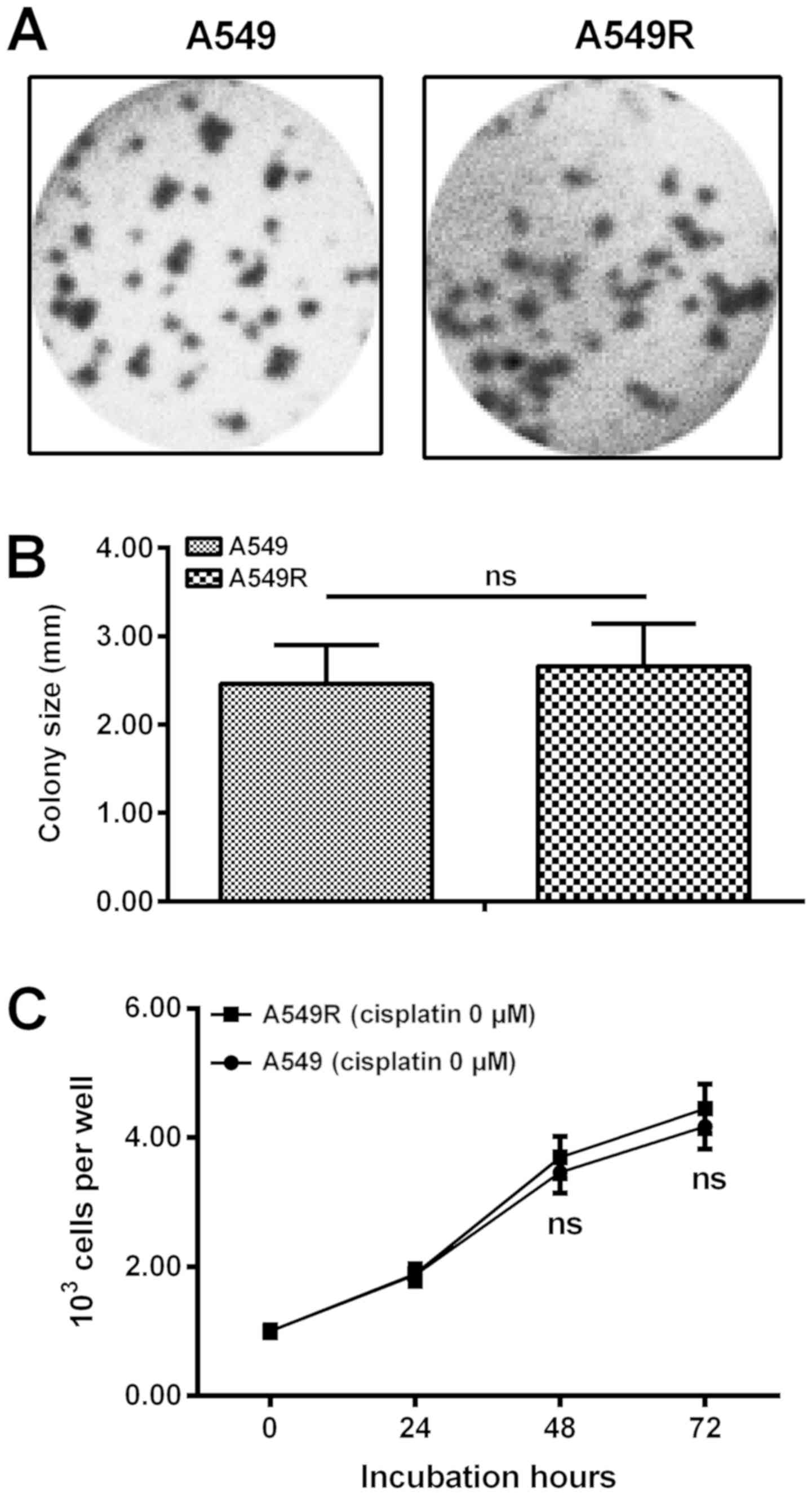
In addition, A549R cells were cultured in DMEM
without cisplatin for an additional 20 passages. It was indicated
in Fig. 2A-C that there was no
marked difference in growth efficiency between A549R and A549
cells.
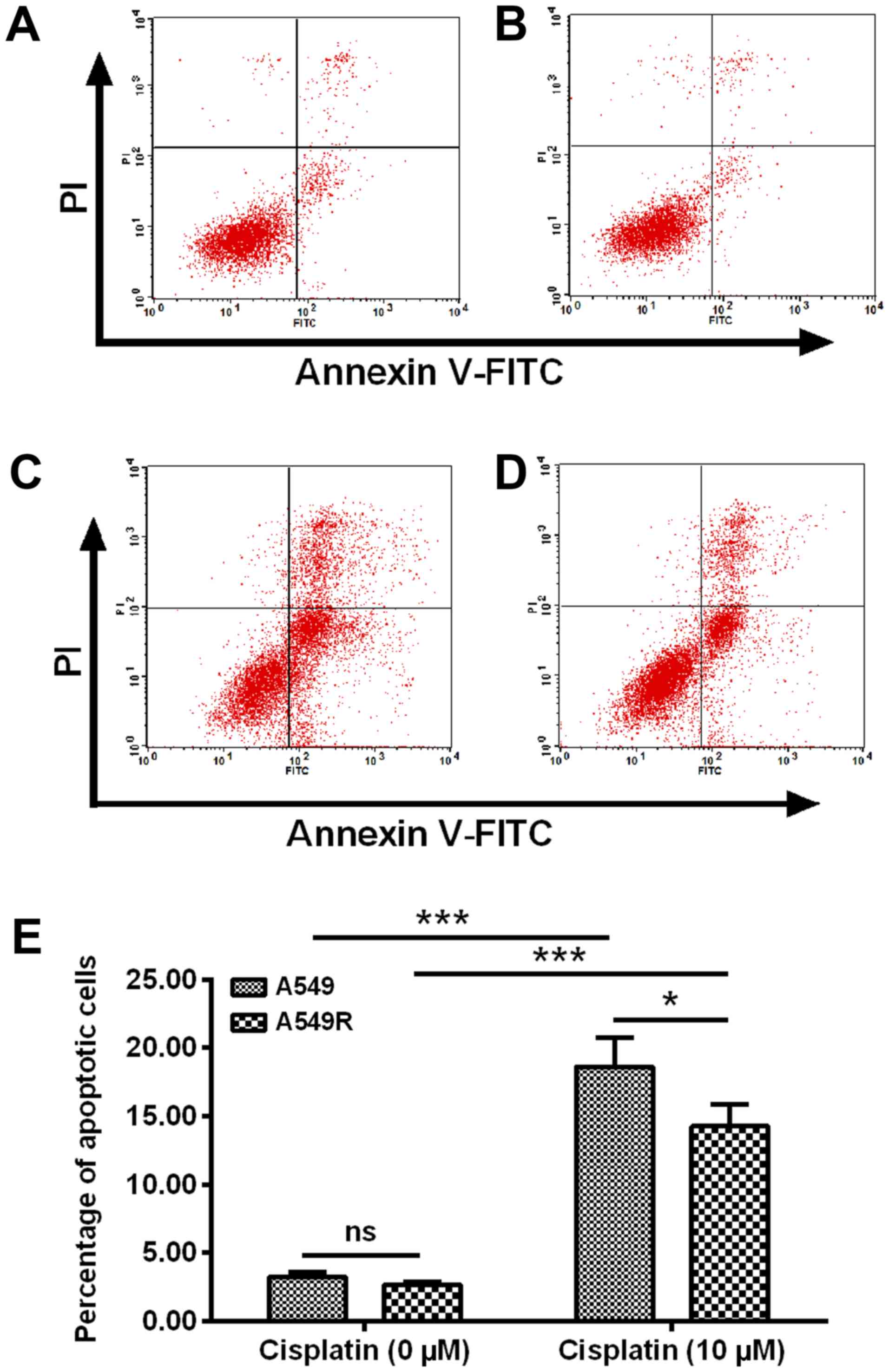
Reduced apoptosis induction by cisplatin
in the cisplatin-resistant A549R cells
To confirm the difference in the sensitivity to
cisplatin between A549R and A549 cells, apoptosis induction of
either A549R or A549 cells, post-treatment with 10 µM
cisplatin for 24 h was examined by flow cytometry analysis
following staining with the Annexin V-FITC/PI Apoptosis Detection
kit. As presented in Fig. 3A-D, in
contrast to the A549 (Fig. 3A) or
A549R (Fig. 3B) cells without
cisplatin treatment, treatment with 10 µM cisplatin for 24 h
induced significantly high levels of apoptosis in A549 and A549R
cells (P<0.001; Fig. 3C-E).
Furthermore, there were less apoptotic cells in the A549R group
(Fig. 3D and E) than in the A549
group following 10 µM cisplatin treatment (P<0.05;
Fig. 3C and E). Therefore, these
results confirmed resistance in A549R cells to cisplatin.
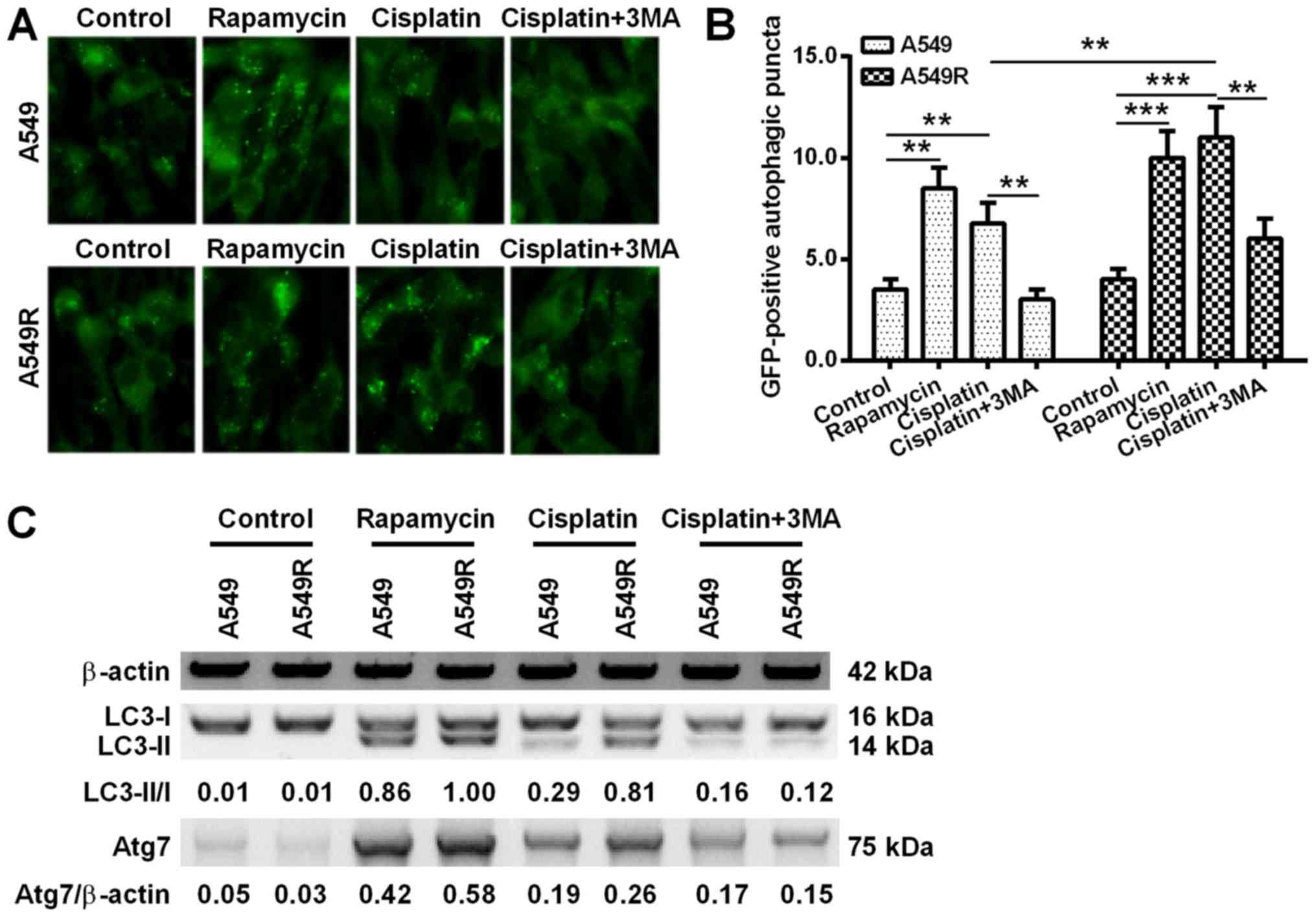
Autophagy induction by cisplatin in A549R
cells
Autophagy has been supported by more studies as one
of mechanisms underlining the chemoresistance of lung cancer cells
(19,20). Firstly in the present study,
autophagy-specific acidic vesicular organelles (AVOs) in A549R or
A549 cells were observed under a fluorescence microscope with a
GFP-LC3 reporter. When compared with the blank A549 or A549R cells,
10 µM rapamycin induced significantly high levels of AVOs
(P<0.01 or P<0.001; Fig. 4A and
B). Notably, the 10 µM cisplatin treatment also induced
significant levels of AVOs in A549 and A549R cells (P<0.01 or
P<0.001). In addition, this induction could be inhibited by the
autophagy inhibitor 3MA in the two types of cells (P<0.01).
Furthermore, more AVOs were induced by cisplatin in A549R cells,
than in A549 cells (P<0.01; Fig.
4B).
Western blotting was also performed to examine the
expression of autophagy-associated genes in the cisplatin-treated
A549 or A549R cells. Fig. 4C
demonstrated that rapamycin and cisplatin induced a high level of
LC3-I to LC3-II conversion and a high expression of Atg7 in A549
and A549R cells, both of which were inhibited by 3MA treatment. In
addition, a greater LC3-II/LC3-I ratio and increased Atg7
expression were observed in the cisplatin-treated A549R cells when
compared with the cisplatin-treated A549 cells.
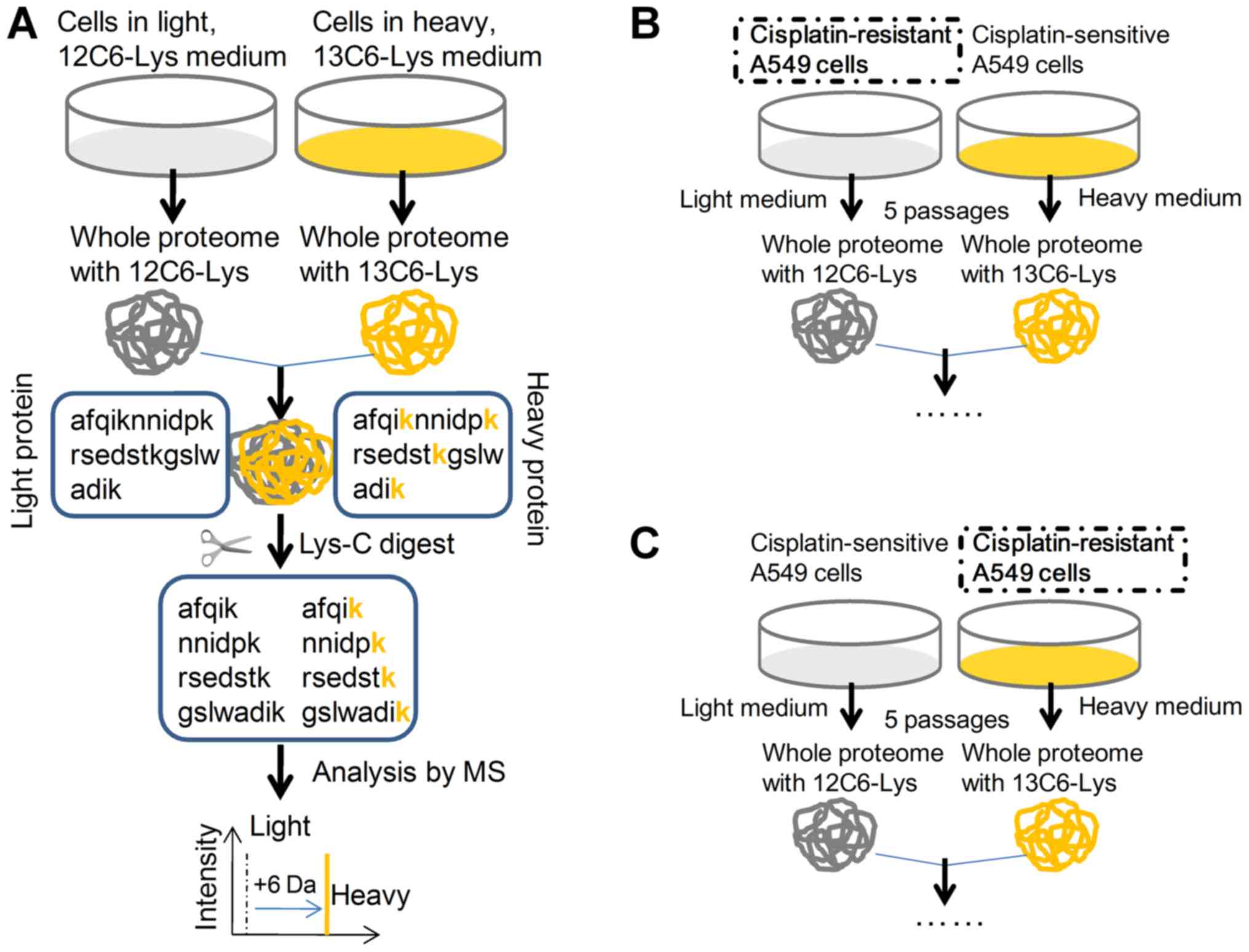
General proteomics information by SILAC
in the cisplatin-treated A549R cells
To recognize the discriminating protein profile
underlining cisplatin resistance in A549R cells, a SILAC method was
adopted to quantify the cellular response to cisplatin treatment in
either A549 or A549R cells. The general technological process of
SILAC is presented in Fig. 5A. The
12C6H14N2O2-Lysine-HCL
(L-labeled) A549R cells (Fig. 5B)
or the
13C6H14N2O2-Lysine-HCL
(H-labeled) A549R cells (Fig. 5C)
were respectively utilized to quantify the responsive protein
profile to cisplatin, with H-labeled or L-labeled A549 cells as
control. To examine the quality of each procedure, cellular
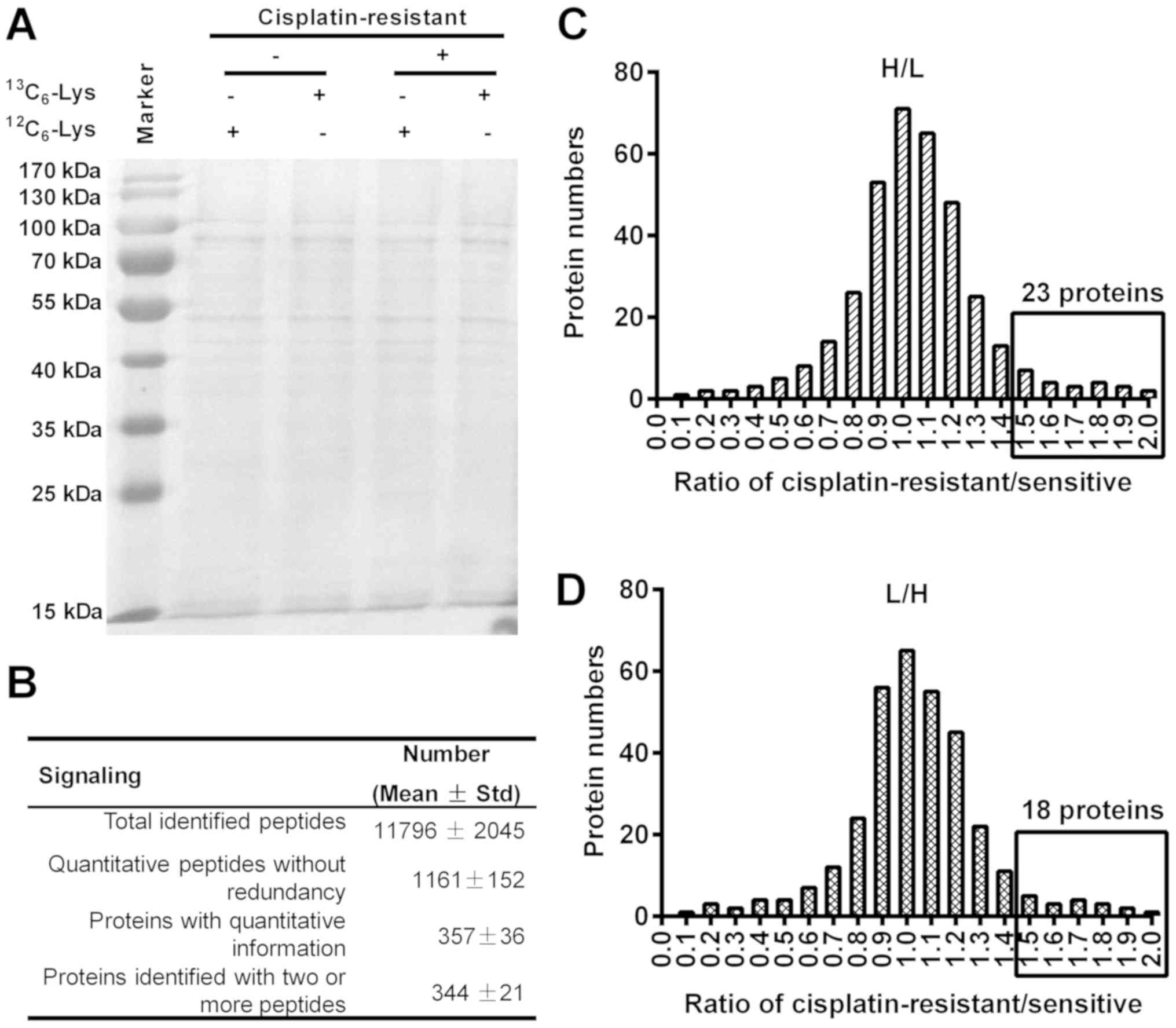
proteins were separated by 12% SDS-PAGE. As shown in Fig. 6A, protein bands were equally
distributed in the H- or L-labeled A549R or A549 cells. The general
difference in proteomics between A549R and A549 cells were
summarized in Fig. 6B: Total of
1,161±152 quantitative peptides, and 357±36 proteins were induced
by cisplatin (10 µM) between A549R and A549 cells.
Upregulation of anti-apoptosis and
autophagy-associated proteins in cisplatin-treated A549R cells via
SILAC screening
Among the upregulated proteins in the H-labeled
A549R cells, there were 23 proteins with expression that was
>1.5-fold greater than in the L-labeled A549 cells (Fig. 6C). In particular, 15 proteins,
including glucose-regulated protein, 78 kDa (GRP78), heat shock
protein 71 (HSP71), heterogeneous nuclear ribonucleoprotein A1
(ROA1) and pre-mRNA processing factor 19 (PRP19), had increased
expression by >2-fold in the H-labeled A549R cells when compared
with the L-labeled A549 cells (Table
I). In another repeated experiment with L-labeled A549R cells
and H-labeled A549 cells, there were 18 proteins recognized also
with expression that was >1.5 fold greater (Fig. 6D; Table II). GO analysis indicated that the
majority of the upregulated proteins were involved in
anti-apoptosis, DNA repair and autophagy (Tables I and II). In addition, the two repeated
experiments demonstrated that 15 proteins [GRP78, HSP71, PRP19,
polypyrimidine tract binding protein 1 (PTBP1), translationally
controlled tumor protein (TCTP), Cathepsin D (CATD), Cytochrome
c (CYC), thioredoxin domain containing 5 (TXND5), MutS
homolog 6 (MSH6), Annexin A2 (ANXA2), RCA2 and Cyclin dependent
kinase inhibitor 1A interacting protein (BCCIP), MSH2, protein
phosphatase 2A 55 kDa regulatory subunit Bα (PP2AB), Rho
glyceraldehyde-3-phosphate-dissociation inhibitor 1 (GDIR1) and
ANXA4)] were repeatedly upregulated by >1.5-fold greater in H-
and L-labeled A549R cells (Tables
I and II).
 | Table IProteins with >1.5-fold change
(H/L) in expression levels in A549R cells when compared with A549
cells. |
Table I
Proteins with >1.5-fold change
(H/L) in expression levels in A549R cells when compared with A549
cells.
| Accession no. | Uniprot ID | Protein name | Fold (H/L) | GO term name in
biological pathway |
|---|
|
P11021* | GRP78 | 78 kDa
glucose-regulated protein (bip) | 3.57 | Anti-apoptosis,
calcium ion binding, enzyme binding, misfolded protein binding |
|
Q9UMS4* | PRP19 | Pre-mRNA-processing
factor 19 | 3.28 | DNA repair,
ubiquitin-ubiquitin ligase activity |
|
P13693* | TCTP |
Translationally-controlled tumor protein
(TCTP) | 3.12 | Anti-apoptosis in
response to DNA damage |
|
P08107* | HSP71 | Heat shock 70 kDa
protein 1A/1B | 2.75 | Anti-apoptosis,
cellular response to oxidative stress, negative regulation of cell
death |
| P09651 | ROA1 | hnRNP core protein
A1 (hnRNP A1) | 2.7 | mRNA processing,
negative regulation of telomere maintenance via telomerase |
|
P26599* | PTBP1 | Polypyrimidine
tract-binding protein 1 (hnRNP I) | 2.63 | mRNA processing,
mRNA splicing, via spliceosome |
| P08758 | ANXA5 | Annexin A5 (Annexin
V) | 2.58 | Anti-apoptosis,
calcium-dependent phospholipid binding |
|
P07339* | CATD | Cathepsin D | 2.56 | Autophagic vacuole
assembly, protein catabolic process, proteolysis |
| Q04760 | LGUL | Lactoylglutathione
lyase (Aldoketomutase) | 2.43 | Anti-apoptosis,
regulation of transcription by RNA polymerase II |
| Q01081 | U2AF1 | Splicing factor
U2AF 35 kDa subunit | 2.37 | mRNA processing,
mRNA splicing, via spliceosome, RNA export from nucleus |
|
P99999* | CYC | Cytochrome C | 2.31 | Cellular
respiration, cellular response to oxidative stress, intrinsic
apoptotic signaling pathway |
|
Q8NBS9* | TXND5 | Thioredoxin
domain-containing protein 5 | 2.25 | Anti-apoptosis,
protein folding, response to endoplasmic reticulum stress |
| P14625 | ENPL | Heat shock protein
90 kDa β member 1 (GRP-94) | 2.12 | Anti-apoptosis, RNA
binding, unfolded protein binding |
|
P52701* | MSH6 | DNA mismatch repair
protein Msh6 (hmsh6) | 2.1 | DNA damage
response, methylated histone binding, mismatched DNA binding |
|
P07355* | ANXA2 | Annexin A2 (Annexin
II) | 2.01 | Skeletal system
development, phosphatidylinositol-4,5-bisphosphate binding,
phospholipase A2 inhibitor activity |
|
Q9P287* | BCCIP | BRCA2 and
CDKN1A-interacting protein | 1.94 | DNA repair,
regulation of cyclin-dependent protein serine/threonine kinase
activity |
|
P43246* | MSH2 | DNA mismatch repair
protein Msh2 (hmsh2) | 1.83 | Mismatch repair,
intrinsic apoptotic signaling pathway in response to DNA damage by
p53 class mediator |
| Q16531 | DDB1 | DNA damage-binding
protein 1 (DDB p127 subunit) | 1.74 | Nucleotide-excision
repair, DNA damage response, detection of DNA damage |
| P13073 | COX41 | Cytochrome C
oxidase subunit 4 isoform 1 (COX IV-1) | 1.71 | Response to
nutrient, mitochondrial electron transport, Cytochrome C to
oxygen |
|
P62714* | PP2AB |
Serine/threonine-protein phosphatase 2A
catalytic subunit β isoform (PP2A-β) | 1.69 | Protein amino acid
dephosphorylation, apoptotic mitochondrial changes, negative
regulation of Ras protein signal, response to endoplasmic reticulum
stress |
|
P52565* | GDIR1 | Rho
GDP-dissociation inhibitor 1 (Rho GDI 1) | 1.67 | Anti-apoptosis,
regulation of Rho protein signal transduction and of small GTPase
mediated signal transduction |
| P62805 | H4 | Histone H4 | 1.56 | Negative regulation
of megakaryocyte differentiation |
|
P09525* | ANXA4 | Annexin A4 (Annexin
IV) | 1.51 | Anti-apoptosis,
negative regulation of NF-κB transcription factor activity |
| Q15833 | STXB2 | Syntaxin-binding
protein 2 | 0.66 | Cellular response
to interferon-gamma, protein transport |
|
Q9NZJ7* | MTCH1 | Mitochondrial
carrier homolog 1 | 0.66 | Activation of
cysteine-type endopeptidase activity, apoptotic process |
| O60218 | AK1BA | Aldo-keto reductase
family 1 member B10 | 0.64 | Aldo-keto reductase
(NADP) activity, geranylgeranyl reductase activity |
|
Q0WX57* | U17LO |
Ubiquitin-specific-processing protease
17 | 0.63 | Apoptotic process,
protein deubiquitination involved in ubiquitin-dependent protein
catabolic process |
| P11498 | PYC | Pyruvate
carboxylase, mitochondrial | 0.63 | Biotin binding,
identical protein binding, biotin metabolic process |
| P10909 | CLUS | Clusterin | 0.61 | Chaperone binding,
positive regulation of apoptotic process |
|
Q16513* | PKN2 |
Serine/threonine-protein kinase N2 | 0.59 | Apoptotic process,
cell adhesion, cell cycle and cell division |
| Q9HBU6 | EKI1 | Ethanolamine kinase
1 | 0.56 | ATP binding,
ethanolamine kinase activity, phosphatidylethanolamine biosynthetic
process |
| Q9GZU2 | PEG3 |
Paternally-expressed gene 3 protein | 0.53 | Apoptotic process,
nucleic acid binding |
| O95140 | MFN2 | Mitofusin-2 | 0.53 | GTP binding,
apoptotic process, macroautophagy |
|
O14763* | TR10B | TRAIL receptor
2 | 0.52 | Receptor for the
cytotoxic ligand TNFSF10/TRAIL, the adapter molecule FADD recruits
caspase-8 to the activated receptor |
| Q96S44 | PRPK | EKC/KEOPS complex
subunit TP53RK | 0.47 | ATP binding,p53
binding, protein serine/threonine kinase activity |
| I0J062 | PANO1 | Proapoptotic
nucleolar protein 1 | 0.47 | Positive regulation
of apoptotic process, regulation of protein stability |
|
Q15464* | SHB | SH2
domain-containing adapter protein B | 0.46 | Apoptotic process,
cell differentiation, SH3/SH2 adaptor activity |
| Q96FX8 | PERP | P53 apoptosis
effector related to PMP-22 | 0.45 | Notch signaling
pathway, positive regulation of proteolysis, regulation of
apoptotic process |
| P26447 | S10A4 | Protein
S100-A4 | 0.38 | Epithelial to
mesenchymal transition, positive regulation of I-κB kinase/NF-κB
signaling |
 | Table IIProteins with >1.5-fold change
(L/H) in expression levels in A549R cells when compared with A549
cells. |
Table II
Proteins with >1.5-fold change
(L/H) in expression levels in A549R cells when compared with A549
cells.
| Accession no. | Uniprot ID | Protein name | Fold (L/H) | GO term name
biological pathway |
|---|
|
P11021* | GRP78 | 78 kDa
glucose-regulated protein (bip) | 2.87 | Anti-apoptosis,
calcium ion binding, enzyme binding, misfolded protein binding |
|
P43246* | MSH2 | DNA mismatch repair
protein Msh2 (hmsh2) | 2.75 | Mismatch repair,
intrinsic apoptotic signaling pathway in response to DNA damage by
p53 class mediator |
|
P26599* | PTBP1 | Polypyrimidine
tract-binding protein 1 (hnRNP I) | 2.7 | mRNA processing,
mRNA splicing, via spliceosome |
|
P07339* | CATD | Cathepsin D | 2.67 | Autophagic vacuole
assembly, protein catabolic process, proteolysis |
|
Q8NBS9* | TXND5 | Thioredoxin
domain-containing protein 5 | 2.6 | Anti-apoptosis,
protein folding, response to endoplasmic reticulum stress |
|
P13693* | TCTP |
Translationally-controlled tumor protein
(TCTP) | 2.58 | Anti-apoptosis in
response to DNA damage |
| Q9Y3F4 | STRAP | Serine-threonine
kinase receptor-associated protein | 2.52 | mRNA processing,
negative regulation of pathway-restricted SMAD protein
phosphorylation |
|
Q9UMS4* | PRP19 | Pre-mRNA-processing
factor 19 | 2.46 | DNA repair,
ubiquitin-ubiquitin ligase activity |
| P61224 | RAP1B | Ras-related protein
Rap-1b | 2.03 | Cell proliferation,
positive regulation of ERK1 and ERK2 cascade |
|
P08107* | HSP71 | Heat shock 70 kDa
protein 1A/1B | 1.89 | Anti-apoptosis,
cellular response to oxidative stress, negative regulation of cell
death |
|
P52701* | MSH6 | DNA mismatch repair
protein Msh6 (hmsh6) | 1.76 | DNA damage
response, methylated histone binding, mismatched DNA binding |
|
P09525* | ANXA4 | Annexin A4 (Annexin
IV) | 1.73 | Anti-apoptosis,
negative regulation of NF-κB transcription factor activity |
|
P99999* | CYC | Cytochrome C | 1.72 | Cellular
respiration, cellular response to oxidative stress, intrinsic
apoptotic signaling pathway |
|
P62714* | PP2AB |
Serine/threonine-protein phosphatase 2A
catalytic subunit β isoform (PP2A-β) | 1.69 | Protein amino acid
dephosphorylation, apoptotic mitochondrial changes, negative
regulation of Ras protein signal, response to endoplasmic reticulum
stress |
|
P52565* | GDIR1 | Rho
GDP-dissociation inhibitor 1 (Rho GDI 1) | 1.67 | Anti-apoptosis,
regulation of Rho protein signal transduction and of small GTPase
mediated signal transduction |
| Q01130 | SFRS2 |
Serine/arginine-rich splicing factor 2
(Protein PR264) | 1.58 | mRNA processing,
mitotic cell cycle, mRNA export from nucleus |
|
P07355* | ANXA2 | Annexin A2 (Annexin
II) | 1.53 | Skeletal system
development, phosphatidylinositol-4,5-bisphosphate binding,
phospholipase A2 inhibitor activity |
|
Q9P287* | BCCIP | BRCA2 and
CDKN1A-interacting protein | 1.51 | DNA repair,
regulation of cyclin-dependent protein serine/threonine kinase
activity |
| O00194 | RB27B | Ras-related protein
Rab-27B | 0.66 | GTP binding, myosin
V binding, protein domain specific binding |
|
Q0WX57* | U17LO |
Ubiquitin-specific-17 processing
protease | 0.66 | Apoptotic process,
protein deubiquitination involved in ubiquitin-dependent protein
catabolic process |
| O94804 | STK10 |
Serine/threonine-protein kinase 10 | 0.64 | Regulation of
apoptotic process, regulation of mitotic cell cycle, signal
transduction by protein phosphorylation |
| O60656 | UD19 |
UDP-glucuronosyltransferase 1-9 | 0.62 |
Glucuronosyltransferase activity, negative
regulation of cellular glucuronidation |
|
Q16513* | PKN2 |
Serine/threonine-protein kinase N2 | 0.62 | Apoptotic process,
cell adhesion, cell cycle and cell division |
| Q16222 | UAP1 |
UDP-N-acetylhexosamine
pyrophosphorylase | 0.6 | Carbohydrate
binding, UDP-N-acetylglucosamine diphosphorylase activity |
| Q16539 | MK14 | Mitogen-activated
protein kinase 14 | 0.59 | Apoptotic process,
DNA damage checkpoint, intracellular signal transduction |
| P62308 | RUXG | Small nuclear
ribonucleoprotein G | 0.58 | RNA binding,
histone mRNA metabolic process, mRNA splicing, RNA splicing |
| Q07812 | BAX | Apoptosis regulator
BAX | 0.58 | Apoptotic
mitochondrial changes, apoptotic process, DNA damage response |
|
Q9NZJ7* | MTCH1 | Mitochondrial
carrier homolog 1 | 0.56 | Activation of
cysteine-type endopeptidase activity, apoptotic process |
|
Q15464* | SHB | SH2
domain-containing adapter protein B | 0.55 | Apoptotic process,
cell differentiation, SH3/SH2 adaptor activity |
|
O14763* | TR10B | TRAIL receptor
2 | 0.53 | Receptor for the
cytotoxic ligand TNFSF10/TRAIL, the adapter molecule FADD recruits
caspase-8 to the activated receptor |
| Q16890 | TPD53 | Tumor protein
D53 | 0.49 | G2/M transition of
mitotic cell cycle source, positive regulation of apoptotic
signaling pathway and of JNK cascade |
| Q9H4P4 | RNF41 | E3
ubiquitin-protein ligase NRDP1 | 0.47 | Autophagy,
extrinsic apoptotic signaling pathway, negative regulation of cell
proliferation |
Downregulation of apoptosis-associated
proteins in cisplatin-treated A549R cells
In addition, there were 26 and 22 proteins that were
downregulated >1.5-fold in the H- and L-labeled A549R cells,
respectively, when compared with the L- and H-labeled A549 cells
(Tables I and II). It was indicated in Table I that there were 16 proteins that
were downregulated by >1.5-fold in H-labeled A549R cells when
compared with L-labeled A549 cells. In another experiment, 14
proteins were revealed to be downregulated in L-labeled A549R cells
when compared with the H-labeled A549 cells (Table II). Notably, the majority of the
downregulated proteins were associated with apoptosis. In
particular, 5 proteins [tumor necrosis factor receptor superfamily
member 10B (TR10B), ubiquitin specific peptidase 17 (U17LO), SHB,
PKN2, MTCH1) were downregulated in H- and L-labeled A549R cells;
all of these proteins were involved in apoptotic processes or
signaling. Therefore, apoptosis-associated proteins were
downregulated in cisplatin-treated A549R cells.
Discussion
Cisplatin-based combinations of cytotoxic
chemotherapy are the primary form of lung cancer chemotherapy as it
significantly improves lung cancer patient outcomes (3,4,21,22).
Approximately 30% of patients with stage IV NSCLC are responsive to
cisplatin-based, two-drug combination treatment, and >95%
patients live >3 years (22,23).
Even for patients with SCLC, their initial response rates to
cisplatin combination are higher, at 50-80%. However, almost all
lung cancers are either initially or ultimately resistant to the
current chemotherapy drugs, including cisplatin (22,23).
In the present study, a NSCLC cell clone, A549, was chosen as a
cell model to evaluate the sensitivity/resistance of lung cancer
cells to cisplatin. Notably, serial passages of A549 cells under
1-2 µM cisplatin treatment gave rise to the
cisplatin-resistant phenotype of A549 cells. A549 colonies with a
larger size were manually enriched via colony forming assay. The
results of growth curve, colony formation and migration assays
confirmed that the A549R cells with the cisplatin-resistant
phenotype grew and migrated more efficiently under cisplatin
treatment than wild-type A549 cells. In addition, cisplatin-induced
apoptosis was significantly decreased in A549R cells when compared
with A549 cells. Taken together, A549R cells were less responsive
to cisplatin.
Marked improvements have been achieved in the past
few decades in our understanding of lung cancer biology (24,25).
The identification of driver oncogenes in lung cancers has led to a
change in cancer treatments. Some studies have provided greater
understanding regarding the mechanisms underlying chemotherapy
sensitivity/resistance and the associated biomarkers of lung cancer
(26-30). Deregulated mesenchymal-epithelial
transition (31) and reduced
apoptosis induction (32) have
been indicated to underlie the chemoresistance in lung cancer.
Autophagy is a self-protective mechanism to guarantee basic energy
supply under nutrition-deficient conditions, such as starvation
(33). The cytoprotective
mechanism of autophagy against chemotherapy has also been
recognized in lung cancer cells (33) and other types of cancers (9,15,34,35).
Thus, autophagy has been highlighted as one of the mechanisms
underlying the chemoresistance of NSCLC. In the present study,
autophagy induction by cisplatin was observed in A549 and A549R
cells, and could be inhibited by the autophagy inhibitor, 3MA.
Notably, a significantly higher level of autophagy was observed in
A549R cells when compared with A549 cells. This implies that
autophagy may contribute to the cisplatin-resistance phenotype of
A549R cells.
Proteomics has been widely utilized to profile,
screen and identify specific phenotype- or genotype-associated
biomarkers (36,37). In recent years, SILAC has stood out
when distinguishing the proteomics from one group to another, such
as in cancer biomarker discovery (11,13)
and in the identification of chemoresistance-associated biomarkers
(16). In the present study, two
rounds of SILAC procedures were performed with paired groups of
H-labeled A549R cells and L-labeled A549 cells, or with paired
groups of L-labeled A549R cells and H-labeled A549 cells. A total
of 1,161±152 quantitative peptides and 357±36 proteins were induced
by cisplatin (10 µM) between A549R and A549 cells. A total
of 344±21 proteins were confirmed by two or more peptides. In
addition, among the 23 proteins with 1.5-fold greater expression in
the H-labeled A549R cells and the 18 proteins with 1.5-fold greater
expression in the L-labeled A549R cells, there were 15 proteins
that were repeated in the 2 rounds of experiments. On the other
hand, there were 17 and 15 proteins that were downregulated in H-
and L-labeled A549R cells, respectively. Particularly, the
downregulation of apoptosis-associated proteins, including TR10B,
U17LO, SHB, PKN2 and MTCH1, was observed in the two types of
labeling experiments. These downregulated proteins may be involved
in mitochondrial dysfunction, the cell response to stress, nuclear
acid damage and finally in apoptosis induction. Exposure of any of
the two proapoptotic domains of MTCH1 on the surface of
mitochondria is sufficient for the induction of apoptosis in a
B-cell lymphoma-2 (Bcl-2)-associated X/Bcl-2
antagonist/killer-independent manner (38). SH2 domain-containing adapter
protein B (SHB) has been indicated to be involved in the Fyn
related Src family tyrosine kinase-SHB signaling pathway, and
regulates cell survival, differentiation and proliferation
(39). The possible roles of
U17LO, PKN2 and TR10B were not clear up until now.
GO analysis indicated that the majority of the
proteins regulated apoptosis (26,28,29,40-43),
DNA damage repairing (43-46) and other biological pathways. The
effect of anti-apoptosis and autophagy promotion was also
identified for these proteins in human lung cancer cells and other
types of cells. GRP78 antagonizes apoptosis and positively
regulates autophagy in human NSCLC cells via the adenosine
monophosphate-activated protein kinase-mammalian target of
rapamycin signaling pathway (28,29).
TCTP also inhibits apoptosis by binding to p53 in lung carcinoma
cells (26,47,48).
The anti-apoptosis effect of HSP71 was also recognized in
azacytidine-treated myeloma cells (49). Given the importance of
anti-apoptosis and autophagy in chemotherapy resistance in cancer,
the present study summarized the involvement of all of the 15
upregulated proteins in H- and L-labeled A549R cells in
anti-apoptosis and/or autophagy promotion (Table III). It was indicated that the
majority of these proteins were closely associated with
anti-apoptosis and/or autophagy promotion in lung cancers or in
other types of cancers. In addition, the majority of proteins that
directly regulate autophagy were upregulated by <1.5-fold,
though autophagy was significantly different in the two groups of
A549 cells, which may be due to the difference in sensitivity among
SILAC and other methods.
 | Table IIIInvolvement of upregulated proteins
in anti-apoptosis and autophagy promotion in human lung cancer and
other types of cells. |
Table III
Involvement of upregulated proteins
in anti-apoptosis and autophagy promotion in human lung cancer and
other types of cells.
| UniProt ID | Anti-apoptosis | Autophagy
promotion |
|---|
| Author, year | Cell type | Ref. | Author, year | Cell type | Ref. |
|---|
| GRP78 | Sun et al,
2012; Ahmad et al, 2014 | Lung cancer | (28,29) | Kim et al,
2012; Xie et al, 2016 | Lung cancer | (27,46) |
| PRP19 | Lu et al,
2007 | Other | (39) | – | NR | / |
| TCTP | Du et al,
2017; Rho et al, 2011 | Lung cancer | (26,43) | Chen et al,
2014; Bonhoure et al, 2017 | Other | (44,47) |
| HSP71 | Tian et al,
2013 | Other | (45) | – | NR | / |
| PTBP1 | Takai et al,
2017 | Other | (48) | Li et al,
2016; Takai et al, 2017 | Other | (33,48) |
| CATD | Wille et al,
2004; Li et al, 2004 | Lung cancer | (37,49) | Oliveira et
al, 2015; Hah et al, 2012 | Other | (50,51) |
| CYC | Chen et al,
2015; Moravcikova et al, 2014; Lee et al, 2015 | Lung cancer | (52-54) | Li et al,
2016; Kaminskyy et al, 2012 | Lung cancer | (38,55) |
| TXND5 | Lee et al,
2010 | Other | (34) | – | NR | / |
| MSH6 | Yu et al,
2017; Habiel et al, 2017 | Lung cancer | (30,56) | Knizhnik et
al, 2013; Zanotto-Filo et al, 2015 | Other | (40,57) |
| ANXA2 | Dassah et
al, 2014; Wang et al, 2017 | Lung cancer | (36,58) | Wang et al,
2017; Chen et al, 2017 | Lung cancer | (58,59) |
| BCCIP | Xu et al,
2017 | Lung cancer | (60) | – | NR | / |
| MSH2 | Zhang et al,
2016; Terry et al, 2015 | Lung cancer | (41,61) | Zeng et al,
2007; Zeng et al, 2007 | Other | (42,62) |
| PP2AB | Huang et al,
2004; Shtrichman et al, 2000 | Other | (63,64) | Banreti et
al, 2012; Ogura et al, 2010 | Other | (65,66) |
| GDIR1 | – | NR | / | – | NR | / |
| ANXA4 | Yao et al,
2016; Nagappan et al, 2016 | Lung cancer | (67,68) | – | NR | / |
However, the detailed signaling pathways underlying
such chemoresistance in A549R cells were not clear. In particular,
though proteins such as GRP78, HSP71, PRP19, PTBP1, TCTP, CATD,
CYC, TXND5, MSH6, ANXA2, BCCIP, MSH2, PP2AB, GDIR1 and ANXA4 were
significantly deregulated in A549R cells, the association of each
molecule with autophagy or directly with chemoresistance requires
further investigation.
In conclusion, the present study isolated a
cisplatin-resistant human lung cancer A549 cell clone, with reduced
apoptosis and high levels of autophagy, in response to cisplatin
treatment. SILAC proteomics recognized the high expression of GRP78
and other proteins that were associated with anti-apoptosis and/or
autophagy promotion in cisplatin-resistant A549R cells.
Funding
The present study was supported by grants from the
National Nature Science Foundation of China (grant no. 80151459),
the Development Project from Science and Technology Department of
Jilin Province (grant no. 140520020JH) and the Thirteen Five
Science and Technology Research Project of Jilin Province
Department of Education (grant no. 2016-467).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZW and GL designed the experiments. ZW, GL and JJ
performed the experiments. ZW conducted the statistical analysis.
GL wrote the manuscript. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Fidler MM, Soerjomataram I and Bray F: A
global view on cancer incidence and national levels of the human
development index. Int J Cancer. 139:2436–2446. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shen C, Wang X, Tian L, Zhou Y, Chen D, Du
H, Wang W, Liu L and Che G: 'Different trend' in multiple primary
lung cancer and intrapulmonary metastasis. Eur J Med Res.
20:172015. View Article : Google Scholar
|
|
3
|
Kalemkerian GP: Advances in
pharmacotherapy of small cell lung cancer. Expert Opin
Pharmacother. 15:2385–2396. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scagliotti GV, Parikh P, von Pawel J,
Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U,
Digumarti R, Zukin M, et al: Phase III study comparing cisplatin
plus gemcitabine with cisplatin plus pemetrexed in
chemotherapy-naive patients with advanced-stage non-small-cell lung
cancer. J Clin Oncol. 26:3543–3551. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Socinski MA, Smit EF, Lorigan P, Konduri
K, Reck M, Szczesna A, Blakely J, Serwatowski P, Karaseva NA,
Ciuleanu T, et al: Phase III study of pemetrexed plus carboplatin
compared with etoposide plus carboplatin in chemotherapy-naive
patients with extensive-stage small-cell lung cancer. J Clin Oncol.
27:4787–4792. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim ES: Chemotherapy resistance in lung
cancer. Adv Exp Med Biol. 893:189–209. 2016. View Article : Google Scholar
|
|
7
|
Willers H, Azzoli CG, Santivasi WL and Xia
F: Basic mechanisms of therapeutic resistance to radiation and
chemotherapy in lung cancer. Cancer J. 19:200–207. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zembutsu H, Ohnishi Y, Tsunoda T, Furukawa
Y, Katagiri T, Ueyama Y, Tamaoki N, Nomura T, Kitahara O, Yanagawa
R, et al: Genome-wide cDNA microarray screening to correlate gene
expression profiles with sensitivity of 85 human cancer xenografts
to anticancer drugs. Cancer Res. 62:518–527. 2002.PubMed/NCBI
|
|
9
|
Kihara C, Tsunoda T, Tanaka T, Yamana H,
Furukawa Y, Ono K, Kitahara O, Zembutsu H, Yanagawa R, Hirata K, et
al: Prediction of sensitivity of esophageal tumors to adjuvant
chemotherapy by cDNA microarray analysis of gene-expression
profiles. Cancer Res. 61:6474–6479. 2001.PubMed/NCBI
|
|
10
|
Beltran H, Yelensky R, Frampton GM, Park
K, Downing SR, MacDonald TY, Jarosz M, Lipson D, Tagawa ST, Nanus
DM, et al: Targeted next-generation sequencing of advanced prostate
cancer identifies potential therapeutic targets and disease
heterogeneity. Eur Urol. 63:920–926. 2013. View Article : Google Scholar :
|
|
11
|
Hoedt E, Zhang G and Neubert TA: Stable
isotope labeling by amino acids in cell culture (SILAC) for
quantitative proteomics. Adv Exp Med Biol. 806:93–106. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Patella F, Neilson LJ, Athineos D, Erami
Z, Anderson KI, Blyth K, Ryan KM and Zanivan S: In-depth proteomics
identifies a role for autophagy in controlling reactive oxygen
species mediated endothelial permeability. J Proteome Res.
15:2187–2197. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lanucara F and Eyers CE: Mass
spectrometric-based quantitative proteomics using SILAC. Methods
Enzymol. 500:133–150. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yeh CC, Hsu CH, Shao YY, Ho WC, Tsai MH,
Feng WC and Chow LP: Integrated stable isotope labeling by amino
acids in cell culture (SILAC) and isobaric tags for relative and
absolute quantitation (iTRAQ) quantitative proteomic analysis
identifies galectin-1 as a potential biomarker for predicting
dorafenib resistance in liver cancer. Mol Cell Proteomics.
14:1527–1545. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu X, Zahari MS, Renuse S, Nirujogi RS,
Kim MS, Manda SS, Stearns V, Gabrielson E, Sukumar S and Pandey A:
Phosphoproteomic analysisidentifies focal adhesion kinase2 (FAK2)
as a potential therapeutic target for tamoxifen resistance in
breast cancer. Mol Cell Proteomics. 14:2887–2900. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu H, Dephoure N, Sun H, Zhang H, Fan F,
Liu J, Ning X, Dai S, Liu B, Gao M, et al: Proteomic profiling of
paclitaxel treated cells identifies a novel mechanism of drug
resistance mediated by PDCD4. J Proteome Res. 14:2480–2491. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bosse K, Haneder S, Arlt C, Ihling CH,
Seufferlein T and Sinz A: Mass spectrometry-based secretome
analysis of non-small cell lung cancer cell lines. Proteomics.
16:2801–2814. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun P, Feng LX, Zhang DM, Liu M, Liu W, Mi
T, Wu WY, Jiang BH, Yang M, Hu LH, et al: Bufalin derivative BF211
inhibits proteasome activity in human lung cancer cells in vitro by
inhibiting β1 subunit expression and disrupting proteasome
assembly. Acta Pharmacol Sin. 37:908–918. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu G, Pei F, Yang F, Li L, Amin AD, Liu
S, Buchan JR and Cho WC: Role of autophagy and apoptosis in
non-small-cell lung cancer. Int J Mol Sci. 18:182017.
|
|
20
|
Lee JG, Shin JH, Shim HS, Lee CY, Kim DJ,
Kim YS and Chung KY: Autophagy contributes to the chemo-resistance
of non-small cell lung cancer in hypoxic conditions. Respir Res.
16:1382015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu M, Ma S, Liu M, Hou Y, Liang B, Su X
and Liu X: Synergistic killing of lung cancer cells by cisplatin
and radiation via autophagy and apoptosis. Oncol Lett. 7:1903–1910.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pisters KM, Evans WK, Azzoli CG, Kris MG,
Smith CA, Desch CE, Somerfield MR, Brouwers MC, Darling G, Ellis
PM, et al Cancer Care Ontario; American Society of Clinical
Oncology: Cancer Care Ontario and American Society of Clinical
Oncology adjuvant chemotherapy and adjuvant radiation therapy for
stages I-IIIA resectable non small-cell lung cancer guideline. J
Clin Oncol. 25:5506–5518. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Paz-Ares L, Mezger J, Ciuleanu TE, Fischer
JR, von Pawel J, Provencio M, Kazarnowicz A, Losonczy G, de Castro
G Jr, Szczesna A, et al INSPIRE investigators: Necitumumab plus
pemetrexed and cisplatin as first-line therapy in patients with
stage IV non-squamous non-small-cell lung cancer (INSPIRE): An
open-label, randomised, controlled phase 3 study. Lancet Oncol.
16:328–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lemjabbar-Alaoui H, Hassan OU, Yang YW and
Buchanan P: Lung cancer: Biology and treatment options. Biochim
Biophys Acta. 1856:189–210. 2015.PubMed/NCBI
|
|
25
|
Sánchez-Céspedes M: Lung cancer biology: A
genetic and genomic perspective. Clin Transl Oncol. 11:263–269.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Du J, Yang P, Kong F and Liu H: Aberrant
expression of translationally controlled tumor protein (TCTP) can
lead to radioactive susceptibility and chemosensitivity in lung
cancer cells. Oncotarget. 8:101922–101935. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim KM, Yu TK, Chu HH, Park HS, Jang KY,
Moon WS, Kang MJ, Lee DG, Kim MH, Lee JH, et al: Expression of ER
stress and autophagy-related molecules in human non-small cell lung
cancer and premalignant lesions. Int J Cancer. 131:E362–E370. 2012.
View Article : Google Scholar
|
|
28
|
Sun Q, Hua J, Wang Q, Xu W, Zhang J, Zhang
J, Kang J and Li M: Expressions of GRP78 and Bax associate with
differentiation, metastasis, and apoptosis in non-small cell lung
cancer. Mol Biol Rep. 39:6753–6761. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ahmad M, Hahn IF and Chatterjee S: GRP78
up-regulation leads to hypersensitization to cisplatin in A549 lung
cancer cells. Anticancer Res. 34:3493–3500. 2014.PubMed/NCBI
|
|
30
|
Yu W, Lu W, Chen G, Cheng F, Su H, Chen Y,
Liu M and Pang X: Inhibition of histone deacetylases sensitizes EGF
receptor-TK inhibitor-resistant non-small-cell lung cancer cells to
erlotinib in vitro and in vivo. Br J Pharmacol. 174:3608–3622.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mortimore GE, Miotto G, Venerando R and
Kadowaki M: Autophagy. Subcell Biochem. 27:93–135. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Notte A, Ninane N, Arnould T and Michiels
C: Hypoxia counteracts taxol-induced apoptosis in MDA-MB-231 breast
cancer cells: Role of autophagy and JNK activation. Cell Death Dis.
4:e6382013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li C, Zhao Z, Zhou Z and Liu R: Linc-ROR
confers gemcitabine resistance to pancreatic cancer cells via
inducing autophagy and modulating the miR-124/PTBP1/PKM2 axis.
Cancer Chemother Pharmacol. 78:1199–1207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee WL, Wen TN, Shiau JY and Shyur LF:
Differential proteomic profiling identifies novel molecular targets
of paclitaxel and phytoagent deoxyelephantopin against mammary
adenocar-cinoma cells. J Proteome Res. 9:237–253. 2010. View Article : Google Scholar
|
|
35
|
Liang S, Xu Z, Xu X, Zhao X, Huang C and
Wei Y: Quantitative proteomics for cancer biomarker discovery. Comb
Chem High Throughput Screen. 15:221–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dassah M, Almeida D, Hahn R, Bonaldo P,
Worgall S and Hajjar KA: Annexin A2 mediates secretion of collagen
VI, pulmonary elasticity and apoptosis of bronchial epithelial
cells. J Cell Sci. 127:828–844. 2014. View Article : Google Scholar :
|
|
37
|
Wille A, Gerber A, Heimburg A, Reisenauer
A, Peters C, Saftig P, Reinheckel T, Welte T and Bühling F:
Cathepsin L is involved in cathepsin D processing and regulation of
apoptosis in A549 human lung epithelial cells. Biol Chem.
385:665–670. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li YR, Li S, Ho CT, Chang YH, Tan KT,
Chung TW, Wang BY, Chen YK and Lin CC: Tangeretin derivative,
5-acetyloxy-6,7,8,4'-tetramethoxyflavone induces G2/M arrest,
apoptosis and autophagy in human non-small cell lung cancer cells
in vitro and in vivo. Cancer Biol Ther. 17:48–64. 2016. View Article : Google Scholar
|
|
39
|
Lu X and Legerski RJ: The Prp19/Pso4 core
complex undergoes ubiquitylation and structural alterations in
response to DNA damage. Biochem Biophys Res Commun. 354:968–974.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Knizhnik AV, Roos WP, Nikolova T, Quiros
S, Tomaszowski KH, Christmann M and Kaina B: Survival and death
strategies in glioma cells: Autophagy, senescence and apoptosis
triggered by a single type of temozolomide-induced DNA damage. PLoS
One. 8:e556652013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang M, Hu C, Tong D, Xiang S, Williams
K, Bai W, Li GM, Bepler G and Zhang X: Ubiquitin-specific peptidase
10 (USP10) deubiquitinates and stabilizes MutS homolog 2 (MSH2) to
regulate cellular sensitivity to DNA damage. J Biol Chem.
291:10783–10791. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zeng X and Kinsella TJ: A novel role for
DNA mismatch repair and the autophagic processing of chemotherapy
drugs in human tumor cells. Autophagy. 3:368–370. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rho SB, Lee JH, Park MS, Byun HJ, Kang S,
Seo SS, Kim JY and Park SY: Anti-apoptotic protein TCTP controls
the stability of the tumor suppressor p53. FEBS Lett. 585:29–35.
2011. View Article : Google Scholar
|
|
44
|
Chen K, Huang C, Yuan J, Cheng H and Zhou
R: Long-term artificial selection reveals a role of TCTP in
autophagy in mammalian cells. Mol Biol Evol. 31:2194–2211. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tian E, Tang H, Xu R, Liu C, Deng H and
Wang Q: Azacytidine induces necrosis of multiple myeloma cells
through oxidative stress. Proteome Sci. 11:242013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xie WY, Zhou XD, Yang J, Chen LX and Ran
DH: Inhibition of autophagy enhances heat-induced apoptosis in
human non-small cell lung cancer cells through ER stress pathways.
Arch Biochem Biophys. 607:55–66. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bonhoure A, Vallentin A, Martin M,
Senff-Ribeiro A, Amson R, Telerman A and Vidal M: Acetylation of
translationally controlled tumor protein promotes its degradation
through chaperone-mediated autophagy. Eur J Cell Biol. 96:83–98.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Takai T, Yoshikawa Y, Inamoto T, Minami K,
Taniguchi K, Sugito N, Kuranaga Y, Shinohara H, Kumazaki M, Tsujino
T, et al: A Novel combination RNAi toward Warburg effect by
replacement with miR-145 and silencing of PTBP1 induces apoptotic
cell death in bladder cancer cells. Int J Mol Sci. 18:182017.
View Article : Google Scholar
|
|
49
|
Li X, Rayford H, Shu R, Zhuang J and Uhal
BD: Essential role for cathepsin D in bleomycin-induced apoptosis
of alveolar epithelial cells. Am J Physiol Lung Cell Mol Physiol.
287:L46–L51. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Oliveira CS, Pereira H, Alves S, Castro L,
Baltazar F, Chaves SR, Preto A and Côrte-Real M: Cathepsin D
protects colorectal cancer cells from acetate-induced apoptosis
through autophagy-independent degradation of damaged mitochondria.
Cell Death Dis. 6:e17882015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hah YS, Noh HS, Ha JH, Ahn JS, Hahm JR,
Cho HY and Kim DR: Cathepsin D inhibits oxidative stress-induced
cell death via activation of autophagy in cancer cells. Cancer
Lett. 323:208–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen H, Liang ZW, Wang ZH, Zhang JP, Hu B,
Xing XB and Cai WB: Akt activation and inhibition of cytochrome C
release: mechanistic insights into leptin-promoted survival of type
II Alveolar Epithelial Cells. J Cell Biochem. 116:2313–2324. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Moravcikova E, Krepela E, Prochazka J,
Benkova K and Pauk N: Differential sensitivity to apoptosome
apparatus activation in non-small cell lung carcinoma and the lung.
Int J Oncol. 44:1443–1454. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lee J, Yeganeh B, Ermini L and Post M:
Sphingolipids as cell fate regulators in lung development and
disease. Apoptosis. 20:740–757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kaminskyy VO, Piskunova T, Zborovskaya IB,
Tchevkina EM and Zhivotovsky B: Suppression of basal autophagy
reduces lung cancer cell proliferation and enhances
caspase-dependent and -independent apoptosis by stimulating ROS
formation. Autophagy. 8:1032–1044. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Habiel DM, Camelo A, Espindola M, Burwell
T, Hanna R, Miranda E, Carruthers A, Bell M, Coelho AL, Liu H, et
al: Divergent roles for clusterin in lung injury and repair. Sci
Rep. 7:154442017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zanotto-Filho A, Braganhol E, Klafke K,
Figueiró F, Terra SR, Paludo FJ, Morrone M, Bristot IJ, Battastini
AM, Forcelini CM, et al: Autophagy inhibition improves the efficacy
of curcumin/temozolomide combination therapy in glioblastomas.
Cancer Lett. 358:220–231. 2015. View Article : Google Scholar
|
|
58
|
Wang K, Zhang T, Lei Y, Li X, Jiang J, Lan
J, Liu Y, Chen H, Gao W, Xie N, et al: Identification of ANXA2
(annexin A2) as a specific bleomycin target to induce pulmonary
fibrosis by impeding TFEB-mediated autophagic flux. Autophagy.
14:269–282. 2018. View Article : Google Scholar :
|
|
59
|
Chen YD, Fang YT, Cheng YL, Lin CF, Hsu
LJ, Wang SY, Anderson R, Chang CP and Lin YS: Exophagy of annexin
A2 via RAB11, RAB8A and RAB27A in IFN-γ-stimulated lung epithelial
cells. Sci Rep. 7:56762017. View Article : Google Scholar
|
|
60
|
Xu XT, Hu WT, Zhou JY and Tu Y: Celecoxib
enhances the radiosensitivity of HCT116 cells in a COX-2
independent manner by up-regulating BCCIP. Am J Transl Res.
9:1088–1100. 2017.PubMed/NCBI
|
|
61
|
Terry MR, Arya R, Mukhopadhyay A, Berrett
KC, Clair PM, Witt B, Salama ME, Bhutkar A and Oliver TG: Caspase-2
impacts lung tumorigenesis and chemotherapy response in vivo. Cell
Death Differ. 22:719–730. 2015. View Article : Google Scholar :
|
|
62
|
Zeng X, Yan T, Schupp JE, Seo Y and
Kinsella TJ: DNA mismatch repair initiates 6-thioguanine - induced
autophagy through p53 activation in human tumor cells. Clin Cancer
Res. 13:1315–1321. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Huang S, Shu L, Easton J, Harwood FC,
Germain GS, Ichijo H and Houghton PJ: Inhibition of mammalian
target of rapamycin activates apoptosis signal-regulating kinase 1
signaling by suppressing protein phosphatase 5 activity. J Biol
Chem. 279:36490–36496. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shtrichman R, Sharf R and Kleinberger T:
Adenovirus E4orf4 protein interacts with both Balpha and B'
subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is
mediated only by the interaction with Balpha. Oncogene.
19:3757–3765. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bánréti Á, Lukácsovich T, Csikós G,
Erdélyi M and Sass M: PP2A regulates autophagy in two alternative
ways in Drosophila. Autophagy. 8:623–636. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ogura K, Okada T, Mitani S, Gengyo-Ando K,
Baillie DL, Kohara Y and Goshima Y: Protein phosphatase 2A
cooperates with the autophagy-related kinase UNC-51 to regulate
axon guidance in Caenorhabditis elegans. Development.
137:1657–1667. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yao H, Sun C, Hu Z and Wang W: The role of
Annexin A4 in cancer. Front Biosci. 21:949–957. 2016. View Article : Google Scholar
|
|
68
|
Nagappan A, Venkatarame Gowda Saralamma V,
Hong GE, Lee HJ, Shin SC, Kim EH, Lee WS and Kim GS: Proteomic
analysis of selective cytotoxic anticancer properties of flavonoids
isolated from Citrus platymamma on A549 human lung cancer cells.
Mol Med Rep. 14:3814–3822. 2016. View Article : Google Scholar : PubMed/NCBI
|