Introduction
Colorectal cancer (CRC) is one of the most common
malignancy types and the fourth most common cause of tumor
associated mortality globally in 2008 (1). Despite improvements in the diagnosis
and treatment of CRC, the efficacy of surgery and chemotherapy
remains unsatisfactory, and the 5 year survival rate of patients
with CRC with metastasis remains <10% in 2012 globally (2-4).
Therefore, further investigation on the molecular mechanisms of CRC
progression is urgently required, which may help to elucidate novel
therapeutic strategies for CRC.
Immunoglobulin like transcript (ILT) 4, which is
also termed lymphocyte immunoglobulin like receptor 2, belongs to
the ILT family. It is an immune inhibitory receptor expressed in
macrophages, monocytes, myeloid and dendritic cells (DCs) (5,6).
ILT4 binds to classical [(human leukocyte antigen A) HLA A and B]
and non classical (HLA-G, E and F) major histocompatibility complex
(MHC) class I molecules, and has been identified to induce
inhibitory signaling via immunoreceptor tyrosine based inhibitory
motifs, which is considered to recruit protein tyrosine phosphatase
(SHP) 1 or SHP 2 (7,8). Our previous studies determined that
ILT4 was aberrantly expressed in breast cancer and non small cell
lung cancer (NSCLC), and its expression was correlated with poor
prognosis (9-11). However, the expression of ILT4 has
not been investigated in CRC to date.
HLA-G is a non classical MHC class I molecule with
four membrane bound (HLA-G 1-4) and three soluble (HLA-G 5-7)
isoforms (11). Binding with the
receptors ILT2 and ILT4, HLA-G serves a crucial role in regulating
immune activities (12-14). Additionally, HLA-G expression has
been demonstrated to be involved in viral infection, organ
transplantation, inflammatory reactions, autoimmune diseases and
cancer, primarily serving an immune inhibitory function. Up to now,
numerous researchers reported that the presence of HLA-G in
numerous cancer types, including NSCLC and leukemia, is correlated
with poor prognosis (15-17). ILT4/HLA-G has been investigated as
an inhibitory axis in simian immunodeficiency virus and human
immunodeficiency virus infections, recurrent implantation failure
and systemic lupus erythematosus (18-20).
In a previous study, the co expression of ILT4/HLA-G in NSCLC and
its correlation with a reduced overall survival (OS) time in
patients with NSCLC was revealed, and also its activation of
extracellular signal regulated kinase (ERK) signaling was
demonstrated.
The present study investigated the co expression of
ILT4/HLA-G in human CRC tissues, and analyzed the association
between ILT4/HLA-G and the clinicopathological characteristics and
survival time of patients with CRC. The co expression of ILT4/HLA-G
was detected in CRC cell lines, in addition to its effect on cell
proliferation, invasion and migration; furthermore, the potential
signaling mechanism of protein kinase B (AKT) and ERK activation
underlying ILT4/HLA-G induced CRC progression was investigated.
Materials and methods
Patients
A total of 88 tumor tissues were obtained from
patients with CRC who underwent surgery between January 2013 and
December 2014 at Jinan Central Hospital Affiliated to Shandong
University (Jinan, China). Prior to the surgery, the patients had
not received chemotherapy, radiotherapy or immunotherapy. Among the
patients, 77.27% (68/88) were male and 22.73% (20/88) were female.
Patients were aged from 20-90 years old with a mean age of 67.17
years. Follow up data were summarized on April 14th 2018, with a
median follow-up time of 60 months. Tumor tissues were classified
according to the American Joint Committee on Cancer Cancer Staging
Manual (21). The present study
was supported by the Institutional Review Boards of Jinan Central
Hospital Affiliated to Shandong University, and written informed
consent was obtained from all patients.
Immunohistochemistry
Human CRC tissues and adjacent normal tissues were
fixed in 10% formalin (Phygene, Fuzhou, China; cat. no. PH0996) for
24 h at room temperature. Subsequently, it was dehydrated with
different concentrations of ethanol (70, 85, 95 and 100%) and
xylene, embedded in paraffin and cut to a thickness of 4 μm.
When the sections had been de-paraffinized by xylene and rehydrated
by different concentrations of ethanol (100, 95 and 75%), antigen
retrieval was performed by heating the specimens in Tris EDTA
buffer (Wuhan Sanying Biotechnology, Wuhan, China; cat. no.
B600011) for 10 min in a microwave oven (Guangdong Galanz Group
Co., Ltd., Guangdong, China; P70D20TL D4) at medium heat. Following
naturally cooling the sections, they were incubated in 3%
H2O2 at room temperature for 10 min and
blocked in 10% goat serum (OriGene Technologies, Inc., Rockville,
MD, USA; cat. no. ZLI 9021) at room temperature for 1 h. The
sections were incubated with rabbit polyclonal anti ILT4 anti body
(1:100; OriGene Technologies, Inc.; cat. no. TA349368) and mouse
monoclonal anti HLA-G antibody (1:200; Abcam, Cambridge, UK; cat.
no. ab52455) at 4°C overnight, followed by detection with an
Elivision Plus Polymer
Horseradish Peroxidase (mouse/rabbit) IHC kit (25
min) and streptavidin conjugated peroxidase (25 min). The sections
were visualized using 3,3′ diaminobenzidine solution (Fuzhou Maixin
Biotech Co., Ltd., Fuzhou, China) and counterstained with
hematoxylin at room temperature. Normal mouse IgG (1:20,000; Abcam;
cat. no. ab205719) and rabbit IgG (1:10,000; OriGene Technologies,
Inc.; cat. no. 17502-0.2mG) were used instead of the primary
antibody as a negative control.
A total of two independent researchers analyzed the
immunohistochemistry results. The stained cell percentages in at
least five fields were recorded at ×400 magnification under a light
microscope (NIKON ECLIPSE TI SR; Nikon Corporation, Tokyo, Japan).
The sections were scored as positive or negative by a previously
described method (11).
Cell lines and cell culture
The human colon epithelial cell line FHC was
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). The HCT1116, HT29, SW480 and SW620 cell lines
were obtained from the Institutes of Biochemistry and Cell Biology
(Shanghai, China), and originated from the ATCC. All cells were
cultured in RPMI 1640 medium (HyClone; GE Healthcare Life Sciences,
Logan, UT, USA) containing 10% fetal bovine serum (FBS; HyClone; GE
Healthcare Life Sciences). All cells were cultured in a humidified
atmosphere containing 5% CO2 at 37°C.
Cell transfection
Downregulation of ILT4 expression was performed by
transfecting plasmid pGPU6/GFP/Neo short hairpin (sh)ILT4-1
(shILT4; Shanghai GenePharma Co., Ltd., Shanghai, China) in to
HCT116 and SW620 cells. Non targeting plasmid [sh negative control
(NC); Shanghai GenePharma Co., Ltd.] was used as a negative
control. Plasmid Pez lv105 ILT4 (ILT4) (GeneCopoeia, Inc.,
Rockville, MD, USA) was added to HT29 and SW480 cells to upregulate
the level of ILT4, plasmid Pez lv105 (NC; GeneCopoeia, Inc.) was
used as negative control. When the cells had reached 40-60%
confluence in 6-well plates, plasmids and X-treme GENE HP reagents
(Roche Diagnostics, Basel, Switzerland) were added to the cells at
a ratio of 2 μg/6 μg according to the manufacturer’s
protocols. The level of mRNA were detected by reverse transcription
quantitative polymerase chain reaction (RT qPCR) after 24 h and the
level of protein was detected by western blot assays after 48 h
following transfection. The interference sequences of ILT4 are
listed in Table I. The
overexpression sequences of ILT4 are listed in Fig. S1.
 | Table IInterference sequences of ILT4. |
Table I
Interference sequences of ILT4.
| Name | Sequence
(5′-3′) |
|---|
| shILT4 |
GAAGAAGAACACCCACAATGC |
| shNC |
GTTCTCCGAACGTGTCACGT |
Protein extraction and western
blotting
Total proteins were extracted from CRC and FHC
cells. Following washing in ice cold PBS for 1 min, cell lysates
were lysed in radioimmunoprecipitation assay lysis buffer
containing a protease and phosphatase inhibitor cocktail (Thermo
Fisher Scientific, Inc., Waltham, MA, USA). Proteins were
quantified using a Pierce bicinchoninic acid assay (Thermo Fisher
Scientific, Inc.). A mass of 30 μg protein/lane was
separated via 10% SDS PAGE and transferred onto polyvinylidene
fluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA) using
semidry transfer apparatus. Following blocking in TBS with 0.1%
Tween 20 and 5% non fat milk for 1 h at room temperature, the PVDF
membranes were incubated with mouse monoclonal anti HLA-G antibody
(1:1,000; Abcam; cat. no. ab52455), rabbit polyclonal anti ILT4
antibody (1:500; Abcam; cat. no. ab128349), rabbit monoclonal anti
phospho AKT antibody (1:1,000; Cell Signaling Technology, Inc.,
Danvers, MA, USA; cat. no. 4060), rabbit monoclonal anti AKT
antibody (1:1,000; Epitomics; Abcam; cat. no. 2957-1), rabbit
monoclonal anti phospho ERK antibody (1:1,000; Abcam; cat. no.
ab201015) and rabbit monoclonal anti ERK antibody (1:1,000; Cell
Signaling Technology, Inc.; cat. no. 4695) at 4°C overnight. The
blots were incubated in horseradish peroxidase conjugated secondary
goat anti mouse (1:10,000; Wuhan Sanying Biotechnology; cat. no.
SA00001-1) or goat anti rabbit antibodies (1:10,000; Wuhan Sanying
Biotechnology; cat. no. SA00001-2) for 1 h at room temperature.
Finally, the blots were developed in Enhanced Chemiluminescence
reagent (EMD Millipore) and exposed using a ChemiDoc™ XRS+ system
(Bio Rad Laboratories, Inc., Hercules, CA, USA). GAPDH (1:10,000;
Wuhan Sanying Biotechnology; cat. no. 10494-1 AP) antibody was used
as the loading control. Image J 1.8.0 (National Institutes of
Health, Bethesda, MD, USA) was employed to quantify protein
levels.
RT-qPCR analysis
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The cDNAs
were synthesized from 2 μg total RNA using a Thermo
Scientific RevertAid First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc.). RT-qPCR was performed using a Fast Start
Universal SYBR® Green RT PCR kit (Roche Diagnostics) and
ABI 7500 Fast Real Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.; 50°C for 2 min, 95°C for 10 min and 95°C
for 15 sec for 40 cycles). The forward and reverse primer sequences
are listed in Table II. The mRNAs
were normalized to GAPDH. The 2-∆∆Cq method was used to
evaluate the relative expression levels of the genes (22).
 | Table IIPrimers used in reverse transcription
quantitative polymerase chain reaction. |
Table II
Primers used in reverse transcription
quantitative polymerase chain reaction.
| Name | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| ILT4 |
GCATCTTGGATTACACGGAT |
CTGACAGCCATATCGCCCTG |
| HLA-G |
GGCAGCCTATGACATTCCCC |
GTCTTGGAGCCTTCTCCCTC |
| GAPDH |
AGAAGGCTGGGGCTCATTTG |
AGGGGCCATCCACAGTCTTC |
Treatment of HLA-G fusion protein and
anti-ILT4 blocking antibody
HT29 cells were plated at a concentration of
1×105 cells/ml in 6 well plates, each well containing 2
ml RPMI 1640 medium with 10% FBS, and cultured at 37°C. When the
cells had reached 60-70% confluence in 6 well plates, HLA-G fusion
protein (Wuhan Sanying Biotechnology; cat. no. Ag10839) at
concentrations of 10, 20, 50, 100, 200 and 500 ng/ml was added
according to the manufacturer’s protocols. Each well contained 2 ml
RPMI 1640 medium with 10% FBS. The cells were incu bated at 37°C,
and then analyzed at 24 h with an RT-qPCR assay and at 48 h by
western blotting according to the afore mentioned protocols.
HT29 cells were treated with 0.5 μg/ml anti
ILT4 blocking antibody (R&D Systems, Inc., Minneapolis, MN,
USA; cat. no. MAB2078) in 6-well plates at 37°C, using 0.5
μg/ml IgG (R&D Systems, Inc.; cat. no. 1-001 A) and PBS
as negative controls. After 12 h, cells were treated with 200 ng/ml
HLA-G fusion protein at 37°C, and after 48 h the total protein
levels were detected by western blotting, according to the
aforementioned protocol. The peptide sequence of the HLA-G fusion
protein was as follows: SHSMRYFSAAVSRPS RGE PRFIAMGYVD
DTQFVRFDSDSACPRMEPRAPWVEREGPEYWEEETRN
TKAHAQTDRMNLQTLRGYYNQSEASSHTLQWMIGC
DLGSDGRLLRGYEQYAYDGKDYLALNEDLRSWTAAD
TAAQISKRKCEAANVAEQRRAYLEGTCVEWLHRYLEN
GKEMLQRADPPKTHVTHHPVFDYEATLRCWALGFYP
AEIILTWQRDGEDQTQDVELVETKPAGDGTFQKWAAV VVPSGEEQRYTCHVQHEG
LPEPLMLRWKQSSLPTIPI (26-308aa encoded by BC021708).
Cell proliferation assay
Cells were plated in 96 well culture plates at a
concentration of 3×103 cells/well, each well containing
100 μl RPMI-1640 medium with 10% FBS. After incubation for
6, 24, 48, 72 and 96 h at 37°C, cells were treated with 10
μl Cell Counting Kit-8 (Dojindo Molecular Technologies,
Inc., Kumamoto, Japan) solution and incubated at 37°C for 4 h, and
subsequently measured at 450 nm. All assays were performed at least
three times.
Wound-healing assay
CRC cells (HCT1116, HT29, SW480 and SW620) were
plated at a concentration of 1×105 cells/ml in 6 well
plates, each well containing 2 ml RPMI 1640 medium with 10% FBS,
and cultured at 37°C for 24 h. Subsequently, the cells were
scratched with a sterile 200 μl pipette tip. PBS was used to
wash the cell debris. Images were acquired at 0 and 24 h in the
same location under a light microscope (Carl Zeiss AG, Oberkochen,
Germany; magnification, ×40). The scratch wound widths were
calculated in three random fields of view.
Transwell migration assay
Cell migration was assessed using Transwell insert
chambers with a pore size of 8 μm (Corning Inc., Corning,
NY, USA). Approximately 1×105 cells in RPMI 1640 without
FBS were cultured in the upper chamber. RPMI 1640 medium
supplemented with 10% FBS was added to the bottom chamber as a
chemoattractant. Subsequently, the cells were incubated at 37°C for
24 h. The migrated cells were fixed with 20% methanol for 20 min at
37°C and stained with 0.1% crystal violet (Invitrogen; Thermo
Fisher Scientific, Inc.) for 15 min at 37°C. The number of cells in
three fields of each sample was counted under a light microscope
(Carl Zeiss AG; magnification, ×100). Cell numbers were calculated
from five random fields.
Matrigel invasion assay
Cell invasion was determined using Matrigel invasion
chambers (BD Biosciences; Becton, Dickinson and Company, Franklin
Lakes, NJ, USA). The upper chambers were covered with Matrigel,
which was diluted in cold PBS and incubated at 37°C for 1 h.
Approximately 1×105 cells were suspended in RPMI 1640
medium without FBS and transferred into the upper chambers. RPMI
1640 medium supplemented with 10% FBS was added to the bottom
chamber as a chemoattractant. Subsequently, the cells were
incubated at 37°C for 24 h. The migrated cells were fixed with 20%
methanol for 20 min at 37°C and stained with 0.1% crystal violet
for 15 min at 37°C. The number of cells in three fields of each
sample was counted under a light microscope (Carl Zeiss AG;
magnification, ×100). Cell numbers were calculated from five random
fields.
Statistical analysis
All data are presented as the mean ± standard
deviation. SPSS version 18.0 (SPSS, Inc., Chicago, IL, USA) and
GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) were
selected to perform the statistical analysis. OS curves were
generated via the by Kaplan Meier method and compared by means of
the log rank test. The associations between ILT4, HLA-G and
clinicopathological characteristics of patients with CRC were
analyzed with the χ2 test. The correlation between ILT4
and HLA-G expression in CRC tissues was analyzed by Spearman’s
correlation coefficient. The differences in characteristics between
two groups were examined by Student’s t test. The differences in
characteristics between three or more groups were examined by one
way analysis of variance followed by Tukey’s post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Co-expression of ILT4 and HLA-G is
detected in human CRC tissues
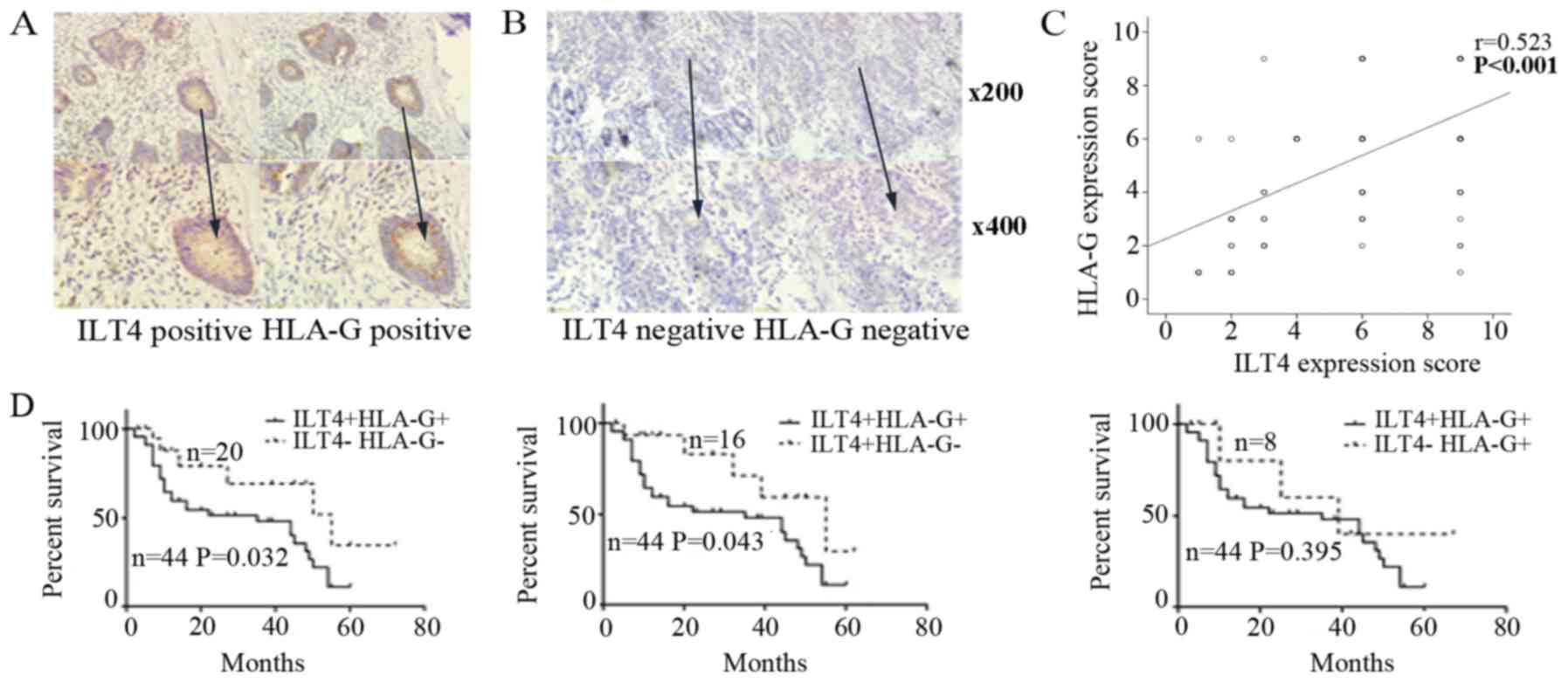
ILT4 and HLA-G positive expression was detected in
the cell cytoplasm and in some cancer stromal cells (Fig. 1A). In adjacent normal colorectal
tissues, ILT4 and HLA-G expression was too weak to be observed
(Fig. 1B). In total, 68.18%
(60/88) of the tissue samples exhibited overexpression of ILT4 and
59.09% (52/88) of the tissue samples exhibited overexpression of
HLA-G. There was a notable positive correlation between ILT4 and
HLA-G expression, and positive ILT4 expression had an increased
probability of being observed in HLA-G positive tissues, compared
with HLA-G negative tissues (P<0.001; Fig. 1C).
Co-expression levels of ILT4/HLA-G are
associated with the clinicopathological factors and prognosis of
patients with CRC
Firstly, the correlation of ILT4 and HLA-G
expression was analyzed with various clinicopathological
characteristics. ILT4 expression was positively correlated with
males and increased lymph node metastasis (Table III; P=0.002 and P=0.002,
respectively). Additionally, increased expression of HLA-G was
correlated with increased CRC Tumor Node Metastasis (TNM) staging
(P=0.030).
 | Table IIIAssociation between the expression of
ILT4 and HLA-G and clinicopathological characteristics in human CRC
tissues. |
Table III
Association between the expression of
ILT4 and HLA-G and clinicopathological characteristics in human CRC
tissues.
| | ILT4
| | | HLA-G
| | |
|---|
| Clinical
parameters | N | + | − | χ2 | P1 | + | − | χ2 | P2 |
|---|
| Age (year) | | | | | | | | | |
| ≤65 | 36 | 24 | 12 | 0.064 | 0.800 | 22 | 14 | 0.103 | 0.748 |
| >65 | 52 | 36 | 16 | | | 30 | 22 | | |
| Sex | | | | | | | | | |
| Male | 68 | 52 | 16 | 9.475 | 0.002b | 42 | 26 | 0.885 | 0.347 |
| Female | 20 | 8 | 12 | | | 10 | 10 | | |
| Primary tumor
localization | | | | | | | | | |
| Right sided | 24 | 16 | 8 | 0.040 | 0.980 | 15 | 9 | 0.159 | 0.924 |
| Left sided | 38 | 26 | 12 | | | 22 | 16 | | |
| Rectum | 26 | 18 | 8 | | | 15 | 11 | | |
| Tumor size
(cm) | | | | | | | | | |
| ≤5 | 52 | 37 | 15 | 0.518 | 0.472 | 33 | 19 | 1.004 | 0.316 |
| >5 | 36 | 23 | 13 | | | 19 | 17 | | |
| Tumor
differentiation | | | | | | | | | |
| Mediate
low/low | 41 | 27 | 14 | 0.192 | 0.661 | 21 | 20 | 1.968 | 0.161 |
| High/mediate | 47 | 33 | 14 | | | 31 | 16 | | |
| Lymph node
metastasis | | | | | | | | | |
| − | 48 | 26 | 22 | 9.561 | 0.002b | 28 | 20 | 0.025 | 0.874 |
| + | 40 | 34 | 6 | | | 24 | 16 | | |
| TNM stage
groupings | | | | | | | | | |
| I/II | 44 | 28 | 16 | 0.838 | 0.360 | 21 | 23 | 4.701 | 0.030a |
| III/IV | 44 | 32 | 12 | | | 31 | 13 | | |
According to ILT4 and HLA-G expression levels,
patients were divided into 4 groups to further examine the
association between ILT4/HLA-G co expression and various
clinicopathological factors. As presented in Table IV, compared with the ILT4 /HLA-G
group, ILT4/HLA-G co expression in the ILT4+/HLA-G+ group was
significantly associated with males (P=0.025) and increased lymph
node metastasis (P=0.041). Additionally, compared with the
ILT4+/HLA-G group, ILT4/HLA-G co expression was associated with
advanced TNM staging (P=0.001). Older age (P=0.042), males
(P=0.001), increased lymph node metastasis (P=0.038) and advanced
TNM staging (P=0.030) were associated with ILT4/HLA-G co
expression, compared with the ILT4 /HLA-G+ group.
 | Table IVAssociation between the co expression
of ILT4/HLA-G and clinicopathological characteristics in human CRC
tissues. |
Table IV
Association between the co expression
of ILT4/HLA-G and clinicopathological characteristics in human CRC
tissues.
| ILT4+/HLA-G+ | ILT4 /HLA-G−
| ILT4+/HLA-G−
| ILT4 /HLA-G+
|
|---|
| Clinical
parameters | N | n | χ2 | P1 | n | χ2 | P2 | n | χ2 | P3 |
|---|
| Age (year) | | | | | | | | | | |
| ≤65 | 16 | 6 | 0.247 | 0.619 | 8 | 0.909 | 0.340 | 6 | 4.140 | 0.042a |
| >65 | 28 | 14 | | | 8 | | | 2 | | |
| Sex | | | | | | | | | | |
| Male | 39 | 13 | 5.042 | 0.025a | 13 | 0.554 | 0.457 | 3 | 11.396 | 0.001b |
| Female | 5 | 7 | | | 3 | | | 5 | | |
| Primary tumor
localization | | | | | | | | | | |
| Right sided | 11 | 4 | 0.283 | 0.868 | 5 | 1.316 | 0.518 | 4 | 4.326 | 0.115 |
| Left sided | 18 | 8 | | | 8 | | | 4 | | |
| Rectum | 15 | 8 | | | 3 | | | 0 | | |
| Tumor size
(cm) | | | | | | | | | | |
| ≤5 | 29 | 11 | 0.698 | 0.403 | 8 | 1.256 | 0.262 | 4 | 0.739 | 0.390 |
| >5 | 15 | 9 | | | 8 | | | 4 | | |
| Tumor
differentiation | | | | | | | | | | |
| Mediate
low/low | 17 | 10 | 0.728 | 0.394 | 10 | 2.700 | 0.100 | 4 | 0.363 | 0.547 |
| High/mediate | 27 | 10 | | | 6 | | | 4 | | |
| Lymph node
metastasis | | | | | | | | | | |
| − | 21 | 15 | 4.156 | 0.041a | 5 | 1.297 | 0.255 | 7 | 4.309 | 0.038a |
| + | 23 | 5 | | | 11 | | | 1 | | |
| TNM stage
groupings | | | | | | | | | | |
| I/II | 15 | 10 | 1.462 | 0.227 | 13 | 10.484 | 0.001b | 6 | 4.705 | 0.030a |
| III/IV | 29 | 10 | | | 3 | | | 2 | | |
As depicted in Fig.
1D, patients with ILT4+/HLA-G+ had a decreased survival time,
compared with the ILT4 /HLA-G (P=0.032) and ILT4+/HLA-G (P=0.043)
groups. However, no significant difference in OS was observed,
compared with the ILT4 /HLA-G+ group (P=0.395).
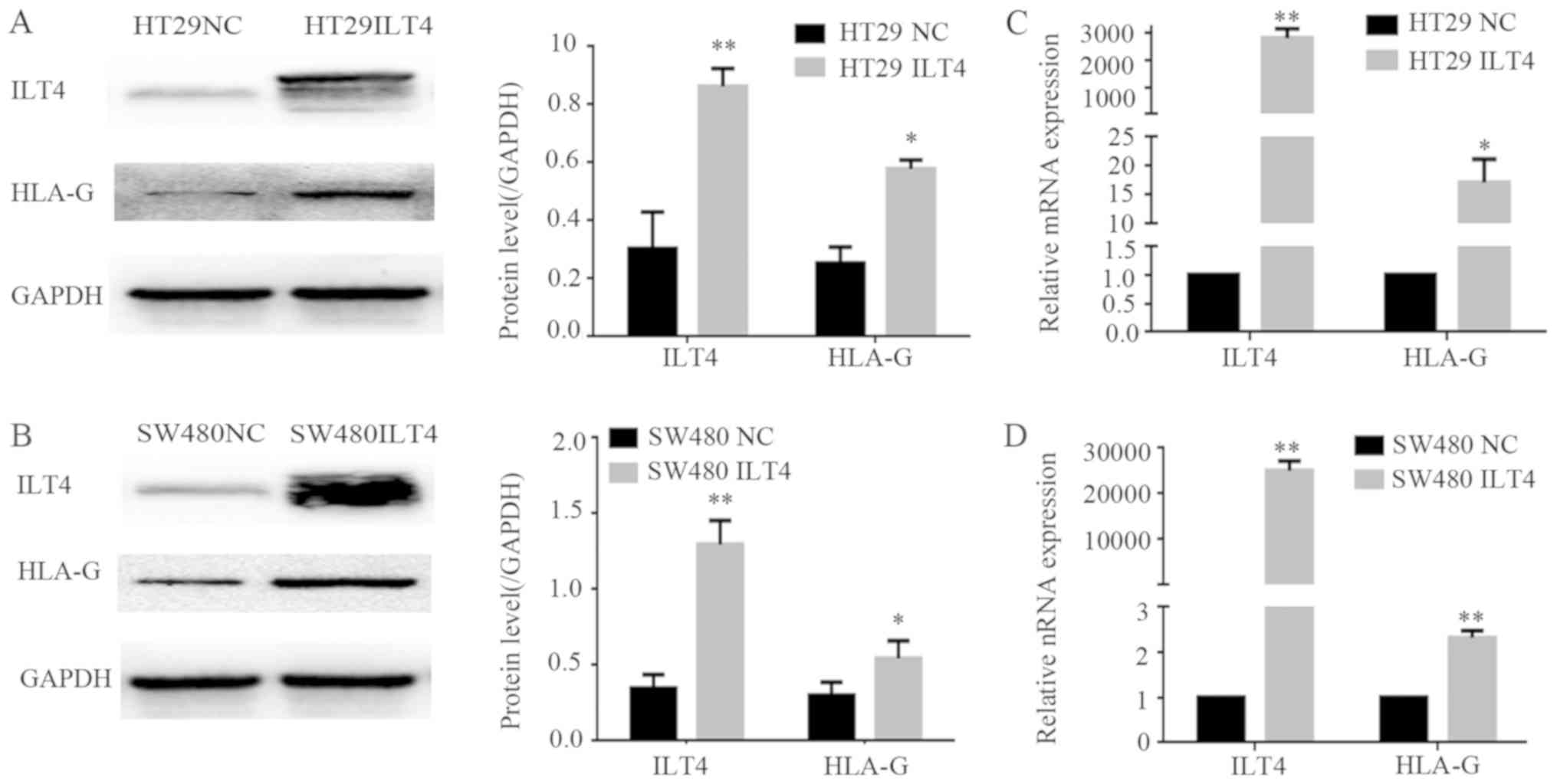
Co expression of ILT4 and HLA-G is also detected in
CRC cells. The expression levels of ILT4 and HLA-G were examined in
FHC and CRC cells (HCT1116, HT29, SW480 and SW620 cells) by western
blotting (Fig. 2A) and RT qPCR
(Fig. 2B and 2C). CRC cells exhibited increased levels
of ILT4 and HLA-G expression, compared with normal cells.
Additionally, increased levels of ILT4 and HLA-G were detected in
HCT116 cells, and reduced levels in HT29 cells, indicating that the
proteins ILT4 and HLA-G may be co expressed in CRC cells.
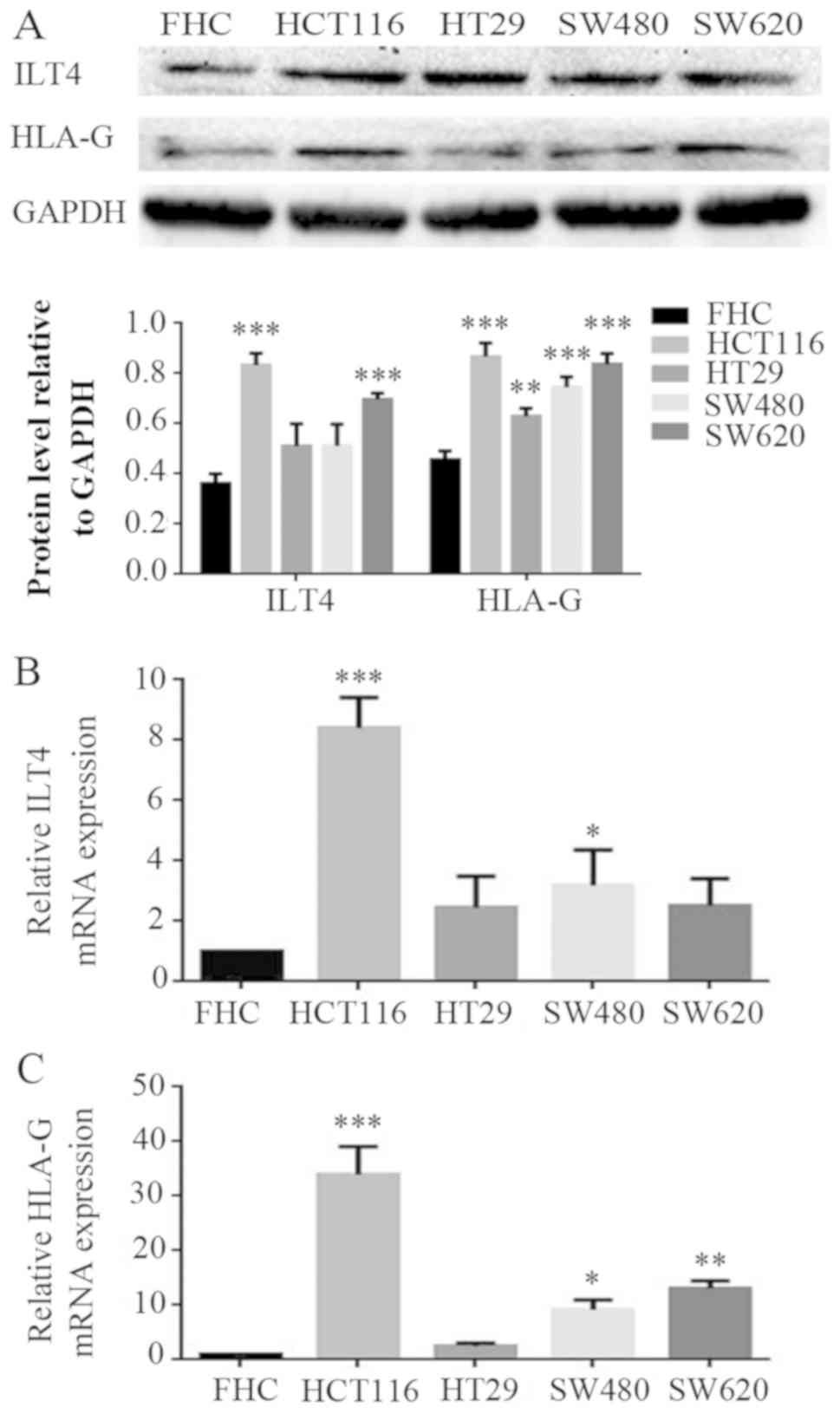 | Figure 2Co expression of ILT4 and HLA-G in
human CRC cells. (A) CRC cells (HCT116, HT29, SW480 and SW620
cells) exhibited increased pro tein levels of ILT4 and HLA-G
expression, compared with normal cells (FHC). (B) CRC cells
(HCT116, HT29, SW480 and SW620 cells) exhibited increased mRNA
levels of ILT4 expression, compared with normal cells (FHC). (C)
CRC cells (HCT116, HT29, SW480 and SW620 cells) exhibited increased
mRNA levels of HLA-G expression, compared with normal cells (FHC).
GAPDH served as a loading control. The results are presented as the
mean ± standard deviation of three independent experiments.
*P<0.05, **P<0.01 and ***P<0.001 vs.
the corresponding expression level of FHC. CRC, colorectal cancer;
HLA-G, human leukocyte antigen G; ILT4, immunoglob ulin like
transcript 4. |
Interference of ILT4 expression affects
the levels of HLA-G and regulates CRC cell proliferation, invasion
and migration
To investigate the regulatory effects of ILT4 on the
expression of HLA-G in CRC cells, ILT4 was overexpressed in HT29
and SW480 cells via an ILT4 plasmid (ILT4 vector), and ILT4
expression was inhibited in HCT116 and SW620 cells via ILT4 shRNA
(shILT 4). Western blotting and RT qPCR assays were performed to
examine HLA-G expression. The overexpression of ILT4 in HT29
(Fig. 3A and B) and SW480 cells
(Fig. 3C and D) upregulated the
expression of HLA-G.
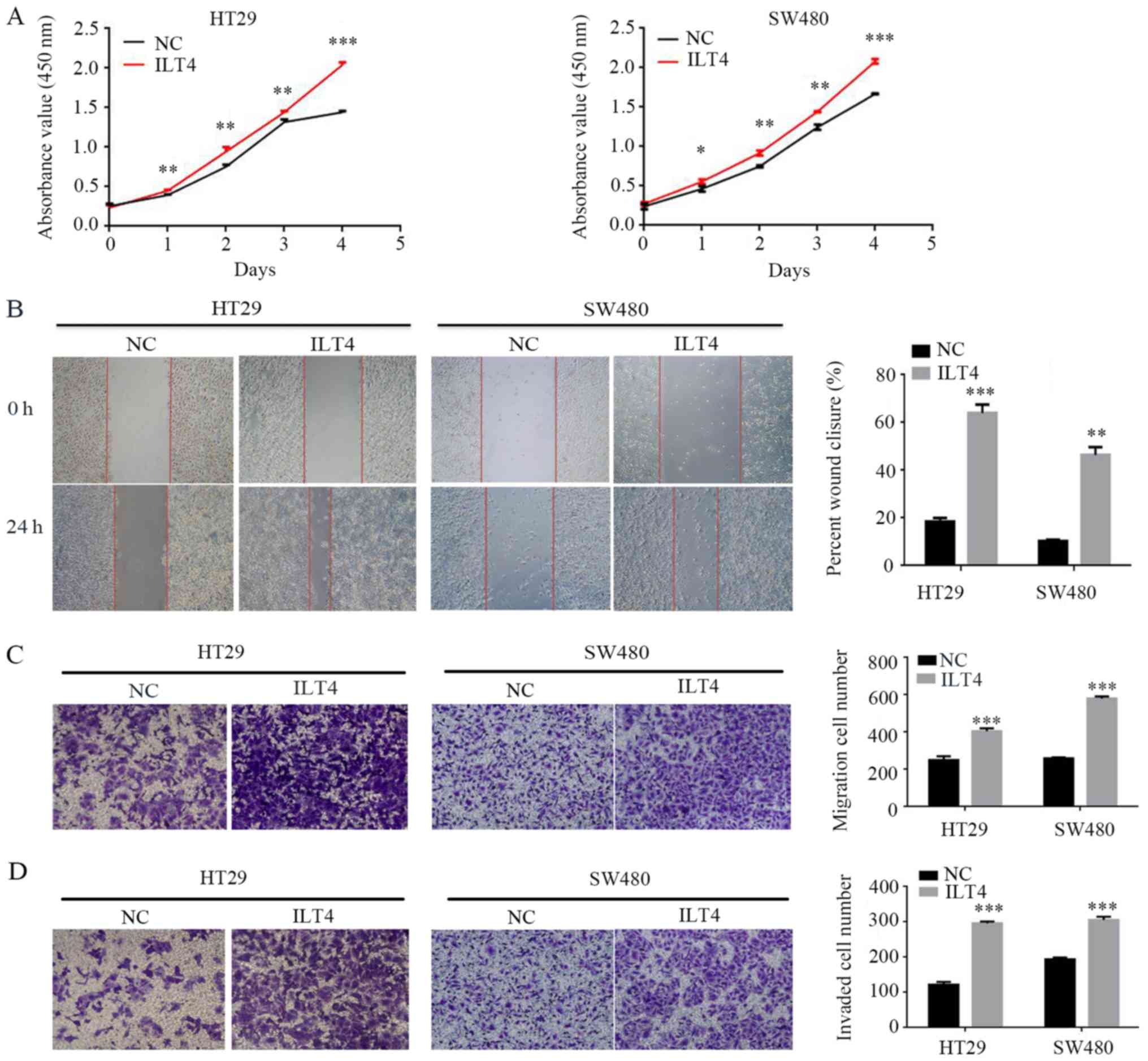
Simultaneously, the present study assessed whether
ILT4 influenced CRC cell proliferation, migration and invasion. The
results demonstrated that ILT4 overexpressing HT29 and SW480 cells
had a notably increased capacity for proliferation, migration and
invasion, compared with the negative control (Fig. 4).
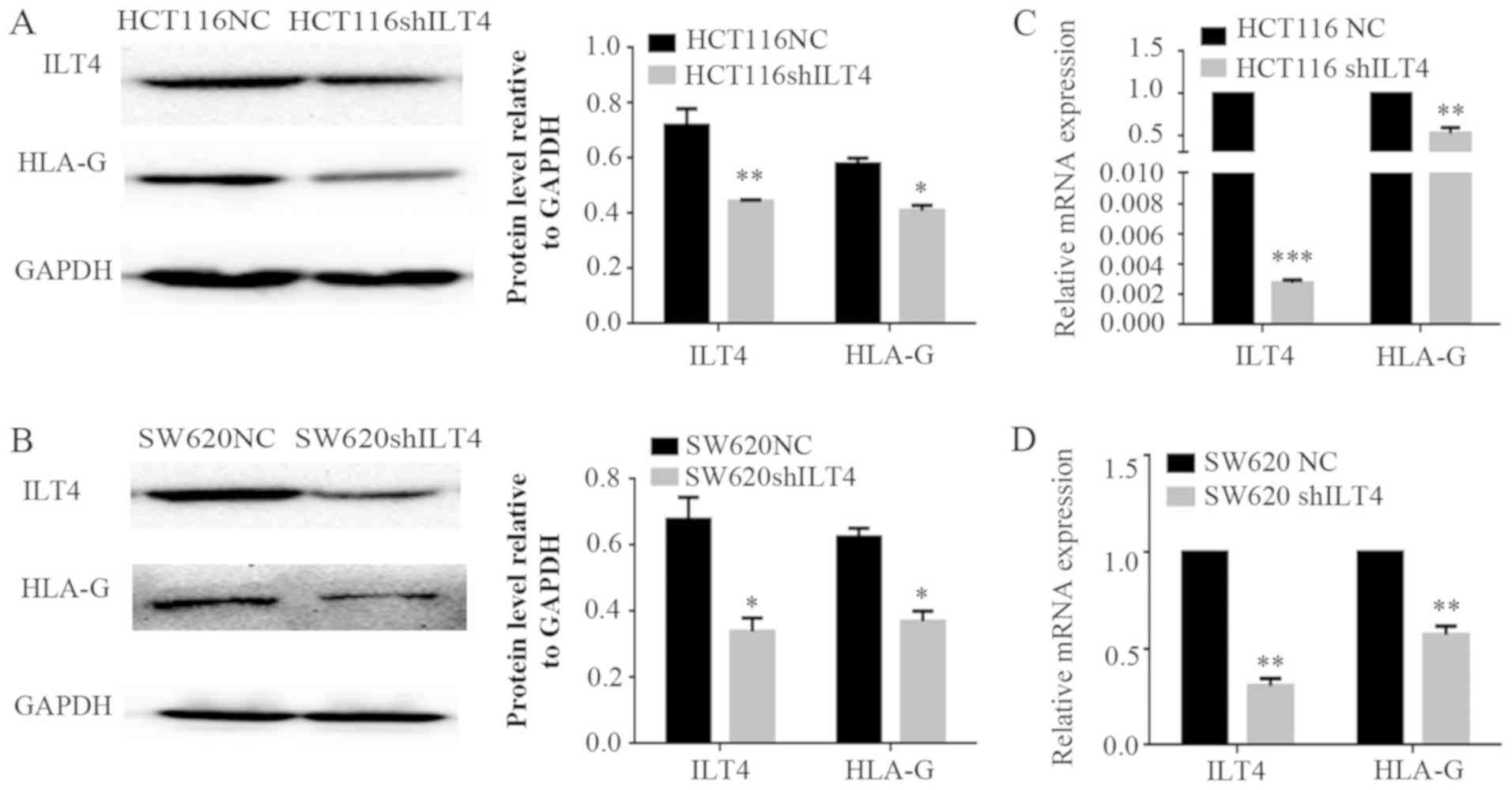
Similarly, HLA-G expression was notably
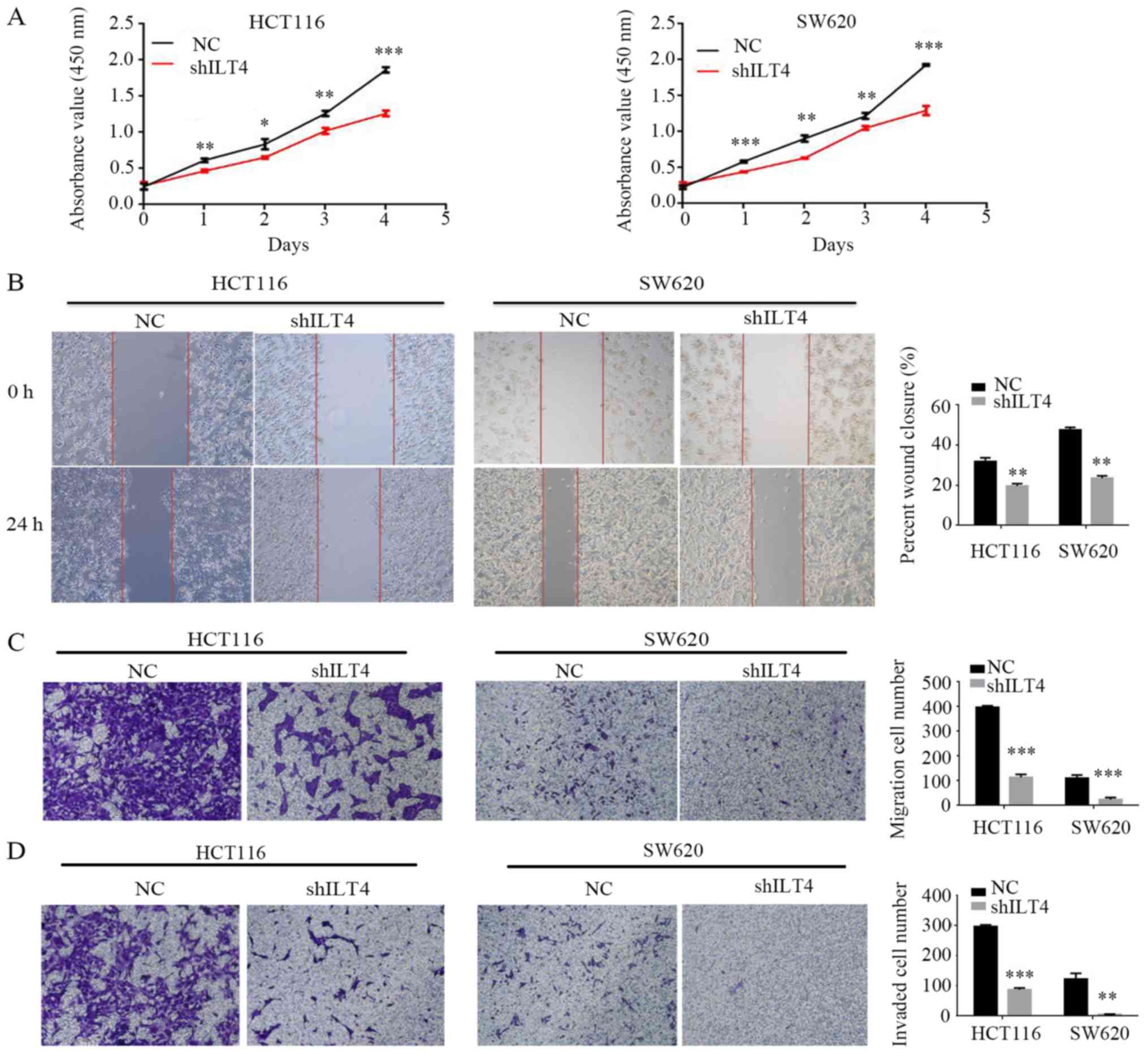
downregulated when ILT4 expression was reduced in HCT116 (Fig. 5A and B) and SW620 cells (Fig. 5C and D). Additionally,
downregulation of ILT4 notably reduced the proliferation, migration
and invasion of HCT116 and SW620 cells (Fig. 6). These results indicated that ILT4
promotes an increased proliferative, migratory and invasive
phenotype in CRC cells.
HLA-G fusion protein upregulates the ILT4
level in CRC cells in a dose-dependent manner
The effects of HLA-G expression on ILT4 expression
were assessed in CRC cell lines. HT29 cells, which express a low
level of ILT4/HLA-G, were treated with different concentrations
(10, 20, 50, 100, 200 and 500 ng/ml) of HLA-G fusion protein. In
the range of 10-200 ng/ml concentration, HLA-G fusion protein
notably upregulated the expression of ILT4 at the mRNA and protein
levels in a dose dependent manner (Fig. 7).
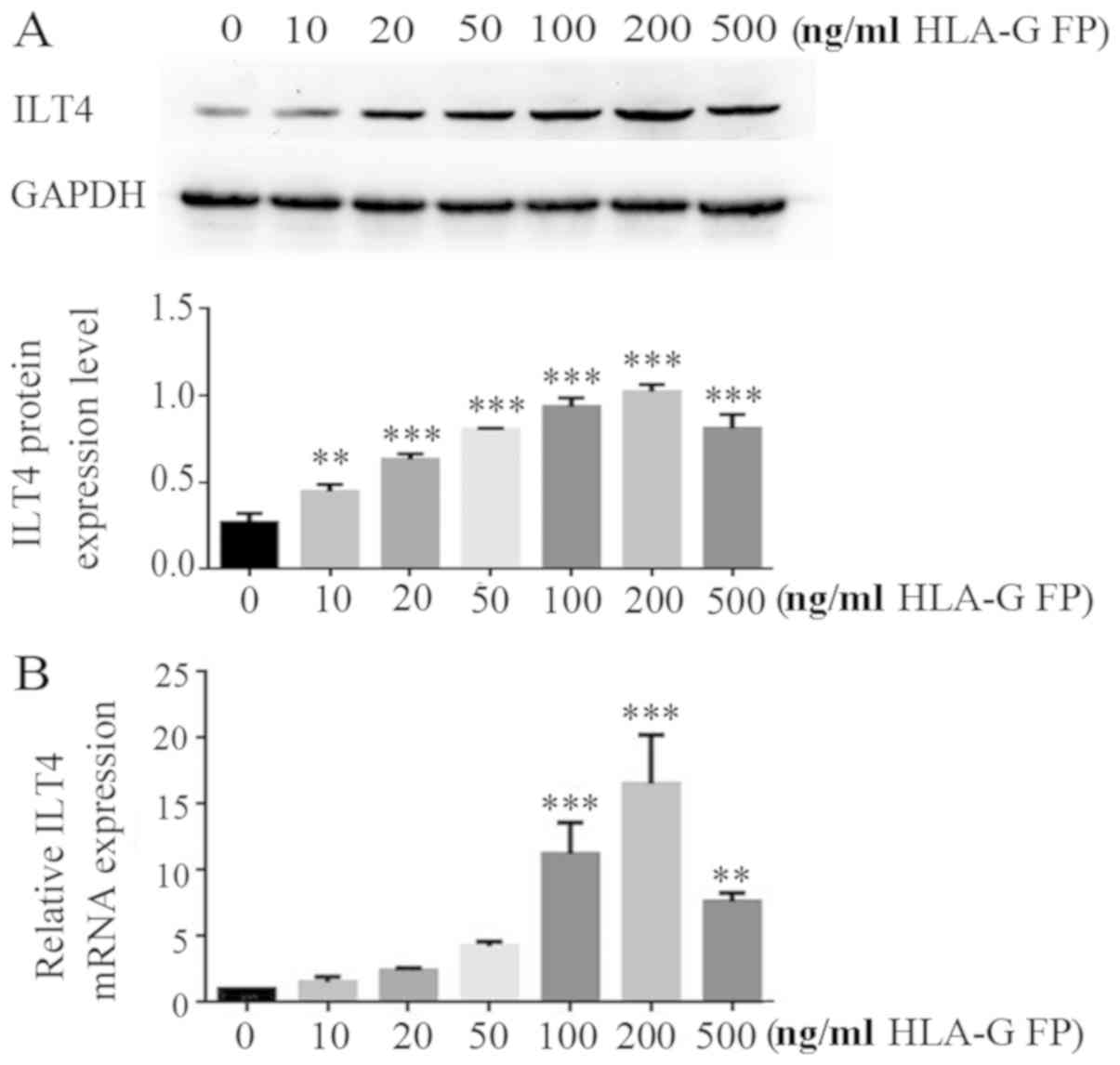 | Figure 7HLA-G FP upregulates ILT4 expression
in CRC cells in a dose dependent manner. (A) Protein expression of
ILT4 in HT29 cells following adding different concentrations (10,
20, 50, 100, 200 and 500 ng/ml) of HLA-G FP was increased, compared
with cells with 0 ng/ml HLA-G FP. (B) The expression of ILT4 mRNA
in HT29 cells following adding different concentrations (10, 20,
50, 100, 200 and 500 ng/ml) of HLA-G FP was increased, compared
with cells with 0 ng/ml HLA-G FP. The results are presented as the
mean ± standard deviation from three independent experiments.
**P<0.01 and ***P<0.001 vs. cells with
0 ng/ml HLA-G FP. HLA-G, human leukocyte antigen G; ILT4,
immunoglobulin like transcript 4; FP, fusion protein. |
HLA-G fusion protein induces the
activation of AKT and ERK protein and promotes the proliferation,
migration and invasion of CRC cells through binding with ILT4
To identify the underlying molecular mechanisms of
ILT4/HLA-G in CRC progression, HT29 cells were treated with 200
ng/ml HLA-G fusion protein, and the expression levels of ERK1/2 and
AKT were analyzed. The results demonstrated that the levels of
phospho ERK1/2 and phospho AKT were notably enhanced, while the
total ERK and AKT levels did not change significantly (Fig. 8A). Simultaneously, following
stimulation by HLA-G, HT29 cells exhibited an increased capacity
for proliferation, migration and invasion (Fig. 8B E).
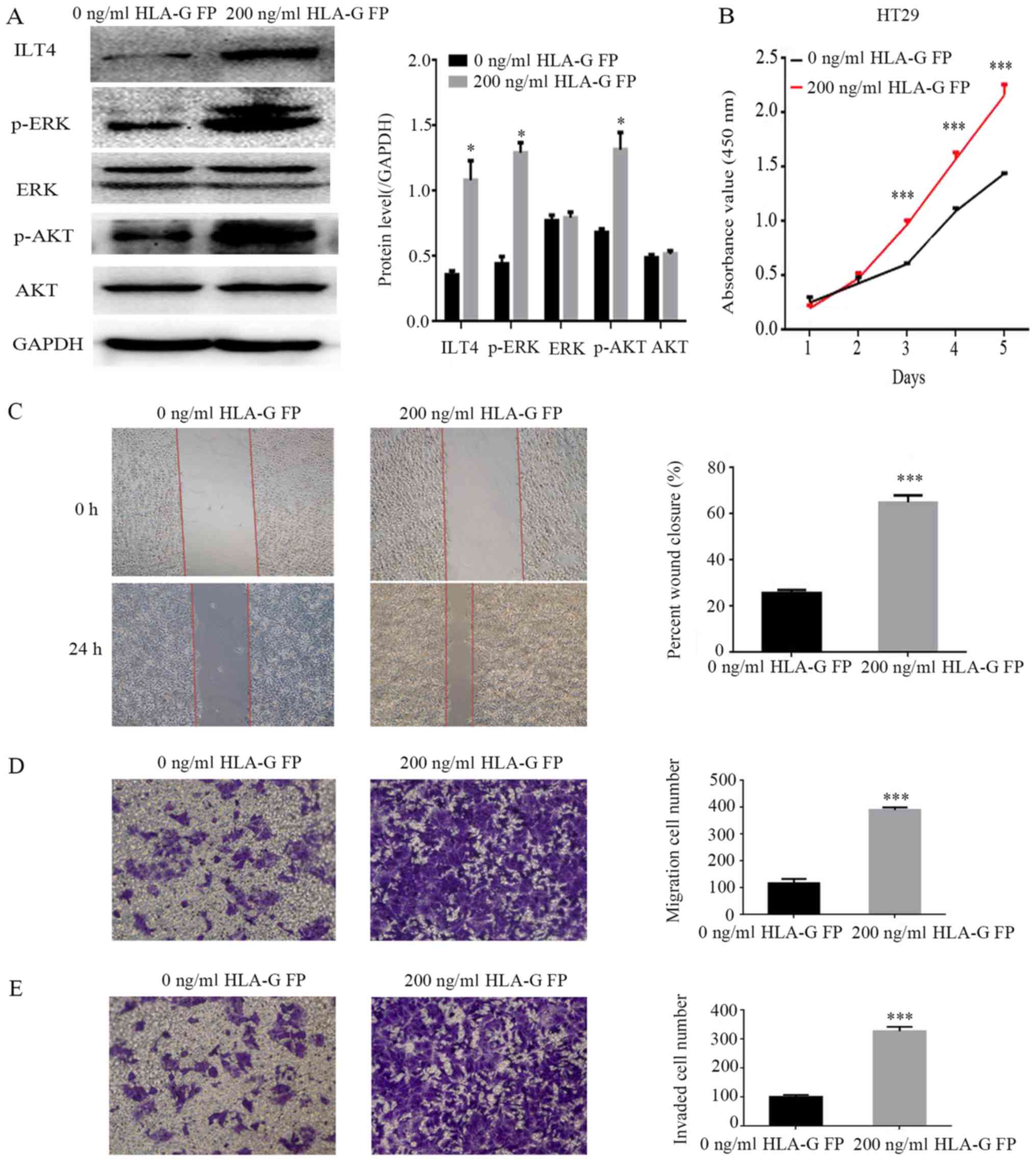 | Figure 8Addition of HLA-G FP activates AKT
and ERK signaling, and promotes the proliferation, migration and
invasion of HT29 cells. (A) Protein expres sion of ILT4, AKT, p
AKT, ERK, p ERK and GAPDH in HT29 following the addition of HLA-G
FP. PBS served as a negative control. (B) The effects of HLA-G FP
on proliferation were evaluated by Cell Counting Kit 8 assays. The
effects of HLA-G FP on migration were detected by (C) wound healing
assay (magnification, ×40) and (D) Matrigel invasion assay
(magnification, ×100). (E) The effects of HLA-G FP on invasion were
measured by Matrigel invasion assays. Magnification, ×100. The
results are presented as the mean ± standard deviation from three
independent experiments. *P<0.05 and
***P<0.001 vs. cells with 0 ng/ml HLA-G FP. HLA-G,
human leukocyte antigen G; ILT4, immunoglobulin like transcript 4;
FP, fusion protein; p , phospho ; ERK, extracellular signal
regulated kinase; AKT, protein kinase B. |
The present study sought to block the ILT4
expression in HT29 cells via anti ILT4 antibody to identify its
function in the cellular malignant behaviors promoted by HLA-G.
Notably, the upregulation of phospho ERK1/2 and phospho AKT
expression induced by HLA-G was no longer significantly altered
(Fig. 9A), and the cell
proliferation, invasion and migration of HT29 were notably reduced
(Fig. 9B E).
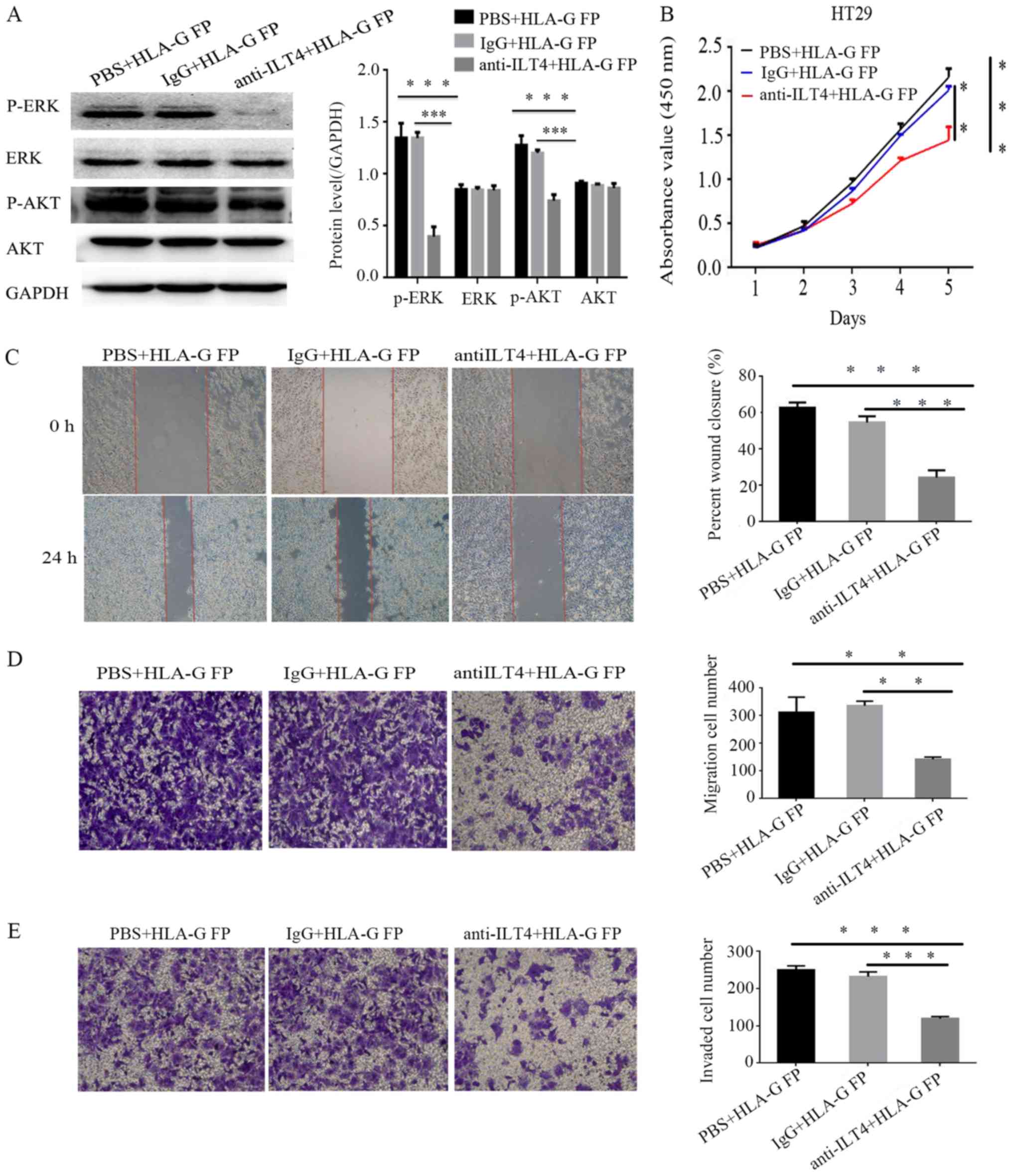 | Figure 9Anti ILT4 antibody can reduce the
signal activation and malignant cellular behaviors promoted by
HLA-G. (A) The protein expression of AKT, p AKT, ERK, p ERK and
GAPDH in HT29 cells following treatment with anti ILT4 antibody and
HLA-G FP. (B) Following blocking ILT4, the effects of HLA-G FP on
proliferation were evaluated by Cell Counting Kit-8 assays, the
effects on migration were measured by (C) wound-healing assay
(magnification, ×40) and (D) Transwell migration assay
(magnification, ×100), and the effects on invasion were measured by
(E) Matrigel invasion assays (magnification, ×100). The same volume
of IgG and PBS were used as negative controls. The results are
presented as the mean ± standard deviation from three independent
experiments. **P<0.01 and ***P<0.001
vs. cells with PBS+HLA-G FP. HLA-G, human leukocyte antigen G;
ILT4, immunoglobulin like transcript 4; FP, fusion protein; p ,
phospho ; ERK, extracellular signal regulated kinase; AKT, protein
kinase B. |
Discussion
ILT4 is a predominantly immunosuppressive molecule
and the majority of researchers have focused on its function on DCs
(23-26). Our previous studies identified ILT4
overexpression in breast cancer and NSCLC tissues (9-11).
However, the expression level of ILT4 in human CRCs and its
underlying role in CRC tumorigenesis have not been reported. HLA-G
has been reported to be able to upregulate the expression of ILT2,
ILT3 and ILT4 in antigen presenting cells (13), and our previous study demonstrated
the interaction of HLA-G with ILT4 in NSCLC (11). Previously, researchers have
investigated the HLA-G/ILT interaction as an immune checkpoint, and
it has been deduced that the use of anti HLA-G or anti ILT
antibodies may be considered as a novel immunotherapy strategy
(16,26-28).
Studies have also detected the expression of HLA-G in CRC, but its
associated mechanisms remain to be investigated (29,30).
The present study demonstrated the high expression
of ILT4 and HLA-G in human CRC tissues, and the co expression of
the two proteins was associated with the pathological
characteristics (age, sex, lymph node metastasis and TNM staging)
and survival time of patients with CRC. Furthermore, the expression
of ILT4 in men is increased, compared with women. Compared with
other groups, patients with CRC in the ILT4+/HLA-G+ group had
larger tumor sizes, increased TNM stages, increased lymph node
metastasis and reduced survival times, which indicated that ILT4
and HLA-G could be prognostic factors to predict poor clinical
response and survival time in patients with CRC. A number of
studies analyzed the association between HLA-G and the OS of
patients with CRC, but have not reported consistent conclusions
(29,31). The present data, analyzed by Kaplan
Meier, demonstrated that the high expression of HLA-G and ILT4 was
statistically associated with the survival time of patients with
CRC.
Additionally, the expression of ILT4/HLA-G and their
interaction in CRC cell lines were detected, at the mRNA and
protein levels. The interference of ILT4 expression was positively
associated with HLA-G expression; however, the administration of
HLA-G fusion protein increased the expression of ILT4 in a dose
dependent manner. The present study confirmed that the interaction
between ILT4/HLA-G in CRC cells may be mediated in an autocrine
manner, which has been demonstrated in NSCLC (11). Furthermore, it was elucidated that
the ILT4/HLA-G interaction notably enhanced cell proliferation,
migration and invasion, which confirmed the potential of ILT4/HLA-G
overexpression as a tumor indicator. Additionally, the present
study sought to reveal the underlying molecular mechanism of
ILT4/HLA-G in tumor promotion. It was demonstrated that HLA-G
induced the activation of AKT and ERK1/2 signaling by binding with
ILT4, accounting for the promoted cell proliferation, invasion and
migration of HT29 cells.
In conclusion, the present study identified the ILT4
and HLA-G overexpression in CRC tissue samples and its association
with poor prognosis in patients with CRC. Furthermore, it was
demonstrated that the ILT4/HLA-G interaction facilitated the cell
proliferation, invasion and migration of CRC by activating AKT and
ERK signaling. Therefore, it may be considered that ILT4 HLA-G may
function as a beneficial checkpoint in the prevention and treatment
of CRC. Further research is necessary to provide in vivo data and
determine other factors involved in ILT4/HLA-G signaling.
Supplementary Materials
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81372334), the
Natural Science Foundation of Shandong Province (grant no.
ZR2010HM105) and the Clinical Medical Innovation Project of Jinan
City (grant no. 201805064).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
ZC, LW and YH performed the cell experiments and
wrote this original manuscript. WG and XW analyzed the clinical
data and collected the human CRC tissue samples. RG and MZ
performed the statistical analysis. YS and SY designed the research
and modified the manuscript. All authors read the final manuscript
and approved the publication.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Jinan Central Hospital Affiliated to Shandong University (Jinan,
China) and written informed consent was obtained from all
patients.
Patient consent for publication
All patients involved in this study gave consent for
the publica tion of the clinical and pathological data.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Abbreviations:
|
CRC
|
colorectal cancer
|
|
HLA-G
|
human leukocyte antigen G
|
|
ILT4
|
immunoglobulin like transcript 4
|
|
AKT
|
protein kinase B
|
|
ERK
|
extracellular signal regulated
kinase
|
|
DC
|
dendritic cell
|
|
MHC
|
major histocompatibility complex
|
|
SHP
|
protein tyrosine phosphatase
|
|
NSCLC
|
non small cell lung cancer
|
|
OS
|
overall survival
|
|
RT qPCR
|
reverse transcription quantitative
polymerase chain reaction
|
References
|
1
|
Liang R, Lin Y, Yuan CL, Liu ZH, Li YQ,
Luo XL, Ye JZ and Ye HH: High expression of estrogen related
receptor α is signifi cantly associated with poor prognosis in
patients with colorectal cancer. Oncol Lett. 15:5933–5939.
2018.PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
3
|
Petera J, Dušek L, Sirák I, Soumarova R
and Jarkovsky J: Cancer in the elderly in the Czech Republic. Eur J
Cancer Care (Engl). 24:163–178. 2015. View Article : Google Scholar
|
|
4
|
Pitule P, Vycital O, Bruha J, Novak P,
Hosek P, Treska V, Hlavata I, Soucek P, Kralickova M and Liska V:
Differential expression and prognostic role of selected genes in
colorectal cancer patients. Anticancer Res. 33:4855–4865.
2013.PubMed/NCBI
|
|
5
|
Colonna M, Nakajima H and Cella M: A
family of inhibitory and activating Ig like receptors that modulate
function of lymphoid and myeloid cells. Semin Immunol. 12:121–127.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Borges L and Cosman D: LIRs/ILTs/MIRs,
inhibitory and stimulatory Ig superfamily receptors expressed in
myeloid and lymphoid cells. Cytokine Growth Factor Rev. 11:209–217.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shiroishi M, Tsumoto K, Amano K,
Shirakihara Y, Colonna M, Braud VM, Allan DS, Makadzange A, Rowland
Jones S, Willcox B, et al: Human inhibitory receptors Ig like
transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I
binding and bind preferentially to HLA-G. Proc Natl Acad Sci USA.
100:8856–8861. 2003. View Article : Google Scholar
|
|
8
|
Nakajima H, Asai A, Okada A, Ping L,
Hamajima F, Sata T and Isobe K: Transcriptional regulation of ILT
family receptors. J Immunol. 171:6611–6620. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun Y, Liu J, Gao P, Wang Y and Liu C:
Expression of Ig like transcript 4 inhibitory receptor in human non
small cell lung cancer. Chest. 134:783–788. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu J, Wang L, Gao W, Li L, Cui X, Yang H,
Lin W, Dang Q, Zhang N and Sun Y: Inhibitory receptor
immunoglobulin like transcript 4 was highly expressed in primary
ductal and lobular breast cancer and significantly correlated with
IL 10. Diagn Pathol. 9:852014. View Article : Google Scholar
|
|
11
|
Zhang Y, Zhao J, Qiu L, Zhang P, Li J,
Yang D, Wei X, Han Y, Nie S and Sun Y: Co expression of ILT4/HLA-G
in human non small cell lung cancer correlates with poor prognosis
and ILT4 HLA-G interaction activates ERK signaling. Tumour Biol.
37:11187–11198. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Djurisic S and Hviid TV: HLA Class Ib
Molecules and Immune Cells in Pregnancy and Preeclampsia. Front
Immunol. 5:6522014. View Article : Google Scholar
|
|
13
|
LeMaoult J, Zafaranloo K, Le Danff C and
Carosella ED: HLA-G up regulates ILT2, ILT3, ILT4, and KIR2DL4 in
antigen presenting cells, NK cells, and T cells. FASEB J.
19:662–664. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Köstlin N, Ostermeir AL, Spring B, Schwarz
J, Marmé A, Walter CB, Poets CF and Gille C: HLA-G promotes myeloid
derived suppressor cell accumulation and suppressive activity
during human pregnancy through engagement of the receptor ILT4. Eur
J Immunol. 47:374–384. 2017. View Article : Google Scholar
|
|
15
|
Rouas Freiss N, Moreau P, LeMaoult J and
Carosella ED: The dual role of HLA-G in cancer. J Immunol Res.
2014:3597482014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rouas Freiss N, LeMaoult J, Verine J,
Tronik Le, Roux D, Culine S, Hennequin C, Desgrandchamps F and
Carosella ED: Intratumor heterogeneity of immune checkpoints in
primary renal cell cancer: Focus on HLA-G/ILT2/ILT4.
OncoImmunology. 6:e13420232017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Özgül Özdemir RB, Özdemir AT, Oltulu F,
Kurt K, Yiğittürk G and Kırmaz C: A comparison of cancer stem cell
markers and nonclassical major histocompatibility complex antigens
in colorectal tumor and noncancerous tissues. Ann Diagn Pathol.
25:60–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alaoui L, Palomino G, Zurawski S, Zurawski
G, Coindre S, Dereuddre Bosquet N, Lecuroux C, Goujard C, Vaslin B,
Bourgeois C, et al: Early SIV and HIV infection promotes the
LILRB2/MHC I inhibitory axis in cDCs. Cell Mol Life Sci.
75:1871–1887. 2018. View Article : Google Scholar
|
|
19
|
Nowak I, Wilczyńska K, Wilczyński JR,
Malinowski A, Radwan P, Radwan M and Kuśnierczyk P: KIR, LILRB and
their Ligands’ Genes as Potential Biomarkers in Recurrent
Implantation Failure. Arch Immunol Ther Exp (Warsz). 65:391–399.
2017. View Article : Google Scholar
|
|
20
|
Guerra de Blas PC, Villaseñor Talavera YS,
Cruz González DJ, Ba randa L, Doníz Padilla L, Abud Mendoza C,
González Amaro R and Monsiváis Urenda AE: Analysis of the
Expression and Function of Immunoglobulin Like Transcript 4 (ILT4,
LILRB2) in Dendritic Cells from Patients with Systemic Lupus
Erythematosus. J Immunol Res. 2016:41630942016. View Article : Google Scholar
|
|
21
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real time quantitative PCR and
the 2(Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Gao A, Sun Y and Peng G: ILT4 functions as
a potential checkpoint molecule for tumor immunotherapy. Biochim
Biophys Acta Rev Cancer. 1869:278–285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Castellaneta A, Mazariegos GV, Nayyar N,
Zeevi A and Thomson AW: HLA-G level on monocytoid dendritic cells
correlates with regulatory T cell Foxp3 expression in liver
transplant tolerance. Transplantation. 91:1132–1140. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang S, Ristich V, Arase H, Dausset J,
Carosella ED and Horuzsko A: Modulation of dendritic cell
differentiation by HLA-G and ILT4 requires the IL 6 STAT3 signaling
pathway. Proc Natl Acad Sci USA. 105:8357–8362. 2008. View Article : Google Scholar
|
|
26
|
Ristich V, Zhang W, Liang S and Horuzsko
A: Mechanisms of prolongation of allograft survival by HLA-G/ILT4
modified dendritic cells. Hum Immunol. 68:264–271. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Carosella ED, Rouas Freiss N, Tronik Le,
Roux D, Moreau P and LeMaoult J: HLA-G: An Immune Checkpoint
Molecule. Adv Immunol. 127:33–144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Apps R, Gardner L and Moffett A: A
critical look at HLA-G. Trends Immunol. 29:313–321. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zeestraten EC, Reimers MS, Saadatmand S,
Goossens Beumer IJ, Dekker JW, Liefers GJ, van den Elsen PJ, van de
Velde CJ and Kuppen PJ: Combined analysis of HLA class I, HLA E and
HLA-G predicts prognosis in colon cancer patients. Br J Cancer.
110:459–468. 2014. View Article : Google Scholar
|
|
30
|
Guo ZY, Lv YG, Wang L, Shi SJ, Yang F,
Zheng GX, Wen WH and Yang AG: Predictive value of HLA-G and HLA E
in the prognosis of colorectal cancer patients. Cell Immunol.
293:10–16. 2015. View Article : Google Scholar
|
|
31
|
Reimers MS, Engels CC, Putter H, Morreau
H, Liefers GJ, van de Velde CJ and Kuppen PJ: Prognostic value of
HLA class I, HLA E, HLA-G and Tregs in rectal cancer: A
retrospective cohort study. BMC Cancer. 14:4862014. View Article : Google Scholar
|