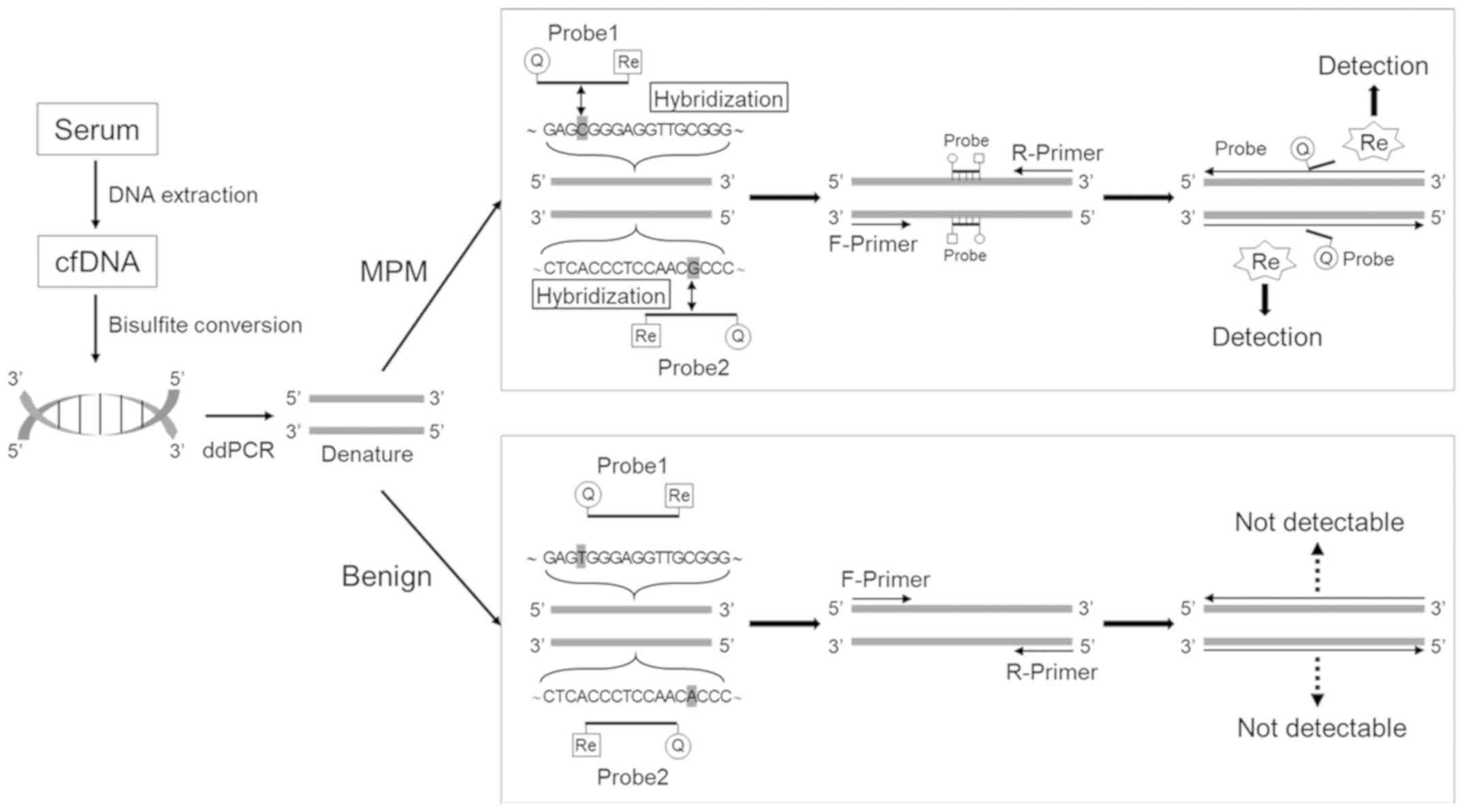
Introduction
Malignant pleural mesothelioma (MPM) is a rare and
highly aggressive tumor arising from the pleura or other
mesothelial surfaces and is most commonly associated with asbestos
exposure, which is known as a major risk factor. Although asbestos
use is now prohibited in Western countries, the incidence of MPM is
not expected to decrease in the near future due to the long
incubation period between asbestos exposure and the onset of MPM
(1,2). Moreover, asbestos continues to be
used in many developing and emerging economies, such as countries
in Southeast Asia, suggesting the possibility of future epidemics
of MPM. In the majority of cases, MPM is only diagnosed at an
advanced disease stage; therefore, the development of a novel
diagnostic approach is warranted (3,4).
Recently, the concept of a ‘liquid biopsy’ for the
diagnosis and monitoring of diseases has attracted attention.
Several studies have suggested that the individual genetic profiles
of cancers are highly heterogeneous and that these profiles can
even change during the course of the disease, particularly in
response to treatment (5,6). At present, the molecular profiles of
patients with solid tumors are generally established using
surgically resected or biopsy specimens. However, the use of tissue
biopsies is limited by their invasiveness, making it difficult to
grasp chronological alterations in molecular profiles and
potentially missing some genomic changes. A liquid biopsy
originally referred to an analysis of the genomic profiles of
circulating tumor cells (7), and
this method has attracted particular interest among experts in the
field of clinical oncology. This definition has now been extended
to include various tumor components, such as circulating cell-free
RNA (cfRNA), circulating cell-free DNA (cfDNA), circulating
cell-free tumor DNA (ctDNA) and exosomes, and this technique
enables clinicians to repeatedly and non-invasively explore
real-time changes in the genomic profiles of human cancers.
MicroRNAs (miRNAs or miRs) are a group of small
noncoding, endogenous, single-stranded RNAs that play an essential
role in the regulation of gene expression. A number of studies have
reported that the aberrant hypermethylation of CpG islands in the
promoter regions is closely related to the silencing of
tumor-suppressive miRs in several types of cancer (8-11).
We previously identified that among several miRs, the epigenetic
silencing of miR-34b/c by aberrant methylation in the promoter
region plays an important role in the tumorigenesis of MPM
(12). miR-34s have been
discovered to be direct transcriptional targets of p53, and to
constitute a part of the p53 tumor suppressor network regulating
cell cycle arrest, apoptosis and senescence (13,14).
As regards the application of miRs as biomarkers, Suzuki et
al reported that the aberrant methylation of miR-34b/c in
biopsy specimens was a predictive marker of metachronous gastric
cancer (15). Wu et al also
reported that the detection of methylation in the promoter regions
of miR-34s using stool DNA was useful as a screening biomarker for
colorectal cancer (16).
Additionally, we have previously revealed that the degree of
miR-34b/c methylation in serum-circulating DNA is associated with
the development of MPM (17).
Although the origins of ctDNA differ, these previous studies
suggest the possibility that the methylation of the miR-34b/c
promoter is a promising biomarker.
In our previous study in 2011, we compared the
degree of methylation using MPM cell lines, MPM tissues and
nonmalignant mesothelial primary cultures that were established
from pleural effusions of cancer-free patients, and we have shown
that the promoter of miR-34b/c is highly methylated in MPM
(12). Based on these findings,
the aim of the present study was to apply the miR-34b/c methylation
specifically observed in MPM to the diagnosis and prediction of the
disease progression. For this purpose, in this study, we
established a novel assay with which to detect DNA methylation in
the blood using droplet digital PCR (ddPCR) technology, enabling
the highly sensitive and quantitative detection of target genes
(18). In ddPCR, the input DNA is
distributed among approximately 20,000 droplets, and each droplet
contains 1 or fewer copies of the target or background DNA; this
makes it possible to detect 0.001% of the target gene from the
background DNA (19-21). Our established assay was then
verified using serum samples from patients with MPM, pleural plaque
patients and healthy volunteers.
Materials and methods
Clinical samples and cell lines
We collected >1 ml peripheral blood sample from
35 cases of MPM, 29 cases of pleural plaque (PP) and 10 healthy
volunteers (HV) at the Okayama University Hospital (Okayama,
Japan), Okayama Rosai Hospital (Okayama, Japan), or the National
Hospital Organization, Yamaguchi Ube Medical Center (Yamaguchi,
Japan), between January, 2005 and January, 2015. The details are
described in Table I. The age of
all the healthy volunteers was >20 years and healthy individuals
who were not any current medications were recruited. None of the
participants had a medical history of cancer other than MPM, and
all the blood samples were collected before any type of treatment.
The blood samples were immediately centrifuged at 5,000 × g for 5
min, and the separated serum samples were stored at −80°C at the
respective institutions. In addition, 3 surgically resected MPM
specimens obtained from the National Hospital Organization,
Yamaguchi Ube Medical Center were also subjected to the methylation
assay. All the tissues were frozen in liquid nitrogen immediately
after surgery and stored at -80°C. This study was conducted with
the approval of the Institutional Review Board/Ethical Committee of
Okayama University; each of the participants provided written
informed consent for the sample collection. All the experiments
were performed in accordance with the Declaration of Helsinki.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| A, Patients in
group A |
|---|
|
|---|
| Characteristic | MPM (n=13) | PP (n=20) |
|---|
| Median (range),
years | 71 (51-90) | 69.5 (65-72) |
| Sex,
male/female | 9/4 | 20/0 |
| Smoking
history | | |
| Never | 3 | 5 |
|
Former/current | 10 | 15 |
| Histological
subtypes | | |
| Epithelioid | 6 | N/A |
| Biphasic | 4 | N/A |
| Sarcomatoid | 3 | N/A |
| Clinical stage | | |
| I | 3 | N/A |
| II | 2 | N/A |
| III | 5 | N/A |
| IV | 1 | N/A |
| Unknown | 2 | N/A |
|
| B, Patients in
group B |
|
| Characteristic | MPM (n=22) | PP (n=9) | HV (n=10) |
|
| Median (range),
years | 61.5 (49-86) | 77 (60-86) | 31 (25-37) |
| Sex,
male/female | 19/3 | 9/0 | 10/0 |
| Smoking
history | | | |
| Never | 6 | 3 | 8 |
|
Former/current | 16 | 6 | 2 |
| Histological
subtypes | | | |
| Epithelioid | 15 | N/A | N/A |
| Biphasic | 5 | N/A | N/A |
| Sarcomatoid | 2 | N/A | N/A |
| Clinical stage | | | |
| I | 8 | N/A | N/A |
| II | 5 | N/A | N/A |
| III | 5 | N/A | N/A |
| IV | 4 | N/A | N/A |
We also used two human MPM cell lines [NCI-H28
(H28), NCI-H2052 (H2052)] and one human normal mesothelial cell
line (MeT-5A) as positive and negative controls, respectively. The
H28 and H2052 cells were obtained as kind gifts from Dr Adi F.
Gazdar (Hamon Center for Therapeutic Oncology Research and
Department of Pathology, University of Texas Southwestern Medical
Center at Dallas, Dallas, TX, USA). The MeT-5A cell line was
purchased from the American Type Culture Collection (ATCC,
Manassas, VA, USA). For the cell lines that had been stored
long-term in liquid nitrogen, a DNA fingerprinting analysis by
short tandem repeat profiling (the PowerPlex 1.2 System, Promega,
Madison, WI, USA) was performed for cell authentication. The cells
were maintained in RPMI-1640 medium (Sigma Chemical Co., St. Louis,
MO, USA) supplemented with 10% FBS and cultured in a humidified
incubator under 5% CO2 at 37°C, and the samples were
routinely tested for mycoplasma using the Venor GeM OneStep kit
(Minerva Biolabs, Berlin, Germany).
DNA extraction, bisulfite conversion, and
bisulfite DNA sequencing
We extracted DNA from the serum samples using the
QIAamp Circulating Nucleic Acid kit (Qiagen, Venlo, The
Netherlands). The DNA concentrations were quantified using the
Qubit 2.0 Fluorometer and Qubit dsDNA HS or BR assay kit (Thermo
Fisher Scientific, Waltham, MA, USA). DNA was also extracted from
the MPM tissues using the phenol-chloroform method. DNA was
extracted from the cell lines using the DNeasy Blood and Tissue kit
(Qiagen). Genomic DNA was subjected to bisulfite conversion using
the Epitect Bisulfite kit (Qiagen), and the methylation status of
miR-34b/c was determined using bisulfite DNA sequencing as
previously described (12,17). The raw sequence chromatograms were
analyzed using Chromas Lite software version 2.6.5 (available at
http://technelysium.com.au/wp/chromas/). The degree of
methylation was determined by comparing the intensity of the
sequencing electropherogram of cytosine with that of thymine at
each of the CpG sites. Based on the electropherograms, we
quantified the relative ratios between the heights of each of the
waves, as described previously (Fig.
1A) (22), and classified the
degree of methylation into three groups, as follows:
Low-methylated, degree of methylation <20%; moderately
methylated, degree of methylation between 20 and 70%; and highly
methylated, degree of methylation >70%.
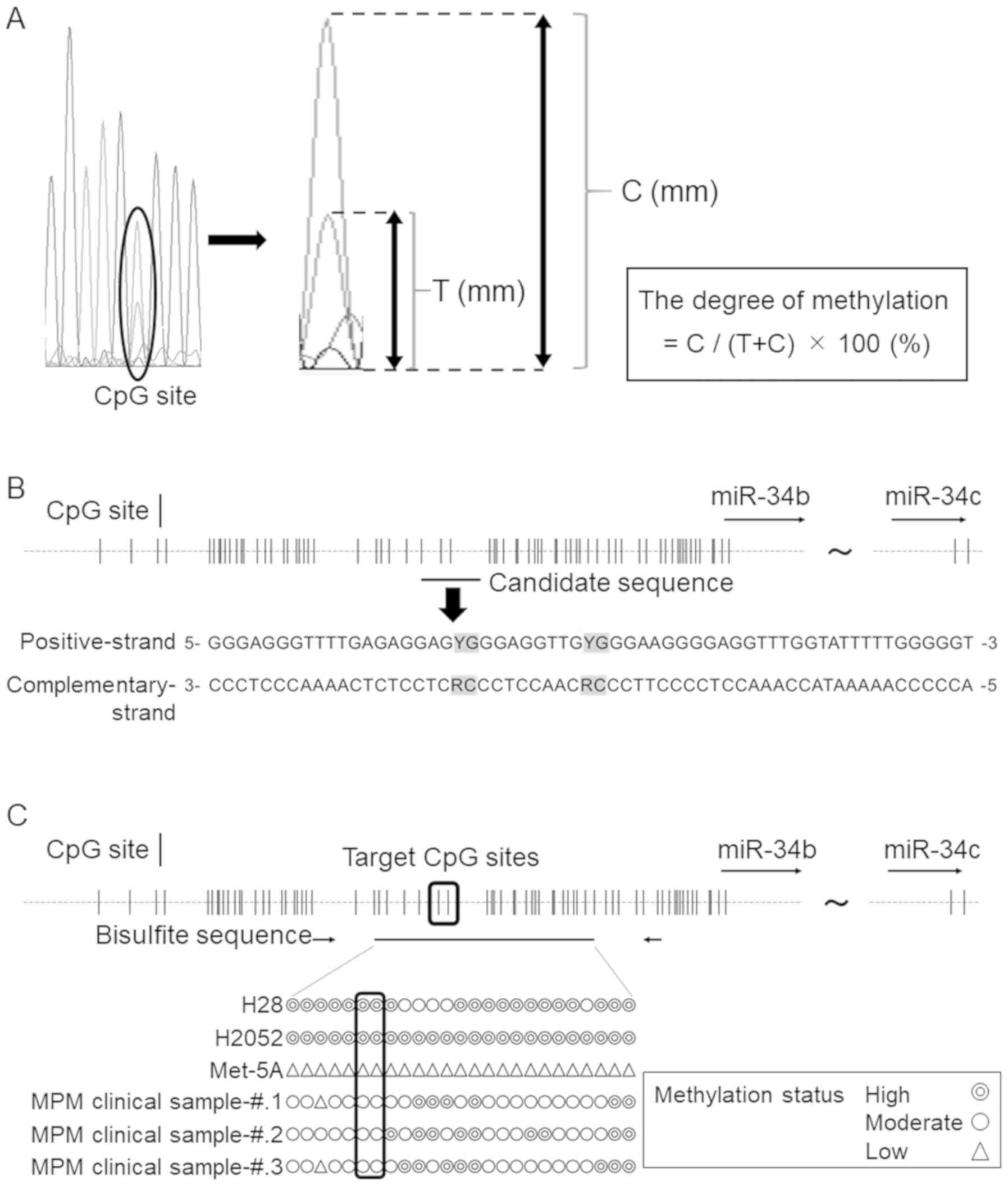 | Figure 1Optimal probe and primer design. (A)
Method for calculating the degree of methylation from sequencing
electropherograms. C, cytosine (methylated allele); T, thymine
(unmethylated allele). (B) Schema of the miR-34b/c promoter region.
CpG sites included in the selected sequence are highlighted in
gray. G, guanine; A, adenine; T, thymine; C, cytosine; Y,
pyrimidine; R, purine. (C) Methylation statuses of 2 MPM cell
lines, 1 normal mesothelial cell line, and 3 MPM tissue specimens.
Double circles represents a highly methylated status, a single
circle represents a moderately methylated status, and a triangle
represents a low methylation status. The target CpG sites and the
methylation status are surrounded by the black line. MPM, malignant
pleural mesothelioma. |
Primers and TaqMan probes
The sequences of the primers and TaqMan probes used
in this study were designed based on the nucleotide sequence
submitted to GenBank (GenBank accession numbers NR 029839.1 for
miR-34b and NR 035765.1 for miR-34c). The melting temperature (Tm)
of each primer was calculated using the Oligo Calculator
(http://mbcf149.dfci.harvard.edu/docs/oligocalc.html).
The primers were synthesized by Invitrogen (Thermo Fisher
Scientific, Yokohama, Japan). The primers, including the mixed-base
and TaqMan probes containing the locked nucleic acids (LNAs) were
designed using the IDT Biophysics software (https://www.idtdna.com/pages/tools) and were
synthesized by Integra ted DNA Technologies KK (Tokyo, Japan).
ddPCR assay for miR-34b/c methylation
detection
ddPCR was performed using the QX200 Droplet Digital
PCR system (Bio-Rad, Hercules, CA, USA). EpiTect Control DNAs
(methylated or unmethylated and bisulfite-converted human DNA,
QIAGEN) were used for the assay validation. The total volume of the
PCR mixture used for the assay was 22 µl, containing 10
µl of ddPCR Supermix for Probes (No dUTP) (Bio-Rad), 1
µM of each primer, 0.25 µM of each probe and 200
µM of dNTP. As for the amount of DNA, 10 µl of cfDNA
extracted from the serum was used, while a total of 5 ng of DNA
(methylated and bisulfite-converted human control DNA) was applied
for the validation of the assay. The PCR conditions were described
in detail in our previous study (23). The annealing temperatures were
optimized by gradient PCR. The PCR products were then subjected to
analysis with the QX-200 droplet reader and QuantaSoft analysis
software (Version 1.7.4.0917) (Bio-Rad). The former measures the
fluorescence value of each droplet, and the latter measures the
number of positive and negative droplets in each sample and
calculates the fraction of positive droplets by a Poisson
algorithm. QuantaSoft analysis software cannot display the
fluorescence intensity of each droplet and standard deviation.
Statistical analysis
All in vitro experiments were performed at
least 3 times. Data are represented as the means ± standard
deviation. The concentrations of the target alleles were calculated
using QuantaSoft software (Bio-Rad) based on Poisson’s
distribution. The receiver-operating characteristic (ROC) curve
analysis was performed using JMP® 9.0.0 for Windows (SAS
Institute, Inc., Cary, NC, USA). A one-way ANOVA followed by
Bonferroni’s multiple comparisons test was conducted using GraphPad
Prism, version 7 (GraphPad Software, San Diego, CA, USA).
Probability values (P-values)<0.05 was considered to indicate
statistically significant differences.
Results
Appropriate sequences for primer
design
First, we examined candidate sequences suitable for
the primer and TaqMan probe design based on some key points, as
follows: i) Multiple CpG sites were included in the target sequence
to increase the sensitivity; ii) CpG sites were not included in the
primer sequences; and iii) the frequency of single nucleotide
polymorphisms (SNPs) was relatively low in the target sequences. In
addition, we made the amplicon size as small as possible to
increase the sensitivity of ctDNA detection, as described
previously (23). One of the
candidate sequences is shown in Fig.
1B, and the SNPs in this target region, as provided by the NCBI
dbSNP database (https://www.ncbi.nlm.nih.gov/projects/SNP/), are
listed in supplementary Table SI.
The possible frequency of SNPs in this region was ≤0.02%, which
reinforced the validity of this sequence. To confirm the
methylation status of the two CpG sites included in this sequence,
we performed bisulfite DNA sequencing. The results revealed that
both CpG sites were moderately or highly methylated in both the MPM
cell lines and the MPM clinical specimens, but not in the normal
mesothelial cell line (Fig. 1C).
Based on these results, we designed the primers as shown in
Table II. Validation of the
primer sets was performed to identify possible non-specific
reactions, and we confirmed the specificity of the primers (data
not shown).
 | Table IISequences of primers and probes. |
Table II
Sequences of primers and probes.
| Oligo name | Oligo sequences 5′
to 3′ | Tm (°C) | Product size
(bp) | Match-mismatch Tm
difference (°C) |
|---|
| Primers | MPM-Fw |
GGGAGGGTTTTGAGAGGAG | 62.54 | 60 | NA |
| MPM-Rv |
ACCCCCAAAAATACCAAACC | 63.28 | | NA |
| MSP-Fw |
AGAGAGTTAGTTTTAGGGTTTGGG | 61.5 | 358 | NA |
| MSP-Rv |
CCTCRAACCCCATTTCAC | 62.95 | | NA |
| Probes | Probe-P | FAM/AC+CT
C+CC+GCT/IABLFQ | 65.41 | NA | 21.03 |
| Probe-C1 |
FAM/TTG+CGGG+AAGGGG/IABLFQ | 64.07 | NA | 14.75 |
| Probe-C2 |
FAM/TG+CGG+G+A+AGG/IABLFQ | 63.23 | NA | 17.83 |
| Probe-C3 |
FAM/AGGTT+G+C+GGGAAG/IABLFQ | 63.56 | NA | 11.85 |
| Probe-C4 |
FAM/TG+CGGGAAGGGGAG/IABLFQ | 64.65 | NA | 13.29 |
Probe design and assay validation
Herein, we present a schema representing the
principle on which our methylation detection assay was based
(Fig. 2). The two CpG sites were
detected separately by two TaqMan probes with the same fluorescent
dye, and thus we examined the optimal probe design. As both CpG
sites in this sequence were located close to each other, we
designed one probe based on the sequence of the positive strand
(Probe-P), and the other based on the sequence of the complementary
strand (Probe-C). In addition, in order to obtain a sufficient
match-mismatch Tm difference, the probes were fabricated using
LNAs. Based on these concepts, we designed several probe sets
(Probe-P, Probe-C#1-4) (Table
II). To verify the validity of these probes, and to consider
the optimum annealing temperatures, gradient PCR was conducted
within the range of 51°C to 61°C. As a result, Probe-P and
Probe-C#1 had a higher fluorescence, compared with the other probes
(Fig. 3A and B). Determining
whether the two probes would function properly without competition
was also important for successful methylation detection. Therefore,
to test the interaction between Probe-P and Probe-C#1, we performed
the same experiment using the two probes in combination. We found
that the fluorescence intensity was enhanced when the probes were
used in combination, suggesting that the probes functioned
cooperatively (Fig. 3C and D), and
the optimal annealing temperature was determined to be 53.1°C.
Lastly, we confirmed whether this assay could correctly distinguish
between methylated and unmethylated DNA. As shown in Fig. 3E and F, the number of droplets with
a fluorescence intensity >3,000 was noticeably larger in the
methylated DNA group.
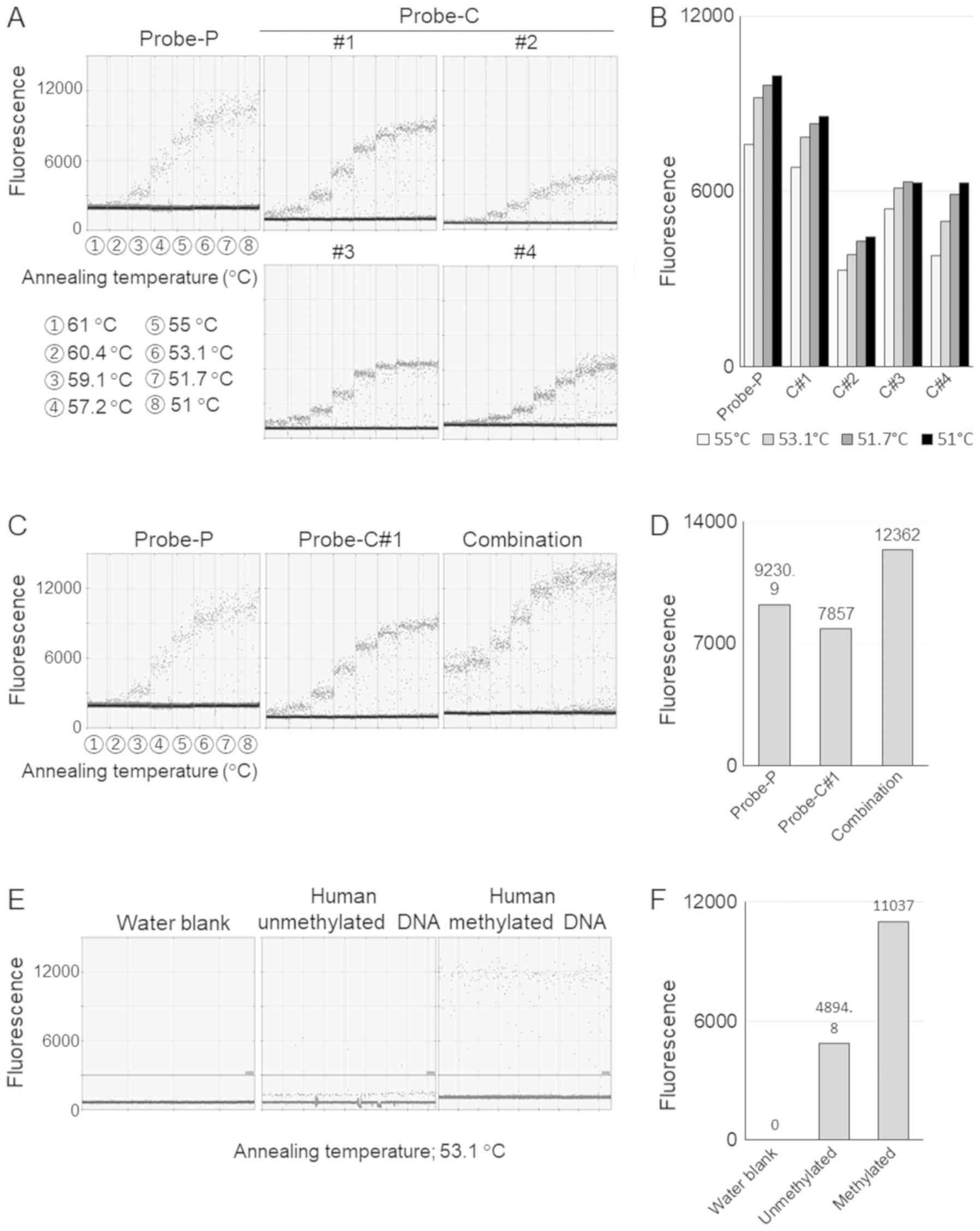 | Figure 3Validation of the established assay.
(A) Validation of probes. Gradient PCR was conducted within an
annealing temperature range of 51 to 61°C; the numbered circles
indicate the following temperatures: 1, 61°C; 2, 60.4°C; 3, 59.1°C;
4, 57.2°C; 5, 55°C; 6, 53.1°C; 7, 51.7°C; and 8, 51°C. Probe-P
represents the probe designed based on the sequence of the positive
strand, and Probe-C represents the probe designed based on the
sequence of the complementary strand. (B) The mean fluorescence
values of droplets with a fluorescence intensity of over 3,000. (C)
Verification of combined use of the probes. The annealing
temperature was ranged from 51 to 61°C; the numbered circles
indicate the following temperatures: 1, 61°C; 2, 60.4°C; 3, 59.1°C;
4, 57.2°C; 5, 55°C; 6, 53.1°C; 7, 51.7°C; and 8, 51°C. The use of
Probe-P and Probe-C#1 in combination was associated with an
enhanced fluorescence intensity, compared with that of each probe
alone (right panel). (D) The mean fluorescence values of droplets
with a fluorescence intensity of over 3,000 at annealing
temperature of 53.1°C. (E) Verification of established assay using
methylated and non-methylated human DNA. (F) The mean fluorescence
values of droplets with a fluorescence intensity of over 3,000 at
annealing temperature of 53.1°C. |
Clinical application of the established
assay
We then evaluated the feasibility of the clinical
application of this assay. We divided the serum samples (35 cases
of MPM, 29 cases of PP and 10 HVs) into group A (n=33) and group B
(n=41) according to their collection site: Samples obtained from
the Okayama Rosai Hospital were classified as group A, while those
obtained from the other two institutions were classified as group
B. The characteristics of the patients in the 2 groups are
summarized in Table I. The median
concentration of cfDNA extracted from the serum was 1.47
ng/µl for the MPM cases and 1.44 ng/µl for the
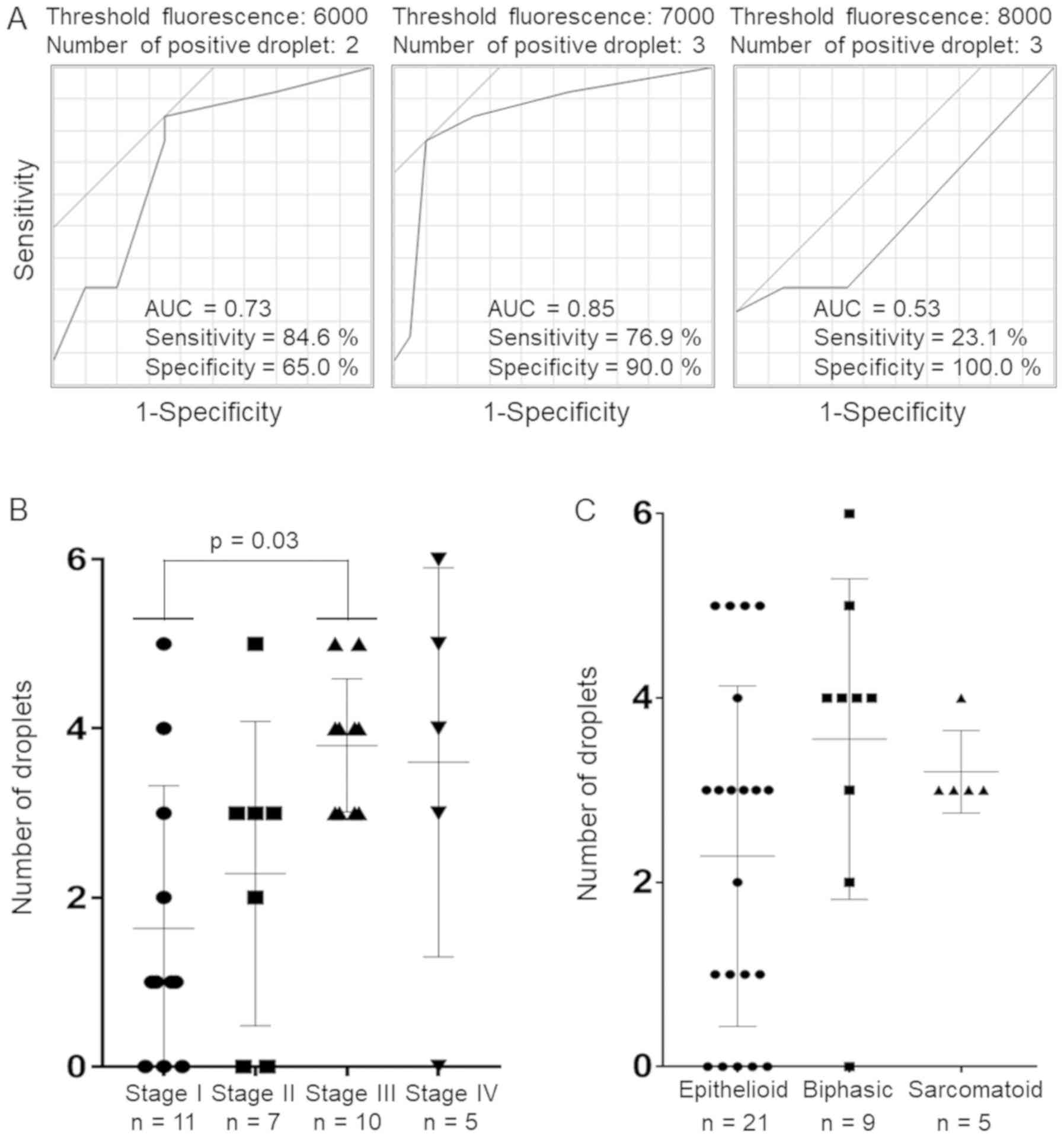
others. Firstly, to determine the positive criterion, we conducted
an ROC curve analysis comparing the MPM cases with other
non-malignant cases using samples from group A (Fig. 4A and Table SII). The results indicated that
the presence of at least 3 droplets with a fluorescence of over a
threshold value of 7,000 was the optimal cut-off for the diagnosis
of MPM, with a sensitivity of 76.9% and a specificity of 90%.
Subsequently, we evaluated the validity of this criterion. The
results are shown in Table III.
The sensitivity and specificity for the diagnosis of MPM in the
group B samples were 59.1 and 100%, respectively, while those for
the entire cohort were 65.7 and 94.9%, respectively, indicating a
moderate sensitivity and a high specificity. In addition, when we
focused on the diagnosis of only stage II or more advanced MPM, the
sensitivity increased to 81.8%. Actually, there were significant
differences in the number of droplets with fluorescence values of
at least 7,000 per case among stages (one-way ANOVA, P= 0.02)
(Bonferroni’s post hoc test; stage I vs. stage II, P>0.99; stage
I vs. stage III, P=0.03; stage I vs. stage IV, P>0.18; stage II
vs. stage III, P=0.39; stage II vs. stage IV, P>0.99; stage III
vs. stage IV, P>0.99), suggesting that the methylated allele
frequency may be associated with the stage of MPM progression
(Fig. 4B). On the other hand,
methylation was not detected in one case with clinical stage IV
MPM. We also assessed whether the histological subtypes were
associated with the methylated allele frequency. However, no
significant difference in the methylated allele frequency was
observed among the histological subtypes (one-way ANOVA, P=0.16)
(Bonferroni’s post-hoc test; epithelioid vs. biphasic, P=0.21;
epithelioid vs. sarcomatoid, P=0.87; biphasic vs. sarcomatoid,
P>0.99) (Fig. 4C).
 | Table IIIAssay sensitivity and specificity of
each group. |
Table III
Assay sensitivity and specificity of
each group.
| MPM | PP or HV |
|---|
| Group A (n=33) | | |
| Positive | 10 | 2 |
| Negative | 3 | 18 |
| Sensitivity, 76.9%;
specificity, 90.0% | | |
| Group B (n=41) | | |
| Positive | 13 | 0 |
| Negative | 9 | 19 |
| Sensitivity, 59.1%;
specificity, 100% | | |
| Entire cohort
(n=74) | | |
| Positive | 23 | 2 |
| Negative | 12 | 37 |
| Sensitivity, 65.7%;
specificity, 94.9% | | |
| Stage II or more
advanced MPM (n=61) | | |
| Positive | 18 | 2 |
| Negative | 4 | 37 |
| Sensitivity, 81.8%;
specificity, 94.9% | | |
Discussion
In this study, we established a TaqMan-based ddPCR
assay for the detection of the methylation of the miR-34b/c
promoter region in circulating DNA. The design of the two probes,
one from the positive strand and the other from the complementary
strand, allowed the successful detection of the methylation of the
two CpG sites located close to each other, with an overall
specificity of 94.9%. Although the sensitivity of our assay was
limited to 65.7%, when the analysis was focused on the detection of
stage II or more advanced cases of MPM, the sensitivity increased
to 81.8%, and there was a tendency that the methylated allele
frequency was higher in more advanced MPM. These findings suggest
that the methylation status may be positively associated with the
stage of MPM progression and that it may be useful for predicting
tumor progression. As for the association between the methylation
status of tumor suppressor genes and the disease progression,
Jezkova et al also reported that the hypermethylation of
RASSF1A and PITX2, which are known for the tumor suppressor gene in
breast cancer, is significantly associated with tumor stage in
breast cancer patients (24). On
the other hand, Guo et al mentioned that there was no
significant difference in the methylation status of HOXD10, which
functions as a tumor suppressor in hepatocellular carcinoma (HCC),
between the HCC patients with stage I and II and those with stage
III and stage IV (25). Thus,
whether the degree of the promoter methylation can predict the
tumor progression may depend on the type of cancer and gene. In
addition, in our series, methylation was not detected in one case
despite the patient having clinical stage IV MPM; therefore,
further studies of the tumor characteristics that may be
particularly related to the degree of methylation are required.
It is well known that both chemotherapy and
radiotherapy induce DNA methylation changes, and chemotherapy or
radiation-induced alterations in DNA methylation result in changes
in the biological response to the treatment. Recently, Flanagan
et al reported that platinum-based chemotherapy induces DNA
methylation changes in blood DNA, and the methylation levels in
blood DNA at the time of relapse can reflect the clinical outcome
of cancer patients (26). Sun
et al also reported that the promoter methylation level of
RASSF1A was affected by oxaliplatin-based chemotherapy, and the
methylation status in blood DNA can be used to predict the outcome
of patients with colorectal cancer (27). Thus, the influences of treatments
on methylation statuses are a very important issue that should be
examined in the future, and miR-34b/c is no exception. Therefore,
the samples that were used in the present study were collected
before any treatment was administered.
Several circulating biomarkers have been reported
for the diagnosis of MPM, including the soluble mesothelin-related
peptides, osteopontin, fibulin-3 and miRs (28,29).
As for protein markers, while they exhibit excellent specificity,
their poor sensitivity reduces their diagnostic usefulness
(30-32). As regards circulating miRs,
although some miRs exhibit diagnostic potential for MPM, there are
problems, such as their origin (whether they are derived from tumor
cells or hematopoietic cells is still controversial) that need to
be resolved, and the majority of the analyses of cfRNA in the blood
remain exploratory (21). On the
other hand, few studies have reported the usefulness of a
diagnostic method targeting the degree of methylation of DNA, not
the miR or protein itself, for MPM. Several studies have reported
the existence of a strong association between the methylation
status in tumor tissue samples and that in ctDNA from blood
samples; therefore, targeting ctDNA methylation is reasonable
(33-36). As suggested by previous studies, a
combination of various approaches could be useful to increase the
sensitivity, and targeting circulating methylated DNA may be a
worthwhile addition (28,29).
Whereas we used a SYBR-Green-based real-time MSP
assay (48 wells/sample) in our previous study, we adopted a
TaqMan-based ddPCR assay (20,000 droplets/well) in the present
study to improve the specificity and accuracy of the detection of
methylated DNA from amongst a large amount of background DNA. As a
result, the specificity of the assay was improved to 94.9%,
compared with that in our previous study. On the other hand, the
sensitivity of the established assay was limited to 65.7%. The
median dosage of cfDNA in this study was approximately 15
ng/sample, corresponding to 4,500 haploid genome equivalents.
Considering the capability of ddPCR, it is possible to process
larger amounts of cfDNA. Increasing the dosage of DNA may lead to
an improvement in sensitivity. Recently, cfDNA in body fluids other
than blood, such as urine or stool, has also attracted attention as
useful biomarkers of cancer (37,38).
The collection of these samples offers the advantage of being truly
non-invasive and allowing large sample volumes to be collected,
which may compensate for the disadvantage of the rather limited
amount of cfDNA in the blood. In addition, the concentration of
ctDNA is one of the key factors for successful cancer detection
using a liquid biopsy, and it is well known that the proportion of
ctDNA in cfDNA varies among patients depending on the tumor
localization, size, vascularization, and clearance, ranging from
<0.005 to 90% in several types of cancer (39-42).
However, the association between ctDNA and total cfDNA in MPM
remains unclear; therefore, further investigation of this issue
using liquid biopsies in patients with MPM will be our next
task.
This study had some limitations. First, the sample
size was too small to enable a definitive conclusion, and the
groups in this study were not matched for background
characteristics, such as age and sex. Considering the rarity of
MPM, large clinical trials would be preferable. Second, plasma
samples are more suitable than serum samples for cfDNA analyses due
to the lower background level of wild-type DNA in the former
(21,43). Therefore, our established assay
should be validated using plasma samples. These factors could have
introduced some bias to our results.
In conclusion, in this study, we established a novel
detection system for the promoter methylation of miR-34b/c using
ddPCR. Our findings suggest the possibility that miR-34b/c
methylation in ctDNA could be a promising circulating biomarker for
the prediction of disease progression in MPM.
Supplementary Materials
Funding
This study was supported by a Management Expenses
Grants.
Availability of data and materials
The datasets analyzed during the current study are
available from the corresponding author on reasonable request.
Authors’ contributions
HS, JS, HY, KS and SToyooka conceived and designed
experiments. HS, STanaka, HTo and KN conducted the experiments. HS
and STomida analyzed data and prepared the figures. KA, NF, HTa, KO
and TK contributed to the sample collection. HS, JS and SToyooka
wrote the manuscript. All authors discussed the results and
commented on the manuscript. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
This study was conducted with the approval of the
Institutional Review Board/Ethical Committee of Okayama University;
each of the participants provided written informed consent for the
sample collection. All the experiments were performed in accordance
with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
MPM
|
malignant pleural mesothelioma
|
|
cfRNA
|
circulating cell-free RNA
|
|
cfDNA
|
circulating cell-free DNA
|
|
ctDNA
|
circulating cell-free tumor DNA
|
|
miR or miRNA
|
microRNA
|
|
ddPCR
|
droplet digital PCR
|
|
PP
|
pleural plaque
|
|
HV
|
healthy volunteers
|
|
LNA
|
locked nucleic acids
|
|
SNPs
|
single nucleotide polymorphisms
|
Acknowledgments
The authors would like to thank Dr Takehiro
Matsubara (Biobank, Okayama University Graduate School of Medicine,
Dentistry and Pharmaceutical Sciences, Okayama, Japan), Ms. Yoko
Kojima (Research Center for Asbestos-related Disease, Okayama Rosai
Hospital), and Ms. Fumiko Isobe (Department of General Thoracic
Surgery and Breast and Endocrinological Surgery, Okayama University
Graduate School of Medicine, Dentistry and Pharmaceutical Sciences,
Okayama, Japan) for their technical support.
References
|
1
|
Henley SJ, Larson TC, Wu M, Antao VC,
Lewis M, Pinheiro GA and Eheman C: Mesothelioma incidence in 50
states and the District of Columbia, United States, 2003-2008. Int
J Occup Environ Health. 19:1–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scherpereel A: Malignant pleural
mesothelioma: new treatments, new hopes? Eur Respir J.
49:17003192017. View Article : Google Scholar
|
|
3
|
Goudar RK: Review of pemetrexed in
combination with cisplatin for the treatment of malignant pleural
mesothelioma. Ther Clin Risk Manag. 4:205–211. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Toyooka S, Kishimoto T and Date H:
Advances in the molecular biology of malignant mesothelioma. Acta
Med Okayama. 62:1–7. 2008.PubMed/NCBI
|
|
5
|
Russo M, Siravegna G, Blaszkowsky LS,
Corti G, Crisafulli G, Ahronian LG, Mussolin B, Kwak EL, Buscarino
M, Lazzari L, et al: Tumor heterogeneity and lesion-specific
response to targeted therapy in colorectal cancer. Cancer Discov.
6:147–153. 2016. View Article : Google Scholar :
|
|
6
|
Murtaza M, Dawson SJ, Pogrebniak K, Rueda
OM, Provenzano E, Grant J, Chin SF, Tsui DW, Marass F, Gale D, et
al: Multifocal clonal evolution characterized using circulating
tumour DNA in a case of metastatic breast cancer. Nat Commun.
6:87602015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alix-Panabières C, Schwarzenbach H and
Pantel K: Circulating tumor cells and circulating tumor DNA. Annu
Rev Med. 63:199–215. 2012. View Article : Google Scholar
|
|
8
|
Loginov VI, Pronina IV, Burdennyy AM,
Filippova EA, Kazubskaya TP, Kushlinsky DN, Utkin DO, Khodyrev DS,
Kushlinskii NE, Dmitriev AA, et al: Novel miRNA genes deregulated
by aberrant methylation in ovarian carcinoma are involved in
metastasis. Gene. 662:28–36. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tian Y, Wei W, Li L and Yang R:
Down-regulation of miR-148a promotes metastasis by DNA methylation
and is associated with prognosis of skin cancer by targeting TGIF2.
Med Sci Monit. 21:3798–3805. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Toyota M, Suzuki H, Sasaki Y, Maruyama R,
Imai K, Shinomura Y and Tokino T: Epigenetic silencing of
microRNA-34b/c and B-cell translocation gene 4 is associated with
CpG island meth-ylation in colorectal cancer. Cancer Res.
68:4123–4132. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kubo T, Toyooka S, Tsukuda K, Sakaguchi M,
Fukazawa T, Soh J, Asano H, Ueno T, Muraoka T, Yamamoto H, et al:
Epigenetic silencing of microRNA-34b/c plays an important role in
the pathogenesis of malignant pleural mesothelioma. Clin Cancer
Res. 17:4965–4974. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Corney DC, Flesken-Nikitin A, Godwin AK,
Wang W and Nikitin AY: MicroRNA-34b and microRNA-34c are targets of
p53 and cooperate in control of cell proliferation and
adhesion-independent growth. Cancer Res. 67:8433–8438. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Suzuki R, Yamamoto E, Nojima M, Maruyama
R, Yamano HO, Yoshikawa K, Kimura T, Harada T, Ashida M, Niinuma T,
et al: Aberrant methylation of microRNA-34b/c is a predictive
marker of metachronous gastric cancer risk. J Gastroenterol.
49:1135–1144. 2014. View Article : Google Scholar :
|
|
16
|
Wu XD, Song YC, Cao PL, Zhang H, Guo Q,
Yan R, Diao DM, Cheng Y and Dang CX: Detection of miR-34a and
miR-34b/c in stool sample as potential screening biomarkers for
noninvasive diagnosis of colorectal cancer. Med Oncol. 31:8942014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Muraoka T, Soh J, Toyooka S, Aoe K,
Fujimoto N, Hashida S, Maki Y, Tanaka N, Shien K, Furukawa M, et
al: The degree of microRNA-34b/c methylation in serum-circulating
DNA is associated with malignant pleural mesothelioma. Lung Cancer.
82:485–490. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Oxnard GR, Paweletz CP, Kuang Y, Mach SL,
O’Connell A, Messineo MM, Luke JJ, Butaney M, Kirschmeier P,
Jackman DM, et al: Noninvasive detection of response and resistance
in EGFR-mutant lung cancer using quantitative next-generation
genotyping of cell-free plasma DNA. Clin Cancer Res. 20:1698–1705.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hindson BJ, Ness KD, Masquelier DA,
Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY,
Hiddessen AL, Legler TC, et al: High-throughput droplet digital PCR
system for absolute quantitation of DNA copy number. Anal Chem.
83:8604–8610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sanmamed MF, Fernández-Landázuri S,
Rodríguez C, Zárate R, Lozano MD, Zubiri L, Perez-Gracia JL,
Martín-Algarra S and González A: Quantitative cell-free circulating
BRAFV600E mutation analysis by use of droplet digital PCR in the
follow-up of patients with melanoma being treated with BRAF
inhibitors. Clin Chem. 61:297–304. 2015. View Article : Google Scholar
|
|
21
|
Siravegna G, Marsoni S, Siena S and
Bardelli A: Integrating liquid biopsies into the management of
cancer. Nat Rev Clin Oncol. 14:531–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Soh J, Okumura N, Lockwood WW, Yamamoto H,
Shigematsu H, Zhang W, Chari R, Shames DS, Tang X, MacAulay C, et
al: Oncogene mutations, copy number gains and mutant allele
specific imbalance (MASI) frequently occur together in tumor cells.
PLoS One. 4:e74642009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suzawa K, Yamamoto H, Ohashi K, Hashida S,
Tomida S, Kubo T, Maki Y, Soh J, Tsukuda K, Kiura K, et al: Optimal
method for quantitative detection of plasma EGFR T790M mutation
using droplet digital PCR system. Oncol Rep. 37:3100–3106. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jezkova E, Kajo K, Zubor P, Grendar M,
Malicherova B, Mendelova A, Dokus K, Lasabova Z, Plank L and Danko
J: Methylation in promoter regions of PITX2 and RASSF1A genes in
association with clinicopathological features in breast cancer
patients. Tumour Biol. 37:15707–15718. 2016. View Article : Google Scholar
|
|
25
|
Guo Y, Peng Y, Gao D, Zhang M, Yang W,
Linghu E, Herman JG, Fuks F, Dong G and Guo M: Silencing HOXD10 by
promoter region hypermethylation activates ERK signaling in
hepato-cellular carcinoma. Clin Epigenetics. 9:1162017. View Article : Google Scholar
|
|
26
|
Flanagan JM, Wilson A, Koo C, Masrour N,
Gallon J, Loomis E, Flower K, Wilhelm-Benartzi C, Hergovich A,
Cunnea P, et al: Platinum-based chemotherapy induces methylation
changes in blood DNA associated with overall survival in patients
with ovarian cancer. Clin Cancer Res. 23:2213–2222. 2017.
View Article : Google Scholar
|
|
27
|
Sun X, Yuan W, Hao F and Zhuang W:
Promoter methylation of RASSF1A indicates prognosis for patients
with stage II and III colorectal cancer treated with
oxaliplatin-based chemotherapy. Med Sci Monit. 23:5389–5395. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cristaudo A, Bonotti A, Guglielmi G,
Fallahi P and Foddis R: Serum mesothelin and other biomarkers: What
have we learned in the last decade? J Thorac Dis. 10(Suppl 2):
S353–S359. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bruno R, Alì G and Fontanini G: Molecular
markers and new diagnostic methods to differentiate malignant from
benign meso-thelial pleural proliferations: A literature review. J
Thorac Dis. 10(Suppl 2): S342–S352. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu ZD, Liu XF, Liu XC, Ding CM and Hu CJ:
Diagnostic accuracy of osteopontin for malignant pleural
mesothelioma: A systematic review and meta-analysis. Clin Chimica
Acta. 433:44–48. 2014. View Article : Google Scholar
|
|
31
|
Creaney J, Dick IM, Meniawy TM, Leong SL,
Leon JS, Demelker Y, Segal A, Musk AW, Lee YC, Skates SJ, et al:
Comparison of fibulin-3 and mesothelin as markers in malignant
mesothelioma. Thorax. 69:895–902. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
van Zandwijk N, Clarke C, Henderson D,
Musk AW, Fong K, Nowak A, Loneragan R, McCaughan B, Boyer M, Feigen
M, et al: Guidelines for the diagnosis and treatment of malignant
pleural mesothelioma. J Thorac Dis. 5:E254–E307. 2013.
|
|
33
|
Xu RH, Wei W, Krawczyk M, Wang W, Luo H,
Flagg K, Yi S, Shi W, Quan Q, Li K, et al: Circulating tumour DNA
methylation markers for diagnosis and prognosis of hepatocellular
carcinoma. Nat Mater. 16:1155–1161. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pishvaian MJ, Joseph Bender R, Matrisian
LM, Rahib L, Hendifar A, Hoos WA, Mikhail S, Chung V, Picozzi V,
Heartwell C, et al: A pilot study evaluating concordance between
blood-based and patient-matched tumor molecular testing within
pancreatic cancer patients participating in the Know Your Tumor
(KYT) initiative. Oncotarget. 8:83446–83456. 2016.
|
|
35
|
Liggett T, Melnikov A, Yi QL, Replogle C,
Brand R, Kaul K, Talamonti M, Abrams RA and Levenson V:
Differential methylation of cell-free circulating DNA among
patients with pancreatic cancer versus chronic pancreatitis.
Cancer. 116:1674–1680. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hoque MO, Feng Q, Toure P, Dem A,
Critchlow CW, Hawes SE, Wood T, Jeronimo C, Rosenbaum E, Stern J,
et al: Detection of aberrant methylation of four genes in plasma
DNA for the detection of breast cancer. J Clin Oncol. 24:4262–4269.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Worm Ørntoft MB: Review of blood-based
colorectal cancer screening: how far are circulating cell-free DNA
methylation markers from clinical implementation? Clin Colorectal
Cancer. 17:e415–e433. 2018. View Article : Google Scholar
|
|
38
|
Stewart CM, Kothari PD, Mouliere F, Mair
R, Somnay S, Benayed R, Zehir A, Weigelt B, Dawson SJ, Arcila ME,
et al: The value of cell-free DNA for molecular pathology. J
Pathol. 244:616–627. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Diehl F, Schmidt K, Choti MA, Romans K,
Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al:
Circulating mutant DNA to assess tumor dynamics. Nat Med.
14:985–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Thierry AR, Mouliere F, El Messaoudi S,
Mollevi C, Lopez-Crapez E, Rolet F, Gillet B, Gongora C, Dechelotte
P, Robert B, et al: Clinical validation of the detection of KRAS
and BRAF mutations from circulating tumor DNA. Nat Med. 20:430–435.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zeng H, He B, Yi C and Peng J: Liquid
biopsies: DNA methylation analyses in circulating cell-free DNA. J
Genet Genomics. 45:185–192. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jung M, Klotzek S, Lewandowski M,
Fleischhacker M and Jung K: Changes in concentration of DNA in
serum and plasma during storage of blood samples. Clin Chem.
49:1028–1029. 2003. View Article : Google Scholar : PubMed/NCBI
|