Introduction
Lung cancer is one of the most common malignancies
globally, and is responsible for >1,700,000 estimated
mortalities in 2018 globally (1).
Lung adenocarcinoma (LA) is the most commonly occurring
histological type of lung cancer, accounting for ~40% of all lung
cancer cases in China in 2010 (2).
An investigation of the underlying molecular mechanisms and
clinical features of LA has resulted in the development of novel
markers and therapies for the diagnosis and treatment of different
stages of LA (3,4). However, numerous patients with LA are
diagnosed at the advanced disease stage, due to a lack of effective
approaches; therefore, the 5-year overall survival rate for
patients with LA remains low (3).
Currently, low-dose computed tomography is the recommended method
of choice for screening early-stage lung cancer in high-risk
populations, based on a previously published clinical study
(5). However, potential false
positive tests, over-diagnosis and psychosocial consequences limit
its clinical applicability (6).
The investigation of classical protein biomarkers, including
carcinoembryonic antigen and cytokeratin 19 fragments, are commonly
recommended in LA diagnosis and management (7). However, the sensitivity of these
tumor markers is dependent on the prevalence rates of lung cancer,
and the cut-off value differs in different populations (8,9).
Therefore, reliable and effective biomarkers in detecting
early-stage LA are urgently required.
Extracellular vesicles (EVs), which are released
from cells, are small membrane particles, ranging from 40 to 1,000
nm in diameter, and contain complex molecules, including nucleic
acids, proteins and lipids (10).
EVs are heterogeneous in size, density, subcellular origin and
function, and are classified into the main two subtypes of exosomes
and microvesicles (11). EVs,
particularly exosomes, are produced by almost all types of cells in
the human body, and are secreted into various human body fluids,
including blood, saliva, pleural effusion, urine and breast milk
(12). A previously published
study has demonstrated that abnormal cell-cell communication
contributes to carcinogenesis (13); furthermore, the microenvironment of
tumors is essential for their rapid proliferation and metastasis to
lymph nodes and organs (14). EVs
fulfill roles in tumor occurrence, development and survival
(15). Previous studies indicated
that abnormal microRNA (miRNA) levels identified in EVs from
patients with lung cancer, as well as cancer cell-delivered miRNAs
or proteins through EVs, promote neighboring cell proliferation and
migration (16,17). miRNAs form a series of regulators
that repress gene expression following transcription by binding
directly to the 3′-untranslated regions (UTRs) of target mRNAs
(18,19). For cells, the levels of endogenous
miRNAs may be changed when the microenvironment is altered
(19-21). Furthermore, altered miRNA profiles
have been implicated in human diseases, ranging from psychiatric
disorders to malignant tumors (22-24).
It is increasingly evident that miRNAs may be sorted into EVs,
particularly exosomes, in which they are protected from degradation
by ribonucleases; additionally, miRNAs serve important roles in
tumorigenesis. However, an assessment of the roles of miRNAs in EVs
remains in its infancy, and additional, more functional studies are
required in order to delineate the complex roles of dysregulated
miRNAs.
In the present study, patients with LA were
assessed, and a three-stage study was developed, which may be used
to examine relative plasma EV miRNA levels for the diagnosis of
early-stage LA. The quantities of plasma EV miRNAs were detected on
the basis of high-throughput sequencing and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Subsequently, the identified plasma EV miRNAs were verified in LA
tissue samples, and compared with those in peripheral plasma
samples. Additionally, an EV miRNA with altered levels was
identified in the patients with LA as a potential, specific and
minimally invasive biomarker for early clinical diagnosis of LA.
Furthermore, subsequent functional studies demonstrated that
miR-505-5p in EVs could be delivered into LA cells, inhibiting cell
apoptosis and promoting cell proliferation by targeting tumor
protein P53-regulated apoptosis-inducing protein 1 (TP53AIP1).
Materials and methods
Participants and samples
A total of 153 patients with LA (age range, 41-78
years; mean, 65.29 years; males, 88; and females, 65) and 75
healthy participants (age range, 43-78 years; mean, 57.35 years;
males, 35; and females, 40) were recruited at Fuxing Hospital,
Capital Medical University (Beijing, China) between February 2017
and May 2018. All the cases of LA were histopathologically
confirmed, and diagnosed as stage I/II. The basis for clinical
staging of LA was the eighth edition of the Union for International
Cancer Control Tumor-Node-Metastasis system (25). Healthy controls involved in the
present study were members of a healthy population undergoing
routine medical examination at Fuxing Hospital, Capital Medical
University. Healthy participants who had a history of acute or
chronic pneumonia were excluded, as well as those with increased
levels of serum tumor markers, or an abnormal chest imaging
examination that had identified features such as small nodules,
fibrous stripes or ground glass opacity in the lung.
The present study was approved by the Human Basic
and Clinical Research Ethics Committee of Fuxing Hospital (approval
no. 2018FXHEC-KY-19). All participants provided written informed
consent prior to sampling.
The present study was divided into three stages,
including the screening (5 patients with LA vs. 5 controls),
validation (40 patients with LA vs. 20 controls) and testing (108
patients with LA vs. 50 controls) stages. The level of miR-505-5p
and miR-382-3p were also examined in different clinical samples,
including plasma EV, raw plasma, and LA and adjacent non-tumor
tissue samples.
Blood samples (10 ml) from patients with LA and the
healthy controls prior to initial treatment were collected in
commercially available ethylenediaminetetraacetic acid-treated
tubes. Cells were removed from the plasma samples by centrifugation
for 10 min at 1,000 × g 4°C using a refrigerated centrifuge (Becton
Dickinson; Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
Platelets were depleted by centrifugation for 15 min at 2,000 × g
4°C from the plasma samples. The supernatant obtained following
these two centrifugation steps was the standard plasma. The samples
were separated out into 0.5 ml aliquots and stored at −80°C. A
total of 40 paired LA and adjacent non-tumor tissue samples were
collected from surgical patients who had not received preoperative
chemoradiotherapy and stored in liquid nitrogen (−196°C).
Isolation of plasma EVs
EVs were extracted from plasma samples using a
PuroExo® exosome isolation kit (101Bio; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), following the manufacturer’s
protocol. Briefly, debris was removed from the plasma samples by
centrifugation for 15 min at 3,000 × g at 4°C. The supernatant was
diluted by the addition of 500 µl sample buffer (provided in
the kit) to the supernatant, prior to mixing with 250 µl
prepared mixture reagent (provided in the kit). After vortex-mixing
for 10 sec at 25°C, the mixture was incubated for 30 min at 4°C
followed by centrifugation at 3,000 × g at 4°C for 2 min, and the
samples were then divided into 3 layers. The middle, ‘fluffy’ layer
was transferred to a fresh tube and centrifuged at 5,000 × g at 4°C
for 3 min. Subsequently, a new 3-layer separation appeared and only
the middle ‘fluffy’ layer was retained. A total of 500 µl
PBS was added to the fluffy pellets, and the resuspended solution
was then centrifuged 5,000 × g at 4°C for 3 min. The supernatant
was placed into a PuroExo® column and centrifuged for 5
min at 3,000 × g at 4°C. The flow-through comprised the isolated
pure exosomes.
Transmission electron microscopy
A solution of 2% paraformaldehyde was prepared to
fix the EVs at 4°C for 10 min, and subsequently 5 µl EV
suspension was transferred to each carbon-coated 200 mesh copper
grid. Adsorption was allowed to occur for 5 min at 4°C. The grids
were washed twice with PBS, followed by 8 washes with deionized
water. The grids were counterstained with uranyl acetate (2%) for 1
min at 4°C, and subsequently air-dried. Negative staining of EVs
was observed on the basis of their shape, structure and size using
a Philips CM12 transmission electron microscope at ×80,000
magnification (Philips Research China, Ltd., Shanghai, China).
Nanoparticle-tracking analysis
Size distribution and concentration of the EVs were
measured using ZetaView® nanoparticle-tracking analysis
instrumentation (Particle Metrix, Meerbusch, Germany). Briefly,
after isolation from the plasma, diluted EVs samples were added to
the measurement system. Particle size distribution was calculated
on the basis of the Brownian motion of individual particles,
according to the Stokes-Einstein equation (26).
RNA extraction and RT-qPCR
Total RNA from plasma EVs or the plasma of patients
with LA and the healthy controls, or frozen tissues of patients
with LA, or cell lines (including A549, H1299 and 293T cells) was
extracted using a miRNeasy Mini kit (Qiagen, Inc., Valencia, CA,
USA). For EVs and plasma RNA extraction, Cel-miR-39 was added into
the samples prior to extraction as external control. RT-qPCR was
applied to detect the relative expression levels of selected
miRNAs. RNA was reverse-transcribed using an Applied Biosystems™
TaqMan® MicroRNA Reverse Transcription kit and amplified
with Applied Biosystems TaqMan Universal PCR Master mix, together
with miRNA-specific Applied Biosystems TaqMan MGB probes (labeled
by fluorescein and minor groove binder; Applied Biosystems; Thermo
Fisher Scientific, Inc.), according to the manufacturer’s protocol.
The quantity of RNA and its purity were measured on a Nanodrop
Technologies ND-2000 spectrophotometer (Thermo Fisher Scientific,
Inc.). Only samples with the appropriate absorbance measurements
(A260/A280 of ~2.0, and
A260/A230 of 1.9-2.2) were considered for
inclusion in the present study. The reaction conditions were as
follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec
and 60°C for 1 min. In order to compare the miRNA levels between
different samples, the 2-ΔΔCq method was used (27). Primers sequences are detailed in
Table I. Plasma EVs and plasma
samples were normalized against Cel-miR-39, whereas cell lines and
frozen tissues were normalized to U6. A total of three triplicate
experiments were performed in each case.
 | Table IPrimers sequences. |
Table I
Primers sequences.
| Primer name | Sequences |
|---|
| UP-R |
5′-GTGCAGGGTCCGAGGT-3′ |
| cel39-F |
5′-ACTCACCGGGTGTAAATCA-3′ |
| miR-16-F |
5′-GTCCAATATTACTGTGCTG-3′ |
| miR-495-F |
5′-GCAAACAAACATGGTGCAC-3′ |
| miR-194-F |
5′-GTTGTAACAGCAACTCCAT-3′ |
| miR-486-3F |
5′-TACGGGGCAGCTCAGTAC-3′ |
| miR-486-5F |
5′-TATCCTGTACTGAGCTGC-3′ |
| miR-505-F |
5′-ATGGGAGCCAGGAAGTATT-3′ |
| miR-126-F |
5′-CGTCGTACCGTGAGTAA-3′ |
| miR-382-F |
5′-CTAATCATTCACGGACAA-3′ |
| miR-410-F |
5′-GCAATATAACACAGATG-3′ |
| miR-411-F |
5′-CGTACGCTATACGGTCTACTA-3′ |
| cel39-RT |
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCAAGCTG-3′ |
| miR-16-RT |
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTAAAGCA-3′ |
| miR-495-RT |
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAAGAAGT-3′ |
| miR-194-RT |
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTCCACAT-3′ |
| miR-486-3RT |
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACATCCTGT-3′ |
| miR-486-5RT |
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCTCGGGG-3′ |
| miR-505-RT |
5′-GTTGGCTCTGGTGCAGGGTCCGA
GGTATTCGCACCAGAGCCAACACATCAA-3′ |
| miR-126-RT |
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCGCATTA-3′ |
| miR-382-RT |
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAAGTGT-3′ |
| miR-410-RT |
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACAGGC-3′ |
| miR-411-RT |
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCGTACG-3′ |
According to the high-through sequencing results,
the top 5 upregulated miRNAs and top 5 downregulated miRNAs were
firstly detected in the plasma EVs from a cohort consists of 40
patients with LA vs. 20 controls. Subsequently, four
significantly-altered miRNAs were selected for the testing stage,
for validation in 108 patients with LA and 50 matched controls.
High-throughput sequencing
A total amount of 1 µg total RNA per sample
was used as input material for the small RNA library.
NEBNext® Multiplex Small RNA Library Prep Set for
Illumina® (New England BioLabs, Inc., Ipswich, MA, USA)
was used to generate the sequencing libraries. The clustering of
index-coded samples was performed on a cBot Cluster Generation
system with TruSeq SR Cluster kit, v3-cBot-HS (Illumina Inc., San
Diego, CA, USA), according to the manufacturer’s protocol.
Subsequently, the library was subjected to an Illumina HiSeq
2500/2000 platform for sequencing, and 50 bp single-end reads were
generated. Raw data were first processed through custom Perl and
Python scripts. Clean data (clean reads) were read by removing
reads containing poly-N with 5′-adapter contaminants, without 3′
adapter or the insert tag, or containing poly-A or -T or -G or -C
from raw data, as well as low-quality reads. Q20, Q30 and the GC
content of raw data were then calculated. Subsequently, a certain
range (18-40 nucleotides) of length from clean reads was selected
for downstream analyses. After unpaired Student’s t-test analysis,
the top 5 upregulated miRNAs and top 5 downregulated miRNAs in the
EVs from patients with LA were selected for further assessment.
MicroRNA targets prediction and gene
ontology analysis
The direct targets of miR-505-5p were predicted
using TargetScan 7.0 (http://www.targetscan.org/vert_70/). The top 1,000
predicted target genes were subjected to Gene Ontology analysis.
The Gene Ontology analysis was processed with DAVID Bioinformatics
Resources 6.8 (https://david.ncifcrf.gov/).
Cell culture
A549, H1299 and 293T cells purchased from the China
Infrastructure of Cell Line Resources (Institute of Basic Medical
Sciences, Chinese Academy of Medical Sciences, Beijing, China) were
cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented
with 10% HyClone™ fetal bovine serum (FBS) (Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA), 100 U/ml penicillin and 100 U/ml
streptomycin at 37°C in a humid atmosphere containing 5%
CO2. For the cell proliferation and apoptosis assay
experiments, exosome-depleted FBS (Systems Biosciences, Palo Alto,
CA, USA) was used for cell culture.
Cell transfection
A549 and H1299 cells (1×105) were seeded into 6-well
plates 1 day prior to transfection. The cells were transfected with
25 nM miR-505 mimic or inhibitor (Shanghai GenePharma Co., Ltd.,
Shanghai, China) with Invitrogen Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer’s protocol. After 48 h following transfection, the
transfection efficiency was determined by RT-qPCR, as
aforementioned. The miR-505 mimic is a double strand RNA oligo,
with sequence scrambled double strand RNA oligo as control.
Furthermore, the miR-505 inhibitor is a single strand RNA oligo,
with sequence scrambled single strand RNA oligo as control. The
sequences for miR-505 mimic, inhibitor and corresponding control
are: miR-505 mimic, 5′-GGGAGCCAGGAAGUAUUGAUGU-3′; mimic control,
5′-AGUGUUGGACUAAGCGGAGGUA-3′; miR-505 inhibitor,
5′-ACAUCAAUACUUCCUGGCUCCC-3′; and inhibitor control,
5′-UAACACGUCUAUACGCCCA-3′.
Co-culture of cells and exosomes
A549 and H1299 cells (5×103) were seeded
into 96-well plates in DMEM supplemented with exosome-depleted FBS,
100 U/ml penicillin and 100 U/ml streptomycin, and the cells were
incubated at 37°C overnight. EVs isolated from the plasma samples
of 10 patients with LA and 10 control subjects were added into the
cells at a final concentration of 100 µg/ml for 48 h at 37°C
for co-cultivation.
Dual luciferase assay
A 221 bp fragment of the 3′-UTR from TP53AIP1 was
cloned downstream of the firefly luciferase gene into a pmirGLO
plasmid (Promega Corporation, Madison, WI, USA) to generate the
reporter vector. miRNA mimic, miRNA inhibitor and sequence
scrambled single- and double-strand control oligos were purchased
from Shanghai GenePharma Co., Ltd. 293T cells were seeded into
48-well plates at 5×104 cells/well, attached overnight
for luciferase reporter assays. The reporter vector was
co-transfected into cells with miRNA mimic or inhibitor using
Invitrogen Lipofectamine 2000. At 2 days after transfection, the
cells were lysed, and the luciferase activity was analyzed using a
Dual-Luciferase® Reporter Assay System (Promega
Corporation). Experiments were performed in triplicate, and the
results were analyzed as the relative luciferase activity (firefly
luciferase/Renilla luciferase).
Immunoblotting
Protein samples were quantified using a Pierce BCA
Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.),
according to the manufacturer’s protocols. Protein samples were
heated with SDS sample buffer (0.25 M Tris-HCl, pH 6.8, 0.5 M DTT,
10% SDS, 50% glycerol and 0.5% bromophenol blue) at 95°C for 5 min,
A total of 30 µg were loaded per lane and then separated on
15% SDS-PAGE and electroblotted onto polyvinylidene fluoride
membranes (Amersham; GE Healthcare Life Sciences, Little Chalfont,
UK). Subsequently, the membranes were incubated with anti-TP53AIP1
rabbit polyclonal antibody at a dilution of 1:1,000 (cat. no.
ab217785; Abcam, Cambridge, MA, USA) and anti-β-actin mouse
monoclonal antibody at a dilution of 1:3,000 (cat. no. sc-47778;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C.
Immunoreactive bands were detected by incubation with horseradish
peroxidase-conjugated goat anti-rabbit (cat. no. ab205718) or
rabbit anti-mouse IgG (cat. no. ab6728; Abcam) at a dilution of
1:5,000 for 2 h at room temperature. Detection by chemiluminescence
was performed using a Pierce enhanced chemiluminescence kit
(Pierce; Thermo Fisher Scientific, Inc.).
Cell proliferation assay
The extent of cell proliferation was measured
according to cell viability, which was evaluated using an MTT
assay. Briefly, A549 and H1299 cells (5×103) were seeded
into 96-well plates in DMEM and incubated at 37°C overnight. The
cells were transfected with miR-505-inhibitor for 24 h using the
same protocol as aforementioned. EVs of patients with LA or healthy
control subjects were added in each well (final concentration, 100
µg/l) 24 h after transfection, and the cells were
subsequently incubated at 37°C for 48 h. Aliquots (20 µl) of
MTT solution (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) were added to each well after co-cultivation, and the
cells were further incubated at 37°C for 4 h. The absorbance was
read at 570 nm on a 96-well plate reader following the addition of
dimethyl sulfoxide.
Cell apoptosis assay
Cell apoptosis was assessed using flow cytometric
analysis, following Annexin V-fluorescein isothiocyanate (FITC) and
propidium iodide staining using a FITC Annexin V Apoptosis
Detection kit (BioLegend, Inc., San Diego, CA, USA). Results were
analyzed with a flow cytometer and FlowJo software (v10.4.1; Tree
Star, Inc., Ashland, OR, USA).
Statistical analysis
Statistical analysis was performed using SPSS
software version 19.0 (IBM Corp., Armonk, NY, USA) and data are
presented as the mean ± standard. Luciferase activity, and the cell
apoptosis and proliferation data were analyzed by paired Student’s
t-test. The miRNA expression levels were evaluated by unpaired
Student’s t-test. Clinical characteristics were evaluated by the
χ2 test. Receiver operating characteristic (ROC) curves
were employed to measure the optimal cut-off value for each
selected miRNA. The score for each miRNA was considered to be 1 for
expression levels above the cutoff value; otherwise, it was set as
0. The area under the ROC curve (AUC) was used to evaluate the
diagnostic value of each selected miRNA in LA. Fisher’s exact test
was used for gene-enrichment analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of altered miRNAs in
patients with LA
To identify altered levels of EV miRNAs in plasma
from patients with early-stage LA, peripheral blood samples were
collected from 153 patients with LA and 75 healthy participants.
The present study was divided into three stages, including the
screening, validation and testing stages. The clinical
characteristics are detailed in Table
II.
 | Table IICharacteristics of the 153 patients
with lung adenocar-cinoma and 75 healthy controls included in the
present study. |
Table II
Characteristics of the 153 patients
with lung adenocar-cinoma and 75 healthy controls included in the
present study.
| Study
stage/variable | Case (%) | Control (%) | P-value |
|---|
| Screening (cohort,
n=10) | | | |
| Sex | | | 1.000 |
| Male | 3 (60) | 2 (40) | |
| Female | 2 (40) | 3 (60) | |
| Age (mean ±
SD) | 62.00±8.37 | 58.40±15.77 | 0.664 |
| Smoking | | | 1.000 |
| No | 2 (40) | 3 (60) | |
| Yes | 3 (60) | 2 (40) | |
| Validation cohort
(n=60) | | | |
| Sex | | | 0.409 |
| Male | 24 (60) | 9 (45) | |
| Female | 16 (40) | 11 (55) | |
| Age (mean ±
SD) | 64.06±9.27 | 60.40±11.07 | 0.221 |
| Smoking | | | 0.370 |
| No | 26 (65%) | 16 (80%) | |
| Yes | 14 (35%) | 4 (20%) | |
| Testing cohort
(n=158) | | | |
| Sex | | | 0.411 |
| Male | 61 (60) | 24 (45) | |
| Female | 47 (40) | 26 (55) | |
| Age (mean ±
SD) | 63.60±8.96 | 61.35±10.92 | 0.439 |
| Smoking | | | 0.684 |
| No | 70 (64.8) | 30 (60) | |
| Yes | 38 (35.2) | 20 (40) | |
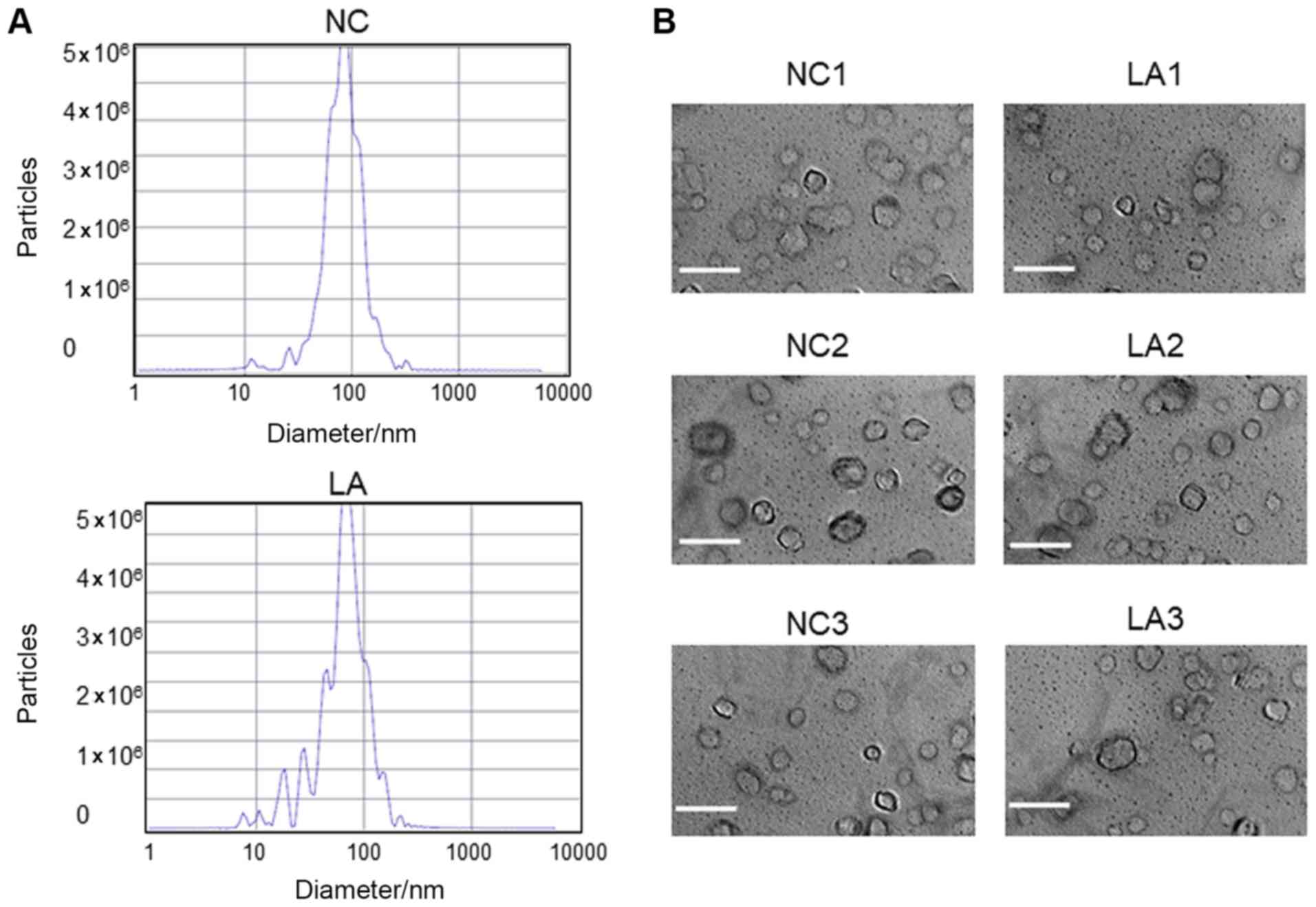
Plasma EVs were assessed by transmission electron
microscopy and nanoparticle-tracking analysis. The results obtained
revealed that the majority of the vesicles had the typical exosome
size (28) (30-200 nm in diameter;
Fig. 1A) and a bilayer membrane
structure (Fig. 1B).
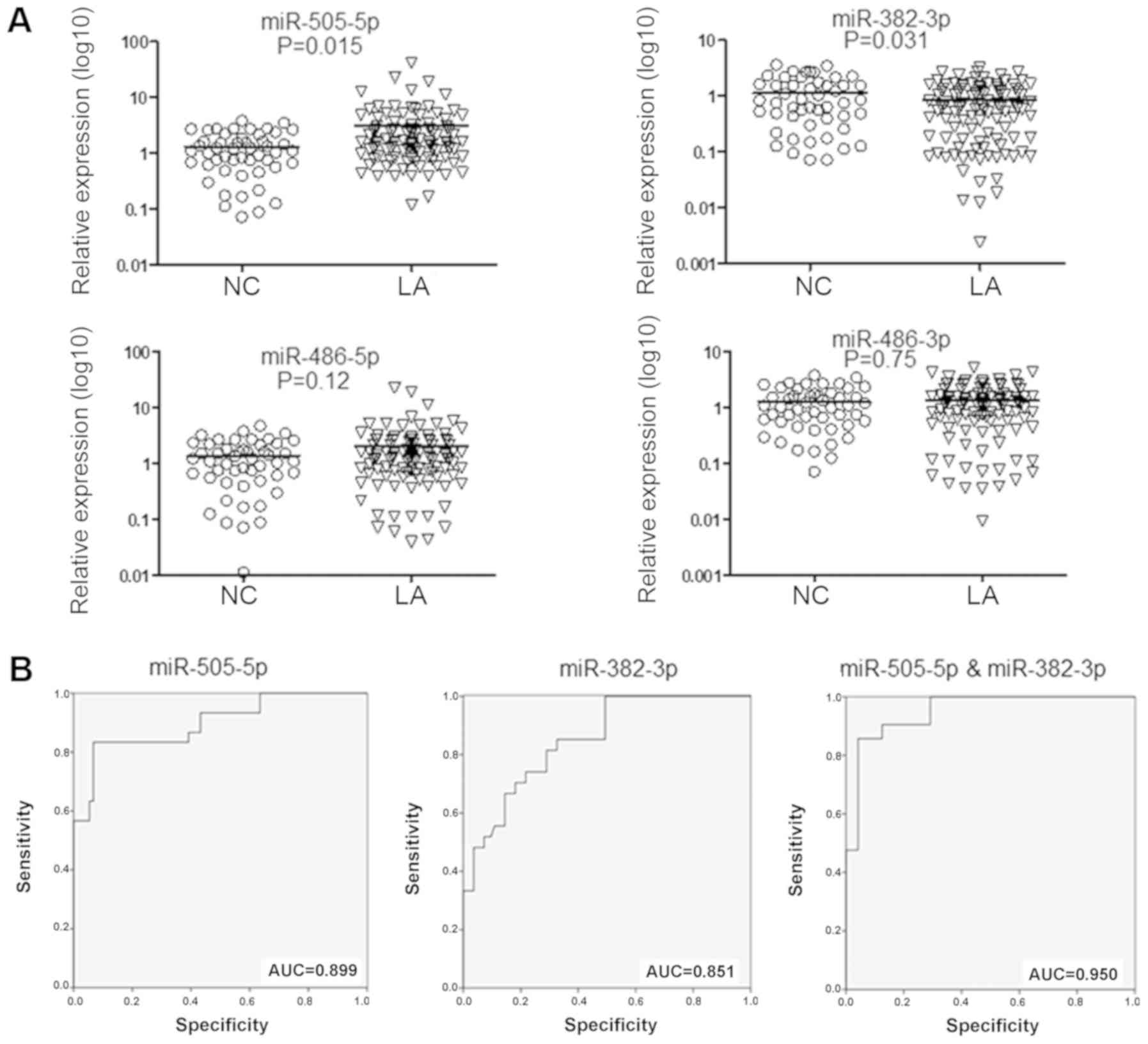
During the screening stage, the expression profile
miRNAs was assessed based on high-throughput sequencing in 5
patients with LA and 5 matched healthy controls. The top 5
upregulated miRNAs (miR-486-3p, miR-194-5p, miR-486-5p, miR-16-3p
and miR-505-5p) and top 5 downregulated miRNAs (miR-411-5p,
miR-126-3p, miR-495-3p, miR-382-3p and miR-410-3p) (Fig. 2A) were selected for further
assessment. In the validation stage, the 10 aforementioned miRNAs
selected as candidate miRNAs were assessed using RT-qPCR, with a
cohort consisting of 40 patients with LA and 20 matched healthy
controls. As depicted in Fig. 2B,
the levels of miR-486-3p, miR-486-5p and miR-505-5p in EVs were
significantly increased in LA patients, compared with the control
subjects (P=0.015, P=0.011 and P=0.031, respectively). Furthermore,
the plasma EV levels of miR-382-3p were significantly reduced in
patients with LA, compared with control subjects (P=0.039).
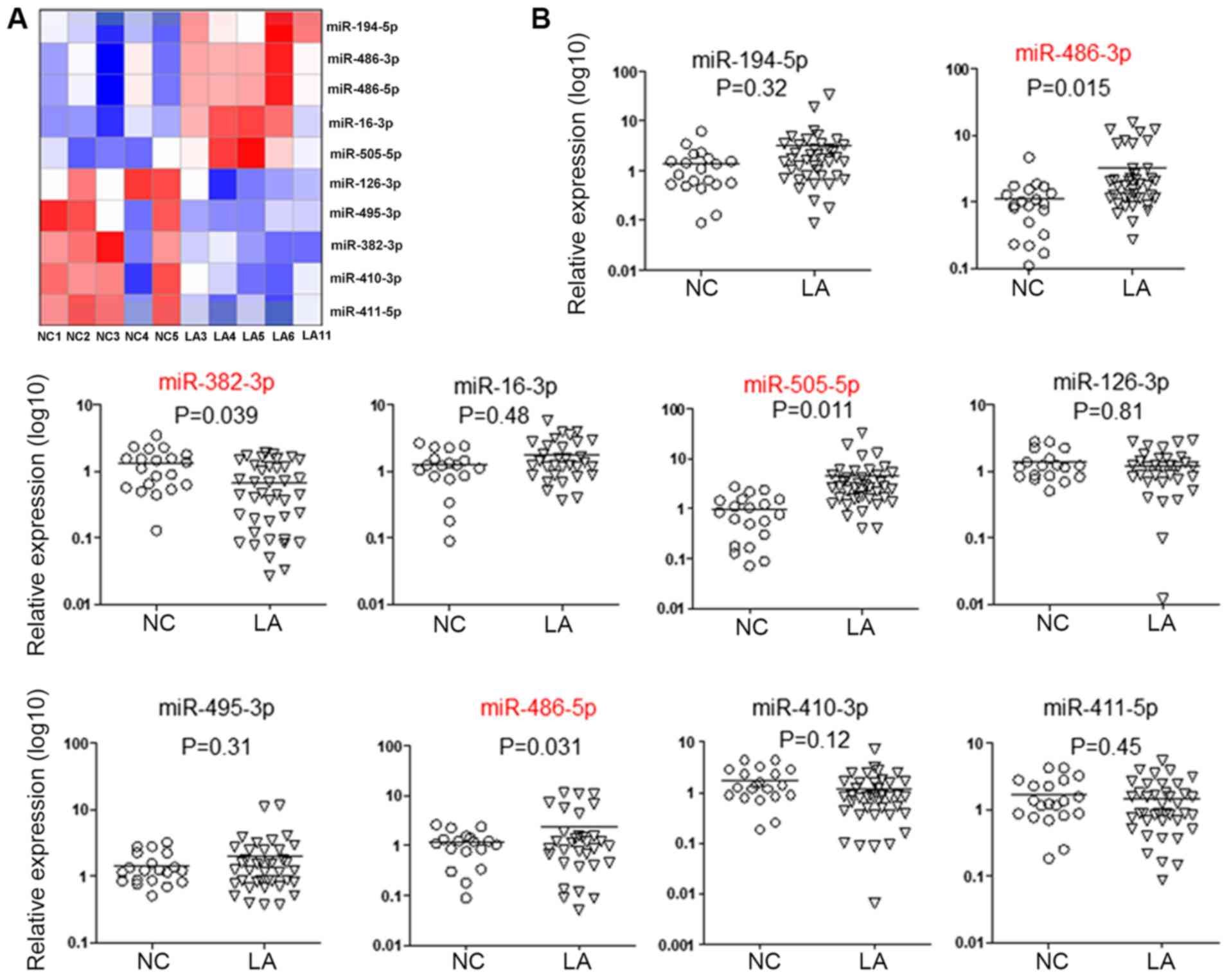 | Figure 2Altered miRNA levels in extracellular
vesicles from patients with LA. (A) Hierarchical cluster analysis.
Heat map illustrating miRNA samples from 5 patients with LA and 5
healthy NCs detected by high-throughput sequencing on an Illumina
HiSeq 2500/2000 platform. The top 5 upregulated (miR-194-5p,
miR-486-3p, miR-486-5p, miR-16-3p and miR-505-5p) and top 5
downregulated (miR-126-3p, miR-495-3p, miR-382-3p, miR-410-3p and
miR-411-5p) miRNAs were selected. (B) The above 10 selected miRNAs
were confirmed by reverse transcription-quantitative polymerase
chain reaction in a cohort of 40 patients with LA and 20 NC
subjects. Results were normalized to the level of cel-miR-39, which
was added into the samples as external control prior to RNA
extraction. The levels of miR-505-5ps, miR-386-5p, miR-386-3p and
miR-382-3p were significantly altered. P<0.05 was considered to
be statistically significant (unpaired Student’s t-test). LA, lung
adenocarcinoma; miRNA, microRNA; NC, normal control. |
Subsequently, these four miRNAs were selected for
the testing stage, for validation in 108 patients with LA and 50
matched controls. As depicted in Fig.
3A, the miR-505-5p levels were significantly elevated in
patients with LA (P=0.015), whereas the levels of miR-382-3p were
significantly reduced (P=0.031), compared with control
subjects.
Diagnostic values of plasma EV
miRNAs
To assess the performances of the two aforementioned
EV miRNAs in distinguishing patients with LA from control subjects,
optimal cut-off values for miR-505-5p and miR-382-3p in EVs were
determined based on ROC curves in the testing cohort. As depicted
in Fig. 3B, miR-505-5p had an AUC
of 0.899 (95% confidence interval, 0.826-0.974), with a sensitivity
and specificity of 83.3 and 93.3%, respectively; whereas,
miR-382-3p exhibited an AUC of 0.851 (95% confidence interval,
0.773-0.928), with a sensitivity and specificity of 81.5 and 71.1%,
respectively. Additionally, the two miRNAs in combination exhibited
an increased ability to distinguish the two experimental groups,
compared with either miRNA alone, with an AUC of 0.950 (95%
confidence interval, 0.906-0.995; sensitivity, 85.7%; and
specificity, 95.8%).
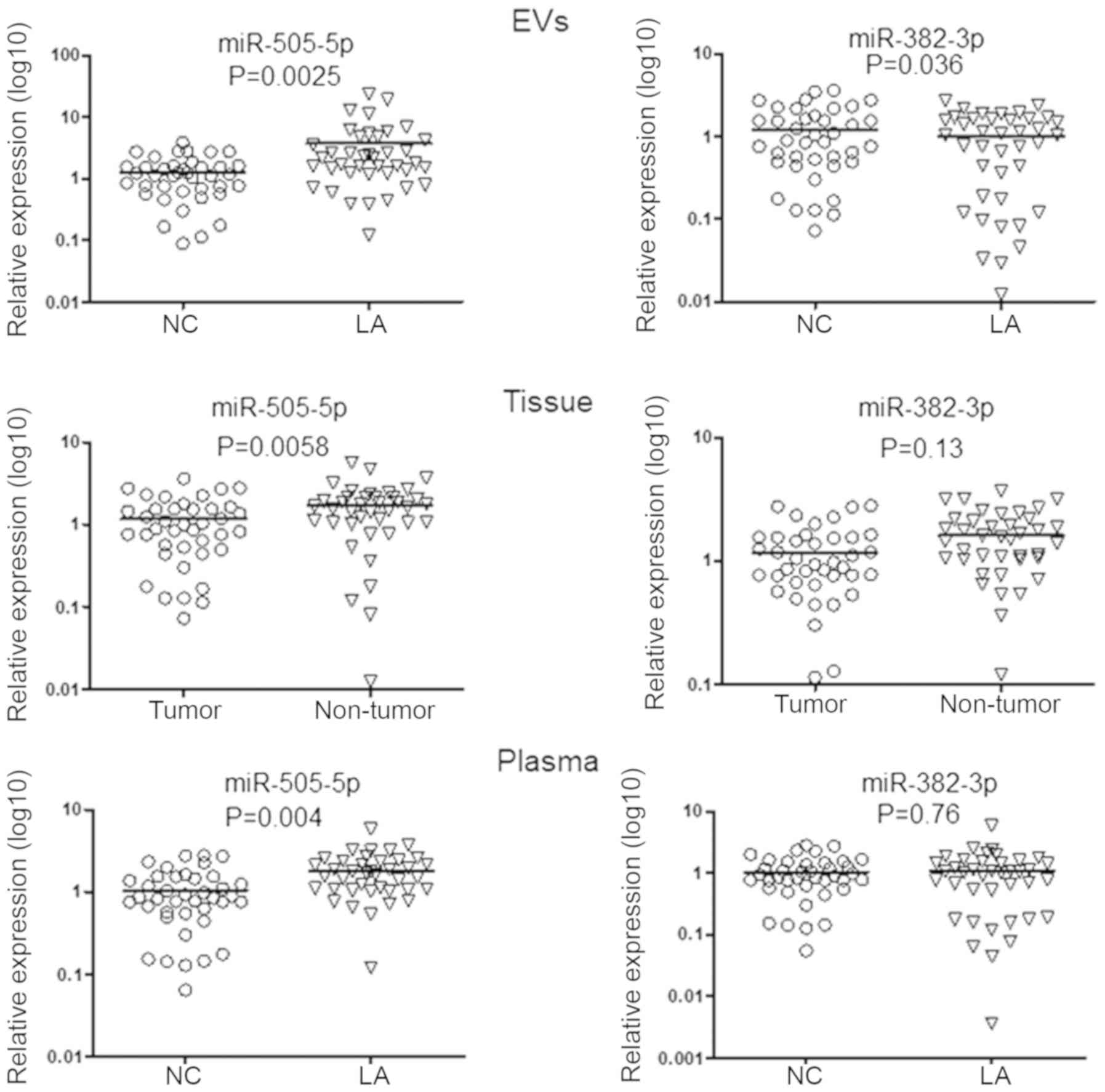
Verification of miRNAs in tissue samples
and plasma
To examine the expression pattern consistency of
miR-505-5p and miR-382-3p in different clinical samples, the levels
of miR-505-5p and miR-382-3p in plasma EV and raw plasma from a
cohort comprising of 40 patients with LA and 40 controls were
analyzed. Furthermore, the levels of these two miRNAs were also
detected in 40 paired LA and adjacent non-tumor tissue samples.
As depicted in Fig.
4, the levels of miR-505-5p were statistically increased in
plasma EVs. Furthermore, the levels of miR-505-5p were
significantly increased in tumor samples, compared with the
corresponding adjacent normal tissues, whereas the levels of
miR-382-3p revealed no statistical differences between tumor and
adjacent normal tissue specimens. Furthermore, only the levels of
miR-505-5p in the plasma of patients with LA were significantly
increased, compared with the control group.
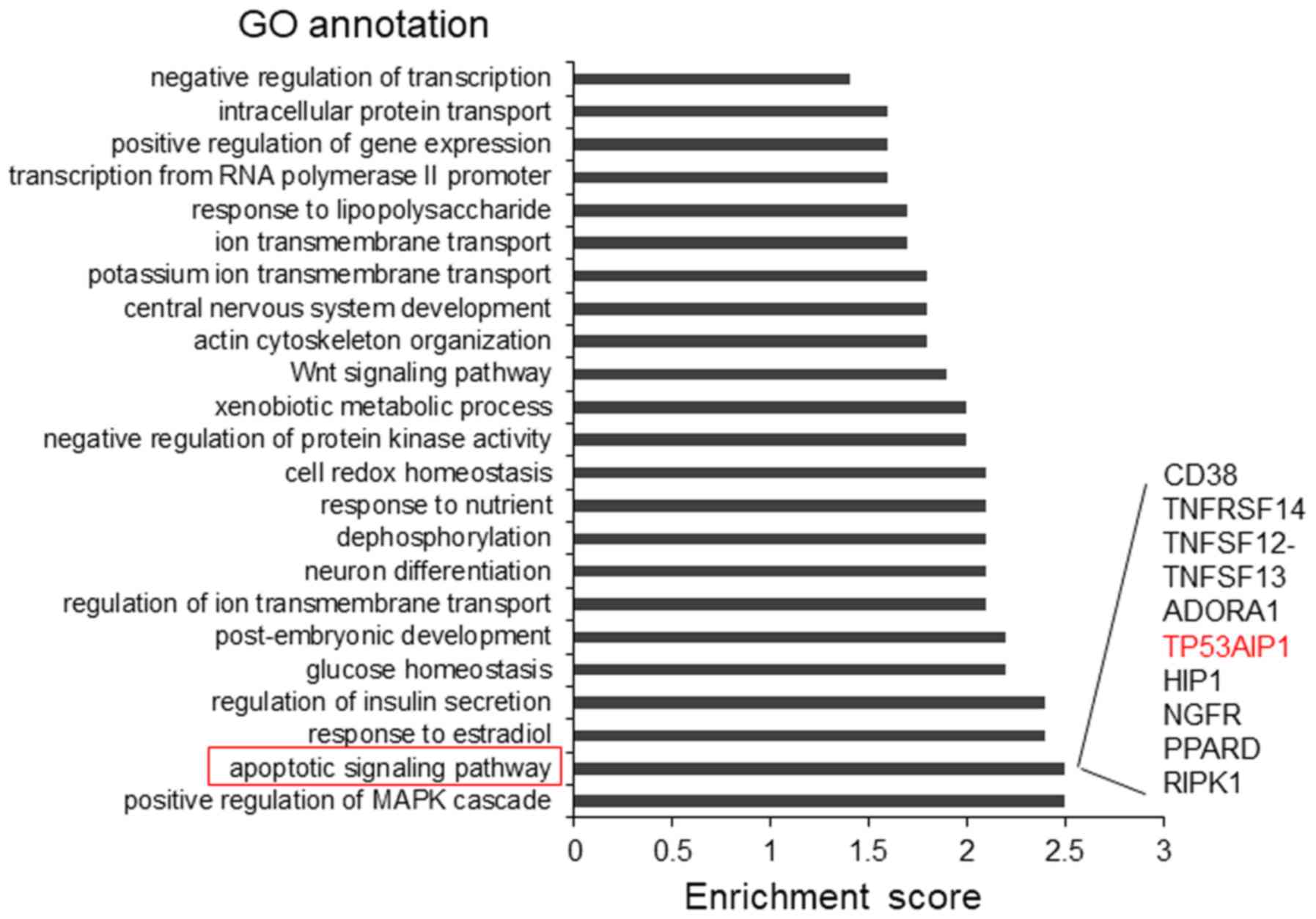
Investigation of miR-505-5p function in
lung cancer
To investigate miR-505-5p function in LA
pathogenesis, the direct targets of miR-505-5p were first predicted
with the TargetScan online bioinformatics tool. The top 1,000
predicted target genes were subjected to Gene Ontology analysis,
and 9 of them were identified to be potentially involved in the
cell apoptotic signaling pathway (Fig.
5), including TP53AIP1.
TP53AIP1 is a p53-inducible gene that serves an
important role in mediating p53-dependent apoptosis (29). Although TP53AIP1 is predominantly
expressed in the thymus, it is also detectable in the lungs, and
reduced levels of TP53AIP1 in lung cancer tissues is considered to
be a predictor of poor prognosis in patients with non-small cell
lung cancer (NSCLC) (30,31). Therefore, a 221 bp fragment of the
TP53AIP1 3′-UTR was subsequently cloned downstream of the firefly
luciferase gene in the pmirGLO plasmid; additionally, a plasmid
with 4 nucleotide mutations was also generated as the mutant
reporter vector (Fig. 6A). As
depicted in Fig. 6B, relative
luciferase activity was significantly downregulated in cells
treated with the miR-505-5p mimic, compared with control, whereas
the luciferase activity was upregulated following transfection with
the miR-505-5p inhibitor, compared with inhibitor control. By
contrast, for the mutant reporter vector, which featured the 4
nucleotide mutations, relative luciferase activity was not
significantly reduced by the miR-505-5p mimic, indicating that
miR-505-5p inhibited luciferase expression by targeting the 3′-UTR
of TP53AIP1. Subsequently, the miR-505-5p mimic and inhibitor were
transfected into the human NSCLC cell lines, A549 and H1299 for 72
h. TP53AIP1 protein levels then were assessed using immunoblotting.
As depicted in Fig. 6C, TP53AIP1
protein expression was downregulated by the miR-505-5p mimic, and
upregulated by the miR-505-5p inhibitor. Furthermore, the
overexpression and knockdown, respectively, of miR-505-5p in
miR-505-5p mimic- or inhibitor-transfected A549 and H1299 cells
were confirmed by RT-qPCR (Fig.
6D). These results revealed that miR-505-5p inhibited
endogenous TP53AIP1 expression by targeting its mRNA 3′-UTR. To
understand the biological function of miR-505-5p, the miR-505-5p
mimic or the inhibitor was transfected into A549 and H1299 cells. A
significant increase in the viability of the
miR-505-5p-overexpressed cells and a significant decrease in the
viability of the miR-505-5p knockdown cells were observed (Fig. 6E). Furthermore, cell apoptosis was
significantly suppressed in the presence of the miR-505-5p mimic
and significantly promoted by the miR-505-5p inhibitor in A549 and
H1299 cells, indicating that miR-505-5p functions as an oncogene in
lung cancer cells (Fig. 6F).
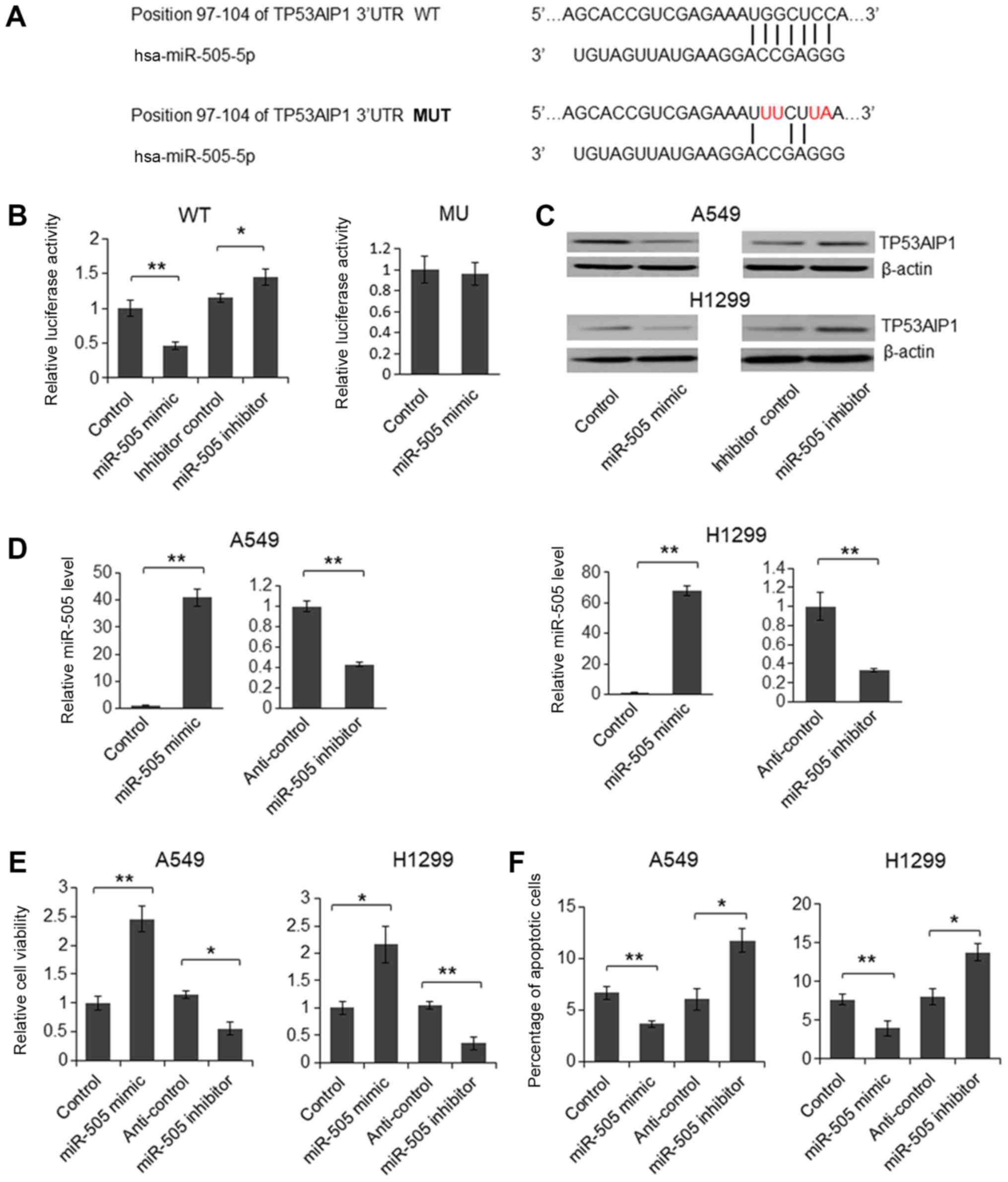 | Figure 6TP53AIP1 is a direct target of
miR-505-5p. (A) The predicted target site of miR-505-5p in the
TP53AIP1 3′-UTR, and the 4 nucleotide mutant sequence (red
letters), are depicted. Results from the (B) dual luciferase assay
and (C) immunoblotting are also depicted. (D) A reverse
transcription-quantitative polymerase chain reaction assay was
performed to determine the miR-505-5p level in miR-505 mimic- or
inhibitor-transfected cells 72 h after transfection. (E) An MTT
assay was performed to determine cell viability, while (F) flow
cytometric analysis was performed to determine the percentage of
apoptotic cells. Paired Student’s t-test was used to analyze the
results. Two-tailed P<0.05 was considered to indicate a
statistically significant difference. *P<0.05 and
**P<0.01, compared with the controls. TP53AIP1, tumor protein
P53-regulated apoptosis-inducing protein 1; LA, lung
adenocarcinoma; miRNA, microRNA; WT, wild-type; MUT, mutated; UTR,
untranslated region. |
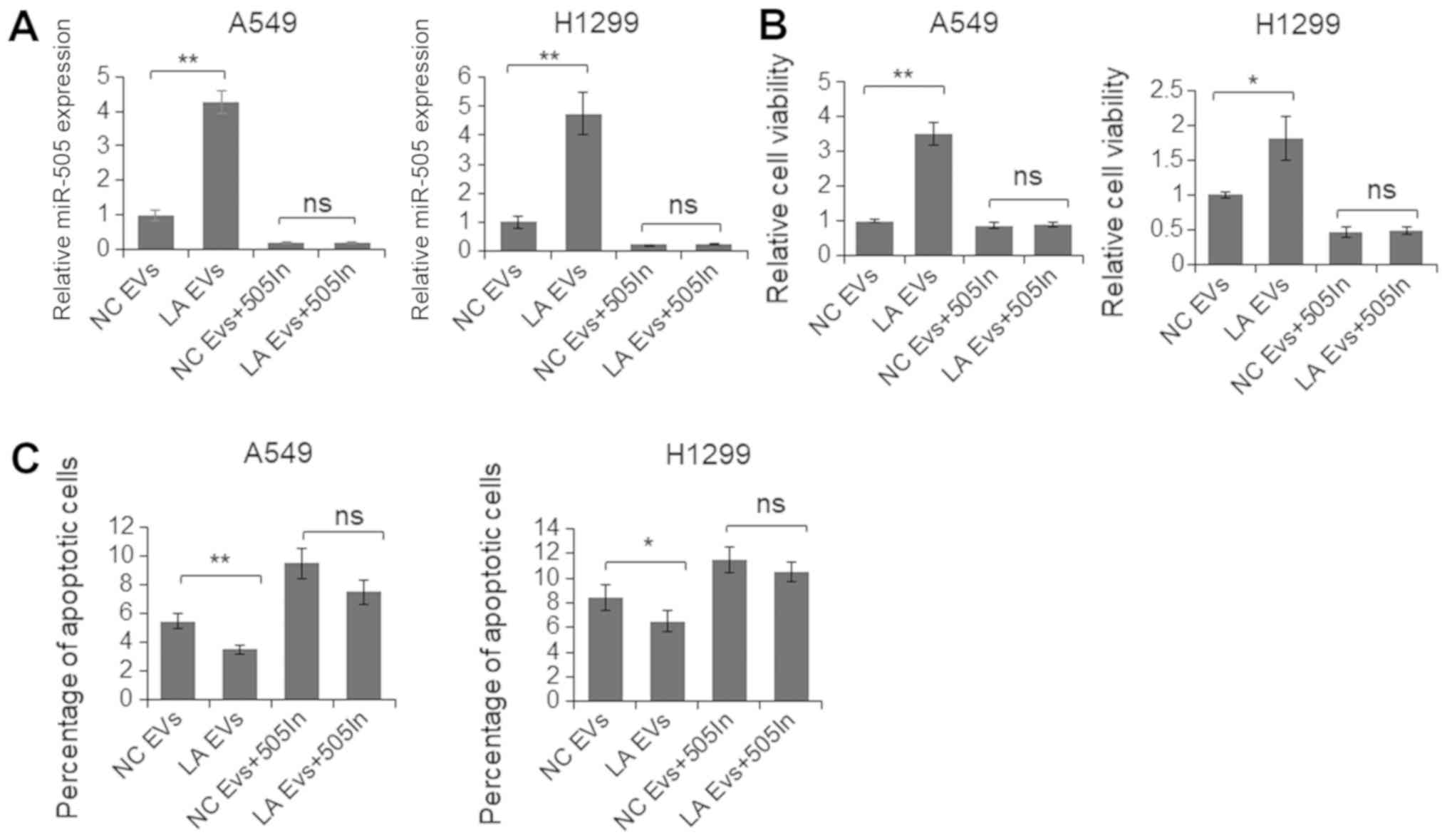
To further assess the functional role of an
increased level of miR-505-5p in EVs, EVs were isolated from the
plasma samples of 10 patients with LA and 10 control subjects. A549
and H1299 cells were co-cultured with the plasma EVs of the
patients and the controls, separately, for 48 h, and the levels of
cell viability and apoptosis were determined. As depicted in
Fig. 7A, following co-culture with
EVs from the patients with LA, the miR-505-5p levels in A549 and
H1299 cells were significantly increased. Additionally, cell
viability was significantly increased, and the apoptotic rate was
significantly reduced (Fig. 7B and
C). By contrast, when cells were first treated with the
miR-505-5p inhibitor and subsequently co-cultured with EVs, the
resultant miR-505-5p levels were revealed to be similar, and cell
viability and apoptosis were not significantly altered, indicating
further that miR-505-5p delivered by the EVs was the key regulator
of cell viability and apoptosis.
Discussion
Tumor-derived EVs containing specific proteins and
RNAs are attracting increasing interest as promising cancer
biomarkers (32). These EVs are
easily obtained from biological fluids, are highly stable and bear
the characteristics of their parental cells. These molecules
fulfill a variety of associated roles in key physiological and
pathological activities, including the transportation of
oncoproteins and tumor-specific miRNA molecules throughout the body
(33). As such, they are
considered as ideal biomarkers for cancer diagnosis and prognosis
due to their high sensitivity and minimal invasiveness (34,35).
However, data regarding plasma EVs in LA are relatively rare, and
are frequently conflicting (36-39,27-30).
In the present study, the presence of altered levels of miRNAs in
plasma EVs was firstly screened from 5 patients with LA and 5
healthy controls. A total of 10 candidate miRNAs were further
examined in a larger population comprising 148 patients with LA and
70 control subjects. The level of miR-505-5p was revealed to be
increased, whereas that of miR-382-3p was reduced in the plasma EVs
derived from the patients with LA. After performing statistical
analysis, the levels of miR-505-5p and miR-382-3p were capable of
being used to distinguish patients with LA from healthy controls,
and had the potential to become a biomarker for early-stage LA
diagnosis.
The human miR-505 gene is first transcribed into an
84 bp pre-miR-505, which is further processed into mature miRNAs,
including miR-505-3p and miR-505-5p (40). miR-505-3p is the predominant miRNA,
having a relative increased level, compared with miR-505-5p in
different human cells (40). It
has been reported that miR-505-3p functions as a tumor suppressor
in endometrial cancer, osteosarcoma and hepatoma by targeting
transforming growth factor-α and high mobility group box 1
(41-43). However, the function of miR-505-5p
has yet to be fully elucidated. In the present study, it was
revealed that only the miR-505-5p level was altered in the plasma
and tumor tissues of patients. The role of miR-505-5p in lung
cancer cell lines was subsequently examined, and it was
demonstrated that miR-505-5p functions as an oncogene by promoting
cell proliferation via the targeting of TP53AIP1. To the best of
our knowledge, the present study is the first published study in
which an investigation of the role of miR-505-5p in LA has
partially uncovered the mechanism of miR-505-5p during the
tumorigenesis of LA, also providing a novel biomarker for LA
treatment.
Chen et al (44) demonstrated that miR-382 expression
is decreased in NSCLC tissue samples, particularly in patients with
stage III/V cancer and metastatic tumors (44). However, in the present study,
altered miR-382 levels in the tumor samples from patients with LA
were not identified. This difference may be attributed to having a
different cohort of research subjects. The present study was
focused only on patients with early-stage LA, whereas that of Chen
et al (44) detected the
level of miR-382 in tumors from all patients with NSCLC. These
contrasting results also indicate that miR-382 may have the
potential for staging lung cancer, a possibility that should be
investigated further in patients with different types and stages of
NSCLC.
A number of groups have identified certain candidate
miRNAs that have the potential for LA diagnosis, although whether
these miRNAs are functional remains unclear (45-47).
Therefore, functional studies were performed to assess the role of
miR-505-5p in EVs. Following co-culture, it was confirmed that
miR-505-5p in EVs could be delivered into cancer cells.
Furthermore, the EV-delivered miR-505-5p retained its functional
properties, promoting proliferation and repressing cell apoptosis,
indicating that EV miR-505-5p serves an important role in cell-cell
communication.
A few limitations of the present study should be
acknowledged. Firstly, the data included the Chinese Han
population, and the roles of miR-505-5p and miR-382-3p in other
populations require further evaluation. Secondly, although the
miR-505-5p inhibitor almost rescued the function of EVs following
co-culture, proteins and other miRNAs in EVs may also have
contributed to the obtained data.
In conclusion, miR-505-5p in EVs was identified as a
candidate molecule for early-stage LA detection, and its function
in association with LA tumorigenesis has been partially uncovered.
These observations provide novel insights into LA development,
early diagnosis and treatment.
Funding
The present study was supported by Open Research
Fund of Key Laboratory of Cell Proliferation and Regulation
Biology, Ministry of Education, College of Life Sciences, Beijing
Normal University (Beijing, China).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
GA and HF designed the experiment. YL, YH, YJ and YW
collected the clinical samples. HF, HL and YG extracted the
exosomes. HF conducted the functional study. HL and YG participated
in the clinical data acquisition, analysis and interpretation. GA
and HF analyzed the results and prepared the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Human Basic
and Clinical Research Ethics Committee of Fuxing Hospital (approval
no. 2018FXHEC-KY-19). All participants provided written informed
consent prior to sampling.
Patients consent for publication
All the patients involved in the present study
provided written informed consent for the publication of any
associated data and accompanying images.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
LA
|
lung adenocarcinoma
|
|
NSCLC
|
non-small cell lung cancer
|
|
EV
|
extracellular vesicle
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
3′-UTR
|
3′-untranslated region
|
|
miRNA
|
microRNA
|
Acknowledgments
Not applicable.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.
|
|
2
|
Zhou C: Lung cancer molecular epidemiology
in China: Recent trends. Transl Lung Cancer Res. 3:270–279.
2014.
|
|
3
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: a heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014.
|
|
4
|
Li Q, Liu M, Ma F, Luo Y, Cai R, Wang L,
Xu N and Xu B: Circulating miR-19a and miR-205 in serum may predict
the sensitivity of luminal A subtype of breast cancer patients to
neoadjuvant chemotherapy with epirubicin plus paclitaxel. PLoS One.
9:e1048702014.
|
|
5
|
Bach PB, Mirkin JN, Oliver TK, Azzoli CG,
Berry DA, Brawley OW, Byers T, Colditz GA, Gould MK, Jett JR, et
al: Benefits and harms of CT screening for lung cancer: a
systematic review. JAMA. 307:2418–2429. 2012.
|
|
6
|
Saghir Z, Dirksen A, Ashraf H, Bach KS,
Brodersen J, Clementsen PF, Døssing M, Hansen H, Kofoed KF, Larsen
KR, et al: Lung cancer screening with low dose CT requires careful
consideration. Ugeskr Laeger. 176:pii: V061403412014.In Danish.
|
|
7
|
Holdenrieder S, Wehnl B, Hettwer K, Simon
K, Uhlig S and Dayyani F: Carcinoembryonic antigen and
cytokeratin-19 fragments for assessment of therapy response in
non-small cell lung cancer: A systematic review and meta-analysis.
Br J Cancer. 116:1037–1045. 2017.
|
|
8
|
Okamura K, Takayama K, Izumi M, Harada T,
Furuyama K and Nakanishi Y: Diagnostic value of CEA and CYFRA 21-1
tumor markers in primary lung cancer. Lung cancer. 80:45–49.
2013.
|
|
9
|
Chen F, Wang XY, Han XH, Wang H and Qi J:
Diagnostic value of Cyfra21-1, SCC and CEA for differentiation of
early-stage NSCLC from benign lung disease. Int J Clin Exp Med.
8:11295–11300. 2015.
|
|
10
|
Reck M and Rabe KF: Precision Diagnosis
and Treatment for Advanced Non-Small-Cell Lung Cancer. N Engl J
Med. 377:849–861. 2017.
|
|
11
|
Zaborowski MP, Balaj L, Breakefield XO and
Lai CP: Extracellular Vesicles: Composition, Biological Relevance,
and Methods of Study. Bioscience. 65:783–797. 2015.
|
|
12
|
Vlassov AV, Magdaleno S, Setterquist R and
Conrad R: Exosomes: current knowledge of their composition,
biological functions, and diagnostic and therapeutic potentials.
Biochim Biophys Acta. 1820:940–948. 2012.
|
|
13
|
Trinchieri G: Cancer and inflammation: an
old intuition with rapidly evolving new concepts. Annu Rev Immunol.
30:677–706. 2012.
|
|
14
|
Pereira ER, Jones D, Jung K and Padera TP:
The lymph node microenvironment and its role in the progression of
metastatic cancer. Semin Cell Dev Biol. 38:98–105. 2015.
|
|
15
|
O’Loghlen A: Role for extracellular
vesicles in the tumour micro-environment. Philos Trans R Soc Lond B
Biol Sci. 373:3732018.
|
|
16
|
Wang Y, Yi J, Chen X, Zhang Y, Xu M and
Yang Z: The regulation of cancer cell migration by lung cancer
cell-derived exosomes through TGF-β and IL-10. Oncol Lett.
11:1527–1530. 2016.
|
|
17
|
Tang Y, Cui Y, Li Z, Jiao Z, Zhang Y, He
Y, Chen G, Zhou Q, Wang W, Zhou X, et al: Radiation-induced
miR-208a increases the proliferation and radioresistance by
targeting p21 in human lung cancer cells. J Exp Clin Cancer Res.
35:72016.
|
|
18
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005.
|
|
19
|
Ma J and Li X: MicroRNAs are involved in
the toxicity of microcystins. Toxin Rev. 36:112017.
|
|
20
|
Ma J, Li Y, Yao L and Li X: Analysis of
MicroRNA Expression Profiling Involved in MC-LR-Induced
Cytotoxicity by High-Throughput Sequencing. Toxins (Basel).
9:92017.
|
|
21
|
Zhao Y, Xie P and Fan H: Genomic profiling
of microRNAs and proteomics reveals an early molecular alteration
associated with tumorigenesis induced by MC-LR in mice. Environ Sci
Technol. 46:34–41. 2012.
|
|
22
|
Maes OC, Chertkow HM, Wang E and Schipper
HM: MicroRNA: Implications for Alzheimer Disease and other Human
CNS Disorders. Curr Genomics. 10:154–168. 2009.
|
|
23
|
Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F,
Xie X, Zhou C and Lu W: Suppressed miR-424 expression via
upregulation of target gene Chk1 contributes to the progression of
cervical cancer. Oncogene. 32:976–987. 2013.
|
|
24
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
|
|
25
|
Detterbeck FC, Boffa DJ, Kim AW and Tanoue
LT: The Eighth Edition Lung Cancer Stage Classification. Chest.
151:193–203. 2017.
|
|
26
|
Coglitore D, Edwardson SP, Macko P,
Patterson EA and Whelan M: Transition from fractional to classical
Stokes-Einstein behaviour in simple fluids. R Soc Open Sci.
4:1705072017.
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001.
|
|
28
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013.
|
|
29
|
Okamura S, Arakawa H, Tanaka T, Nakanishi
H, Ng CC, Taya Y, Monden M and Nakamura Y: p53DINP1, a
p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell.
8:85–94. 2001.
|
|
30
|
Yamashita S, Chujo M, Miyawaki M, Tokuishi
K, Anami K, Yamamoto S and Kawahara K: Combination of p53AIP1 and
survivin expression is a powerful prognostic marker in non-small
cell lung cancer. J Exp Clin Cancer Res. 28:222009.
|
|
31
|
Yamashita SI, Masuda Y, Yoshida N,
Matsuzaki H, Kurizaki T, Haga Y, Ikei S, Miyawaki M, Kawano Y,
Chujyo M and Kawahara K: p53AIP1 expression can be a prognostic
marker in non-small cell lung cancer. Clin Oncol (R Coll Radiol).
20:148–151. 2008.
|
|
32
|
Verma M, Lam TK, Hebert E and Divi RL:
Extracellular vesicles: Potential applications in cancer diagnosis,
prognosis, and epidemiology. BMC Clin Pathol. 15:62015.
|
|
33
|
Liu S, Zhan Y, Luo J, Feng J, Lu J, Zheng
H, Wen Q and Fan S: Roles of exosomes in the carcinogenesis and
clinical therapy of non-small cell lung cancer. Biomed
Pharmacother. 111:338–346. 2018.
|
|
34
|
Bonjoch L, Gironella M, Iovanna JL and
Closa D: REG3β modifies cell tumor function by impairing
extracellular vesicle uptake. Sci Rep. 7:31432017.
|
|
35
|
Mrowczynski OD, Madhankumar AB, Slagle
Webb B, Lee SY, Zacharia BE and Connor JR: HFE genotype affects
exosome phenotype in cancer. Biochim Biophys Acta Gen Subj.
1861:1921–1928. 2017.
|
|
36
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: a novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008.
|
|
37
|
Lin Q, Mao W, Shu Y, Lin F, Liu S, Shen H,
Gao W, Li S and Shen D: A cluster of specified microRNAs in
peripheral blood as biomarkers for metastatic non-small-cell lung
cancer by stem-loop RT-PCR. J Cancer Res Clin Oncol. 138:85–93.
2012.
|
|
38
|
Wang ZX, Bian HB, Wang JR, Cheng ZX, Wang
KM and De W: Prognostic significance of serum miRNA-21 expression
in human non-small cell lung cancer. J Surg Oncol. 104:847–851.
2011.
|
|
39
|
Silva J, Garcia V, Zaballos A, Provencio
M, Lombardía L, Almonacid L, García JM, Domínguez G, Peña C, Diaz
R, et al: Vesicle-related microRNAs in plasma of nonsmall cell lung
cancer patients and correlation with survival. Eur Respir J.
37:617–623. 2011.
|
|
40
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42(D1): D68–D73. 2014.
|
|
41
|
Chen S, Sun KX, Liu BL, Zong ZH and Zhao
Y: MicroRNA-505 functions as a tumor suppressor in endometrial
cancer by targeting TGF-α. Mol Cancer. 15:112016.
|
|
42
|
Liu YJ, Li W, Chang F, Liu JN, Lin JX and
Chen DX: MicroRNA-505 is downregulated in human osteosarcoma and
regulates cell proliferation, migration and invasion. Oncol Rep.
39:491–500. 2018.
|
|
43
|
Lu L, Qiu C, Li D, Bai G, Liang J and Yang
Q: MicroRNA-505 suppresses proliferation and invasion in hepatoma
cells by directly targeting high-mobility group box 1. Life Sci.
157:12–18. 2016.
|
|
44
|
Chen T, Ren H, Thakur A, Yang T, Li Y,
Zhang S, Wang T and Chen M: miR-382 inhibits tumor progression by
targeting SETD8 in non-small cell lung cancer. Biomed Pharmacother.
86:248–253. 2017.
|
|
45
|
Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R,
Cheng W, Wang F, Qi LW, Chen Y, et al: A six-microRNA panel in
plasma was identified as a potential biomarker for lung
adenocarcinoma diagnosis. Oncotarget. 8:6513–6525. 2017.
|
|
46
|
Feng M, Zhao J, Wang L and Liu J:
Upregulated Expression of Serum Exosomal microRNAs as Diagnostic
Biomarkers of Lung Adenocarcinoma. Ann Clin Lab Sci. 48:712–718.
2018.
|
|
47
|
Maemura K, Watanabe K, Ando T, Hiyama N,
Sakatani T, Amano Y, Kage H, Nakajima J, Yatomi Y, Nagase T, et al:
Altered editing level of microRNAs is a potential biomarker in lung
adenocarcinoma. Cancer Sci. 109:3326–3335. 2018.
|