Introduction
The Ci county of Hebei Province is one of the
high-incidence regions of esophageal cancer in China. Over the past
40 years, the incidence and mortality rate of esophageal cancer
have significantly decreased with the application of primary and
secondary prevention measures. However, in 2011, the age-adjusted
esophageal cancer incidence and mortality rates were
106.74/105 and 103.07/105, respectively, in
men, and 75.41/105 and 53.52/105,
respectively, in women (1).
Therefore, esophageal cancer remains a major public health concern
in this region. A number of environmental and genetic factors
chronically interact, leading to esophageal cancer development.
Genome-wide association studies (GWAS) is a powerful tool for
identifying genetic markers for various diseases with
high-throughput genotyping technology (2). Recent GWAS identified susceptibility
loci for esophageal squamous cell carcinoma (ESCC), the most common
histological type of esophageal cancer, in the phospholipase C ε-1
gene (PLCE1) (3–5). Subsequently, several independent
studies were conducted to validate the association between
PLCE1 single-nucleotide polymorphisms (SNPs) and ESCC, with
controversial results (6–12). PLCE1 is a member of the phospholipase
C family, a group of proteins able to convert phosphoinositol
4,5-bisphosphate to two second messengers, inositol
1,4,5-triphosphate (IP3) and diacylglycerol (DAG). IP3 and DAG
participate in Ca2+ immobilization and protein kinase C
activation, respectively (13). It
is noteworthy that PLCE1 may be regulated by multiple signaling
inputs from both Ras family GTPases and G proteins (14). Hence, PLCE1 plays a crucial role in
regulating cell growth, differentiation, apoptosis and angiogenesis
(15). It was previously
demonstrated that PLCE1 was involved in the development and
progression of various cancers, including esophageal cancer
(3,10,16–22). In
addition, the expression of PLCE1 was associated with the survival
of patients with esophageal cancer (23,24). In
fact, identifying applicable biomarkers for ESCC prognosis may be
helpful in improving the outcome of ESCC patients. Our previous
studies indicated that the PLCE1 rs2274223 and rs11599672
SNPs were associated with susceptibility to ESCC in a
high-incidence population from the Ci county of Hebei province in
northern China (25). To the best of
our knowledge, whether these two SNPs may be used as predictive
biomarkers for the prognosis of ESCC patients in this
high-incidence region has not been determined to date.
Materials and methods
Study subjects
The survival information of 207 ESCC patients was
collected. All the study subjects were ethnically homogeneous (of
Han descent) and permanent residents of the Ci county, and they
were recruited during an endoscopic screening campaign between 2008
and 2012. The patients had histologically confirmed ESCC.
Information on the sex, age, smoking habits and family history of
upper gastrointestinal cancer (UGIC) was obtained from the cancer
patients by two professional interviewers directly after blood
sampling. Smokers were defined as those who formerly or currently
smoked no less than five cigarettes per day for at least 2 years.
Individuals who had at least one first-degree relative or at least
two second-degree relatives who had esophageal/cardiac/gastric
cancer were defined as having a family history of UGIC. The present
study was approved by the Ethics Committee of the Fourth Hospital
of Hebei Medical University. Informed consent forms were obtained
from all recruited subjects.
DNA extraction
Venous blood (5 ml) from each subject was collected
in Vacutainer tubes containing EDTA and stored at 4°C. After
sampling, genomic DNA was extracted within 1 week by proteinase K
(Merck, Darmstadt, Germany) digestion, followed by a salting out
procedure according to the method published by Miller et al
(26).
Polymorphism genotyping
The genotypes of PLCE1 polymorphisms were
determined by the Shanghai Generay Biotech Co., Ltd. (Shanghai,
China) using the polymerase chain reaction-ligase detection
reaction method, which has been described in detail in our previous
study (25).
Statistical analysis
Statistical analysis was performed using the SPSS
version 22.0 software package (IBM Corp., Armonk, NY, USA).
P<0.05 was considered significant for all statistical analyses.
Survival time was calculated from the date of ESCC diagnosis to the
date of death or the last follow-up. The associations of survival
time with demographic characteristics and PLCE1 SNPs were
estimated using the Kaplan-Meier method and the log-rank test.
Univariate or multivariate Cox regression analysis was fitted to
estimate the crude hazard ratios (HRs), adjusted HRs and 95%
confidence intervals (CIs).
Results
Patient characteristics
The mean age of the 207 ESCC patients was 60.3±7.9
years. Sex, age, smoking status and UGIC family history were not
found to be associated with the survival time of the ESCC patients
(Table I).
 | Table I.ESCC patient characteristics and
survival. |
Table I.
ESCC patient characteristics and
survival.
| Characteristics | Patients, n (%) | Deaths, n (%) | MST (months) | Log-rank P-value | HR (95% CI) |
|---|
| Sex |
|
|
|
|
|
| Male | 141 (68.1) | 60 (42.6) | 42.2 |
| 1.000 |
|
Female | 66
(31.9) | 2
(39.4) | 45.3 | 0.593 | 0.883
(0.557–1.399) |
| Age, years |
|
|
|
|
|
| ≤60 | 106 (51.2) | 44 (41.5) | 43.5 |
| 1.000 |
|
>60 | 101 (48.8) | 42 (41.6) | 42.9 | 0.943 | 1.015
(0.665–1.550) |
| Smoking status |
|
|
|
|
|
|
Non-smoker | 92
(44.4) | 34 (37.0) | 45.2 |
| 1.000 |
|
Smoker | 115 (55.6) | 52 (45.2) | 41.6 | 0.282 | 1.265
(0.821–1.949) |
| Family history of
UGIC |
|
|
|
|
|
|
Negative | 129 (62.3) | 59 (45.7) | 41.7 |
| 1.000 |
|
Positive | 78
(37.7) | 27 (34.6) | 45.8 | 0.159 | 0.723
(0.459–1.141) |
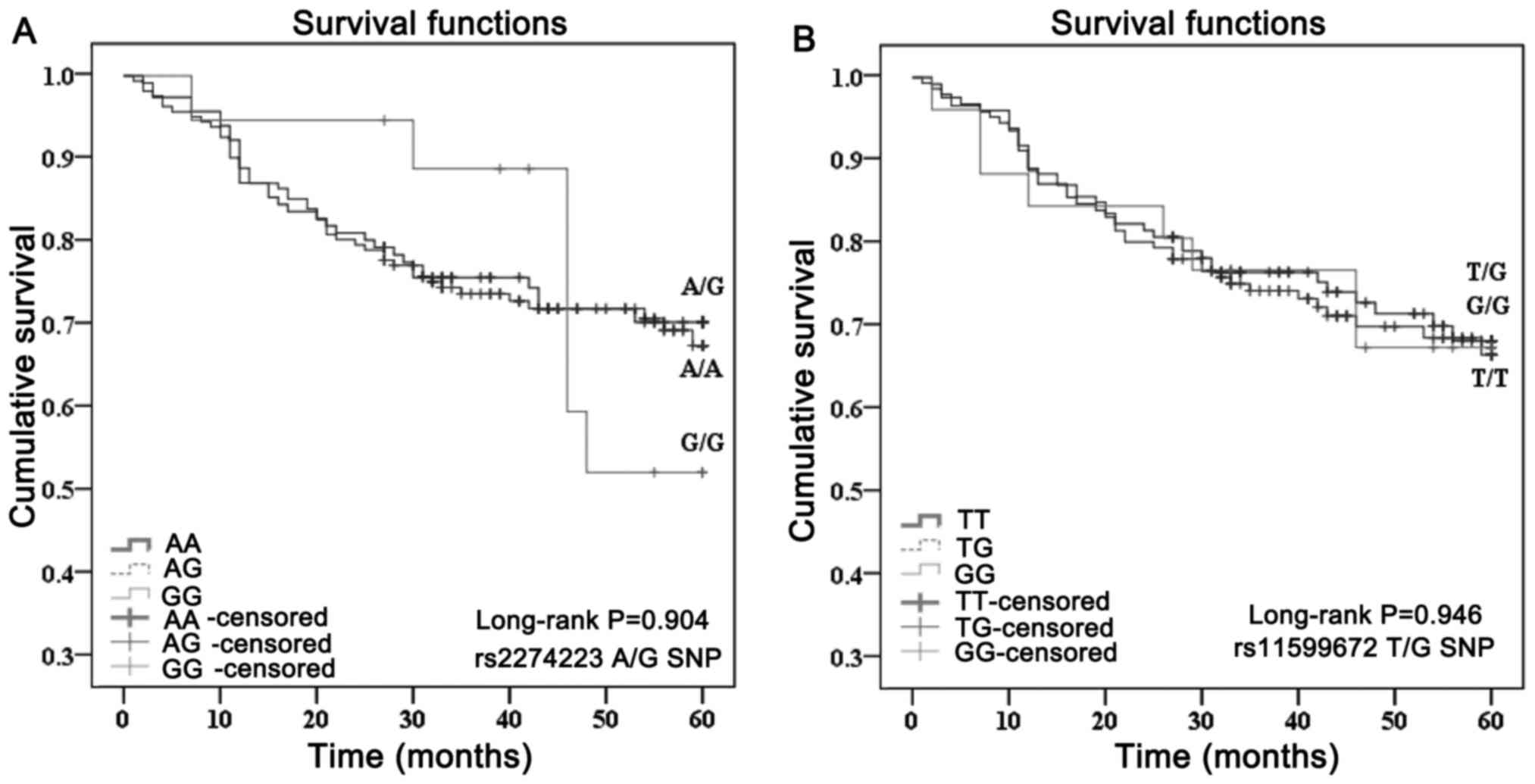
Effect of different SNP genotypes on
survival
The mean survival time of rs2274223 SNP A/A, A/G and
G/G genotype carriers was 42.9, 43.4 and 46.3 months, respectively.
Compared with the A/A genotype, the A/G and G/G genotypes did not
modify the mortality risk of ESCC patients (HR=0.924 and 0.986, 95%
CI: 0.585–1.459 and 0.439–2.216, respectively). For rs11599672 SNP
T/T, T/G and G/G genotype carriers, the mean survival time was
42.8, 43.8 and 42.7 months, respectively. Compared with the T/T
genotype, the T/G and G/G genotypes did not significantly affect
the mortality risk of ESCC patients (HR=0.902 and 0.993, 95% CI:
0.575–1.414 and 0.436–1.994, respectively) (Table II, Fig.
1).
 | Table II.PLCE1 SNPs and survival of
ESCC patients. |
Table II.
PLCE1 SNPs and survival of
ESCC patients.
| SNP | Patients, n
(%) | Deaths, n (%) | MST (months) | Log-rank
P-value | HR (95%
CI)a |
|---|
| rs2274223 A/G |
|
|
|
|
|
|
A/A | 113 (54.6) | 47 (41.6) | 42.9 |
| 1.000 |
|
A/G | 81 (39.1) | 32 (39.5) | 43.4 |
| 0.924
(0.585–1.459) |
|
G/G | 13 (6.3) | 7
(53.8) | 46.3 | 0.904 | 0.986
(0.439–2.216) |
| rs11599672 T/G |
|
|
|
|
|
|
T/T | 102 (49.3) | 43 (42.2) | 42.8 |
| 1.000 |
|
T/G | 87 (42.0) | 35 (40.2) | 43.8 |
| 0.902
(0.575–1.414) |
|
G/G | 118 (8.7) | 8
(44.4) | 42.7 | 0.946 | 0.993
(0.436–1.994) |
Discussion
The TNM staging system, which is based on tumor
depth (T), presence and number of regional nodes with metastatic
disease (N) and presence or absence of distant metastasis (M) is
used to predict the prognosis of cancer patients. However, the
overall survival of the patients with similar TNM stage varies,
which may be attributed to overlooking the biological or molecular
characteristics of each individual tumor. Therefore, it is
necessary to detect the alterations in the patients' genomic,
epigenetic and/or proteomic profile, and even in single markers,
and to identify useful biomarkers for the prognosis of cancer
patients (27). In the present
study, it was investigated whether the PLCE1 rs2274223 and
rs11599672 SNPs may be used to predict the outcome of ESCC
patients; however, no correlation was observed between these two
SNPs and the survival of ESCC patients.
Two studies demonstrated that the PLCE1
rs2274223 SNP was associated with the risk of ESCC (3,10).
Subsequently, Wang et al investigated the PLCE1 protein
expression of ESCC tissues with different genotypes of rs2274223
SNP and found a higher expression level in ESCC tissues with the
A/G genotype compared with those with the A/A genotype (28). Similar results were observed in the
study by Hu et al (10).
However, it remains controversial whether PLCE1 acts as an oncogene
or a tumor suppressor in ESCC development and progression. Hu et
al (10) and Li et al
(24) reported decreased mRNA in
ESCC tumors compared with that in the adjacent normal esophagus,
but no difference in protein expression. By contrast, ESCC tissues
exhibited a higher PLCE1 protein expression compared with normal
tissues in other studies (3,23,28–30).
This disparity may be attributed to differences in sample size,
genetic background or experimental methods. It is worth mentioning
that downregulation of PLCE1 inhibited ESCC cell proliferation and
promoted cell apoptosis in vitro (23,29).
As regards the association of PLCE1 expression with
the survival of ESCC patients, the results were inconsistent.
Specifically, one study reported that high PLCE1 protein expression
in ESCC was associated with poor survival (23), while increased tumor/normal-fold
change of mRNA and protein expression in ESCC was associated with
improved survival in another study (24). To the best of our knowledge, the
present study is the first to evaluate the association between the
rs2274223 SNP and survival of ESCC patients of Han nationality, and
found no correlation, which is similar to the results in the
northern Indian population (7). The
PLCE1 rs2274223 SNP, a non-synonymous SNP causing an amino
acid change from His to Arg in the 26th exon, is located at the
calcium-binding domain, indicating its crucial functional
significance. However, data analysis by bioinformatics software
suggests rs2274223 SNP to be benign and tolerated and the variation
between wild-type and mutated structures negligible, which may
partly explain the null results (7).
Interestingly, the same SNP played a different role in ESCC
susceptibility and prognosis of ESCC patients. The possible reason
for this discrepancy may be the complex structure of PLCE1 and its
involvement in various signaling pathways, such as nuclear
factor-κB and Ras-mitogen-activated protein kinase (31).
The PLCE1 rs11599672 SNP is situated within
the transcription factor-binding site, and the substitution of T to
G may alter the transcriptional activity of PLCE1. Our
previous study indicated that a family history of UGIC increased
the risk of ESCC in subjects with the T/T genotype. In addition,
the rs11599672 SNP G/G genotype was found to be associated with a
decreased risk of non-oropharyngeal tumors in a non-Hispanic white
population (32). However, there was
no association between the rs11599672 SNP and the prognosis of ESCC
patients in the Ci county, which is a high-incidence region. To
date, the functional role of rs11599672 SNP has not been
determined. Therefore, defining the function of the rs11599672 SNP,
such as the association of PLCE1 expression with different
genotypes of rs11599672 SNP, may be useful in providing mechanistic
evidence for the findings.
In conclusion, the PLCE1 rs227423 and
rs11599672 SNPs cannot be used as predictive markers for survival
of ESCC cases in a high-incidence region of northern China. Further
investigation is required to determine the association of other
SNPs in PLCE1 with the prognosis of ESCC patients.
Acknowledgements
The present study was supported by the Hebei
Province Medical Scientific Research Key Project (grant no.
20110146). The authors would like to thank Zhi-feng Chen for
helping with sample collection and acknowledge Guo-hui Song and Lei
Wang for their help with data collection.
Glossary
Abbreviations
Abbreviations:
|
GWAS
|
genome-wide association study
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
SNP
|
single-nucleotide polymorphism
|
|
IP3
|
inositol 1,4,5-triphosphate
|
|
DAG
|
diacylglycerol
|
|
UGIC
|
upper gastrointestinal cancer
|
References
|
1
|
He Y, Wu Y, Song G, Li Y, Liang D, Jin J,
Wen D and Shan B: Incidence and mortality rate of esophageal cancer
has decreased during past 40 years in Hebei Province, China. Chin J
Cancer Res. 27:562–571. 2015.PubMed/NCBI
|
|
2
|
Manolio TA: Genomewide association studies
and assessment of the risk of disease. N Engl J Med. 363:166–176.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang LD, Zhou FY, Li XM, Sun LD, Song X,
Jin Y, Li JM, Kong GQ, Qi H, Cui J, et al: Genome-wide association
study of esophageal squamous cell carcinoma in Chinese subjects
identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet.
42:759–763. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abnet CC, Freedman ND, Hu N, Wang Z, Yu K,
Shu XO, Yuan JM, Zheng W, Dawsey SM, Dong LM, et al: A shared
susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma
and esophageal squamous cell carcinoma. Nat Genet. 42:764–767.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu C, Hu Z, He Z, Jia W, Wang F, Zhou Y,
Liu Z, Zhan Q, Liu Y, Yu D, et al: Genome-wide association study
identifies three new susceptibility loci for esophageal
squamous-cell carcinoma in Chinese populations. Nat Genet.
43:679–684. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Duan F, Xie W, Cui L, Wang P, Song C, Qu
H, Wang K, Zhang J and Dai L: Novel functional variants locus in
PLCE1 and susceptibility to esophageal squamous cell carcinoma:
Based on published genome-wide association studies in a central
Chinese population. Cancer Epidemiol. 37:647–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Umar M, Upadhyay R, Kumar S, Ghoshal UC
and Mittal B: Role of novel and GWAS originated PLCE1 genetic
variants in susceptibility and prognosis of esophageal cancer
patients in northern Indian population. Tumour Biol.
35:11667–11676. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cui XB, Chen YZ, Pang XL, Liu W, Hu JM, Li
SG, Yang L, Zhang WJ, Liu CX, Cao YW, et al: Multiple polymorphisms
within the PLCE1 are associated with esophageal cancer via
promoting the gene expression in a Chinese Kazakh population. Gene.
530:315–322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu H, Ding G, Zhang W, Liu C, Chen Y, Chen
S and Jiang P: Replication study of PLCE1 and C20orf54 polymorphism
and risk of esophageal cancer in a Chinese population. Mol Biol
Rep. 39:9105–9111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu H, Yang J, Sun Y, Yang Y, Qian J, Jin
L, Wang M, Bi R, Zhang R, Zhu M, et al: Putatively functional PLCE1
variants and susceptibility to esophageal squamous cell carcinoma
(ESCC): A case-control study in eastern Chinese populations. Ann
Surg Oncol. 19:2403–2410. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Palmer AJ, Lochhead P, Hold GL, Rabkin CS,
Chow WH, Lissowska J, Vaughan TL, Berry S, Gammon M, Risch H and
El-Omar EM: Genetic variation in C20orf54, PLCE1 and MUC1 and the
risk of upper gastrointestinal cancers in Caucasian populations.
Eur J Cancer Prev. 21:541–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jia X, Liu P, Zhang M, Feng T, Tang H,
Tang Z, Zhao H and Jin T: Genetic variants at 6p21, 10q23, 16q21
and 22q12 are associated with esophageal cancer risk in a Chinese
Han population. Int J Clin Exp Med. 8:19381–19387. 2015.PubMed/NCBI
|
|
13
|
Rhee SG: Regulation of
phosphoinositide-specific phospholipase C. Annu Rev Biochem.
70:281–312. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smrcka AV, Brown JH and Holz GG: Role of
phospholipase Cε in physiological phosphoinositide signaling
networks. Cell Signal. 24:1333–1343. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Citro S, Malik S, Oestreich EA,
Radeff-Huang J, Kelley GG, Smrcka AV and Brown JH: Phospholipase
Cepsilon is a nexus for Rho and Rap-mediated G protein-coupled
receptor-induced astrocyte proliferation. Proc Natl Acad Sci USA.
104:pp. 15543–15548. 2007; View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang M, Zhang R, He J, Qiu L, Li J, Wang
Y, Sun M, Yang Y, Wang J, Yang J, et al: Potentially functional
variants of PLCE1 identified by GWASs contribute to gastric
adenocarcinoma susceptibility in an eastern Chinese population.
PLoS One. 7:e319322012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen J, Wang W, Zhang T, Ji J, Qian Q, Lu
L, Fu H, Jin W and Cui D: Differential expression of phospholipase
C epsilon 1 is associated with chronic atrophic gastritis and
gastric cancer. PLoS One. 7:e475632012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sorli SC, Bunney TD, Sugden PH, Paterson
HF and Katan M: Signaling properties and expression in normal and
tumor tissues of two phospholipase C epsilon splice variants.
Oncogene. 24:90–100. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang X, Zbou C, Qiu G, Fan J, Tang H and
Peng Z: Screening of new tumor suppressor genes in sporadic
colorectal cancer patients. Hepatogastroenterology. 55:2039–2044.
2008.PubMed/NCBI
|
|
20
|
Ling Y, Chunli L, Xiaohou W and Qiaoling
Z: Involvement of the PLCε/PKCα pathway in human BIU-87 bladder
cancer cell proliferation. Cell Biol Int. 35:1031–1036. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Yan L, Zhao Y, Ou L, Wu X and Luo
C: Knockdown of phospholipase C-epsilon by short-hairpin
RNA-mediated gene silencing induces apoptosis in human bladder
cancer cell lines. Cancer Biother Radiopharm. 28:233–239. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du HF, Ou LP, Yang X, Song XD, Fan YR, Tan
B, Luo CL and Wu XH: A new PKCα/β/TBX3/E-cadherin pathway is
involved in PLCε-regulated invasion and migration in human bladder
cancer cells. Cell Signal. 26:580–593. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cui XB, Li S, Li TT, Peng H, Jin TT, Zhang
SM, Liu CX, Yang L, Shen YY, Li SG, et al: Targeting oncogenic
PLCE1 by miR-145 impairs tumor proliferation and metastasis of
esophageal squamous cell carcinoma. Oncotarget. 7:1777–1795. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li WQ, Hu N, Burton VH, Yang HH, Su H,
Conway CM, Wang L, Wang C, Ding T, Xu Y, et al: PLCE1 mRNA and
protein expression and survival of patients with esophageal
squamous cell carcinoma and gastric adenocarcinoma. Cancer
Epidemiol Biomarkers Prev. 23:1579–1588. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou RM, Li Y, Wang N, Liu BC, Chen ZF and
Zuo LF: PLC-ε1 gene polymorphisms significantly enhance the risk of
esophageal squamous cell carcinoma in individuals with a family
history of upper gastrointestinal cancers. Arch Med Res.
43:578–584. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miller SA, Dykes DD and Polesky HF: A
simple salting out procedure for extracting DNA from human
nucleated cells. Nucleic Acids Res. 16:12151988. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Plum PS, Bollschweiler E, Hölscher AH and
Warnecke-Eberz U: Novel diagnostic and prognostic biomarkers in
esophageal cancer. Expert Opin Med Diagn. 7:557–571. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang LD, Bi X, Song X, Pohl NM, Cheng Y,
Zhou Y, Shears S, Ansong E, Xing M, Wang S, et al: A sequence
variant in the phospholipase C epsilon C2 domain is associated with
esophageal carcinoma and esophagitis. Mol Carcinog. 52 Suppl
1:E80–E86. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao L, Wei ZB, Yang CQ, Chen JJ, Li D, Ji
AF and Ma L: Effects of PLCE1 gene silencing by RNA interference on
cell cycling and apoptosis in esophageal carcinoma cells. Asian Pac
J Cancer Prev. 15:5437–5442. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen YZ, Cui XB, Hu JM, Zhang WJ, Li SG,
Yang L, Shen XH, Liu CX, Pan QF, Yu SY, et al: Overexpression of
PLCE1 in Kazakh esophageal squamous cell carcinoma: Implications in
cancer metastasis and aggressiveness. APMIS. 121:908–918. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tyutyunnykova A, Telegeev G and Dubrovska
A: The controversial role of phospholipase C epsilon (PLCε) in
cancer development and progression. J Cancer. 8:716–729. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma H, Wang LE, Liu Z, Sturgis EM and Wei
Q: Association between novel PLCE1 variants identified in published
esophageal cancer genome-wide association studies and risk of
squamous cell carcinoma of the head and neck. BMC Cancer.
11:2582011. View Article : Google Scholar : PubMed/NCBI
|