Introduction
Breast cancer is classified into several intrinsic
subtypes based on gene expression profiles (1). Different subtypes, including luminal
A, luminal B, human epidermal growth factor receptor 2 (HER2) and
basal-like breast cancer, have been indicated to express different
biological characteristics (2).
Pathological examination of breast cancer samples is used to detect
the expression of estrogen receptor (ER), progesterone receptor
(PgR), HER2 and Ki67, in order to select suitable therapeutic
strategies in the clinic (3).
Triple-negative breast cancer (TNBC), which is
character-ized by the absence of ER, PgR, and HER2, exhibits a
relatively aggressive phenotype, therapeutic resistance and is
associated with a poor prognosis (4). TNBC is associated with phenotypes of
cancer stem cells (CSCs) (5,6) and
epithelial-mesenchymal transition (EMT), which is characterized by
the downregulation of epithelial markers and mesenchymal phenotypes
with high migration ability (7).
Carboxypeptidase A4 (CPA4) catalyzes the
release of carboxy-terminal amino acids (8), and its overexpression has been
associated with cancer progression in several types of cancer
(9-14). Furthermore, CPA4 has been indicated
to be secreted in higher amounts from breast cancer cells compared
with noncancerous mammary epithelial cells (15). However, few studies have addressed
the association between CPA4 expression and clinicopathological
factors in patients with breast cancer, including TNBC.
The purpose of the present study was to clarify the
significance of CPA4 expression and function in breast cancer. To
this end, immunohistochemical analysis was performed to evaluate
the association between CPA4 expression and clinicopathological
markers, including ER, PgR, HER2, CD44, CD24, aldehyde
dehydrogenase 1 (ALDH1), E-cadherin, EGFR, cytokeratin 5/6 (CK5/6),
claudins, vimentin and androgen receptor (AR) in 221 breast cancer
cases. In addition, the in vitro effects of small
interfering RNA (siRNA)-mediated CPA4 knockdown on the viability
and migration ability of human TNBC cell lines was examined.
Materials and methods
Cell lines
In the present study, cell lines representative of
cancer types were selected as described previously (16): MCF7 and T47D for luminal A type;
BT474 for luminal B type; and MDA-MB468, MDA-MB231, HCC70 and
HCC1143 for TNBC. The human breast cancer cell lines were obtained
from the Riken Cell Bank (MCF7; Riken BioResource Research Center,
Tsukuba, Japan) and from the American Type Culture Collection
(T47D, BT474, MDA-MB468, MDA-MB231, HCC70 and HCC1143; Manassas,
VA, USA). Cells were cultured in RPMI-1640 medium (Wako Pure
Chemical Industries, Ltd., Osaka, Japan) supplemented with 100 U/ml
of penicillin, 100 U/ml of streptomycin and 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a
humidified atmosphere containing 5% CO2 at 37°C.
Data mining for new molecular target
therapy candidates in TNBC
Four genes were identified as candidates for
molecular target therapy in TNBC using whole transcriptome analysis
and a lentiviral shRNA-screening library. For transcriptome
analysis, total RNA was prepared from cell lines (MCF7, BT474,
T47D, MDA-MB468, HCC70 and HCC1143) using NucleoSpin RNA (Takara
Bio, Inc., Shiga, Japan). The quality of the RNA was assessed using
the RNA integrity number (RIN) obtained by the Agilent RNA6000 Pico
Kit and the Agilent 2100 Bioanalyzer (both Agilent Technologies,
Santa Clara, CA, USA). Samples used for transcriptome analysis had,
on average, an RIN value of 9.4 and a minimum RIN value of 8.9. The
library was prepared using the TruSeq RNA Sample Prep Kit v2
(Illumina, Inc., San Diego, CA, USA) from 1 µg of total RNA
according to the manufacturer's protocol. The resulting libraries
were subjected to single-end sequencing of 76-bp reads using the
NextSeq 500 High Output v1 Kit on the Illumina NextSeq 500 system
(both from Illumina, Inc.). Data processing and analyses were
performed using the TopHat version 2 alignment, cufflinks assembly
and differential expression apps (all from Illumina, Inc.).
Briefly, the reads were filtered, trimmed and aligned in the UCSC
reference human genome 19 (hg19) using the Tophat2 (v2.0.7) and
Bowtie1 (0.12.9) pipelines. The transcripts were assembled using
Cufflinks 2.1.1, and differentially expressed transcripts were
detected using Cuffdiff 2.1.1. Genes with a false discovery
rate-adjusted q-value of <0.05 and log2-fold change
(TNBC/non-TNBC) of >5 were defined as significantly upregulated
genes in TNBC cells.
MISSION LentiPlex Human Pooled shRNA Library
(SHPH01-1SET; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) is a
genome-wide shRNA library that covers ~15,000 genes, with 80,000
shRNA clones. In order to identify shRNAs that targeted genes able
to specifically destroy TNBC cell lines compared with non-TNBC cell
lines, a mixture of lentiviral particles (SHPH01-1SET;
Sigma-Aldrich; Merck KGaA) was introduced to the breast cancer cell
lines of non-TNBC (MCF7, T47D and BT474) and TNBC (MDA-MB468, HCC70
and HCC1143) subtypes at multiplicity of infections that yielded
30-50% infected cells according to the manufacturer's protocol. The
infected cells were subsequently selected using puromycin for 7
days, and genomic DNA with integrated shRNA was isolated from the
cell lines. Thirty cycles of polymerase chain reaction (PCR) was
performed using KAPA HiFi HotStart ReadyMix (2X) (KAPA Biosystems;
Roche Diagnostics, Indianapolis, IN, USA). The following primers
were used for the PCR of the shRNA vector:
5′-ATTTCTTGGCTTTATATATCTTGTGGAAAG-3′ (sense) and reverse primer,
5′-TGTGGATGAATACTGCCATTTGT CTC-3′ (antisense). The PCR were
performed on 3-µg genomic DNAs. PCR conditions consisted of
initial denaturation at 95°C for 3 min, followed by 30 cycles of
denaturation at 98°C for 20 sec, annealing at 60°C for 15 sec and
elongation at 72°C for 30 sec. Sequencing adaptors (BIOO
Scientific, Austin, TX, USA) were ligated to the PCR amplicons with
8 cycles of PCR and Illumina sequencing was performed for 150
cycles on an Illumina NextSeq500 sequencer with 10% PhiX control
DNA (Illumina, Inc.). Data processing was performed in accordance
with the manufacturer's protocol. Briefly, FASTQ files were used to
trim and align the adapter sequences to the reference shRNA
sequences using Bowtie2. The Bowtie output files were converted to
count files through a python script 'countBowtieHits.py' and
'RTable.py', which were obtained from the manufacturer's protocol.
TCC-edgeR was used for normalization and statistical analysis as
described previously (17).
The Reference Expression dataset (RefEx: http://refex.dbcls.jp) was used to examine the
expression levels of CPA4 in several noncancerous tissues using RNA
sequencing methods (http://refex.dbcls.jp). Specific low-expression genes
in 40 normal organs were picked up as maximum fragments per
kilobase of transcript per million mapped reads of <1.0. The
present transcriptome data were submitted to a public repository,
the Gene Expression Omnibus (accession no. GSE113653).
The Cancer Genome Atlas database (cBioPortal Breast
Cancer: METABRIC, Nature 2012 and Nat Commun 2016: http://www.cbioportal.org) was used to validate the
prognostic significance of CPA4 in patients with breast cancer.
Patients and samples
Tumor specimens from 221 patients with primary
breast cancer who underwent surgery for excision of a primary tumor
between January 1999 and October 2010 at the Gunma University
Hospital (Maebashi, Japan) were retrospectively analyzed. The
median follow-up period for survivors was 118 months. Some data
from patients with ductal carcinoma in situ, preoperative
chemotherapy, preoperative hormone therapy and male breast
carcinoma from the present tissue microarray preparations were
excluded. Tumor staging was based on the Union for International
Cancer Control TNM classification (seventh edition) (18). Nuclear grades (NGs) were defined as
the sum of scores for the nuclear atypia, as described previously
(19). The present research
conformed to the tenets of the Declaration of Helsinki and to the
guidelines of the Gunma University Ethical Review Board for Medical
Research Involving Human Subjects (approval no. 1297).
Immunohistochemistry (IHC)
For tissue microarray, clinical formalin-fixed
paraffin-embedded samples were stored in the archives of the
Clinical Department of Pathology, Gunma University Hospital
(Maebashi, Japan). For each patient, one paraffin block containing
representative non-necrotic tumor areas was selected. Breast cancer
tissue cores (2.0-mm diameter per tumor) were punched out from the
representative areas near the invasive front and transferred into
the paired recipient paraffin block using a tissue array instrument
(Beecher Instruments, Inc., Silver Spring, MD, USA).
For IHC, a 4-µm section was cut from the
sample paraffin blocks. Each section was mounted on a silane-coated
glass slide, deparaffinized and soaked for 30 min at room
temperature in 0.3% H2O2/methanol to block
endogenous peroxidases. The sections were subsequently heated in
boiled 10 mM citrate buffer (pH 6.0) at 98°C for 30 min.
Non-specific binding sites were blocked by incubating with 0.25%
Casein/1% bovine serum albumin (Code 81-003-3; EMD Millipore,
Kankakee, IL, USA) for 30 min at room temperature. A rabbit
polyclonal anti-CPA4 antibody (HPA021030; Sigma-Aldrich; Merck
KGaA) was applied at a dilution of 1:400 for 24 h at 4°C. A MAX-PO
secondary antibody from the Histofine Simple Stain MAX-PO (Multi)
Kit (Nichirei Corporation, Tokyo, Japan) was used for 30 min at
room temperature according to the manufacturer's instructions. The
chromogen 3,3-diaminobenzidine tetrahydrochloride (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) was applied as a
0.02% solution containing 0.005% H2O2 in 50 mM Tris-HCl buffer (pH
7.6). The sections were lightly counterstained with Mayer's
hematoxylin and mounted. Negative controls were established by
omitting the primary antibody. Other IHC was performed using the
following primary antibodies: Anti-ER (ready to use; SP1), anti-PgR
(ready to use; 1E2), anti-HER2 (ready to use; 4B5), anti-Ki67
(ready to use; 30-9) (Ventana Medical Systems, Inc., Tucson, AZ,
USA), anti-EGFR (ready to use; 31G7; Nichirei Corporation),
anti-CK5/6 (1:50; 5/16 B4; Dako; Agilent Technologies, Inc., Santa
Clara, CA, USA), anti-E-cadherin (ready to use; 36; Ventana Medical
Systems, Inc.) anti-ALDH1 (1:400; 46/ALDH; BD Biosciences, San
Jose, CA, USA), anti-CD44 (1:50; DF1485; Dako; Agilent
Technologies, Inc.), anti-CD24 (1:40; SN3b; Thermo Fisher
Scientific, Inc.), AR (1:200; AR441; Thermo Fisher Scientific,
Inc.), Claudin1 (1:50; 1449527A; Invitrogen; Thermo Fisher
Scientific, Inc.), Claudin3 (1:100; GR253635-1; Abcam, Cambridge,
UK), Claudin4 (1:50; 3E2C1; Invitrogen; Thermo Fisher Scientific,
Inc.), Claudin7 (1:50; GR212894-5; Abcam) and Vimentin (ready to
use; V9; Dako; Agilent Technologies, Inc.). A Ventana BenchMark XT
system (Roche Diagnostics, Basel, Switzerland) was used for ER,
PgR, HER2, Ki67 and E-cadherin. All sections were examined under a
BX43 light microscope (Olympus Corporation, Tokyo, Japan).
Immunohistochemical evaluation and
intrinsic subtype
CPA4 expression was deemed positive in cells with
stained cytoplasms. In addition, the cutoff value for CPA4
positivity was 20% (14). The
cutoff value for ER and PgR positivity was 1%. HER2 expression was
scored according to the American Society of Clinical
Oncology/College of American Pathologists guideline (20). Ki67 labeling index was used to
calculate the percentage of cells with high nuclear expression in
~1,000 cells per sample, as described previously (21). EGFR expression was scored in the
same manner as HER2 expression; scores of 0 and 1+ were considered
as negative, and 2+ and 3+ as positive. A cutoff value of 10% for
CK5/6, ALDH1, CD44, CD24, claudin and AR was used (19,22).
The cutoff value for high E-cadherin expression was set as >70%
(23).
Based on the IHC results, the breast cancer subtypes
were defined as follows: Luminal A-like (ER or PgR+ and HER2
0/1+/2+), luminal B-like (ER or PgR+ and HER2 3+), HER2-like (ER-,
PgR- and HER2 3+) and TNBC-like (ER-, PgR- and HER2 0/1+/2+).
Fluorescent IHC
The sections were prepared, and endogenous
peroxidase was blocked as described above. Antigen retrieval was
performed by heating samples in boiled Immunosaver Antigen
Activator (Nisshin EM Co., Ltd., Tokyo, Japan) at 98°C for 45 min
and stripping was performed by heating in boiled 10 mM citrate
buffer (pH 6.0) at 98°C 15 min in a microwave. Nonspecific binding
sites were blocked as described above (19). The sections were incubated with
primary antibodies CPA4 (1:400) and E-cadherin (1:500) overnight at
4°C or for 1 h at room temperature, respectively. The secondary
antibody was used as described for the protocol for IHC. Multiplex
covalent labeling was performed (CPA4; Fluorescein, E-cadherin;
Cyanine 3) with tyramide signal amplification (Opal 3-Plex Kit;
PerkinElmer, Inc., Waltham, MA, USA) according to the
manufacturer's protocol. All sections were counterstained with
4′,6-diamidino-2-phenylin-dole at room temperature (Opal 3-Plex
Kit) and examined under an All-in-One BZ-X710 fluorescence
microscope (Keyence Corporation, Osaka, Japan).
CPA4 small interfering RNA (siRNA)
transfection
CPA4-specific siRNAs #1
(5′-CCAAAGAACAUCUGAGAUGtt-3′) and siRNA #2
(5′-CAGCAAAUCUUGUAGGGAUtt-3′) and negative-control siRNA were
purchased from GeneDesign, Inc. (Osaka, Japan) and Bonac
Corporation (Fukuoka, Japan). MDA-MB231 and MDA-MB468 at a density
of 1×106 cells/well were seeded in 100 µl of Opti
MEM Reduced Serum Medium (Invitrogen; Thermo Fisher Scientific,
Inc.). In total, 20 nM of CPA4-specific siRNAs 1 and 2, and
scrambled siRNA (as a negative control) were used to treat cells;
siRNAs was transfected using an electroporator CUY-21 EDIT II (Bex
Co., Ltd., Tokyo, Japan) according to the manufacturer′s
instructions. The subsequent experiments were performed following
24 h of transfection.
Western blot analysis
The proteins were extracted using lysis buffer [10
mM Tris-HCl (pH 7.5), 1 mM ethylene-diaminetetraacetic acid, 10%
glycerol, 0.5% NP-40 detergent, 400 mM NaCl, 4 µg/ml of
aprotinin, phenylmethylsulfonyl fluoride and dithiothreitol]. The
BCA protein assay was used for the protein determination. Total
protein (10 µg loaded per lane) was separated using SDS-PAGE
(on a 10% polyacrylamide gel), at 300 mA for 90 min and transferred
on a nitrocellulose membrane (Invitrogen; Thermo Fisher Scientific,
Inc.). Blocking of the membrane was performed using 4% skimmed milk
for 60 min at room temperature. The protein expression levels of
CPA4, E-cadherin and β-actin were assessed using western blot
analysis. These proteins were detected using specific antibodies to
CPA4 (1:200; HPA021030; Sigma-Aldrich; Merck KGaA), E-cadherin
(1:1,000; M106; Takara Bio, Inc.), and β-actin (1:1,000; #3700;
Cell Signaling Technology, Inc.). The membrane was incubated in
primary antibodies for overnight at 4°C. β-actin served as a
loading control. ECL anti-mouse or anti-rabbit IgG,
peroxidase-linked whole antibody was used for secondary antibody
(GE Healthcare Life Sciences, Little Chalfont, UK). The signals
were detected using the ECL Western Blotting Detection System and
an Image Quant LAS 4000 machine (GE Healthcare Life Sciences).
Proliferation assay
Cell proliferation analyses were performed using
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan). Breast cancer cell line cells were plated onto
96-well plates in 100 µl of RPMI-1640 medium containing 10%
FBS, at ~3,000 cells per well following siRNA transfection.
MDA-MB231 and MDA-MB468 cells were evaluated at 48 and 72 h
post-CPA4 siRNA treatment, respectively. For quantifying cell
proliferation, 10 µl of CCK-8 solution was added to each
well and incubated at 37°C for 2 h. Following this, the absorbance
of each well at 450 nm was detected using a plate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Migration assay
Cell migration was examined using MDA-MB231 and
MDA-MB468 cells transfected with a negative control or CPA4 siRNAs.
The transfected breast cancer cells were grown in 6-well plates
until 100% confluence, and a uniform straight wound was produced in
the monolayer in each well using a pipette tip. Wells were washed
with phosphate-buffered saline to remove all cell debris Cells were
cultured in an atmosphere containing 5% CO2 at 37°C.
Closure or filling of the wound was evaluated at 3 and 24 h in
MDA-MB231 and MDA-MB468 plates, respectively.
Statistical analysis
Statistical analyses were performed using the
Wilcoxon test for continuous variables, and the χ2 test
and one-way analysis of variance for categorical variables.
Survival curves were generated according to the Kaplan-Meier
method. The differences between survival curves were examined using
the log-rank test. Cox proportional hazard regression was used to
examine the independent prognostic contribution of standard
prognostic variables, including CPA4 expression, age, tumor factor,
NG, lymph node metastasis (LNM), venous invasion and adjuvant
therapy. P<0.05 was considered to indicate a statistically
significant difference. JMP software version 12 (SAS Institute,
Cary, NC, USA) was used to analyze all statistical analyses.
Results
Identification of CPA4 as a novel
therapeutic target according to the shRNA library and transcriptome
analysis in breast cancer cells
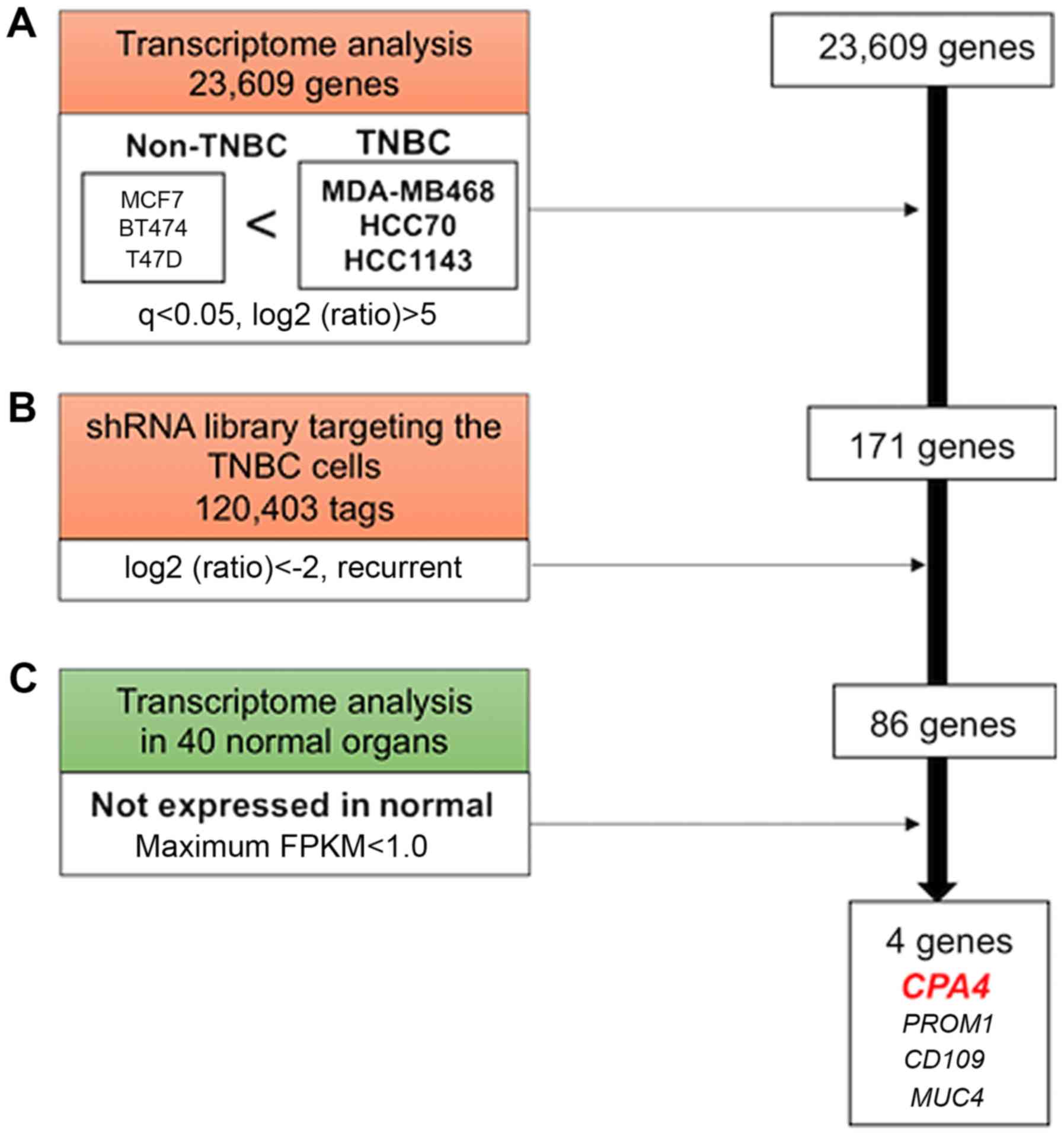
Among 23,609 genes, 171 that were highly expressed
in TNBC compared with their expression in non-TNBC cell lines were
selected for transcriptome analysis (Fig. 1A). Following this, 86 of the 171
genes, in which suppression was indicated to have inhibited the
viability of TNBC cell lines according to the shRNA library, were
selected (Fig. 1B). Finally, 4
genes that were not expressed in normal organs according to a
public database were selected (Fig.
1C). It was deduced that these 4 candidates were associated
with TNBC viability because they were highly expressed in the TNBC
cells compared with non-TNBC cells and normal organs. In the
present study, CPA4 was investigated in order to discover a
novel molecular target against TNBC.
Immunohistochemical analysis of CPA4
expression in breast cancer
Cytoplasmic expression of CPA4 in normal breast
tissues was decreased compared with breast cancer tissues (Fig. 2). Out of the 221 breast cancer
samples, 153 specimens (69.2%) were assigned to the low CPA4
expression group (Fig. 2B) and 68
specimens (30.8%) were assigned to the high CPA4 expression group
(Fig. 2C and D).
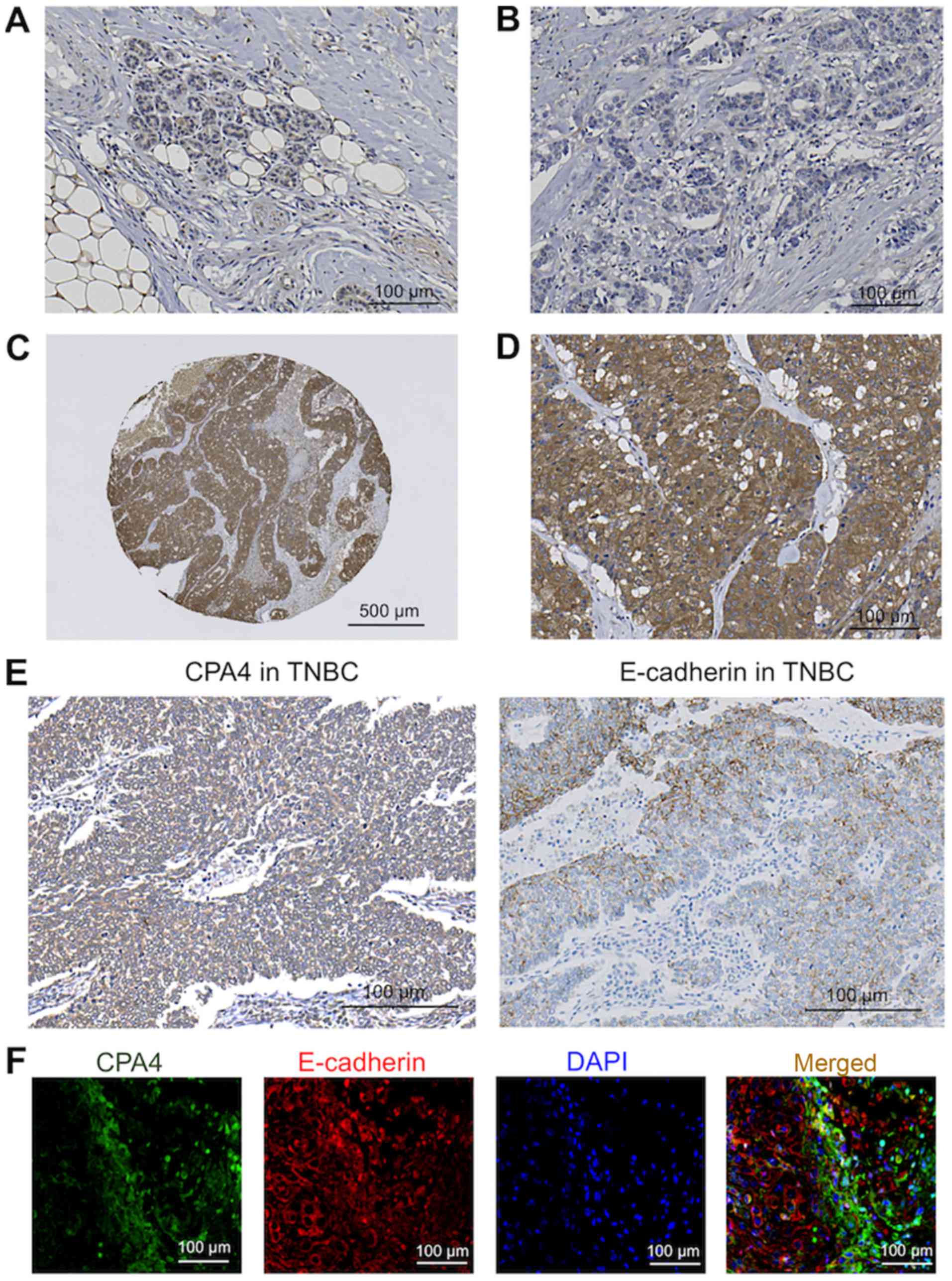 | Figure 2Immunohistochemical CPA4 and
E-cadherin analyses in representative breast tissue samples. (A)
Low CPA4 expression in normal breast tissue. Scale bar, 100
µm. (B) Low CPA4 expression in breast cancer tissue. Scale
bar, 100 µm. (C) High CPA4 expression in breast cancer
tissue (low-power field). Scale bar, 500 µm. (D) High CPA4
expression in breast cancer tissue (high-power field). Scale bar,
100 µm. (E) High CPA4 expression (left panel) and low
E-cadherin expression (right panel) in serial TNBC sections using
immunohistochemical analysis. Scale bar, 100 µm. (F)
Fluorescent immunohistochemical analyses of CPA4 and E-cadherin
expression in the representative TNBC tissue. TNBC tissues were
immunostained with antibodies against CPA4 (green) and E-cadherin
(red). This section was counterstained with DAPI (blue). Scale bar,
100 µm. TNBC, triple-negative breast cancer; CPA4,
carboxypeptidase A4; DAPI, 4′,6-diamidino-2-phenylindole. |
Association between CPA4 expression and
breast cancer clinicopathological features
The association between CPA4 expression and
clinicopathological characteristics of the patients were indicated
(Table I). Notably, no association
was identified between CPA4 expression and clinicopathological
characteristics in the 221 breast cancer cases. Furthermore, the
association between CPA4 expression levels with breast CSCs markers
(ALDH1, CD44 and CD24), EMT markers (vimentin and E-cadherin) and
the AR was examined using immunohistochemical staining. An
association between high CPA4 expression and high ALDH1 expression
was noted (Table II, P=0.027).
However, only a few cases exhibited high ALDH1 expression.
 | Table IPatient characteristics and CPA4
expression in 221 breast cancer cases. |
Table I
Patient characteristics and CPA4
expression in 221 breast cancer cases.
| Clinical
factors | Total breast cancer
cohort (n=221)
| P-value |
|---|
Low CPA4
(n=153) | High CPA4
(n=68) |
|---|
| Age (years) | 55.0±12.49 | 56.7±12.52 | 0.371 |
| Subtype | | | |
| Luminal A | 89 | 37 | 0.435 |
| Luminal B | 4 | 0 | |
| HER2 | 14 | 9 | |
| TNBC | 46 | 22 | |
| ER | | | |
| Negative | 62 | 31 | 0.482 |
| Positive | 91 | 37 | |
| PgR | | | |
| Negative | 80 | 44 | 0.086 |
| Positive | 73 | 24 | |
| HER2 | | | |
| Score 0, 1+,
2+ | 135 | 59 | 0.758 |
| Score 3+ | 18 | 9 | |
| EGFR | | | |
| Score 0, 1+ | 139 | 64 | 0.412 |
| Score 2+, 3+ | 14 | 4 | |
| CK5/6 | | | |
| Negative | 145 | 66 | 0.45 |
| Positive | 8 | 2 | |
| Ki67 labeling
index | 21.2±24.22 | 13.8±15.50 | 0.068 |
| Tumor size | | | |
| <2 cm | 63 | 29 | 0.838 |
| ≥2 cm | 90 | 39 | |
| Tumor stage | | | |
| 1 | 70 | 35 | 0.881 |
| 2 | 74 | 29 | |
| 3 | 7 | 3 | |
| 4 | 2 | 1 | |
| Lymph node
metastasis | | | |
| Negative | 89 | 48 | 0.079 |
| Positive | 64 | 20 | |
| Metastasis | | | |
| Negative | 152 | 68 | 0.504 |
| Positive | 1 | 0 | |
| Stage (UICC, 7th
Ed) | | | |
| I | 49 | 28 | 0.353 |
| II | 73 | 32 | |
| III | 30 | 8 | |
| IV | 1 | 0 | |
| Lymphatic
invasion | | | |
| Negative | 46 | 25 | 0.325 |
| Positive | 107 | 43 | |
| Venous
invasion | | | |
| Negative | 103 | 51 | 0.252 |
| Positive | 50 | 17 | |
| NG | | | |
| NG1 | 33 | 12 | 0.428 |
| NG2 | 39 | 23 | |
| NG3 | 81 | 33 | |
 | Table IICPA4 expression in stem cell markers,
epithelial-mesenchymal transition markers and AR of 221 breast
cancer cases. |
Table II
CPA4 expression in stem cell markers,
epithelial-mesenchymal transition markers and AR of 221 breast
cancer cases.
| Clinical
factors | Total breast cancer
cohort (n=221)
| P-value |
|---|
Low CPA4
(n=153) | High CPA4
(n=68) |
|---|
| ALDH1 | | | |
| Low | 147 | 60 | 0.027a |
| High | 6 | 8 | |
| CD44/CD24 | | | |
| High/low | 83 | 43 | 0.213 |
| Others | 70 | 25 | |
| Claudin1 | | | |
| Negative | 107 | 40 | 0.106 |
| Positive | 46 | 28 | |
| Claudin3 | | | |
| Negative | 112 | 46 | 0.206 |
| Positive | 37 | 22 | |
| Unknown | 4 | | |
| Claudin4 | | | |
| Negative | 73 | 23 | 0.055 |
| Positive | 80 | 45 | |
| Claudin7 | | | |
| Negative | 82 | 33 | 0.511 |
| Positive | 68 | 32 | |
| Unknown | 3 | 3 | |
| E-cadherin | | | |
| Low | 95 | 41 | 0.800 |
| High | 58 | 27 | |
| Vimentin | | | |
| Negative | 121 | 55 | 0.463 |
| Positive | 29 | 10 | |
| Unknown | 3 | 3 | |
| AR | | | |
| Negative | 66 | 26 | 0.755 |
| Positive | 82 | 39 | |
| Unknown | 5 | 3 | |
Clinicopathological factors were significantly
different in 22/68 TNBC cases in the high CPA4 expression group
(Tables III and IV). TNBC with high CPA4 expression was
associated with low Ki67 expression and the expression of CD44/CD24
(Table III, P=0.011; and
Table IV, P=0.016, respectively).
The association between CPA4 and E-cadherin, a representative
epithelial marker (7), was also
assessed. High CPA4 expression was associated with low E-cadherin
expression (Table IV, P=0.016).
The association between high CPA4 and low E-cadherin expression was
validated in representative TNBC sections using IHC (Fig. 2E and F). No other significant
differences in the other evaluated factors were indicated.
 | Table IIIPatient characteristics and CPA4
expression in 68 TNBC cases. |
Table III
Patient characteristics and CPA4
expression in 68 TNBC cases.
| Clinical
factors | Total TNBC cohort
(n=68)
| P-value |
|---|
Low CPA4
(n=46) | High CPA4
(n=22) |
|---|
| Age (years) | 56.3±11.4 | 61.5±14.9 | 0.119 |
| Basal-like
type | | | |
| Basal | 15 | 5 | 0.403 |
| Non-basal | 31 | 17 | |
| Ki67 labeling
index | 41.5±30.35 | 22.4±22.6 | 0.011a |
| Tumor size | | | |
| <2 cm | 21 | 8 | 0.469 |
| ≥2 cm | 25 | 14 | |
| Tumor stage | | | |
| 1 | 23 | 10 | 0.532 |
| 2 | 21 | 12 | |
| 3 | 2 | 0 | |
| 4 | 0 | 0 | |
| Lymph node
metastasis | | | |
| Negative | 28 | 17 | 0.181 |
| Positive | 18 | 5 | |
| Metastasis | | | |
| Negative | 46 | 22 | N.D. |
| Positive | 0 | 0 | |
| Stage (UICC, 7th
Ed) | | | |
| I | 17 | 8 | 0.302 |
| II | 21 | 13 | |
| III | 8 | 1 | |
| IV | 0 | 0 | |
| Lymphatic
invasion | | | |
| Negative | 16 | 11 | 0.230 |
| Positive | 30 | 11 | |
| Venous
invasion | | | |
| Negative | 30 | 15 | 0.809 |
| Positive | 16 | 7 | |
| NG | | | |
| NG1 | 3 | 3 | 0.567 |
| NG2 | 5 | 3 | |
| NG3 | 38 | 16 | |
 | Table IVCPA4 expression in stem cell markers,
epithelial-mesenchymal transition markers and AR of 68 TNBC
cases. |
Table IV
CPA4 expression in stem cell markers,
epithelial-mesenchymal transition markers and AR of 68 TNBC
cases.
| Clinical
factors | Total TNBC cohort
(n=68)
| P-value |
|---|
Low CPA4
(n=46) | High CPA4
(n=22) |
|---|
| ALDH1 | | | |
| Low | 41 | 16 | 0.086 |
| High | 5 | 6 | |
| CD44/CD24 | | | |
| High/low | 15 | 14 | 0.016a |
| Others | 31 | 8 | |
| Claudin1 | | | |
| Negative | 18 | 8 | 0.826 |
| Positive | 28 | 14 | |
| Claudin3 | | | |
| Negative | 26 | 12 | 0.760 |
| Positive | 19 | 10 | |
| Unknown | 1 | | |
| Claudin4 | | | |
| Negative | 7 | 6 | 0.237 |
| Positive | 39 | 16 | |
| Claudin7 | | | |
| Negative | 20 | 10 | 0.782 |
| Positive | 25 | 12 | |
| Unknown | 1 | | |
| E-cadherin | | | |
| Low | 17 | 15 | 0.016a |
| High | 29 | 7 | |
| Vimentin | | | |
| Negative | 26 | 14 | 0.256 |
| Positive | 20 | 7 | |
| Unknown | | 1 | |
| AR | | | |
| Negative | 33 | 14 | 0.741 |
| Positive | 12 | 7 | |
| Unknown | 1 | 1 | |
Prognostic significance of CPA4
expression in patients with breast cancer
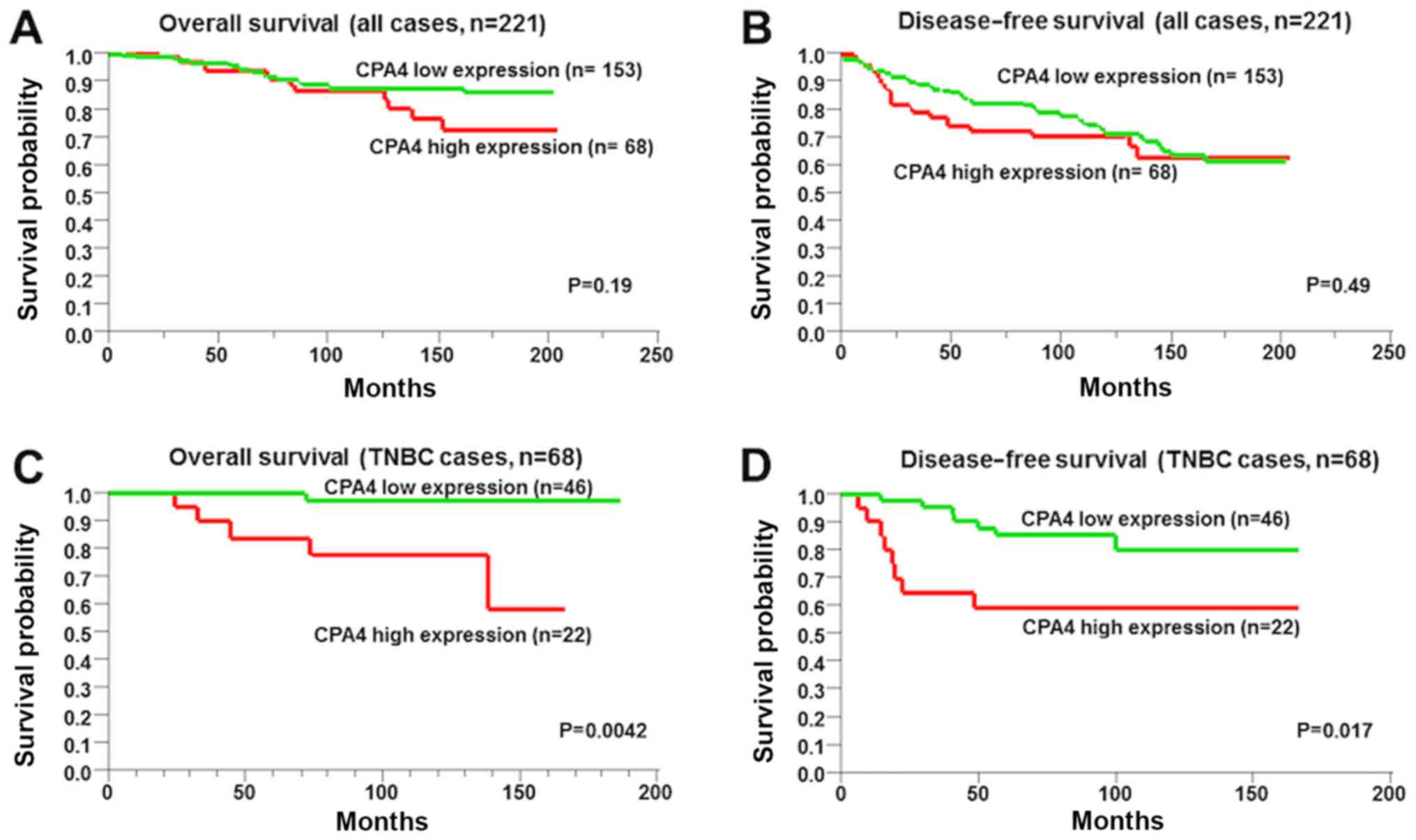
The overall and disease-free survival were not
significantly associated with CPA4 expression (Fig. 3A and B; P=0.19 and P=0.49,
respectively). However, the overall and disease-free survival
intervals of high-CPA4-expressing TNBC cells were worse compared
with those of low-CPA4-expressing TNBC cells (Fig. 3C and D; P=0.004 and P=0.017,
respectively).
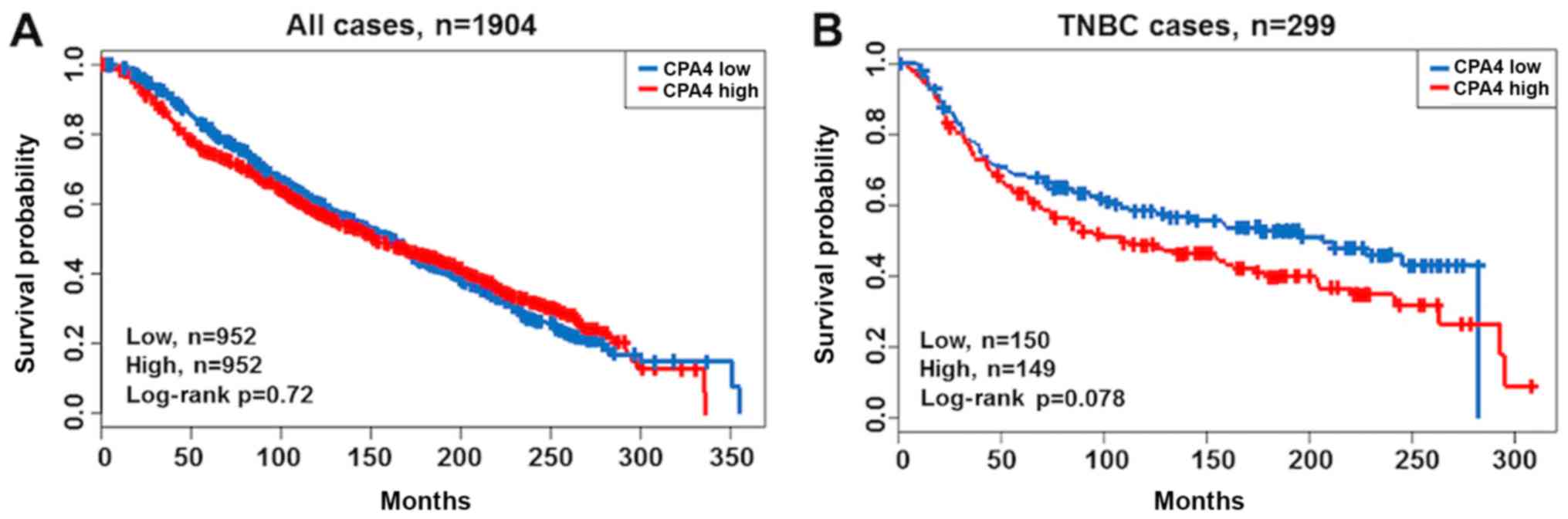
Using a public database cohort, the prognostic
significance of CPA4 expression on overall survival was validated
(Fig. 4). As expected, the
patients with TNBC and high CPA4 expression exhibited poorer
prognoses compared with those with low CPA4 expression; however,
the differences were not significant (Fig. 4A and B; P=0.720 and P=0.078,
respectively).
The association between CPA4 expression and other
factors was also explored, including the NG and LNM (Fig. 5). Among NG1-2 and LNM-negative
cases, the overall and disease-free survival were not significantly
associated with CPA4 expression (Fig.
5A, B, E and F; P=0.39, P=0.66, P=0.29 and P=0.16,
respectively). However, among NG3 cases and LNM-positive cases, the
overall and disease-free survival intervals of highly expressed
CPA4 TNBC cells were worse than those of low CPA4 TNBC cells
(Fig. 5C, D, G and 5H; P=0.0002, P=0.019, P=0.0004 and
P=0.015, respectively). To clarify the prognostic significance of
CPA4, multivariate analysis was performed for survival, and results
indicated that high CPA4 expression was an independent prognostic
factor for poor survival (Table V;
P=0.001).
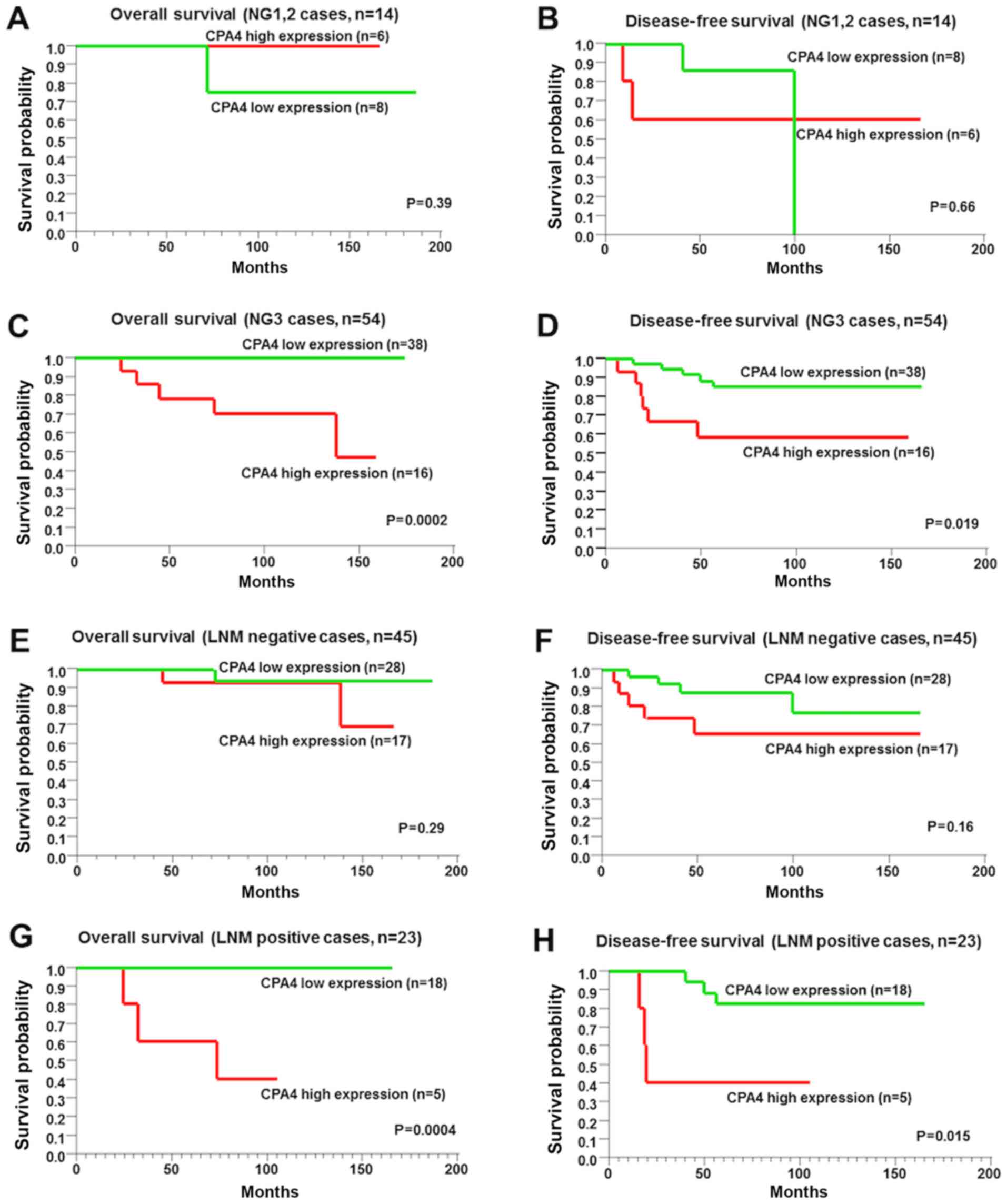 | Figure 5Kaplan-Meier curves analysis of CPA4
expression in 68 patients with TNBC, with or without progression of
NG and LNM. (A) Overall survival in patients with NG1-2 and high or
low CPA4 expression (n=14, P=0.39). (B) Disease-free survival in
patients with NG1-2 and high or low CPA4 expression (n=14, P=0.66).
(C) Overall survival in patients with NG3 and high or low CPA4
expression (n=54, P=0.0002). (D) Disease-free survival in patients
with NG3 and high or low CPA4 expression (n=54, P=0.019). (E)
Overall survival in LNM-negative patients with high or low CPA4
expression (n=45, P=0.29).(F) Disease-free survival in LNM-negative
patients with high or low CPA4 expression (n=45, P=0.16). (G)
Overall survival in LNM-positive patients with high or low CPA4
expression (n=23, P=0.0004). (H) Disease-free survival in
LNM-positive patients with high or low CPA4 expression (n=23,
P=0.015). NG, nuclear grade; LNM, lymph node metastasis; CPA4,
carboxypeptidase A4. |
 | Table VCox univariate/multivariate
regression analysis of variables associated with overall survival
in patients with TNBC. |
Table V
Cox univariate/multivariate
regression analysis of variables associated with overall survival
in patients with TNBC.
| Clinicopathologic
variables | Univariate analysis
| Multivariate
analysis
|
|---|
| RR | 95% Cl | P-value | RR | 95% a | P-value |
|---|
| CPA4 expression
(Low vs. high) | 3.43 | 1.38-15.02 | 0.007a | 30.38 | 3.44-1071.53 | 0.001a |
| Age (<58 vs.
58≤) | 1.63 | 0.71-4.43 | 0.250 | 155 | 0.09-29.95 | 0.750 |
| Tumor factor CT1
vs.T2-T3) | 1.4 | 0.62-3.76 | 0.420 | 2.99 | 0.23-103.06 | 0.430 |
| Nuclear grade
(NG1-2 vs.NG3) | 1.02 | 0.41-4.48 | 0.970 | 0.37 | 0.01-10.92 | 0520 |
| Lymph node
metastasis (Absent vs. present) | 1.32 | 0.56-3.08 | 0.510 | 46.02 | 152-18844.18 | 0.020a |
| Lymphatic invasion
(Absent vs. present) | 0.81 | 0.35-1.89 | 0.610 | _ | _ | _ |
| Venous invasion
(Absent vs. present) | 1.03 | 0.38-2.33 | 0.950 | 053 | 0.02-10.29 | 0.690 |
| Adjuvant therapy
(Absent vs. present) | 051 | 0.22-1.37 | 0.160 | 0.04 | 0.0006-0.77 | 0.030a |
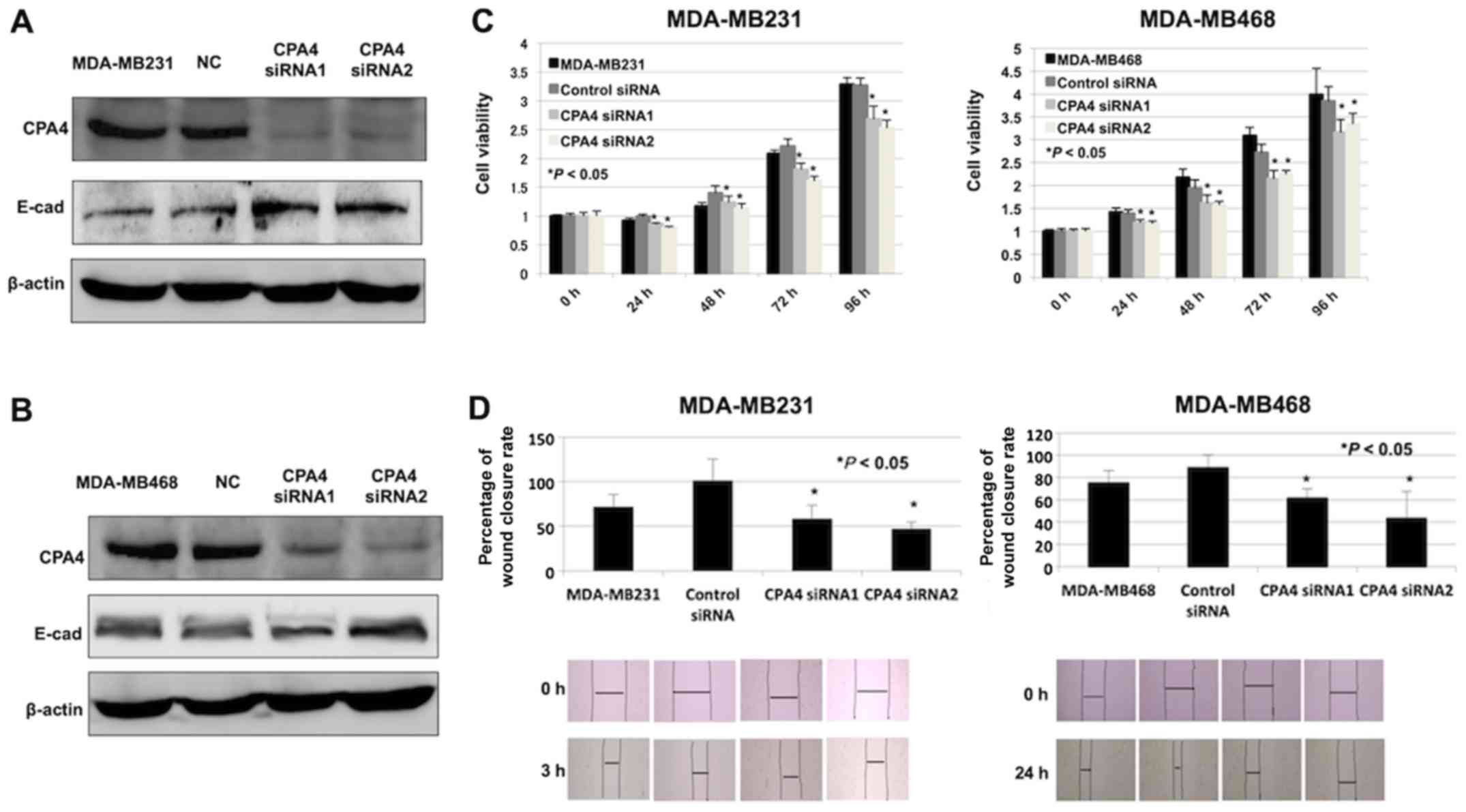
Cell viability and migration inhibition
in CPA4-knockdown TNBC cells
The role of CPA4 on cell viability and migration
ability was assessed using RNA interference. Western blot analysis
was performed to validate the CPA4 knockdown experiments in TNBC
cell lines treated with specific CPA4 siRNAs (Fig. 6A and B). In addition, it was
identified that cell viability in the CPA4 siRNA group was
significantly inhibited compared with that in the negative-control
group (Fig. 6C, P<0.05).
Furthermore, CPA4 knockdown suppressed cell migration in comparison
with the negative-control cells (Fig.
6D, P<0.05).
Suppression of E-cadherin in
CPA4-suppressed TNBC cells
The association between CPA4 and the expression of
E-cadherin was examined. The expression of E-cadherin was increased
in CPA4 siRNA groups (Fig. 6A and
B). These data were consistent with the inverse association
identified between CPA4 and E-cadherin expression in clinical TNBC
samples.
Discussion
Lehmann et al (24) indicated that TNBC can be classified
according to the gene expression profiles. The subtypes include
basal-like1, basal-like2, immunomodulatory, mesen-chymal-like,
mesenchymal stem-like, and luminal androgen receptor (LAR) types.
Among these subtypes, the present analyses indicated that high CPA4
expression in TNBC was independent of the following
characteristics: Basal-like subtypes with high proliferation
ability and LAR subtypes with LAR accumulation. Furthermore, the
present results indicated E-cadherin upregulation and
reduced-migration ability following CPA4 knockdown. The CPA4
accumulation in TNBC tissues were identified to be associated with
low E-cadherin expression, high CSC marker expression and a poor
prognosis. Furthermore, the data indicated that CPA4 may be a
regulator of EMT and CSCs in TNBC cells. Therefore, it was
suggested that CPA4 in TNBC may be associated with the
mesenchymal-like or mesenchymal stem-like subtypes, with CSC and/or
EMT phenotypes. Tanco et al (8) reported that proneurotensin, a
precursor of neurotensin, is a substrate of CPA4 using kinetic
analysis. Their study revealed that neuro-tensin production was
regulated by CPA4 enzyme activity. Notably, neurotensin has also
been reported to be associated with EMT induction (25). From these observations, it was
suggested that CPA4-mediated EMT may be induced via neurotensin
activation via CPA4 enzymes.
The present study was performed to identify
therapeutic targets for TNBC, as patients with TNBC lack treatment
options available that are readily available options for patients
with other types of breast cancer, including hormone therapy and
molecularly targeted therapies (26). From the present data, CPA4
expression in normal tissues, including mammary glands and non-TNBC
tissues, was significantly decreased compared with that in TNBC
tissues. Furthermore, in the present cohort, CPA4 expression in
patients with breast cancer was not significantly associated with
patient prognosis. However, patients with TNBC and high CPA4
expression had a poorer prognosis compared with those with low CPA4
expression. To the best of our knowledge, the present study is the
first to report that high expression of CPA4 may be a specific
predictor of poor prognosis for patients with TNBC.
Suppression of CPA4 significantly reduced the cell
viability in TNBC cell lines the present study, and previous data
has also suggested that CPA4 inhibitors may successfully inhibit
cancer cell viability in patients with TNBC (27). The carboxypeptidase inhibitor
Sabellastarte magnifica (SmCI) has been reported to suppress
the metallo-carboxypeptidase activity of CPA4 by forming a complex
with CPA4 (27). SmCI inhibits
metallo-carboxypeptidases and serine proteases, such as trypsin and
elastase (28). Serine protease is
a known promising molecular target in TNBC cells (29). Thus, these findings suggest that
targeting CPA4 using inhibitors, such as SmCI, in patients with
TBNC may be effective in reducing cancer cell viability via the
inhibition of metallo-carboxypeptidases and serine proteases.
Therefore, further studies are required before SmCI or other
candidate drug trials can be implemented for future clinical
applications.
In conclusion, the present results indicated that
high CPA4 expression is a powerful marker for poor prognosis and
aggressive phenotypes, such as EMT, in TNBC. The present results
suggest that targeting CPA4 in TNBC may be a promising therapeutic
strategy for controlling aggressive phenotypes in refractory
TNBC.
Funding
This study was supported by Grants-in-Aid for
Scientific Research from the Japan Society for the Promotion of
Science (JSPS; grant nos. 15K08339, 15K10085, 17K19893 and
18K07665).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TH, AK, TY, HK and TO contributed to the study
design and wrote the manuscript. AY, TF, SO, SK, RKI, NG, MN, TA
and KS performed sample collection and data analysis. RKI performed
transcriptome analysis.
Ethics approval and consent to
participate
This research conformed to the tenets of the
Declaration of Helsinki and to the guidelines of the Gunma
University Ethical Review Board for Medical Research Involving
Human Subjects (approval no. 1297). Patient consent was
obtained.
Patient consent for publication
Patient consent was obtained.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carey LA, Perou CM, Livasy CA, Dressler
LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S,
et al: Race, breast cancer subtypes, and survival in the Carolina
Breast Cancer Study. JAMA. 295:2492–2502. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goldhirsch A, Winer EP, Coates AS, Gelber
RD, Piccart-Gebhart M, Thürlimann B and Senn HJ; Panel members:
Personalizing the treatment of women with early breast cancer:
highlights of the St Gallen International Expert Consensus on the
Primary Therapy of Early Breast Cancer 2013. Ann Oncol.
24:2206–2223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Giatromanolaki A, Sivridis E, Fiska A and
Koukourakis MI: The CD44+/CD24- phenotype
relates to 'triple-negative' state and unfavorable prognosis in
breast cancer patients. Med Oncol. 28:745–752. 2011. View Article : Google Scholar
|
|
6
|
Idowu MO, Kmieciak M, Dumur C, Burton RS,
Grimes MM, Powers CN and Manjili MH: CD44(+)/CD24(-/low) cancer
stem/progenitor cells are more abundant in triple-negative invasive
breast carcinoma phenotype and are associated with poor outcome.
Hum Pathol. 43:364–373. 2012. View Article : Google Scholar
|
|
7
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tanco S, Zhang X, Morano C, Avilés FX,
Lorenzo J and Fricker LD: Characterization of the substrate
specificity of human carboxypeptidase A4 and implications for a
role in extracellular peptide processing. J Biol Chem.
285:18385–18396. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun L, Cao J, Guo C, Burnett J, Yang Z,
Ran Y and Sun D: Associations of carboxypeptidase 4 with ALDH1A1
expression and their prognostic value in esophageal squamous cell
carcinoma. Dis Esophagus. 30:1–5. 2017. View Article : Google Scholar
|
|
10
|
Sun L, Guo C, Burnett J, Pan J, Yang Z,
Ran Y and Sun D: Association between expression of carboxypeptidase
4 and stem cell markers and their clinical significance in liver
cancer development. J Cancer. 8:111–116. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun L, Guo C, Yuan H, Burnett J, Pan J,
Yang Z, Ran Y, Myers I and Sun D: Overexpression of
carboxypeptidase A4 (CPA4) is associated with poor prognosis in
patients with gastric cancer. Am J Transl Res. 8:5071–5075.
2016.PubMed/NCBI
|
|
12
|
Sun L, Guo C, Burnett J, Yang Z, Ran Y and
Sun D: Serum carboxypeptidaseA4 levels predict liver metastasis in
colorectal carcinoma. Oncotarget. 7:78688–78697. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun L, Wang Y, Yuan H, Burnett J, Pan J,
Yang Z, Ran Y, Myers I and Sun D: CPA4 is a novel diagnostic and
prognostic marker for human non-small-cell lung cancer. J Cancer.
7:1197–1204. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun L, Burnett J, Guo C, Xie Y, Pan J,
Yang Z, Ran Y and Sun D: CPA4 is a promising diagnostic serum
biomarker for pancreatic cancer. Am J Cancer Res. 6:91–96.
2015.
|
|
15
|
Tan AA, Phang WM, Gopinath SC, Hashim OH,
Kiew LV and Chen Y: Revealing Glycoproteins in the Secretome of
MCF-7 Human Breast Cancer Cells. BioMed Res Int. 2015:4532892015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Finn RS, Dering J, Conklin D, Kalous O,
Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, et al: PD
0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially
inhibits proliferation of luminal estrogen receptor-positive human
breast cancer cell lines in vitro. Breast Cancer Res. 11:R772009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun J, Nishiyama T, Shimizu K and Kadota
K: TCC: An R package for comparing tag count data with robust
normalization strategies. BMC Bioinformatics. 14:2192013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sobin LH, Gospodarowicz MK and Wittekind
C; International Union against Cancer: TNM Classification of
Malignant Tumours. Wiley-Blackwell; West Sussex, UK; Hoboken, NJ:
2010
|
|
19
|
Handa T, Katayama A, Yokobori T, Yamane A,
Horiguchi J, Kawabata-Iwakawa R, Rokudai S, Bao P, Gombodorj N,
Altan B, et al: Caspase14 expression is associated with triple
negative phenotypes and cancer stem cell marker expression in
breast cancer patients. J Surg Oncol. 116:706–715. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al American Society of Clinical Oncology; College
of American Pathologists: Recommendations for human epidermal
growth factor receptor 2 testing in breast cancer: American Society
of Clinical Oncology/College of American Pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dowsett M, Nielsen TO, A'Hern R, Bartlett
J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T, et
al International Ki-67 in Breast Cancer Working Group: Assessment
of Ki67 in breast cancer: Recommendations from the International
Ki67 in Breast Cancer working group. J Natl Cancer Inst.
103:1656–1664. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Obayashi S, Horiguchi J, Higuchi T,
Katayama A, Handa T, Altan B, Bai T, Bao P, Bao H, Yokobori T, et
al: Stathmin1 expression is associated with aggressive phenotypes
and cancer stem cell marker expression in breast cancer patients.
Int J Oncol. 51:781–790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Senol S, Sayar I, Ceyran AB, Ibiloglu I,
Akalin I, Firat U, Kosemetin D, Engin Zerk P and Aydin A: Stromal
clues in endometrial carcinoma: Loss of expression of β-catenin,
epithelial-mesenchymal transition regulators, and
estrogen-Progesterone Receptor. Int J Gynecol Pathol. 35:238–248.
2016. View Article : Google Scholar :
|
|
24
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750–2767.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ye Y, Liu P, Wang Y, Li H, Wei F, Cheng Y,
Han L and Yu J: Neurotensin, a novel messenger to cross-link
inflammation and tumor invasion via epithelial-mesenchymal
transition pathway. Int Rev Immunol. 35:340–350. 2016.PubMed/NCBI
|
|
26
|
Johnston SR: New strategies in estrogen
receptor-positive breast cancer. Clin Cancer Res. 16:1979–1987.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Alonso del Rivero M, Reytor ML, Trejo SA,
Chávez MA, Avilés FX and Reverter D: A noncanonical mechanism of
carboxypeptidase inhibition revealed by the crystal structure of
the Tri-Kunitz SmCI in complex with human CPA4. Structure.
21:1118–1126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alonso-del-Rivero M, Trejo SA, Reytor ML,
Rodriguez-de-la-Vega M, Delfin J, Diaz J, González-González Y,
Canals F, Chavez MA and Aviles FX: Tri-domain bifunctional
inhibitor of metallocarboxypeptidases A and serine proteases
isolated from marine annelid Sabellastarte magnifica. J Biol Chem.
287:15427–15438. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Al-Awadhi FH, Salvador LA, Law BK, Paul VJ
and Luesch H: Kempopeptin C, a novel marine-derived serine protease
inhibitor targeting invasive breast cancer. Mar Drugs. 15:152017.
View Article : Google Scholar
|