Introduction
The post-translational β-O-linkage of
N-acetylglucosamine (O-GlcNAc) to cellular proteins represents one
signaling pathway, which has been implicated in the pathophysiology
of cardiovascular disease (1).
O-GlcNAc transferase (OGT), which adds O-GlcNAc to proteins, has
been demonstrated to be required for cell division and
embryogenesis (2). Cardiomyocytes
ablation of OGT induces heart failure and OGT is required as part
of the endogenous compensatory response to infarct-induced heart
failure (3). In developed
countries, the single most prevalent cause of mortality and
morbidity is congestive heart failure (CHF) (4). CHF accounts for ~5,000,000
hospitalizations annually in developed nations, which results in
the treatment of CHF being a major expenditure in the health care
budgets of all developed nations, and is also becoming an
increasing expense for those of numerous developed nations. CHF is
known to occur as a common result of a wide range of etiologies;
however, the propensity to develop CHF varies significantly
following a given insult (4,5).
This is considered to be due to responses, including inherited
variation in myocardial and vascular responses. However, the
factors underlying the ultimate development of CHF remain to be
fully elucidated. The clinical management of CHF is enabled by the
identification of circulating biomarkers, including brain
natriuretic peptide (BNP) (6,7).
However, there remains a requirement for the identification of
novel circulating biomarkers as objective indicators of CHF, which
can be readily and reliably measured.
MicroRNAs (miRNAs) were first identified in mammals
as a large class of evolutionarily conserved small, non-coding
RNAs. These are involved in the regulation of gene expression at
the post-transcriptional level through targeted interactions with
the 3′-untranslated region (UTR) of messenger RNA (mRNA)
transcripts (8). miRNAs are
important regulators of a wide range of biological processes by
affecting protein translation (8).
In addition, miRNAs can be efficiently suppressed for prolonged
periods using antisense technologies, which has been of increasing
interest due to its potential therapeutic use in cardiovascular
diseases through the targeted inhibition of specific miRNAs
(9). A study in 2008 revealed that
miRNAs were also present in blood, where they were detected in
plasma, platelets, erythrocytes and nucleated blood cells. Plasma
miRNAs were found to remain stable, even under conditions of
stress, including boiling, low or high pH, long-term storage at
room temperature and multiple freeze-thaw cycles (10–12).
These characteristics suggest that miRNAs may be a suitable
biomarker for disease diagnosis.
In previous years, miR-423-5p has been investigated
widely as a potential biomarker for heart diseases. Array
investigations have reported that miR-423-5p is upregulated in
human failing myocardium (13).
Tijsen et al (14)
demonstrated that circulating levels of miR-423-5p are increased in
patients with clinical heart failure, defined by the Framingham
criteria and elevated pro-brain (pro)BNP levels. This indicated
that the levels of miR-423-5p levels were associated with proBNP
and ejection fraction in this patient group (14). However, the association between
miR-423-5p and CHF and the underlying mechanism remain to be
elucidated. Therefore, elucidating the direct target of miR-423-5p
and the associated pathways in cardiomyocytes is essential for
understanding the association between miR-423-5p and heart disease,
and to determine why miR-423-5p fails as a biomarker for systemic
ventricular and left ventricular remodeling (15,16).
The present study aimed to examine the role of
miR-423-5p in the development of CHF through investigating the
expression of miR-423-5p in the plasma of patients with CHF and
healthy volunteers. The expression levels of miR-423-5p, proBNP,
OGT and the downstream targets of OGT, and apoptosis in the
cardiomyocytes were examined.
Materials and methods
Clinical study
The present study was approved by the ethics
committee of Sichuan Academy of Medical Sciences & Sichuan
Provinicial People’s Hospital (Chengdu, China). Human plasma
samples were obtained with informed consent under a general waiver
by the Academic Medical Center institutional review board for the
proper secondary use of human material. Plasma samples were
obtained from the patients and healthy volunteers at the People’s
Hospital of Sichuan Province and experiments were performed on
samples at the Sichuan Academy of Medical Science. Written informed
consent was obtained from the patients and volunteers.
Definition of CHF diagnosis
The subjects were classified as CHF cases in
accordance with the Framingham criteria for CHF diagnosis (17) and if they had circulating proBNP
levels of >1,000 pg/ml. The subjects were classified as non-CHF
cases if the clinical diagnosis excluded CHF and if circulating
proBNP levels were below the age-associated cut-off points. In
total, 51 of the 74 subjects screened for CHF fulfilled the
criteria.
Cell culture and transfection
Mouse cardiomyocytes were obtained from Shanghai
Cell Bank (Shanghai, China). The cardiomyocytes were cultured in
Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal
bovine serum (FBS; Gibco-BRL, Carlsbad, CA, USA). The cells were
maintained in a humidified atmosphere containing 5% CO2
at 37°C. Cell transfection was performed using FuGENE®
HD Transfection Reagent (Roche Diagnostics, Indianapolis, IN, USA),
according to the manufacturer’s instructions. Briefly, the
cardiomyocytes were seeded into six-well plates at a density of
2×105 cells/well and cultured for 24 h to reach 70–80%
confluence. Plasmids (2 μg), encoding mouse (m)-miR-423-5p
and miR-Ctrl, were diluted in 100 μl media without serum.
Subsequently, 5 μl FuGENE® HD Transfection
Reagent was added to tubes containing the diluted DNA, mixed,
incubated with the transfection complex for 15 min at room
temperature and then added to the six-well plates. A total of 24 or
48 h post-transfection the cells were collected for total mRNA and
protein extraction. miR-Ctrl was transfected for 48 h. Medium (DMEM
containing 10% FBS) alone was used as the blank agent.
Cardiomyoctes was placed in a six-well plate at a density of
2×105 cells/well. After 24 h, puromycin (Beyotime
Biotechnology, Shanghai, China) was used to treat cardiomyoctes at
0, 5, 10, 20 or 40 μM. The cell vitality was observed in the
following 48–72 h The terminal concentration was found to be 20
μM. Consequently, after plasmid transfection for 48 h, 20
μM puromycin was added to treat cardiomyoctes. The surviving
cells were selected as the stable cells which expressed the
plasmid.
Bioinformatics analysis
To analyze the potential targets of miR-423-5p,
online miRNA databases were used. TargetScan (http://www.targetscan.org/) and microRNA.org (http://www.microrna.org/microrna/getGeneForm.do) were
used to predict the potential targets of miR-423-5p, and O-GlcNAc
was identified as a target. Further study was performed to
determine whether O-GlcNAc is the direct target of miR-423-5p.
Luciferase assays
The miR-423-5p binding site was synthesized and
cloned into an Ambion pMIR-REPORT vector (Ambion Life Technologies,
Carlsbad, CA, USA) to generate pMiRluc-423-5p. Briefly, each miRNA
and plasmid oligonucleotide was annealed at 90°C for 3 min, cooled
to 37°C, and incubated for 1 h. The annealed dsDNA oligonucleotides
were ligated between HindIII and SacI (Fermentas,
Thermo Scientific, Waltham, MA, USA) sites on the pMIR-REPORT
vector. The resulting recombinant plasmids were named
pMIR-miR-423-5p and pMIR-miR-Ctrl. The two constructs were verified
by DNA sequencing (Invitrogen Life Technologies, Carlsbad, CA,
USA). Plasmids were extracted using EndoFree Plasmid Giga kits
(Qiagen GmbH, Hilden, Germany) from DH5α (Genewiz, Suzhou, China)
Escherichia coli transformants and stored at −20°C until further
use. The concentration was determined by measuring the A260/A280
ratio using a Thermo ND 2000 spectrophotometer (Thermo Scientific).
The 3′-UTRs of O-GlcNAc transferase (OGT) containing miR-423-5p
binding sites were amplified and cloned into the same vector to
generate pMiRluc-OGT. The reporter was co-transfected with a
cytomegalovirus β-galactosidase vector using FuGENE HD® (Roche
Diagnostics). Luciferase activity was measured 4 h later using a
Luciferase Reporter assay (Promega Corporation, Madison, WI, USA).
Values were normalized to the activity of β-galactosidase.
26S proteasome activity assay
The function of the 26S proteasome was assayed, as
described previously (17).
Briefly, the cells were seeded at a density of 2×105
cells/well and cultured for 24 h, until they had reached 70–80%
confluence. The cells were then washed with phosphate-buffered
saline (ZsBio, Beijing, China), and then with buffer I, containing
I50 mM Tris (pH 7.4), 2 mM dithiothreitol, 5 mM MgCl2
and 2 mM adenosine triphosphate (ATP)], and pelleted by
centrifugation (Mini Centrifuge #166-0623; Bio-Rad Laboratories,
Inc., Hercules, CA, USA) at 200 × g for 5 min. Glass beads
(Beyotime Biotechnology) and homogenization buffer, containing 50
mM Tris (pH 7.4), 1 mM dithiothreitol, 5 mM MgCl2, 2 mM
ATP and 250 mM sucrose, were added and vortexed (MixPlus; Aibenseng
Bio, Hefei, China) for 1 min. The beads and cell debris were
removed by centrifugation at 1,000 × g for 5 min and then at 10,000
× g for 20 min. The protein concentrations were determined using a
bicinchoninic acid assay (Pierce Biotechnology, Inc., Rockford, IL,
USA), according to the manufacturer’s instructions. The protein
from each sample (100 μg) was diluted with buffer I to a
final volume of 1,000 μl, and
Suc-LLVY-7-amido-4-methylcoumarin fluorogenic proteasome substrate
(chymotrypsin-like; Sigma-Aldrich, St., Louis, MO, USA) was added
to a final concentration of 80 μM in 1% dimethyl sulfoxide
(Sigma-Aldrich). In order to access the 20S function, the buffer I
was replaced with an ATP-free buffer containing 20 mM HEPES (pH
7.8) 0.5 mM EDTA and 0.03% SDS (18). Cleavage activity was monitored
continuously by detection of free 7-amido-4-methylcoumarin (AMC)
using a fluorescence plate reader (Gemini; Molecular Devices,
Sunnyvale, CA, USA) at 380/460 nm and 37°C. As controls, the AMC (2
μM) was incubated with the drugs in buffer I without the
cell extracts, and measurements of proteasome function were
corrected when necessary.
Quantitative polymerase chain reaction
(qPCR)
Total RNA was extracted from each experimental group
using TRIzol® eagent (Invitrogen Life Technologies,
Carlsbad, CA, USA), according to the manufacturer’s instructions.
RNA concentration was assessed spectrophotometrically at 260 nm
(Thermo ND 2000; Thermo Scientific). Reverse transcription was
performed on the isolated total RNA using a Reverse Transcription
kit (Takara Bio Inc., Otsu, Japan), and PCR was carried out using a
Real Time PCR kit (Takara Bio, Inc.). Reverse transcription was
performed at 65°C for 5 min, 30°C for 10 min, 42°C for 10–30 min
and 92°C for 3 min. PCR conditions were as follows: Denaturation at
94°C for 2 min; amplification for 30 cycles at 94°C for 0.5 min,
annealing at 60°C for 0.5 min, and extension at 72°C for 1 min;
followed by a terminal elongation step at 72°C for 10 min, and was
perfomed on a Bio-Rad CF×96 thermal cycler (Bio-Rad Laboratories,
Inc.). U6 was amplified as an internal control and the Ct value of
each PCR product was calculated, and the fold change was analyzed.
The m-miR-423-5p, h-miR-423-5p, m-U6 and h-U6 primers were supplied
by Riob Bio Technology (Guangzhou, China); the sequences were not
supplied due to the rules of the company.
Detection of miR-423-5p downstream
targets using western blot analysis
The cardiomyocytes were transfected with
m-miR-423-5p or miR-Ctrl for 24 or 48 h, as described above,
following which the total protein was collected. The cells were
lysed on ice for 30 min using radioimmunoprecipitation assay lysis
buffer, containing 50 mM Tris-HCl (pH 7.4), 1% NP-40, 0.25%
Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl
fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1
μg/ml pepstatin, 1 mM Na3VO4 and 1 mM
NaF. The proteins (20 μg) were separated by
SDS-polyacrylamide gel electrophoresis (SDS-PAGE; Beyotime
Biotechnology) and electronically transferred onto a polyvinylidene
difluoride membrane (Millipore, Bedford, MA, USA). Following
blocking with 5% skim milk powder in tris-buffered saline with
Tween, the membranes were incubated with recommended dilution
primary antibodies against OGT (rabbit polyclonal; 1:800 for 1 h at
37°C; cat. no. sc-32921; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA), phosphorylated (p)-adenosine monophosphate
(AMP)-activated protein kinase (AMPK; cat. no. 4188; 1:1,000), AMPK
(cat. no. 2532; 1:1,000) p53 (cat. no. 2527; 1:1,200) and caspase 3
(cat. no. 9662; 1:600) for 1 h at 37°C (all purchased from Cell
Signaling Technology, Inc., Danvers, MA, USA), and GAPDH (cat. no.
sc-25778; 1:5,000; Santa Cruz Biotechnology, Inc.). This was
followed by incubation with peroxidase-conjugated secondary
antibodies (Abcam, Cambridge, MA, USA). The peroxidase-labeled
bands were visualized using an enhanced chemiluminescence kit
(Pierce Biotechnology, Inc.). The ratio of OGT, p-AMPK, AMPK, p53
and caspase 3/GAPDH were calculated using densitometry.
Terminal deoxynucleotidyl
transferase-deoxyuridine triphosphate nick-end labeling (TUNEL)
assay
In order to detect apoptotic cells in cardiomyocytes
following transfection with miR-423-5p and miR-Ctrl, a TUNEL assay
was performed using a DeadEndTM Fluorometric TUNEL system (Promega
Corporation), according to the manufacturer’s instructions. Cell
nuclei with dark green fluorescent staining were defined as
TUNEL-positive nuclei, which were visualized using a fluorescence
microscope (DT×500; Nikon Corporation, Tokyo, Japan). In order to
quantify the TUNEL-positive cells, the number of green
fluorescence-positive cells were counted in randomly selected
fields at ×200 magnification. The cell nuclei were then
counter-stained with 4′,6-diamidino-2-phenylindole (Beyotime
Biotechnology).
Statistical analysis
Statistical comparisons of all the results were
analyzed using a one-way analysis of variance. Statistical analyses
were performed using SPSS version 19.0 (IBM SPSS, Armonk, NY, USA).
Values are expressed as the mean ± standard error of the mean.
P<0.05 was considered to indicate a statistically significant
difference.
Results
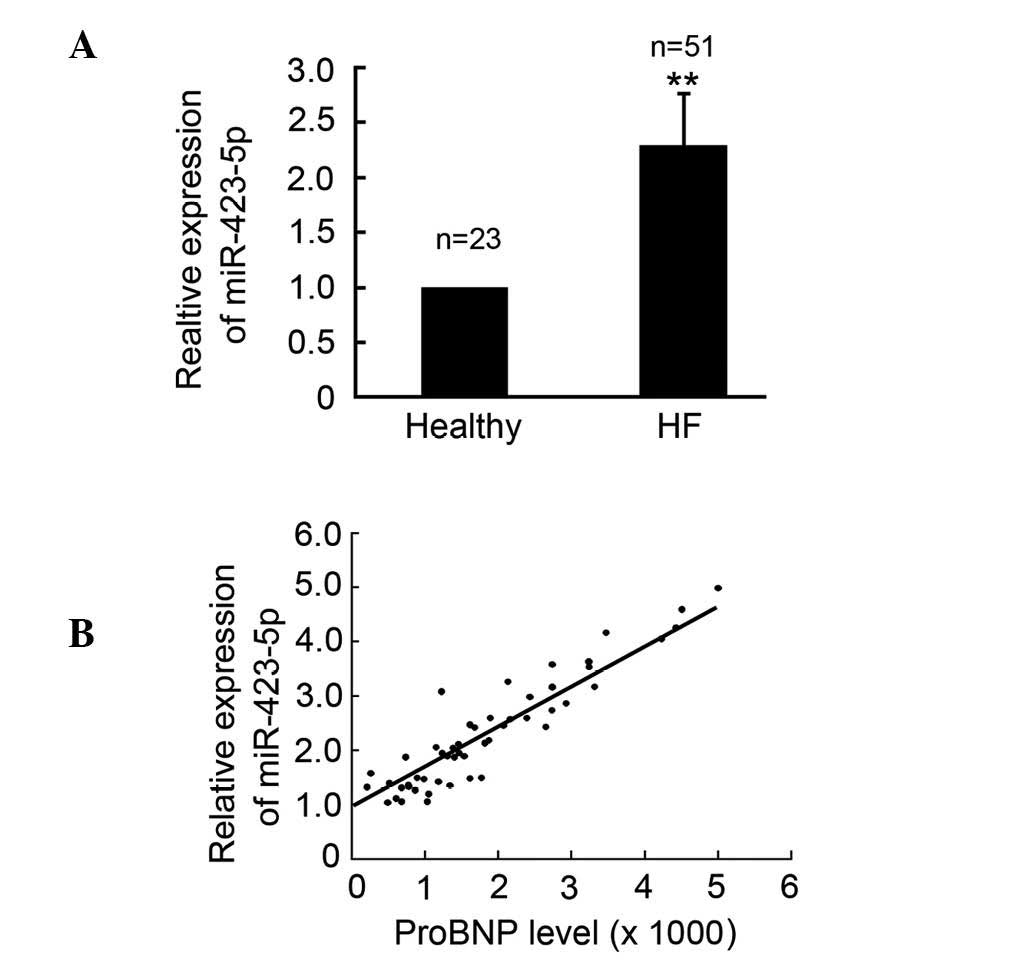
miR-423-5p is upregulated in patients
with CHF
In order to investigate whether the expression of
miR-423-5p was altered in patients with CHF, plasma samples from
patients and healthy controls were collected and qPCR was performed
to determine the expression levels of circulating miR-423-5p in the
patients with CHF and the healthy controls. The results
demonstrated that the expression of miR-423-5p was upregulated in
the patients, with CHF compared with the healthy controls (Fig. 1A). The circulating protein levels
of the proBNP biomarker of CHF were also determined and the results
indicated that the circulating levels of miR-423-5p were positively
correlated with the expression of proBNP and the CHF classification
(Fig. 1B).
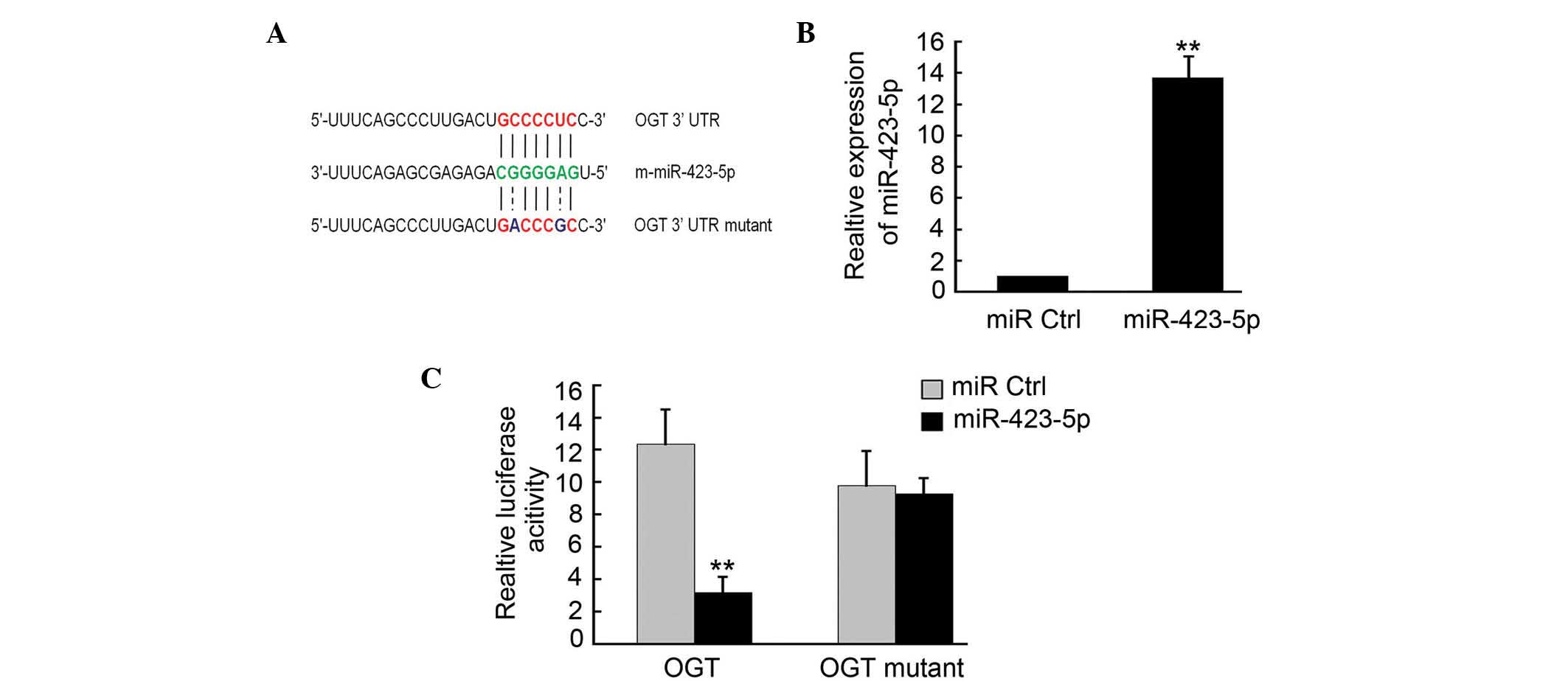
OGT is a target of m-miR-423-5p
To determine the targets of m-miR-423-5p, a number
of potential target proteins in a database library were screened to
identify potential miRNA binding seed sequences within the 3′-UTR.
Using bioinformatic analyses (Targetscan, microRNA.org and
microRNASeq), it was found that m-OGT was a potential candidate
target for m-miR-423-5p (Fig. 2A).
In order to verify this, m-OGT-3′UTR containing an miR-423-5p
binding site were cloned downstream of the luciferase open reading
frame. The m-OGT-3′UTR mutant, which contained a mutated
m-miR-423-5p binding site, was also introduced into the luciferase
construct. The plasmid expressing m-miR-423-5p was transfected into
the cardiomyoctes, and puromycin (Beyotime Biotechnology) was used
to select the stable expression cells. qPCR analysis confirmed that
m-miR-423-5p was upregulated in these cells (Fig. 2B). The luciferase-OGT-3′UTR and
luciferase-OGT-3′UTR mutant constructs were then transfected into
the 293 cells stably expressing m-miR-423-5p. Following 4–6 h
transfection, the expression of luciferase in the OGT-3′UTR
constructs was significantly downregulated compared with that of
the miR-Ctrl, whereas luciferase activity was unaffected in the
cells transfected with the mutant construct (Fig. 2C).
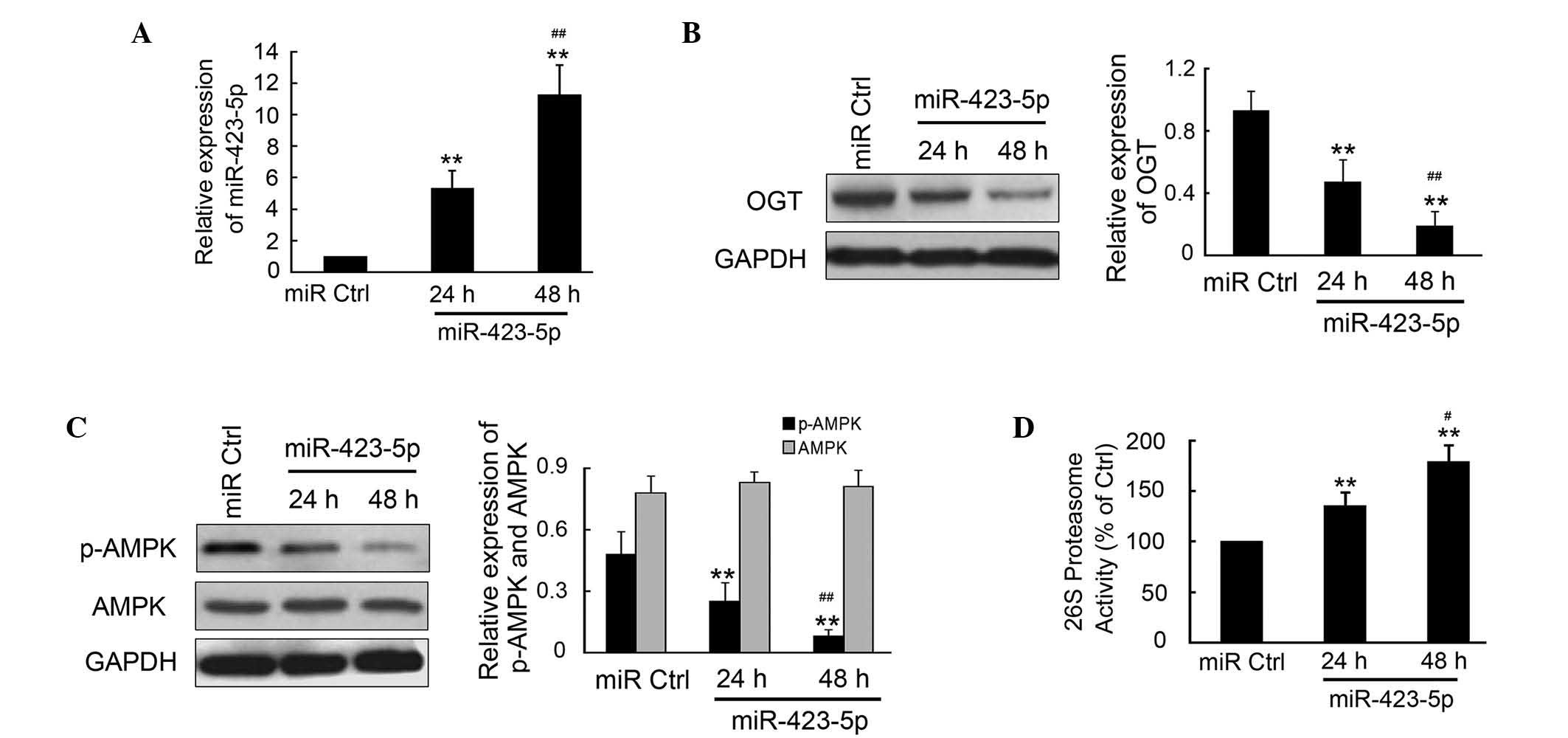
m-miR-423-5p regulates the expression
levels of OGT and downstream targets of OGT in cardiomyocytes
The effects of m-miR-423-5p transfection were
investigated in cardiomyocytes from mice due to the limited use of
human cardiomyocytes in investigations. The plasmid encoding
m-miR-423-5p was transfected into the mouse cardiomyocytes and
incubated for 24 or 48 h. The cells were then collected and qPCR
was performed to determine the expression of levels of
m-miR-423-5p. m-miR-423-5p was significantly upregulated at 24 and
48 h post-transfection compared with the miR-Ctrl (Fig. 3A). There was significantly
increased expressed of miR-423-5p at 48 h compared with that at 24
h (P<0.01). Western blotting and densitometric analysis
indicated that the OGT in the cardiomyocytes was significantly
inhibited by m-miR-423-5p (Fig.
3B). The activities of AMPK and the 26S proteasome, as the
downstream targets of OGT, were also determined. The results
demonstrated that the phosphorylation of AMPK was inhibited by
m-miR-423-5p, which was comparable with the reduction in OGT
(Fig. 3C). However, no significant
change was observed in the expression of total AMPK. Furthermore,
the activities of the 26S proteasome increased significantly by
1.35±0.12- and 1.79±0.16-fold 24 and 48 h post-transfection with
m-miR-423-5p, respectively, compared with miR-Ctrl.
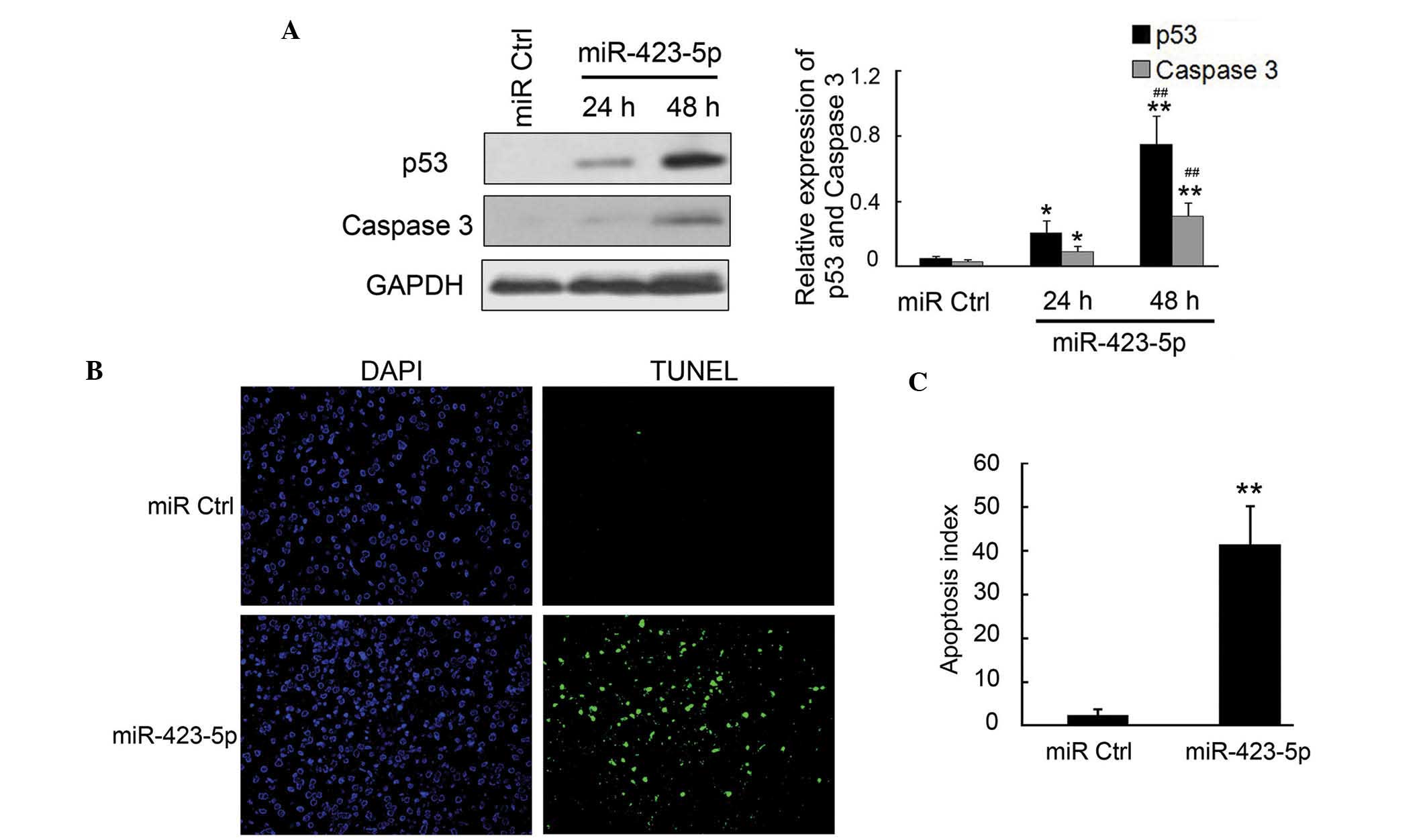
m-mir-423-5p induces apoptosis in
cardiomyocytes through upregulation of p53 and caspase-3
Cardiomyocyte apoptosis has been previously reported
as the primary cause of CHF (19);
therefore, the present study examined the apoptosis and the
expression of apoptosis-associated proteins using TUNEL and western
blot analysis. The results of the western blot analysis revealed
that the pro-apoptotic proteins p53 and caspase-3, which are the
downstream targets of AMPK, were significantly upregulated by
m-miR-423-5p in the cardiomyocytes (Fig. 4A and B). The TUNEL assays indicated
that the apoptotic rate was significantly increased in the
m-miR-423-5p-transfected cardiomyocytes, compared with the miR-Ctrl
group (Fig. 4C and D). Overall,
these results demonstrated that the increased expression of
m-miR-423-5p in cardiomyocytes significantly induced cell apoptosis
via the upregulation of p53 and caspase-3.
Discussion
The results of the present study demonstrated that
the expression of miR-423-5p was positively correlated with CHF
diagnosis and the plasma levels of proBNP. In addition, the results
indicated that OGT was a target of miR-423-5p; therefore, it was
hypothesized that miR-423-5p directly targeted the OGT 3′-UTR,
inhibiting the expression of OGT and inducing apoptosis in the
cardiomyocytes. Furthermore, the inhibition of OGT by miR-423-5p
significantly decreased the expression of p-AMPK and upregulated
the activity of the 26S proteasome and the protein expression
levels of pro-apoptotic p53 and caspase-3. The TUNEL analysis also
revealed increased apoptosis in the cardiomyocytes following
transfection with miR-423-5p.
Previous evidence suggests that circulating miRNAs
may be used as stable blood-based biomarkers in cancer (12). In particular, the expression levels
of miR-1 and miR-208 cardiac miRNAs are elevated in plasma
following myocardial injury, which has been suggested to occur due
to the release of these miRNAs from the damaged cardiac cells
(20,21). The results of a previous array
study reported that miR-423-5p is upregulated in the failing human
myocardium (13) and circulating
levels of miR-423-5p have been associated with proBNP and ejection
fractions in this patient groups (14). However, the use of miR-423-5p as a
biomarker for systemic ventricular and left ventricular remodeling
has been unsuccessful (15,16).
The results of the present study demonstrated that circulating
levels of miR-423-5p were upregulated in theCHF patients and were
associated with the experssion of proBNP. In addition, the
expression levels of miR-423-5p in the plasma may also provide
novel biomarker for CHF. However, further clinical investigation is
required in order to confirm this hypothesis.
The post-translational β-O-linkage of
N-acetylglucosamine (O-GlcNAc) to cellular proteins is an important
signaling pathway, which has been implicated in the pathophysiology
of cardiovascular disease (22).
OGT enables the addition of O-GlcNAc to proteins and is required
for cell division and embryogenesis (23). Cardiomyocyte ablation of OGT
induces heart failure, and OGT is required for the endogenous
compensatory response to infarct-induced heart failure (24). AMPK is present in all mammalian
cells and is a sensor of cellular energy (25) and cellular redox status (26). As a downstream target of OGT, the
reduction of p-AMPK by OGT in endothelial cells upregulates
proteasome activity (27). The
results of the present study indicated that OGT was a target of
miR-423-5p; and that the expression of miR-423-5p in mouse
cardiomyocytes significantly inhibited the expression of OGT and
phosphorylation of the downstream target, AMPK. Of note, 26S
proteasome activity and the expression levels of the p53 and
caspase-3 pro-apoptotic proteins were markedly increased.
Furthermore, according to the molecular changes observed in the
TUNEL assay, the apoptotic rate of cells was increased in
cardiomyocytes transfected with miR-423-5p.
References
|
1
|
Dassanayaka S and Jones SP: O-GlcNAc and
the cardiovascular system. Pharmacol Ther. 142:62–71. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shaf R, Iyer SP, Ellies LG, O’Donnell N,
Marek KW, Chui D, et al: The O-GlcNAc transferase gene resides on
the X chromosome and is essential for embryonic stem cell viability
and mouse ontogeny. Proc Natl Acad Sci USA. 97:5735–5739. 2000.
View Article : Google Scholar
|
|
3
|
Watson LJ, Facundo HT, Ngoh GA, Ameen M,
Brainard RE, Lemma KM, et al: O-linked beta-N-acetylglucosamine
transferase is indispensable in the failing heart. Proc Natl Acad
Sci USA. 107:17797–17802. 2010. View Article : Google Scholar
|
|
4
|
Mann DL and Bristow MR: Mechanisms and
models in heart failure: the biomechanical model and beyond.
Circulation. 111:2837–2849. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Seidman JG and Seidman C: The genetic
basis for cardiomyopathy: from mutation identification to
mechanistic paradigms. Cell. 104:557–567. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Januzzi JL, van Kimmenade R, Lainchbury J,
Bayes-Genis A, Ordonez-Llanos J, Santalo-Bel M, et al: NT-proBNP
testing for diagnosis and short-term prognosis in acute
destabilized heart failure: an international pooled analysis of
1256 patients: the International Collaborative of NT-proBNP Study.
Eur Heart J. 27:330–337. 2006. View Article : Google Scholar
|
|
7
|
van Kimmenade RR, Pinto YM and Januzzi JL
Jr: Importance and interpretation of intermediate (gray zone)
amino-terminal pro-B-type natriuretic peptide concentrations. Am J
Cardiol. 101:39–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Rooij E, Marshall WS and Olson EN:
Toward microRNA-based therapeutics for heart disease: the sense in
antisense. Circ Res. 103:919–928. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
et al: Characterization of microRNAs in serum: a novel class of
biomarkers for diagnosis of cancer and other diseases. Cell Res.
18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lawrie CH, Gal S, Dunlop HM, Pushkaran B,
Liggins AP, Pulford K, et al: Detection of elevated levels of
tumour-associated microRNAs in serum of patients with diffuse large
B-cell lymphoma. Br J Haematol. 141:672–675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, et al: Circulating microRNAs as
stable blood-based markers for cancer detection. Proc Natl Acad Sci
USA. 105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thum T, Galuppo P, Wolf C, Fiedler J,
Kneitz S, van Laake LW, et al: MicroRNAs in the human heart: a clue
to fetal gene reprogramming in heart failure. Circulation.
116:258–267. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tijsen AJ, Creemers EE, Moerland PD, de
Windt LJ, van der Wal AC, Kok WE, et al: MiR423-5p as a circulating
biomarker for heart failure. Circ Res. 106:1035–1039. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bauters C, Kumarswamy R, Holzmann A,
Bretthauer J, Anker SD, Pinet F, et al: Circulating miR-133a and
miR-423-5p fail as biomarkers for left ventricular remodeling after
myocardial infarction. Int J Cardiol. 168:1837–1840. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tutarel O, Dangwal S, Bretthauer J,
Westhoff-Bleck M, Roentgen P, Anker SD, et al: Circulating
miR-423_5p fails as a biomarker for systemic ventricular function
in adults after atrial repair for transposition of the great
arteries. Int J Cardiol. 167:63–66. 2013. View Article : Google Scholar
|
|
17
|
Fekete MR, McBride WH and Pajonk F:
Anthracyclines, proteasome activity and multi-drug-resistance. BMC
Cancer. 5:1142005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rosamond WD, Chang PP, Baggett C, et al:
Classification of heart failure in the atherosclerosis risk in
communities (ARIC) study: A comparsion of diagnostic criteria. Circ
Heart Fail. 5:152–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang C, Liu Z, Liu K and Yang P:
Mechanisms of Ghrelin anti-heart failure: Inhibition of Ang
II-induced cardiomyocyte apoptosis by down-regulating AT1R
expression. PLoS One. 9:e857852014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J,
et al: Circulating microRNA-1 as a potential novel biomarker for
acute myocardial infarction. Biochem Biophys Res Commun. 391:73–77.
2010. View Article : Google Scholar
|
|
21
|
Ji X, Takahashi R, Hiura Y, Hirokawa G,
Fukushima Y and Iwai N: Plasma miR-208 as a biomarker of myocardial
injury. Clin Chem. 55:1944–1949. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dassanayaka S and Jones SP: O-GlcNAc and
the cardiovascular system. Pharmacol Ther. 142:62–71. 2014.
View Article : Google Scholar :
|
|
23
|
Shaf R, Iyer SP, Ellies LG, O’Donnell N,
Marek KW, Chui D, et al: The O-GlcNAc transferase gene resides on
the X chromosome and is essential for embryonic stem cell viability
and mouse ontogeny. Proc Natl Acad Sci USA. 97:5735–5739. 2000.
View Article : Google Scholar
|
|
24
|
Watson LJ, Facundo HT, Ngoh GA, Ameen M,
Brainard RE, Lemma KM, et al: O-linked beta-N-acetylglucosamine
transferase is indispensable in the failing heart. Proc Natl Acad
Sci USA. 107:17797–17802. 2010. View Article : Google Scholar
|
|
25
|
Hardie DG: AMPK and Raptor: matching cell
growth to energy supply. Mol Cell. 30:263–265. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zou MH and Wu Y: AMP-activated protein
kinase activation as a strategy for protecting vascular endothelial
function. Clin Exp Pharmacol Physiol. 35:535–545. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu J, Wang S, Viollet B and Zou MH:
Regulation of the proteasome by AMPK in endothelial cells: the role
of O-GlcNAc transferase (OGT). PloS one. 7:e367172012. View Article : Google Scholar : PubMed/NCBI
|