Signals from the overhead pacemaker of the circadian
clock, the SCN, mediate the oscillation on a cellular level through
clock gene expression and feedback (1). A disruption in these signaling pathways
may have a crucial influence on the organism affected. Circadian
genes may be involved in regulating cancer-related pathways,
including cell proliferation, DNA damage response, and apoptosis
(2). Cancer-related genes like
c-myc and p53 exhibit a circadian rhythm in
vivo (3,4). Oncogenic activity such as excessive
cell proliferation, loss of DNA damage control and increased tumor
development has been detected in mice with a loss of functioning
circadian genes (4). The lifestyle
in the twenty-first century has changed due to more
industrialization of society, which has altered the endogenous
circadian rhythm in ~50% of the world's population. This, among
other reasons, has led to increased development of cancer
throughout the world (5). There are
studies showing the effect of dysfunctional circadian machinery in
humans, for example mutations, non-standard expression, and
translocation of clock genes, which has led to different cancer
types including breast, colorectal, gastric, kidney, lung,
prostate, pancreatic, and oral cancer (6). The circadian clock and the cell cycle
share some common features in molecular pathways and theoretical
stages. It has been hypothesized that clock genes have a crucial
role in the cell cycle and with this role they are highly involved
in tumorigenesis (4). The underlying
molecular mechanisms and the role of clock genes in oral
carcinogenesis is elusive. The aim of this review is to summarize
the current state of knowledge and to provide insight to guide
future research on involvement of clock genes in oral cancer.
The circadian clock is an endogenous timekeeping
system shared by most organisms. Although there are some
differences between species, the underlying molecular mechanisms of
the circadian clock are very similar (7). The ability to adapt to a continuously
changing environment is an essential key to selective advantage for
living creatures to survive and thrive. The circadian clock system
is one of these adapting abilities, which organisms have acquired
in order to synchronize their daily behavior and internal
mechanisms with the most profound environmental signal: The
circadian light cycle of 24 h. Body temperature, feeding, hormonal
levels, and the sleep-wake cycle all varies synchronized with
light-dark cycle (8). Another great
ability of this system is adjustment to the 24 h cycle showing a
crucial plastic capacity (9). There
is a hypothesis called ‘escape from UV’, which is based on the S
phase of the cell cycle, which is during night-time. It suggests
that ancient lifeforms have adapted to the environment by limiting
this UV-sensitive phase of the cycle to nighttime in order to avoid
DNA-damage (10).
The circadian clock system consists of almost as
many individual clocks as there are cells. It is based on different
levels and controls the whole rhythmicity of the organism (11). In mammals there is a central
pacemaker in the suprachiasmatic nucleus (SCN) of the hypothalamus
consisting of ~15,000 neurons (12).
The input to this central pacemaker comes through different
pathways. The primarily input, i.e. light, is registered in the
retina by a subset of melanopsin-expressing retinal ganglions and
accesses the SCN via the retino-hypothalamic tract (RHT). From the
SCN there are output pathways leading to the whole body (11,13,14).
These feeding pathways are regulated by interlocked
transcription-translation feedback loops (TTFLs) (15), where the clock gene family exerts an
important role. In Drosophila and zebrafish, light has a
direct influence on the circadian behavior of the peripheral cells
(12,16), whereas in mammals the clock genes in
peripheral tissues are not light sensitive. Here, they maintain and
regulate TTFLs in almost every cell of the body by feeding pathways
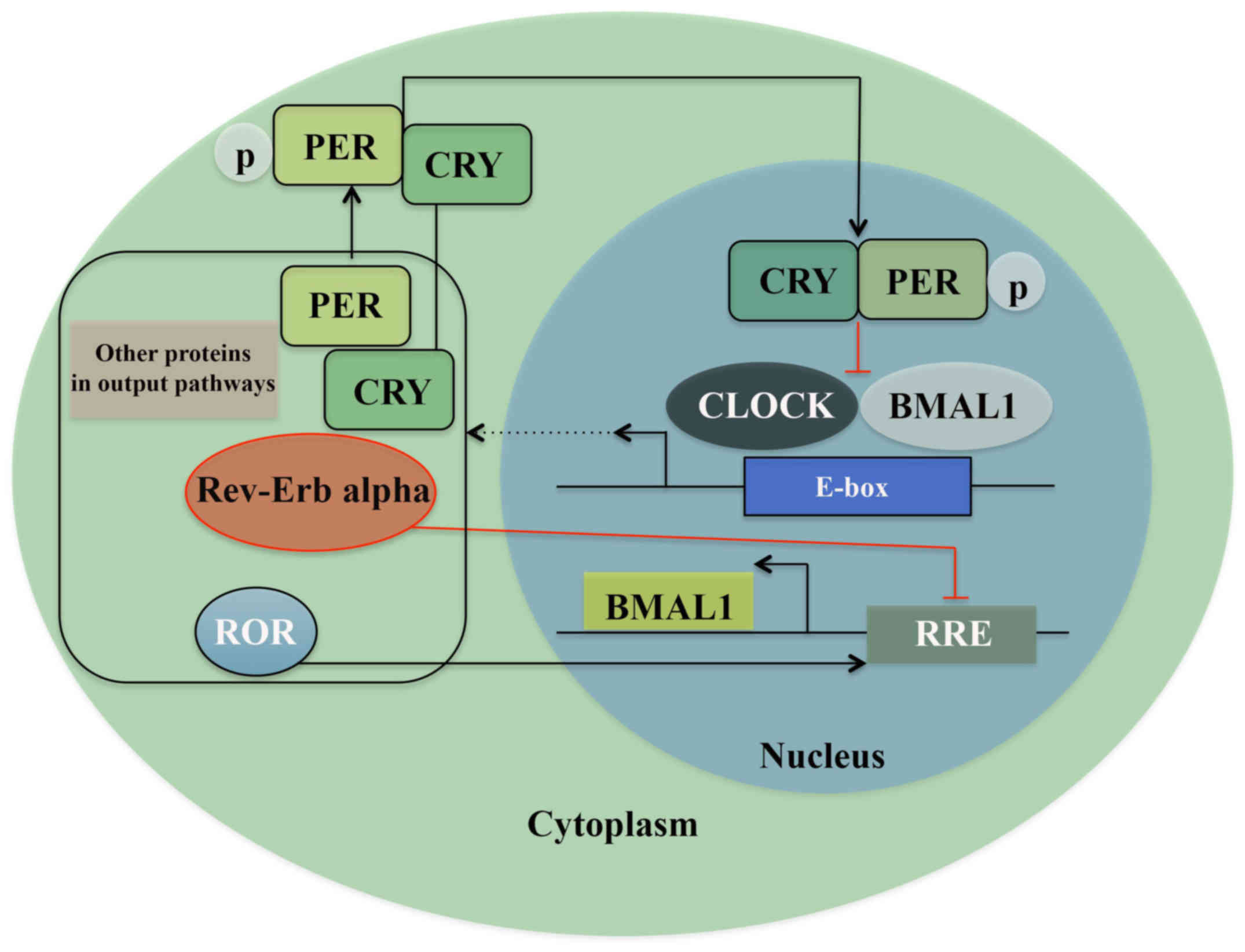
from the SCN and other molecular processes (11). One of these transcription/translation
feedback loops consists of the heterodimeric transcription complex:
CLOCK/BMAL1, which in the morning binds to the E-boxes in the
promoter region of genes expressing Period proteins (PER1, PER2,
and PER3) and Cryptochrome proteins (CRY1 and CRY2). When these
proteins accumulate, and reach an acute concentration in cytoplasm,
factors like Skp1-Cullin-F-box protein (SCF) E3 ubiquitin ligase
complexes, caseinkinase 1ε/δ (CK1ε/δ), and AMP kinase (AMPK) lead
to the formation of the PER/CRY complex. These protein complexes
translocate into the nucleus and reduce the activity of CLOCK/BMAL1
by direct protein-protein interaction by night, i.e., a negative
feedback loop. The robustness of this feedback loop is ensured by a
secondary mechanism where two subfamilies of nuclear hormone
receptors Rev-erb and Ror, regulate the transcription of Bmal1 and
thereby directly regulate the core feedback loop (Fig. 1) (17–19).
Further, chromatin remodeling and posttranslational modifications
ensure the regulation required for maintenance of the circadian
rhythm (8,20,21). A
study based on systematic mathematical and computational analysis
of the biological rhythms has revealed that oscillations are
created by the negative feedback signals, whereas the frequency of
these oscillations is adjusted by the positive feedback signals
without altering the amplitude of the oscillation (22). Another study has shown that the SCN
plays a more significant role in synchronizing the peripheral
clocks than regulating their oscillation, which suggests a more
cell-independent model of the system (23,24).
Accordingly, most circadian genes, except for clock and
CKIε, have a rhythmic expression in periods of 24 h. The
clock/Bmal1 complex regulates the transcription of many other genes
in addition to clock genes. A circadian oscillation is observed in
the transcription of >10% of mammalian genes, and naturally the
clock gene family exhibits an important role in many physiological
functions such as food intake, body temperature, metabolism and
synthesis and release of hormones (25–27).
Circadian regulators, being directly involved in the circadian
machinery, are suggested to control cell cycle. For example,
CLOCK/BMAL1 regulates cell cycle gene Wee1 being important in the
G2/M phase, 12c-myc in the G0/G1 phase and Cyclin D1, which is
important in the G1/S phase (3).
Moreover, an interaction is detected between PER1 and checkpoint
proteins such as ATM and Chk2 and 17 (28).
It has been suggested that disease caused by
circadian rhythm alterations is due to gene dosage changes and
failure in controlling gene dosages in TTFLs (29). Alterations and disruption of the
circadian clock are a more common problem nowadays due to the
industrialization of our society where artificial lighting, working
night shifts, and rapid long-distance travelling through several
time zones are common features. This is speculated to be directly
linked to the increasingly higher risk of acquiring a number of
health problems and diseases including cancer (30). Epidemiologic studies of circadian
clock alterations have suggested a link between cardiovascular,
metabolic, gastrointestinal, and mental disorders as well as
numerous cancer forms such as breast, ovarian, lung, pancreatic,
prostate, colorectal, and endometrial cancers, non-Hodgkin's
lymphoma (NHL), osteosarcoma, acute myeloid leukemia (AML), head
and neck squamous cell carcinoma and hepatocellular carcinoma
(31–45). The risk of acquiring cancer alters
with the frequency and duration of disruption of the endogenous
circadian clock. (40,46–49).
The hypothesis of artificial lighting altering the
circadian clock and leading to a higher cancer risk is further
strengthened by the findings in visually impaired individuals that
were not affected by light input and depended on other inputs for
regulating their endogenous clock. Studies have shown a lower
cancer risk for these individuals than others in the same
environment (50–52). In 2007, research and evidence
gathered resulted in classifying ‘shiftwork that involves circadian
disruption’ as a probable carcinogen by the International Agency
for Research in Cancer (44).
The disruption of the circadian clock has not only
been shown to increase the risk of disease, but also to affect the
prognosis and treatment outcome of patients. Studies have
demonstrated that variation in circadian cortisol value in blood
and sleeping patterns of the patients with metastatic breast,
colorectal, or lung cancer are linked to overall survival of these
patients (53–59). Although these findings suggest that
the circadian clock undergoes significant changes in human
tumorigenesis, the direct links between aberrant circadian clock
gene expression and human malignancies, including oral and head and
neck carcinomas, remain largely elusive. The present review focuses
on the role of clock genes in oral squamous cell cancer.
The molecular process in how clock genes expression
levels prevent or enhance tumorigenesis is not yet fully
understood; however, several studies have registered a correlation
between different expression levels of each clock gene and
different cancer types. NPAS2 has shown a significant association
with a lack of metastasis and survival prognosis in breast cancer
patients (60–62). Similar results were presented in
colorectal cancer, and decreased expression of NPAS2 was strongly
correlated to tumor size, TNM stage and metastasis rate (63). In ERα-positive breast cancer tissue
the high expression of the clock gene was reduced through a
knockout technique and a reduction in proliferation was observed.
In contrast, administration of estrogen resulted in increased
expression of clock and the proliferation of breast cancer
cells (64). Also, in colorectal
cancer a higher expression of clock is registered in
diseased tissue (65). These
findings present a diagnostic value for both genes and the adverse
effects of two different clock genes where a high level of NPAS2
expression is correlated to better prognosis whereas a high level
of clock expression correlates to increased proliferation of
cancerous tissue.
A lower expression level of PER1, PER2, and PER3 has
been registered in diseased tissue compared to normal adjacent
tissue in breast cancer, prostate cancer, colorectal cancer,
pancreatic ductal adenocarcinoma, gastric cancer, kidney cancer and
non-small-cell lung cancer (61,66–71).
PER1 and PER2 have shown a tumor-suppressing effect in a number of
studies. Higher expression of PER2 in breast cancer tissue
correlated with a lack of metastasis (61). In a study where the PER2 expression
was downregulated, a substantial increase in tumor growth rate and
higher proliferation in diseased cells were observed both in
vivo and in vitro (72).
In gastric cancer tissue a suppressing role of PER1 and PER2 on
tumor progression and metastasis was registered and a low
expression of PER1 and PER2 was correlated with poorer prognosis
(64). Overexpression of PER1 showed
a great inhibition of growth and stimulated apoptosis in prostate
cancer cell lines (71). A
correlation between decreased levels of PER1 and a lower survival
rate and liver metastasis in gastric cancer patients has also been
detected (65,66).
A link has been discovered between CRY2 and breast
cancer progression and prognosis where lower expression levels were
registered in diseased tissue (73).
Moreover, the CRY1 gene was associated with fatal prostate
cancer (74). Results from an animal
study with Bmal1 knockout mice showed that circadian
behavior in total darkness came to a complete stop (75). Research has registered lower
expression of BMAL1 in diseased tissue compared to normal adjacent
tissue in patients with colorectal cancer, pancreatic cancer, and
pancreatic ductal adenocarcinoma (67,76,77). A
knockdown of Bmal1 in pancreatic cancer cell lines led to
increased cell proliferation and decreased apoptosis (77). In vivo and in vitro
studies of colorectal cancer patients show that a higher level of
BMAL1 expression correlates to less tumor cell proliferation and
higher survival (76). Collectively,
these data open avenues for novel diagnostic and therapeutic models
for different cancer forms and prove the need for, and importance
of, further research on clock genes.
Worldwide, almost 300,000 people are annually
diagnosed with oral cancer, which makes it the 10th most common
type of cancer (78). Oral cancer
incidence and mortality rates vary widely across the world, and the
highest rates are generally registered in a few developing
countries, i.e., Sri Lanka, India, Pakistan, and Bangladesh
(79). The etiology of oral cancer
is multifactorial. The main risk factors are tobacco use and
alcohol consumption with combined multiplicative effects possibly
leading to DNA damage or mutations. Human papilloma virus infection
and genetic polymorphism can also be mentioned as risk factors
(80–82). Men are overrepresented in this
patient group and high age and lower socioeconomic status may have
an impact (83).
Oral squamous cell carcinoma (OSCC), one type of
oral cancer, is the eighth most common cancer worldwide (84,85).
Over 90% of oral malignancies are squamous cell carcinomas and its
variants (45,86). This cancer type usually emerges from
the tongue, floor of the mouth, buccal mucosa, gingiva and hard
palate. Cancer located in the tongue is associated with poorer
prognosis (87,88).
Clock genes have been detected in healthy oral
mucosa and their diurnal oscillations are mapped (89,90).
Rhythmical oscillation of the genes and their different peaks has
been shown to occur simultaneously with different phases of the
cell-cycle. PER1 peaked simultaneously as p53, which is a G1-marker
and an important gene in oncogenesis. BMAL1 peaked simultaneously
with the M-phase marker cyclinβ1 (90). Studies have confirmed that cyclinβ1
and p53 are targets of human clock genes where loss of BMAL1
reduces the expression of p53 along with PER1, PER2, and PER3. It
has also been hypothesized that p53 is involved in regulating PER2
expression by blocking the CLOCK/BMAL1 complex from binding to a
promotor region (77,91–93).
This further supports the theory that there is a connection between
clock gene activity and the cell cycle (2).
Clock genes have a clear role in cancer development,
prognosis, and therapy. From the perspective of biological rhythms,
focusing on clock genes may provide novel ideas and methods for a
better understanding of the occurrence and development of tumors,
and for individualized treatment of cancer. So far, the results
suggest that the PER1 gene may be used as a marker to determine
clinical staging and the metastatic risk, and as a novel target for
the prevention and treatment of oral cancer. (94–96)
However, future studies are warranted in order to concentrate on
the translational and post-translational levels and to illustrate
the molecular function and the regulatory effects in the clock gene
network and the tumor-suppression mechanisms of PER1, providing new
and effective molecular targets for the treatment of oral
cancer.
Investigation of the role of PER2 in OSCC cell line
Tca8113 cells, showed a lower level of expression than in healthy
tissue. The expression of PER2 was down regulated, and cell cycle,
cell proliferation and apoptosis was analyzed using flow cytometry
and RT-qPCR. The down-regulation of PER2 expression had a great
effect on the CDK/CKI cell cycle network and altered the expression
levels of many factors including decreasing p53. A significantly
higher cell proliferation and lower apoptosis were observed
(101). Tan et al
investigated the circadian pattern of PER2 and various cell cycle
genes in golden hamsters. PER2 and P53 had a decreased level while
Cyclin D1, CDK1, and Cyclin B1 levels increased during cancer
development (102).
Reports show that PER1 has a pro-apoptotic role in
many cancer types, for example in human colon cancer and prostate
cancer (71,99). But it has also been reported that
PER1 has an anti-apoptotic role in pancreatic and hepatocellular
cancer cells (103). A
pro-apoptotic role of PER1 is suggested in OSCCs, but in contrast,
in the gingival cancer cell line CA9-22, PER1 had an increased
level of expression in cancer cells compared to healthy gingival
cells, while PER3 had a decreased level of expression in diseased
cells compared to normal cells. An anti-apoptotic role was observed
for PER1 and pro-apoptotic role for PER3 (104). These results emphasize the
importance of the variation in cancer cell properties and indicate
that the same gene may play a substantially different role in
different parts of the body and that thorough research is crucial
for obtaining useful results concerning cancer diagnosis and
therapeutic methods.
We are grateful to the Department of Oral Biology,
University of Oslo and the Department of Medical Biochemistry, Oslo
University Hospital for the funding.
|
1
|
Moore RY: The suprachiasmatic nucleus and
the circadian timing system. Prog Mol Biol Transl Sci. 119:1–28.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Soták M, Sumová A and Pácha J: Cross-talk
between the circadian clock and the cell cycle in cancer. Ann Med.
46:221–232. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fu L, Pelicano H, Liu J, Huang P and Lee
C: The circadian gene Period2 plays an important role in tumor
suppression and DNA damage response in vivo. Cell. 111:41–50. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hunt T and Sassone-Corsi P: Riding tandem:
Circadian clocks and the cell cycle. Cell. 129:461–464. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hrushesky WJ, Grutsch J, Wood P, Yang X,
Oh EY, Ansell C, Kidder S, Ferrans C, Quiton DF, Reynolds J, et al:
Circadian clock manipulation for cancer prevention and control and
the relief of cancer symptoms. Integr Cancer Ther. 8:387–397. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gery S and Koeffler HP: Circadian rhythms
and cancer. Cell Cycle. 9:1097–1103. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roenneberg T and Merrow M: Circadian
clocks-the fall and rise of physiology. Nat Rev Mol Cell Biol.
6:965–971. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Grimaldi B, Nakahata Y, Kaluzova M,
Masubuchi S and Sassone-Corsi P: Chromatin remodeling, metabolism
and circadian clocks: The interplay of CLOCK and SIRT1. Int J
Biochem Cell Biol. 41:81–86. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aschoff J, Fatranská M, Giedke H, Doerr P,
Stamm D and Wisser H: Human circadian rhythms in continuous
darkness: Entrainment by social cues. Science. 171:213–215. 1971.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rosato E and Kyriacou CP: Origins of
circadian rhythmicity. J Biol Rhythms. 17:506–511. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schibler U and Sassone-Corsi P: A web of
circadian pacemakers. Cell. 111:919–922. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Whitmore D, Foulkes NS and Sassone-Corsi
P: Light acts directly on organs and cells in culture to set the
vertebrate circadian clock. Nature. 404:87–91. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reppert SM and Weaver DR: Coordination of
circadian timing in mammals. Nature. 418:935–941. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Golombek DA and Rosenstein RE: Physiology
of circadian entrainment. Physiol Rev. 90:1063–1102. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shearman LP, Sriram S, Weaver DR, Maywood
ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings
MH, et al: Interacting molecular loops in the mammalian circadian
clock. Science. 288:1013–1019. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Giebultowicz JM: Peripheral clocks and
their role in circadian timing: Insights from insects. Philos Trans
R Soc Lond B Biol Sci. 356:1791–1799. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Preitner N, Damiola F, Lopez-Molina L,
Zakany J, Duboule D, Albrecht U and Schibler U: The orphan nuclear
receptor REV-ERBalpha controls circadian transcription within the
positive limb of the mammalian circadian oscillator. Cell.
110:251–260. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cho H, Zhao X, Hatori M, Yu RT, Barish GD,
Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al:
Regulation of circadian behaviour and metabolism by REV-ERB-α and
REV-ERB-β. Nature. 485:123–127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sato TK, Panda S, Miraglia LJ, Reyes TM,
Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA and Hogenesch
JB: A functional genomics strategy reveals Rora as a component of
the mammalian circadian clock. Neuron. 43:527–537. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Padmanabhan K, Robles MS, Westerling T and
Weitz CJ: Feedback regulation of transcriptional termination by the
mammalian circadian clock PERIOD complex. Science. 337:599–602.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duong HA, Robles MS, Knutti D and Weitz
CJ: A molecular mechanism for circadian clock negative feedback.
Science. 332:1436–1439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Novák B and Tyson JJ: Design principles of
biochemical oscillators. Nat Rev Mol Cell Biol. 9:981–991. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nishide SY, Honma S, Nakajima Y, Ikeda M,
Baba K, Ohmiya Y and Honma K: New reporter system for Per1 and
Bmal1 expressions revealed self-sustained circadian rhythms in
peripheral tissues. Genes Cells. 11:1173–1182. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoo SH, Yamazaki S, Lowrey PL, Shimomura
K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al:
PERIOD2: LUCIFERASE real-time reporting of circadian dynamics
reveals persistent circadian oscillations in mouse peripheral
tissues. Proc Natl Acad Sci USA. 101:pp. 5339–5346. 2004;
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Akhtar RA, Reddy AB, Maywood ES, Clayton
JD, King VM, Smith AG, Gant TW, Hastings MH and Kyriacou CP:
Circadian cycling of the mouse liver transcriptome, as revealed by
cDNA microarray, is driven by the suprachiasmatic nucleus. Curr
Biol. 12:540–550. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duffield GE, Best JD, Meurers BH, Bittner
A, Loros JJ and Dunlap JC: Circadian programs of transcriptional
activation, signaling, and protein turnover revealed by microarray
analysis of mammalian cells. Curr Biol. 12:551–557. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ceriani MF, Hogenesch JB, Yanovsky M,
Panda S, Straume M and Kay SA: Genome-wide expression analysis in
Drosophila reveals genes controlling circadian behavior. J
Neurosci. 22:9305–9319. 2002.PubMed/NCBI
|
|
28
|
Unsal-Kaçmaz K, Mullen TE, Kaufmann WK and
Sancar A: Coupling of human circadian and cell cycles by the
timeless protein. Mol Cell Biol. 25:3109–3116. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee HM, Chen R, Kim H, Etchegaray JP,
Weaver DR and Lee C: The period of the circadian oscillator is
primarily determined by the balance between casein kinase 1 and
protein phosphatase 1. Proc Natl Acad Sci USA. 108:pp. 16451–16456.
2011; View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Costa G, Haus E and Stevens R: Shift work
and cancer-considerations on rationale, mechanisms and
epidemiology. Scand J Work Environ Health. 36:163–179. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schernhammer ES, Laden F, Speizer FE,
Willett WC, Hunter DJ, Kawachi I, Fuchs CS and Colditz GA:
Night-shift work and risk of colorectal cancer in the nurses'
health study. J Natl Cancer Inst. 95:825–828. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kubo T, Ozasa K, Mikami K, Wakai K, Fujino
Y, Watanabe Y, Miki T, Nakao M, Hayashi K, Suzuki K, et al:
Prospective cohort study of the risk of prostate cancer among
rotating-shift workers: Findings from the Japan collaborative
cohort study. Am J Epidemiol. 164:549–555. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Viswanathan AN, Hankinson SE and
Schernhammer ES: Night shift work and the risk of endometrial
cancer. Cancer Res. 67:10618–10622. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lahti TA, Partonen T, Kyyrönen P,
Kauppinen T and Pukkala E: Night-time work predisposes to
non-Hodgkin lymphoma. Int J Cancer. 123:2148–2151. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stevens RG: Artificial lighting in the
industrialized world: Circadian disruption and breast cancer.
Cancer Causes Control. 17:501–507. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Touitou Y, Bogdan A, Lévi F, Benavides M
and Auzéby A: Disruption of the circadian patterns of serum
cortisol in breast and ovarian cancer patients: Relationships with
tumour marker antigens. Br J Cancer. 74:1248–1252. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Panzer A: Melatonin in osteosarcoma: An
effective drug? Med Hypotheses. 48:523–525. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Skibola CF, Holly EA, Forrest MS, Hubbard
A, Bracci PM, Skibola DR, Hegedus C and Smith MT: Body mass index,
leptin and leptin receptor polymorphisms, and non-hodgkin lymphoma.
Cancer Epidemiol Biomarkers Prev. 13:779–786. 2004.PubMed/NCBI
|
|
39
|
Kloog I, Haim A, Stevens RG and Portnov
BA: Global co-distribution of light at night (LAN) and cancers of
prostate, colon, and lung in men. Chronobiol Int. 26:108–125. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bhatti P, Mirick DK and Davis S: Invited
commentary: Shift work and cancer. Am J Epidemiol. 176:760–765.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Buzzelli G, Dattolo P, Pinzani M, Brocchi
A, Romano S and Gentilini P: Circulating growth hormone and
insulin-like growth factor-I in nonalcoholic liver cirrhosis with
or without superimposed hepatocarcinoma: Evidence of an altered
circadian rhythm. Am J Gastroenterol. 88:1744–1748. 1993.PubMed/NCBI
|
|
42
|
Rafnsson V, Tulinius H, Jónasson JG and
Hrafnkelsson J: Risk of breast cancer in female flight attendants:
A population-based study (Iceland). Cancer Causes Control.
12:95–101. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Scheer FA, Hilton MF, Mantzoros CS and
Shea SA: Adverse metabolic and cardiovascular consequences of
circadian misalignment. Proc Natl Acad Sci USA. 106:pp. 4453–4458.
2009; View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Straif K, Baan R, Grosse Y, Secretan B,
Ghissassi FE, Bouvard V, Altieri A, Benbrahim-Tallaa L and Cogliano
V; WHO, ; International Agency for Research on Cancer Monograph
Working Group, : Carcinogenicity of shift-work, painting and
fire-fighting. Lancet Oncol. 8:1065–1066. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hsu CM, Lin SF, Lu CT, Lin PM and Yang MY:
Altered expression of circadian clock genes in head and neck
squamous cell carcinoma. Tumour Biol. 33:149–155. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Haus EL and Smolensky MH: Shift work and
cancer risk: Potential mechanistic roles of circadian disruption,
light at night and sleep deprivation. Sleep Med Rev. 17:273–284.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schernhammer ES, Laden F, Speizer FE,
Willett WC, Hunter DJ, Kawachi I and Colditz GA: Rotating night
shifts and risk of breast cancer in women participating in the
nurses' health study. J Natl Cancer Inst. 93:1563–1568. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Stevens RG, Hansen J, Costa G, Haus E,
Kauppinen T, Aronson KJ, Castaño-Vinyals G, Davis S, Frings-Dresen
MH, Fritschi L, et al: Considerations of circadian impact for
defining ‘shift work’ in cancer studies: IARC Working Group Report.
Occup Environ Med. 68:154–162. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pukkala E, Aspholm R, Auvinen A, Eliasch
H, Gundestrup M, Haldorsen T, Hammar N, Hrafnkelsson J, Kyyrönen P,
Linnersjö A, et al: Incidence of cancer among Nordic airline pilots
over five decades: Occupational cohort study. BMJ. 325:5672002.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Feychting M, Osterlund B and Ahlbom A:
Reduced cancer incidence among the blind. Epidemiology. 9:490–494.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kliukiene J, Tynes T and Andersen A: Risk
of breast cancer among Norwegian women with visual impairment. Br J
Cancer. 84:397–399. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Verkasalo PK, Pukkala E, Stevens RG, Ojamo
M and Rudanko SL: Inverse association between breast cancer
incidence and degree of visual impairment in Finland. Br J Cancer.
80:1459–1460. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rich T, Innominato PF, Boerner J, Mormont
MC, Iacobelli S, Baron B, Jasmin C and Lévi F: Elevated serum
cytokines correlated with altered behavior, serum cortisol rhythm,
and dampened 24-hour rest-activity patterns in patients with
metastatic colorectal cancer. Clin Cancer Res. 11:1757–1764. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sephton SE, Sapolsky RM, Kraemer HC and
Spiegel D: Diurnal cortisol rhythm as a predictor of breast cancer
survival. J Natl Cancer Inst. 92:994–1000. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kim KS, Kim YC, Oh IJ, Kim SS, Choi JY and
Ahn RS: Association of worse prognosis with an aberrant diurnal
cortisol rhythm in patients with advanced lung cancer. Chronobiol
Int. 29:1109–1120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sephton SE, Lush E, Dedert EA, Floyd AR,
Rebholz WN, Dhabhar FS, Spiegel D and Salmon P: Diurnal cortisol
rhythm as a predictor of lung cancer survival. Brain Behav Immun.
30 Suppl:S163–S170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Innominato PF, Giacchetti S, Bjarnason GA,
Focan C, Garufi C, Coudert B, Iacobelli S, Tampellini M, Durando X,
Mormont MC, et al: Prediction of overall survival through circadian
rest-activity monitoring during chemotherapy for metastatic
colorectal cancer. Int J Cancer. 131:2684–2692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu HS, Davis JE and Natavio T: Fatigue and
disrupted sleep-wake patterns in patients with cancer: A shared
mechanism. Clin J Oncol Nurs. 16:E56–E68. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Innominato PF, Focan C, Gorlia T, Moreau
T, Garufi C, Waterhouse J, Giacchetti S, Coudert B, Iacobelli S,
Genet D, et al: Circadian rhythm in rest and activity: A biological
correlate of quality of life and a predictor of survival in
patients with metastatic colorectal cancer. Cancer Res.
69:4700–4707. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yi C, Mu L, de la Longrais IA, Sochirca O,
Arisio R, Yu H, Hoffman AE, Zhu Y and Katsaro D: The circadian gene
NPAS2 is a novel prognostic biomarker for breast cancer. Breast
Cancer Res Treat. 120:663–669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cadenas C, van de Sandt L, Edlund K, Lohr
M, Hellwig B, Marchan R, Schmidt M, Rahnenführer J, Oster H and
Hengstler JG: Loss of circadian clock gene expression is associated
with tumor progression in breast cancer. Cell Cycle. 13:3282–3291.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhu Y, Stevens RG, Leaderer D, Hoffman A,
Holford T, Zhang Y, Brown HN and Zheng T: Non-synonymous
polymorphisms in the circadian gene NPAS2 and breast cancer risk.
Breast Cancer Res Treat. 107:421–425. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xue X, Liu F, Han Y, Li P, Yuan B, Wang X,
Chen Y, Kuang Y, Zhi Q and Zhao H: Silencing NPAS2 promotes cell
growth and invasion in DLD-1 cells and correlated with poor
prognosis of colorectal cancer. Biochem Biophys Res Commun.
450:1058–1062. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xiao L, Chang AK, Zang MX, Bi H, Li S,
Wang M, Xing X and Wu H: Induction of the CLOCK gene by E2-ERα
signaling promotes the proliferation of breast cancer cells. PLoS
One. 9:e958782014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Oshima T, Takenoshita S, Akaike M,
Kunisaki C, Fujii S, Nozaki A, Numata K, Shiozawa M, Rino Y, Tanaka
K, et al: Expression of circadian genes correlates with liver
metastasis and outcomes in colorectal cancer. Oncol Rep.
25:1439–1446. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mazzoccoli G, Panza A, Valvano MR, Palumbo
O, Carella M, Pazienza V, Biscaglia G, Tavano F, Di Sebastiano P,
Andriulli A and Piepoli A: Clock gene expression levels and
relationship with clinical and pathological features in colorectal
cancer patients. Chronobiol Int. 28:841–851. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Relles D, Sendecki J, Chipitsyna G, Hyslop
T, Yeo CJ and Arafat HA: Circadian gene expression and
clinicopathologic correlates in pancreatic cancer. J Gastrointest
Surg. 17:443–450. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hu ML, Yeh KT, Lin PM, Hsu CM, Hsiao HH,
Liu YC, Lin HY, Lin SF and Yang MY: Deregulated expression of
circadian clock genes in gastric cancer. BMC Gastroenterol.
14:672014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mazzoccoli G, Piepoli A, Carella M, Panza
A, Pazienza V, Benegiamo G, Palumbo O and Ranieri E: Altered
expression of the clock gene machinery in kidney cancer patients.
Biomed Pharmacother. 66:175–179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu B, Xu K, Jiang Y and Li X: Aberrant
expression of Per1, Per2 and Per3 and their prognostic relevance in
non-small cell lung cancer. Int J Clin Exp Pathol. 7:7863–7871.
2014.PubMed/NCBI
|
|
71
|
Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu
J and Koeffler HP: A role for the clock gene per1 in prostate
cancer. Cancer Res. 69:7619–7625. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yang X, Wood PA, Oh EY, Du-Quiton J,
Ansell CM and Hrushesky WJ: Down regulation of circadian clock gene
Period 2 accelerates breast cancer growth by altering its daily
growth rhythm. Breast Cancer Res Treat. 117:423–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Mao Y, Fu A, Hoffman AE, Jacobs DI, Jin M,
Chen K and Zhu Y: The circadian gene CRY2 is associated with breast
cancer aggressiveness possibly via epigenomic modifications. Tumour
Biol. 36:3533–3539. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Markt SC, Valdimarsdottir UA, Shui IM,
Sigurdardottir LG, Rider JR, Tamimi RM, Batista JL, Haneuse S,
Flynn-Evans E, Lockley SW, et al: Circadian clock genes and risk of
fatal prostate cancer. Cancer Causes Control. 26:25–33. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Bunger MK, Wilsbacher LD, Moran SM,
Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS and
Bradfield CA: Mop3 is an essential component of the master
circadian pacemaker in mammals. Cell. 103:1009–1017. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zeng ZL, Luo HY, Yang J, Wu WJ, Chen DL,
Huang P and Xu RH: Overexpression of the circadian clock gene Bmal1
increases sensitivity to oxaliplatin in colorectal cancer. Clin
Cancer Res. 20:1042–1052. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Jiang W, Zhao S, Jiang X, Zhang E, Hu G,
Hu B, Zheng P, Xiao J, Lu Z, Lu Y, et al: The circadian clock gene
Bmal1 acts as a potential anti-oncogene in pancreatic cancer by
activating the p53 tumor suppressor pathway. Cancer Lett.
371:314–325. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Sanderson RJ, de Boer MF, Damhuis RA,
Meeuwis CA and Knegt PP: The influence of alcohol and smoking on
the incidence of oral and oropharyngeal cancer in women. Clin
Otolaryngol Allied Sci. 22:444–448. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Scully C and Field J: Genetic aberrations
in squamous cell carcinoma of the head and neck (SCCHN), with
reference to oral carcinoma (review). Int J Oncol. 10:5–21.
1997.PubMed/NCBI
|
|
82
|
Yuan H, Li H, Ma H, Niu Y, Wu Y, Zhang S,
Hu Z, Shen H and Chen N: Genetic polymorphisms in key DNA repair
genes and risk of head and neck cancer in a Chinese population. Exp
Ther Med. 3:719–724. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang J, Gao F, Yang AK, Chen WK, Chen SW,
Li H, Zhang X, Yang ZY, Chen XL and Song M: Epidemiologic
characteristics of oral cancer: Single-center analysis of 4097
patients from the Sun Yat-sen University Cancer Center. Chin J
Cancer. 35:242016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Amit M, Yen TC, Liao CT, Chaturvedi P,
Agarwal JP, Kowalski LP, Ebrahimi A, Clark JR, Kreppel M, Zöller J,
et al: Improvement in survival of patients with oral cavity
squamous cell carcinoma: An international collaborative study.
Cancer. 119:4242–4248. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sand L, Wallström M and Hirsch JM:
Smokeless tobacco, viruses and oral cancer. Oral Health Dent Manag.
13:372–378. 2014.PubMed/NCBI
|
|
87
|
Listl S, Jansen L, Stenzinger A, Freier K,
Emrich K, Holleczek B, Katalinic A, Gondos A and Brenner H; GEKID
Cancer Survival Working Group, : Survival of patients with oral
cavity cancer in Germany. PLoS One. 8:e534152013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Engholm G, Ferlay J, Christensen N, Bray
F, Gjerstorff ML, Klint A, Køtlum JE, Olafsdóttir E, Pukkala E and
Storm HH: NORDCAN-a Nordic tool for cancer information, planning,
quality control and research. Acta Oncol. 49:725–736. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zieker D, Jenne I, Koenigsrainer I,
Zdichavsky M, Nieselt K, Buck K, Zieker J, Beckert S, Glatzle J,
Spanagel R, et al: Circadian expression of clock- and tumor
suppressor genes in human oral mucosa. Cell Physiol Biochem.
26:155–166. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Bjarnason GA, Jordan RC, Wood PA, Li Q,
Lincoln DW, Sothern RB, Hrushesky WJ and Ben-David Y: Circadian
expression of clock genes in human oral mucosa and skin:
Association with specific cell-cycle phases. Am J Pathol.
158:1793–1801. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Mullenders J, Fabius AW, Madiredjo M,
Bernards R and Beijersbergen RL: A large scale shRNA barcode screen
identifies the circadian clock component ARNTL as putative
regulator of the p53 tumor suppressor pathway. PLoS One.
4:e47982009. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Miki T, Matsumoto T, Zhao Z and Lee CC:
p53 regulates Period2 expression and the circadian clock. Nat
Commun. 4:24442013. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zeng ZL, Wu MW, Sun J, Sun YL, Cai YC,
Huang YJ and Xian LJ: Effects of the biological clock gene Bmal1 on
tumour growth and anti-cancer drug activity. J Biochem.
148:319–326. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chen R, Yang K, Zhao NB, Zhao D, Chen D,
Zhao CR and Tang H: Abnormal expression of PER1 circadian-clock
gene in oral squamous cell carcinoma. Onco Targets Ther. 5:403–407.
2012.PubMed/NCBI
|
|
95
|
Li HX, Fu XJ, Yang K, Chen D, Tang H and
Zhao Q: The clock gene PER1 suppresses expression of tumor-related
genes in human oral squamous cell carcinoma. Oncotarget.
7:20574–20583. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhao Q, Zheng G, Yang K, Ao YR, Su XL, Li
Y and Lv XQ: The clock gene PER1 plays an important role in
regulating the clock gene network in human oral squamous cell
carcinoma cells. Oncotarget. 7:70290–70302. 2016.PubMed/NCBI
|
|
97
|
Zhao N, Tang H, Yang K and Chen D:
Circadian rhythm characteristics of oral squamous cell carcinoma
growth in an orthotopic xenograft model. Onco Targets Ther.
6:41–46. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Ye H, Yang K, Tan XM, Fu XJ and Li HX:
Daily rhythm variations of the clock gene PER1 and cancer-related
genes during various stages of carcinogenesis in a golden hamster
model of buccal mucosa carcinoma. Onco Targets Ther. 8:1419–1426.
2015.PubMed/NCBI
|
|
99
|
Gery S, Komatsu N, Baldjyan L, Yu A, Koo D
and Koeffler HP: The circadian gene per1 plays an important role in
cell growth and DNA damage control in human cancer cells. Mol Cell.
22:375–382. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Fu XJ, Li HX, Yang K, Chen D and Tang H:
The important tumor suppressor role of PER1 in regulating the
cyclin-CDK-CKI network in SCC15 human oral squamous cell carcinoma
cells. Onco Targets Ther. 9:2237–2245. 2016.PubMed/NCBI
|
|
101
|
Wang Q, Ao Y, Yang K, Tang H and Chen D:
Circadian clock gene Per2 plays an important role in cell
proliferation, apoptosis and cell cycle progression in human oral
squamous cell carcinoma. Oncol Rep. 35:3387–3394. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Tan XM, Ye H, Yang K, Chen D, Wang QQ,
Tang H and Zhao NB: Circadian variations of clock gene Per2 and
cell cycle genes in different stages of carcinogenesis in golden
hamster buccal mucosa. Sci Rep. 5:99972015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Sato F, Nagata C, Liu Y, Suzuki T, Kondo
J, Morohashi S, Imaizumi T, Kato Y and Kijima H: PERIOD1 is an
anti-apoptotic factor in human pancreatic and hepatic cancer cells.
J Biochem. 146:833–838. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Sato F, Wu Y, Bhawal UK, Liu Y, Imaizumi
T, Morohashi S, Kato Y and Kijima H: PERIOD1 (PER1) has
anti-apoptotic effects, and PER3 has pro-apoptotic effects during
cisplatin (CDDP) treatment in human gingival cancer CA9-22 cells.
Eur J Cancer. 47:1747–1758. 2011. View Article : Google Scholar : PubMed/NCBI
|