Introduction
MicroRNAs (miRNAs) are a class of single-stranded
non-coding RNAs that regulate target gene expression, predominantly
by base pairing to the 3′-untranslated (3′-UTR) region of their
target mRNAs (1). miRNAs have been
linked to carcinogenesis due to their apparent proximity to
chromosomal breakpoints and aberrant expression levels in various
malignancies. Furthermore, approximately half of miRNA genes are
located in cancer-correlated genomic regions or fragile sites
(2). A number of studies have
provided evidence that miRNAs are associated with cancer
development and progression, including in gastric (3–5),
lung (6), breast (7,8) and
hepatocellular carcinoma (9).
miRNAs that are upregulated or downregulated in various cancer
types are respectively referred to as oncogenic or tumor-suppressor
miRNAs (10–12).
MiRNAs have been identified to be associated with
the carcinogenesis of human gastric cancer, which ranks as the
fourth most common cancer and the second leading cause of
cancer-associated mortality worldwide (13,14).
miRNAs including miR-150, miR-27a, miR-21, miR-106b and miR-25 are
upregulated in gastric cancer, and as oncogenes, they promote
proliferation and migration of gastric cancer cells (15–18).
Other miRNAs are downregulated in gastric cancer, including
miR-218, miR-9, miR-143 and miR-145, and function as possible
tumor-suppressor genes to inhibit cell proliferation, migration and
metastasis (19–21). However, a number of studies have
identified no further evidence for their causative role in gastric
carcinogenesis. The mechanisms and functions of the majority of
these miRNAs remains to be elucidated.
Recently, we performed miRNA microarray profiling in
primary gastric cancer tissues and their adjacent non-tumor tissues
and identified a number of miRNAs that were dysregulated in gastric
cancer (22). The expression
pattern of 6 miRNAs (miR-663, miR-21, miR-25, miR-106a, miR-106b
and miR-375) in gastric cancer was consistent with other
independent studies (18,23–25).
For example, we identified that downregulated miR-375 in the
majority of gastric cancer tissues was able to suppress gastric
cancer cell proliferation and viability, which is confirmed by the
data from Moriyama et al (25). In this screening, we also provide
evidence that miR-125a-5p, a miRNA with unknown function, appeared
to be one of the most downregulated miRNAs in gastric cancer
tissues. In the present study, it was confirmed that miR-125a-5p
expression was reduced in >80% of gastric cancer tissues and we
demonstrated that ectopic expression of miR-125a-5p inhibited the
proliferation, migration and invasion of gastric cancer cells,
suggesting a tumor-suppressive role of miR-125a-5p in gastric
carcinogenesis. Furthermore, miR-125a-5p targets E2F3 via its
3′-UTR region, indicating that E2F3 may be a potential downstream
target of miR-125a-5p in gastric cancer progression.
Materials and methods
Tissues and cell lines
The clinical characteristics of all 51 patients in
our study are listed in Table I.
Gastric tissues were obtained from 51 gastric cancer patients
undergoing gastric resection at the Sir Run Run Shaw Hospital
(Hangzhou, Zhejiang, China). Written consent was obtained prior to
surgery, as described previously (22). The study was approved by the Ethics
Comittee of Zhejiang University School of Medicine (Hangzhou,
China). The tumor tissues and adjacent non-tumor tissues were
quickly separated into two sections following resection. One was
immediately frozen in liquid nitrogen for RNA isolation and another
was fixed in formalin for pathological examination. Final
pathological diagnosis was independently made by at least two
professional pathologists.
 | Table IClinical and pathological
characteristics of included patient samples and miR-125a-5p
expression. |
Table I
Clinical and pathological
characteristics of included patient samples and miR-125a-5p
expression.
| Expression of
miR-125a-5p | |
|---|
|
| |
|---|
| Characteristic | Upregulated
(n=9) | Downregulated
(n=42) | P-value |
|---|
| Gender |
| Male | 8 | 30 | 0.28 |
| Female | 1 | 12 | |
| Age (years) |
| ≤65 | 4 | 23 | 0.52 |
| >65 | 5 | 19 | |
| Lauren’s
classification |
| Intestinal | 1 | 21 | 0.06 |
| Diffuse | 4 | 7 | |
| Others | 4 | 14 | |
|
Differentiation |
| Well | 1 | 14 | 0.23 |
| Moderate | 2 | 11 | |
| Low | 6 | 17 | |
| Invasion depth |
| T1 | 0 | 4 | 0.26 |
| T2 | 2 | 2 | |
| T3 | 7 | 35 | |
| T4 | 0 | 1 | |
| Lymph node
metastasis |
| Absent | 3 | 10 | 0.55 |
| Present | 6 | 32 | |
| Distant
metastasis |
| M0 | 6 | 39 | 0.03a |
| M1 | 3 | 3 | |
| UICC stage |
| I | 1 | 5 | 0.82 |
| II | 1 | 6 | |
| III | 4 | 23 | |
| IV | 3 | 8 | |
The source and culture conditions of 6 gastric
cancer cell lines (NCI-N87, AGS, MGC-803, HGC-27, SGC-7901 and
BGC-823 cells) and one non-malignant gastric epithelial cell line
(GES-1 cells) were previously described (22,26).
RNA extraction and quantitative real-time
PCR
Total RNA, including low molecular weight RNA from
gastric tissue samples and gastric epithelial cells, were isolated
using the MirVana™ miRNA Isolation kit (Ambion, Foster City, CA,
USA) according to the manufacturer’s instructions. Expression of
miR-125a-5p was analyzed using a TaqMan microRNA assay kit (Applied
Biosystems, Grand Island, NY, USA). Briefly, for each sample, 10 ng
total RNA was reversely transcribed using a TaqMan microRNA reverse
transcript kit (PN: 4366597) and a miR-125a-5p specific reverse
transcript primer (Applied Biosystems, Foster City, CA, USA).
Quantitative real-time PCR was then performed using TaqMan 2X PCR
master mix (Applied Biosystems) on an Applied Biosystems 7500
system. All of the miR-125a-5p threshold cycles (Ct values) were
normalized to snRNA U6 (RNU6B, Applied Biosystems) and an
endogenous control. The normalized values (ΔCt) from gastric cancer
tissues were subsequently compared with that of their adjacent
non-tumor tissues.
Cell proliferation assay
AGS and MGC-803 cells (4,000 cells/well) were
transfected with miR-125a-5p precursor (pre-miR-125a-5p) or a
negative control (Applied Biosystems) in 96-well plates using
siPORT™ NeoFX™ transfection agent (Ambion) following the
manufacturer’s instructions. At 24, 48 and 72 h post-transfection,
cells were imaged by phase contrast microscopy (Olympus IX81,
Olympus Corporation, Tokyo, Japan) and counted manually using a
hemocytometer. MTT assays were performed as described previously
(26). Each group was conducted in
triplicate and all experiments were repeated at least three times
independently.
Transwell migration analysis
AGS and MGC-803 cells (80,000 cells/well) were
transfected with pre-miR-125a-5p or negative control using siPORT
NeoFX transfection agent in 24-well plates, following the
manufacturer’s instructions Following transfection (24 h), the
cells were placed in a serum-free medium for another 12 h,
chemotactic assays were then conducted in 24-well Transwell inserts
with 8 μm pore size (Corning Costar Corp.). The cells (30,000) were
suspended in 100 μl corresponding culture medium without fetal
bovine serum (FBS) and loaded into the top chamber of Transwell
insert for the migration assay. The bottom chamber was placed with
600 μl medium containing 20% FBS. Migration of cells was allowed to
proceed for 12 h at 37°C. The cells that migrated into the bottom
chamber were then fixed, stained with 4′,6-diamidino-2-phenylindole
(DAPI) for 2 min, visualized under phase contrast microscope and
photographed. Total number of migrated cells in nine randomly
selected fields was counted by IPP 6.0 (Image-Pro Plus 6.0)
software (Media Cybernetics, Inc. Rockville, MD, USA). All
experiments were independently repeated at least three times.
Scratch-wound healing assay
AGS and MGC-803 cells were transfected as indicated
and allowed to grow to confluence. The cells were then cultured in
the corresponding medium without serum for 12 h and scratched with
a pipette tip. Wound areas were marked and photographed at 0 and 24
h using a phase-contrast microscope (Olympus IX81, Olympus
Corporation), respectively. The rate of cell migration was
evaluated by photographing and quantifying the migrated distance of
cells moved from the wound edge toward the wound center using IPP
6.0 software (Media Cybernetics, Inc.). All experiments were
repeated three times.
Matrigel invasion assay
For the Matrigel invasion assay, each 24-well insert
with 8 μm pore size was pre-coated with 50 μl (1 μg/μl) Matrigel
(BD Biosciences, Bedford, MA, USA). AGS and MGC-803 cells were
transfected with miR-125a-5p precursor or negative control as
previously described. Following transfection (24 h), 50,000 cells
were plated in 100 μl serum-free medium in the upper
Matrigel-coated chamber. The bottom chamber was placed with 600 μl
medium containing 20% FBS. Following incubation for 12 h at 37°C,
the cells on the upper chamber were removed. Then, the cells in the
bottom chamber were fixed, stained with DAPI for 2 min, visualized
under phase contrast microscope and photographed. The total number
of invasive cells in nine randomly selected fields was counted by
IPP software. All experiments were independently repeated at least
three times.
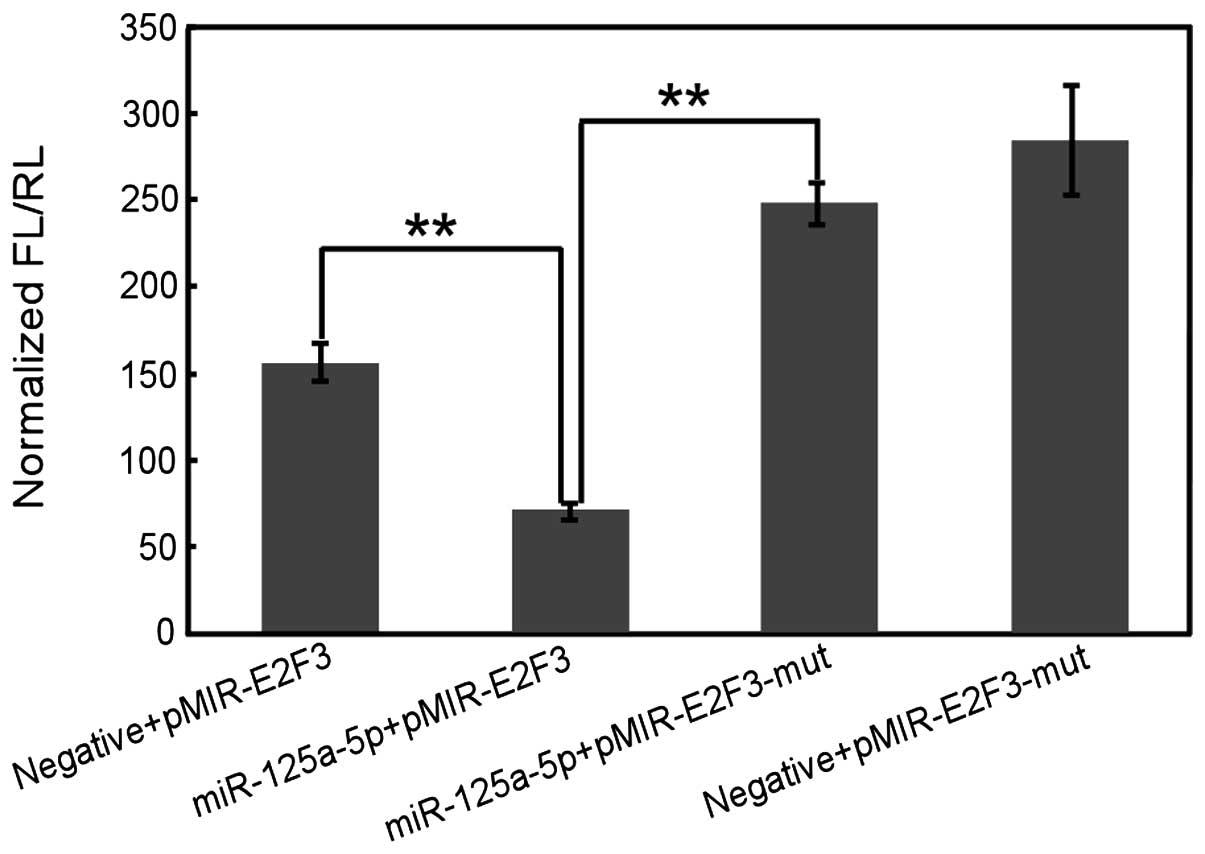
Luciferase reporter assays
The E2F3 3′-UTR, containing the predicted
miR-125a-5p binding site, was amplified and cloned into pMIR-REPORT
vector (pMIR-E2F3) containing Firefly luciferase (Applied
Biosystems). The binding site was also mutant and cloned into
pMIR-REPORT vector (pMIR-E2F3-mut). Cells were cotransfected with
pMIR-E2F3 or pMIR-E2F3-mut together with miR-125a-5p precursor or a
negative control using siPORT amine transfection agent (Ambion).
The pRL-TK vector containing Renilla luciferase was
cotransfected as a reference control. Luciferase was measured by
using dual-luciferase reporter assay (Promega Corporation, Madison,
WI, USA).
Statistical analysis
Data are represented as the mean ± standard error
(SE) of at least three independent experiments. Student’s t-test
was performed to determine statistical significance. P<0.05 was
considered to indicate a statistically significant result.
Results
Downregulation of miR-125a-5p in primary
gastric cancer tissues and cell lines
To investigate the expression profile of miRNAs in
gastric cancer, an miRNA microarray was utilized to analyze five
primary gastric cancer tissues compared with their matched
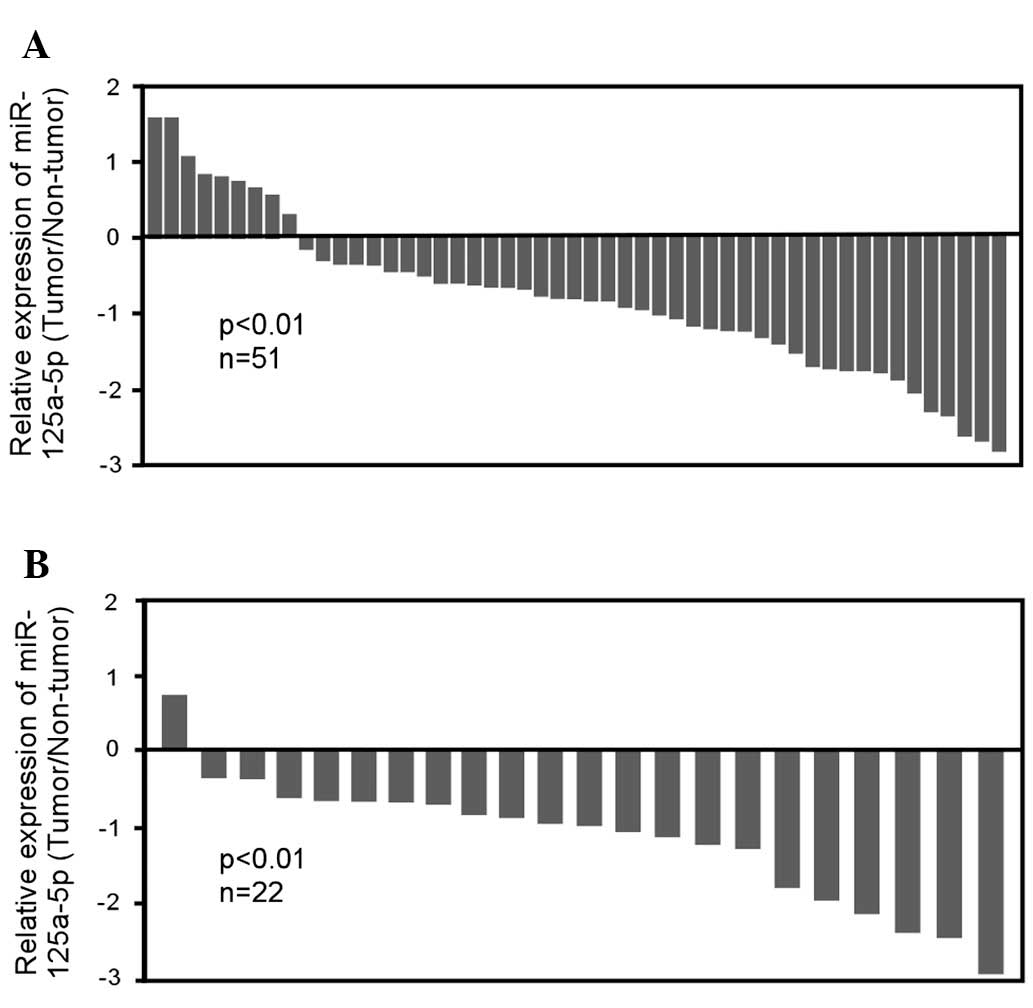
non-tumor tissues. It was identified that miR-125a-5p expression
was markedly reduced in gastric cancer tissue. qRT-PCR analysis
confirmed that miR-125a-5p was significantly downregulated in ~82%
of gastric tumor tissues (42 of 51 patients, P<0.01), with
2.3-fold reduction relative to their adjacent non-tumor tissues
(Fig. 1A). Of note, the reduction
of miR-125a-5p expression was demonstrated in 95.5% of
intestinal-type gastric cancer (21 of 22 patients, P<0.01)
relative to their matched non-tumor tissues (Fig. 1B), whereas there was no significant
change in miR-125a-5p expression in diffused-type and
undetermined-type gastric cancer (data not shown). To further
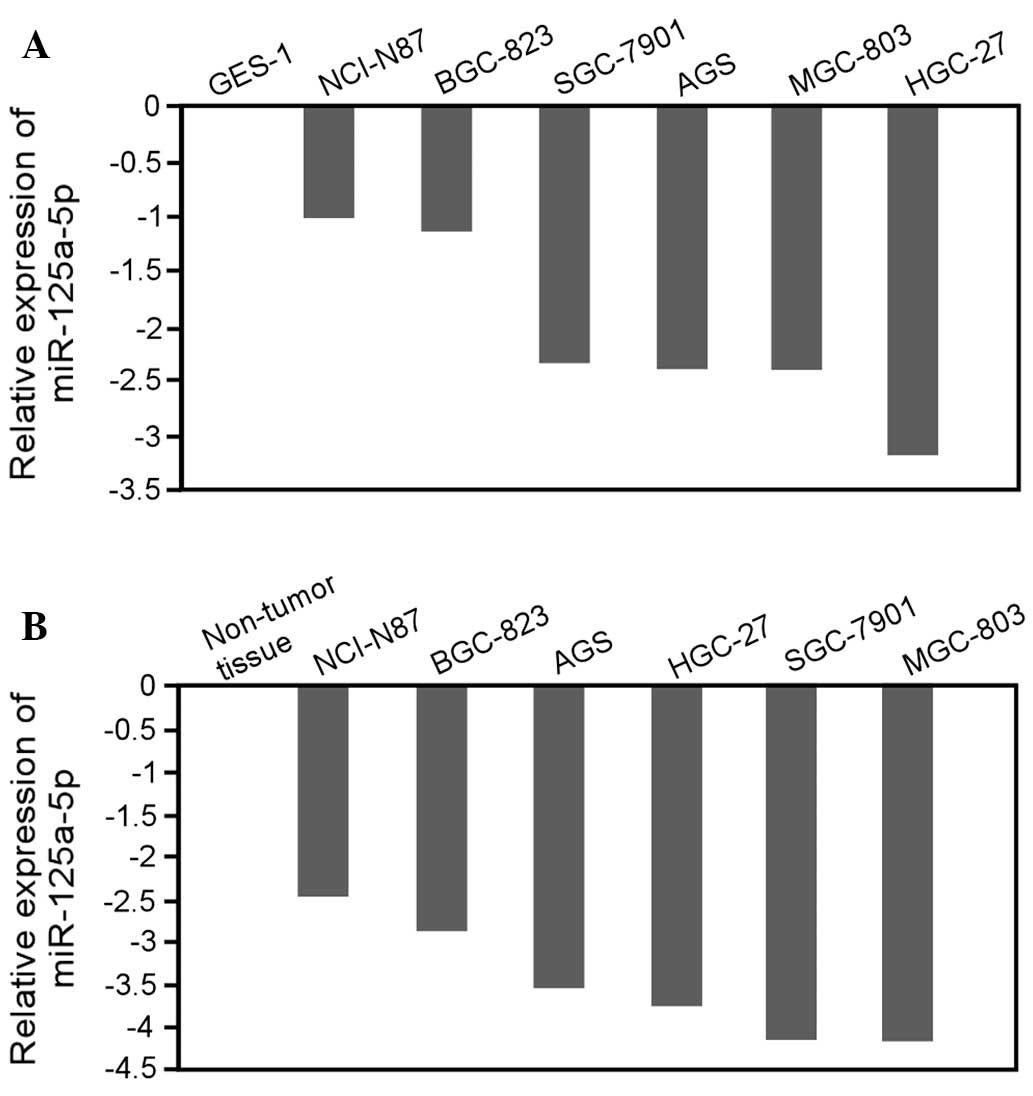
evaluate the correlation between miR-125a-5p expression and gastric
cancer, we detected the expression of miR-125a-5p in six gastric
cancer cell lines (AGS, BGC-823, HGC-27, MGC-803, NCI-N87 and
SGC-7901 cells). The data revealed that the expression level of
miR-125a-5p was significantly decreased in all cell lines derived
from gastric cancers with various differentiation degrees, compared
with the non-malignant gastric cell line GES-1 (Fig. 2A) or non-tumor stomach tissue
(Fig. 2B). Taken together, these
results indicate that miR-125a-5p is frequently downregulated in
gastric cancer and therefore may be associated with gastric cancer
progression.
Reduction of miR-125a-5p expression in
gastric cancer is associated with distant metastasis
The correlation between the expression level of
miR-125a-5p and the clinicopathological characteristics of gastric
cancer are listed in Table I. A
statistically significant association between miR-125a-5p
expression level and distant metastasis was observed (P<0.05).
However, no significant correlation was observed between
miR-125a-5p expression level and gender, age, Lauren’s
classification, differentiation, node metastasis, invasion depth or
stage. More clinical gastric cancer samples were required to
further evaluate the correlation between miR-125a-5p expression and
gastric cancer clinicopathological characteristics.
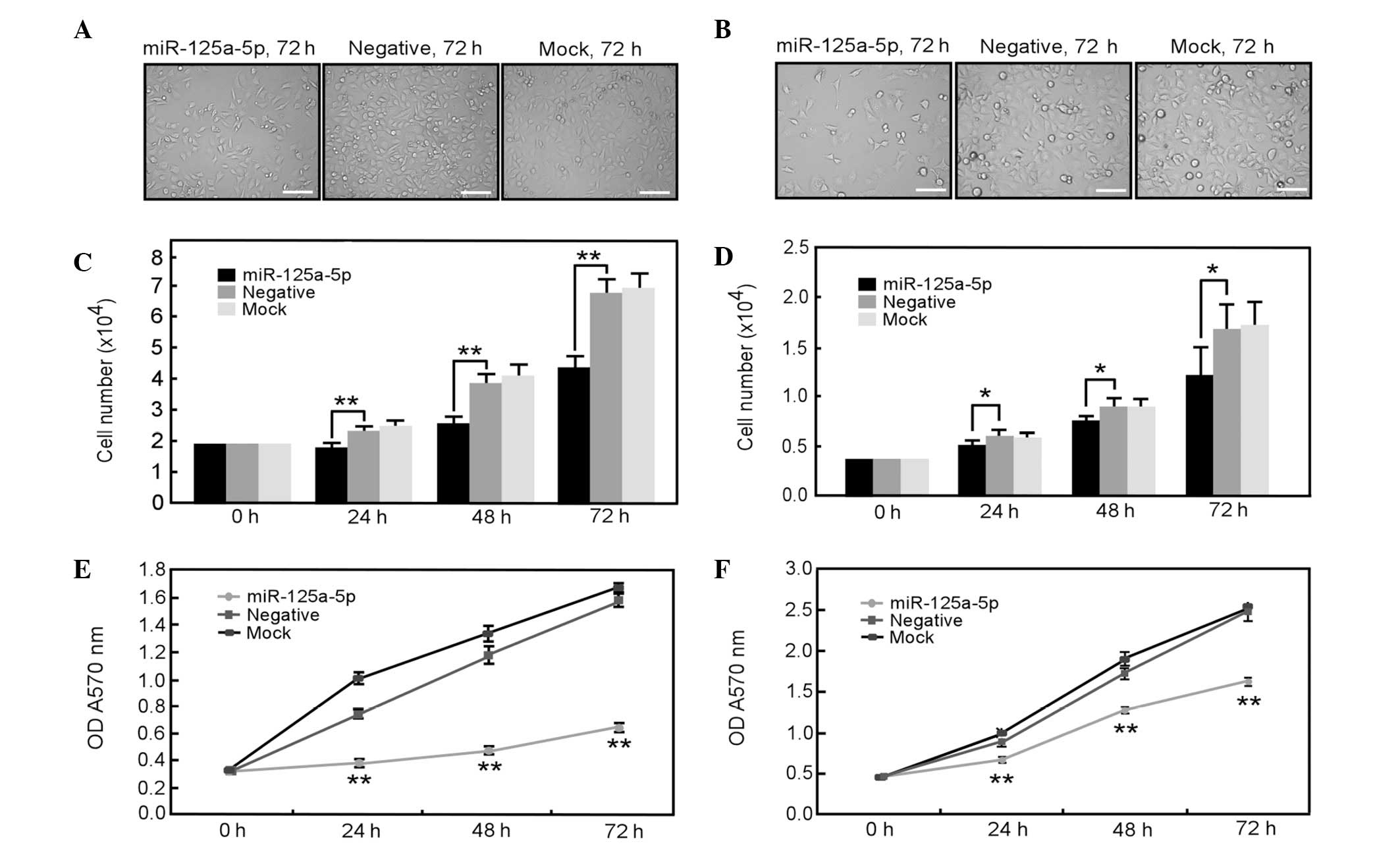
Overexpression of miR-125a-5p inhibits
gastric cancer cell proliferation
To examine the potential role of miR-125a-5p in
malignant phenotypes of gastric cancer cells, first, the effect of
miR-125a-5p overexpression on the proliferation of AGS and MGC-803
gastric cancer cells with low endogenous expression of miR-125a-5p
was investigated. All of the results from phase contrast
microscopy, direct cell count and MTT assays clearly demonstrated
that overexpression of miR-125a-5p significantly inhibited the
proliferation of AGS and MGC-803 cells (Fig. 3). Therefore, these results indicate
that miR-125a-5p may have an important function in the
proliferation of gastric cancer cells.
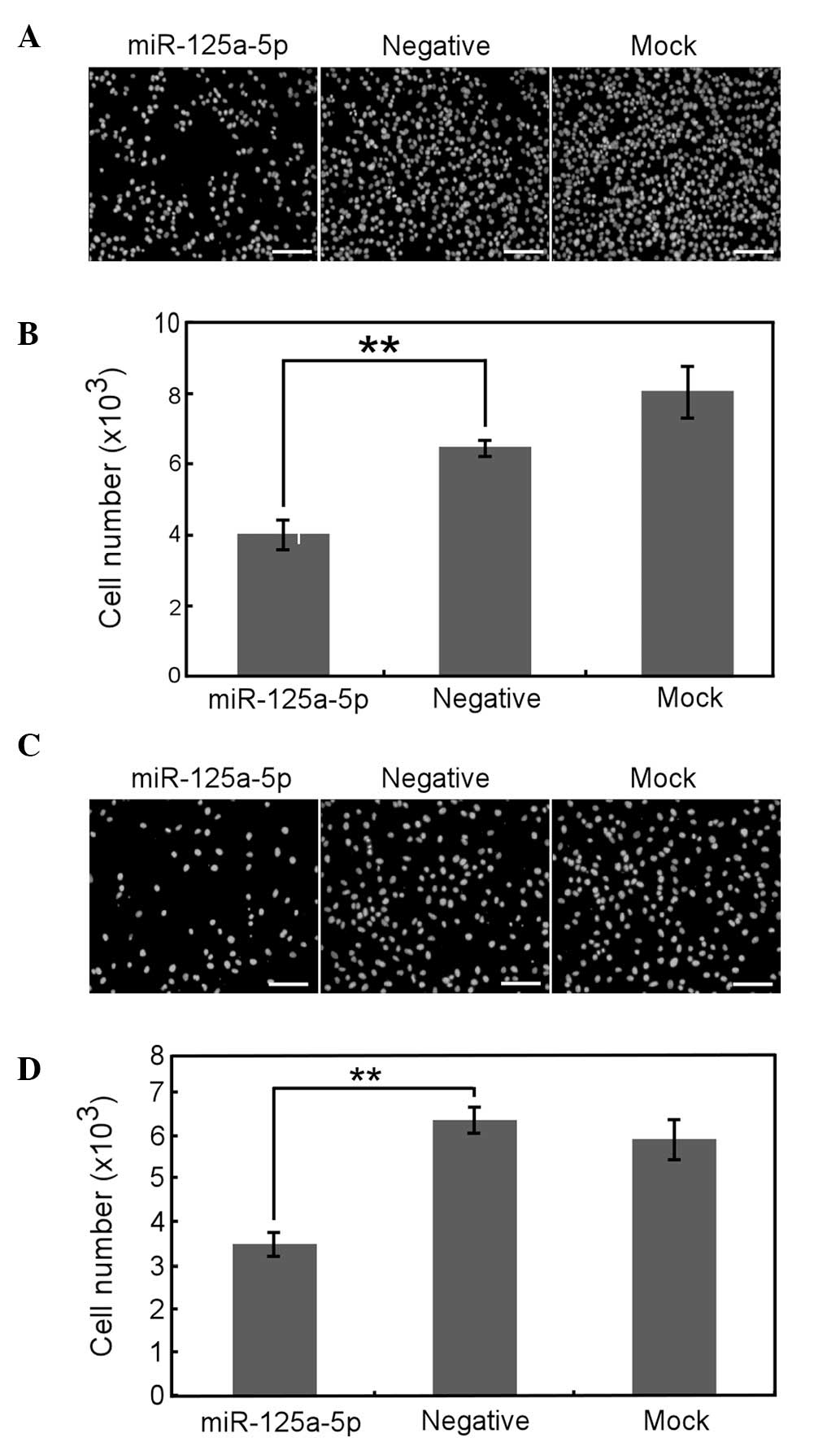
Overexpression of miR-125a-5p inhibits
gastric cancer cell migration
The ability of cell migration and invasion are
associated with cancer metastasis, so the role of miR-125a-5p in
the migration and invasion of gastric cancer cells was
investigated. The Transwell experiment demonstrated that
miR-125a-5p overexpression greatly inhibited the migration of AGS
(Fig. 4A and B) and MGC-803 cells
(Fig. 4E and F). Furthermore,
scratch wound healing assays revealed that the velocity of AGS
(Fig. 4C and D) and MGC-803
(Fig. 4G and H) cell migration
toward the wound area was also significantly reduced by
overexpression of miR-125a-5p. Therefore, these data imply that
miR-125a-5p may be important in gastric cancer cell migration.
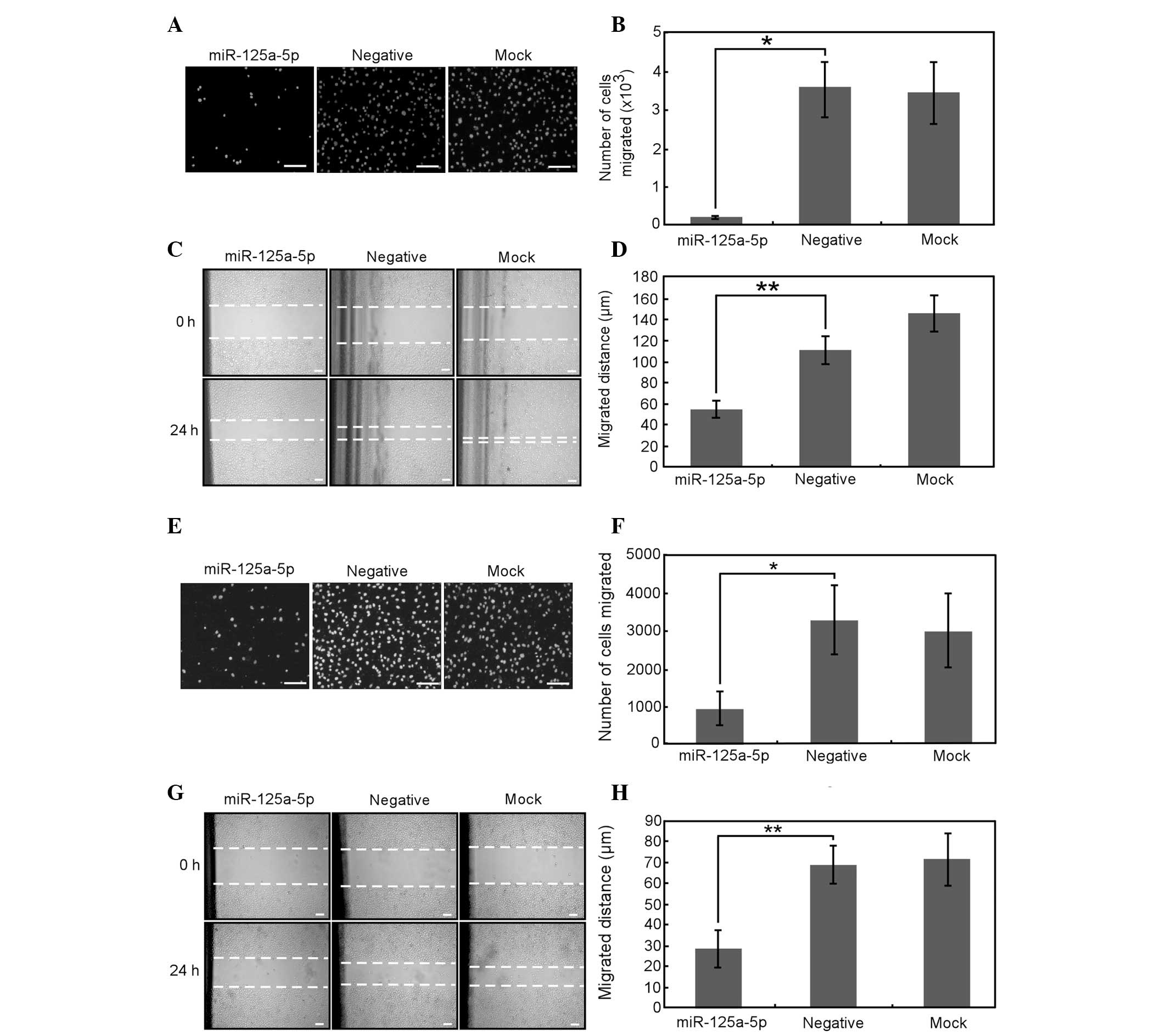 | Figure 4Overexpression of miR-125a-5p inhibits
the migration of AGS and MGC-803 cells. (A–D) AGS and (E–H) MGC-803
cells transfected with miR-125a-5p precursor, negative control or
neither of the above for 24 h were subjected to (A, B, E and F)
Transwell and (C, D, G and H) scratch-wound healing analysis. (A
and E) The cells that migrated to the bottom chamber were fixed and
stained with DAPI. (B and F) Total number of migrated cells from
nine randomly chosen fields was counted by IPP 6.0 software. (C and
G) The cell migration to the wounded area was photographed by
microscopy at 0 and 24 h post-wounding. The dotted lines indicate
the areas lacking cells. (D and H) The rate of migration was
examined by measuring the distance of cells moved from the wound
edge toward the center following scratching. Data are presented as
the mean ± SE from at least three independent experiments. Bars, 50
μm. *P<0.05 and **P<0.01. cp,[ared woth
the negative control. DAPI, 4′,6-diamidino-2-phenylindole; SE,
standard error. |
Overexpression of miR-125a-5p inhibits
gastric cancer cell invasion
Cell invasion into the surrounding tissue is a
crucial, early step in gastric cancer development. Therefore, to
determine whether miR-125a-5p affects the invasion ability of
gastric cancer cells, we employed a Matrigel invasion assay. It was
identified that miR-125a-5p overexpression significantly inhibited
the invasion ability of AGS and MGC-803 cells (Fig. 5). These results indicate that
downregulation of miR-125a-5p in gastric cancer may be sufficient
to promote gastric cancer invasion. In the two cell lines, ectopic
expression of miR-125a-5p did result in a 2- to 4-fold decrease in
cell migration or invasiveness.
miR-125a-5p regulates E2F3 through
3′-UTR
To assess the mechanism of miR-125a-5p function in
gastric cancer cell proliferation, migration and invasion, by using
prediction tools including miRanda (mirSVR; http://www.microrna.org/microrna/home.do), TargetScan
and Pictar algorithms, the potential targets of miR-125a-5p were
screened. Among the hundreds of targets that were predicted, E2F3
was further studied as a potential target. E2F3 is regarded as an
oncogene because it is amplified in various human tumor types. To
obtain direct evidence that E2F3 is a potential target of
miR-125a-5p, we examined whether the predicted binding sites of
miR-125a-5p in the 3′-UTR of E2F3 mRNA were responsible for its
regulation. The 3′-UTR of E2F3 downstream of a luciferase reporter
gene (pMIR-E2F3) was cloned, and this vector was co-transfected
with an miR-125a-5p precursor or its negative control into gastric
cancer cells. The luciferase activity of cells transfected with
miR-125a-5p precursor was significantly reduced compared with the
negative control (P<0.01; Fig.
6). Furthermore, mutation of the putative binding site clearly
abrogated the repression of luciferase activity caused by
miR-125a-5p overexpression. These data suggest that miR-125a-5p may
inhibit E2F3 expression through 3′-UTR at the post-transcriptional
level. Further investigations are required to examine the
association between the expression of miR-125a-5p and E2F3 in
gastric cancer tissues.
Discussion
Recent studies have established the presence of
miRNA expression signatures in gastric cancer, but our
understanding of the function of aberrant miRNAs in gastric
carcinoma progression remains in its infancy. Our presented data
demonstrate a potential role for miR-125a-5p in gastric cancer. In
our previous study, we aimed to examine the miRNA expression
profile in gastric cancer by using miRNA microarray techniques, and
it was identified that miR-125a-5p is one of the most downregulated
miRNAs in gastric cancer. In the present study, it was revealed
that miR-125a-5p was significantly downregulated in >80% of
gastric cancer samples compared with their adjacent non-tumor
tissues obtained from patients undergoing gastric resection, which
was further confirmed by the data from gastric cancer cell lines.
Restoration of miR-125a-5p expression substantially inhibited
proliferation, migration and invasion activities of gastric cancer
cells. These results suggest a potential role of miR-125a-5p in
gastric carcinogenesis.
miR-125a-5p has been identified to be downregulated
in a number of malignancies, including lung, head and neck, gastric
and esophageal and hepatocellular carcinogenesis (27–31),
which are consistent with the results of our study. In vitro
assays revealed that HER2 is a direct target of miR-125a-5p, which
potently suppresses the proliferation of gastric cancer cells
(28,31). Whereas, limited data is available
identifying the targets of miR-125a-5p involved in gastric cancer
cells migration and invasion.
As part of our investigations into how the
downregulation of miR-125a-5p expression affects gastric cancer
progression, a number of the prediction algorithms were employed,
including PicTar, TargetScan and miRanda (32–34),
to search putative targets of miR-125a-5p. Since the overexpression
of miR-125a-5p inhibits cell proliferation, migration and invasion,
it is possible to expect that the target genes of miR-125a-5p may
have oncogenic characteristics. Among the hundreds of targets that
were predicted, E2F3 is regarded as an oncogene because it is
amplified in various human tumor types, including lung, bladder,
prostate, colon and breast cancer, and is involved in cancer cells
apoptosis and proliferation (35–39).
Furthermore, E2F3 amplification and overexpression is demonstrated
to be associated with cell migration and invasion with
comprehensive mechanisms (40–42).
For example, the data of Oeggerli’s et al study concluded
that E2F3 was frequently amplified and overexpressed in invasively
growing bladder cancer, which indicated an important role of E2F3
in cancer progression (42). In
the present study, using the Luciferase assay, direct evidence was
obtained that E2F3 is potentially a direct target of miR-125a-5p.
These data demonstrated that miR-125a-5p significantly inhibited
the luciferase activity of pMIR-E2F3 which contained the 3′-UTR of
E2F3 but had no effect on pMIR-E2F3-mut, which accommodated the
mutant 3′-UTR of E2F3. Together, it was hypothesized that the
downregulation of miR-125a-5p results in overexpression of E2F3
which subsequently contributes to gastric cancer progression.
Additionally, these data reveal that miR-125a-5p is
significantly downregulated in the majority of the intestinal-type
gastric cancer samples (21 of 22 patients; P<0.05), implying a
highly possible association between the expression level of
miR-125a-5p and the Lauren’s classification of gastric cancer. A
statistically significant association between miR-125a-5p
expression level and distant metastasis was also observed
(P<0.05). However, no significant correlation was observed
between the expression level of miR-125a-5p and gender, age,
Lauren’s classification, differentiation, node metastasis, invasion
depth or stage. To confirm the association between miR-125a-5p
expression and the clinicopathological characteristics of gastric
cancer, future studies should employ a larger sample, to further
analyze the association of miR-125a-5p expression and the clinical
outcomes of gastric cancer.
The identification of the expression levels and
tumor-suppressive function of miR-125a-5p in gastric cancer types,
provides a new window of therapeutic opportunity. The development
of modified miRNAs with longer half-time and higher efficiency has
produced favorable anticancer outcomes in experimental models,
including the locked nucleic acid-modified oligonucleotides and the
antisense oligonucleotides termed ‘antagomirs’ (43–45).
Therefore, enforced expression of miR-125a-5p utilizing approaches
such as transfection of miR-125a-carrying viruses or synthetic
miR-125a-5p oligonucleotides, will be required for future study in
gastric carcinoma pathology.
Acknowledgements
This study was supported by Natural Scientific
Foundation of China (nos. 81402429, 30771107 and 30901714), and the
Natural Scientific Foundation of Zhejiang Province, China (nos.
LQ14H160003, Z2100247 and Y2100106).
References
|
1
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar
|
|
3
|
Li X, Zhang Y, Zhang Y, Ding J, Wu K and
Fan D: Survival prediction of gastric cancer by a seven-microRNA
signature. Gut. 59:579–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kong KL, Kwong DL, Chan TH, et al:
MicroRNA-375 inhibits tumour growth and metastasis in oesophageal
squamous cell carcinoma through repressing insulin-like growth
factor 1 receptor. Gut. 61:33–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hsu KW, Wang AM, Ping YH, et al:
Down-regulation of tumor suppressor MBP-1 by microRNA-363 in
gastric carcinogenesis. Carcinogenesis. Oct 4–2013.(Epub ahead of
print).
|
|
6
|
Peng Y, Dai Y, Hitchcock C, et al: Insulin
growth factor signaling is regulated by microRNA-486, an
underexpressed microRNA in lung cancer. Proc Natl Acad Sci USA.
110:15043–15048. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang N, Wang X, Huo Q, et al:
MicroRNA-30a suppresses breast tumor growth and metastasis by
targeting metadherin. Oncogene. Jul 15–2013.(Epub ahead of
print).
|
|
8
|
Volinia S, Galasso M, Sana ME, et al:
Breast cancer signatures for invasiveness and prognosis defined by
deep sequencing of microRNA. Proc Natl Acad Sci USA. 109:3024–3029.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li D, Liu X, Lin L, et al: MicroRNA-99a
inhibits hepatocellular carcinoma growth and correlates with
prognosis of patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Osada H and Takahashi T: let-7 and
miR-17–92: small-sized major players in lung cancer development.
Cancer Sci. 102:9–17. 2011.
|
|
11
|
Aqeilan RI, Calin GA and Croce CM: miR-15a
and miR-16-1 in cancer: discovery, function and future
perspectives. Cell Death Differ. 17:215–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wiklund ED, Bramsen JB, Hulf T, et al:
Coordinated epigenetic repression of the miR-200 family and miR-205
in invasive bladder cancer. Int J Cancer. 128:1327–1334. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
14
|
Hohenberger P and Gretschel S: Gastric
cancer. Lancet. 362:305–315. 2003. View Article : Google Scholar
|
|
15
|
Wu Q, Jin H, Yang Z, et al: MiR-150
promotes gastric cancer proliferation by negatively regulating the
pro-apoptotic gene EGR2. Biochem Biophys Res Commun. 392:340–345.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu T, Tang H, Lang Y, Liu M and Li X:
MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by
targeting prohibitin. Cancer Lett. 273:233–242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chan SH, Wu CW, Li AF, Chi CW and Lin WC:
miR-21 microRNA expression in human gastric carcinomas and its
clinical association. Anticancer Res. 28:907–911. 2008.PubMed/NCBI
|
|
18
|
Kim YK, Yu J, Han TS, et al: Functional
links between clustered microRNAs: suppression of cell-cycle
inhibitors by microRNA clusters in gastric cancer. Nucleic Acids
Res. 37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao C, Zhang Z, Liu W, Xiao S, Gu W and Lu
H: Reduced microRNA-218 expression is associated with high nuclear
factor kappa B activation in gastric cancer. Cancer. 116:41–49.
2010.PubMed/NCBI
|
|
20
|
Luo H, Zhang H, Zhang Z, et al:
Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp
Clin Cancer Res. 28:822009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Takagi T, Iio A, Nakagawa Y, Naoe T,
Tanigawa N and Akao Y: Decreased expression of microRNA-143 and
-145 in human gastric cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ding L, Xu Y, Zhang W, et al: MiR-375
frequently downregulated in gastric cancer inhibits cell
proliferation by targeting JAK2. Cell Res. 20:784–793. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pan J, Hu H, Zhou Z, et al:
Tumor-suppressive mir-663 gene induces mitotic catastrophe growth
arrest in human gastric cancer cells. Oncol Rep. 24:105–112.
2010.PubMed/NCBI
|
|
24
|
Zhang Z, Li Z, Gao C, et al: miR-21 plays
a pivotal role in gastric cancer pathogenesis and progression. Lab
Invest. 88:1358–1366. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsukamoto Y, Nakada C, Noguchi T, et al:
MicroRNA-375 is downregulated in gastric carcinomas and regulates
cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res.
70:2339–2349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Du Y, Xu Y, Ding L, et al: Down-regulation
of miR-141 in gastric cancer and its involvement in cell growth. J
Gastroenterol. 44:556–561. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang L, Huang Q, Chang J, Wang E and Qiu
X: MicroRNA HSA-miR-125a-5p induces apoptosis by activating p53 in
lung cancer cells. Exp Lung Res. 37:387–398. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nishida N, Mimori K, Fabbri M, et al:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar
|
|
29
|
Odar K, Boštjančič E, Gale N, Glavač D and
Zidar N: Differential expression of microRNAs miR-21, miR-31,
miR-203, miR-125a-5p and miR-125b and proteins PTEN and p63 in
verrucous carcinoma of the head and neck. Histopathology.
61:257–265. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim JK, Noh JH, Jung KH, et al: Sirtuin7
oncogenic potential in human hepatocellular carcinoma and its
regulation by the tumor suppressors MiR-125a-5p and MiR-125b.
Hepatology. 57:1055–1067. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fassan M, Pizzi M, Realdon S, et al: The
HER2-miR125a5p/miR125b loop in gastric and esophageal
carcinogenesis. Hum Pathol. 44:1804–1810. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Krek A, Grun D, Poy MN, et al:
Combinatorial microRNA target predictions. Nat Genet. 37:495–500.
2005. View
Article : Google Scholar
|
|
33
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar
|
|
35
|
Cooper CS, Nicholson AG, Foster C, et al:
Nuclear overexpression of the E2F3 transcription factor in human
lung cancer. Lung Cancer. 54:155–162. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Feber A, Clark J, Goodwin G, et al:
Amplification and overexpression of E2F3 in human bladder cancer.
Oncogene. 23:1627–1630. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Foster CS, Falconer A, Dodson AR, et al:
Transcription factor E2F3 overexpressed in prostate cancer
independently predicts clinical outcome. Oncogene. 23:5871–5879.
2004. View Article : Google Scholar
|
|
38
|
Fang Y, Gu X, Li Z, Xiang J and Chen Z:
miR-449b inhibits the proliferation of SW1116 colon cancer stem
cells through downregulation of CCND1 and E2F3 expression. Oncol
Rep. 30:399–406. 2013.PubMed/NCBI
|
|
39
|
Vimala K, Sundarraj S, Sujitha MV and
Kannan S: Curtailing overexpression of E2F3 in breast cancer using
siRNA (E2F3)-based gene silencing. Arch Med Res. 43:415–422. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
McClellan KA, Ruzhynsky VA, Douda DN, et
al: Unique requirement for Rb/E2F3 in neuronal migration: evidence
for cell cycle-independent functions. Mol Cell Biol. 27:4825–4843.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ziebold U, Reza T, Caron A and Lees JA:
E2F3 contributes both to the inappropriate proliferation and to the
apoptosis arising in Rb mutant embryos. Genes Dev. 15:386–391.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Oeggerli M, Tomovska S, Schraml P, et al:
E2F3 amplification and overexpression is associated with invasive
tumor growth and rapid tumor cell proliferation in urinary bladder
cancer. Oncogene. 23:5616–5623. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ørom UA, Kauppinen S and Lund AH:
LNA-modified oligonucleotides mediate specific inhibition of
microRNA function. Gene. 372:137–141. 2006.PubMed/NCBI
|
|
44
|
Surdziel E, Eder M and Scherr M:
Lentivirus-mediated antagomir expression. Methods Mol Biol.
667:237–248. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Velu CS and Grimes HL: Utilizing antagomiR
(antisense microRNA) to knock down microRNA in murine bone marrow
cells. Methods Mol Biol. 928:185–195. 2012.PubMed/NCBI
|