Introduction
Pancreatic cancer is a deadly disease and it is
usually diagnosed at an advanced stage. Chemotherapy plays a key
role in the treatment of pancreatic cancer, often producing a
temporary clinical benefit (1,2). Genomic
aberrations in breast cancer 1 (BRCA1) and BRCA2
genes, which are components of a common DNA repair pathway, are
inherited in an autosomal dominant pattern (3,4). A
number of studies have demonstrated that BRCA mutations,
particularly those in BRCA2, increase the risk of developing
pancreatic adenocarcinoma (5). It
has been suggested that DNA-damaging agents, such as platinum salts
or poly(ADP-ribose) polymerase (PARP) inhibitors, may be used in
patients with pancreatic cancer carrying BRCA mutations
(6,7). In the present study, genomic evolution
was investigated in a patient with stage IV pancreatic cancer
bearing a germline BRCA2 mutation.
Case report
A 60-year-old male patient carrying a known
deleterious BRCA2 mutation (1153insT) presented with
locally advanced pancreatic cancer. The patient underwent surgery,
and the subsequent pathological analysis revealed pancreatic
adenocarcinoma. Two months later, prior to commencing adjuvant
chemotherapy, the patient developed liver metastasis. He received a
cisplatin-based regimen and rapidly achieved a complete response
that lasted for 18 months. When relapse occurred in the liver, the
patient resumed the same protocol, achieving a partial response for
an additional 20 months. At that point, disease progression was
detected in the brain. The patient received multidisciplinary
treatment that included resection of one lesion and stereotactic
radiosurgery (SRS) for the second lesion. Subsequently, he received
irinotecan and bevacizumab, which resulted in a response that
lasted for 7 months.
The patient provided written informed consent, in
accordance with the Hadassah Institutional Review Board-approved
protocol.
DNA isolation
Formalin-fixed, paraffin-embedded (FFPE) tumor
tissues were assessed by a board-certified pathologist. The regions
of tumor tissue were marked and the DNA was extracted using a
QIAamp DNA FFPE Tissue kit (Qiagen, Solana Beach, CA, USA). DNA was
extracted from the blood using a DNeasy Blood and Tissue kit
(Qiagen), according to the manufacturer's instructions.
Massive parallel sequencing
The clinical samples were screened for mutations in
50 cancer-associated genes using an Ion AmpliSeq™ Cancer Hotspot
Panel v2, and a panel spanning the coding sequences of an
additional 22 cancer-associated genes. DNA extraction and
sequencing were performed as previously described (8).
The blood, primary tumor tissue (prior to any
treatment) and the brain metastatic tissue were examined using a
targeted deep sequencing assay with a mean 1,320-fold coverage. The
known deleterious BRCA2 mutation (1153insT) was
identified in all three samples, while a BRCA1 mutation
(NM_007294.3:c.4535G>T; NP_009225.1:p.Ser1512Ile;
rs1800744) and a checkpoint kinase 2 (CHEK2) mutation
(NM_001005735.1:c.1399T>C; NP_001005735.1:p.Tyr467His) were
identified only in the pancreatic tumor tissue and brain
metastasis. The pancreatic and brain samples shared other somatic
genetic aberrations (Table I), but
no significant mutational differences were detected between the
two. This suggests that the isolated relapse in the brain was due
to pharmacological sanctuary rather than further genomic
alternations. The patient survived ~7 years with metastatic disease
until succumbing to his illness.
 | Table I.Somatic mutations detected in primary
tumor and metastases. |
Table I.
Somatic mutations detected in primary
tumor and metastases.
|
|
|
| Allele frequency
(%) |
|---|
|
|
|
|
|
|---|
| Mutation | RNA | Protein | Pancreas
(primary) | Brain
(metastasis) |
|---|
| BRCA1 |
NM_007294.3:c.4535G>T |
NP_009225.1:p.Ser1512Ile | 47 | 57 |
| P53 |
c.831T>ANM_001126112.2 |
NP_000537.3:p.C277* | 33 | 46 |
| K-RAS |
NM_004985.4:c.35G>C |
NP_004976.2:p.Gly12Asp | 44 | 32 |
| CHEK2 |
NM_001005735.1:c.1399T>C |
NP_001005735.1:p.Tyr467His | 50 | 57 |
Discussion
In order to identify biomarkers for response and
resistance to platinum-based therapy in exceptional responder,
parallel genomic molecular characterization of the primary and
metastatic tumors was conducted. Genetic sequencing indicated that
selective therapeutic pressure did not lead to any significant
genomic alternation in the brain metastasis, which suggests that
the isolated relapse in the brain was instead due to
pharmacological sanctuary at this site. This hypothesis was
confirmed by the clinical observation that the patient also
responded well to SRS and irinotecan administered to treat the
brain recurrence, suggesting that the therapeutic sensitivity to
DNA-damaging agents was retained.
Moreover, a BRCA1 gene mutation was
identified in both the primary tumor tissue and the brain
metastasis, in addition to the germline BRCA2 mutation. Such
dual BRCA1/2 loss of function may be the cause of the
exceptional clinical sensitivity to DNA-damaging agents throughout
the treatment of this patient.
In light of the role of BRCA in DNA repair,
it is suggested that BRCA1 or BRCA2 mutations result
in increased sensitivity to DNA-damaging agents (9,10).
Indeed, mounting evidence suggests a better response to PARP
inhibitors or cisplatin in BRCA-associated malignancies
(11–16). However, over time,
BRCA-deficient tumors become resistant and disease
progression may occur.
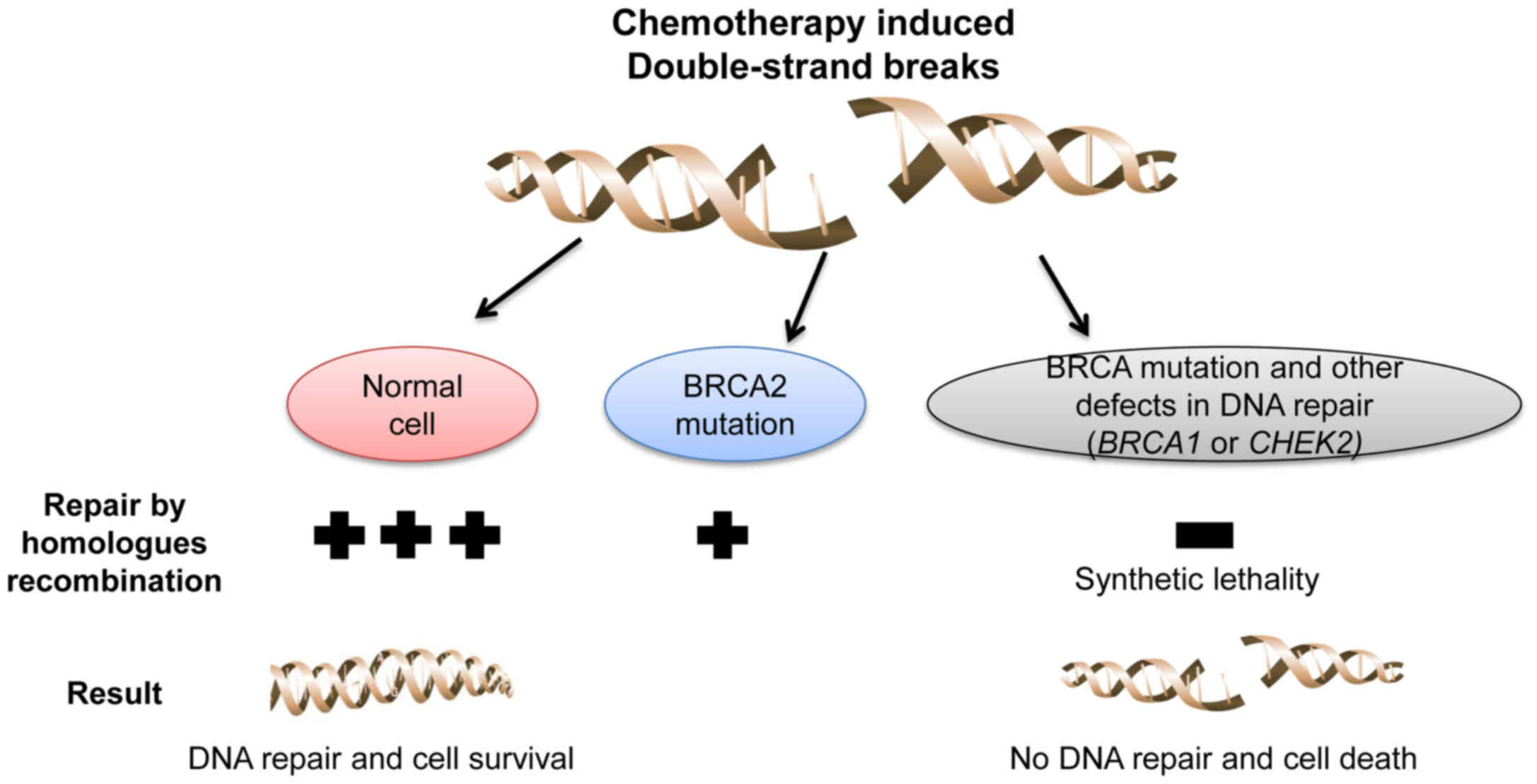
The ‘synthetic lethality’ concept has provided new
opportunities for drug development (17). For example, in cancer cells with loss
of function of BRCA, treatment with PARP inhibitors leads to
an accumulation of single-strand breaks that subsequently develop
into double-strand breaks, which cannot be fixed by homologous
recombination (18,19).
A mutation in the CHEK2 gene in the primary
tumor and brain metastatic tissues was also detected. CHEK2
is an important regulator of cellular response to DNA damage, and
has been identified as tumor-suppressor gene in various human
malignancies (20,21). This CHEK2 mutation may also
contribute to the defects in DNA repair mechanisms in the described
tumor.
To conclude, the findings of the present study
suggest that loss of BRCA1, BRCA2 and CHEK2 function
may result in greater sensitivity of cancer cells to DNA-damaging
agents compared with the loss of function of only one of these
genes (Fig. 1). If such a strategy
becomes pharmacologically applicable, it may represent a novel
synthetic effective approach to the treatment of pancreatic cancer,
as well as other malignancies.
Acknowledgements
We would like to thank Tamar H. for her
assistance.
Funding
AS is supported by a Clinical Research Career
Development Award from the Israel Cancer Research Fund grants
(16-116-CRCDA) and from the Israeli cancer research association
(2017–0140).
Availability of data and materials
Not applicable
Authors' contributions
AS, AZ, TP and AH conceived of the study and
participated in its design and coordination, analyzed and
interpreted the data and wrote the manuscript. MM and SC performed
technical work.
Ethics approval and consent to
participate
The patient provided written informed consent for
the genetic research studies performed in accordance with protocols
approved by the Institutional Review Board.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gresham GK, Wells GA, Gill S, Cameron C
and Jonker DJ: Chemotherapy regimens for advanced pancreatic
cancer: A systematic review and network meta-analysis. BMC Cancer.
14:4712014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heinemann V, Haas M and Boeck S: Systemic
treatment of advanced pancreatic cancer. Cancer Treat Rev.
38:843–853. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Roy R, Chun J and Powell SN: BRCA1 and
BRCA2: Different roles in a common pathway of genome protection.
Nat Rev Cancer. 12:68–78. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mavaddat N, Peock S, Frost D, Ellis S,
Platte R, Fineberg E, Evans DG, Izatt L, Eeles RA, Adlard J, et al:
Cancer risks for BRCA1 and BRCA2 mutation carriers: Results from
prospective analysis of EMBRACE. J Natl Cancer Inst. 105:812–822.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hahn SA, Greenhalf B, Ellis I, Sina-Frey
M, Rieder H, Korte B, Gerdes B, Kress R, Ziegler A, Raeburn JA, et
al: BRCA2 germline mutations in familial pancreatic carcinoma. J
Natl Cancer Inst. 95:214–221. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sonnenblick A, Kadouri L, Appelbaum L,
Peretz T, Sagi M, Goldberg Y and Hubert A: Complete remission, in
BRCA2 mutation carrier with metastatic pancreatic adenocarcinoma,
treated with cisplatin based therapy. Cancer Biol Ther. 12:165–168.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lowery MA, Kelsen DP, Stadler ZK, Yu KH,
Janjigian YY, Ludwig E, D'Adamo DR, Salo-Mullen E, Robson ME, Allen
PJ, et al: An emerging entity: Pancreatic adenocarcinoma associated
with a known BRCA mutation: Clinical descriptors, treatment
implications and future directions. Oncologist. 16:1397–1402. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zick A, Peretz T, Lotem M, Hubert A, Katz
D, Temper M, Rottenberg Y, Uziely B, Nechushtan H, Meirovitz A, et
al: Treatment inferred from mutations identified using massive
parallel sequencing leads to clinical benefit in some heavily
pretreated cancer patients. Medicine (Baltimore). 96:e69312017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Muggia F, Safra T and Dubeau L: BRCA
genes: Lessons learned from experimental and clinical cancer. Ann
Oncol. 22 Suppl 1:i7–i10. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sonnenblick A, de Azambuja E, Azim HA Jr
and Piccart M: An update on PARP inhibitors-moving to the adjuvant
setting. Nat Rev Clin Oncol. 12:27–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Silver DP, Richardson AL, Eklund AC, Wang
ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, et
al: Efficacy of neoadjuvant cisplatin in triple-negative breast
cancer. J Clin Oncol. 28:1145–1153. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fong PC, Boss DS, Yap TA, Tutt A, Wu P,
Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et
al: Inhibition of Poly(ADP-Ribose) polymerase in tumors from BRCA
mutation carriers. N Engl J Med. 361:123–134. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ledermann J, Harter P, Gourley C,
Friedlander M, Vergote I, Rustin G, Scott CL, Meier W,
Shapira-Frommer R, Safra T, et al: Olaparib maintenance therapy in
patients with platinum-sensitive relapsed serous ovarian cancer: A
preplanned retrospective analysis of outcomes by BRCA status in a
randomised phase 2 trial. Lancet Oncol. 15:852–861. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kwa M, Edwards S, Downey A, Reich E,
Wallach R, Curtin J and Muggia F: Ovarian cancer in BRCA mutation
carriers: Improved outcome after intraperitoneal (IP) cisplatin.
Ann Surg Oncol. 21:1468–1473. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Golan T, Kanji ZS, Epelbaum R, Devaud N,
Dagan E, Holter S, Aderka D, Paluch-Shimon S, Kaufman B,
Gershoni-Baruch R, et al: Overall survival and clinical
characteristics of pancreatic cancer in BRCA mutation carriers. Br
J Cancer. 111:1132–1138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fong PC, Yap TA, Boss DS, Carden CP,
Mergui-Roelvink M, Gourley C, De Greve J, Lubinski J, Shanley S,
Messiou C, et al: Poly(ADP)-ribose polymerase inhibition: frequent
durable responses in BRCA carrier ovarian cancer correlating with
platinum-free interval. J Clin Oncol. 28:2512–2519. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kaelin WG Jr: The concept of synthetic
lethality in the context of anticancer therapy. Nat Rev Cancer.
5:689–698. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Farmer H, McCabe N, Lord CJ, Tutt AN,
Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I,
Knights C, et al: Targeting the DNA repair defect in BRCA mutant
cells as a therapeutic strategy. Nature. 434:917–921. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bryant HE, Schultz N, Thomas HD, Parker
KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ and Helleday T:
Specific killing of BRCA2-deficient tumours with inhibitors of
poly(ADP-ribose) polymerase. Nature. 434:913–917. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nevanlinna H and Bartek J: The CHEK2 gene
and inherited breast cancer susceptibility. Oncogene. 25:5912–5919.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bartek J and Lukas J: Chk1 and Chk2
kinases in checkpoint control and cancer. Cancer Cell. 3:421–429.
2003. View Article : Google Scholar : PubMed/NCBI
|