Introduction
Osteosarcoma is an aggressive malignant tumor of
bone, which is mainly observed in children and adolescents
(1). Up to 70% of patients with
osteosarcoma are treated with multimodal therapies that include
chemotherapy [such as doxorubicin (DOX), methotrexate and
cisplatin] and surgical excision (2,3).
However, over the past three decades, survival rates have not
substantially improved. Furthermore, there is a lack of effective
therapeutic options for patients suffering from a recurrence and
metastasis of the disease. The lack of options is mainly due to the
complicated etiologies of the disease, which are responsible for an
extremely complex genomic organization that results from chromosome
aberrations, chromothripsis, kataegis, and genome instability with
a high rate of copy number alterations and structural variations
(4,5). In addition, accumulating evidence
indicates that there are epigenetic changes in osteosarcoma, mainly
through histone modifications and DNA methylation (6). This aberrance induces significant
alterations in the expression levels of various oncogenic proteins
and tumor suppressors in osteosarcoma (7-10).
The intricate heterogeneity introduces difficulties for the
identification of key oncogene-driven pathways, which may be
potential targets for effective treatment of osteosarcoma.
While genomics provide an insight into the molecule
profile of cancer, which indicates key players in the tumorigenesis
and progression of the disease, proteomics connects the genomic
information with disease phenotypes. Defects at the genomic level
affect cellular function and regulation, which are revealed at the
protein level. Recent advances in current proteomic techniques and
bioinformatics allow detailed and more complete views of biological
networks relating to disease.
Cancer patient tissue is an ideal biological
specimen for studying disease etiology at the genomic and proteomic
levels as it most accurately represents the subject. In addition,
the method of fresh freezing is a compatible preservation procedure
for subsequent proteomic analysis. Therefore, in the present study,
proteomics analysis using freshly frozen biopsy tissues from
patients with osteosarcoma was performed. Due to the
pathophysiology of osteosarcoma, one of limitations in a proteomics
study is that there are no adequate comparable control tissues. To
date, the majority of proteomic studies of osteosarcoma tissues
have been performed using clinical specimens from patients compared
with benign bone tumors (11,12).
No consensus representative normal tissues have been defined. In
the present study, soft tissue callus was used as a non-cancerous
control tissue. Soft callus is formed during the normal fracture
repair process primarily from periosteal cells, which are the major
source of osteoblasts and chondrocytes (13,14).
Bone callus has been demonstrated to contain the highest density of
active osteoblasts at 3 months after a fracture, and then the
density decreases over time (15).
Thus, soft callus is a good source of non-cancerous tissue
containing high numbers of osteoblastic cells that can be used as a
control in a proteomics study of osteosarcoma.
The aim of the present study was to obtain novel
information regarding osteosarcoma mechanisms by studying protein
patterns directly from clinical tissues together with
bioinformatics tools. The findings suggest disease-relevant
pathways that may serve as novel targets for the treatment of
osteosarcoma.
Materials and methods
Patients and tissue samples
Chemonaïve osteosarcoma tissues were obtained from
patients diagnosed and treated at Maharaj Nakorn Chiang Mai
Hospital (Chiang Mai, Thailand). Soft tissue callus was collected
from donors who were treated at the Trauma Unit, Maharaj Nakorn
Chiang Mai Hospital. All tissue samples were freshly frozen and
stored at −80°C until use. Patients, or parents in the case of
minors, and volunteers gave written consent to be included in the
study. This study was approved by the Research Ethics Committee of
the Faculty of Medicine, Chiang Mai University (Chiang Mai,
Thailand). All clinicopathological parameters were retrieved from
hospital records and pathology reports (Table I).
 | Table ICharacteristics of osteosarcoma
patients and donors. |
Table I
Characteristics of osteosarcoma
patients and donors.
| Sample ID | Sex | Age at
diagnosis | Site | Enneking stage | Neoadjuvant
chemotherapy | % tumor
necrosis | Lung
metastasis |
|---|
| Osteosarcoma |
| OS1 | Male | 24 | Distal femur | IIB | DOX/Ifos | 80 | Yes |
| OS2 | Female | 15 | Proximal
humerus | IIB | - | 10 | No |
| OS3 | Male | 12 | Proximal tibia | IIB | MTX NA | No | |
| OS4 | Female | 5 | Distal femur | IIB | DOX/CIS/MTX | 10 | Yes |
| OS5 | Female | 9 | Distal femur | IIB | DOX/Carbo | 90 | No |
| OS6 | Female | 15 | Distal femur | IIB | DOX/CIS | 90 | No |
| OS7 | Female | 11 | Distal femur | IIB | ICE | 95 | No |
| OS8 | Female | 5 | Distal femur | IIB | DOX/CIS/MTX | 10 | Yes |
| OS9 | Male | 15 | Distal femur | IIB | DOX/CIS | 42 | Yes |
| OS10 | Female | 61 | Distal femur | IIB | DOX/CIS | 35 | Yes |
| OS11 | Male | 12 | Proximal tibia | III | DOX/CIS/MTX | 95 | Yes |
| OS12 | Female | 54 | Ilium | III | DOX/CIS | 50 | Yes |
| OS13 | Female | 13 | Proximal
humerus | III | - | NA | Yes |
| Soft callus and
osteoblast |
| Cal1 | Female | 58 | Distal femur | - | - | - | - |
| Cal2 | Female | 36 | Distal femur | - | - | - | - |
| Cal3 | Female | 64 | Distal femur | - | - | - | - |
| Cal4 | Male | 28 | Distal femur | - | - | - | - |
| Cal5 | Male | 19 | Tibia | - | - | - | - |
| Cal6 | Male | 26 | Proximal femur | - | - | - | - |
| Cal7 | Male | 17 | Shaft femur | - | - | - | - |
| Cal8 | Male | 32 | Shaft femur | - | - | - | - |
Two-dimensional electrophoresis (2DE) of
proteins from osteosarcoma and soft callus tissues
Osteosarcoma and soft callus tissues were lysed in
radioimmunoprecipitation assay buffer supplemented with 1% protease
inhibitor (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany),
homogenized and incubated on ice for 30 min. The supernatant was
collected by centrifugation at 15,000 × g at 4°C for 20 min. Crude
tissue lysates were further prepared for 2DE analysis using a 2-D
Clean-Up kit, according to the manufacturer's protocol (GE
Healthcare Life Sciences, Uppsala, Sweden). Protein concentration
was determined using Bradford assay. The protein pellets were
resuspended in 2D lysis buffer. Individual samples (60 µg
protein) were applied by overnight in-gel rehydration of 7 cm pH
3-10 nonlinear gradient IPG strips (GE Healthcare, Chicago, IL,
USA). IEF was performed at 7,000 Vh, 55 mA per gel strip using an
Ettan IPGphor 3 (GE Healthcare). The IPG strips were equilibrated
and separated in 14% SDS-PAGE followed by SYPRO Ruby staining
(Thermo Fisher Scientific, Inc., Waltham, MA, USA), as previously
described (16). Gels were scanned
and visualized using Ettan DIGE Imager (GE Healthcare).
Spot detection and analysis
To minimize contamination by the highly abundant
protein present in the tissue samples, the protein spots on the 2DE
gels were aligned to human serum protein patterns, as previously
reported (17). Major highly
abundant proteins, including α and γ Ig heavy chain, serum albumin,
hemoglobin β chain, complement factor B and transferrin, were
observed in every 2DE gel. The volume of abundant proteins present
was subtracted from the total volume of all protein spots on
individual gels. The gels were then analyzed using ImageMaster 2D
Platinum 7.0 software (GE Healthcare). The relative volume (%) of a
spot was used to compare differences in the expression of each
protein between the osteosarcoma and soft callus tissues. In
addition to strict control of sample preparation and the
experimental procedure of the first- and second-dimension
separation, statistical analyses were applied in order to minimize
experimental variations and increase the reliability and overall
reproducibility of the 2DE results. Each gel of the osteosarcoma
tissues (4 cases) was independently compared to each gel from the
soft tissue callus (4 cases), generating 16 match sets in total.
Only spots that were consistently upregulated or downregulated in
≥80% of all match sets and significantly different by ≥1.5-fold
(P<0.05, Student's t-test) were subjected to protein
identification.
In-gel digestion
The protein spots from the osteosarcoma tissues were
excised and subjected to in-gel trypsin digestion, as previously
described (16). Briefly, the
excised gels were destained with 50% ACN in 0.1 M
NH4HCO3, reduced with 10 mM DTT, alkylated in
100 mM iodoacetamide and digested with trypsin (Promega
Corporation, Madison, WI, USA) at 37°C overnight. The digested
peptides were dried and collected for protein identification by
liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Protein identification by LC-MS/MS
The digested peptides were identified using nanoflow
liquid chromatography coupled with nano ESI MS/MS (Q-TOF micro)
(both from Waters Corporation, Milford, MA, USA). The MS/MS spectra
were processed using MassLynx 4.0 software (Waters Corporation) and
converted to PKL files using ProteinLynx 2.2 software (Waters
Corporation). Protein identification was performed using the Mascot
search engine with NCBInr version 20130630 sequence and SwissProt
databases (http://www.matrixscience.com). The search parameters
were set as follows: Peptide mass tolerance, 1.2 Da; MS/MS ion mass
tolerance, 0.2 Da; allowance, 1 missed cleavage; enzyme, trypsin;
and limit of peptide charges, 1+, 2+ and
3+. Proteins identified with P≤0.05 were considered as
promising hits. Proteins with a molecular weight and pI consistent
with the spots on the 2DE gels were considered to be positively
identified.
Gene ontology (GO) terms enrichment and
pathway analysis
Functional enrichment analysis of the upregulated
proteins in osteosarcoma was performed using WebGestalt (18). ClueGO (version 2.3.3) and CluePedia
(version 1.3.3), a plugin for Cytoscape version 3.4, were used for
functionally grouped network analysis (19,20).
Primary cell extraction and
characterization
Primary osteoblastic cells were isolated from bone
grafts of patients who had been diagnosed with non-cancerous
orthopedic conditions and required the use of autologous bone
grafts as substitution procedures. Primary osteoblasts used in this
study were derived from a set of bone graft specimens previously
reported (6). Primary osteosarcoma
cells were extracted from chemonaïve biopsy tissues of patients
with osteosarcoma. Extraction, culturing and characterization of
the primary cells were performed according to previously described
protocols (21). Briefly, the
primary cells were isolated by incubating the tissues in 5 mg/ml
collagenase type I solution (Gibco; Thermo Fisher Scientific, Inc.)
at 37°C for 18 h. The cells were pelleted by centrifugation and
cultured in freshly prepared Dulbecco's modified Eagle's medium
(DMEM) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified 5% CO2
incubator. All primary cells were characterized for doubling time
and osteogenicity. Cancer markers, including matrix
metallopeptidase (MMP)-9 and collagen type X, were determined by
reverse transcription-quantitative polymerase chain reaction,
according to a previously described protocol (21).
Cell culture
Osteosarcoma and osteoblastic cell lines used in
this study include MNNG/HOS (CLS 300289; Cell Lines Service, GmbH,
Eppelheim, Germany), U2OS (CLS-300364; Cell Lines Service, GmbH),
143B (CRL-8303; ATCC, Manassas, VA, USA), MG-63 (CRL-1427; ATCC),
Saos-2 (HTB-85; ATCC) and normal human osteoblast cell line
hFOB1.19 (CRL-11372; ATCC). MNNG/HOS was cultured in Roswell Park
Memorial Institute (RPMI)-1640 medium supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.). MG-63 was cultured in DMEM
supplemented with 10% FBS. 143B cells was cultured in DMEM
supplemented with 10% FBS and 0.015 mg/ml 5-bromo-2′-deoxyuridine
(Merck KGaA). U2OS, Saos-2 and hFOB1.19 were cultured in F-12
Medium supplemented with 10% FBS. All the cells were maintained at
37°C in humidified 5% CO2 incubator.
Western blot analysis and total protein
staining
Crude proteins were extracted from osteosarcoma
cells and tissue samples, as well as osteoblastic cells and soft
callus tissues, with RIPA buffer supplemented with 1% protease
inhibitor cocktail (Amresco, LLC, Solon, OH, USA). Protein
concentration was determined by Bradford assay. Extracted proteins
(10–15 µg) were separated in 10% SDS-PAGE and transferred to
polyvinylidene difluoride (PVDF) membranes (Immobilon-P; EMD
Millipore, Billerica, MA, USA). Protein bands on the blots of
tissue samples were stained using 0.15% (w/v) Ponceau S dye
(Sigma-Aldrich; Merck KGaA) in 1% acetic acid. The protein images
were used to control protein loading by quantifying the whole lane
of protein bands. The membranes were destained in TBS/T (TBS, 0.1%
Tween-20) for 5 min. After blocking with 10% skimmed milk in TBS/T,
membranes were incubated with antibodies specific to GRP78
(1:1,000; cat. no. ab108615), GRP94 (1:1,000; cat. no. ab108606),
prelamin-A/C (1:5,000; cat. no. ab169532), and calreticulin
(1:2,000; cat. no. ab22683) (all from Abcam, Cambridge, UK) at 4°C
overnight. Membranes were then washed with TBS/T, and incubated
with secondary antibody conjugated with horseradish peroxidase
(HRP; 1:5,000; cat. no. ab6721; Abcam) at room temperature for 1 h.
Bands on immunoblots were detected using ECL-Advance Western
Blotting Detection kit (GE Healthcare).
RT-qPCR
RNA was extracted from osteosarcoma and osteoblastic
cells using Ilustra RNAspin Mini kit (GE Healthcare), and cDNA was
synthesized from total RNA using a Bioline SensiFAST cDNA synthesis
kit (Bioline, London, UK). qPCR was performed in Chromo4 Real-Time
PCR Detection system (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) using a Bioline SensiFAST SYBR No-ROX kit (Bioline). To
compare the mRNA expression of each group, β-actin was used as an
internal control. The primers used were: sliced X-box binding
protein 1 (XBP1s), forward, 5′-TGCTGAGTCCGCAGCAGGTG-3′ and reverse,
5′-GCTGGCAGGCTCTGGGGAAG-3′ (22);
β-actin, forward, 5′-TTCAACACCCCAGCCATGT-3′ and reverse, 5′-TGGTAC
GGCCAGAGGCGTACAG-3′; MMP-9, forward, 5′-TGAGAAC CAATCTCACCGACAG-3′
and reverse, 5′-TGCCACCCGAG TGTAACCAT-3′; collagen type X, forward,
5′-AGGAATGCC TGTGTCTGCTT-3′ and reverse, 5′-ACAGGCCTACCCAA
ACATGA-3′; collagen type I, forward, 5′-CAGCCGCTTCACC TACAGC-3′ and
reverse, 5′-TTTTGTATTCAATCACTGTCTT GCC-3′; osteonectin, forward,
5′-TCCACAGTACCGGATT CTCTCT-3′ and reverse, 5′-TCTATGTTAGCACCTTGTCTC
CAG-3′; and bone sialoprotein, forward, 5′-GCAGTAGTGAC
TCATCCGAAGAA-3′ and reverse, 5′-GCCTCAGAGTCT TCATCTTCATTC-3′. The
thermal profile was set according to previous report as follows:
initial incubation step at 95°C for 3 min, followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing at 62°C for 15 sec, and
extension at 72°C for 30 sec (22). Relative fold changes in mRNA
expression were calculated using the 2−ΔΔCq method
(23).
Statistical analysis
Data are presented as means ± standard deviation
with three independent replications. Statistical analyses were
performed using GraphPad Prism version 8.0 (GraphPad Software,
Inc., La Jolla, CA, USA). The significance of the differential
expression of UPR-related proteins and XBP1s in multiple cells and
tissues was tested using one-way analysis of variance followed by
Bonferroni's test. The difference between two groups was tested
using Student's t-test for parametric tests and the Mann-Whitney U
test for nonparametric tests. P<0.05 was considered to indicate
a statistically significant difference.
Results
Histological evaluation of osteosarcoma
tissue and soft tissue callus
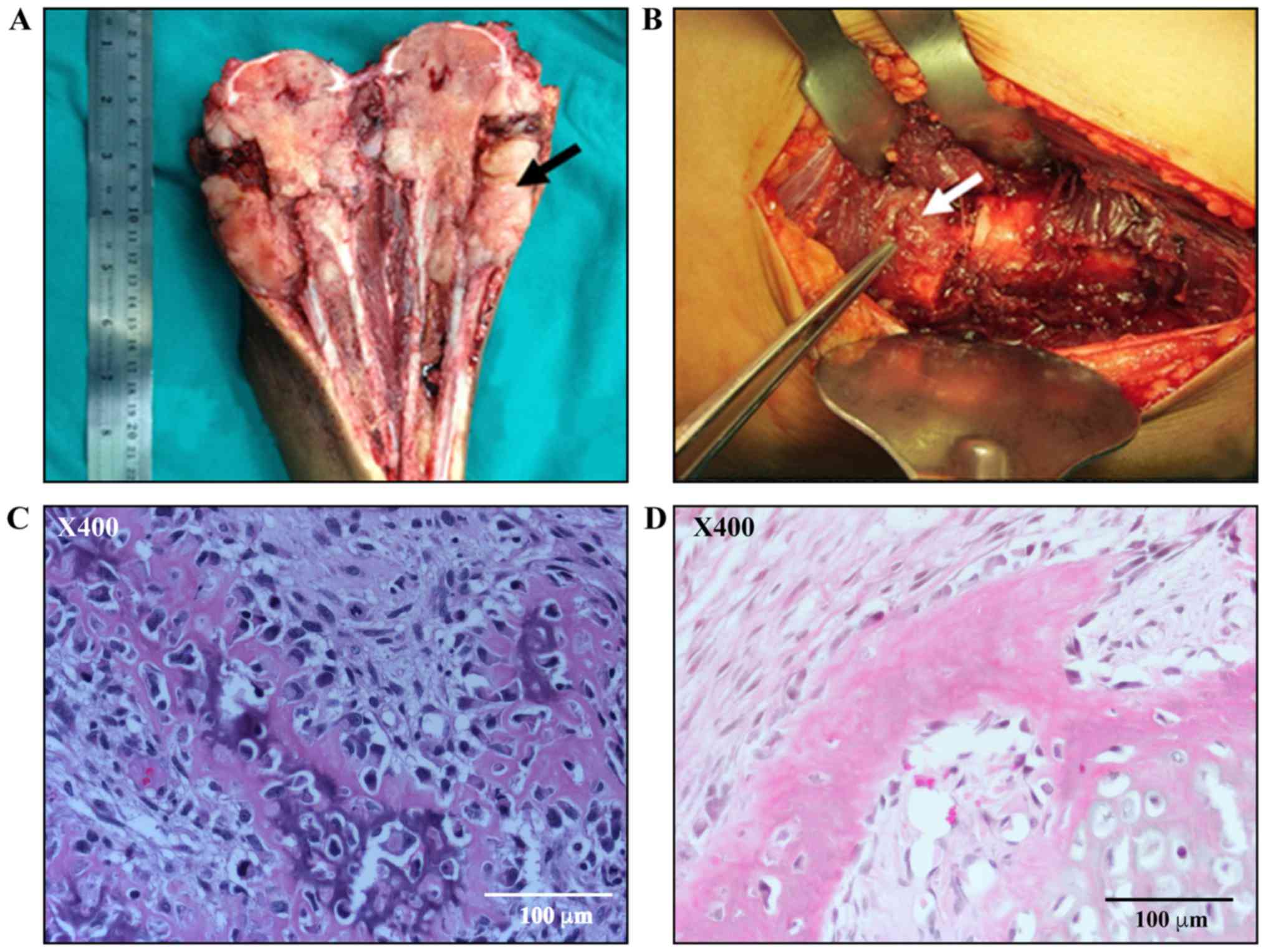
A representative image of the osteosarcoma tissue
from the proximal tibias of patients who had undergone amputation
is shown in Fig. 1A. The white
fresh tissue extruding from destructive metaphyseal was harvested.
Soft callus was derived from the fracture site of donors who were
treated at the Trauma Unit. Soft callus appears as soft fresh
tissue covering the fracture site, as illustrated in Fig. 1B. Formalin-fixed paraffin-embedded
sections of osteosarcoma tissue and soft tissue callus were stained
with hematoxylin and eosin and evaluated by an experienced
musculoskeletal pathologist. Histologically, the osteosarcoma
tissue exhibited high cellularity and consisted of malignant plump
spindle cells with osteoid matrix production (Fig. 1C). The staining of the soft callus
demonstrated a high content of osteoblastic cells (Fig. 1D).
Identification of differentially
expressed proteins in osteosarcoma tissues in comparison to soft
tissue callus using 2DE and LC-MS/MS
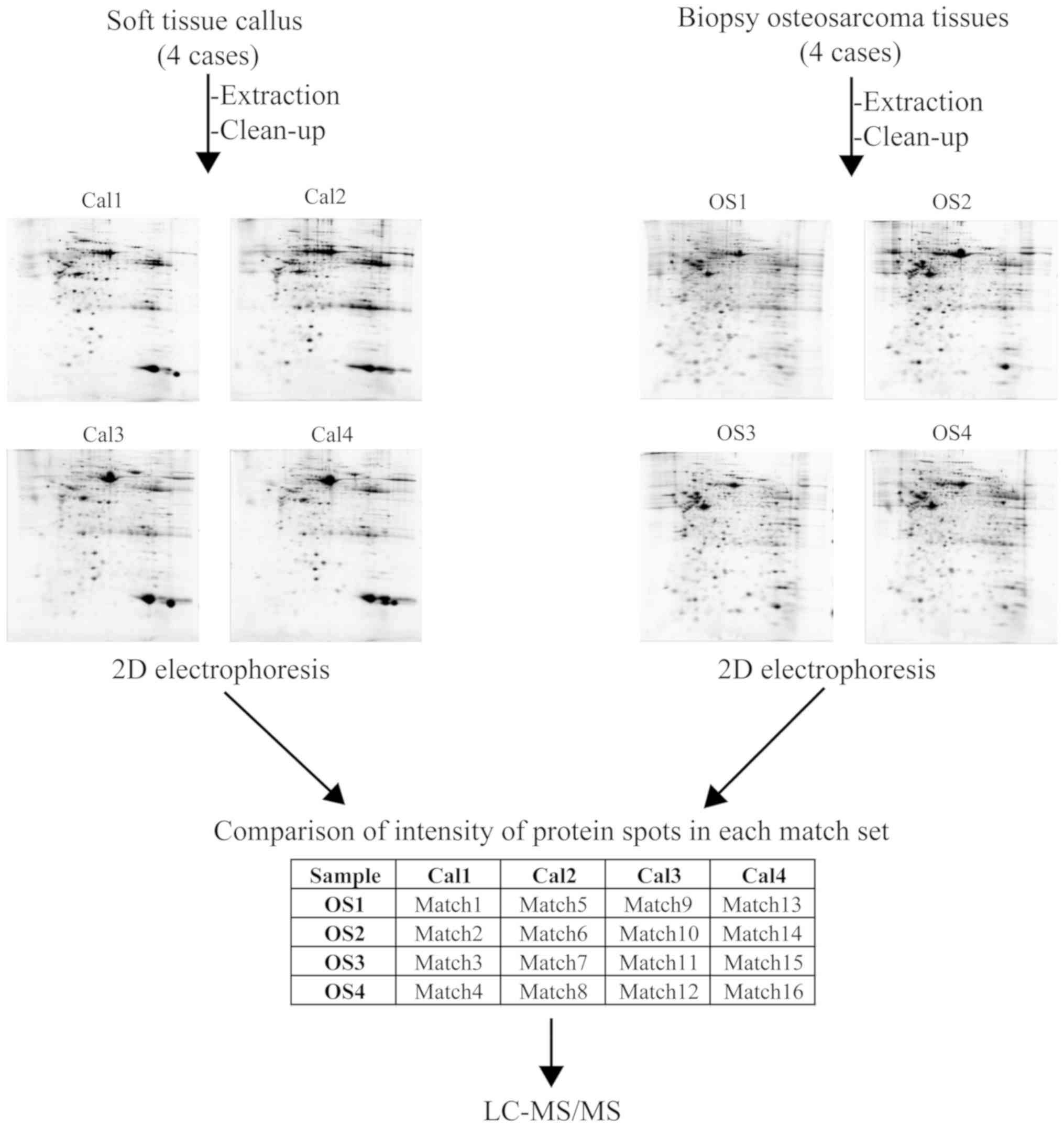
In the protein profiling of the chemonaïve tissues
derived from patients with osteosarcoma (4 cases; OS1-4) and soft
tissue callus from donors (4 cases; Cal1-4), 2DE was used to
analyze the protein extracted from the individual samples. The
characteristics of all patients and donors are presented in
Table I. The protein spots on the
individual 2DE gels from the osteosarcoma tissues and soft tissue
callus were matched, generating a total of 16 match sets (Fig. 2).
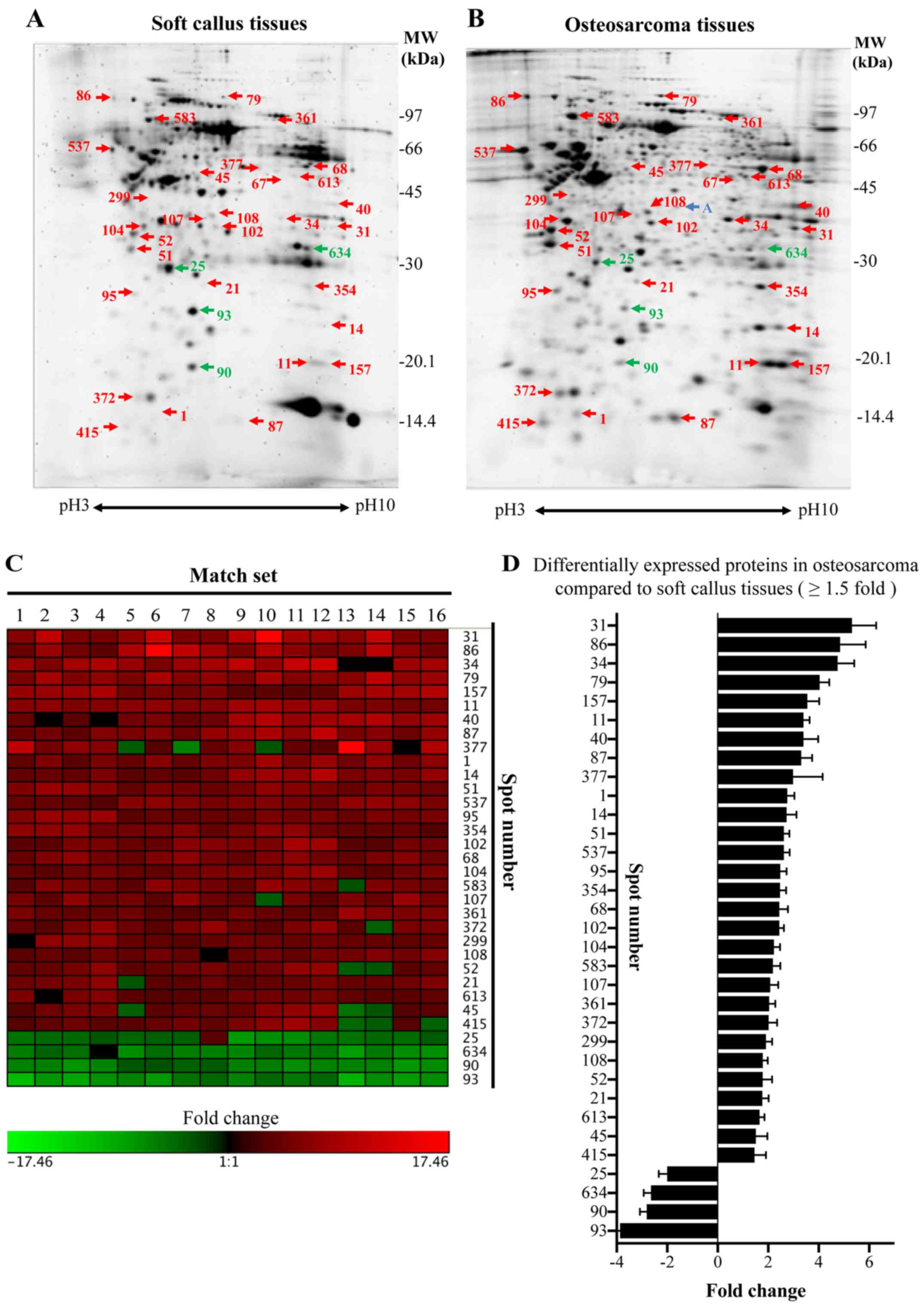
A total of 329 protein spots were perfectly matched
between the osteosarcoma and soft callus tissues. Overall, 28
proteins were upregulated and 4 proteins were downregulated in
osteosarcoma compared with soft tissue callus (≥1.5-fold change,
P<0.05; Fig. 3A and B). One
protein spot was specifically present in the osteosarcoma tissues
but not in the soft callus tissues. The differences between the
volume of each protein spot in the osteosarcoma and soft callus
tissues were mostly consistent among the match sets (Fig. 3C). The average fold changes of each
protein spot are illustrated in Fig.
3D. All protein spots with ≥1.5-fold change were cut, digested
with trypsin, and subjected to LC-MS/MS analysis. The protein
identification is presented in Table
II.
 | Table IIIdentification of differentially
expressed proteins in osteosarcoma tissues compared with soft
callus tissues. |
Table II
Identification of differentially
expressed proteins in osteosarcoma tissues compared with soft
callus tissues.
| Spot no.a | Accession no. | Name | Gene | MW/pIb | Scorec | Peptide
matchd | Coveragee(%) | Regulation | Fold changef (mean±standard error) |
|---|
| 1 | P58546 | Myotrophin | MTPN | 12.89/5.27 | 71 | 3 | 14 | Up | ↑2.76±0.28 |
| 11 | P62937 | Peptidyl-prolyl
cis-trans isomerase A, Cyclophilin | PPIA | 18.00/7.68 | 93 | 3 | 10 | Up | ↑3.40±0.24 |
| 14 | P30086 |
Phosphatidylethanolamine-binding protein
1 | PEBP1 | 21.04/7.01 | 45 | 2 | 8 | Up | ↑2.73±0.39 |
| 21 | P28070 | Proteasome subunit
β type-4 | PSMB4 | 29.18/5.72 | 79 | 2 | 7 | Up | ↑1.77±0.25 |
| 31 | P00338 | L-lactate
dehydrogenase A chain | LDHA | 36.66/8.44 | 69 | 4 | 11 | Up | ↑5.32±0.96 |
| 34 | P04083 | Annexin A1 | ANXA1 | 38.69/6.57 | 82 | 2 | 6 | Up | ↑4.75±0.66 |
| 40 | P04075 |
Fructose-bisphosphate aldolase A | ALDOA | 39.40/8.30 | 71 | 5 | 11 | Up | ↑3.40±0.58 |
| 45 | P31930 | Cytochrome b-c1
complex subunit 1 | UQCRC1 | 52.61/5.94 | 72 | 4 | 7 | Up | ↑1.51±0.46 |
| | | | | | | | | |
| 51 | Q04917 | 14-3-3 protein
eta | YWHAH | 28.20/4.76 | 101 | 5 | 17 | Up | ↑2.62±0.22 |
| 52 | P67936 | Tropomyosin α-4
chain | TPM4 | 28.50/4.67 | 124 | 9 | 26 | Up | ↑1.78±0.37 |
| 68 | P06733 | α-enolase | ENO1 | 47.14/7.01 | 177 | 9 | 20 | Up | ↑2.44±0.18 |
| 79 | Q14697 | Neutral
α-glucosidase AB | GANAB
106.81/5.74 | | 97 | 6 | 5 | Up | ↑4.04±0.38 |
| 86 | P14625 | Endoplasmin | HSP90B1 | 92.41/4.76 | 43 | 2 | 2 | Up | ↑4.85±1.01 |
| 87 | P31949 | Protein
S100-A11 | S100A11 | 11.73/6.56 | 50 | 2 | 17 | Up | ↑3.31±0.43 |
| 95 | P28072 | Proteasome subunit
β type-6 | PSMB6 | 25.34/4.80 | 70 | 1 | 10 | Up | ↑2.48±0.25 |
| 102 | P09525 | Annexin A4 | ANXA4 | 35.86/5.84 | 71 | 5 | 12 | Up | ↑2.44±0.34 |
| 104 | P08758 | Annexin A5 | ANXA5 | 35.91/4.94 | 57 | 4 | 10 | Up | ↑2.23±0.24 |
| 107 | P47755 | F-actin-capping
protein subunit α-2 | CAPZA2 | 32.93/5.57 | 74 | 3 | 10 | Up | ↑2.08±0.32 |
| 108 | P07195 | L-lactate
dehydrogenase B chain | LDHB | 36.62/5.71 | 253 | 10 | 29 | Up | ↑1.79±0.19 |
| 157 | P17742 | Peptidyl-prolyl
cis-trans isomerase A, Cyclophilin A | PPIA | 18.00/7.68 | 43 | 1 | 5 | Up | ↑3.55±0.47 |
| 299 | Q9Y3F4 | Serine-threonine
kinase receptor-associated protein | STRAP | 38.41/4.98 | 69 | 5 | 14 | Up | ↑1.91±0.25 |
| 354 | P51157 | Ras-related protein
Rab-28 | RAB28 | 24.83/5.70 | 49 | 1 | 4 | Up | ↑2.47±0.24 |
| 361 | P02545 | Prelamin-A/C | LMNA | 74.10/6.57 | 255 | 12 | 14 | Up | ↑2.04±0.24 |
| 372 | P10599 | Thioredoxin | TXN | 11.73/4.82 | 84 | 2 | 20 | Up | ↑2.02±0.33 |
| 377 | P26641 | Elongation factor
1-γ | EEF1G | 50.09/6.25 | 51 | 3 | 5 | | ↑2.99±1.17 |
| 415 | Q9H299 | SH3 domain binding
glutamic acid-rich-like protein 3 | SH3BGRL3 | 10.43/4.82 | 46 | 1 | 10 | Up | ↑1.46±0.45 |
| 537 | P27797 | Calreticulin | CALR | 48.11/4.29 | 68 | 5 | 12 | Up | ↑2.62±0.23 |
| 583 | P11021 | 78 kDa
glucose-regulated protein | HSPA5 | 72.29/5.07 | 64 | 9 | 13 | Up | ↑2.19±0.29 |
| 613 | O75874 | Isocitrate
dehydrogenase [NADP] cytoplasmic | IDH1 | 46.63/6.53 | 60 | 3 | 7 | Up | ↑1.66±0.19 |
| 25 | P02647 | Apolipoprotein
A-I | APOA1 | 30.76/5.56 | 147 | 10 | 32 | Down | ↑2.00±0.33 |
| 90 | P02766 | Transthyretin | TTR | 15.88/5.52 | 42 | 1 | 8 | Down | ↑2.81±0.27 |
| 93 | P32119 |
Peroxiredoxin-2 | PRDX2 | 21.88/5.66 | 113 | 7 | 32 | Down | ↑3.87±0.45 |
| 634 | Q15404 | Ras suppressor
protein 1 | RSU1 | 31.52/8.57 | 64 | 1 | 3 | Down | ↑2.65±0.29 |
| A | P05388 | 60S acidic
ribosomal protein P0 | RPLP0
34.254/5.71 | | 48 | 1 | 3 | Present in
osteosarcoma | - |
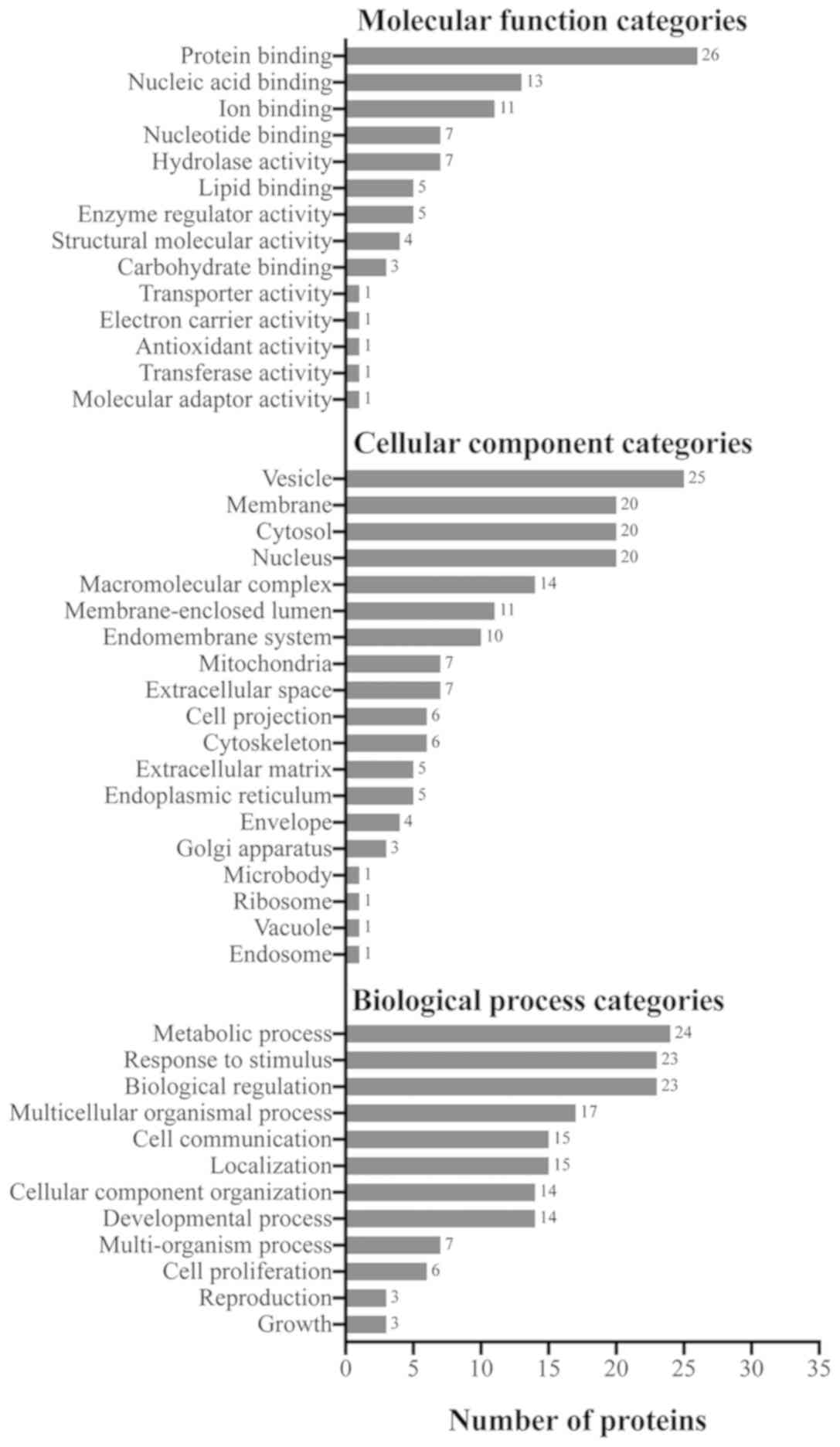
Enrichment analysis of GO terms of the
upregulated proteins in osteosarcoma
The WebGestalt web tool was used to determine the
enriched biological processes, cellular components and molecular
functions of the upregulated proteins in osteosarcoma. Of the 29
upregulated proteins, the majority were involved in metabolic
processes, biological regulation, and response to stimulus
(Fig. 4). Major localizations of
the upregulated proteins included the secretory vesicle, nucleus,
cytosol and membrane (Fig. 4).
Furthermore, the majority of the observed proteins appear to have
functions in protein binding (Fig.
4).
Pathway analysis of the upregulated
proteins in osteosarcoma
To further investigate the functionally grouped
networks of upregulated proteins in osteosarcoma, the ClueGO and
the CluePedia plugins of Cytoscape were used to identify the
enriched pathways. ClueGO was used to integrate biological
processes, molecular functions, Reactome, Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathways and WikiPathways, and to
visualize a functionally grouped network. CluePedia was used in
conjunction with ClueGO for comprehensive analysis of pathways
relating to protein-protein interactions and functions. With the κ
score level set at ≥0.3, 22 terms were found to be connected by 65
edges. Only the statistically significant (P<0.05) enrichment
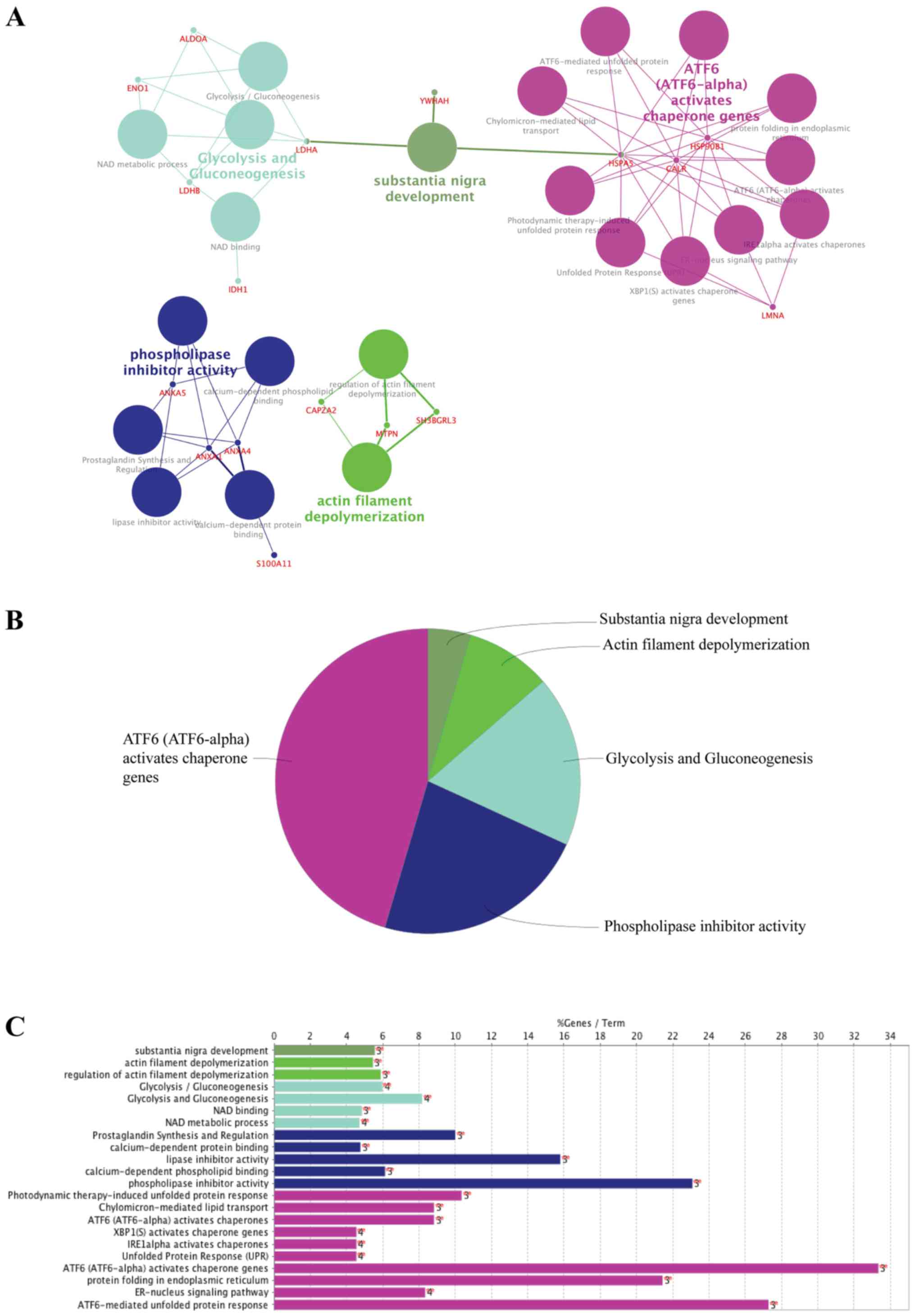
terms are presented in Fig. 5A.
The most significant functional groups included 'activating
transcription factor 6 (ATF6; ATF6-alpha) activates chaperone
genes', 'phospholipase inhibitor activity', 'glycolysis and
gluconeogenesis', 'actin filament depolymerization' and
'substantial nigra development' (Fig.
5B). All GO and pathway terms along with the % of genes
associated with the upregulated proteins in osteosarcoma are
presented in Fig. 5C.
Expression of important markers of the
unfolded protein response (UPR) pathway is elevated in osteosarcoma
cells
The results from the proteomics study demonstrated
elevated levels of a number of proteins involved in the UPR
pathway. The expression profiles of these proteins, including 78
kDa glucose-regulated protein (GRP78), endoplasmin (GRP94),
calreticulin (ERp60) and prelamin-A/C, were validated in
osteosarcoma cell lines and in the patient-derived osteosarcoma
cells using western blot analysis. As illustrated in Fig. 6A, higher expression levels of
GRP78, GRP94 and prelamin-A/C were observed in all osteosarcoma
cell lines examined, including MNNG/HOS, 143B, U2OS, Saos-2 and
MG-63, compared with the osteoblastic cells hFOB 1.19. A similar
trend was also observed in the primary osteosarcoma cells; GRP78,
GRP94 and prelamin-A/C were significantly upregulated in the
primary osteosarcoma cells (6 cases) compared with the primary
osteoblastic cells (5 cases), as illustrated in Fig. 6B. The expression levels of
calreticulin were slightly altered in all of the osteosarcoma cell
lines and primary cells, but these results were not as
significant.
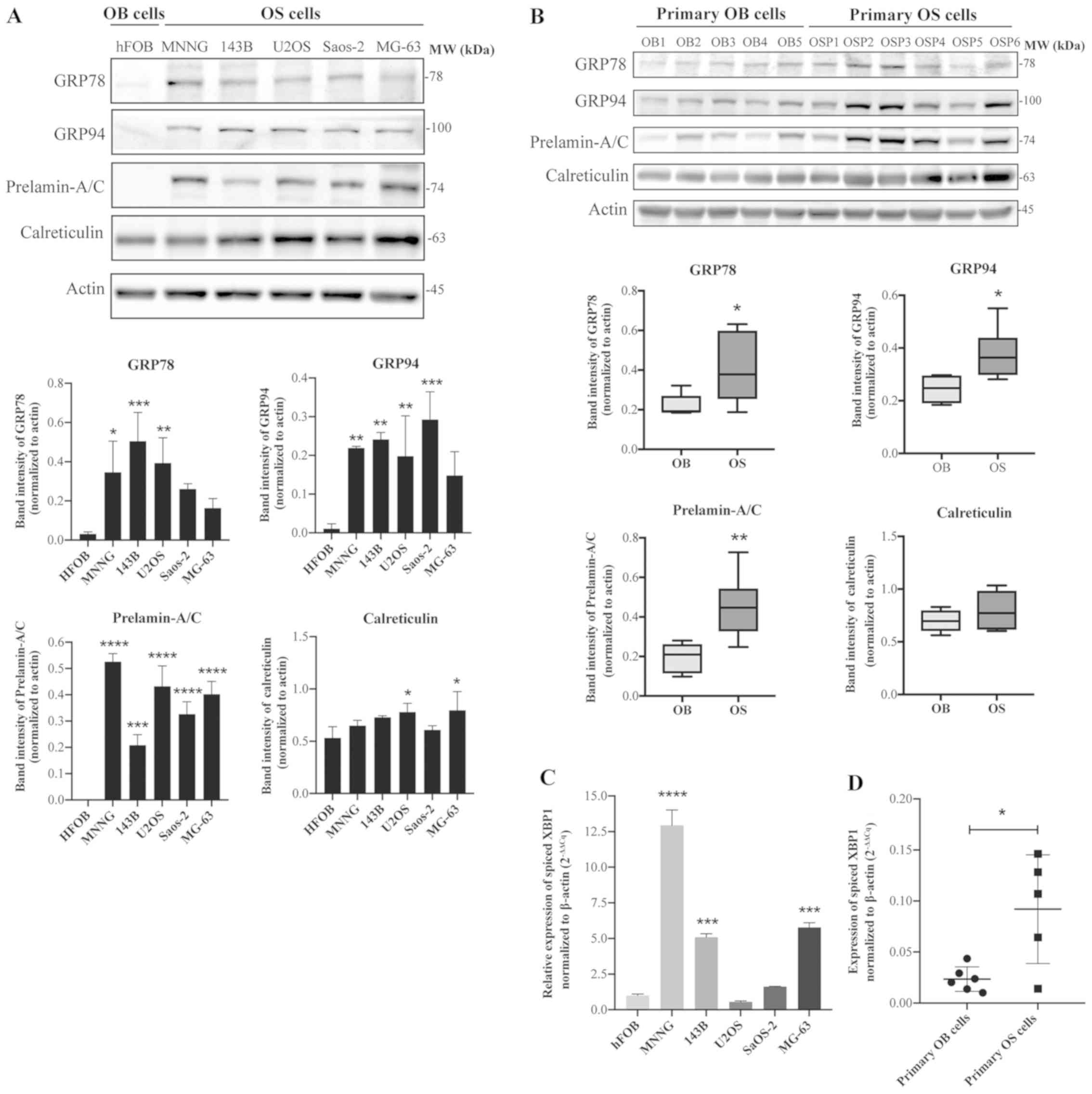 | Figure 6Expression of unfolded protein
response-related proteins is upregulated in osteosarcoma cells.
Immunoblots of GRP78, GRP94, calreticulin and prelamin-A/C in (A)
osteosarcoma cell lines (MNNG/HOS, 143B, U2OS, Saos-2 and MG-63)
compared with the osteoblastic cells (hFOB 1.19), and (B) primary
osteosarcoma cells (OSP1-6) compared with primary osteoblastic
cells (OB1-5). (C) Reverse transcription-quantitative polymerase
chain reaction analysis of spliced XBP1 expression levels in
osteoblastic cell line vs. osteosarcoma cell lines and (D) primary
osteoblastic cells vs. primary osteosarcoma cells. Data are
presented as means ± standard deviation from three independent
experiments. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 compared to
osteoblasts. GRP78, 78 kDa glucose-related protein; GRP94,
endoplasmin; OSP, primary osteosarcoma cell; OB, primary
osteoblastic cell; XBP1, X-box binding protein 1. |
Furthermore, to examine the induction of ER stress
and the UPR pathway in osteosarcoma cells, the expression levels of
the spliced form of XBP1 mRNA (XBP1s) were measured using RT-qPCR
and specific primers to XBP1s mRNA (22). The results demonstrated that XBP1s
expression levels were significantly upregulated in most
osteosarcoma cells compared with osteoblastic cells (Fig. 6C). Similar results were observed
with the patient samples, with primary osteosarcoma cells
exhibiting a significantly elevated expression of XBP1s compared
with primary osteoblastic cells (Fig.
6D).
Association of UPR-related protein
expression levels with clinical outcomes in osteosarcoma
The expression of the four identified proteins was
further validated in independent sets of tissues samples that were
not used in the original discovery set of the proteomics study. In
addition, the expression patterns of these proteins were further
investigated in osteosarcoma tissues from patients with different
stages of disease, as follows: patients with stage IIB who were
sensitive to chemotherapy or good responders (GR; tumor necrosis
≥90%); patients with stage IIB who responded poorly to chemotherapy
(PR; tumor necrosis <90%); and patients with stage III and
initial metastasis (Met). The results demonstrated that the
expression of GRP94 was upregulated in the tissues of patients with
non-metastasis (stage IIB) and metastasis (stage III) compared with
the soft callus controls (Fig. 7A and
B). Calreticulin was significantly overexpressed in the
patients with stage II. Prelamin-A/C had a tendency towards
overexpression in the osteosarcoma tissues. The expression of GRP78
did not differ in the patients with osteosarcoma compared with the
soft callus when the patients were classified according to the
Enneking staging system. Notably, by considering the expression of
GRP78 protein in relation to the chemosensitivity of patients who
had received the same chemotherapy regimen (DOX and platinum-based
chemotherapy), the results revealed that GRP78 was markedly
upregulated in the PR group compared with the soft callus and the
GR groups (Fig. 7C).
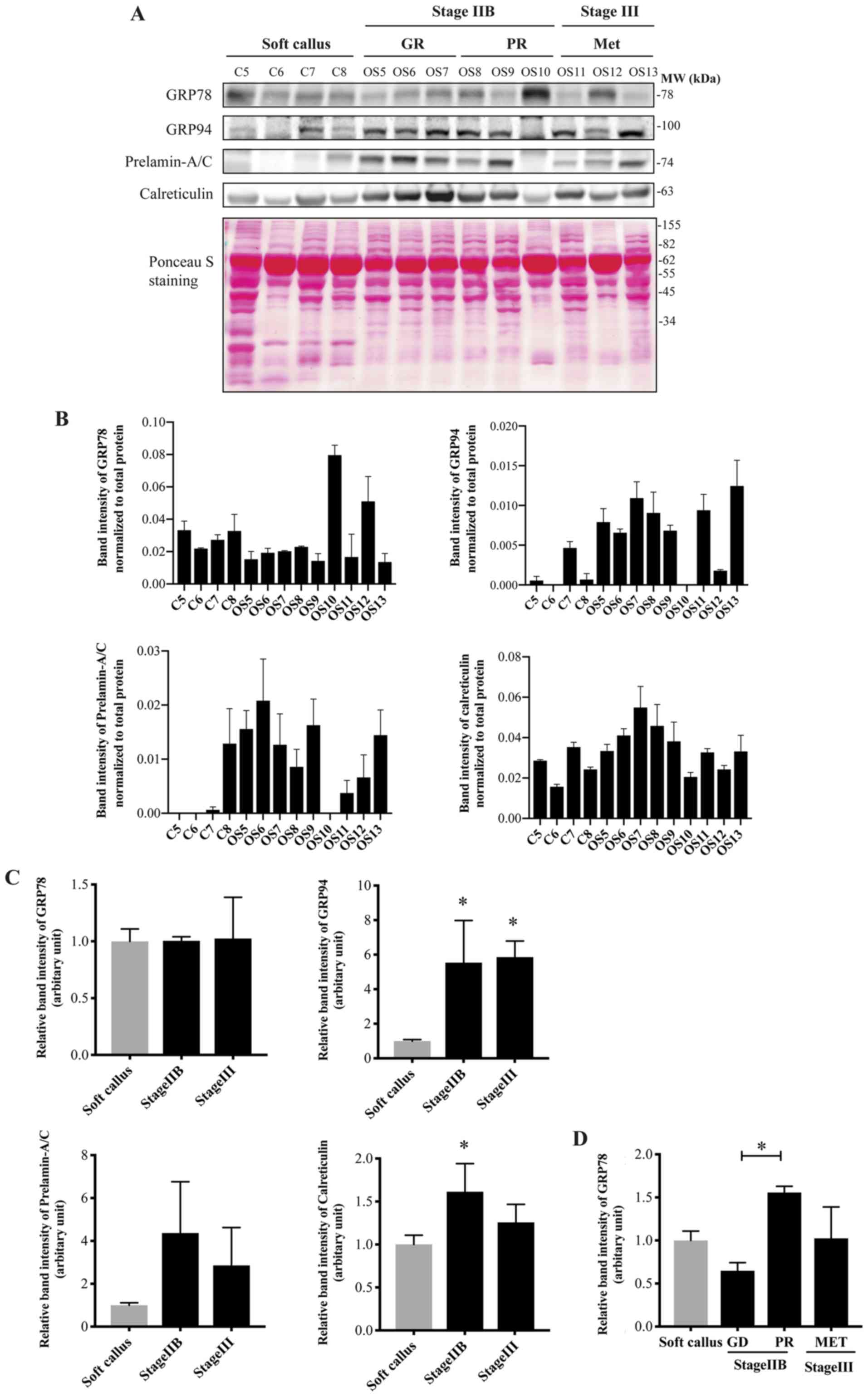 | Figure 7Expression of UPR-related proteins in
soft callus (C5-C8) and osteosarcoma tissues (OS5-OS13), including
patients with stage IIB (6 cases) and stage III (3 cases) disease.
(A) Western blotting of GRP78, GRP94, prelamin-A/C and calreticulin
in soft callus and osteosarcoma tissues. Total proteins, used as a
loading control, were evaluated using Ponceau S staining. (B) Bar
graphs demonstrating the levels of the UPR-related proteins
normalized to total protein in individual samples. (C and D)
Quantification and statistical analysis of the levels of the tested
proteins in the soft callus and osteosarcoma tissues. Data are
presented as means ± standard deviation from three independent
experiments. *P<0.05 compared to the soft callus, or
as indicated by brackets. UPR, unfolded protein response; GRP78, 78
kDa glucose-related protein; GRP94, endoplasmin; GR, good
responders; PR, poor responders; Met, metastatic; C, soft callus
control; OS, osteosarcoma. |
Discussion
Fresh frozen tissue is the most biologically
representative clinical specimen and is compatible with proteomics
analysis. In the present study, upregulated proteins in biopsy
tissues from patients with osteosarcoma were identified using 2DE
and LC-MS/MS. Soft tissue callus was used to represent
non-cancerous tissue. The formation of soft callus involves
chondroblasts and osteoblasts and commonly occurs during the
fracture repair process (24,25).
In the present study, histological evaluation of osteoblastic
morphology in the soft callus was conducted. The results revealed
high osteoblastic cell content in the soft callus, suggesting that
soft callus is a relevant source of osteoblasts that can represent
non-cancerous tissues for proteomics study of osteosarcoma.
The results of the 2DE and LC-MS/MS in the present
study revealed significant changes in the expression levels of
various proteins from the osteosarcoma tissues compared with the
soft callus. Using bioinformatics tools, the significantly enriched
network groups identified in osteosarcoma included 'ATF6
(ATF6-alpha) activates chaperone genes', 'phospholipase inhibitor
activity', 'glycolysis and gluconeogenesis', 'actin filament
depolymerization' and 'substantial nigra development'. Among these
functional groups, the majority of the upregulated proteins,
including GRP78 (also known as BiP or HSP70 family protein 5),
GRP94 (also known as heat shock protein 90 kDa β member 1) and
calreticulin (also known as ERp60 or GRP60), were related to the
'ATF6 (ATF6-alpha) activates chaperone genes' group, which is one
of the three major endoplasmic reticulum (ER) stress sensors
controlling the UPR pathway. In addition, elevated levels of
prelamin-A/C were observed, which is a protein related to the XBP1
arm of the UPR pathway. The findings from the proteomics study were
further validated by investigation of the levels of these
UPR-related proteins in various types of samples, including
osteosarcoma cell lines, patient-derived primary cells and an
independent set of tissue samples. These results substantiated the
data from the proteomics study and demonstrated that all of the
proteins were upregulated in osteosarcoma.
The present study revealed overexpression of three
key ER chaperone proteins, including two classical UPR markers,
GRP78 and GRP94, in addition to a well-characterized chaperone,
calreticulin. Accumulating evidence has demonstrated that elevated
expression of these proteins in a wide range of cancer types
results from activation of the UPR (26). Under intrinsic perturbations and
hostile environments, cancer cells restore protein homeostasis
(proteostasis), which is one of the hallmarks of cancer, in order
to promote cancer cell transformation and progression (27,28).
The intrinsic factors include oncogenic activation, alteration in
chromosome number, genomic instability, high secretion levels of
proteins, and high levels of protein production (29,30).
Among the extrinsic factors are hypoxia, nutrient deprivation,
acidosis, altered cell metabolism, hyperproliferation and
hypoglycemia (31). These factors
induce ER stress, which is the result of an accumulation of
misfolded proteins in the ER lumen (32). In response to ER stress, cancer
cells activate the UPR, an adaptive mechanism to recover the ER
protein-folding environment and proteostasis that assist cancer
cell survival (31). A large
amount of evidence suggests that activation of the UPR serves
important roles in numerous aspects of cancer development and
progression, including cell survival, angiogenesis, metastasis,
resistance to treatment, adaptation, transformation and tumor
dormancy (33). A diagram of the
UPR pathway is provided in Fig.
8.
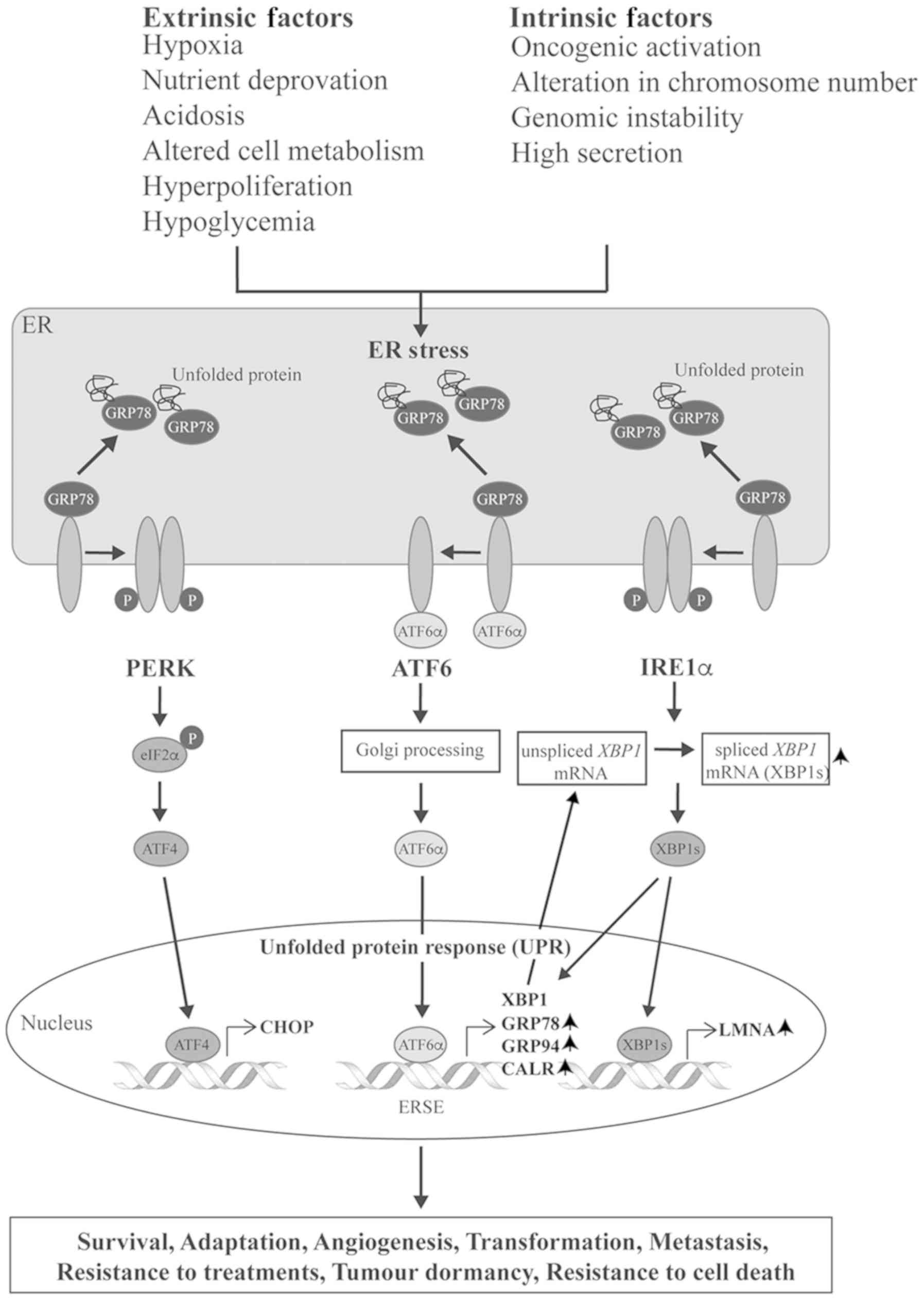 | Figure 8Diagram demonstrating the unfolded
protein response pathway and downstream chaperone proteins.
Upwards-pointing arrows next to protein names indicate upregulated
proteins in osteosarcoma. ER, endoplasmic reticulum; GRP78, 78 kDa
glucose-related protein; P, phosphorylated; PERK, PRKR-like ER
kinase; ATF, activating transcription factor; IRE,
inositol-requiring enzyme; CHOP, C/EBP homologous protein; XBP1,
X-box binding protein 1; GRP94, endoplasmin; CALR, calreticulin;
LMNA, lamin A/C. |
GRP78 is the ER master chaperone, which acts as a
major regulator of the UPR (34).
In non-stressed conditions, GRP78 inactivates the UPR by binding to
all three ER stress inducers, PRKR-like ER kinase (PERK),
inositol-requiring enzyme 1 (IRE1) and ATF6 (35). Upon ER stress, GRP78 activates the
UPR pathways by dissociating from the stress inducers and then
binding to misfolded proteins, which accumulate in the ER (Fig. 8). Free ATF6 subsequently triggers
downstream signaling to increase ER protein folding capacity and
ER-associated protein degradation through the transcription of
various related genes that encode for the chaperone proteins,
including GRP78, GRP94, calreticulin and XBP1 (34,36–38).
As a consequence, during tumorigenesis, expression of GRP78 and
other ER chaperones is enhanced by the UPR. Notably, Luo et
al (39) demonstrated that
inhibition of GRP78 upregulates ATF4 and the pro-apoptotic factor
C/EBP homologous protein (CHOP) and sensitizes osteosarcoma cells
to bortezomib, an FDA-approved proteasome inhibitor. Luo et
al (39) also reported that
upon ER stress induction, elevated expression of GRP78 protects
against osteosarcoma cell death through diminishing the activity of
CHOP by promoting ubiquitinating degradation of CHOP protein.
Furthermore, the present study revealed an
upregulation of the spliced form of XBP1 mRNA, XBP1s, in
osteosarcoma cells compared with osteoblastic cells. This finding
confirmed that the ATF6 pathway was induced in osteosarcoma,
because it is well known that expression of XBP1 mRNA is a
downstream target of ATF6 and XBP1s (Fig. 8) (38). Significant levels of XBP1 mRNA are
essential for subsequent splicing processes activated by the
IRE1α-dependent pathway (40). The
spliced form of XBP1 is a more stable and potent transcription
activator than the un-spliced XBP1 (XBP1u). As a consequence, XBP1s
effectively induces transcription of various UPR-related genes and
chaperones.
Not only was GRP78 overexpressed in osteosarcoma in
the present study, elevated levels of GRP78 were observed in
chemonaïve tissues of patients with osteosarcoma who responded
poorly to chemotherapy compared with good responders, suggesting an
association of GRP78 with intrinsic drug resistance. Accumulating
evidence has demonstrated an association between GRP78 and
pre-existing resistance mechanisms in various types of cancer. For
example, a high expression level of GRP78 in chemonaïve cancer
tissues has been identified to be associated with subsequent
development of chemoresistance (41,42).
Furthermore, an enhanced level of GRP78 has been demonstrated to be
strongly correlated with an acquired chemoresistance mechanism in
cancer (43,44). Notably, Xia et al (45) demonstrated the role of GRP78 in
acquired chemoresistance of osteosarcoma cell lines; DOX treatment
induced P-glycoprotein (P-gp) and GRP78 expression, as well as
activation of the serine/threonine kinase Akt, both in vitro
and in vivo. The results also revealed an important role of
GRP78 in full Akt activity, in which GRP78 activates Akt and
enhances Akt-mediated P-gp expression in response to DOX treatment.
In addition, DOX-resistant osteosarcoma lines that express higher
levels of P-gp markedly stimulate the resistance mechanism.
Overexpression of proteins involved with the UPR
pathway, particularly in the ATF6 arm, suggest that the activation
of ATF6 signaling may contribute to oncogenesis events in
osteosarcoma. Schewe and Aguirre-Ghiso (46) reported a potential role of ATF6 in
tumor dormancy and chemoresistance. They demonstrated that
transcription factor ATF6α is an essential survival factor
exclusively for quiescent cells. Silencing of the ATF6 gene reduces
survival and resistance to rapamycin in dormant squamous carcinoma
cells in vivo. The findings of the present study suggest
that upregulation of important proteins in the ATF6 pathway may
contribute to chemoresistance and tumor dormancy phenotypes in
osteosarcoma cells. These findings also suggest a possible strategy
for targeting UPR components for the treatment of osteosarcoma.
One of the most promising strategies is to use an
agent that augments ER stress, shifting it from an adaptive to an
apoptotic mechanism. Prolonged activation of UPR signals can
trigger pro-apoptotic events that eliminate damaged cells.
Proteasome inhibitors are key players in this respect, since the
attenuation of proteasome activity effectively induces apoptosis in
UPR-dependent tumors through various mechanisms (29). Prevention of proteasomal
degradation results in an overwhelming accumulation of misfolded
proteins in the cytosol, which induces ER stress, overcomes the UPR
adaptive mechanism, and ultimately triggers apoptosis of cells. The
antitumor effect of bortezomib, a FDA-approved proteasome inhibitor
for the treatment of multiple myeloma, has been tested in
osteosarcoma (47). Bortezomib was
demonstrated to inhibit cell growth through the apoptosis pathway
both in vitro and in vivo. A phase II clinical trial
of bortezomib was conducted in patients with advanced or metastatic
sarcoma, including adult patients with osteosarcoma (NCT00027716).
Direct inhibition of GRP78 is another promising approach. In a
phase I clinical trial, the GRP78 monoclonal immunoglobulin M
antibody PAT-SM6 was used as a single agent in relapsed or
refractory multiple myeloma (48).
The results revealed that PAT-SM6 was well tolerated with modest
clinical activity in the patients.
Limitations of the present study are the rarity of
this type of tumor and the small number of patients. Future work
will need to be performed in a larger cohort of osteosarcoma
patients to validate these findings of association of UPR-related
proteins and chemo-resistance.
In conclusion, the present study identified a number
of pathways that are aberrantly regulated in osteosarcoma. The most
significant one is an activation of the adaptive mechanism, the
UPR, induced by ER stress. This finding suggests a potential role
for therapeutic agents that target the UPR pathway in the treatment
of osteosarcoma, inducing a shift to pro-apoptotic events and/or
acting on specific key proteins.
Funding
This study was supported by the National Science and
Technology Development Agency (grant no. P-15-50265), the Faculty
of Medicine Chiang Mai University, the National Research University
fund, and the Musculoskeletal Science and Translational Research
Center.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
DP and PC designed the experiments. JSe performed
the histological evaluation of osteosarcoma tissue and soft tissue
callus. PS and PD performed the experiments in 2DE. NS performed
the experiments in western blotting analysis. PT and JK performed
primary cell extraction and characterization. DC performed the
LC-MS/MS. PS and PT analyzed the proteomics data. PC performed
network analysis, generated the data, and assembled the figures and
tables. CS and JSv reviewed and edited the manuscript. DP and PC
wrote the manuscript. All authors have reviewed and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The protocol involving human clinical samples was
approved by the Ethics Committee of the Faculty of Medicine, Chiang
Mai University. Written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would also like to express their sincere
thanks to Dr G. Lamar Robert, and Associate Professor Dr Chongchit
Sripun Robert, for critical review of our manuscript, and Areerak
Phanphaisarn for help with for statistical analysis. Part of this
study was presented at the 9th Asia-Oceania Human Proteome
Organization (AOHUPO) Conference (2018), as a poster presentation
with interim findings.
References
|
1
|
Settakorn J, Lekawanvijit S,
Arpornchayanon O, Rangdaeng S, Vanitanakom P, Kongkarnka S,
Cheepsattayakorn R, Ya-In C and Thorner PS: Spectrum of bone tumors
in Chiang Mai University Hospital, Thailand according to WHO
classification 2002: A study of 1,001 cases. J Med Assoc Thai.
89:780–787. 2006.PubMed/NCBI
|
|
2
|
Friebele JC, Peck J, Pan X, Abdel-Rasoul M
and Mayerson JL: Osteosarcoma: A meta-analysis and review of the
literature. Am J Orthop (Belle Mead NJ). 44:547–553. 2015.
|
|
3
|
Whelan JS, Bielack SS, Marina N, Smeland
S, Jovic G, Hook JM, Krailo M, Anninga J, Butterfass-Bahloul T,
Böhling T, et al: EURAMOS collaborators: EURAMOS-1, an
international randomised study for osteosarcoma: Results from
pre-randomisation treatment. Ann Oncol. 26:407–414. 2015.
View Article : Google Scholar
|
|
4
|
Chen X, Bahrami A, Pappo A, Easton J,
Dalton J, Hedlund E, Ellison D, Shurtleff S, Wu G, Wei L, et al St
Jude Children's Research Hospital-Washington University Pediatric
Cancer Genome Project: Recurrent somatic structural variations
contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep.
7:104–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Botter SM, Neri D and Fuchs B: Recent
advances in osteosarcoma. Curr Opin Pharmacol. 16:15–23. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chaiyawat P, Pruksakorn D, Phanphaisarn A,
Teeyakasem P, Klangjorhor J and Settakorn J: Expression patterns of
class I histone deacetylases in osteosarcoma: A novel prognostic
marker with potential therapeutic implications. Mod Pathol.
31:264–274. 2018. View Article : Google Scholar :
|
|
7
|
Xie C, Wu B, Chen B, Shi Q, Guo J, Fan Z
and Huang Y: Histone deacetylase inhibitor sodium butyrate
suppresses proliferation and promotes apoptosis in osteosarcoma
cells by regulation of the MDM2-p53 signaling. Onco Targets Ther.
9:4005–4013. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Watanabe K, Okamoto K and Yonehara S:
Sensitization of osteo-sarcoma cells to death receptor-mediated
apoptosis by HDAC inhibitors through downregulation of cellular
FLIP. Cell Death Differ. 12:10–18. 2005. View Article : Google Scholar
|
|
9
|
Rao-Bindal K, Koshkina NV, Stewart J and
Kleinerman ES: The histone deacetylase inhibitor, MS-275
(entinostat), downregulates c-FLIP, sensitizes osteosarcoma cells
to FasL, and induces the regression of osteosarcoma lung
metastases. Curr Cancer Drug Targets. 13:411–422. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu S, Denman CJ, Cobanoglu ZS, Kiany S,
Lau CC, Gottschalk SM, Hughes DP, Kleinerman ES and Lee DA: The
narrow-spectrum HDAC inhibitor entinostat enhances NKG2D expression
without NK cell toxicity, leading to enhanced recognition of cancer
cells. Pharm Res. 32:779–792. 2015. View Article : Google Scholar
|
|
11
|
Li Y, Liang Q, Wen YQ, Chen LL, Wang LT,
Liu YL, Luo CQ, Liang HZ, Li MT and Li Z: Comparative proteomics
analysis of human osteosarcomas and benign tumor of bone. Cancer
Genet Cytogenet. 198:97–106. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rao UN, Hood BL, Jones-Laughner JM, Sun M
and Conrads TP: Distinct profiles of oxidative stress-related and
matrix proteins in adult bone and soft tissue osteosarcoma and
desmoid tumors: A proteomics study. Hum Pathol. 44:725–733. 2013.
View Article : Google Scholar
|
|
13
|
Einhorn TA and Gerstenfeld LC: Fracture
healing: Mechanisms and interventions. Nat Rev Rheumatol. 11:45–54.
2015. View Article : Google Scholar :
|
|
14
|
Murao H, Yamamoto K, Matsuda S and Akiyama
H: Periosteal cells are a major source of soft callus in bone
fracture. J Bone Miner Metab. 31:390–398. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han W, He W, Yang W, Li J, Yang Z, Lu X,
Qin A and Qian Y: The osteogenic potential of human bone callus.
Sci Rep. 6:363302016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Srisomsap C, Sawangareetrakul P,
Subhasitanont P, Panichakul T, Keeratichamroen S, Lirdprapamongkol
K, Chokchaichamnankit D, Sirisinha S and Svasti J: Proteomic
analysis of cholangiocarcinoma cell line. Proteomics. 4:1135–1144.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Herosimczyk A, Dejeans N, Sayd T, Ozgo M,
Skrzypczak WF and Mazur A: Plasma proteome analysis: 2D gels and
chips. J Physiol Pharmacol. 57(Suppl 7): 81–93. 2006.
|
|
18
|
Zhang B, Kirov S and Snoddy J: WebGestalt:
An integrated system for exploring gene sets in various biological
contexts. Nucleic Acids Res. 33:W741–W748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lotia S, Montojo J, Dong Y, Bader GD and
Pico AR: Cytoscape app store. Bioinformatic s. 29:1350–1351. 2013.
View Article : Google Scholar
|
|
21
|
Pruksakorn D, Teeyakasem P, Klangjorhor J,
Chaiyawat P, Settakorn J, Diskul-Na-Ayudthaya P, Chokchaichamnankit
D, Pothacharoen P and Srisomsap C: Overexpression of KH-type
splicing regulatory protein regulates proliferation, migration, and
implantation ability of osteosarcoma. Int J Oncol. 49:903–912.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
van Schadewijk A, van't Wout EF, Stolk J
and Hiemstra PS: A quantitative method for detection of spliced
X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic
reticulum (ER) stress. Cell Stress Chaperones. 17:275–279. 2012.
View Article : Google Scholar :
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Schindeler A, McDonald MM, Bokko P and
Little DG: Bone remodeling during fracture repair: The cellular
picture. Semin Cell Dev Biol. 19:459–466. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mörike M, Schulz M, Nerlich A, Koschnik M,
Teller WM, Vetter U and Brenner RE: Expression of osteoblastic
markers in cultured human bone and fracture callus cells. J Mol Med
(Berl). 73:571–575. 1995. View Article : Google Scholar
|
|
26
|
Luo B and Lee AS: The critical roles of
endoplasmic reticulum chaperones and unfolded protein response in
tumorigenesis and anticancer therapies. Oncogene. 32:805–818. 2013.
View Article : Google Scholar
|
|
27
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Obacz J, Avril T, Le Reste PJ, Urra H,
Quillien V, Hetz C and Chevet E: Endoplasmic reticulum proteostasis
in glioblastoma-From molecular mechanisms to therapeutic
perspectives. Sci Signal. 10:102017. View Article : Google Scholar
|
|
29
|
Wang M and Kaufman RJ: The impact of the
endoplasmic reticulum protein-folding environment on cancer
development. Nat Rev Cancer. 14:581–597. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dejeans N, Manié S, Hetz C, Bard F, Hupp
T, Agostinis P, Samali A and Chevet E: Addicted to secrete - novel
concepts and targets in cancer therapy. Trends Mol Med. 20:242–250.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma Y and Hendershot LM: The role of the
unfolded protein response in tumour development: Friend or foe? Nat
Rev Cancer. 4:966–977. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hetz C: The unfolded protein response:
Controlling cell fate decisions under ER stress and beyond. Nat Rev
Mol Cell Biol. 13:89–102. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dufey E, Urra H and Hetz C: ER
proteostasis addiction in cancer biology:. Novel concepts Semin
Cancer Biol. 33:40–47. 2015. View Article : Google Scholar
|
|
34
|
Lee AS: Glucose-regulated proteins in
cancer: Molecular mechanisms and therapeutic potential. Nat Rev
Cancer. 14:263–276. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang M, Wey S, Zhang Y, Ye R and Lee AS:
Role of the unfolded protein response regulator GRP78/BiP in
development, cancer, and neurological disorders. Antioxid Redox
Signal. 11:2307–2316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chang SC, Erwin AE and Lee AS:
Glucose-regulated protein (GRP94 and GRP78) genes share common
regulatory domains and are coordinately regulated by common
trans-acting factors. Mol Cell Biol. 9:2153–2162. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamamoto K, Sato T, Matsui T, Sato M,
Okada T, Yoshida H, Harada A and Mori K: Transcriptional induction
of mammalian ER quality control proteins is mediated by single or
combined action of ATF6alpha and XBP1. Dev Cell. 13:365–376. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yoshida H, Matsui T, Yamamoto A, Okada T
and Mori K: XBP1 mRNA is induced by ATF6 and spliced by IRE1 in
response to ER stress to produce a highly active transcription
factor. Cell. 107:881–891. 2001. View Article : Google Scholar
|
|
39
|
Luo J, Xia Y, Luo J, Li J, Zhang C, Zhang
H, Ma T, Yang L and Kong L: GRP78 inhibition enhances ATF4-induced
cell death by the deubiquitination and stabilization of CHOP in
human osteosarcoma. Cancer Lett. 410:112–123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee K, Tirasophon W, Shen X, Michalak M,
Prywes R, Okada T, Yoshida H, Mori K and Kaufman RJ: IRE1-mediated
unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge
to regulate XBP1 in signaling the unfolded protein response. Genes
Dev. 16:452–466. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee E, Nichols P, Spicer D, Groshen S, Yu
MC and Lee AS: GRP78 as a novel predictor of responsiveness to
chemotherapy in breast cancer. Cancer Res. 66:7849–7853. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mhaidat NM, Alzoubi KH, Almomani N and
Khabour OF: Expression of glucose regulated protein 78 (GRP78)
determines colorectal cancer response to chemotherapy. Cancer
Biomark. 15:197–203. 2015. View Article : Google Scholar
|
|
43
|
Gifford JB, Huang W, Zeleniak AE, Hindoyan
A, Wu H, Donahue TR and Hill R: Expression of GRP78, master
regulator of the unfolded protein response, increases
chemoresistance in pancreatic ductal adenocarcinoma. Mol Cancer
Ther. 15:1043–1052. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li W, Wang W, Dong H, Li Y, Li L, Han L,
Han Z, Wang S, Ma D and Wang H: Cisplatin-induced senescence in
ovarian cancer cells is mediated by GRP78. Oncol Rep. 31:2525–2534.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xia YZ, Yang L, Xue GM, Zhang C, Guo C,
Yang YW, Li SS, Zhang LY, Guo QL and Kong LY: Combining GRP78
suppression and MK2206-induced Akt inhibition decreases
doxorubicin-induced P-glycoprotein expression and mitigates
chemoresistance in human osteosarcoma. Oncotarget. 7:56371–56382.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schewe DM and Aguirre-Ghiso JA:
ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor
cells in vivo. Proc Natl Acad Sci USA. 105:10519–10524. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shapovalov Y, Benavidez D, Zuch D and
Eliseev RA: Proteasome inhibition with bortezomib suppresses growth
and induces apoptosis in osteosarcoma. Int J Cancer. 127:67–76.
2010. View Article : Google Scholar
|
|
48
|
Rasche L, Duell J, Castro IC, Dubljevic V,
Chatterjee M, Knop S, Hensel F, Rosenwald A, Einsele H, Topp MS, et
al: GRP78-directed immunotherapy in relapsed or refractory multiple
myeloma - results from a phase 1 trial with the monoclonal
immunoglobulin M antibody PAT-SM6. Haematologica. 100:377–384.
2015. View Article : Google Scholar : PubMed/NCBI
|