|
1
|
Raza K: The Michael Mason prize: early
rheumatoid arthritis - the window narrows. Rheumatology (Oxford).
49:406–410. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nell V, Machold K, Eberl G, Stamm T,
Uffmann M and Smolen J: Benefit of very early referral and very
early therapy with disease-modifying anti-rheumatic drugs in
patients with early rheumatoid arthritis. Rheumatology (Oxford).
43:906–914. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bigelow RL, Jen EY, Delehedde M, Chari NS
and McDonnell TJ: Sonic hedgehog induces epidermal growth factor
dependent matrix infiltration in HaCaT keratinocytes. J Invest
Dermatol. 124:457–465. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iwasaki A and Medzhitov R: Regulation of
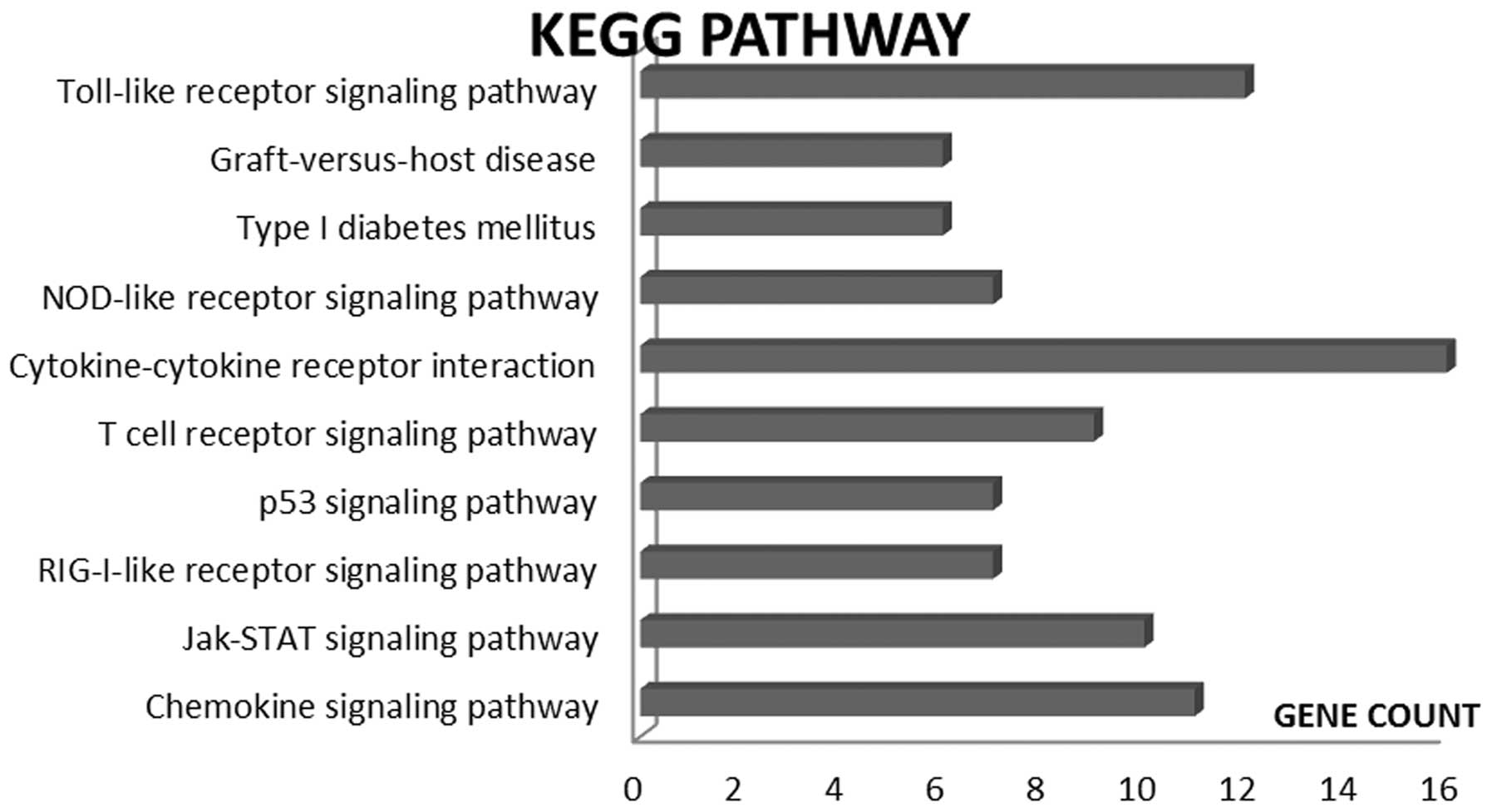
adaptive immunity by the innate immune system. Science.
327:291–295. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hoebe K, Janssen E and Beutler B: The
interface between innate and adaptive immunity. Nat Immunol.
5:971–974. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wijbrandts CA, Vergunst CE, Haringman JJ,
Gerlag DM, Smeets TJ and Tak PP: Absence of changes in the number
of synovial sublining macrophages after ineffective treatment for
rheumatoid arthritis: implications for use of synovial sublining
macrophages as a biomarker. Arthritis Rheum. 56:3869–3871. 2007.
View Article : Google Scholar
|
|
7
|
Firestein GS, Nguyen K, Aupperle KR, Yeo
M, Boyle DL and Zvaifler NJ: Apoptosis in rheumatoid arthritis: p53
overexpression in rheumatoid arthritis synovium. Am J Pathol.
149:2143–2151. 1996.PubMed/NCBI
|
|
8
|
Yamanishi Y, Boyle DL, Pinkoski MJ, et al:
Regulation of joint destruction and inflammation by p53 in
collagen-induced arthritis. Am J Pathol. 160:123–130. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Salvador G, Sanmarti R, Garcia-Peiró A,
Rodríguez-Cros J, Muñoz-Gómez J and Cañete J: p53 expression in
rheumatoid and psoriatic arthritis synovial tissue and association
with joint damage. Ann Rheum Dis. 64:183–187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McGovern DP, Hysi P, Ahmad T, et al:
Association between a complex insertion/deletion polymorphism in
NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum
Mol Genet. 14:1245–1250. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Porcelli S, Yockey CE, Brenner MB and Balk
SP: Analysis of T cell antigen receptor (TCR) expression by human
peripheral blood CD4-8-alpha/beta T cells demonstrates preferential
use of several V beta genes and an invariant TCR alpha chain. J Exp
Med. 178:1–16. 1993. View Article : Google Scholar
|
|
12
|
Sen M: Wnt signalling in rheumatoid
arthritis. Rheumatology (Oxford). 44:708–713. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deon D, Ahmed S, Tai K, et al: Cross-talk
between IL-1 and IL-6 signaling pathways in rheumatoid arthritis
synovial fibroblasts. J Immunol. 167:5395–5403. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feldmann M and Maini RN; Lasker Clinical
Medical Research Award. TNF defined as a therapeutic target for
rheumatoid arthritis and other autoimmune diseases. Nat Med.
9:1245–1250. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nakano K, Whitaker JW, Boyle DL, Wang W
and Firestein GS: DNA methylome signature in rheumatoid arthritis.
Ann Rheum Dis. 72:110–117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee HK, Hsu AK, Sajdak J, Qin J and
Pavlidis P: Coexpression analysis of human genes across many
microarray data sets. Genome Res. 14:1085–1094. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stuart JM, Segal E, Koller D and Kim SK: A
gene-coexpression network for global discovery of conserved genetic
modules. Science. 302:249–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee H-M, Sugino H, Aoki C, et al: Abnormal
networks of immune response-related molecules in bone marrow cells
from patients with rheumatoid arthritis as revealed by DNA
microarray analysis. Arthritis Res Ther. 13:R892011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Irizarry RA, Hobbs B, Collin F, et al:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar
|
|
20
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu H, Liu B-H, Ye ZQ, Li C, Li YX and Li
YY: Link-based quantitative methods to identify differentially
coexpressed genes and gene pairs. BMC Bioinformatics. 12:3152011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu BH, Yu H, Tu K, Li C, Li YX and Li YY:
DCGL: an R package for identifying differentially coexpressed genes
and links from gene expression microarray data. Bioinformatics.
26:2637–2638. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang da W, Sherman BT, Tan Q, Collins JR,
Alvord G, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID Gene Functional Classification Tool: a novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007.
|
|
25
|
Wingender E, Chen X, Hehl R, et al:
TRANSFAC: an integrated system for gene expression regulation.
Nucleic Acids Res. 28:316–319. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
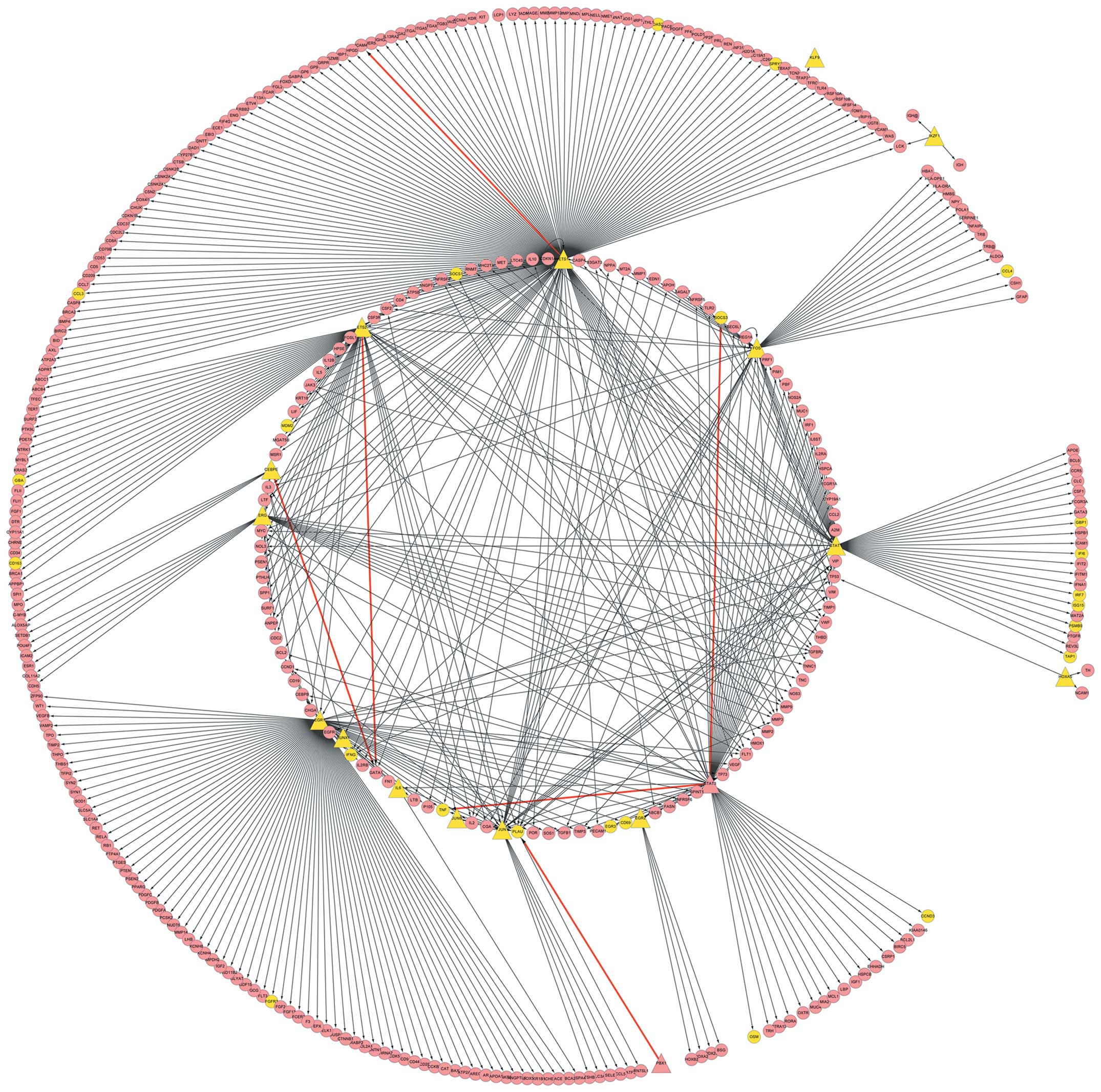
Smoot ME, Ono K, Ruscheinski J, Wang P-L
and Ideker T: Cytoscape 2.8: new features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang ML, Huang ZZ and Zheng Y: Estimates
and prediction on incidence, mortality and prevalence of breast
cancer in China, 2008. Zhonghua Liu Xing Bing Xue Za Zhi.
33:1049–1051. 2012.(In Chinese).
|
|
28
|
Hu F, Mu R, Zhu J, et al: Hypoxia and
hypoxia-inducible factor-1α provoke toll-like receptor
signalling-induced inflammation in rheumatoid arthritis. Ann Rheum
Dis. May 3–2013.(Epub ahead of print).
|
|
29
|
Xue F, Zhang C, He Z, Ding L and Xiao H:
Analysis of critical molecules and signaling pathways in
osteoarthritis and rheumatoid arthritis. Mol Med Rep. 7:603–607.
2013.PubMed/NCBI
|
|
30
|
Plantinga TS, Fransen J, Knevel R, et al:
Role of NOD1 polymorphism in susceptibility and clinical
progression of rheumatoid arthritis. Rheumatology (Oxford).
52:806–814. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Alexander WS: Suppressors of cytokine
signalling (SOCS) in the immune system. Nat Rev Immunol. 2:410–416.
2002.PubMed/NCBI
|
|
32
|
Kubo M, Hanada T and Yoshimura A:
Suppressors of cytokine signaling and immunity. Nat Immunol.
4:1169–1176. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Egan PJ, Lawlor KE, Alexander WS and Wicks
IP: Suppressor of cytokine signaling-1 regulates acute inflammatory
arthritis and T cell activation. J Clin Invest. 111:915–924. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Hooge AS, van de Loo FA, Koenders MI,
et al: Local activation of STAT-1 and STAT-3 in the inflamed
synovium during zymosan-induced arthritis: exacerbation of joint
inflammation in STAT-1 gene-knockout mice. Arthritis Rheum.
50:2014–2023. 2004.PubMed/NCBI
|
|
35
|
Isomäki P, Alanärä T, Isohanni P, et al:
The expression of SOCS is altered in rheumatoid arthritis.
Rheumatology (Oxford). 46:1538–1546. 2007.PubMed/NCBI
|
|
36
|
Nakajima K, Yamanaka Y, Nakae K, et al: A
central role for Stat3 in IL-6-induced regulation of growth and
differentiation in M1 leukemia cells. EMBO J. 15:3651–3658.
1996.PubMed/NCBI
|
|
37
|
Feldmann M, Brennan FM and Maini RN: Role
of cytokines in rheumatoid arthritis. Annu Rev Immunol. 14:397–440.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Elliott MJ, Maini R, Feldmann M, et al:
Randomised double-blind comparison of chimeric monoclonal antibody
to tumour necrosis factor alpha (cA2) versus placebo in rheumatoid
arthritis. Lancet. 344:1105–1110. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Elliott MJ, Maini R, Feldmann M, et al:
Repeated therapy with monoclonal antibody to tumour necrosis factor
alpha (cA2) in patients with rheumatoid arthritis. Lancet.
344:1125–1127. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mihara M, Moriya Y, Kishimoto T and Ohsugi
Y: Interleukin-6 (IL-6) induces the proliferation of synovial
fibroblastic cells in the presence of soluble IL-6 receptor. Br J
Rheumatol. 34:321–325. 1995. View Article : Google Scholar
|
|
41
|
Tamura T, Udagawa N, Takahashi N, et al:
Soluble interleukin-6 receptor triggers osteoclast formation by
interleukin 6. Proc Natl Acad Sci USA. 90:11924–11928. 1993.
View Article : Google Scholar : PubMed/NCBI
|